Abstract
Pancreatic ductal adenocarcinoma (PDAC) is the fourth and fifth leading cause of cancer death for each gender in developed countries. With lack of effective treatment and screening scheme available for the general population, the mortality rate is expected to increase over the next several decades in contrast to the other major malignancies such as lung, breast, prostate and colorectal cancers. Endoscopic ultrasound, with its highest level of detection capacity of smaller pancreatic lesions, is the commonly employed and preferred clinical imaging-based PDAC detection method. Various molecular biomarkers have been investigated for characterization of the disease, but none are shown to be useful or validated for clinical utilization for early detection. As seen from studies of a small subset of familial or genetically high-risk PDAC groups, the higher yield and utility of imaging-based screening methods are demonstrated for these groups. Multiple recent studies on the unique cancer metabolism including PDAC, demonstrate the potential for utility of the metabolites as the discriminant markers for this disease. In order to generate an early PDAC detection screening strategy available for a wider population, we propose to expand the population of higher risk PDAC group with combination clinical and metabolomics parameters.
Keywords: Pancreatic cancer, Endoscopic ultrasound, Metabolomics, Early detection, Biomarkers
Core tip: This is a summary of current pancreatic cancer cohort early detection studies and a potential approach being considered for future application. This is an area that requires heightened efforts as lack of effective treatment and screening scheme for wider population is leading this particular disease to be the second lethal cancer by 2030.
INTRODUCTION
Currently, pancreatic ductal adenocarcinoma (PDAC) is the fourth major cause of cancer mortality in the United States[1]. It is predicted that 46420 new cases and 39590 deaths would result from pancreatic cancer in the United States in 2014[2]. Worldwide, there were 277668 new cases and 266029 deaths from this cancer in 2008[3]. In comparison to other major malignancies such as breast, colon, lung and prostate cancers with their respective 89%, 64%, 16%, 99% 5-year survival rate, PDAC at 6% is conspicuously low[2]. For PDAC, the only curative option is surgical resection, which is applicable in only 10%-15% of patients due to the common discovery of late stage at diagnosis[4]. In fact, PDAC is notorious for late stage discovery as evidenced by the low percentage of localized disease at diagnosis, compared to other malignancies: breast (61%), colon (40%), lung (16%), ovarian (19%), prostate (91%), and pancreatic cancer (7%)[5]. With the existing effective screening methods, the decreasing trends of cancer death rate are seen in major malignancies such as breast, prostate and colorectal cancer. In contrast, it is estimated that PDAC is expected to be surfacing as the second leading cause of cancer death by 2030[6].
With the distinct contribution of late-stage discovery and general lack of effective medical therapy, a critical approach in reversing the poor outcome of pancreatic cancer is to develop an early detection scheme for the tumor. In support of this, we see the trend that despite the poor prognosis of the disease, for those who have undergone curative resection with negative margins, the 5-year survival rate is 22% in contrast to 2% for the advanced-stage with distant metastasis[7,8]. An earlier diagnosis with tumor less than 2 cm (T1) is associated with a better 5-year survival of 58% compared to 17% for stage IIB PDAC[9]. Ariyama et al[10] reported complete survival of 79 patients with less than 1 cm tumors after surgical resection. Furthermore, as a recent report indicates, the estimated time from the transformation to pre-metastatic growths of pancreatic cancer is approximately 15 years[11]; there is a wide potential window of opportunity to apply developing technologies in early detection of this cancer.
In this article, we will review the recent studies on the PDAC early detection approaches and ongoing research endeavors in developing early detection schemes for this devastating disease, with specific attention to application of combined clinical and metabolomics parameters.
PDAC EARLY DETECTION AND DIAGNOSIS - IMAGING-BASED TESTS
Over the past few decades, endoscopic ultrasound (EUS) has proven itself to be a superior imaging study for detection of a small or early-stage pancreatic neoplasm as compared to other modalities such as transabdominal ultrasound, computed tomography (CT), magnetic resonance imaging (MRI), positron emission tomography scans and angiography[12-14]. Yasuda et al[15] and Rösch et al[16] had initially demonstrated the superiority of EUS in detection of small pancreatic lesions. More recently, DeWitt et al[17] had verified the superiority of EUS as compared to multi-detector CT scan. In another study, Khashab et al[18] demonstrated that the sensitivity of EUS in detecting a pancreatic mass was significantly greater than that of CT images, and particularly for pancreatic neuroendocrine tumors, which commonly consist of smaller lesions. In addition, EUS detected CT-negative tumors in more than 90% of the cases. As an additional diagnostic modality, EUS-guided fine needle aspiration (FNA) provides success rates of 90%-95%, with an overall sensitivity and specificity of 85%-90% and 98%-99%, respectively[19-22]. Thus, the utility and the advantage of EUS enable visualization and targeting of small pancreatic masses. Lesions of 5 mm or less could be visualized and sampled, which might not have been accessible or identifiable by other imaging modalities[23].
DIAGNOSTIC MOLECULAR MARKERS AND PANCREATIC CANCER
In order to enhance the diagnostic accuracy of PDAC, molecular markers on EUS-FNA samples have been evaluated in recent years. Utilities of DNA mutations and loss of heterozygosity are being reported as potential surrogate markers of the cancer[24,25]. In a recent study, Takahashi et al[26] assessed k-ras point mutations in PDAC and chronic focal pancreatitis samples obtained by EUS-FNA[27,28]. The study revealed the presence of point mutations of k-ras in 74% of patients with PDAC compared to no mutations in chronic focal pancreatitis. In another study, Tada et al[29] reported a high k-ras gene mutation rate in 20 of 26 cases of EUS-FNA specimens (77%) and in 12 of 19 cases of pancreatic juice (63%) in PDAC. However, the presence of k-ras mutations in a benign condition such as chronic pancreatitis and premalignant lesions such as intraductal papillary mucinous neoplasm (IPMN) in addition to lack of such mutations in 20% of PDAC limit the usage of this test solely as a diagnostic or a detection tool. Other studies analyzing p53 by immunohistochemistry[30], telomerase activity with a ribonucleoprotein enzyme[31], and a broad panel of microsatellite allele loss markers demonstrated similar results[32]. In the presence of inconclusive EUS-FNA cytology, molecular markers could potentially complement EUS-FNA cytology results to help establish the diagnosis of malignancy.
SELECT POPULATION-BASED RESEARCH FOR EARLY DETECTION SCHEME DEVELOPMENT
PDAC screening in high-risk individuals
Currently, a general population-screening program for PDAC is not cost-effective because of low relative disease incidence and non-availability of simple, cheap, highly accurate non-invasive tests. The main goal of the screening is to identify clinically significant precursor or early stage PDAC. However, since overwhelming majority of premalignant and small PDAC lesions is asymptomatic, we do not have a definite surrogate marker to identify a subset population for screening. Consequently, as one of the approaches in investigating the risks, research has focused on identification of a subset of individuals with a higher-risk for PDAC development in order to elucidate the genetic predilection. Up to 10% of PDAC patients have a familial/genetic basis and they have increased risk of developing both pancreatic and extra-pancreatic malignancies[33-37]. Classic categorization of high-risk patients are based on the highly associated genetic risks defined as those who have significant family history of the cancer or have an inherited PDAC syndrome with a known genetic abnormality (Table 1).
Table 1.
