Abstract
DEAD-box RNA helicase Dbp4 is required for 18S rRNA synthesis: cellular depletion of Dbp4 impairs the early cleavage reactions of the pre-rRNA and causes U14 small nucleolar RNA (snoRNA) to remain associated with pre-rRNA. Immunoprecipitation experiments (IPs) carried out with whole-cell extracts (WCEs) revealed that hemagglutinin (HA)-tagged Dbp4 is associated with U3 snoRNA but not with U14 snoRNA. IPs with WCEs also showed association with the U3-specific protein Mpp10, which suggests that Dbp4 interacts with the functionally active U3 RNP; this particle, called the small-subunit (SSU) processome, can be observed at the 5′ end of nascent pre-rRNA. Electron microscopy analyses indicated that depletion of Dbp4 compromised SSU processome formation and cotranscriptional cleavage of the pre-rRNA. Sucrose density gradient analyses revealed that depletion of U3 snoRNA or the Mpp10 protein inhibited the release of U14 snoRNA from pre-rRNA, just as was seen with Dbp4-depleted cells, indicating that alteration of SSU processome components has significant consequences for U14 snoRNA dynamics. We also found that the C-terminal extension flanking the catalytic core of Dbp4 plays an important role in the release of U14 snoRNA from pre-rRNA.
INTRODUCTION
Ribosome biogenesis in the nucleoli of eukaryotic cells begins with the transcription of large rRNA precursors (pre-rRNAs), which are matured and assembled into the small (40S) and large (60S) ribosomal subunits. It is well established that a key step in the making of ribosomes is the production of mature rRNAs, the functional components of ribosomes (1, 2).
In Saccharomyces cerevisiae, RNA polymerase I synthesizes a long precursor of 35S that is processed into the 18S, 5.8S, and 25S rRNAs, while the 5S rRNA is independently transcribed by RNA polymerase III (1, 3). The 35S pre-rRNA is subjected to an orderly maturation process that requires more than 200 trans-acting factors (4–6). In addition, tens of small nucleolar RNAs (snoRNAs) base pair with pre-rRNAs and direct the site-specific posttranscriptional modification of rRNAs (7). The snoRNAs are grouped in two large families, called C/D and H/ACA, and they assemble into RNPs with specific proteins to carry out 2′-O-ribose methylation and pseudouridylation, the conversion of uridines into pseudouridines (Ψ) (7–9).
In contrast to the plethora of snoRNAs guiding posttranscriptional modifications, very few snoRNAs are required for the endonucleolytic cleavages that remove spacer sequences from pre-rRNAs. The U3, U14, and snR30 snoRNAs are essential for the early cleavage reactions that lead to the production of 18S rRNA; snR10 is also implicated in these cleavages but is not essential for growth (1, 10–12). U3 (C/D class) and snR30 (H/ACA class) are involved in the three early processing reactions, i.e., the cleavages at sites A0, A1, and A2 (for details on the processing pathway, see reference 13). U14 and snR10 are unique in that they have dual functions: they are involved in pre-rRNA processing at sites A1 and A2 but not at A0, and U14 targets the 2′-O-methylation of C414 in 18S rRNA, while snR10 directs the formation of Ψ2923 in 25S rRNA (14). Another essential snoRNA is RRP2, the RNA component of RNase MRP; this RNA does not belong to the C/D or H/ACA class but is required for cleavage at site A3 and for the production of 5.8SS rRNA (1, 13).
The small-subunit (SSU) processome is a very large RNP of ∼80S that forms the terminal knob observed at the 5′ end of a nascent pre-rRNA transcript (15, 16). The SSU processome is composed of U3 snoRNA and more than 70 proteins, most of which are specific for U3 and are required for its function (13, 15, 17–20). At present, it is not known whether U14 snoRNP contains specific proteins required for its function. Yeast U14 snoRNA has an extra stem-loop structure called the Y domain, which is essential for growth (10). In a search for a protein(s) that interacts with the Y domain, Liang et al. (21) identified Dbp4 in a multicopy suppressor screen; growth defects caused by deleterious mutations in the Y domain of U14 could be suppressed by overexpression of Dbp4, a phylogenetically conserved DEAD-box RNA helicase (21, 22).
RNA helicases are viewed as molecular motors that rearrange RNA structures or RNA-protein complexes in an energy-dependent fashion (23). These enzymes are characterized by signature motifs that form a central, catalytic domain of about 350 amino acids. The catalytic core is flanked by N- and C-terminal extensions that differ in length and amino acid composition; these regions are thought to be important for substrate recognition and for the function of an individual helicase (24, 25). Of the 45 putative RNA helicases identified in yeast (26), 20 are required for ribosome biogenesis (27, 28): this reflects the complexity of the RNA-RNA and RNA-protein rearrangements that are needed for the production of functional ribosomes.
The DEAD-box RNA helicase Dbp4 is essential for viability, indicating that its function cannot be complemented by another helicase (21). A role for Dbp4 in ribosome biogenesis was demonstrated by Kos and Tollervey (29): they showed that depletion of Dbp4 impaired early cleavage reactions at sites A0, A1, and A2 of the pre-rRNA, a phenotype that is normally seen with cells depleted of U3 snoRNA or U3-specific proteins (30). Cellular depletion of Dbp4 also impaired the release of U14 snoRNA from pre-rRNA, suggesting that Dbp4 is the RNA helicase that unwinds the U14 snoRNA–pre-rRNA duplex (29). We showed recently that Dbp4 is associated with U14 snoRNA in a complex that sediments at about 50S in sucrose density gradients (31). The human homologue of Dpb4 is also present in a complex of 50S but is associated with U3 snoRNA and not with U14 (32), indicating that the Dbp4 complex of 50S is different in yeast and humans (31).
Here we report that Dbp4 is associated with U3 snoRNA and the U3-specific protein Mpp10, which are components of the SSU processome. Electron microscopy (EM) analyses of chromatin spreads indicated that cellular depletion of Dbp4 inhibited SSU processome formation. We also show that trapping of U14 snoRNA on pre-rRNA is observed not only in Dbp4-depleted cells but also upon depletion of the U3 snoRNA or Mpp10 protein. Moreover, we found that the C-terminal extension of Dbp4 plays an important role in the release of U14 snoRNA from high-molecular-weight complexes and is necessary for the association of Dbp4 with U14 in the 50S complex.
MATERIALS AND METHODS
Yeast strains and media.
The yeast strains used in this study are listed in Table 1. The strains were usually grown in rich YP medium (1% yeast extract, 2% peptone) supplemented with either 2% dextrose (YPD) or 2% galactose (YPGal). For the selection of auxotrophic markers, cells were grown in synthetic medium (0.17% yeast nitrogen base) supplemented with the appropriate dropout mix (Clontech) and 2% galactose or glucose, as required. Media in culture plates included 2% Bacto agar. Yeast strains expressing hemagglutinin (HA)-tagged proteins were generated as described elsewhere (33) by using appropriate oligonucleotides (oligonucleotide sequences are available on request): cells were plated onto nonselective YPD or YPGal agar plates, as required, and were then replica plated onto selective agar plates containing 200 μg/ml Geneticin (Gibco). The DBP4-HA strain was further engineered into a depletion strain by replacing its promoter with the inducible GAL1 promoter as described by Longtine et al. (34). The GAL::DBP4-HA depletion strain was used for complementation assays with constructs that were constitutively expressed from the single-copy plasmid pCM188 (35). Plasmid-encoded Dbp4 contained a Myc tag at its C terminus.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| YPH499 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 | 40 |
| DBP4-HA | Same as YPH499 except DBP4::DBP4-3HA-kanMX6 | This study |
| GAL::DBP4-HA | Same as DBP4-HA except TRP1::PGAL1-DBP4-3HA | This study |
| NOP1-HA | Same as YPH499 except NOP1::NOP1-3HA-kanMX6 | 15 |
| YS626 | MATa leu2 ura3 trp1 his3 HIS3::GAL1::U14 | 21 |
| YSS1 | Same as YS626 except DBP4::DBP4-3HA-kanMX6 | This study |
| JH84 | MATα leu2-3,12 ura3-52 his3-Δ ade2-1 can1-100 u3aΔ UASGAL-U3A::URA3 U3B::LEU2 | 47 |
| YSS2 | Same as GAL::U3 except DBP4::DBP4-3HA-kanMX6 | This study |
| YPH258 | MATa ura3-52 lys2-801 ade2-101 his3-Δ200 leu2-Δ1 | 40 |
| GAL::MPP10 | Same as YPH258 except mpp10::HIS3 ADE2 URA3::PGAL1-10-MPP10 | 37 |
| YSS3 | Same as GAL::MPP10 except DBP4::DBP4-3HA-kanMX6 | This study |
Immunoprecipitations.
Immunoprecipitation experiments (IPs) with whole-cell extracts (WCEs) were carried out essentially as described previously (15). For each IP, 4 mg of protein A–Sepharose CL-4B (GE Healthcare) was first saturated with a mouse anti-HA monoclonal antibody (MAb 12CA5): binding was carried out overnight on a nutator at 4°C in TMN100 buffer (25 mM Tris-HCl [pH 7.6], 10 mM MgCl2, 100 mM NaCl, 0.1% Nonidet P-40, 1 mM dithiothreitol [DTT]). The “HA beads” were then washed three times with 1 ml TMN100 and were subsequently incubated with 500 μl of WCEs on a nutator at 4°C for 1 h. WCEs were prepared from exponentially growing cells (A600, 0.5 to 0.7), and the equivalent of 5 A600 units of cells was used for each IP. Cells were disrupted with glass beads (Sigma) in TMN100 buffer containing Complete protease inhibitor cocktail (Roche). After vigorous vortexing (seven times for 45 s each time, with intervals of 45 s, on ice), lysates were cleared by centrifugation at 4°C in a microcentrifuge (5 min at 18,000 × g). After incubation, the beads were washed 5 times with 1 ml TMN100, TMN200, or TMN400 (the number after “TMN” indicates the millimolar concentration of NaCl). IPs with complexes isolated from sucrose gradient fractions were carried out as described in reference 31. Coimmunoprecipitated RNAs were recovered by phenol-chloroform extraction, followed by ethanol precipitation in the presence of 40 μg glycogen (Roche). For protein analyses, the beads were mixed with 2× SDS loading buffer, and the proteins were eluted by incubation at 90°C for 5 min.
