Abstract
The modular assembly (MA) method of generating engineered zinc finger proteins (ZFPs) was the first practical method for creating custom DNA-binding proteins. As such, MA has enabled a vast exploration of sequence-specific methods and reagents, ushering in the modern era of zinc finger-based applications that are described in this volume. The first zinc finger nuclease to cleave an endogenous site was created using MA, as was the first artificial transcription factor to enter phase II clinical trials. In recent years, other excellent methods have been developed that improved the affinity and specificity of the engineered ZFPs. However, MA is still used widely for many applications. This chapter will describe methods and give guidance for the creation of ZFPs using MA. Such ZFPs might be useful as starting materials to perform other methods described in this volume. Here, we also describe a single-strand annealing recombination assay for the initial testing of zinc finger nucleases.
Keywords: Zinc finger nuclease, single-strand annealing, homologous recombination, zinc finger protein engineering methods, modular assembly
1. Introduction
ZFPs are among the most common type of DNA-binding protein found in eukaryotes, and often comprise arrays of 8–10 fingers (1). The 1991 crystal structure of the three-finger ZFP Zif268 bound to DNA (2) revealed that each finger used amino acids in positions −1, 3 and 6 of its α-helix to contact a 3-bp subsite on the DNA. This seemingly simple mode of recognition inspired an early effort to create custom-DNA binding proteins (3). Through a combination of rational design and combinatorial methods such as phage display, individual zinc fingers were optimized to recognize one of the 64 possible 3-bp DNA subsites (4–8). In principle, these fingers could be assembled in any order to create multi-finger ZFPs to any desired sequence. Since each finger recognized an independent 3-bp subsite, ZFPs containing six fingers were expected to recognize 18 bp of DNA, a site sufficiently large to specify a unique locus in the human genome (9). The construction of ZFPs in this manner is referred to as modular assembly (MA).
Three large sets of MA fingers have been described. A widely-used and well-characterized set was developed by Carlos Barbas of The Scripps Research Institute that covers all 3-bp subsites of the type GNN, most ANN, many CNN, and some TNN (see Table 1) (4, 5, 7). Sangamo Biosciences developed a set that covers GNN subsites, often with slightly different protein sequences depending on whether the finger would be used as finger 1, 2, or 3 in a three-finger protein (see Table 2) (6). This set is not widely used by Sangamo Bioscience, and several recent studies found this set was less likely than others to generate highly active ZFNs (10, 11). ToolGen, another company, developed a set that was based on selections of zinc fingers that occurred naturally in the human genome, as opposed to synthetic variants of Zif268 (8). The final set contains more than 50 full-length fingers that collectively recognize about 25 subsites (see Tables 3 and 4).
Table 1.
The Barbas set of zinc finger modules
| DNAa | Fingerb | DNA | Finger | DNA | Finger | RAc | DNA | Finger |
|---|---|---|---|---|---|---|---|---|
| AAA | QRANLRA | CAA | QSGNLTE | GAA | QSSNLVR | 0.5 | TAA | |
| AAC | DSGNLRV + | CAC | SKKALTE + | GAC | DPGNLVR | 3 | TAC | |
| AAGk | RKDNLKN | CAGk | RADNLTE + | GAGk | RSDNLVR | 1 | TAGk | REDNLHT + |
| AAT | TTGNLTV | CAT | TSGNLTE − | GAT | TSGNLVR | 3 | TAT | |
| ACA | SPADLTR + | CCA | TSHSLTE + | GCA | QSGDLRRd | 2 | TCA | |
| ACC | DKKDLTR | CCC | SKKHLAE − | GCC | DCRDLAR | 80 | TCC | |
| ACGk | RTDTLRD | CCGk | RNDTLTE + | GCGk | RSDDLVR | 9 | TCG | |
| ACT | THLDLIR + | CCT | TKNSLTE + | GCT | TSGELVRd | 65 | TCT | |
| AGA | QLAHLRA | CGA | QSGHLTE | GGA | QRAHLER | 3 | TGA | QAGHLAS |
| AGC | CGC | HTGHLLE + | GGC | DPGHLVR + | 40 | TGC | ||
| AGGk | RSDHLTN | CGGk | RSDKLTE | GGGk | RSDKLVR | 6 | TGGk | RSDHLTT |
| AGT | HRTTLTN − | CGT | SRRTCRA + | GGT | TSGHLVR | 15 | TGT | |
| ATA | QKSSLIA | CTA | QNSTLTE | GTA | QSSSLVR | 2.5e | TTA | |
| ATC | CTC | GTC | DPGALVR | 40 | TTC | |||
| ATGk | RRDELNV − | CTGk | RNDALTE − | GTGk | RSDELVR | 15 | TTG | |
| ATT | HKNALQN | CTT | TTGALTE − | GTT | TSGSLVR | 5 | TTT |
The 3-bp subsite to which the finger was designed. A “k” following the triplet indicates that this neighboring subsite should begin with either a G or T (K in the IUPAC nomenclature). This is due to TSO interactions, as described in the I introduction.
The DNA-recognition portion of the finger, consisting of amino acids −1, 1, 2, 3, 4, 5, and 6 with respect to the α-helix (based on (4, 5, 7)). These amino acids should be inserted into the appropriate positions in an Sp1C or Zif268 zinc-finger scaffold (see Fig. 1C and 1D). Also indicated is whether the finger was recommended favorably (+) or unfavorably (−) by the large-scale ToolGen study (28). Note that many fingers were not evaluated.
The relative affinity of each finger in nM, as determined in a context as the middle finger of the three-finger Zif268 DNA-binding domain. Note that this information is only currently available for the GNN set of fingers (based on (7)).
As described in the introduction, QSGDLRR also binds the subsite 5’-GCT-3’ with a relative affinity of 10, which is six-fold stronger than the more 5’-GCT-3’-specific finger, TSGELVR.
Table 2.
The Sangamo set of zinc finger modules
| DNAa | Finger 1b | Finger 2 | Finger 3 |
|---|---|---|---|
| GAA | QRSNLVR | QSGNLAR | QSGNLAR |
| GAC | DRSNLTR | DRSNLTR | DRSNLTR |
| GAGk | RSDNLAR | RSDNLAR | RSDNLTR |
| GAT | QSSNLAR | TSGNLVR | TSANLSR |
| GCA | QSGSLTR | QSGDLTR | QSGDLTR |
| GCC | ERGTLAR | DRSDLTR | DRSDLTR |
| GCGk | RSDDLTR | RSDDLQR | RSDDLTR |
| GCT | QSSDLTR | QSSDLTR | QSSDLQR |
| GGA | QSGHLAR | QSGHLQR | QSGHLQR |
| GGC | DRSHLTR | DRSHLAR | DRSHLAR |
| GGGk | RSDHLAR | RSDHLTR | RSDHLSR |
| GGT | QSSHLTR | TSGHLVR | TSGHLVR |
| GTA | QSGALTR | QSGALAR | QRASLTR |
| GTC | DRSALAR | DRSALAR | DRSALAR |
| GTGk | RSDALRT | RSDALSR | RSDALTR |
| GTT | TTSALTR | TSGALTR | QSSALTR |
The 3-bp subsite to which the finger was designed. A “k” following the triplet indicates that this neighboring subsite should begin with either a G or T (K in the IUPAC nomenclature). This is due to TSO interactions, as described in the Introduction.
The DNA-recognition portion of the finger, consisting of amino acids −1, 1, 2, 3, 4, 5, and 6 with respect to the α-helix (based on (6)). Note that this set often recommends a slightly different protein sequence depending on whether the finger would be used as finger 1, 2, or 3 in a three-finger protein. These amino acids should be inserted into the appropriate positions in an Sp1C or Zif268 zinc-finger scaffold (see Fig. 1C and 1D).
Table 3.