Pancreatic ductal adenocarcinoma related genetic syndromes
| Syndrome | Inheritance | Gene mutation | Risk of PDAC |
| Peutz-Jeghers syndrome[38] | AD | STK11/LKB1 | SIR = 132 |
| Hereditary pancreatitis[39-41] | AD | PRSS1 | OR = 69.9 |
| SPINK1 | |||
| Familial atypical multiple mole melanoma syndrome[42-44] | AD | CDKN2A | SIR = 13-38 |
| Hereditary breast-ovarian cancer syndrome[45-51] | AD | BRCA2 | BRCA2: OR = 3.5-10-fold increased risk |
| BRCA1 | BRCA1: OR = 2.26 times average population | ||
| Lynch syndrome[52] | AD | MLH1, MSH2, MSH6 or PMS2 | SIR = up to 8.6 |
| Cystic fibrosis[53] | Autosomal recessive | CFTR | OR = 5.3-6.6 |
AD: Autosomal dominant; PDAC: Pancreatic ductal adenocarcinoma; SIR: Standardized incidence ratio.
Familial pancreatic cancer: Familial pancreatic cancer (FPC) cohort (cancer in two or more first-degree relatives (FDRs) or in three or more affected family members - including one first-degree relative) is considered a high-risk and a candidate for screening program[47,54,55]. Currently, the genetic foundation for FPC is not fully understood. Various investigations have demonstrated the presence of a germline mutation in the BRCA2 gene[47-49], association of BRCA1[46,56], paladin gene mutation[57] as well as other genes such as apolipoprotein A4, CEA, keratin 19, stratifin, trefoil factor, and S100A6[58,59] in FPC, and more recently identification of PALB2[60], as a pancreatic cancer susceptibility gene. These facts suggest that multiple and heterogeneous factors are likely at play for the genesis of PDAC in this subset.
Analysis of the PDAC kindred data from Johns Hopkins’ National Familial Pancreas Tumor Registry has shown that the risk of PDAC in individuals with two afflicted FDRs is 6.4% and the lifetime risk is 8%-12%; for persons with three afflicted FDRs, the relative and lifetime risks for PDAC increase to 32% and 16%-32%, respectively[36]. Brune et al[61] in their recent article reported a higher risk of PDAC among FPC kindred with a younger onset (age less than 50). Rulyak et al[62] in another study found smoking as a significant risk factor in FPC cohort, especially among males and those under age 50. This factor increases the cancer risk by 2.0-3.7 times and lowers the onset age by 10 years. A risk assessment software tool, PancPRO, has been generated and is available for calculating the risk for individuals with familial pancreatic cancer (http://www4.utsouthwestern.edu/breasthealth/cagene/default.asp)[35].
Screening modalities and the current screening programs: Many of the screening programs have used additional investigational biomarker to complement imaging tests to identify the early lesions. A commonly used marker, CA19-9, is neither sensitive nor specific independently for early PDAC or precursor detection. Kim et al[63] in their study found only 0.9% positive-predictive value using the standard cut-off value (37 U/mL). Other biomarkers investigated recently include MIC-1, CEACAM-1, SPan1, DUPAN, Alpha4GNT, and PAM4, but none is validated for routine clinical use[64]. In another approach, elevated fasting-glucose level has been demonstrated to be associated with sporadic PDAC[65] and is currently utilized by an European registry in high-risk people with mutational analysis of pancreatic juice along with p16 promoter methylation status.
Several international screening programs exist for PDAC in high-risk individuals. “Cancer of the Pancreas Screening Study” (CAPS study), led by John Hopkins University, is the largest screening program that involves 24 American Centers of Excellence. To date, three studies, CAPS 1, CAPS 2 and CAPS 3, have been completed (Table 2).
Table 2.
Results of screening programs for pancreatic cancer in high-risk groups n (%)
| Study | CAPS1 | CAPS2 | CAPS3 | U of Washington | FaPaCa | Dutch Study |
| Diagnostic yield | 2 (5.3) | 8 (10) | 92 (43) | 10 (13) | 1 (1.3) | 10 (23) |
Positive finding of abnormal imaging such as mass (solid, cyst) or abnormal duct in the cases. CAPS: Cancer of the Pancreas Screening Study; FaPaCa: Familial Pancreatic Cancer Study.
In the CAPS 1, thirty-eight patients including 31 from a kindred with > 3 affected with PDAC, 6 with 2 affected relatives, and 1 patient with Peutz-Jeghers syndrome (PJS) were studied. Screening protocol with EUS revealed six pancreatic masses: 1 invasive PDAC, 1 benign IPMN, 2 serous cystadenomas, and 2 non-neoplastic lesions. The yield of screening was 5.3% in this study[66]. In the CAPS 2, seventy-eight high-risk patients were studied[67]. In 8 patients, the screening found pancreatic neoplasia, confirmed by surgery or FNA: 6 benign IPMNs, 1 IPMN that progressed to invasive PDAC, and another had high-grade pancreatic intraepithelial neoplasia (PanIN-3). The CAPS 3 was a multicenter prospective cohort study involving annual EUS, MRI screening with assays of DNA and protein markers in serum and pancreatic juice. Over 200 patients were enrolled over a three-year period. The study results on the detection modality comparison demonstrated that the EUS has the highest rate of detection of early neoplastic changes in up to 42.6% of the asymptomatic high-risk group[68].
In another study from the University of Washington, high-risk familial cohorts were screened with EUS, beginning 10 years prior to the earliest index PDAC case. If EUS was normal, they underwent a repeat EUS in 2-3 years. With abnormal EUS findings, they were referred for ERCP and if abnormalities were noted, patients were offered surgical intervention[69]. Among 75 screened subjects, 15 had gone to surgery for abnormal EUS and ERCP findings. All surgical cases revealed premalignant lesions: PanIN-3 in 10 and PanIN-2 in five cases[70]. The study gave a yield of 13% (10 out of 75) for detecting PanIN-3 lesions. A single patient developed unresectable PDAC during the surveillance.
In a German PDAC screening program (FaPaCa), 76 patients were followed using annual EUS, MRCP, and laboratory assays (CDKN2a and BRCA2 genetic analysis, CA19-9 and CEA). Any appreciable lesion was evaluated with EUS. With an abnormal finding, the patient underwent surgical exploration and if malignancy was detected, total pancreatectomy was performed. In 10 cases, lesions were seen on EUS as compared to only seven detected by MR scan. Out of the seven MRCP-positive cases, six underwent resections and the histology showed one PanIN-3, one PanIN-2, one PanIN-1, and three with other benign lesions. This resulted in a diagnostic yield of 1.3% for PanIN-3 detection[71]. Another study from the Netherlands in 44 high-risk subjects demonstrated a 7% detection rate for asymptomatic PDAC and a 16% for premalignant lesions[72].
The International CAPS Consortium have recently met and reported a suggested guideline for current PDAC screening based on the risk[73]. A consensus (≥ 75% agreement by the participants) was reached that the following groups should be offered screening (only to individuals who are surgical candidate): (1) FDRs of the cancer patients from a familial pancreatic cancer cohort with at least two affected FDRs; (2) patients with Peutz-Jeghers syndrome; and (3) p16, BRCA2 and hereditary non-polyposis colorectal cancer mutation carriers with at least single affected FDR. The initial screening should include EUS and/or MRI. However, consensus was not reached on the beginning and the end age of screening/surveillance and the interval of the examination. Their conclusions also included requirement for further studies, and the clinical management should occur at high-volume centers with multidisciplinary teams.
FUTURE OF PANCREATIC CANCER SCREENING
Current screening programs have demonstrated that the EUS evaluation can detect premalignant lesions and early cancers in certain small subset of high-risk groups. However, as the overwhelming majority of PDAC cases involve patients who develop the disease sporadically without a recognized genetic abnormality, the application of this modality for PDAC detection screening is very limited for the general adult population.