3′-end labeling of RNAs.
RNAs were labeled at their 3′ ends essentially as described previously (36). RNAs were mixed with T4 RNA ligase reaction buffer (50 mM HEPES [pH 8.3], 10 mM MgCl2, 50 μM ATP, 3.3 mM DTT, 10% [vol/vol] dimethyl sulfoxide [DMSO], 40 U RNasin [Promega], 10 μCi [5′-32P]pCp, and 20 U T4 RNA ligase [New England BioLabs]). The reaction mixture (30 μl) was incubated at 4°C for 16 h, and an equal volume of stop solution (USB) was added to the reaction. Samples were heat denatured for 3 min at 90°C and were immediately put on ice. Aliquots of 6 μl were separated on 8% polyacrylamide sequencing gels.
Northern blotting.
Small RNAs recovered in IPs or from sucrose gradient fractions were extracted with phenol-chloroform, precipitated with ethanol, separated on a denaturing polyacrylamide (8%) gel, transferred to a nylon membrane, cross-linked under UV light, and hybridized with specific 5′-end-labeled antisense oligonucleotides (sequences are available on request) (31). Hybridization was carried out for 16 h at 37°C. Following two washes in 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) containing 0.1% SDS and one wash in 0.5× SSPE containing 0.1% SDS, the membranes were exposed to a phosphor screen and were analyzed with a Molecular Imager F/X system (Bio-Rad).
Western blotting.
Proteins recovered after immunoprecipitations or from sucrose gradient fractions were separated by SDS-PAGE and were transferred to a polyvinylidene difluoride (PVDF) membrane (Immobilon-P; Millipore). The blots were incubated for 1 h with one of the following primary antibodies: mouse anti-HA monoclonal antibody 12CA5, mouse anti-Myc monoclonal antibody 9E10, rabbit anti-Mpp10 polyclonal antibodies (37), rabbit anti-Tsr1 polyclonal antibodies (38), and rabbit anti-Rpl3 and anti-Rpl30 polyclonal antibodies. Note that anti-Rpl30 (formerly designated L32) also recognizes Rps2 (formerly designated S4) (39). After three washes, the blots were incubated with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies (GE Healthcare) according to the manufacturer's recommendations. The blots were washed and were revealed using the ECL-Plus Western blotting detection system (GE Healthcare).
Sucrose density gradient analyses.
Prior to the harvesting of cells, cultures were incubated with cycloheximide (100 μg/ml; Sigma), and cycloheximide was maintained in all subsequent steps. Cells (30 A600 units) were harvested by centrifugation, washed with cold water, and resuspended in 0.5 ml of TMK100 buffer (the same as TMN100 except that KCl was substituted for NaCl) containing Complete protease inhibitor cocktail (Roche) and cycloheximide. Cellular extracts were prepared with glass beads (Sigma) by vigorous vortexing (seven times for 45 s each time, with intervals of 45 s, on ice), and lysates were cleared by centrifugation at 4°C in a microcentrifuge (5 min at 2,500 × g). Fifteen A260 units of extract was loaded onto linear 7-to-47% (wt/vol) sucrose gradients prepared in TMK100. The gradients were spun in a SW41 rotor at 39,000 rpm for 165 min. Fractions were collected using an Isco density gradient fractionator equipped with a UA-6 detector with constant monitoring of the absorbance at 254 nm to follow the presence of 40S and 60S ribosomal subunits, 80S ribosomes, and polysomes. Each fraction was separated into two aliquots: an aliquot of 200 μl was used for RNA analyses by Northern blot hybridization (see above), and 300 μl was subjected to trichloroacetic acid (TCA) precipitation before analysis of proteins by Western blotting.
Cellular depletion of individual components.
Conditional strains were first grown to exponential phase (A600 ≈ 0.5) at 30°C in liquid YPGal medium and were then transferred to prewarmed YPD medium. When required, cultures were diluted with prewarmed YPD to maintain exponential growth. Cellular growth was monitored at different time points by measuring the A600.
Electron microscopy.
Miller chromatin spreading was performed as described in reference 16. Briefly, 1 ml of the appropriate yeast culture was added to 5 mg Zymolyase for 4 min and was centrifuged briefly, and the pellet was resuspended in 1 ml 0.025% Triton (pH 9.2). After thorough mixing, the solution was added to 6 ml 11 mM KCl (pH 7) and was allowed to disperse for about 40 min with swirling. One-tenth volume of 0.1 M sucrose–10% formalin (pH 8.65) was added, and grids were made 15 min later.
RESULTS
Dbp4 specifically associates with U3 snoRNA.
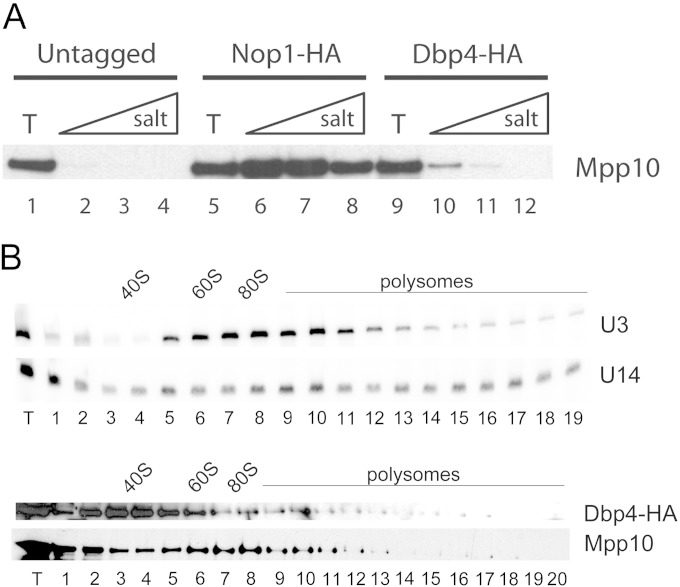
Deleterious mutations in the Y domain of U14 snoRNA can be suppressed by overexpression of DEAD-box protein Dbp4 (21). To examine the possible association of Dbp4 with U14 snoRNA, we carried out immunoprecipitation experiments (IPs) with cellular extracts prepared from the yeast DBP4-HA strain, a derivative of YPH499 (40). The DBP4-HA strain expresses Dbp4 with a triple HA epitope at its C terminus (referred to below as Dbp4-HA); it should be noted that in this strain, Dbp4-HA is not overexpressed, because transcription is under the control of its endogenous promoter (33). No growth defects were observed for the DBP4-HA strain, indicating that the triple HA tag at the C terminus of Dbp4 did not affect its essential function (data not shown).
We conducted IPs with cellular extracts prepared from the DBP4-HA strain and other control strains, and we analyzed the coimmunoprecipitated RNAs. The immunoprecipitates were washed with increasing salt concentrations in order to assess the stability of complexes. In a first series of experiments, RNAs that coimmunoprecipitated with Dbp4-HA were directly labeled at their 3′ ends with [5′-32P]pCp and were separated on a sequencing gel. These experiments revealed a faint band of about 330 nucleotides (nt) that progressively disappeared with increasing salt concentrations (Fig. 1A, lanes 3 to 5). This band comigrated with U3 snoRNA, which gave a very strong signal in control IPs with HA-tagged Nop1, a protein common to all C/D snoRNPs (7) (Fig. 1A, lanes 7 to 9). In marked contrast to the Nop1-HA IPs, no band corresponding to U14 snoRNA (126 nt) was detected in the Dbp4-HA immunoprecipitates (Fig. 1A, compare lanes 3 to 5 with lanes 7 to 9). Note that tRNAs, 5S rRNA, and 5.8S rRNA were also detected in these experiments, but those RNAs are known as sticky, nonspecific contaminants (15, 37, 41). To identify the RNAs that coimmunoprecipitated with Dbp4, we carried out Northern blot hybridization analyses with 32P-labeled oligonucleotides. These experiments showed that HA-tagged Dbp4 is specifically associated with U3 snoRNA (Fig. 1B). The intensity of the signal for U3 decreased with increasing salt concentrations, showing that the association of Dbp4 with U3 was salt sensitive, in contrast with the U3-Nop1 association (Fig. 1B, compare lanes 8 to 10 with lanes 13 to 15). Control experiments with the untagged parental strain did not reveal the presence of U3 in immunoprecipitates (Fig. 1B, lanes 3 to 5), ruling out the possibility that the faint U3 band observed in Dbp4-HA IPs resulted from a nonspecific interaction.
FIG 1.