The ToolGen set of zinc finger modules
| DNAa | Module Nameb |
DNA | Module Name |
DNA | Module Name |
DNA | Module Name |
|
|---|---|---|---|---|---|---|---|---|
| AAA | QSNI + | CAA | QSNI | GAA | CSNR1 | QSNR1 + | TAA | QSNK − |
| QSNK | QSNV1 | CSNR2 | QSNR2 | QSNV2 | ||||
| QSNT | QSNV2 + | ISNR | QSNR3 | |||||
| QSNV1 | QSNV3 | QGNR | QSNR4 | |||||
| QSNV2 | QSNV4 | QSHR5 | QSNV2 | |||||
| QSNV3 | QSNK | QSTV | ||||||
| QSNV4 | ||||||||
| AAC | HSNK | CAC | GAC | CSNR1 + | HSNK − | TAC | ||
| CSNR2 | QSTV | |||||||
| DSNR + | ||||||||
| AAGk | CAGk | GAGk | CSNR1c | QSTVc | TAGk | |||
| CSNR2c | RDER1 | |||||||
| KSNRc | RSNRc − | |||||||
| QFNR c − | SSNRc | |||||||
| AAT | ISNV | CAT | GAT | ISNR + | TAT | VDYK + | ||
| VSNV + | ||||||||
| ACA | CCA | GCA | QSSR1 + | QSTR − | TCA | |||
| QSSR2 | VSTR | |||||||
| QSSR3 | ||||||||
| ACC | CCC | GCC | DSCR + | TCC | ||||
| ACGk | CCGk | GCGk | RDER1 + | RDER4 | TCGk | |||
| RDER2 + | RDER5 | |||||||
| RDER3 | RDER6 | |||||||
| ACT | CCT | GCT | VSTR+ | TCT | ||||
| AGA | QSHR1 | CGA | QSHT − | GGA | QAHR | QSHR5 | TGA | QSHT − |
| QSHR3 | QSHV + | QSHR1 | QTHR1 − | QSHV + | ||||
| QSHT | QTHQ | QSHR2 + | QTHR2 | QTHQ | ||||
| QSHV | QTHR1 | QSHR3 | WSNR | |||||
| QTHQ + | QSHR4 | |||||||
| QTHR1 | ||||||||
| AGC | CGC | GGC | TGC | |||||
| AGGk | RDHT | CGGk | RDHT + | GGGk | RDHR1 + | RDKI | TGGk | RDHT + |
| RDKR + | RDHR2 | RDKR | ||||||
| RDHT | RSHRc + | |||||||
| AGT | CGT | GGT | WSNR + | TGT | ||||
| ATA | QNTQ + | CTA | GTA | QSSR1 + | QSTR − | TTA | ||
| QSSR2 | VSSR | |||||||
| QSSR3 | ||||||||
| ATC | DSAR | CTC | GTC | DSAR + | TTC | |||
| ATGk | CTGk | GTGk | RDER1 + | TTGk | ||||
| RDER2 + | ||||||||
| VSSRc | ||||||||
| ATT | CTT | GTT | HSSR − | TTT | ||||
| VSSR − | ||||||||
The 3-bp subsite to which the finger was designed. A “k” following the triplet indicates that this neighboring subsite should begin with either a G or T (K in the IUPAC nomenclature). This is due to TSO interactions, as described in the Introduction.
The unique ToolGen name for each finger (8). The name corresponds to the amino acids in positions −1, 2, 3, and 6 of the α-helix; however, the full sequence of the finger is provided in Table 4. Also indicated is whether the finger was recommended favorably (+) or unfavorably (−) by the large-scale ToolGen study (28). Note that some fingers were not evaluated.
These fingers do not contain Asp in position 2 of the α-helix. Therefore, they are not expected to have a TSO interaction (i.e., the neighboring subsite could begin with any base).
Table 4.
The ToolGen finger sequences and functional data
| Module Namea |
Finger Sequenceb | DNA-Fullc | Td | DNA-ZFNe | RAf | DNA-RAg | |
|---|---|---|---|---|---|---|---|
| CSNR1 | YKCKQCGKAFG | CPSNLRR HGRTH | GA(ACG) | + | GAC | 2.3 | GAC |
| CSNR2 | YQCNICGKCFS | CNSNLHR HQRTH | GA(ACG) | ||||
| DSAR | YSCGICGKSFS | DSSAKRR HCILH | (AG)TC | + | GTC | 0.4 | GTC |
| DSCR | YTCSDCGKAFR | DKSCLNR HRRTH | GCC | + | GCC | 3.9 | GCC |
| DSNR | YRCKYCDRSFS | DSSNLQR HVRNIH | + | GAC | |||
| HSNK | YKCKECGKAFN | HSSNFNK HHRIH | (AG)AC | − | GAC | 178.6 | GAC |
| HSSR | FKCPVCGKAFR | HSSSLVR HQRTH | GTT | − | GTT | 0.8 | GTT |
| ISNR | YRCKYCDRSFS | ISSNLQR HVRNIH | GA(AT) | + | GAT | 0.1 | GAT |
| ISNV | YECDHCGKAFS | IGSNLNV HRRIH | AAT | ||||
| KSNR | YGCHLCGKAFS | KSSNLRR HEMIH | GAG | 1.7 | GAG | ||
| QAHR | YKCKECGQAFR | QRAHLIR HHKLH | GGA | 8.4 | GGA | ||
| QFNR | YKCHQCGKAFI | QSFNLRR HERTH | GAG | − | GAG | 66.1 | GAG |
| QGNR | FQCNQCGASFT | QKGNLLR HIKLH | GAA | 1.2 | GAA | ||
| QNTQ | YTCSYCGKSFT | QSNTLKQ HTRIH | + | ATA | |||
| QSHR1 | YACHLCGKAFT | QSSHLRR HEKTH | (AG)GA | ||||
| QSHR2 | YKCGQCGKFYS | QVSHLTR HQKIH | GGA | + | GGA | ||
| QSHR3 | YACHLCGKAFT | QCSHLRR HEKTH | (AG)GA | ||||
| QSHR4 | YACHLCAKAFI | QCSHLRR HEKTH | GGA | ||||
| QSHR5 | YVCRECGRGFR | QHSHLVR HKRTH | G(AG)A | 2 | GGA | ||
| QSHT | YKCEECGKAFR | QSSHLTT HKIIH | (ACT)GA | − | (CT)GA | 17.1 | AGA |
| QSHV | YECDHCGKSFS | QSSHLNV HKRTH | (ACT)GA | + | (CT)GA | 64.3 | CGA |
| QSNI | YMCSECGRGFS | QKSNLTI HQRTH | (AC)AA | + | AAA | 51.8 | CAA |
| QSNK | YKCEECGKAFT | QSSNLTK HKKIH | (AGT)AA | − | TAA | 2.7 | GAA |
| QSNR1 | FECKDCGKAFI | QKSNLIR HQRTH | GAA | + | GAA | 1.1 | GAA |
| QSNR2 | YVCRECRRGFS | QKSNLIR HQRTH | GAA | ||||
| QSNR3 | YECEKCGKAFN | QSSNLTR HKKSH | GAA | ||||
| QSNR4 | YECVQCGKSYS | QSSNLFR HQRRH | GAA | ||||
| QSNT | YECVQCGKGFT | QSSNLIT HQRVH | AAA | ||||
| QSNV1 | YECNTCRKTFS | QKSNLIV HQRTH | (AC)AA | ||||
| QSNV2 | YVCSKCGKAFT | QSSNLTV HQKIH | (ACGT)AA | + | CAA | 4.1 | CAA |
| QSNV3 | YKCDECGKNFT | QSSNLIV HKRIH | (AC)AA | ||||
| QSNV4 | YECDVCGKTFT | QKSNLGV HQRTH | (AC)AA | ||||
| QSSR1 | YKCPDCGKSFS | QSSSLIR HQRTH | G(CT)A | + | G(CT)A | 0.8 | GTA |
| QSSR2 | YECQDCGRAFN | QNSSLGR HKRTH | G(CT)A | ||||
| QSSR3 | YECNECGKFFS | QSSSLIR HRRSH | G(CT)A | ||||
| QSTR | YKCEECGKAFN | QSSTLTR HKIVH | G(CT)A | − | G(CT)A | 0.9 | GTA |
| QSTV | YECNECGKAFA | QNSTLRV HQRIH | GA(ACG) | ||||
| QTHQ | YECHDCGKSFR | QSTHLTQ HRRIH | (ACT)GA | + | AGA | 0.6 | CGA |
| QTHR1 | YECHDCGKSFR | QSTHLTR HRRIH | (ACG)GA | − | GGA | 1.6 | GGA |
| QTHR2 | HKCLECGKCFS | QNTHLTR HQRTH | GGA | ||||
| RDER1 | YVCDVEGCTWKFA | RSDELNR HKKRH | G(ACT)Gk | + | G(CT)Gk | 1.4 | GCG |
| RDER2 | YHCDWDGCGWKFA | RSDELTR HYRKH | GCGk | + | G(CT)Gk | ||
| RDER3 | YRCSWEGCEWRFA | RSDELTR HFRKH | GCGk | ||||
| RDER4 | FSCSWKGCERRFA | RSDELSR HRRTH | GCGk | ||||
| RDER5 | FACSWQDCNKKFA | RSDELAR HYRTH | GCGk | ||||
| RDER6 | YHCNWDGCGWKFA | RSDELTR HYRKH | GCGk | ||||
| RDHR1 | FLCQYCAQRFG | RKDHLTR HMKKSH | GGGk | + | GGGk | 0.9 | GGG |
| RDHR2 | CRCNECGKSFS | RRDHLVR HQRTH | GGGk | ||||
| RDHT | FQCKTCQRKFS | RSDHLKT HTRTH | (ACGT)GGk | + | (CT)GGk | 6.8 | AGG |
| RDKI | FACEVCGVRFT | RNDKLKI HMRKH | GGGk | ||||
| RDKR | YVCDVEGCTWKFA | RSDKLNR HKKRH | (AG)GGk | + | AGGk | 4.5 | GGG |
| RSHR | YKCMECGKAFN | RRSHLTR HQRIH | GGG | + | GGG | 0.2 | GGG |
| RSNR | YICRKCGRGFS | RKSNLIR HQRTH | GAG | − | GAG | 0.7 | GAG |
| SSNR | YECKECGKAFS | SGSNFTR HQRIH | GAG | 1.3 | GAG | ||
| VDYK | FHCGYCEKSFS | VKDYLTK HIRTH | + | TAT | |||
| VSNV | YECDHCGKAFS | VSSNLNV HRRIH | AAT | + | AAT | 2.5 | AAT |
| VSSR | YTCKQCGKAFS | VSSSLRR HETTH | GT(AGT) | − | GTT | 2.1 | GTG |
| VSTR | YECNYCGKTFS | VSSTLIR HQRIH | GC(AT) | + | GCT | 14.5 | GCT |
| WSNR | YRCEECGKAFR | WPSNLTR HKRIH | GG(AT) | + | GGT | 1.3 | GGT |
| Zif268 | FACDICGRKFA | RSDERKR HTKIH | GCG | 10 | GCG | ||
The unique ToolGen name for each finger (8). The name corresponds to the amino acids in positions −1, 2, 3, and 6 of the α-helix; however, the full sequence of the finger is provided in the next column.