Select population based approach
Identification of a higher-PDAC-risk group: As the prevalence of PDAC in the general United States population over the age 55 is approximately 68 per 100000, a candidate discriminant test with a specificity of 98% and a sensitivity of 100% would generate 1999 false-positive test results and 68 true-positives[74]. Thus, relying on a single determinant for distinguishing the PDAC early-stage cases from the general population would necessitate a highly accurate test with a specificity of greater than 99%. More practical approach, then, would be to begin with a subset of population with a higher prevalence, and in conjunction with novel surrogate markers to curtail the at-risk subset, we could begin to identify the group with significantly increased PDAC risk for whom the endoscopic/imaging-based screening strategy could be applied.
An initial approach in selection of the screening population is to utilize selective clinical parameters that could be used to curtail the subset of the general population at increased PDAC risk. For instance, based on the epidemiological evidence, such clinical parameters include hyperglycemia or diabetes, which are noted in 50%-80% of pancreatic cancer patients[75-79]. Though not encompassing all PDAC patients, this subset includes a much larger proportion of PDAC patients for whom we may select further for screening. Similarly, patients with a history of chronic pancreatitis or obesity are reported to have increased PDAC risk during their lifetime[80-85]. Recent findings from molecular biology and animal studies investigating effects of diet-induced obesity in a PDAC mouse model demonstrated increased occurrence of pancreatic inflammation and accelerated pancreatic neoplastic changes, supporting the association of obesity and pancreatic inflammation and PDAC risks[86,87]. Considering that millions are being diagnosed with diabetes or glucose intolerance, chronic pancreatitis, or obesity annually in comparison to PDAC, however, further refinement of the screening patient group is critically needed to justify for developing a larger scale screening protocol.
Translational research - application of metabolomics approach
Initially established as a key methodology in the field of inborn metabolic errors and toxicology, metabolomics have developed over the years to examine a much wider array of low-molecular-weight products or intermediates within the biological state of a cell, tissue, organ, or organism. A metabolome represents a physiological readout of the biochemical state in an individual’s body compartment, and provides the functional terminal signals of the genome and proteome, reflecting more closely the current phenotypic state of an individual in response to the environmental stimuli[88]. Thus, metabolomics data has considerable potential in elucidating cancer-development risks, with its additional capacity for providing temporal molecular information to the ongoing changes originating from genetic PDAC risk alone.
With the recent advancement in the technology and resumed interest in the cancer-associated metabolic abnormality[89,90], application of metabolomics in the cancer field has attracted more attention[91]. Cancer-related metabolic reprogramming, Warburg effect, has been known since nearly a century ago in association with various solid tumors including PDAC[92], as cancer cells undergo energetically inefficient glycolysis even in the presence of oxygen in the environment (aerobic glycolysis)[93]. A number of common cancer mutations including Akt1, HIF (hypoxia-inducible factor), and p53 have been shown to support the Warburg effect through glycolysis and down-regulation of metabolite flux through the Krebs cycle[94-101]. In PDAC, increased phosphorylation or activation of Akt1 has also been reported (illuminating on the importance of enzyme functionality)[102] as well as involvement of HIF1 in the tumor growth via effects on glycolytic process[103,104] and membrane-bound glycoprotein (MUC17) regulation[105] - reflective of activation of metabolic pathways. Further evidences of loss-of-function genetic mutations in key mitochondrial metabolic enzymes such as succinate dehydrogenase and fumarate hydratase, isocitrate dehydrogenase, phosphoglycerate dehydrogenase support carcinogenesis and the Warburg effect[106-110]. Other important alternative pathways in cancer metabolism such as glutaminolysis and pyruvate kinase isoform suppression have been shown to accumulate respective upstream intermediates and reduction of associated end products such as NADPH, ribose-5-phosphate and nucleic acids[111-116]. As such, various groups have reported metabolomics biomarker applications for different cancers[117,118].
As a major organ involved in metabolic regulation in a healthy individual, pancreatic disorder such as malignancy is anticipated to influence the normal metabolism, presenting further rationale and interest in elucidating the implication of malignant transformation and PDAC development. Proteomic analysis of the pancreatic cancer cells demonstrated alteration in proteins involved in metabolic pathways including increased expression of glycolytic and reduced Krebs cycle enzymes, and accumulation of key proteins involved in glutamine metabolism, in support of Warburg effect. These in turn play significant role in nucleotide and amino acid biosynthesis required for sustaining the proliferating cancer cells[119]. Applications of sensitive mass spectrometric techniques in metabolomics study of PDAC detection biomarkers have led to identification of a set of small molecules or metabolites (or biochemical intermediates) that are potent discriminants of developing PDAC and the controls (See Figure 1 as an example of metabolomics based analysis, allowing segregation of PDAC from benign cases). Recent reports from our group as well as others have demonstrated that specific candidate metabolites consisting of amino acids, bile acids, and a number of lipids and fatty acids - suspected to be reflective of tumor proliferation as well as many systemic response yet to be determined - were identified as potential discriminant for blood-based PDAC biomarkers[120-123]. As a further supporting data, elucidation of lipids and fatty acids as discriminant factors from PDAC and benign lesions from the cancer tissue and adjacent normal tissue has been reported recently[124].
Figure 1.
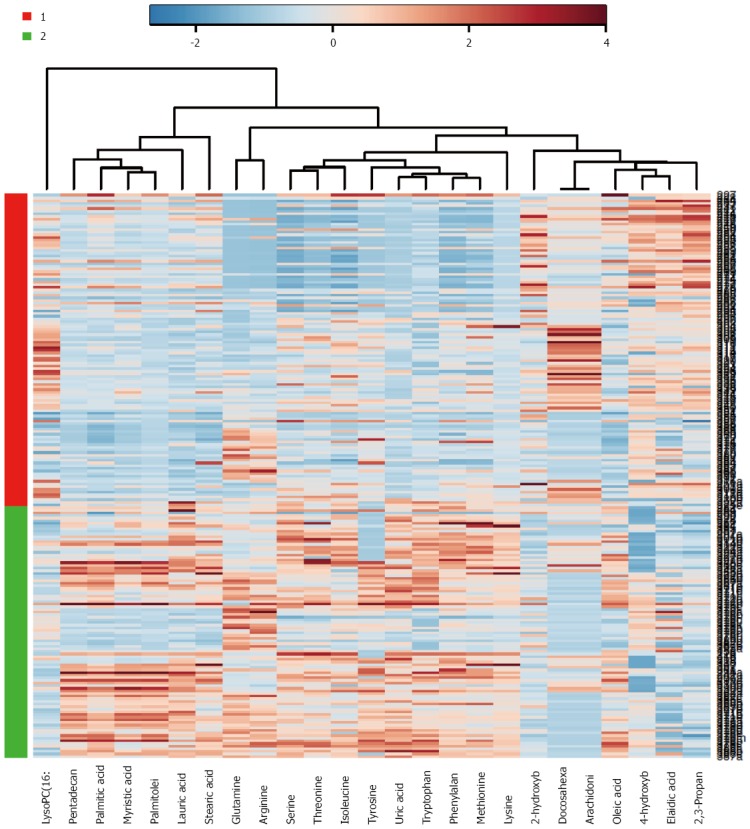
Example of metabolomics based analysis, allowing segregation of pancreatic ductal adenocarcinoma from benign cases. Heat map illustration of discriminant capability of a metabolite set derived from gas chromatography and liquid chromatography/mass spectrometry plasma metabolomics dataset comparing pancreatic ductal adenocarcinoma patients (Red or group 1; n = 110) and benign pancreas (includes benign cysts, chronic pancreatitis, and normal pancreas) (Green or group 2; n = 90). Metabolites are plotted on x-axis, and the cases on the y-axis. Blue color indicates data points with a value smaller than the median of the respective metabolite and the red indicates higher values. This candidate set of metabolites enabled the segregation of the two groups.