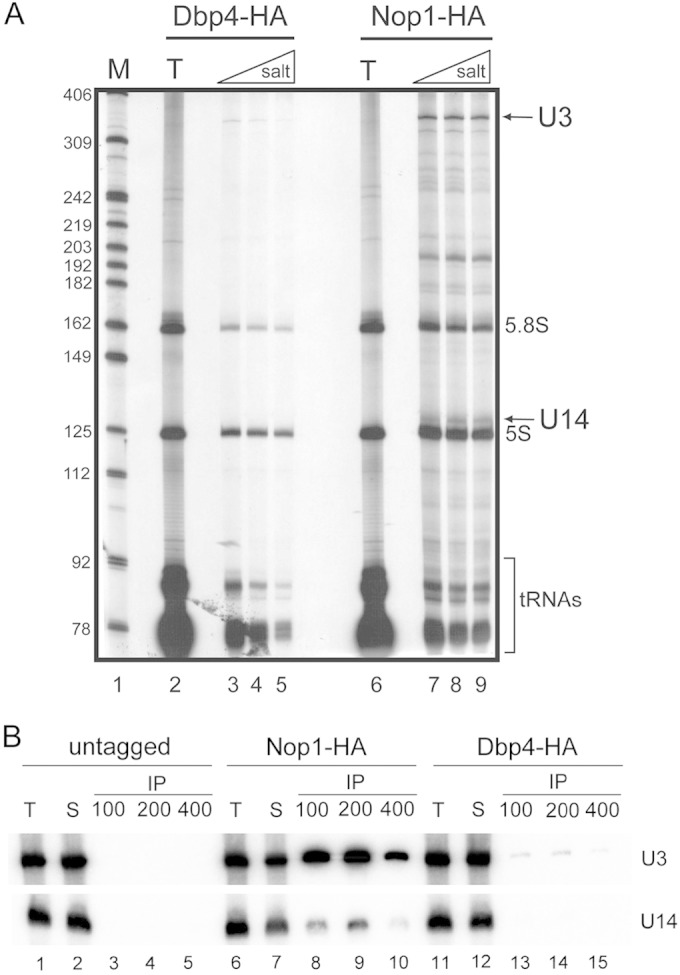
Dbp4 is associated with U3 snoRNA. (A) 3′-end labeling of RNAs. WCEs were prepared from yeast strains expressing HA-tagged Dbp4 (lanes 2 to 5) or HA-tagged Nop1 (lanes 6 to 9). Beads coated with an anti-HA MAb were incubated with WCEs, recovered by centrifugation, and washed with increasing salt concentrations. RNAs were extracted with phenol-chloroform, precipitated with ethanol, labeled at their 3′ ends with [5′-32P]pCp, and separated on a sequencing gel. T lanes, total RNAs from WCEs. DNA molecular size markers (M) are shown in lane 1 (in nucleotides). The predicted positions of some RNAs are indicated on the right. (B) Northern blotting of recovered RNAs. IPs and RNA isolation were conducted as for panel A with WCEs prepared from an untagged yeast strain (lanes 1 to 5) or from yeast strains expressing HA-tagged Nop1 (lanes 6 to 10) or HA-tagged Dbp4 (lanes 11 to 15). Immunoprecipitated RNAs were separated in a denaturing gel, transferred to a nylon membrane, and probed with radiolabeled oligonucleotides complementary to various snoRNAs (indicated on the right). T lanes correspond to total RNAs isolated from WCEs, and S lanes correspond to RNAs from supernatants.
Dbp4 interacts with the SSU processome, the active U3 particle.
U3 is a dynamic RNP that can be detected in alternative states of ∼12S to 15S and ∼80S; the larger form is the functionally active U3 RNP and is designated the SSU processome (15, 42, 43). Since it is an RNA helicase, Dbp4 could be required for assembly of the U3 RNP at an early (∼12S to 15S) or a late (∼80S) stage. To assess the latter possibility, we asked whether Dbp4 coimmunoprecipitated with the U3-specific protein Mpp10, which is an integral constituent of the SSU processome (15, 16, 37). Western blot analyses indicated that Dbp4 was associated with Mpp10 and that this association was salt sensitive (Fig. 2A, lanes 10 to 12). The coimmunoprecipitation (co-IP) of Mpp10 with Dbp4-HA was much less efficient than that with the core component Nop1-HA (Fig. 2A, compare lanes 6 to 8 with lanes 10 to 12), but the signal was well above background levels (Fig. 2A, compare lanes 2 to 4 with lanes 10 to 12), indicating that the co-IP of Mpp10 with Dbp4 was specific and was not due to fortuitous interactions. Sucrose gradient sedimentation analyses indicated that Dbp4-HA was enriched in the lower-density fractions of the gradient, forming a peak at 40S to 50S, as observed elsewhere (31), and that a very small portion of Dbp4-HA cosedimented with Mpp10 and the U3 snoRNA in the 80S region of the gradient (Fig. 2B). These results are consistent with IPs showing that only a fraction of the U3 snoRNA and Mpp10 coimmunoprecipitated with Dbp4-HA (Fig. 1B and 2A). We proposed recently that Dbp4 is an SSU processome component (31); here we showed that the association of Dbp4 with the SSU processome is not stable, which could reflect the transient nature of the interaction between the RNA helicase Dbp4 and the SSU processome.
FIG 2.
Dbp4 associates with Mpp10 but is not enriched in SSU processome fractions. (A) Western blot analysis of immunoprecipitates from IPs conducted as for Fig. 1B. WCEs were prepared from an untagged yeast strain (lanes 1 to 4) or from yeast strains expressing HA-tagged Nop1 (lanes 5 to 8) or HA-tagged Dbp4 (lanes 9 to 12). Proteins were fractionated by SDS-PAGE, transferred to a PVDF membrane, and subjected to immunoblotting with anti-Mpp10 antibodies. (B) Sucrose gradient sedimentation analyses. (Top) Northern blots for U3 and U14 snoRNAs; (bottom) Western blots for Dbp4 and Mpp10 proteins. The positions of 40S and 60S ribosomal subunits, 80S ribosomes, and polysomes are indicated.
Loss of Dbp4 alters the sedimentation of snoRNPs.
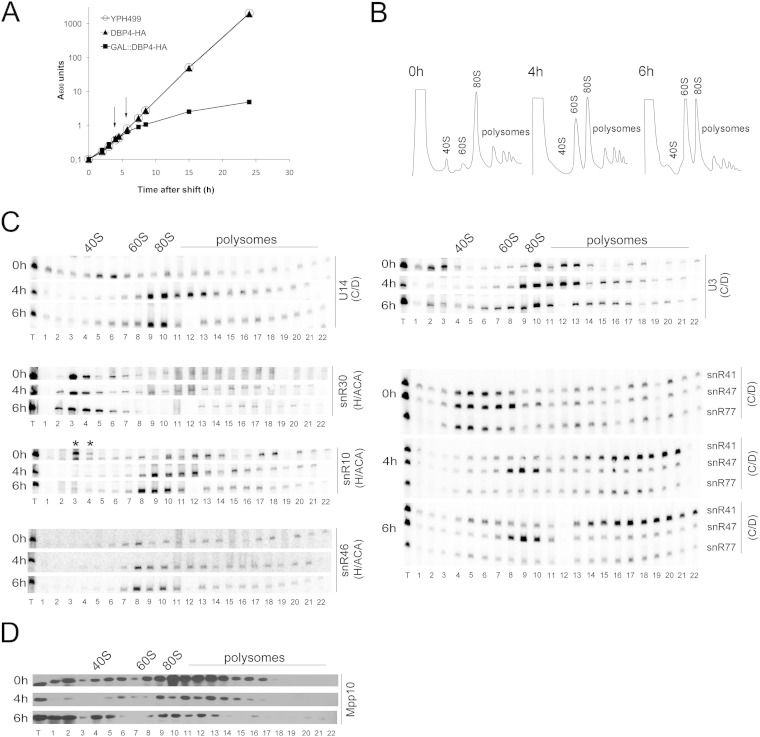
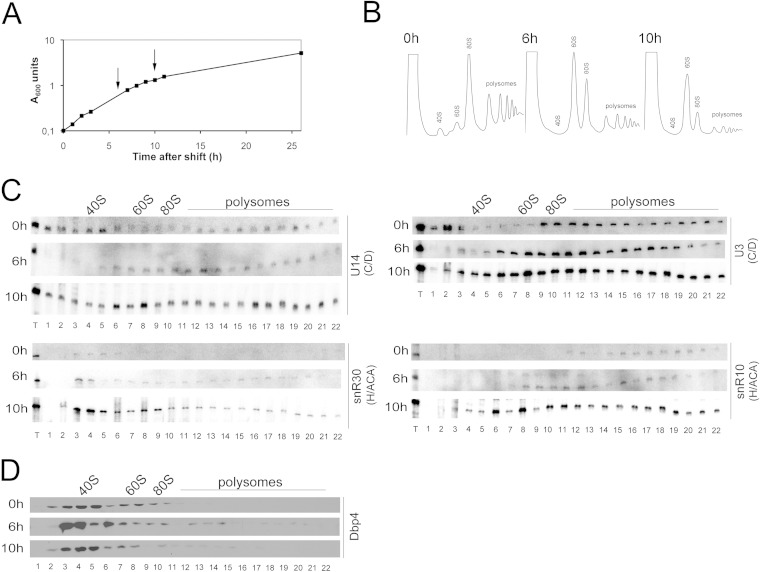
We generated the GAL::DBP4-HA conditional strain, which expresses Dbp4-HA under the control of the GAL1 promoter (34). This promoter is active in galactose-containing medium (YPGal) but is turned off when cells are grown in the presence of glucose (dextrose) (YPD). We found that the growth rate of the GAL::DBP4-HA strain was identical to that of the parental DBP4-HA strain and the wild-type strain YPH499 when the cells were grown in YPGal (data not shown). When exponentially growing cells were shifted from YPGal to YPD, the growth of the GAL::DBP4-HA strain began to slow down within 3 to 4 h after the shift to YPD, whereas the DBP4-HA and YPH499 strains maintained exponential growth rates (Fig. 3A).
FIG 3.
Depletion of Dbp4 affects the sedimentation of U14 and other snoRNAs. (A) Growth curves of the YPH499, DBP4-HA, and GAL::DBP4-HA strains after the shift into glucose-containing medium. The cultures were diluted as necessary to maintain exponential growth. Absorbance at 600 nm was measured over time. Arrows indicate depletion time points that were chosen for subsequent analyses. (B) Sucrose gradient sedimentation profiles of extracts prepared from the GAL::DBP4-HA strain before (0 h) and after (4 h and 6 h) the shift to YPD. The positions of free 40S and 60S subunits, 80S ribosomes, and polysomes are indicated. (C) Northern blot analysis of snoRNAs in sucrose gradient fractions. RNAs were extracted from gradient fractions of the GAL::DBP4-HA strain grown in YPGal (0 h) or in YPD to deplete Dbp4 for 4 h or 6 h (the times of depletion are indicated on the left). We used oligonucleotides complementary to different snoRNAs that belong to the C/D or H/ACA class (indicated on the right). Gradients were fractionated into 22 samples (numbered 1 to 22). T lanes, total RNAs of the extracts. Sedimentation is from left to right. The positions of 40S and 60S subunits, 80S ribosomes, and polysomes are indicated on the top. In the snR10 panel, asterisks indicate that snR10 corresponds to the lower band in fractions 3 and 4 (at 0 h). Note that fraction 12 of the 6-h depletion time was lost. (D) Western blotting of sucrose gradient fractions. The time of growth of the GAL::DBP4-HA strain in YPD medium is indicated on the left of each panel. Samples (numbered 1 to 22) were analyzed with an antibody against Mpp10.
