Abstract
Retroviruses have evolved complex transcriptional enhancers and promoters that allow their replication in a wide range of tissue and cell types. Embryonic stem (ES) cells, however, characteristically suppress transcription of proviruses formed after infection by exogenous retroviruses and also of most members of the vast array of endogenous retroviruses in the genome. These cells have unusual profiles of transcribed genes and are poised to make rapid changes in those profiles upon induction of differentiation. Many of the transcription factors in ES cells control both host and retroviral genes coordinately, such that retroviral expression patterns can serve as markers of ES cell pluripotency. This overlap is not coincidental; retrovirus-derived regulatory sequences are often used to control cellular genes important for pluripotency. These sequences specify the temporal control and perhaps “noisy” control of cellular genes that direct proper cell gene expression in primitive cells and their differentiating progeny. The evidence suggests that the viral elements have been domesticated for host needs, reflecting the wide-ranging exploitation of any and all available DNA sequences in assembling regulatory networks.
INTRODUCTION
Retroviruses are significant pathogens for their hosts. Some members of this large family of viruses can be directly cytopathic, but many others cause disease—most often leukemias—by insertional activation of genes in an infected somatic cell (1, 2). The rare insertion of proviral DNAs into the germ line can cause even more extended damage, creating mutations that are inherited by offspring of the infected individual as a new Mendelian locus (3, 4). Because of these serious consequences, there has been strong pressure for the host to evolve mechanisms to restrict or limit the spread of retroviral genomes, especially in embryonic cells. A vast array of virus restriction factors, often controlled by the interferon system, has been discovered and is currently under intense study (5, 6). In addition, embryonic cells have long been known to exhibit a specific ability to potently suppress the expression of both exogenous (7) and endogenous (8) retroviral sequences. This transcriptional silencing is probably critical to the maintenance of the genetic stability of these cells, limiting inappropriate transcription and further genotypic damage from subsequent retroviral replication. Remarkably, the silencing of retroviruses is closely correlated with the transcriptional regulation of developmentally important genes. Recent findings are revealing that this correlation is not coincidental: retroviral regulatory elements have frequently been coopted by the cell to control genes important in embryogenesis (9). This “domestication” of retroviral elements is an example of the flexible exploitation of DNAs during evolution. We here review and ruminate on some of these findings.
RETROVIRAL SILENCING IN MOUSE ES CELLS
Mouse embryonic stem (ES) cell lines were first generated in 1981 by culturing mouse inner cell mass (ICM) explants on feeder layers and since then have been exploited as a model system to study the characteristics of pluripotency (10). They are unique among primary cells in that they can give rise to all cell types of the body and have a very high self-renewing capacity. Diverse epigenetic and chromatin marks are used in ES cells for the genomic silencing of incoming and endogenous retroviruses. The silencing of the Moloney murine leukemia virus (MMLV) genome in embryonic cells has been studied in great detail (Fig. 1). MMLV can infect and integrate the viral DNA into the genomes of embryonic carcinoma and ES cells, but once integrated, the provirus is transcriptionally silent and cannot produce infectious progeny virions (7). The repression is heavily dependent on a conserved sequence element termed the primer binding site (PBS) (Fig. 1) (11), an 18-nucleotide sequence complementary to the 3′ end of proline tRNA, the tRNA primer used for initiation of reverse transcription by MMLV (12, 13). The zinc finger DNA binding protein ZFP809 was shown to mediate the silencing by binding to the proline PBS sequence of the integrated provirus DNA (14). A well-characterized corepressor, Trim28/Kap-1/Tif1b, interacts with ZFP809 to initiate the epigenetic marking of the provirus (15, 16). Another zinc finger protein, the cofactor yin yang 1 (YY1), can also bind the proviral long terminal repeat (LTR) of many retroviruses to enhance the recruitment of Trim28 (17). This explains why many retroviral vectors utilizing alternative PBS sequences, which are not recognized by ZFP809, are still subject to some transcriptional repression (18, 19). YY1 binding to the proviral LTR is highly efficient and specific during the first 4 days after infection, e.g., during the initiation of the silencing. At this stage, binding of both YY1 and ZFP809 is needed for the effective recruitment of Trim28. In cells lacking YY1 expression, or in differentiated cells where YY1 does not bind Trim28, the downstream recruitment of the epigenetic silencing complex is blocked and the expression level of the provirus is elevated. When tethered to the provirus by ZFP809 and YY1, Trim28 recruits several other key players to establish silencing. Binding of heterochromatin protein 1 (HP1) is absolutely necessary for MMLV silencing (20). Additional factors are required to impose a closed and silenced chromatin structure on the provirus. Other members of the complex identified so far are ErbB3 binding protein 1 (EBP1) (21), the H3K9 trimethylase ESET (22, 23), and the polycomb complex catalytic subunit EZH2 (24). Very likely other factors, as yet unidentified, contribute to the efficient epigenetic silencing observed in ES cells.
FIG 1.