By virtue of simultaneously depicting the multiple metabolite levels, metabolomics approach reveals various biochemical pathways that are uniquely involved in malignant conditions and has led to findings such as abnormalities of glycine and its mitochondrial biosynthetic pathway, as a potential therapeutic target in certain cancers[125]. Moreover, in combination with other systems biology approaches such as transcriptomics and proteomics, further refinement in characterization of cancer development and therapeutic targets as well as identification of potential biomarkers could be realized for PDAC. Since many enzymes in a metabolic network determine metabolites’ level and nonlinear quantitative relationship from the genes to the proteome and metabolome levels exist, a metabolome cannot be easily decomposed to a specific single marker, which will designate the cancer state[126]. Thus, in order to delineate a pathological state such as PDAC, multiple metabolomic features might be required for accurate depiction of a developing cancer. Future studies are anticipated to incorporate cancer systems’ biological knowledge, including metabolomics, for optimal designation of PDAC biomarkers, which would be utilized in conjunction with a clinical-parameter-derived population subset for establishing the PDAC screening population. Subsequently, further validation studies for the PDAC biomarkers need to be performed.
CONCLUSION
Current imaging-based detection and diagnostic methods for PDAC is effectively providing answers to clinical questions raised for patients with signs or symptoms of suspected pancreatic lesions. However, the endoscopic/imaging-based screening schemes are currently limited in applications to early PDAC detection in asymptomatic patients, aside from a small group of known genetically high-risk groups. There is a high demand for developing a method of selecting distinct subsets among the general population for implementing the endoscopic/imaging screening test effectively. Application of combinations of clinical risk parameters/factors with the developing molecular biomarkers from translational science such as metabolomics analysis brings hopes of providing us with early PDAC detection markers, and developing effective early detection screening scheme for the patients in the near future.
Footnotes
Conflict-of-interest: The author has no conflict of interest related to the manuscript.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: July 8, 2014
First decision: August 15, 2014
Article in press: January 8, 2015
P- Reviewer: Falconi M, Gangl A, Hayano K S- Editor: Ma YJ L- Editor: A E- Editor: Ma S
References
- 1.National Cancer Institute. Cancer topics: Pancreatic cancer. Available from: http://www.cancer.gov/cancertopics/types/pancreatic.
- 2.American Cancer Society. Cancer Facts & Figures, 2014. Available from: http://www.cancer.org/research/cancerfactsstatistics/cancerfactsfigures2014/index.
- 3.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 4.Beger HG, Rau B, Gansauge F, Poch B, Link KH. Treatment of pancreatic cancer: challenge of the facts. World J Surg. 2003;27:1075–1084. doi: 10.1007/s00268-003-7165-7. [DOI] [PubMed] [Google Scholar]
- 5.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 6.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 7.Ahmad NA, Lewis JD, Ginsberg GG, Haller DG, Morris JB, Williams NN, Rosato EF, Kochman ML. Long term survival after pancreatic resection for pancreatic adenocarcinoma. Am J Gastroenterol. 2001;96:2609–2615. doi: 10.1111/j.1572-0241.2001.04123.x. [DOI] [PubMed] [Google Scholar]
- 8.Howard TJ, Krug JE, Yu J, Zyromski NJ, Schmidt CM, Jacobson LE, Madura JA, Wiebke EA, Lillemoe KD. A margin-negative R0 resection accomplished with minimal postoperative complications is the surgeon’s contribution to long-term survival in pancreatic cancer. J Gastrointest Surg. 2006;10:1338–1345; discussion 1345-1346. doi: 10.1016/j.gassur.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Egawa S, Takeda K, Fukuyama S, Motoi F, Sunamura M, Matsuno S. Clinicopathological aspects of small pancreatic cancer. Pancreas. 2004;28:235–240. doi: 10.1097/00006676-200404000-00004. [DOI] [PubMed] [Google Scholar]
- 10.Ariyama J, Suyama M, Ogawa K, Ikari T. [Screening of pancreatic neoplasms and the diagnostic rate of small pancreatic neoplasms] Nihon Rinsho. 1986;44:1729–1734. [PubMed] [Google Scholar]
- 11.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rösch T, Braig C, Gain T, Feuerbach S, Siewert JR, Schusdziarra V, Classen M. Staging of pancreatic and ampullary carcinoma by endoscopic ultrasonography. Comparison with conventional sonography, computed tomography, and angiography. Gastroenterology. 1992;102:188–199. doi: 10.1016/0016-5085(92)91800-j. [DOI] [PubMed] [Google Scholar]
- 13.Müller MF, Meyenberger C, Bertschinger P, Schaer R, Marincek B. Pancreatic tumors: evaluation with endoscopic US, CT, and MR imaging. Radiology. 1994;190:745–751. doi: 10.1148/radiology.190.3.8115622. [DOI] [PubMed] [Google Scholar]
- 14.Agarwal B, Abu-Hamda E, Molke KL, Correa AM, Ho L. Endoscopic ultrasound-guided fine needle aspiration and multidetector spiral CT in the diagnosis of pancreatic cancer. Am J Gastroenterol. 2004;99:844–850. doi: 10.1111/j.1572-0241.2004.04177.x. [DOI] [PubMed] [Google Scholar]
- 15.Yasuda K, Mukai H, Fujimoto S, Nakajima M, Kawai K. The diagnosis of pancreatic cancer by endoscopic ultrasonography. Gastrointest Endosc. 1988;34:1–8. doi: 10.1016/s0016-5107(88)71220-1. [DOI] [PubMed] [Google Scholar]
- 16.Rösch T, Lorenz R, Braig C, Feuerbach S, Siewert JR, Schusdziarra V, Classen M. Endoscopic ultrasound in pancreatic tumor diagnosis. Gastrointest Endosc. 1991;37:347–352. doi: 10.1016/s0016-5107(91)70729-3. [DOI] [PubMed] [Google Scholar]
- 17.DeWitt J, Devereaux B, Chriswell M, McGreevy K, Howard T, Imperiale TF, Ciaccia D, Lane KA, Maglinte D, Kopecky K, et al. Comparison of endoscopic ultrasonography and multidetector computed tomography for detecting and staging pancreatic cancer. Ann Intern Med. 2004;141:753–763. doi: 10.7326/0003-4819-141-10-200411160-00006. [DOI] [PubMed] [Google Scholar]
- 18.Khashab MA, Yong E, Lennon AM, Shin EJ, Amateau S, Hruban RH, Olino K, Giday S, Fishman EK, Wolfgang CL, et al. EUS is still superior to multidetector computerized tomography for detection of pancreatic neuroendocrine tumors. Gastrointest Endosc. 2011;73:691–696. doi: 10.1016/j.gie.2010.08.030. [DOI] [PubMed] [Google Scholar]
- 19.Vilmann P, Jacobsen GK, Henriksen FW, Hancke S. Endoscopic ultrasonography with guided fine needle aspiration biopsy in pancreatic disease. Gastrointest Endosc. 1992;38:172–173. doi: 10.1016/s0016-5107(92)70385-x. [DOI] [PubMed] [Google Scholar]