The full sequence of the human zinc finger (based on (28, 44)). These amino acids should be inserted between canonical 5-residue linkers (TGEKP) to create new multi-finger ZFPs (see Fig. 1E). The DNA-recognition portion of the finger, consisting of amino acids −1, 1, 2, 3, 4, 5, and 6 with respect to the α-helix, is separated from the scaffold regions by spaces.
The full set of 3-bp subsites to which the finger bound as determined by a yeast one-hybrid assay (8).
Indicates this finger was recommended favorably (+) or unfavorably (−) by the large-scale ToolGen study (28). Note that some fingers were not evaluated.
The 3-bp subsite to which the finger bound when it performed favorably in the large-scale ToolGen study (28). A “k” following the triplet indicates that this neighboring subsite should begin with either a G or T (K in the IUPAC nomenclature). This is due to TSO interactions, as described in the Introduction.
The relative affinity of each finger in nM, as determined in a context as finger 3 of the threefinger Zif268 DNA-binding domain. Note that these values are based on (8), but have been normalized (multiplied by a factor of 17.86) to set the value of Zif268 equal to 10 nM. This was done for easier comparison with the Barbas relative affinity values, for which Zif268 had a value of 10 nM (7).
The 3-bp subsite used to test the finger in the relative affinity assay (8).
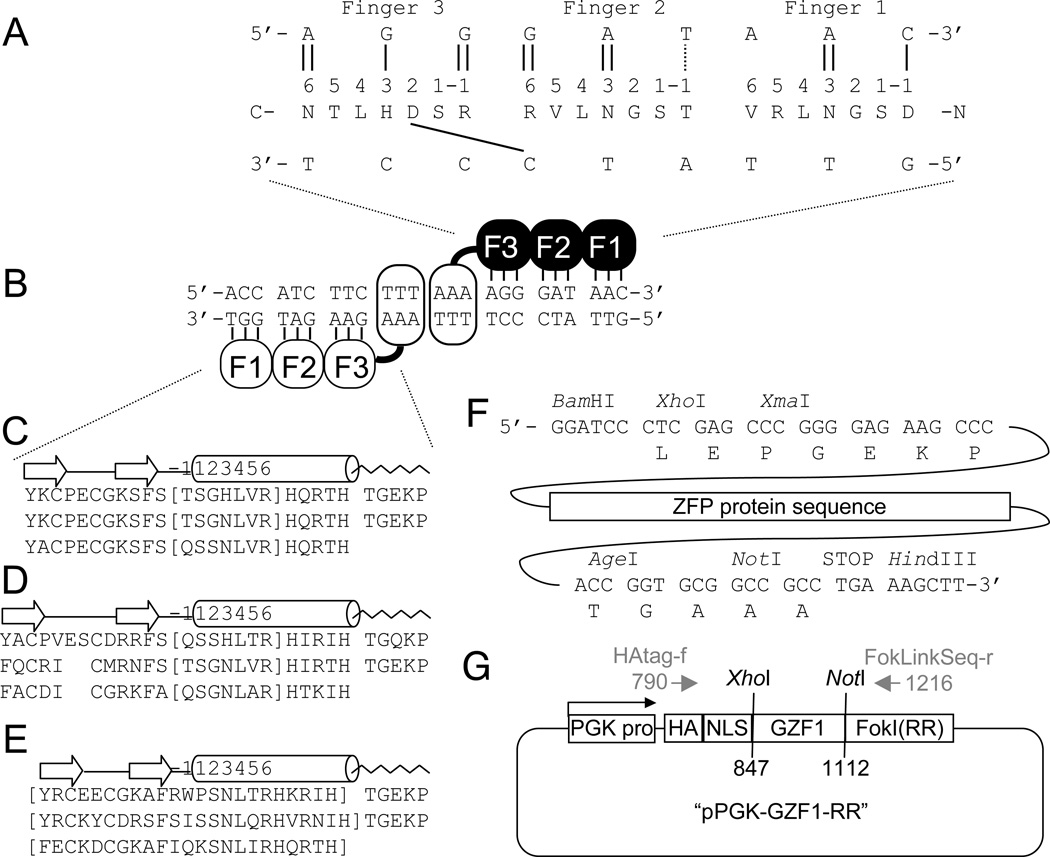
The ability to assemble the fingers in any order is based on the assumption that each finger binds its 3-bp subsite as an independent module. However, evidence soon began to emerge that, in some cases, there were “context-dependent effects”, interactions that occur when two particular fingers are assembled next to each other on a particular DNA sequence. For example, strong structural and biochemical evidence demonstrated that when the residue in position 2 of the α-helix was aspartate, its side chain made energetically important contacts that favored G or T as the first bp of the previous finger’s subsite (see Fig. 1A) (12). We can refer to this type of context-dependant effect, a helical amino acid (aa) specifying a base pair (bp) in the neighboring subsite, as target site overlap (TSO). However, there is only scattered evidence for other examples of TSO interactions. Other types of context-dependent effects, such as sequence-specific irregularities in the DNA structure due to base stacking, may in fact be more prevalent but are difficult to discover or predict.
Fig. 1.
Construction of ZFNs by MA. (A) A schematic diagram of the hydrogen bonding between the ZFP GZF3 and its 9-bp DNA target site. The ZFP binds “anti-parallel” (C-term. to N-term.) with respect to the DNA strand containing the target sequence (5’ to 3’). Vertical lines are presumed hydrogen bonds. The diagonal line represents a Target Site Overlap (TSO) by Finger 3, influencing the 5’ base of the neighboring Finger-2 subsite to be G. Fingers with potential TSO interactions are indicated in Tables 1–4. (B) A heterodimeric ZFN. Each monomer binds in an everted orientation so that the FokI cleavage domains at their C-termini (open ovals) can dimerize and cleave the DNA within the 6-bp spacer (TTTAAA) between the ZFP sites. (C) The sequence of the three-finger ZFP GZF1, created by inserting the Barbas modules (in brackets) into the Sp1C scaffold. Regions corresponding to beta strands (arrows), alpha helix (tube), and the interfinger linker (TGEKP, wavy line) are indicated. (D) The sequence for GZF1 using the Sangamo modules inserted into the Zif268 scaffold. (E) The sequence of GZF1 using the ToolGen modules, which consist of full-length fingers (in brackets) joined by interfinger linkers. Noted that D and E are for illustrative purposes; they have not been evaluated for function. (F) Additional protein sequences must be added to the ZFPs in panels C, D, or E, and must be encoded by the specific DNA sequences indicated in order to allow proper cloning into the ZFP cloning vector. (G) A schematic diagram of the ZFP cloning vector. A PGK promoter (PGK pro), HA tag (HA), SV40 Nuclear Localization Signal (NLS), ZFP (GZF1), and FokI cleavage domain (here shown to contain the obligate heterodimer modification RR), are indicated. PCR primers used for sequencing are shown in gray.
An appreciation of these context-dependent effects led to the development of what can be called “second generation” ZFP engineering methods, in which selections were performed on larger segments of the intended DNA target sequence. Yen Choo and Sir Aaron Klug developed a “bipartite” method (see Chapter 3 of this volume), allowing for rapid selection of essentially 1.5 fingers in the context of each other (13). Sangamo Biosciences eventually acquired a set of two-finger modules created by this method, and has used them as starting materials for their engineered ZFPs (14). Carl Pabo developed a “sequential selection” method, in which each new finger was selected in the context of its neighbor (15). A second method from the Pabo group, dubbed “B2H” in reference to the use of a bacterial two-hybrid system for selections, involved two steps: an enrichment of fingers on out-of-context 3-bp subsites as in classic modular assembly, followed by a final selection of the enriched fingers in the context of the full intended DNA target sequence (16). This method was extremely laborious, but was shown to frequently produce three-finger ZFPs that were superior in both affinity and specificity to three-finger ZFPs assembled by classical MA. Recently, J. Keith Joung developed a more streamlined version of this method called “oligomerized pool engineering” (OPEN) (11). In this method, the first step of the B2H method is performed by the Joung laboratory in high volume. The pools of enriched fingers are then made available to other investigators for the second step of selection on the intended target (see Chapter 2).