To avoid possible secondary effects caused by the depletion of Dbp4, we chose to analyze cellular extracts prepared at early time points after the shift to YPD. Cells were harvested at 4 and 6 h following the shift to YPD (Fig. 3A, arrows), and cellular extracts were analyzed by ultracentrifugation through sucrose density gradients. Continuous monitoring of the absorbance at 254 nm during gradient fractionation generated a sedimentation profile of ribosomal particles (40S and 60S ribosomal subunits, 80S ribosomes, and polysomes). For comparison, extracts were prepared from nondepleted exponentially growing cells (0 h in YPD) and were analyzed in parallel. Note that the 60S peak was less pronounced than the 40S peak in nondepleted cells (this was also seen with extracts from other nondepleted strains and from the parental DBP4-HA strain), and switching cells from galactose- to dextrose-containing medium restored the normal smaller size of the 40S peak (data not shown). Depletion of Dbp4 caused a strong decrease in the amount of free 40S ribosomal subunits and a gradual increase in the amount of free 60S ribosomal subunits over time (Fig. 3B). These results are consistent with the implication of Dbp4 in 18S rRNA production (21, 29). Indeed, impairment of 18S rRNA synthesis leads to a deficit in 40S subunits, which is accompanied by an excess of free 60S subunits, because 80S ribosomes can no longer be formed. Curiously however, the intensity of the 80S peak did not diminish, even 6 h after the shift to YPD. This phenomenon is unique to Dbp4 depletion; it was not observed with the depletion of U14, U3, or Mpp10 (see below).
The distribution of snoRNAs in sucrose density gradients was analyzed by Northern blot hybridization (Fig. 3C). In comparison with that in nondepleted cells (0 h in YPD), the distribution patterns of U3 and U14 snoRNAs changed: U14 moved from the 50S fraction to higher-molecular-weight fractions of the gradient, indicating that U14 was trapped in higher-order complexes. U3 normally sediments as free RNPs (∼12S to 15S) and active particles (∼80S), but during Dbp4 depletion, free RNPs were barely detected, and the bulk of U3 accumulated in high-density regions of the gradient (≥80S). Individual fractions were also analyzed for the presence of the U3-specific protein Mpp10 by Western blotting (Fig. 3D). In depleted cells, the sedimentation profile of Mpp10 was similar to that of U3 snoRNA, except that a larger portion of Mpp10 was found in free form at the top of the gradient; it is known that the Mpp10 complex, which is small and contains Imp3 and Imp4, independently joins the nascent SSU processome (19, 20, 44–46).
We also examined the sedimentation profiles of modification guide RNAs, because depletion of Dbp4 can alter their sedimentation (29). In Dbp4-depleted cells, the box C/D guide snoRNAs snR41 and snR77 showed increased distribution in fractions corresponding to very large complexes (polysome-size fractions), and snR47 moved from the 40S-to-60S region to the 60S-to-80S region of the gradient. In contrast, we did not observe major changes in the distribution of the H/ACA guide snoRNAs snR10 and snR46 or in that of the H/ACA processing snoRNA snR30. Overall, these sedimentation profiles suggest that depletion of Dbp4 can alter the dynamics of snoRNPs, with a more pronounced effect on C/D snoRNPs than on H/ACA snoRNPs.
Effects of depleting U14 snoRNA.
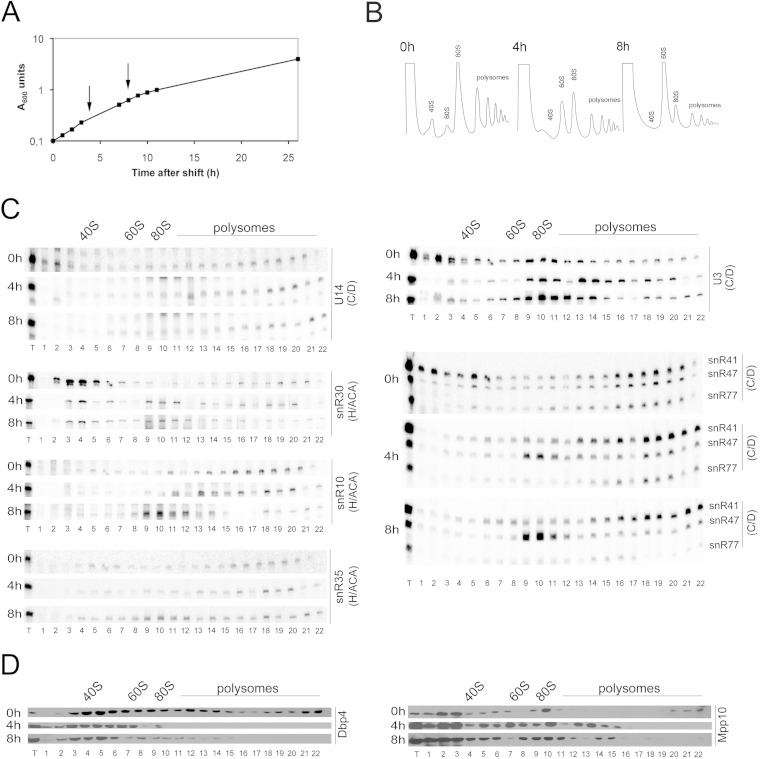
Considering the link between Dbp4 and U14 snoRNA (21, 29), we analyzed the effects of depleting U14. Strain YS626, which expresses U14 under the control of the GAL1 promoter (21), was further engineered to encode HA-tagged Dbp4 under the control of its endogenous promoter (33). This new strain was named YSS1 and was used for our analyses. Exponentially growing cells were transferred from YPGal to YPD, and the growth rate decreased gradually after the shift (Fig. 4A). Early depletion times of 4 and 8 h were chosen so as to avoid secondary effects in further experiments with strain YSS1. We examined the sedimentation profiles of ribosomal particles before and after the shift to YPD (Fig. 4B). In accord with the essential role of U14 in 18S rRNA production, the peak of free 40S ribosomal subunits was almost undetectable upon U14 depletion. As expected, this deficit was accompanied by an increase in free 60S ribosomal subunits, and the overall content of 80S ribosomes and polysomes was decreased due to the impaired balance between the amounts of 40S and 60S ribosomal subunits (Fig. 4B). Gradient fractions were subjected to Northern and Western blot analyses (Fig. 4C and D). Interestingly, U14 depletion caused a change in the sedimentation profile of U3 snoRNA: as seen with Dbp4-depleted cells (Fig. 3), free U3 RNPs were no longer detected in the lower-density region of the gradient, and U3 accumulated in large complexes. Before its depletion, U14 sedimented in low-molecular-weight fractions, but after a few hours in YPD, it was no longer detected in those fractions.
FIG 4.
Effects of depleting U14 snoRNA. (A) Effect of U14 gene repression on cellular growth. Strain YSS1 was grown to exponential phase in YPGal and was shifted to YPD as described for Fig. 3A. (B) Polysome profile of strain YSS1 at 0 h (before depletion) and at 4 h and 8 h after the shift to YPD. The peaks of free 40S and 60S subunits, 80S ribosomes, and polysomes are indicated. (C) Northern blot hybridization analysis of snoRNAs in sucrose gradient fractions (see the legend to Fig. 3C for details). (D) Sedimentation analysis of the Dbp4 and Mpp10 proteins by Western blotting. Sucrose gradient fractions were analyzed before and after the depletion of U14 snoRNA (times in YPD are indicated on the left).
Depletion of U14 also caused a redistribution of box C/D guide snoRNAs, which was similar to that seen upon depletion of Dbp4. Note, however, that these snoRNAs could be detected in large complexes even before depletion, a pattern different from that observed with the GAL::DBP4-HA strain (compare the 0-h results for snR41, snR47, and snR77 in Fig. 3C and 4C). As reported previously, the sedimentation profiles of snoRNAs can differ between strains with different genetic backgrounds (29). Nevertheless, U14 depletion changed the sedimentation profile of these C/D guide snoRNAs in a manner similar to that seen upon Dbp4 depletion. Moreover, U14 depletion affected the sedimentation of the H/ACA processing snoRNA snR30, which largely disappeared from low-molecular-weight fractions and accumulated in the 80S region of the gradient (Fig. 4C). H/ACA guide snoRNAs snR10 and snR35 also accumulated slightly in the 80S region of the gradient upon U14 depletion (Fig. 4C). Western blot analyses showed that U14 depletion had little effect on the distribution of Mpp10, while Dbp4 was no longer detected in high-density fractions (Fig. 4D). Taken together, these results indicate that U14 depletion affects the sedimentation profiles of many nucleolar factors.
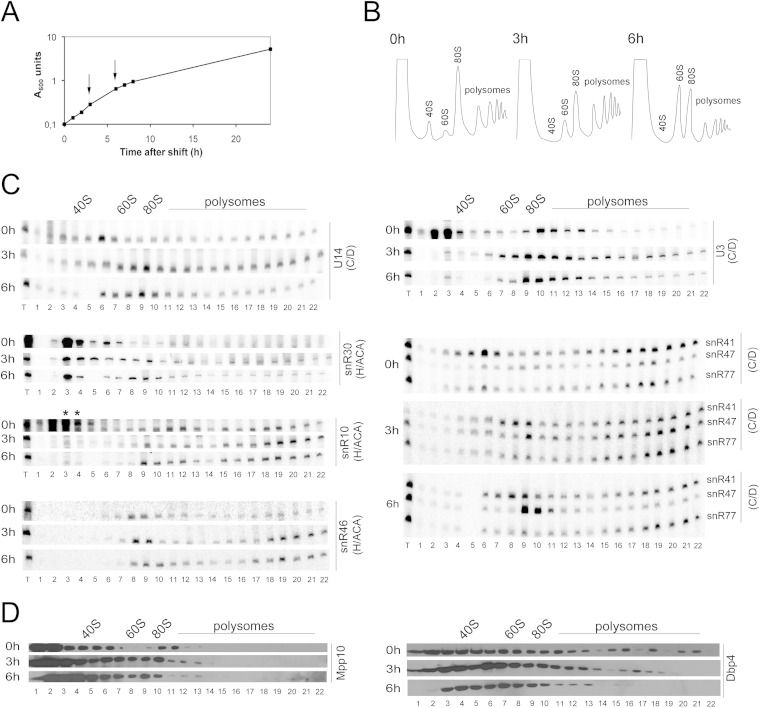
Depletion of U3 snoRNA alters the sedimentation of snoRNAs.