- 20.Hewitt MJ, McPhail MJ, Possamai L, Dhar A, Vlavianos P, Monahan KJ. EUS-guided FNA for diagnosis of solid pancreatic neoplasms: a meta-analysis. Gastrointest Endosc. 2012;75:319–331. doi: 10.1016/j.gie.2011.08.049. [DOI] [PubMed] [Google Scholar]
- 21.Hébert-Magee S, Bae S, Varadarajulu S, Ramesh J, Frost AR, Eloubeidi MA, Eltoum IA. The presence of a cytopathologist increases the diagnostic accuracy of endoscopic ultrasound-guided fine needle aspiration cytology for pancreatic adenocarcinoma: a meta-analysis. Cytopathology. 2013;24:159–171. doi: 10.1111/cyt.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luz LP, Al-Haddad MA, Sey MS, DeWitt JM. Applications of endoscopic ultrasound in pancreatic cancer. World J Gastroenterol. 2014;20:7808–7818. doi: 10.3748/wjg.v20.i24.7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fritscher-Ravens A, Izbicki JR, Sriram PV, Krause C, Knoefel WT, Topalidis T, Jaeckle S, Thonke F, Soehendra N. Endosonography-guided, fine-needle aspiration cytology extending the indication for organ-preserving pancreatic surgery. Am J Gastroenterol. 2000;95:2255–2260. doi: 10.1111/j.1572-0241.2000.02311.x. [DOI] [PubMed] [Google Scholar]
- 24.Pellisé M, Castells A, Ginès A, Solé M, Mora J, Castellví-Bel S, Rodríguez-Moranta F, Fernàndez-Esparrach G, Llach J, Bordas JM, et al. Clinical usefulness of KRAS mutational analysis in the diagnosis of pancreatic adenocarcinoma by means of endosonography-guided fine-needle aspiration biopsy. Aliment Pharmacol Ther. 2003;17:1299–1307. doi: 10.1046/j.1365-2036.2003.01579.x. [DOI] [PubMed] [Google Scholar]
- 25.Goggins M. Molecular markers of early pancreatic cancer. J Clin Oncol. 2005;23:4524–4531. doi: 10.1200/JCO.2005.19.711. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi K, Yamao K, Okubo K, Sawaki A, Mizuno N, Ashida R, Koshikawa T, Ueyama Y, Kasugai K, Hase S, et al. Differential diagnosis of pancreatic cancer and focal pancreatitis by using EUS-guided FNA. Gastrointest Endosc. 2005;61:76–79. doi: 10.1016/s0016-5107(04)02224-2. [DOI] [PubMed] [Google Scholar]
- 27.Maluf-Filho F, Kumar A, Gerhardt R, Kubrusly M, Sakai P, Hondo F, Matuguma SE, Artifon E, Monteiro da Cunha JE, César Machado MC, et al. Kras mutation analysis of fine needle aspirate under EUS guidance facilitates risk stratification of patients with pancreatic mass. J Clin Gastroenterol. 2007;41:906–910. doi: 10.1097/MCG.0b013e31805905e9. [DOI] [PubMed] [Google Scholar]
- 28.Bournet B, Souque A, Senesse P, Assenat E, Barthet M, Lesavre N, Aubert A, O’Toole D, Hammel P, Levy P, et al. Endoscopic ultrasound-guided fine-needle aspiration biopsy coupled with KRAS mutation assay to distinguish pancreatic cancer from pseudotumoral chronic pancreatitis. Endoscopy. 2009;41:552–557. doi: 10.1055/s-0029-1214717. [DOI] [PubMed] [Google Scholar]
- 29.Tada M, Komatsu Y, Kawabe T, Sasahira N, Isayama H, Toda N, Shiratori Y, Omata M. Quantitative analysis of K-ras gene mutation in pancreatic tissue obtained by endoscopic ultrasonography-guided fine needle aspiration: clinical utility for diagnosis of pancreatic tumor. Am J Gastroenterol. 2002;97:2263–2270. doi: 10.1111/j.1572-0241.2002.05980.x. [DOI] [PubMed] [Google Scholar]
- 30.Itoi T, Takei K, Sofuni A, Itokawa F, Tsuchiya T, Kurihara T, Nakamura K, Moriyasu F, Tsuchida A, Kasuya K. Immunohistochemical analysis of p53 and MIB-1 in tissue specimens obtained from endoscopic ultrasonography-guided fine needle aspiration biopsy for the diagnosis of solid pancreatic masses. Oncol Rep. 2005;13:229–234. [PubMed] [Google Scholar]
- 31.Mishra G, Zhao Y, Sweeney J, Pineau BC, Case D, Ho C, Blackstock AW, Geisinger K, Howerton R, Levine E, et al. Determination of qualitative telomerase activity as an adjunct to the diagnosis of pancreatic adenocarcinoma by EUS-guided fine-needle aspiration. Gastrointest Endosc. 2006;63:648–654. doi: 10.1016/j.gie.2005.11.056. [DOI] [PubMed] [Google Scholar]
- 32.Salek C, Benesova L, Zavoral M, Nosek V, Kasperova L, Ryska M, Strnad R, Traboulsi E, Minarik M. Evaluation of clinical relevance of examining K-ras, p16 and p53 mutations along with allelic losses at 9p and 18q in EUS-guided fine needle aspiration samples of patients with chronic pancreatitis and pancreatic cancer. World J Gastroenterol. 2007;13:3714–3720. doi: 10.3748/wjg.v13.i27.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi C, Hruban RH, Klein AP. Familial pancreatic cancer. Arch Pathol Lab Med. 2009;133:365–374. doi: 10.5858/133.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Permuth-Wey J, Egan KM. Family history is a significant risk factor for pancreatic cancer: results from a systematic review and meta-analysis. Fam Cancer. 2009;8:109–117. doi: 10.1007/s10689-008-9214-8. [DOI] [PubMed] [Google Scholar]
- 35.Wang W, Chen S, Brune KA, Hruban RH, Parmigiani G, Klein AP. PancPRO: risk assessment for individuals with a family history of pancreatic cancer. J Clin Oncol. 2007;25:1417–1422. doi: 10.1200/JCO.2006.09.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein AP, Brune KA, Petersen GM, Goggins M, Tersmette AC, Offerhaus GJ, Griffin C, Cameron JL, Yeo CJ, Kern S, et al. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res. 2004;64:2634–2638. doi: 10.1158/0008-5472.can-03-3823. [DOI] [PubMed] [Google Scholar]
- 37.Wang L, Brune KA, Visvanathan K, Laheru D, Herman J, Wolfgang C, Schulick R, Cameron JL, Goggins M, Hruban RH, et al. Elevated cancer mortality in the relatives of patients with pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:2829–2834. doi: 10.1158/1055-9965.EPI-09-0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giardiello FM, Brensinger JD, Tersmette AC, Goodman SN, Petersen GM, Booker SV, Cruz-Correa M, Offerhaus JA. Very high risk of cancer in familial Peutz-Jeghers syndrome. Gastroenterology. 2000;119:1447–1453. doi: 10.1053/gast.2000.20228. [DOI] [PubMed] [Google Scholar]
- 39.Rebours V, Boutron-Ruault MC, Schnee M, Férec C, Maire F, Hammel P, Ruszniewski P, Lévy P. Risk of pancreatic adenocarcinoma in patients with hereditary pancreatitis: a national exhaustive series. Am J Gastroenterol. 2008;103:111–119. doi: 10.1111/j.1572-0241.2007.01597.x. [DOI] [PubMed] [Google Scholar]
- 40.Lowenfels AB, Maisonneuve P, DiMagno EP, Elitsur Y, Gates LK, Perrault J, Whitcomb DC. Hereditary pancreatitis and the risk of pancreatic cancer. International Hereditary Pancreatitis Study Group. J Natl Cancer Inst. 1997;89:442–446. doi: 10.1093/jnci/89.6.442. [DOI] [PubMed] [Google Scholar]
- 41.Raimondi S, Lowenfels AB, Morselli-Labate AM, Maisonneuve P, Pezzilli R. Pancreatic cancer in chronic pancreatitis; aetiology, incidence, and early detection. Best Pract Res Clin Gastroenterol. 2010;24:349–358. doi: 10.1016/j.bpg.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 42.Goldstein AM, Fraser MC, Struewing JP, Hussussian CJ, Ranade K, Zametkin DP, Fontaine LS, Organic SM, Dracopoli NC, Clark WH. Increased risk of pancreatic cancer in melanoma-prone kindreds with p16INK4 mutations. N Engl J Med. 1995;333:970–974. doi: 10.1056/NEJM199510123331504. [DOI] [PubMed] [Google Scholar]