Currently, there are three major sources of ZFPs for investigators interested in engineered zinc finger technology, such as zinc finger nucleases (ZFNs). Sangamo Biosciences uses proprietary methods to create ZFNs that are highly potent and can target an impressively diverse spectrum of DNA sequences. Custom ZFNs from Sangamo are available through Sigma-Aldrich as CompoZr ZFN Technology, albeit at considerable cost (http://www.compozrzfn.com). MA is still one of the most popular methods for engineering ZFPs, and has been referred to in several chapters in this volume (see Chapters 6–11, 17, 23, and 24). Due to its large repertoire of targetable subsites, low cost, and ease of use, MA has been used to produce numerous regulators of endogenous genes, targetable live-cell imaging probes, recombinases, integrases, transposases, and methylases (17–21). Indeed, the first zinc finger nuclease to cleave an endogenous site was created using MA, as was the first artificial transcription factor to enter phase II clinical trial (22, 23). However, many have reported difficulty creating highly active ZFNs using MA. The Zinc Finger Consortium (ZFC), a confederation of academic laboratories, recently found that three-finger MA ZFPs composed of any set of modules could only produce a “positive” binding signal in a B2H reporter assay for 24% of 104 DNA targets (i.e., an “unexpected” failure rate of 76%) (10). The OPEN system has become the method of choice for the ZFC, and has been shown consistently to create high quality ZFNs (11). However, it is important to consider that the current version of OPEN is still a fairly complex procedure and only allows selection of fingers that recognize GNN and a few TNN subsites. If three-finger ZFPs engineered by OPEN were evaluated on the same 104 target sequences as used in the MA study, they would have an “expected” failure rate of 87%. Further advances are therefore required to develop methods for engineering high quality ZFPs that are simultaneously inexpensive, accessible, comprehensive, and robust.
In that regard, several recent reports offer helpful guidance for practitioners of MA. Many of the Barbas set of fingers were selected based on high specificity rather than high affinity. A more recent study by the ZFC showed that three-finger MA ZFPs containing one or more of the four lowest affinity Barbas fingers (GGC, GTC, GCT, GCC) tended to be poor binders in a B2H assay (24). These results argue against the proposition that MA is fundamentally flawed because it fails to address context-dependent effects (25), and suggest that one approach to improving performance might be to replace low affinity fingers with higher affinity variants. An interesting example supporting this approach is TSGELVR, a Barbas finger engineered to recognize 5’-GCT-3’ with high specificity but relatively low affinity (7). QSGDLRR, a Barbas finger designed to recognize 5’-GCA-3’, also binds 5’-GCT-3’ with six-fold higher affinity than TSGELVR. Several successful ZFNs in the literature actually use fingers essentially identical to QSGDLRR to recognize 5’-GCT-3’ (26, 27). Another approach might be to construct ZFPs containing 4, 5, or 6 fingers, which may have increased affinity compared to the three-finger MA ZFPs examined in the ZFC study (10). In support of this approach, a recent large-scale analysis by ToolGen found that ZFN pairs composed of two four-finger MA ZFPs were more frequently active than those composed of three-finger ZFPs (28). The authors also recommended a subset of 37 ToolGen and Barbas modules to produce active ZFNs based on their study (see Table 1 and 3).
In this chapter we describe methods and design considerations for creating ZFPs by MA (see Method 3.1). Once constructed, it is often useful to test some performance characteristics of the ZFPs (i.e., affinity, specificity, or activity). Many such assays are described in other chapters of this volume, including in vitro methods such as EMSA (see Chapters 6 and 16), in vitro ZFN cleavage assays (see Chapter 13), cell-based transcription reporter assays such as B2H or luciferase (see Chapters 2 and 7), and expression assays such as westerns (see Chapters 11 and 16). Here we describe a single-strand annealing (SSA) recombination assay for the initial analysis of ZFNs (see Method 3.2). Once active ZFN pairs are identified, assays at the endogenous site can be performed, such as the Surveyor mutagenesis detection assay (see Chapter 15), the restriction site modification assay (see Chapter 19), the chromatin immunoprecipitation (ChIP) binding assay (see Chapters 11 and 27). Upon achieving activity at the endogenous site, more laborious characterizations of the ZFN are warranted, such as in vitro target site selection assays (SELEX or Bind-n-Seq, (29, 30)), in vivo genome-wide binding site assays (ChIP-seq, see Chapter 27), or other target-specific activity assays (e.g., see Chapter 18).
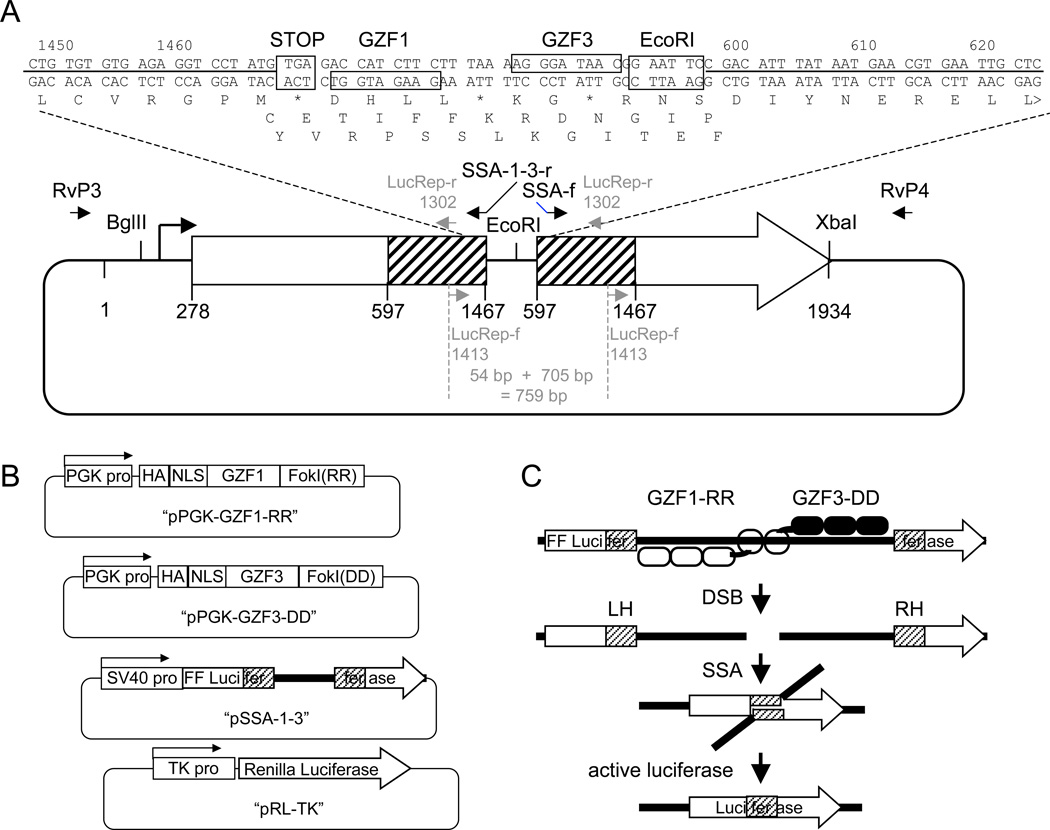
In the SSA recombination assay, a gene encoding firefly luciferase is divided into two segments, separated by a stop codon and a ZFN target site. Both segments contain an 870-bp region of homology in direct repeat orientation (see Fig. 2). A ZFN-induced DSB between the segments allows efficient SSA homologous recombination, resulting in an active luciferase gene. Firefly luciferase activity is therefore proportional to ZFN activity. The assay has several features that make it well suited as an initial test of ZFN activity. First, the assay is performed in the biologically-relevant environment of a human cell (or could be any easily transfected cell type). Adverse events, such as cytotoxicity due to off-target cleavages, will result in the death of transfected cells. Loss of firefly luciferase signal due to toxicity-dependant cell death can be distinguished from poor ZFN activity by the inclusion of a Renilla luciferase control plasmid. Second, there are many special challenges to the success of a ZFN at an endogenous target site, including limited chromatin accessibility, competing endogenous binding factors, and unexpected local DNA structures. If ZFN activity is not observed at an endogenous site, knowing that the ZFN was functional in an SSA assay will help to differentiate complications at the endogenous site from problems with the ZFN. Third, because it is difficult to anticipate the challenges at the endogenous site, a pragmatic approach would be to create 5–10 ZFNs that target different sites in the region of interest. The SSA assay can be easily reconfigured with a new ZFN target site with a single oligonucleotide. Finally, ZFNs are composed of a heterodimer of two ZFP-FokI cleavage domain fusion proteins ((31), and see Fig. 1B). Often only one monomer of the ZFN pair can be the cause of heterodimer failure. It is therefore important that the functional assay identify potential underperforming monomers, which can be easily done by configuring the SSA assay with homodimer ZFN target sites. Once identified, improvements can be directed towards the deficient monomer, such as replacing poorly-binding fingers with stronger variants or adding additional fingers.
Fig. 2.
The SSA assay. (A) A schematic diagram of the SSA reporter plasmid. The sequence at the junction site between the homologous repeats is shown at the top. The GZF1 and GZF3 heterodimer target site is indicated. PCR primers used in construction are shown as black arrows on the plasmid diagram below. PCR primers used for amplification of the junction region and sequencing are shown in gray. (B) Schamatics of the four plasmids that will be transfected into the test cells for the SSA assay. (C) The expressed ZFN pair will bind at the target site and create a double strand break (DSB). The break will be efficiently repaired by the SSA pathway to reconstruct an active firefly luciferase reporter gene.