The U3 snoRNA is the central constituent of the SSU processome. Because U3 and Dbp4 are associated (Fig. 1), we analyzed the consequences of depleting U3 snoRNA. Strain JH84, which conditionally expresses U3 snoRNA from the GAL1 promoter (47), was modified to encode HA-tagged Dbp4 from its endogenous promoter (strain YSS2). As observed with the depletion of other components, blocking the production of U3 snoRNA caused severe growth defects (Fig. 5A). We chose to carry out further experiments with cells depleted of U3 snoRNA for 3 and 6 h. The sedimentation profile of ribosomal particles was similar to that seen for U14-depleted cells, except that the decrease in the 80S peak was less pronounced (compare Fig. 5B and 4B). The sedimentation profile of the U3 snoRNA itself was similar to that seen for cells depleted of Dbp4 or U14 snoRNA (compare Fig. 5C with Fig. 3C and 4C). Surprisingly, depletion of U3 snoRNA also affected the sedimentation profile of U14, which showed increased distribution and abundance in the fractions corresponding to large complexes (Fig. 5C). In fact, the sedimentation profile of U14 snoRNA was very similar to that observed during Dbp4 depletion (Fig. 3C), indicating that depletion of U3 snoRNA also resulted in the trapping of U14 in high-molecular-weight complexes.
FIG 5.
Depletion of U3 snoRNA affects the sedimentation of U14. (A) Cellular growth analysis during U3 snoRNA depletion. The experiment with strain YSS2 was carried out as described for Fig. 3A. (B) Polysome profile of strain YSS2 at 0 h (before depletion) and at 3 h and 6 h after depletion. The peaks of free 40S and 60S subunits, 80S ribosomes, and polysomes are indicated. (C) Analysis of the sedimentation profiles of different snoRNAs by Northern blot hybridization as described for Fig. 3C. (D) Distribution patterns of Dbp4 and Mpp10 in the presence or absence of U3 snoRNA, determined by using anti-HA and anti Mpp10 antibodies, respectively (times in YPD are indicated on the left).
We noted that, as with the U14 depletion strain YSS1 grown in YPGal (Fig. 4), snR41 accumulated in two regions of the gradient in nondepleted cells, one at ∼50S and the other corresponding to high-density fractions (Fig. 5C). Upon U3 depletion, this distribution changed, and snR41 moved from the ∼50S region of the gradient to the 60S-to-80S region, while a good proportion of snR41 was maintained in high-density fractions. There were no major changes in the distribution of guide snoRNAs snR10, snR46, and snR77; however, snR47 largely accumulated in the ∼80S region of the gradient, as seen above during the depletion of Dbp4 (Fig. 3C) and U14 snoRNA (Fig. 4C). Only minor changes were observed in the sedimentation profile of the H/ACA processing snoRNA snR30 (Fig. 5C) and for the Mpp10 and Dbp4 proteins (Fig. 5D).
Effects of Mpp10 depletion.
The U3-specific protein Mpp10 is required for pre-18S rRNA processing and is a genuine SSU processome component (15, 37). We have shown that Mpp10 and Dbp4 are associated in vivo (Fig. 2A). Hence, we analyzed the effect of depleting Mpp10 using the depletion strain YSS3, which expresses Mpp10 under the control of a GAL1-10 promoter (37) and HA-tagged Dbp4 from its endogenous promoter. When exponentially growing cells were shifted from YPGal to YPD, there was a gradual reduction in the growth rate (Fig. 6A), and further experiments were carried out 6 and 10 h after the shift to YPD. The sedimentation profiles of ribosomal particles from Mpp10-depleted cells were very similar to those seen for U14-depleted cells (compare Fig. 4B and 6B). Loss of Mpp10 induced a strong deficit of 80S ribosomes and concomitant accumulation of free 60S subunits. We also examined the sedimentation profiles of snoRNAs involved in processing reactions and that of Dbp4 by Northern and Western blotting (Fig. 6C and D). After 6 h of depletion, U3 and U14 snoRNAs were no longer detected in low-molecular-weight fractions (free RNPs) and accumulated in higher density fractions (∼80S to 90S). After 10 h of depletion, there was a quite uniform distribution of U3 and U14 snoRNAs throughout the gradient. In contrast, there was almost no change in the distribution of snoRNAs snR10 and snR30, or in that of the Dbp4 protein, upon depletion of Mpp10. The effects on snoRNAs are reminiscent of the observations for Dbp4-depleted cells and suggest that Mpp10 depletion affected primarily the distribution of U3 and U14 snoRNAs.
FIG 6.
Depletion of Mpp10 affects the sedimentation of U14. (A) Cellular growth analysis during Mpp10 depletion. Strain YSS3 was grown in YPD (see the legend to Fig. 3A for details). (B) Polysome profile of strain YSS3 at 0 h (before depletion) and at 6 h and 10 h after the shift to YPD. The peaks of free 40S and 60S subunits, 80S ribosomes, and polysomes are indicated. (C) Northern blot hybridization analysis of processing snoRNAs with strain YSS3 (see the legend to Fig. 3C for details). (D) Western blots of Dbp4 with strain YSS3 (times in YPD are indicated on the left).
Dbp4 is required for SSU processome formation.
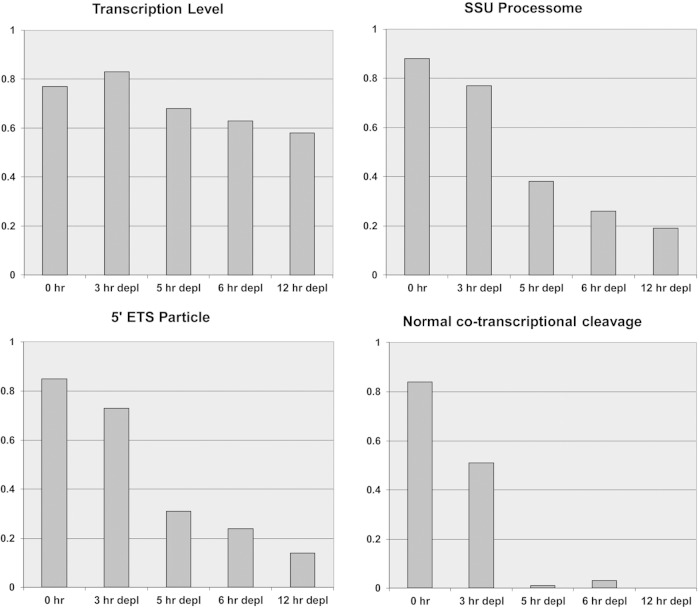
The SSU processome forms at the 5′ end of nascent pre-rRNA and decorates active transcription units, which take the shape of “Christmas trees”; electron microscopy (EM) analyses revealed that depletion of various components of the SSU processome abolished its formation, although the extents of alterations seen by EM differed between various SSU processome components (15, 16, 48). Because Dbp4 depletion leads to pre-18S rRNA processing defects related to SSU processome dysfunction, we carried out EM analyses on chromatin spreads of the GAL::DBP4-HA strain to determine if the absence of Dbp4 could affect the formation of “Christmas trees.” As shown in Fig. 7, there was a marked difference between nondepleted cells (0 h in YPD) and cells depleted of Dbp4 for various times. In nondepleted cells, rRNA genes showed the characteristic pattern in which nascent transcripts are first packaged into SSU processomes and then cotranscriptionally cleaved at site A2 (16). After a few hours of depletion, there were fewer transcripts per gene on average and fewer terminal knobs. The few SSU processomes that formed did so on more-mature transcripts near the 3′ end of the gene (Fig. 7A, small arrows). This phenotype was exacerbated over time and resulted in nearly complete loss of terminal knobs within 12 h of depletion (Fig. 7 and 8). The most striking effect of Dbp4 depletion was seen on normal cotranscriptional cleavage of pre-rRNA, which was nearly abolished within 5 h of depletion (Fig. 8). Taken together, these results demonstrate that Dbp4 is required for cotranscriptional SSU processome formation and function.
FIG 7.
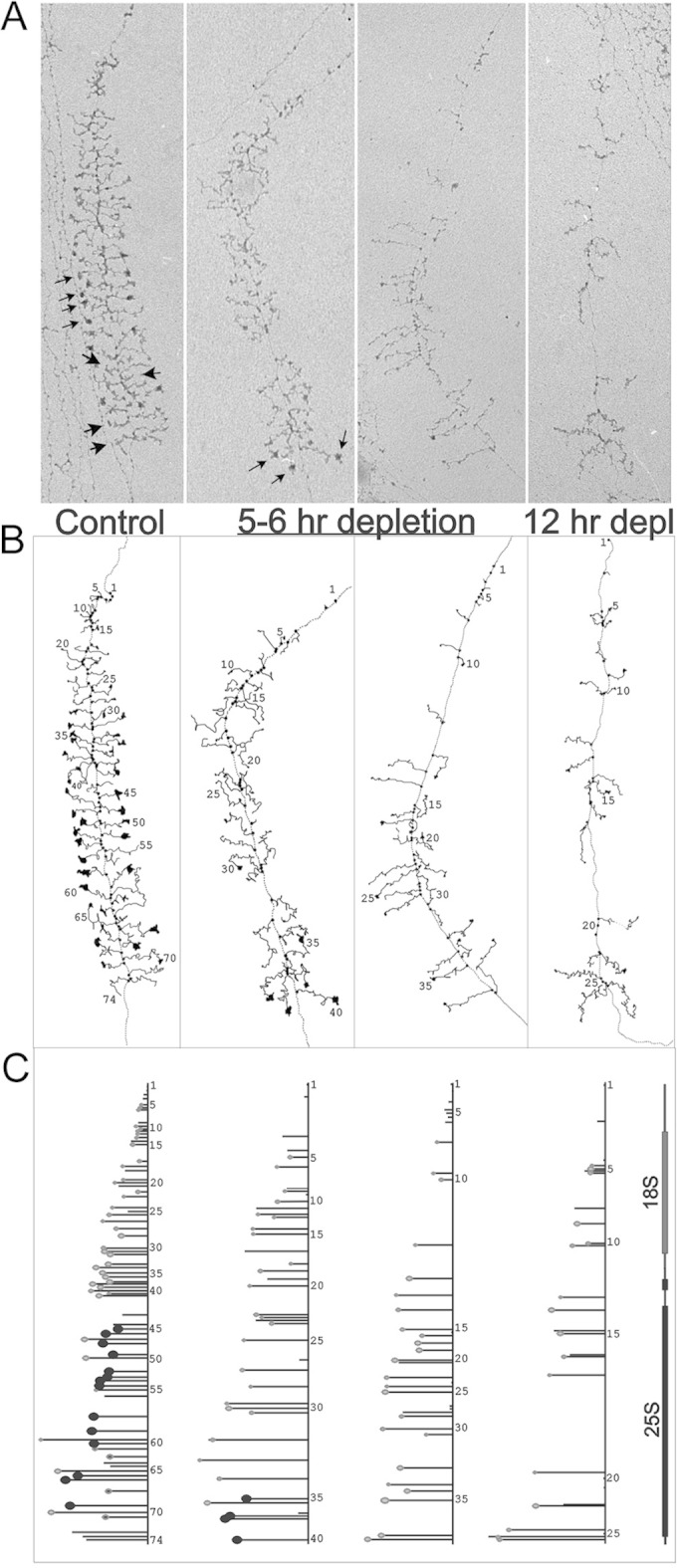
Dbp4 is required for SSU processome formation. (A) Electron micrographs of rRNA genes from the GAL::DBP4-HA strain. Examples of genes from 0 h (nondepleted) and from 5-h, 6-h, and 12-h depletions are displayed. Small arrows indicate SSU processomes, and large arrows indicate transcripts that have been cotranscriptionally cleaved. (B) Interpretive line tracings of the genes. (C) Maps of the genes with the following features highlighted: 5′ external transcribed spacer particles (small and light gray), SSU processomes (larger and black), and LSU particles beginning to form on the control gene only, i.e., transcripts 62, 68, and 71 (small and light gray with a black center) (16).