- 43.Whelan AJ, Bartsch D, Goodfellow PJ. Brief report: a familial syndrome of pancreatic cancer and melanoma with a mutation in the CDKN2 tumor-suppressor gene. N Engl J Med. 1995;333:975–977. doi: 10.1056/NEJM199510123331505. [DOI] [PubMed] [Google Scholar]
- 44.Klein AP. Genetic susceptibility to pancreatic cancer. Mol Carcinog. 2012;51:14–24. doi: 10.1002/mc.20855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Asperen CJ, Brohet RM, Meijers-Heijboer EJ, Hoogerbrugge N, Verhoef S, Vasen HF, Ausems MG, Menko FH, Gomez Garcia EB, Klijn JG, et al. Cancer risks in BRCA2 families: estimates for sites other than breast and ovary. J Med Genet. 2005;42:711–719. doi: 10.1136/jmg.2004.028829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brose MS, Rebbeck TR, Calzone KA, Stopfer JE, Nathanson KL, Weber BL. Cancer risk estimates for BRCA1 mutation carriers identified in a risk evaluation program. J Natl Cancer Inst. 2002;94:1365–1372. doi: 10.1093/jnci/94.18.1365. [DOI] [PubMed] [Google Scholar]
- 47.Couch FJ, Johnson MR, Rabe KG, Brune K, de Andrade M, Goggins M, Rothenmund H, Gallinger S, Klein A, Petersen GM, et al. The prevalence of BRCA2 mutations in familial pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:342–346. doi: 10.1158/1055-9965.EPI-06-0783. [DOI] [PubMed] [Google Scholar]
- 48.Goggins M, Schutte M, Lu J, Moskaluk CA, Weinstein CL, Petersen GM, Yeo CJ, Jackson CE, Lynch HT, Hruban RH, et al. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360–5364. [PubMed] [Google Scholar]
- 49.Hahn SA, Greenhalf B, Ellis I, Sina-Frey M, Rieder H, Korte B, Gerdes B, Kress R, Ziegler A, Raeburn JA, et al. BRCA2 germline mutations in familial pancreatic carcinoma. J Natl Cancer Inst. 2003;95:214–221. doi: 10.1093/jnci/95.3.214. [DOI] [PubMed] [Google Scholar]
- 50.Lal G, Liu G, Schmocker B, Kaurah P, Ozcelik H, Narod SA, Redston M, Gallinger S. Inherited predisposition to pancreatic adenocarcinoma: role of family history and germ-line p16, BRCA1, and BRCA2 mutations. Cancer Res. 2000;60:409–416. [PubMed] [Google Scholar]
- 51.Murphy KM, Brune KA, Griffin C, Sollenberger JE, Petersen GM, Bansal R, Hruban RH, Kern SE. Evaluation of candidate genes MAP2K4, MADH4, ACVR1B, and BRCA2 in familial pancreatic cancer: deleterious BRCA2 mutations in 17% Cancer Res. 2002;62:3789–3793. [PubMed] [Google Scholar]
- 52.Kastrinos F, Mukherjee B, Tayob N, Wang F, Sparr J, Raymond VM, Bandipalliam P, Stoffel EM, Gruber SB, Syngal S. Risk of pancreatic cancer in families with Lynch syndrome. JAMA. 2009;302:1790–1795. doi: 10.1001/jama.2009.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McWilliams R, Highsmith WE, Rabe KG, de Andrade M, Tordsen LA, Holtegaard LM, Petersen GM. Cystic fibrosis transmembrane regulator gene carrier status is a risk factor for young onset pancreatic adenocarcinoma. Gut. 2005;54:1661–1662. doi: 10.1136/gut.2005.074534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greenhalf W, Grocock C, Harcus M, Neoptolemos J. Screening of high-risk families for pancreatic cancer. Pancreatology. 2009;9:215–222. doi: 10.1159/000210262. [DOI] [PubMed] [Google Scholar]
- 55.Brand RE, Lerch MM, Rubinstein WS, Neoptolemos JP, Whitcomb DC, Hruban RH, Brentnall TA, Lynch HT, Canto MI. Advances in counselling and surveillance of patients at risk for pancreatic cancer. Gut. 2007;56:1460–1469. doi: 10.1136/gut.2006.108456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lynch HT, Deters CA, Snyder CL, Lynch JF, Villeneuve P, Silberstein J, Martin H, Narod SA, Brand RE. BRCA1 and pancreatic cancer: pedigree findings and their causal relationships. Cancer Genet Cytogenet. 2005;158:119–125. doi: 10.1016/j.cancergencyto.2004.01.032. [DOI] [PubMed] [Google Scholar]
- 57.Pogue-Geile KL, Chen R, Bronner MP, Crnogorac-Jurcevic T, Moyes KW, Dowen S, Otey CA, Crispin DA, George RD, Whitcomb DC, et al. Palladin mutation causes familial pancreatic cancer and suggests a new cancer mechanism. PLoS Med. 2006;3:e516. doi: 10.1371/journal.pmed.0030516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Earl J, Yan L, Vitone LJ, Risk J, Kemp SJ, McFaul C, Neoptolemos JP, Greenhalf W, Kress R, Sina-Frey M, et al. Evaluation of the 4q32-34 locus in European familial pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:1948–1955. doi: 10.1158/1055-9965.EPI-06-0376. [DOI] [PubMed] [Google Scholar]
- 59.Zervos EE, Tanner SM, Osborne DA, Bloomston M, Rosemurgy AS, Ellison EC, Melvin WS, de la Chapelle A. Differential gene expression in patients genetically predisposed to pancreatic cancer. J Surg Res. 2006;135:317–322. doi: 10.1016/j.jss.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 60.Jones S, Hruban RH, Kamiyama M, Borges M, Zhang X, Parsons DW, Lin JC, Palmisano E, Brune K, Jaffee EM, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brune KA, Lau B, Palmisano E, Canto M, Goggins MG, Hruban RH, Klein AP. Importance of age of onset in pancreatic cancer kindreds. J Natl Cancer Inst. 2010;102:119–126. doi: 10.1093/jnci/djp466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rulyak SJ, Lowenfels AB, Maisonneuve P, Brentnall TA. Risk factors for the development of pancreatic cancer in familial pancreatic cancer kindreds. Gastroenterology. 2003;124:1292–1299. doi: 10.1016/s0016-5085(03)00272-5. [DOI] [PubMed] [Google Scholar]
- 63.Kim JE, Lee KT, Lee JK, Paik SW, Rhee JC, Choi KW. Clinical usefulness of carbohydrate antigen 19-9 as a screening test for pancreatic cancer in an asymptomatic population. J Gastroenterol Hepatol. 2004;19:182–186. doi: 10.1111/j.1440-1746.2004.03219.x. [DOI] [PubMed] [Google Scholar]
- 64.Bussom S, Saif MW. Methods and rationale for the early detection of pancreatic cancer. Highlights from the “2010 ASCO Gastrointestinal Cancers Symposium”. Orlando, FL, USA. January 22-24, 2010. JOP. 2010;11:128–130. [PubMed] [Google Scholar]
- 65.Stolzenberg-Solomon RZ, Graubard BI, Chari S, Limburg P, Taylor PR, Virtamo J, Albanes D. Insulin, glucose, insulin resistance, and pancreatic cancer in male smokers. JAMA. 2005;294:2872–2878. doi: 10.1001/jama.294.22.2872. [DOI] [PubMed] [Google Scholar]
- 66.Canto MI, Goggins M, Yeo CJ, Griffin C, Axilbund JE, Brune K, Ali SZ, Jagannath S, Petersen GM, Fishman EK, et al. Screening for pancreatic neoplasia in high-risk individuals: an EUS-based approach. Clin Gastroenterol Hepatol. 2004;2:606–621. doi: 10.1016/s1542-3565(04)00244-7. [DOI] [PubMed] [Google Scholar]