2. Materials
2.1 The construction of ZFNs by MA
The list of Barbas zinc finger modules (see Table 1).
The list of Sangamo zinc finger modules (see Table 2).
The list of ToolGen zinc finger modules (see Tables 3 and 4).
pPGK-GZF1-wt ZFN-expression plasmid: this plasmid contains a phosphoglycerate kinase promoter driving expression of a gene encoding an HA-tag (YPYDVPDYA), the nuclear localization signal of SV40 large T antigen (PKKKRKV), a three-finger ZFP (GZF1), a 6-aa linker (TGAAAR), and a wildtype FokI endonuclease catalytic domain (32). The expressed ZFN is active as a homodimer. All non-commercial plasmids are available from the authors upon request.
pPGK-GZF1-RR ZFN-expression plasmid: This plasmid is similar to pPGK-GZF1-wt, except the FokI cleavage domain contains a modification that prevents activity as a homodimer. The expressed ZFN is only active as a heterodimer with a DD-modified ZFN (i.e., GZF3-DD; see below).
pPGK-GZF3-DD ZFN-expression plasmid: This plasmid is similar to pPGK-GZF3-wt, except the FokI cleavage domain contains a modification that prevents activity as a homodimer. The expressed ZFN is only active as a heterodimer with an RR-modified ZFN (i.e., GZF1-RR; see above).
- Oligonucleotide primers for sequencing the pPGK ZFN expression plasmid:
- HAtag-f : 5’-ATGTACCCATACGATGTCCCAGACTACG-3’
- FokLinkSeq-r: 5’-TCTATCCTGAGTGGAATTTCTGGC-3’
Standard equipment and reagents for agarose gel electrophoresis.
Restriction endonucleases and appropriate buffers: XholI and NotI.
T4 DNA ligase.
2.2 The SSA recombination assay
2.2.1 Preparation of the SSA reporter plasmid
- Oligonucleotide primers for SSA reporter plasmid construction:
- RvP3-f: 5’-CTAGCAAAATAGGCTGTCCC-3’
- SSA-1-3-r: 5’-CTC GAATTC C – [GTT ATC CCT TTTAAA GAA GAT GGT]-CTCACATAGGACCTCTCACACACAG –3’
- Additional custom SSA-r oligonucleotides, in which the desired ZFN target site replaces the bracketed sequence in SSA-1-3-r.
- Oligonucleotide primers for sequencing the SSA reporter plasmid:
- LucRep-f: 5’-CGAAGGTTGTGGATCTGGATACC-3’
- LucRep-r: 5’-TAGCTGATGTAGTCTCAGTGAGC-3’
pGL3-Control plasmid: This plasmid expresses full-length firefly luciferase under the control of a SV40 promoter and enhancer (Invitrogen). It serves as the template for construction of the SSA reporter plasmid, and can be used as a positive control.
pSSA-1-3 reporter plasmid: This plasmid contains the target site for the ZFN target site for the heterodimer of GZF1 and GZF3, flanked by a split firefly luciferase reporter gene. This is the exemplary plasmid that will be constructed in Method 3.2.1, but it is also used for the construction of new SSA reporter plasmids. This and all other non-commercial plasmid vectors are available from the investigator upon request.
2× Taq-Pro Complete polymerase mix with dNTPs and 2.0 mM MgCl2 (Denville Scientific, Cat. CB4070-4). Thaw the 3-mL solution on ice, aliquot 250-µL volumes into 1.5-µL tubes, and store at −20°C.
Standard equipment and reagents for agarose gel electrophoresis.
PCR purification kit (e.g., QIAquick PCR purification kit, Qiagen).
Restriction endonucleases and appropriate buffers: BglII and EcoRI.
T4 DNA ligase.
2.2.2 Performing the SSA assay
Appropriate ZFN expression plasmids and SSA reporter plasmids (see Note 1).
pPGK-empty vector: This plasmid is similar to pPGK-GZF1-wt but lacks the ZFP and FokI domains. It can be used as a negative control.
pRL-TK vector: This plasmid expresses Renilla luciferase under control of a HSVthymidine kinase promoter. It can be used to assess cytotoxicity.
24-well polystyrene tissue culture plates.
2× Poly-d-Lysine: Poly-d-Lysine (Poly-K; MP Biomedicals, Cat. 102694) is supplied as a 10 mg dessicant that is resuspended in 50 mL of filtered autoclaved water. This 2× stock should be stored at 4°C. Fresh 1× Poly-K is made by diluting the 2× stock in filtered autoclaved water.
HEK 293-T cells (ATCC, Cat. CRL-11268).
75-cm2 tissue culture flasks.
Phosphate Buffered Saline, sterile for tissue culture (e.g., Dulbecco’s PBS, Invitrogen, Cat. 14190).
Trypsin-EDTA (Invitrogen, Cat. 25300-054).
Pen-Strep (Invitrogen, Cat. 15140-122), contains 100 U/mL of penicillin and 100 µg/mL streptomycin. 5-mL aliquots are stored at −20°C.
Bovine Calf Serum (JR Scientific, Cat. WBJR050). 50-mL aliquots are stored at −20°C.
DMEM Complete Medium: DMEM medium (Invitrogen, Cat. 11995) is supplemented with 5 mL of Pen-Strep and 50 mL of Bovine Calf Serum (10% final). This mixture should be stored at 4°C.
Hemocytometer.
OPTI-MEM reduced serum medium (Invitrogen, Cat. 31985). This should be stored at 4°C.
Lipofectamine2000 (1 mg/mL solution, Invitrogen, Cat. 11668-019). This should be stored at 4°C.
1× Reporter Lysis Buffer. Dilute the 5× stock solution (Promega, Cat. E397A) with filtered autoclaved water to make a 1× working solution. The 1× solution can be stored at room temperature.
25× Protease Inhibitor Cocktail: Suspend one Complete Protease Inhibitor Cocktail Tablet without EDTA (Roche, Cat. 11873580001) in 2 mL of filtered autoclaved water. Prepare small aliquots and store at −20°C.
Bright-Glo Luciferase Assay System (Promega, Cat. E2620). Aliquot in 15-mL tubes and store at −80°C. This substrate is light-sensitive, keep it wrapped in aluminum foil.
Renilla Luciferase Assay System (Promega, Cat. E2820). Aliquot in 15-mL tubes and store at −80°C. This substrate is light-sensitive, keep it wrapped in aluminum foil.
Opaque white 96-well plates (Denville, Cat. P9725).
Microplate luminometer (e.g., Veritas Microplate Luminometer, Turner Biosystems).
3. Methods
3.1 The construction of ZFNs by MA
Find 5–10 appropriate target sites within your DNA region of interest (see Note 2).
Verify that the target sites are reasonably unique (not more than 1–2 mismatches) in the target genome. Usually, this can be achieved using BLAST (see Note 3).
Design the ZFP coding region based on the information in Tables 1–4 and Fig. 1. For the Barbas and Sangamo module sets, only the DNA-contacting regions corresponding to positions −1, 1, 2, 3, 4, 5, and 6 of the α-helix are inserted into a Sp1C (see Fig. 1C) or Zif268 (see Fig. 1D) three-finger scaffold. It is not clear if either scaffold has any advantage compared to the other. The ToolGen modules listed in Table 4 are connected with TGEKP linkers to create a finger-specific scaffold (see Fig. 1E and Note 4).
The coding regions can be physically constructed by commercial synthesis (see Note 5). Typically this is rapid and relatively inexpensive, especially for new users. In addition, the codons can be optimized to the target organism. Be sure to request that the DNA sequences are appended to the protein coding region as indicated in Fig. 1F to facilitate cloning into the pPGK-based ZFN expression plasmids (see Fig. 1G).
Digest both the ZFP coding region and a pPGK-based ZFN expression vector with XhoI and NotI. Subclone the ZFP into the expression vector. To create obligate heterodimer ZFN pairs, one ZFP will replace GZF1 in pPGK-GZF1-RR and the other will replace GZF3 in pPGK-GZF3-DD. For homodimer ZFNs, each ZFP will replace GZF1 in pPGK-GZF1-wt.
Verify the new constructs by sequencing using primers HAtag-f and/or FokLinkSeq-r.
3.2 The SSA recombination assay
3.2.1 Preparation of the SSA reporter plasmid
- Prepare the left arm of the split luciferase gene in a 50-µL PCR reaction consisting of the following components (see Fig. 2 and Note 6):
- 19 µL of H2O
- 1.0 µL of 10 ng/µL pGL3-Control template
- 2.5 µL of 5 µM RvP3 primer
- 2.5 µL of 5 µM SSA-1-3-r primer (or other SSA-r primer with a desired ZFN site)
- 25 µL of 2× TaqPro polymerase mix
Check for correct amplification by analyzing 10 µL of the reaction using agarose gel electrophoresis. Purify the amplicon using a PCR purification kit.
Digest both the amplicon and pSSA-1-3 using BglII and EcoRI (see Note 7).
Ligate the amplicon into the pSSA vector using T4 DNA ligase.