FIG 8.
EM analyses of cotranscriptional pre-rRNA processing. Results of semiquantitative analysis of the state of rRNA genes in the GAL::DBP4-HA strain at various times of depletion are shown. (Cells were grown in YPGal and were then switched to YPD for depletion.) Fields of rRNA genes from EM analysis of Miller chromatin spreads were examined, and the following features were given a visual score of 0 to 3: the transcription level (i.e., density of transcripts/gene), the presence of 5′ external transcribed spacer (ETS) particles, the presence of SSU processomes, and normal cotranscriptional cleavage. The scores were then normalized (0 to 1) and were plotted as a bar graph. The number of fields examined ranged from 56 to 133 (average, 83), and each field contained multiple genes. As a general rule, the rRNA genes in a given nucleolus display similar morphologies. The transcription level at 0 h is slightly lower than that at 3 h of depletion because of the transcriptional advantage of growth in glucose.
The C-terminal extension of Dbp4 is required for association with U14.
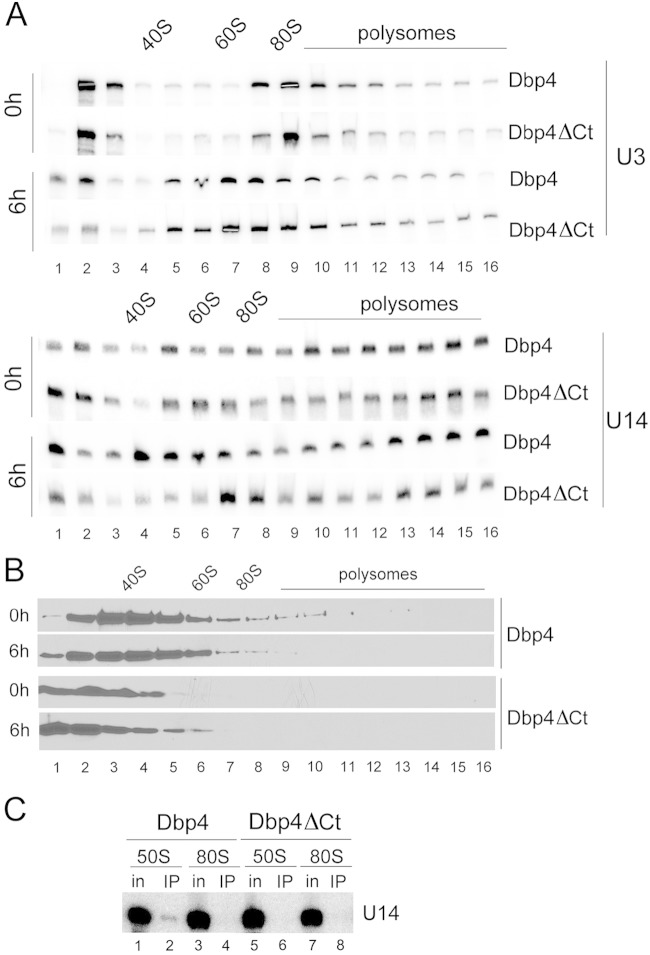
Point mutations in the catalytic core of Dbp4 induced the trapping of U14 snoRNA in high-molecular-weight complexes, suggesting that the ATPase/helicase activity of Dbp4 is required to release U14 from pre-rRNA (29). The extensions flanking the catalytic core of DEAD-box proteins are thought to be important for substrate recognition (24, 25). We therefore tested whether deleting the C-terminal extension of Dbp4 would affect the release of U14 from pre-rRNA. The GAL::DBP4-HA depletion strain was transformed with single-copy plasmids that constitutively express full-length Dbp4 (control) or a truncated version lacking the C-terminal extension (Dbp4ΔCt); these constructs each bear a Myc tag at the C terminus to distinguish them from chromosome-encoded HA-tagged Dbp4. Expression of Myc-tagged Dbp4 restored growth in YPD, but Dbp4ΔCt expression did not (data not shown). Cultures were harvested before depletion or 6 h after the shift to YPD, and cellular extracts were fractionated on sucrose gradients in order to analyze the sedimentation patterns of the U3 and U14 snoRNAs. The distribution of U3 was the same in cells expressing plasmid-borne Dbp4 or Dbp4ΔCt either before (0 h) or after (6 h) depletion (Fig. 9A, top). The distribution of U14 was also similar in nondepleted cells (0 h) expressing plasmid-borne Dbp4 or Dbp4ΔCt (Fig. 9A), but in depleted cells (6 h), there was a marked difference between cells expressing Dbp4ΔCt and those expressing full-length Dbp4 (Fig. 9A, bottom). In fact, the distribution of U14 in cells expressing Dbp4ΔCt was identical to that in cells depleted of Dbp4 (Fig. 3C). These results suggest that, in addition to the helicase activity of Dbp4 (29), the C-terminal extension of Dbp4 is required to release U14 from pre-rRNA.
FIG 9.
Deletion of the C-terminal extension of Dbp4. Depletion strain GAL::DBP4-HA was transformed with a plasmid expressing full-length Dbp4 (Dbp4) or a truncated version lacking its C-terminal extension (DBP4ΔCt). (A and B) Cells were grown in galactose-containing medium (0 h) and were shifted to YPD for 6 h (6 h). Northern blot hybridization (A) and Western blot (B) analyses were carried out as for Fig. 3 except that 16 fractions were collected instead of 22, and immunoblotting was carried out with an anti-Myc MAb. (C) IPs with an anti-Myc MAb were carried out on sucrose gradient fractions corresponding to the 50S and 80S peaks, and U14 snoRNA was detected by Northern blot hybridization (as described in reference 31). in, input.
We examined the sedimentation profiles of plasmid-borne Dbp4 and Dbp4ΔCt by Western blotting. The distributions of full-length Dbp4 were similar in nondepleted and depleted cells, and the same was observed for Dbp4ΔCt; however, in contrast to full-length Dbp4, very little Dbp4ΔCt was detected in high-molecular-weight fractions (Fig. 9B). We also verified that expression of the Myc-tagged Dbp4 constructs did not alter the distribution of chromosome-encoded Dbp4 in nondepleted cells (data not shown).
IPs carried out with WCEs indicated that Dbp4 is not associated with U14 snoRNA (Fig. 1). However, when cellular extracts are fractionated on sucrose gradients and IPs are carried out with 50S fractions, U14 coimmunoprecipitates with Dbp4 (31). We investigated whether the C-terminal extension of Dbp4 is required for its association with U14 snoRNA in the 50S complex (Fig. 9C). IPs were carried out with sucrose gradient fractions of 50S and 80S (SSU processome) as described previously (31), except that extracts were prepared from cells expressing Myc-tagged Dbp4 or Dbp4ΔCt. U14 snoRNA did not coimmunoprecipitate with Dbp4ΔCt in the 50S complex (Fig. 9C, compare lanes 2 and 6). We conclude that the C-terminal extension of Dbp4 is required for its association with U14 snoRNA.
Dbp4-depleted cells accumulate mature LSU RNAs in the 80S peak.