- 67.Canto MI, Goggins M, Hruban RH, Petersen GM, Giardiello FM, Yeo C, Fishman EK, Brune K, Axilbund J, Griffin C, et al. Screening for early pancreatic neoplasia in high-risk individuals: a prospective controlled study. Clin Gastroenterol Hepatol. 2006;4:766–81; quiz 665. doi: 10.1016/j.cgh.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 68.Canto MI, Hruban RH, Fishman EK, Kamel IR, Schulick R, Zhang Z, Topazian M, Takahashi N, Fletcher J, Petersen G, et al. Frequent detection of pancreatic lesions in asymptomatic high-risk individuals. Gastroenterology. 2012;142:796–804; quiz e14-5. doi: 10.1053/j.gastro.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kimmey MB, Bronner MP, Byrd DR, Brentnall TA. Screening and surveillance for hereditary pancreatic cancer. Gastrointest Endosc. 2002;56:S82–S86. doi: 10.1016/s0016-5107(02)70092-8. [DOI] [PubMed] [Google Scholar]
- 70.Carlson C, Greenhalf W, Brentnall TA. Screening of hereditary pancreatic cancer families. In: Berger HG, Bucher M, Kozarek R, Lerch M, Neoptolemos J, et al., editors. The Pancreas: An Integrated Textbook of Basic Science, Medicine and Surgery. Second Edition. Oxford UK: Wiley-Blackwell Publishing; 2008. pp. 636–642. [Google Scholar]
- 71.Langer P, Kann PH, Fendrich V, Habbe N, Schneider M, Sina M, Slater EP, Heverhagen JT, Gress TM, Rothmund M, et al. Five years of prospective screening of high-risk individuals from families with familial pancreatic cancer. Gut. 2009;58:1410–1418. doi: 10.1136/gut.2008.171611. [DOI] [PubMed] [Google Scholar]
- 72.Poley JW, Kluijt I, Gouma DJ, Harinck F, Wagner A, Aalfs C, van Eijck CH, Cats A, Kuipers EJ, Nio Y, et al. The yield of first-time endoscopic ultrasonography in screening individuals at a high risk of developing pancreatic cancer. Am J Gastroenterol. 2009;104:2175–2181. doi: 10.1038/ajg.2009.276. [DOI] [PubMed] [Google Scholar]
- 73.Canto MI, Harinck F, Hruban RH, Offerhaus GJ, Poley JW, Kamel I, Nio Y, Schulick RS, Bassi C, Kluijt I, et al. International Cancer of the Pancreas Screening (CAPS) Consortium summit on the management of patients with increased risk for familial pancreatic cancer. Gut. 2013;62:339–347. doi: 10.1136/gutjnl-2012-303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lennon AM, Wolfgang CL, Canto MI, Klein AP, Herman JM, Goggins M, Fishman EK, Kamel I, Weiss MJ, Diaz LA, et al. The early detection of pancreatic cancer: what will it take to diagnose and treat curable pancreatic neoplasia? Cancer Res. 2014;74:3381–3389. doi: 10.1158/0008-5472.CAN-14-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chari ST, Leibson CL, Rabe KG, Timmons LJ, Ransom J, de Andrade M, Petersen GM. Pancreatic cancer-associated diabetes mellitus: prevalence and temporal association with diagnosis of cancer. Gastroenterology. 2008;134:95–101. doi: 10.1053/j.gastro.2007.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chari ST, Leibson CL, Rabe KG, Ransom J, de Andrade M, Petersen GM. Probability of pancreatic cancer following diabetes: a population-based study. Gastroenterology. 2005;129:504–511. doi: 10.1053/j.gastro.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ogawa Y, Tanaka M, Inoue K, Yamaguchi K, Chijiiwa K, Mizumoto K, Tsutsu N, Nakamura Y. A prospective pancreatographic study of the prevalence of pancreatic carcinoma in patients with diabetes mellitus. Cancer. 2002;94:2344–2349. doi: 10.1002/cncr.10493. [DOI] [PubMed] [Google Scholar]
- 78.Pannala R, Basu A, Petersen GM, Chari ST. New-onset diabetes: a potential clue to the early diagnosis of pancreatic cancer. Lancet Oncol. 2009;10:88–95. doi: 10.1016/S1470-2045(08)70337-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang F, Herrington M, Larsson J, Permert J. The relationship between diabetes and pancreatic cancer. Mol Cancer. 2003;2:4. doi: 10.1186/1476-4598-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lowenfels AB, Maisonneuve P, Cavallini G, Ammann RW, Lankisch PG, Andersen JR, Dimagno EP, Andrén-Sandberg A, Domellöf L. Pancreatitis and the risk of pancreatic cancer. International Pancreatitis Study Group. N Engl J Med. 1993;328:1433–1437. doi: 10.1056/NEJM199305203282001. [DOI] [PubMed] [Google Scholar]
- 81.Arslan AA, Helzlsouer KJ, Kooperberg C, Shu XO, Steplowski E, Bueno-de-Mesquita HB, Fuchs CS, Gross MD, Jacobs EJ, Lacroix AZ, et al. Anthropometric measures, body mass index, and pancreatic cancer: a pooled analysis from the Pancreatic Cancer Cohort Consortium (PanScan) Arch Intern Med. 2010;170:791–802. doi: 10.1001/archinternmed.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li D, Morris JS, Liu J, Hassan MM, Day RS, Bondy ML, Abbruzzese JL. Body mass index and risk, age of onset, and survival in patients with pancreatic cancer. JAMA. 2009;301:2553–2562. doi: 10.1001/jama.2009.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Calle EE, Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat Rev Cancer. 2004;4:579–591. doi: 10.1038/nrc1408. [DOI] [PubMed] [Google Scholar]
- 84.Rapp K, Schroeder J, Klenk J, Stoehr S, Ulmer H, Concin H, Diem G, Oberaigner W, Weiland SK. Obesity and incidence of cancer: a large cohort study of over 145,000 adults in Austria. Br J Cancer. 2005;93:1062–1067. doi: 10.1038/sj.bjc.6602819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Larsson SC, Permert J, Håkansson N, Näslund I, Bergkvist L, Wolk A. Overall obesity, abdominal adiposity, diabetes and cigarette smoking in relation to the risk of pancreatic cancer in two Swedish population-based cohorts. Br J Cancer. 2005;93:1310–1315. doi: 10.1038/sj.bjc.6602868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dawson DW, Hertzer K, Moro A, Donald G, Chang HH, Go VL, Pandol SJ, Lugea A, Gukovskaya AS, Li G, et al. High-fat, high-calorie diet promotes early pancreatic neoplasia in the conditional KrasG12D mouse model. Cancer Prev Res (Phila) 2013;6:1064–1073. doi: 10.1158/1940-6207.CAPR-13-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Philip B, Roland CL, Daniluk J, Liu Y, Chatterjee D, Gomez SB, Ji B, Huang H, Wang H, Fleming JB, et al. A high-fat diet activates oncogenic Kras and COX2 to induce development of pancreatic ductal adenocarcinoma in mice. Gastroenterology. 2013;145:1449–1458. doi: 10.1053/j.gastro.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oliver SG. Functional genomics: lessons from yeast. Philos Trans R Soc Lond B Biol Sci. 2002;357:17–23. doi: 10.1098/rstb.2001.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Garber K. Energy boost: the Warburg effect returns in a new theory of cancer. J Natl Cancer Inst. 2004;96:1805–1806. doi: 10.1093/jnci/96.24.1805. [DOI] [PubMed] [Google Scholar]