- Because the duplicated region also makes sequencing of the final plasmid difficult, the junction of the repeated region (containing the ZFN target site) should be first amplified in the following 50-µL PCR reaction:
- 19 µL of H2O
- 1.0 µL of 10 ng/µL pSSA reporter plasmid
- 2.5 µL of 5 µM LucRep-f primer
- 2.5 µL of 5 µM LucRep-r primer
- 25 µL of 2× TaqPro polymerase mix
Check for correct amplification by analyzing 10 µL of the reaction using agarose gel electrophoresis. Purify the amplicon using a PCR purification kit.
The sequence of the newly constructed SSA reporter plasmid can then be verified by sequencing the PCR amplicon using LucRep-f as the sequencing primer.
3.2.2 Performing the SSA assay
Day 0: Seed the cells in the 24-well plates
-
1
Pretreat the assay tissue culture plates with Poly-K to help loosely-adherent cells such as HEK 293-T to remain bound during washing procedures. Add 300 µL of 1× Poly-K to each well of a 24-well plate. Allow the Poly-K solution to incubate at room temperature (RT) for 30 min. Steps 1–19 should be performed in a biosafety cabinet to maintain sterility and biosafety.
-
2
Prepare the HEK 293-T cells to seed on 24-well plates. Aspirate the medium from the cells growing in a 75-cm2 flask.
-
3
Wash the cells with 5 mL of PBS. Swirl the PBS around to wash away remaining medium, then aspirate the PBS.
-
4
Add 2 mL of Trypsin-EDTA and swirl the flask to ensure the entire surface is covered. Incubate at RT for 1 min.
-
5
Resuspend the cells using 8 mL of DMEM Complete Medium.
-
6
Place 15 µL of the above suspension onto a hemocytometer. Count the number of cells using an inverted microscope. After use, decontaminate the hemocytometer in 10% final bleach solution.
-
7
Aspirate the 1× Poly-K solution from the 24-well plate.
-
8
Add 100,000 – 150,000 cells to each well.
-
9
Incubate for 24 h in a 37°C CO2 incubator before transfection.
Day 1: Transfection
-
10
To prepare the DNA for transfection, add an appropriate amount of DNA (25 ng of Renilla reporter plasmid, 25 ng of SSA reporter plasmid, and 100 ng of each ZFN plasmid) to 250 µL of OPTI-MEM in a 1.5-mL tube, and incubate at RT for 5 min.
-
11
Add 2 µL of Lipofectamine2000 to 250 µL of OPTI-MEM in a separate 1.5-mL tube incubate at RT for 5 min (see Note 8).
-
12
Combine the two and mix gently. Incubate at RT for 20 min.
-
13
To transfect the cells, remove the medium from cells in the 24-well plate from the previous day. Add the DNA-Lipofectmine2000-OPTI-MEM solution to wells.
-
14
Incubate for 4–5 h in a 37°C incubator.
-
15
Add 1 mL of DMEM Complete Medium.
-
16
Incubate the cells in a 37°C incubator, then allow 24 h for ZFN expression and activity (see Note 9).
Day 2: Harvesting
-
17
To preparing the cell lysates, remove the media from wells.
-
18
Wash twice with 0.5 mL of PBS. Carefully aspirate the PBS.
-
19
Supplement the 1× Reporter Lysis Buffer with 1× Protease Inhibitor Cocktail (4 µL of the 25× Protease Inhibitor Cocktail stock for every 100 µL of 1× Reporter Lysis Buffer). Add 100 µL of supplemented Reporter Lysis Buffer to each well of the 24-well plate.
-
20
Allow to incubate on ice for 5 min.
-
21
Harvest the cell lysates by scraping the cells from the bottom of the well with pipette tip. Keep the 24-well plate on ice to prevent any degradation of proteins. Wear appropriate personal protective equipment, such as a lab coat and eye goggles.
-
22
Transfer the lysates to 1.5-mL tubes on ice.
-
23
Centrifuge the tubes at 20,817 × g for 5 min at 4°C to pellet cellular debris. The supernatants can be used directly in the luciferase assays (see Note 10).
-
24
Thaw the Bright-Glo solution. This luciferase substrate is light sensitive so keep it wrapped in aluminum foil.
-
25
For each sample, pipette 20 µL of cell lysate into a well of an opaque white 96-well plate. The opaque walls ensure that the luminescence generated by one sample does not interfere with measurements of the neighboring samples.
-
26
Add 100 µL/well of Bright-Glo Luciferase Assay solution and pipette to mix. Cover the plate with aluminum foil.
-
28
Read the plate using a microplate luminometer. Integrate the signal of each sample for 1 s.
-
29
To read the Renilla luciferase signal, pipette another 20 µL of cell lysate into unused wells of the opaque white 96-well plate. Add 100 µL/well of Renilla Luciferase Assay solution, mix, and read using the microplate luminometer (see Note 11).
-
30
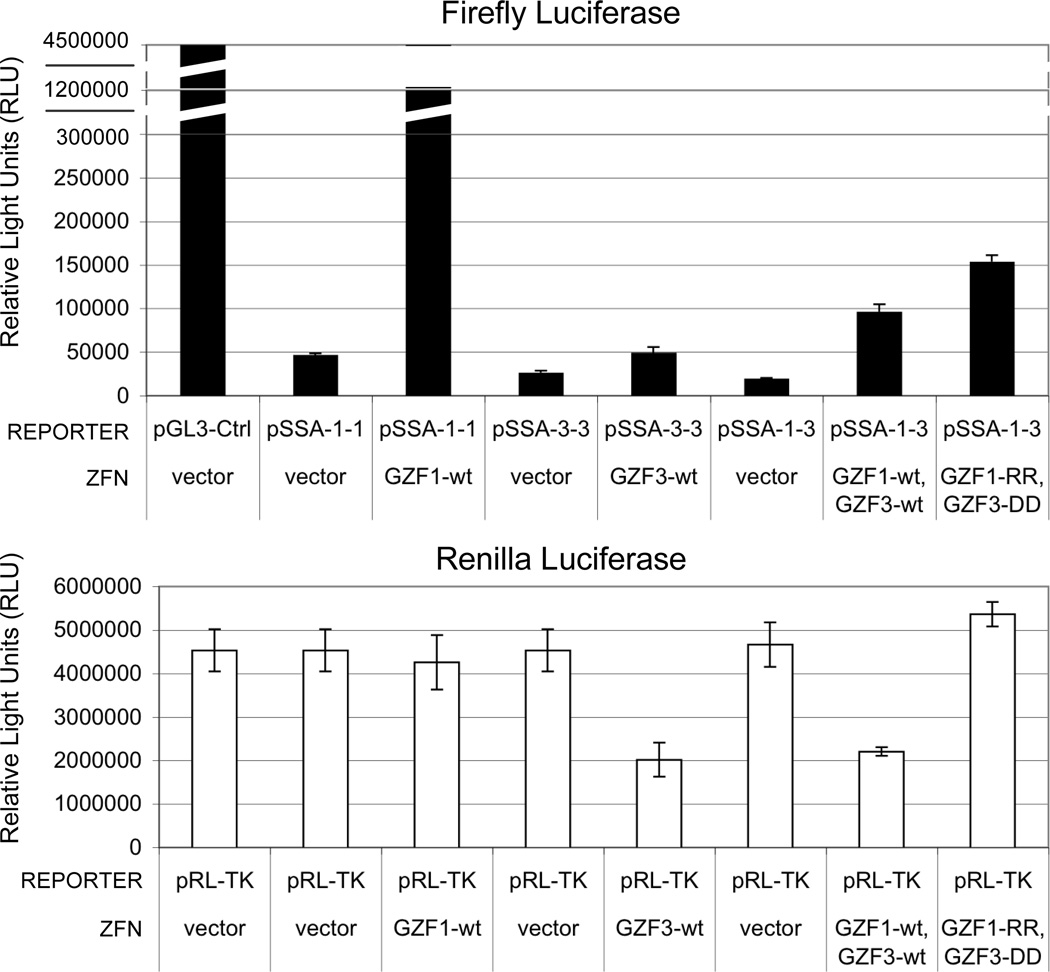
Calculate and plot the average and standard deviation of the firefly and Renilla luciferase signals for each set of duplicates or triplicates (see Fig. 3). The standard deviation of the replicates will be an indication of well-to-well variation, which should be small. The gain in firefly luciferase signal is an indicator of ZFN activity. The loss of Renilla luciferase signal is an indicator of ZFN toxicity, due to significant off-target cleavage activity (typically leading to apoptosis of the transfected cells). Note that a low firefly luciferase signal may be due either to low ZFN activity or high ZFN toxicity. A good example is the ZFN GZF3-wt (see Fig. 3). The presence of this monomer, either as a GZF3-wt homodimer or as a GZF1-wt/GZF3-wt heterodimer, leads to significant toxicity (reduced Renilla signal) and a corresponding apparent loss of ZFN activity (reduced firefly signal). In fact, the nuclease is active, but is causing the transfected cells to die. This conclusion is supported by the obligate heterodimer modifications, GZF1-RR/GZF3-DD. The GZF3 monomer is no longer active by itself, and the heterodimer pair now displays reduced toxicity and increased activity.
Fig. 3.
Exemplary data from an SSA assay. Firefly (SSA recombination due to ZFN activity) and Renilla (constitutively high in the absence of ZFN toxicity) luciferase data is shown for some of the experiments described in Note 1, including the analysis of GZF1-wt and GZF3-wt homodimers, as well as GZF1-wt/GZF3-wt and GZF1-RR/GZF3-DD heterodimers. Luciferase activity was measured 24 h post-transfection. Error bars indicate the standard deviation of triplicate experiments.