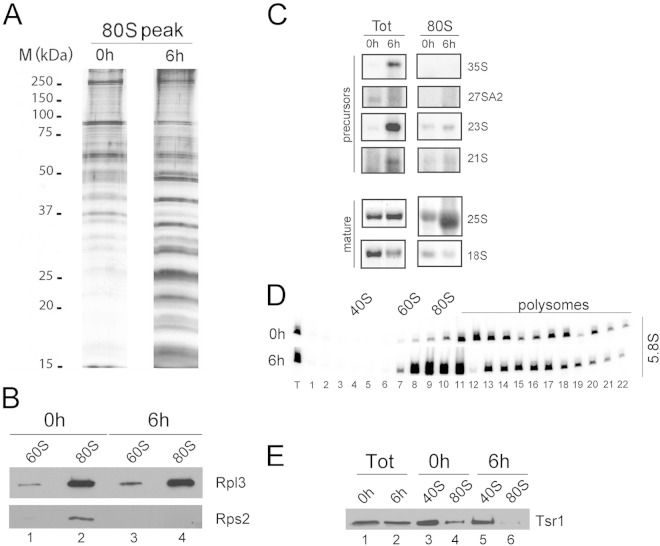
The sucrose gradient sedimentation profile seen with Dbp4-depleted cells was very peculiar: even though production of 40S subunits was impaired in those cells and was accompanied by an excess of free 60S subunits (large subunits [LSU]), there was no diminution of the 80S peak that normally contains free ribosomes (Fig. 3B). Because Dbp4 is required for pre-rRNA processing (29), the unaltered 80S peak seen in Dbp4-depleted cells could result from the accumulation of immature, preribosomal RNPs. We thoroughly examined extracts prepared from cells depleted of Dbp4 for 6 h and found that the 80S peak was enriched in low-molecular-weight proteins compared to the 80S peak of nondepleted cells (Fig. 10A, compare the 0-h and 6-h time points). Western blot analyses with antibodies directed against ribosomal proteins L3 and S2 (also known as uL3 and uS5, respectively [49]) showed that the ratios of Rpl3 to Rps2 in the 80S peak were increased upon depletion of Dbp4 (Fig. 10B). This suggests that the 80S peak of Dbp4-depleted cells is devoid of 40S subunits and does not represent free ribosomes. Northern blot hybridization revealed that the 80S peak of Dbp4-depleted cells was enriched in mature 25S and 5.8S rRNAs (Fig. 10C and D, respectively), whereas the abundances of various pre-rRNA species in the 80S peak were the same in depleted and nondepleted cells (Fig. 10C, compare third and fourth lanes). Furthermore, 7S and 6S pre-rRNAs, which are precursors of 5.8S rRNA (13), did not accumulate in the 80S peaks of Dbp4-depleted cells (data not shown).
FIG 10.
Depletion of Dbp4 leads to accumulation of mature LSU rRNAs in the 80S peak. Extracts were prepared from nondepleted cells (0 h) or cells depleted of Dbp4 for 6 h. (A) Proteins isolated from the 80S peaks of sucrose gradients were first analyzed by SDS-PAGE and then stained with silver nitrate. The sizes of molecular mass markers are given on the left. (B) Analysis of ribosomal proteins. Proteins of the 60S and 80S peaks of the gradients were first fractionated by SDS-PAGE and then transferred to a PVDF membrane. The blot was subjected to immunodetection with anti-Rpl3 (LSU protein) and anti-Rps2 (SSU protein) antibodies. (C) Northern blot hybridization of precursor and mature rRNAs. Total RNA (Tot) and RNAs from the 80S peak were fractionated in a denaturing agarose gel and were transferred to a nylon membrane. The blot was probed for various pre-rRNA species or mature rRNAs (indicated on the right). (D) Northern blot of 5.8S rRNA in fractions of sucrose gradients. The blots shown in Fig. 3C (0 h and 6 h) were probed for mature 5.8S rRNA. (E) Analysis of the pre-40S assembly factor Tsr1. Proteins of the extracts (Tot) and the 40S and 80S peaks of sucrose gradients from nondepleted (0 h) or Dbp4-depleted (6 h) cells were first fractionated by SDS-PAGE and then transferred to a PVDF membrane. The blot was subjected to immunodetection with anti-Tsr1 antibodies.
Alteration of SSU biogenesis can lead to relocation of pre-40S RNPs into 80S-like particles (38, 50). To determine if the 80S peak observed upon Dbp4 depletion contained pre-40S particles, sucrose gradient fractions corresponding to 40S and 80S peaks were isolated from nondepleted and Dbp4-depleted cells and were subjected to Western blot analyses for the pre-40S assembly factor Tsr1 (38). There was no accumulation of Tsr1 in the 80S peak from Dbp4-depleted cells (Fig. 10E, compare lane 6 with lane 4). We conclude that depletion of Dbp4 did not lead to relocation of pre-40S particles in the 80S peak.
DISCUSSION
A role for Dbp4 in ribosome biogenesis was originally proposed on the basis of its genetic interaction with U14 snoRNA, which is essential for 18S rRNA production (1, 10): overexpression of Dbp4 could suppress growth defects caused by mutations in the Y domain of U14, and it was proposed that Dbp4 would also be required for 18S rRNA synthesis (21). It was later shown that cellular depletion of Dbp4 impaired the production of 18S rRNA due to cleavage defects at sites A0, A1, and A2, leading to the accumulation of the aberrant 23S pre-rRNA species (29), a phenotype identical to that seen in cells depleted of U3 snoRNA or U3-specific proteins (1, 15, 37). Dbp4 depletion also caused U14 snoRNA to remain associated with the 35S pre-rRNA, and it was envisioned that the RNA helicase activity of Dbp4 could release U14 from the pre-rRNA (29).
To examine the physical interaction of Dbp4 with U14 snoRNA, we first carried out IPs using whole-cell extracts (WCEs) prepared from a yeast strain that expresses HA-tagged Dbp4; it should be noted that in this strain, Dbp4-HA is placed under the control of its endogenous promoter in order to prevent its overexpression (33). IPs with an anti-HA MAb indicated that Dbp4 is associated with U3 snoRNA and the U3-specific protein Mpp10 (Fig. 1 and 2A). Since U3 and Mpp10 are required for pre-rRNA processing reactions at sites A0, A1, and A2, our results are consistent with the observation that cellular depletion of Dbp4 leads to processing defects at sites A0 to A2 (29). U14 snoRNA did not coimmunoprecipitate with Dbp4 when WCEs were used (Fig. 1B). Although this was also observed in previous experiments (31), our results are different from those of the earlier study by Kos and Tollervey (29), who showed that both U3 and U14 snoRNAs coimmunoprecipitated with Dbp4. This could be due to differences in experimental procedures, such as the difference in the concentration of WCEs, which was about 20-fold higher in the experiments carried out by Kos and Tollervey (29).
U3 snoRNA and Mpp10 are components of the SSU processome, a large RNP that sediments at ∼80S in sucrose gradients (15). As seen in Fig. 1 and 2A, the association of Dbp4 with U3 and Mpp10 was not salt stable. This is in marked contrast to what is generally observed in IPs conducted with genuine SSU processome components (15, 17). The sensitivity to salt concentrations suggests that the interaction of Dbp4 with the SSU processome could depend on electrostatic interactions (51, 52), which is plausible, because Dbp4 contains patches of positively and negatively charged residues (21, 22). Sucrose gradient sedimentation analyses indicated that a large fraction of Dbp4 did not cosediment with U3 and Mpp10 in the ∼80S region of the gradient (Fig. 2B). This observation is in agreement with proteomic studies that failed to identify Dbp4 in SSU processome/90S preribosome preparations (15, 17, 53), and it supports the idea that Dbp4 is not a stable component of the SSU processome. Nevertheless, Dbp4 is required for SSU processome formation and function. EM studies on chromatin spreads revealed that depletion of Dbp4 rapidly caused defects in the appearance of terminal knobs (Fig. 7); however, the most striking defect seen in Dbp4-depleted cells was the strong reduction in pre-rRNA cotranscriptional cleavage, which crashed within a few hours of depletion (Fig. 8). Taken together, our results suggest that Dbp4 is an SSU processome component but that its association with the SSU processome is likely to be transient.
Cellular depletion of Dbp4 leads to retention of U14 snoRNA on the pre-rRNA (29) (Fig. 3C), and it is reasonable to assume that Dbp4 is the RNA helicase that unwinds the U14–pre-rRNA duplex. Here we showed that depletion of two SSU processome components, U3 snoRNA and Mpp10, also led to the trapping of U14 in very large complexes (Fig. 5C and 6C). With extracts from U3- and Mpp10-depleted cells, we were expecting that the sucrose gradient sedimentation profiles of Dbp4 and U14 would be the same, which was not the case. In fact, in marked contrast to the findings for U14 snoRNA, depletion of U3 or Mpp10 did not considerably alter the sedimentation profile of Dbp4 (Fig. 5D and 6D), implying that Dbp4 did not follow U14 in very large complexes.
Depletion of U3, Mpp10, U14, or Dbp4 prevents SSU processome formation (15, 16) (Fig. 7) and leads to the trapping of U14 in large complexes (Fig. 3 to 6). Depletion of Bfr2, another SSU processome component, caused a similar phenotype for U14 snoRNA (31). Therefore, assembly of the SSU processome could be a prerequisite for Dbp4 to “find” U14 and release it from pre-rRNA. If the trapping of U14 is a consequence of impaired SSU processome formation, one could view U14 as a sentinel or sensor of correct SSU processome assembly and function, and the release of U14 by Dbp4 would be a checkpoint in this process. It is not clear what mediates the interaction of Dbp4 with the SSU processome. Although we showed an association of Dbp4 with U3 snoRNA (Fig. 1), a direct interaction of Dbp4 with U3 appears unlikely, because many proteins are probably already present in the particle before Dbp4 interacts with it (see also the model proposed in reference 31). Since Dbp4 contains a highly conserved coiled-coil motif in its C-terminal extension (31), it is probable that Dbp4 would contact the SSU processome through interactions with SSU processome proteins, many of which contain coiled-coil motifs (15). Such interactions could allow the correct positioning of Dbp4 prior to its action on the U14–pre-rRNA duplex.
Complementation assays with a truncated version of Dbp4 that lacks its C-terminal extension (Dbp4ΔCt) revealed that this region is essential for growth (data not shown). Dbp4ΔCt changed the sucrose gradient sedimentation pattern of U14 snoRNA, which was trapped in high-molecular-weight complexes (Fig. 9A). This phenotype was identical to that seen when cells were depleted of Dbp4 (Fig. 3) or when point mutations were introduced into the catalytic core of Dbp4 (29). Therefore, in addition to the ATPase/helicase activity of Dbp4, the C-terminal extension plays a critical role in releasing U14 from pre-rRNA. We know that Dbp4 associates with U14 snoRNA in a complex of about 50S (31). Here we demonstrated that the C-terminal extension is required to maintain the association of Dbp4 with U14 snoRNA in the 50S complex (Fig. 9C).