- 90.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 91.Spratlin JL, Serkova NJ, Eckhardt SG. Clinical applications of metabolomics in oncology: a review. Clin Cancer Res. 2009;15:431–440. doi: 10.1158/1078-0432.CCR-08-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lai IL, Chou CC, Lai PT, Fang CS, Shirley LA, Yan R, Mo X, Bloomston M, Kulp SK, Bekaii-Saab T, et al. Targeting the Warburg effect with a novel glucose transporter inhibitor to overcome gemcitabine resistance in pancreatic cancer cells. Carcinogenesis. 2014;35:2203–2213. doi: 10.1093/carcin/bgu124. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 93.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 94.Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 95.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 96.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 97.Lu CW, Lin SC, Chen KF, Lai YY, Tsai SJ. Induction of pyruvate dehydrogenase kinase-3 by hypoxia-inducible factor-1 promotes metabolic switch and drug resistance. J Biol Chem. 2008;283:28106–28114. doi: 10.1074/jbc.M803508200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mathupala SP, Heese C, Pedersen PL. Glucose catabolism in cancer cells. The type II hexokinase promoter contains functionally active response elements for the tumor suppressor p53. J Biol Chem. 1997;272:22776–22780. doi: 10.1074/jbc.272.36.22776. [DOI] [PubMed] [Google Scholar]
- 99.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 100.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 101.Kondoh H, Lleonart ME, Gil J, Wang J, Degan P, Peters G, Martinez D, Carnero A, Beach D. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65:177–185. [PubMed] [Google Scholar]
- 102.Britton D, Zen Y, Quaglia A, Selzer S, Mitra V, Löβner C, Jung S, Böhm G, Schmid P, Prefot P, et al. Quantification of pancreatic cancer proteome and phosphorylome: indicates molecular events likely contributing to cancer and activity of drug targets. PLoS One. 2014;9:e90948. doi: 10.1371/journal.pone.0090948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Qin Y, Zhu W, Xu W, Zhang B, Shi S, Ji S, Liu J, Long J, Liu C, Liu L, et al. LSD1 sustains pancreatic cancer growth via maintaining HIF1α-dependent glycolytic process. Cancer Lett. 2014;347:225–232. doi: 10.1016/j.canlet.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 104.Chien W, Lee DH, Zheng Y, Wuensche P, Alvarez R, Wen DL, Aribi AM, Thean SM, Doan NB, Said JW, et al. Growth inhibition of pancreatic cancer cells by histone deacetylase inhibitor belinostat through suppression of multiple pathways including HIF, NFkB, and mTOR signaling in vitro and in vivo. Mol Carcinog. 2014;53:722–735. doi: 10.1002/mc.22024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kitamoto S, Yokoyama S, Higashi M, Yamada N, Matsubara S, Takao S, Batra SK, Yonezawa S. Expression of MUC17 is regulated by HIF1α-mediated hypoxic responses and requires a methylation-free hypoxia responsible element in pancreatic cancer. PLoS One. 2012;7:e44108. doi: 10.1371/journal.pone.0044108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 107.Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, Chung YL, Merino M, Trepel J, Zbar B, Toro J, et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8:143–153. doi: 10.1016/j.ccr.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 108.King A, Selak MA, Gottlieb E. Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene. 2006;25:4675–4682. doi: 10.1038/sj.onc.1209594. [DOI] [PubMed] [Google Scholar]
- 109.Pollard PJ, Brière JJ, Alam NA, Barwell J, Barclay E, Wortham NC, Hunt T, Mitchell M, Olpin S, Moat SJ, et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Hum Mol Genet. 2005;14:2231–2239. doi: 10.1093/hmg/ddi227. [DOI] [PubMed] [Google Scholar]
- 110.MacKenzie ED, Selak MA, Tennant DA, Payne LJ, Crosby S, Frederiksen CM, Watson DG, Gottlieb E. Cell-permeating alpha-ketoglutarate derivatives alleviate pseudohypoxia in succinate dehydrogenase-deficient cells. Mol Cell Biol. 2007;27:3282–3289. doi: 10.1128/MCB.01927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dang CV, Kim JW, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer. 2008;8:51–56. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- 113.Dang CV. Rethinking the Warburg effect with Myc micromanaging glutamine metabolism. Cancer Res. 2010;70:859–862. doi: 10.1158/0008-5472.CAN-09-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 115.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, Yang X. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol. 2011;13:310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Denkert C, Bucher E, Hilvo M, Salek R, Orešič M, Griffin J, Brockmöller S, Klauschen F, Loibl S, Barupal DK, et al. Metabolomics of human breast cancer: new approaches for tumor typing and biomarker discovery. Genome Med. 2012;4:37. doi: 10.1186/gm336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vermeersch KA, Styczynski MP. Applications of metabolomics in cancer research. J Carcinog. 2013;12:9. doi: 10.4103/1477-3163.113622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhou W, Capello M, Fredolini C, Racanicchi L, Piemonti L, Liotta LA, Novelli F, Petricoin EF. Proteomic analysis reveals Warburg effect and anomalous metabolism of glutamine in pancreatic cancer cells. J Proteome Res. 2012;11:554–563. doi: 10.1021/pr2009274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Urayama S, Zou W, Brooks K, Tolstikov V. Comprehensive mass spectrometry based metabolic profiling of blood plasma reveals potent discriminatory classifiers of pancreatic cancer. Rapid Commun Mass Spectrom. 2010;24:613–620. doi: 10.1002/rcm.4420. [DOI] [PubMed] [Google Scholar]
- 121.Bathe OF, Shaykhutdinov R, Kopciuk K, Weljie AM, McKay A, Sutherland FR, Dixon E, Dunse N, Sotiropoulos D, Vogel HJ. Feasibility of identifying pancreatic cancer based on serum metabolomics. Cancer Epidemiol Biomarkers Prev. 2011;20:140–147. doi: 10.1158/1055-9965.EPI-10-0712. [DOI] [PubMed] [Google Scholar]
- 122.Kobayashi T, Nishiumi S, Ikeda A, Yoshie T, Sakai A, Matsubara A, Izumi Y, Tsumura H, Tsuda M, Nishisaki H, et al. A novel serum metabolomics-based diagnostic approach to pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2013;22:571–579. doi: 10.1158/1055-9965.EPI-12-1033. [DOI] [PubMed] [Google Scholar]
- 123.Ritchie SA, Akita H, Takemasa I, Eguchi H, Pastural E, Nagano H, Monden M, Doki Y, Mori M, Jin W, et al. Metabolic system alterations in pancreatic cancer patient serum: potential for early detection. BMC Cancer. 2013;13:416. doi: 10.1186/1471-2407-13-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang G, He P, Tan H, Budhu A, Gaedcke J, Ghadimi BM, Ried T, Yfantis HG, Lee DH, Maitra A, et al. Integration of metabolomics and transcriptomics revealed a fatty acid network exerting growth inhibitory effects in human pancreatic cancer. Clin Cancer Res. 2013;19:4983–4993. doi: 10.1158/1078-0432.CCR-13-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.ter Kuile BH, Westerhoff HV. Transcriptome meets metabolome: hierarchical and metabolic regulation of the glycolytic pathway. FEBS Lett. 2001;500:169–171. doi: 10.1016/s0014-5793(01)02613-8. [DOI] [PubMed] [Google Scholar]