Acknowledgments
This chapter is based upon work supported by grant CA103651 from the National Cancer Institute, NIH (DJS).
Footnotes
- pPGK-GZF1-RR, pPGK-GZF3-DD, pSSA-1-3. This set can serve as a standard and should be included routinely in every assay.
- pPGK-empty vector, pSSA-1-3. In the absence of a ZFN-induced double strand break, the reporter plasmid should not produce a luciferase signal. However, mechanical damage to the plasmid, such as occasionally occurs during the preparation of plasmid from bacteria, can cause strand breaks that enable to reporter to recombine in the absence of ZFN cleavage. For this reason, it is important to monitor the background signal of the reporter plasmid. For any preparations that produce a significant signal, we have found that treating the preparation with T4 DNA ligase often restores the integrity of the plasmid and reduces the signal to baseline.
- pPGK-empty vector, pGL3-Control. This sample produces a luciferase signal that would be equivalent to every molecule of SSA reporter plasmid recombining to yield an active luciferase gene immediately upon transfection. Since the efficiency and kinetics of recombination will always be less than this idyllic scenario, the pGL3-Control signal will always be higher than that of a ZFN/SSA reporter sample. The standard ZFN/SSA set described in a. above typically produces a signal that is typically 10–30% of the pGL3-control signal (somewhat less than 10% under the conditions used in Fig. 3).
- A new ZFN pair and the cognate new heterodimer SSA reporter plasmid. The ZFN pair can either have wild type cleavage domains or obligate heterodimer domains with DD and RR modifications (33). Obligate heterodimers generally reduce the cytotoxicity of the ZFN.
- pPGK-empty vector and the new heterodimer SSA reporter plasmid.
- Optionally, it may be useful to test the activity of each monomer of the ZFN pair, especially if the heterodimer activity is low. For testing homodimers, wild type cleavage domains should be used, since the use of obligate heterodimer versions of the domains will cause 2/3 of the homodimer pairs to be inactive. Therefore, testing of the homodimers may require additional subcloning steps for the ZFNs.
- Optionally, pPGK-empty vector and the new homodimer SSA reporter plasmid. Note that each ZFN pair consists of two monomers; therefore testing of the homodimers requires the construction of two new homodimer SSA reporters. h. For each sample a-g listed above, duplicate or triplicate assays should be performed on the same plate. 24-well plates are useful to accommodate the many samples required for an accurate assessment of ZFN activity.
Clearly there are many factors that should be considered in selecting a target site, most of which will be beyond the scope of this methods article. If the intended target is within a living cell, it is often advisable to seek regions with accessible chromatin. In principle, accessibility can be determined experimentally (e.g., in vivo DNaseI footprinting) or extrapolated from existing genomic data (e.g., the UCSC Genome Browser). Targeting regions near actively-transcribed genes is often useful. Depending on the application, chromatin-remodeling drugs can be used to increase chromatin accessibility temporarily (34). However, the most important strategy to ensure a successful outcome is to choose 5–10 different target sites.
The composition of the fingers in the ZFPs is also a critical parameter. Our understanding of the relationship between affinity and the number of fingers in a ZFP is still evolving. Generally, three fingers are the minimum required for biologically relevant binding. Recent studies suggest that fingers with greater relative affinity improve the functional capabilities of three-finger ZFPs (24). The conventional wisdom is that ZFP composed of GNN-binding fingers tend to perform better; however, it is important to remember that not all GNN-binding fingers have high relative affinity. Increasing the number of fingers may be a useful strategy to increase the overall affinity of the ZFP, especially (as is common) if lower affinity fingers are required to bind the target site. In principle, more fingers also provide greater specificity. Even a three-finger ZFP with perfect specificity for a 9-bp site will nominally have about 22,000 perfect binding sites in the human genome. A six-finger ZFP with perfect specificity for an 18-bp site would have 0–1 perfect sites. However, the caveat for multi-finger proteins is that subgroups of three or more fingers may have sufficient affinity by themselves to cause biologically relevant binding (35, 36). If this occurs, the ZFP will use different binding modes to recognize numerous and diverse binding sites. The subgroup phenomenon will be exacerbated by using an excess of fingers with high relative affinity, such as many GNN-binding fingers. The goal should therefore be to create ZFPs with “appropriate” affinity, using fewer but stronger binding fingers, or more but weaker binding fingers.
Some guidance is available to help predict performance. The relative affinities for some Barbas and ToolGen fingers have been measured (see Tables 1 and 4). ToolGen has identified a number of Barbas and ToolGen fingers that occurred frequently in successful ZFNs (see Tables 1 and 3). Also, since G and A bases often form stronger bonds to their contacting amino acids than C and T, a simple count of the purines (including any TSO interactions that are present) can serve as a rough guide to affinity. Two web-based tools are also available. The site http://www.zincfingertools.org/ is run by the Barbas lab and allows easy searching and coding of the Barbas set of modules (37). The site http://bindr.gdcb.iastate.edu/ZiFiT/ is run by the ZFC (38). It allows searching of Barbas, Sangamo, and ToolGen modules. The output of the search is not protein sequence but rather a set of catalog numbers to their library of pre-cloned fingers, which are available from the internet distribution service http://addgene.org/. Both sites provide voluminous predictive data, which in most cases will have similar accuracy to the guidance outlined here. Unfortunately, due to the many parameters and significant weaknesses in our understanding, our ability to predict the behaviors of assembled ZFPs is limited. A robust assay, such as the SSA assay described in this chapter (see Method 3.2), may be the most useful approach to functionally determine the optimal length and composition of the ZFP.
Finally, ZFNs have special target site considerations. The two monomers of the ZFN pair must bind in a tail-to-tail format, such that their C-termini (containing the cleavage domains) face each other. The optimal spacing between the monomer binding sites is 5, 6, or 7 bp. Maximum ZFN activity for a 5, 6, or 7-bp spacer may require different peptide linkers between the last zinc finger and the FokI cleavage domain. The relationship of spacers and linkers is best described in (39). The ZFN constructs described in this chapter are designed to provide maximal activity at target sites with a 6-bp spacer.
Unfortunately, BLAST does not work very well for ZFN target sites, because the 6-bp spacer between the two monomer binding sites can have any sequence without affecting cleavage efficiency. A custom search algorithm would be more appropriate. A C-based program for searching any downloaded genome is available from the author upon request.
Pay very close attention to the order of the fingers with respect to the DNA (see Fig. 1). The most 5’ 3-bp subsite of the DNA sequence to be recognized is bound by the most Cterm finger of the ZFP. The most 3’ 3-bp subsite is bound by the most N-terminal finger (i.e., Finger 1). Failure to correctly order the fingers in this “anti-parallel” arrangement is the most common technical error when designing ZFPs.
For commercial synthesis, we use a Canadian company called BioBasic. At the time of this writing, the cost of synthesis is $0.50/bp and deliver typically takes 3–6 weeks. Of course there are many other assembly methods. Chapters 7 and 12 of this volume describe two oligonucleotide-based assembly methods, and several others have been described (40–42). The ZFC has constructed a library of single fingers cloned into plasmids, and has published detailed methods for their assembly (43). The entire library of 141 fingers is available at a modest cost from Addgene.
It is often difficult to clone small DNA fragments, such as such as a 24-bp ZFN target site. Therefore, we usually amplify the whole “left arm” of the split luciferase gene with the ZFN target site appended, then subclone the amplicon into the SSA reporter vector. The forward primer anneals upstream of the BglII site (RvP3, see Fig. 2). The reverse primer anneals around nucleotide 1467 (in the numbering of the pGL-Control plasmid), and additional contains the ZFN target sequence followed by an EcoRI site (SSA-1-3-r). Since this primer anneals to both parts of the duplicated region, this PCR reaction must use the original pGL3-Control as a template (i.e., not a SSA plasmid).
As an alternative to ligating the “left arm” into an existing pSSA vector, a “right arm” can be prepared, and the left and right arms can be ligated into a digested pGL3-Control plasmid in a three-part ligation. The right arm is prepared by performing a PCR reaction similar to that described in Method 3.2.1.1, using primers SSA-f (5’-GAGGAG GAATTC CGACATTTATAATGAACGTGAATTGCTC-3’) and RvP4-r (5’-GACGATAGTCATGCCCCGCGC-3’) (see Fig. 2). The right arm is digested with EcoRI and XbaI, and a pGL3-Control plasmid is digested with BglII and XbaI.
For HEK 293-T cells, we normally transfect with 1–2 µL of Lipofectamine2000 and approximately 250 ng of DNA when the cells are about 95% confluent. However, these parameters should be re-optimized for different cell types and conditions, as large variations in transfection efficiency can result.
Firefly luciferase signals are often higher after 48 h of incubation, probably due to an increased opportunity for ZFN-induced cleavage and an increased time to express the recombined luciferase.
Alternatively, the cell lysates can be transferred to a clean tube and stored at −80°C until further use. Thaw these lysates on ice prior to use.
Alternatively, the firefly and Renilla luciferase signals can be read sequentially from a single cell lysate sample using the Dual-Luciferase Reporter Assay System (Promega, Cat. E1960). However, this system requires a microplate luminometer with dual injection pumps.