Given that Dbp4 depletion impairs 18S rRNA synthesis (29), it was stunning to observe that the 80S peak did not diminish upon Dbp4 depletion (Fig. 3B). We first suspected that the 80S peak contained 23S pre-rRNA associated with ribosomal proteins and other nucleolar factors, but a detailed analysis indicated that this 80S peak was not enriched in 23S pre-rRNA but rather contained large amounts of mature rRNAs of the LSU (25S and 5.8S) and very little mature 18S rRNA (Fig. 10). To determine whether the 80S-like particles could be pre-40S subunits, we examined the presence of Tsr1, a late pre-40S assembly factor (38, 50). Tsr1 did not accumulate in the 80S peak, suggesting that it did not contain pre-40S particles (Fig. 10E). Taken together, the data suggest that Dbp4-depleted cells accumulate LSU particles containing mature rRNAs; these LSUs likely interact with additional cellular components to form RNPs of ∼80S. To our knowledge, this is the first time such an observation has been reported. Further studies will be required to determine what makes “normal” 60S subunits sediment at 80S.
ACKNOWLEDGMENTS
We are grateful to Susan Baserga, Maurille Fournier, Katrin Karbstein, Jonathan Warner, and Karen Wehner for the generous gift of yeast strains and reagents.
This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada to F.D. (RGPIN 249792) and by grants from the National Science Foundation (MCB-0818818) and NIH (R01GM063952) to A.L.B.
This article is dedicated to the memory of Brianna Maree Mitchell.
REFERENCES
- 1.Venema J, Tollervey D. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet 33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 2.Moore PB, Steitz TA. 2002. The involvement of RNA in ribosome function. Nature 418:229–235. doi: 10.1038/418229a. [DOI] [PubMed] [Google Scholar]
- 3.Kressler D, Linder P, de La Cruz J. 1999. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol Cell Biol 19:7897–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. 2003. Ribosome assembly in eukaryotes. Gene 313:17–42. doi: 10.1016/S0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 5.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kressler D, Hurt E, Bassler J. 2010. Driving ribosome assembly. Biochim Biophys Acta 1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Henras AK, Dez C, Henry Y. 2004. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr Opin Struct Biol 14:335–343. doi: 10.1016/j.sbi.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 8.Kiss T. 2001. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J 20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decatur WA, Fournier MJ. 2003. RNA-guided nucleotide modification of ribosomal and other RNAs. J Biol Chem 278:695–698. doi: 10.1074/jbc.R200023200. [DOI] [PubMed] [Google Scholar]
- 10.Maxwell ES, Fournier MJ. 1995. The small nucleolar RNAs. Annu Rev Biochem 64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 11.King TH, Liu B, McCully RR, Fournier MJ. 2003. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol Cell 11:425–435. doi: 10.1016/S1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- 12.Liang XH, Liu Q, King TH, Fournier MJ. 2010. Strong dependence between functional domains in a dual-function snoRNA infers coupling of rRNA processing and modification events. Nucleic Acids Res 38:3376–3387. doi: 10.1093/nar/gkq043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woolford JL Jr, Baserga SJ. 2013. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195:643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piekna-Przybylska D, Decatur WA, Fournier MJ. 2007. New bioinformatic tools for analysis of nucleotide modifications in eukaryotic rRNA. RNA 13:305–312. doi: 10.1261/rna.373107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, Beyer AL, Hunt DF, Baserga SJ. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL. 2004. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol Cell 16:943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 17.Bernstein KA, Gallagher JE, Mitchell BM, Granneman S, Baserga SJ. 2004. The small-subunit processome is a ribosome assembly intermediate. Eukaryot Cell 3:1619–1626. doi: 10.1128/EC.3.6.1619-1626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phipps KR, Charette J, Baserga SJ. 2011. The small subunit processome in ribosome biogenesis—progress and prospects. Wiley Interdiscip Rev RNA 2:1–21. doi: 10.1002/wrna.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim YH, Charette JM, Baserga SJ. 2011. Assembling a protein-protein interaction map of the SSU processome from existing datasets. PLoS One 6:e17701. doi: 10.1371/journal.pone.0017701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez-Fernandez J, Martin-Marcos P, Dosil M. 2011. Elucidation of the assembly events required for the recruitment of Utp20, Imp4 and Bms1 onto nascent pre-ribosomes. Nucleic Acids Res 39:8105–8121. doi: 10.1093/nar/gkr508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang WQ, Clark JA, Fournier MJ. 1997. The rRNA-processing function of the yeast U14 small nucleolar RNA can be rescued by a conserved RNA helicase-like protein. Mol Cell Biol 17:4124–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia I, Albring MJ, Uhlenbeck OC. 2012. Duplex destabilization by four ribosomal DEAD-box proteins. Biochemistry 51:10109–10118. doi: 10.1021/bi301172s. [DOI] [PubMed] [Google Scholar]
- 23.Jankowsky E. 2011. RNA helicases at work: binding and rearranging. Trends Biochem Sci 36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silverman E, Edwalds-Gilbert G, Lin RJ. 2003. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene 312:1–16. doi: 10.1016/S0378-1119(03)00626-7. [DOI] [PubMed] [Google Scholar]
- 25.Cordin O, Banroques J, Tanner NK, Linder P. 2006. The DEAD-box protein family of RNA helicases. Gene 367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 26.de la Cruz J, Kressler D, Linder P. 1999. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem Sci 24:192–198. doi: 10.1016/S0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- 27.Bleichert F, Baserga SJ. 2007. The long unwinding road of RNA helicases. Mol Cell 27:339–352. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez-Galan O, Garcia-Gomez JJ, de la Cruz J. 2013. Yeast and human RNA helicases involved in ribosome biogenesis: current status and perspectives. Biochim Biophys Acta 1829:775–790. doi: 10.1016/j.bbagrm.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Kos M, Tollervey D. 2005. The putative RNA helicase Dbp4p is required for release of the U14 snoRNA from preribosomes in Saccharomyces cerevisiae. Mol Cell 20:53–64. doi: 10.1016/j.molcel.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 30.Granneman S, Baserga SJ. 2004. Ribosome biogenesis: of knobs and RNA processing. Exp Cell Res 296:43–50. doi: 10.1016/j.yexcr.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 31.Soltanieh S, Lapensée M, Dragon F. 2014. Nucleolar proteins Bfr2 and Enp2 interact with DEAD-box RNA helicase Dbp4 in two different complexes. Nucleic Acids Res 42:3194–3206. doi: 10.1093/nar/gkt1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner AJ, Knox AA, Prieto JL, McStay B, Watkins NJ. 2009. A novel small-subunit processome assembly intermediate that contains the U3 snoRNP, nucleolin, RRP5, and DBP4. Mol Cell Biol 29:3007–3017. doi: 10.1128/MCB.00029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963–972. doi:. [DOI] [PubMed] [Google Scholar]
- 34.Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961. doi:. [DOI] [PubMed] [Google Scholar]
- 35.Gari E, Piedrafita L, Aldea M, Herrero E. 1997. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13:837–848. doi:. [DOI] [PubMed] [Google Scholar]
- 36.Dragon F, Pogacic V, Filipowicz W. 2000. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol Cell Biol 20:3037–3048. doi: 10.1128/MCB.20.9.3037-3048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunbar DA, Wormsley S, Agentis TM, Baserga SJ. 1997. Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol Cell Biol 17:5803–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strunk BS, Novak MN, Young CL, Karbstein K. 2012. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell 150:111–121. doi: 10.1016/j.cell.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vilardell J, Warner JR. 1997. Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol Cell Biol 17:1959–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lemay V, Hossain A, Osheim YN, Beyer AL, Dragon F. 2011. Identification of novel proteins associated with yeast snR30 small nucleolar RNA. Nucleic Acids Res 39:9659–9670. doi: 10.1093/nar/gkr659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fabrizio P, Esser S, Kastner B, Luhrmann R. 1994. Isolation of S. cerevisiae snRNPs: comparison of U1 and U4/U6.U5 to their human counterparts. Science 264:261–265. doi: 10.1126/science.8146658. [DOI] [PubMed] [Google Scholar]
- 43.Billy E, Wegierski T, Nasr F, Filipowicz W. 2000. Rcl1p, the yeast protein similar to the RNA 3′-phosphate cyclase, associates with U3 snoRNP and is required for 18S rRNA biogenesis. EMBO J 19:2115–2126. doi: 10.1093/emboj/19.9.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wehner KA, Gallagher JE, Baserga SJ. 2002. Components of an interdependent unit within the SSU processome regulate and mediate its activity. Mol Cell Biol 22:7258–7267. doi: 10.1128/MCB.22.20.7258-7267.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perez-Fernandez J, Roman A, De Las Rivas J, Bustelo XR, Dosil M. 2007. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol Cell Biol 27:5414–5429. doi: 10.1128/MCB.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dutca LM, Gallagher JE, Baserga SJ. 2011. The initial U3 snoRNA:pre-rRNA base pairing interaction required for pre-18S rRNA folding revealed by in vivo chemical probing. Nucleic Acids Res 39:5164–5180. doi: 10.1093/nar/gkr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samarsky DA, Fournier MJ. 1998. Functional mapping of the U3 small nucleolar RNA from the yeast Saccharomyces cerevisiae. Mol Cell Biol 18:3431–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ. 2004. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev 18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, McAlear MA, Moore PB, Noller HF, Ortega J, Panse VG, Ramakrishnan V, Spahn CM, Steitz TA, Tchorzewski M, Tollervey D, Warren AJ, Williamson JR, Wilson D, Yonath A, Yusupov M. 2014. A new system for naming ribosomal proteins. Curr Opin Struct Biol 24:165–169. doi: 10.1016/j.sbi.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferreira-Cerca S, Kiburu I, Thomson E, LaRonde N, Hurt E. 2014. Dominant Rio1 kinase/ATPase catalytic mutant induces trapping of late pre-40S biogenesis factors in 80S-like ribosomes. Nucleic Acids Res 42:8635–8647. doi: 10.1093/nar/gku542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katsamba PS, Myszka DG, Laird-Offringa IA. 2001. Two functionally distinct steps mediate high affinity binding of U1A protein to U1 hairpin II RNA. J Biol Chem 276:21476–21481. doi: 10.1074/jbc.M101624200. [DOI] [PubMed] [Google Scholar]
- 52.Doetsch M, Schroeder R, Furtig B. 2011. Transient RNA-protein interactions in RNA folding. FEBS J 278:1634–1642. doi: 10.1111/j.1742-4658.2011.08094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schafer T, Kuster B, Tschochner H, Tollervey D, Gavin AC, Hurt E. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol Cell 10:105–115. doi: 10.1016/S1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]