References
- 1.Emerson RO, Thomas JH. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009;5:e1000325. doi: 10.1371/journal.pgen.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 Å. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 3.Segal DJ, Barbas CF., III Custom DNA-binding proteins come of age: polydactyl zinc-finger proteins. Curr. Opin. Biotech. 2001;12:632–637. doi: 10.1016/s0958-1669(01)00272-5. [DOI] [PubMed] [Google Scholar]
- 4.Dreier B, Beerli RR, Segal DJ, Flippin JD, Barbas CF., III Development of zinc finger domains for recognition of the 5'-ANN-3' family of DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem. 2001;276:29466–29478. doi: 10.1074/jbc.M102604200. [DOI] [PubMed] [Google Scholar]
- 5.Dreier B, Fuller RP, Segal DJ, Lund CV, Blancafort P, Huber A, Koksch B, Barbas CF., 3rd Development of zinc finger domains for recognition of the 5'-CNN-3' family DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem. 2005;280:35588–35597. doi: 10.1074/jbc.M506654200. [DOI] [PubMed] [Google Scholar]
- 6.Liu Q, Xia Z, Zhong X, Case CC. Validated zinc finger protein designs for all 16 GNN DNA triplet targets. J Biol Chem. 2002;277:3850–3856. doi: 10.1074/jbc.M110669200. [DOI] [PubMed] [Google Scholar]
- 7.Segal DJ, Dreier B, Beerli RR, Barbas CF., III Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5'-GNN-3' DNA target sequences. Proc Natl Acad Sci U S A. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bae KH, Kwon YD, Shin HC, Hwang MS, Ryu EH, Park KS, Yang HY, Lee DK, Lee Y, Park J, Kwon HS, Kim HW, Yeh BI, Lee HW, Sohn SH, Yoon J, Seol W, Kim JS. Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat Biotechnol. 2003;21:275–280. doi: 10.1038/nbt796. [DOI] [PubMed] [Google Scholar]
- 9.Liu Q, Segal DJ, Ghiara JB, Barbas CF., III Design of polydactyl zincfinger proteins for unique addressing within complex genomes. Proc Natl Acad Sci U S A. 1997;94:5525–5530. doi: 10.1073/pnas.94.11.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, Cathomen T, Voytas DF, Joung JK. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, Unger-Wallace E, Sander JD, Muller-Lerch F, Fu F, Pearlberg J, Gobel C, Dassie JP, Pruett- Miller SM, Porteus MH, Sgroi DC, Iafrate AJ, Dobbs D, McCray PB, Jr, Cathomen T, Voytas DF, Joung JK. Rapid "open-source" engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isalan M, Choo Y, Klug A. Synergy between adjacent zinc fingers in sequence-specific DNA recognition. Proc Natl Acad Sci U S A. 1997;94:5617–5621. doi: 10.1073/pnas.94.11.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isalan M, Klug A, Choo Y. A rapid, generally applicable method to engineer zinc fingers illustrated by targeting the HIV-1 promoter. Nat Biotechnol. 2001;19:656–660. doi: 10.1038/90264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 15.Greisman HA, Pabo CO. A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- 16.Joung JK, Ramm EI, Pabo CO. A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions. Proc Natl Acad Sci U S A. 2000;97:7382–7387. doi: 10.1073/pnas.110149297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carroll D. Progress and prospects: Zinc-finger nucleases as gene therapy agents. Gene Ther. 2008;15:1463–1468. doi: 10.1038/gt.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolb AF, Coates CJ, Kaminski JM, Summers JB, Miller AD, Segal DJ. Site-directed genome modification: nucleic acid and protein modules for targeted integration and gene correction. Trends Biotechnol. 2005;23:399–406. doi: 10.1016/j.tibtech.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 19.Gordley RM, Gersbach CA, Barbas CF., 3rd Synthesis of programmable integrases. Proc Natl Acad Sci U S A. 2009;106:5053–5058. doi: 10.1073/pnas.0812502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camenisch TD, Brilliant MH, Segal DJ. Critical parameters for genome editing using zinc finger nucleases. Mini Rev Med Chem. 2008;8:669–676. doi: 10.2174/138955708784567458. [DOI] [PubMed] [Google Scholar]
- 21.Sera T. Zinc-finger-based artificial transcription factors and their applications. Adv Drug Deliv Rev. 2009;61:513–526. doi: 10.1016/j.addr.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 22.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price SA, Dent C, Duran-Jimenez B, Liang Y, Zhang L, Rebar EJ, Case CC, Gregory PD, Martin TJ, Spratt SK, Tomlinson DR. Gene transfer of an engineered transcription factor promoting expression of VEGF-A protects against experimental diabetic neuropathy. Diabetes. 2006;55:1847–1854. doi: 10.2337/db05-1060. [DOI] [PubMed] [Google Scholar]
- 24.Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. An affinity-based scoring scheme for predicting DNA-binding activities of modularly assembled zinc-finger proteins. Nucleic Acids Res. 2009;37:506–515. doi: 10.1093/nar/gkn962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cathomen T, Joung JK. Zinc-finger nucleases: the next generation emerges. Mol Ther. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- 26.Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, Holmes MC, Guschin D, Waite A, Miller JC, Rebar EJ, Gregory PD, Klug A, Collingwood TN. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci U S A. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minczuk M, Papworth MA, Miller JC, Murphy MP, Klug A. Development of a single-chain, quasi-dimeric zinc-finger nuclease for the selective degradation of mutated human mitochondrial DNA. Nucleic Acids Res. 2008;36:3926–3938. doi: 10.1093/nar/gkn313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segal DJ, Beerli RR, Blancafort P, Dreier B, Effertz K, Huber A, Koksch B, Lund CV, Magnenat L, Valente D, Barbas CF., III Evaluation of a modular strategy for the construction of novel polydactyl zinc finger DNA-binding proteins. Biochemistry. 2003;42:2137–2148. doi: 10.1021/bi026806o. [DOI] [PubMed] [Google Scholar]
- 30.Zykovich A, Korf I, Segal DJ. Bind-n-Seq: high-throughput analysis of in vitro protein-DNA interactions using massively parallel sequencing. Nuc Acids Res. 2009 doi: 10.1093/nar/gkp802. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol. 2001;21:289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alwin S, Gere MB, Guhl E, Effertz K, Barbas CF, 3rd, Segal DJ, Weitzman MD, Cathomen T. Custom Zinc-Finger Nucleases for Use in Human Cells. Mol Ther. 2005;12:610–617. doi: 10.1016/j.ymthe.2005.06.094. [DOI] [PubMed] [Google Scholar]
- 33.Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 34.Beltran AS, Sun X, Lizardi PM, Blancafort P. Reprogramming epigenetic silencing: artificial transcription factors synergize with chromatin remodeling drugs to reactivate the tumor suppressor mammary serine protease inhibitor. Mol Cancer Ther. 2008;7:1080–1090. doi: 10.1158/1535-7163.MCT-07-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imanishi M, Nakamura A, Morisaki T, Futaki S. Positive and negative cooperativity of modularly assembled zinc fingers. Biochem Biophys Res Commun. 2009;387:440–443. doi: 10.1016/j.bbrc.2009.07.059. [DOI] [PubMed] [Google Scholar]
- 37.Mandell JG, Barbas CF., 3rd Zinc Finger Tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34:W516–W523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. Zinc Finger Targeter (ZiFiT): an engineered zinc finger/target site design tool. Nucleic Acids Res. 2007;35:W599–w605. doi: 10.1093/nar/gkm349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Handel EM, Alwin S, Cathomen T. Expanding or restricting the target site repertoire of zinc-finger nucleases: the inter-domain linker as a major determinant of target site selectivity. Mol Ther. 2009;17:104–111. doi: 10.1038/mt.2008.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cathomen T, Segal DJ, Brondani V, Muller-Lerch F. Generation and functional analysis of zinc finger nucleases. Methods Mol Biol. 2008;434:277–290. doi: 10.1007/978-1-60327-248-3_17. [DOI] [PubMed] [Google Scholar]
- 41.Carroll D, Morton JJ, Beumer KJ, Segal DJ. Design, construction and in vitro testing of zinc finger nucleases. Nat Protoc. 2006;1:1329–1341. doi: 10.1038/nprot.2006.231. [DOI] [PubMed] [Google Scholar]
- 42.Mani M, Kandavelou K, Dy FJ, Durai S, Chandrasegaran S. Design, engineering, and characterization of zinc finger nucleases. Biochem Biophys Res Commun. 2005;335:447–457. doi: 10.1016/j.bbrc.2005.07.089. [DOI] [PubMed] [Google Scholar]
- 43.Wright DA, Thibodeau-Beganny S, Sander JD, Winfrey RJ, Hirsh AS, Eichtinger M, Fu F, Porteus MH, Dobbs D, Voytas DF, Joung JK. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat Protoc. 2006;1:1637–1652. doi: 10.1038/nprot.2006.259. [DOI] [PubMed] [Google Scholar]
- 44.Kim JS, Kwon YD, Kim H, Ryu EH, Hwang MS. Zinc finger domains and methods of identifying same. US2007087371. US Patent. 2007