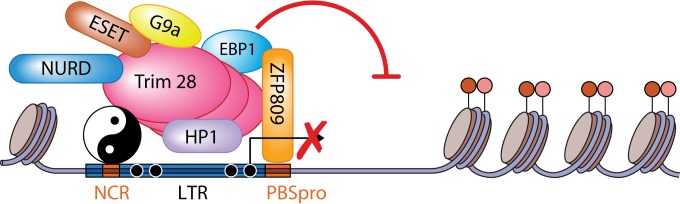
Transcriptional regulation of the integrated provirus in embryos and embryonic cells. Trim28 binding to the DNA binding proteins ZFP809 (14) and YY1 (17), as well as to EBP1 (21) and HP1 (20), results in epigenetic silencing of the newly integrated or endogenous provirus elements. The silencing is mediated by Trim28 recruitment of many factors involved in transcriptional silencing and heterochromatin formation, including the histone methyltransferases ESET (22) and G9a (34) and the NuRD histone deacetylase complex (92). These histone modifiers are responsible for the trimethylation of H3K9 (H3K9Me3), H4K20 (H4K20Me3) (22), and H3K27 (H3K27me3) (24) (red and pink circles) histone tails as well as for subsequent DNA methylation (93). NCR, noncoding region.
Mammalian genomes contain hundreds of endogenous retroviruses (ERVs) (25). In the mouse genome, the ERVs can be divided into class I, II, and III families based on sequence similarities. Trim28 is involved in the silencing of many of these ERVs in pluripotent cells. In cells lacking Trim28, the expression of ERV sequences is highly elevated, and the repressive chromatin marks on the ERV DNAs are not established properly (26). Trim28 mediates the silencing of the murine class I and II ERVs via the recruitment of ESET, which leads to the trimethylation of H3K9 (H3K9Me3) and histone H4 lysine 20 (H4K20Me3) (22, 27, 28). YY1 is responsible, at least in part, for the recruitment of Trim28 to these ERV sequences (17, 29) and thus to the initiation of silencing in ES cells. Another zinc finger protein, ZFP819, was also recently shown to be involved in ERV silencing (30). The murine class III ERVs exhibit different expression patterns; they are highly expressed in early stages after fertilization of the oocyte and are downregulated at the morula and blastula stages. The expression of murine ERV-L (MERV-L), a member of this group, is upregulated in cells lacking the histone demethylase LSD1/KDM1A (31), the pluripotency-specific transcription factor ZFP42/Rex1 (29, 32), the polycomb-associated protein RYBP (33), and the H3K9 trimethyl transferase G9a (34). Although their contribution is less clear, the polycomb group (PcG) complex I and II (PRC1 and -2) proteins—responsible for histone H3K27 trimethylation—were shown to be involved in the silencing of these ERV classes as well (24, 35). Mouse ES lines with null mutations in the Suv39h histone methyltransferase show decreased H3K9me3 levels in the repetitive elements and a marked increase in H3K27me3 enrichment, suggesting potential feedback control between these two modification systems and at least some shared functionality of the two modifications (36). While the exact mechanisms involved in silencing of diverse ERV classes remain to be elucidated, it is clear that chromatin state plays an important role in suppressing the activities of ERVs and other transposable elements.
ERV LTRs AS REGULATORS OF CELLULAR GENES
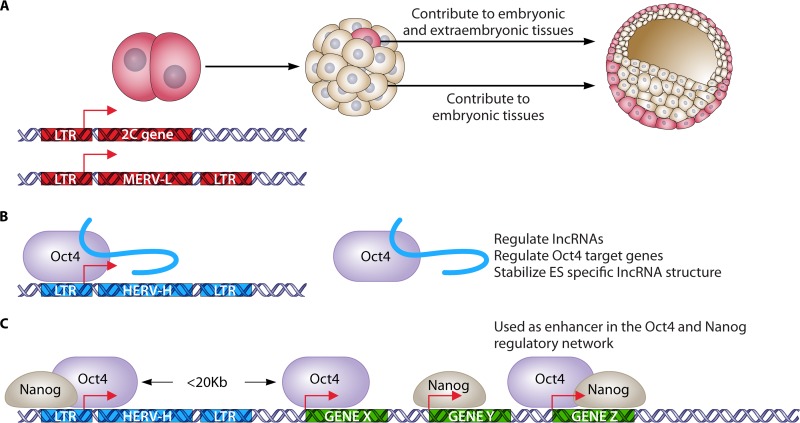
When Barbara McClintock first discovered transposable elements in maize more than 50 years ago, she called them “controlling elements” because they altered gene expression (37). We now appreciate that many ERVs similarly affect cellular gene expression by contributing to the activities of nearby promoters and enhancers. The accumulating impact of retroviral infections on the evolution of the genome has led to the rewiring of major genetic networks in both mouse and human stem cells. This realization stems from the many examples of ERV integration events that do not cause harmful mutations but provide additional regulatory sequences that change the regulation of genes and genetic networks in useful ways. Some of the known ways that ERVs perform useful functions are diagrammed in Fig. 2. ERV promoters can drive the expression of tissue-specific genes, can express long noncoding RNAs (lncRNAs) that autoregulate and transregulate cellular genes, and provide enhancer elements that are used by cellular genes.
FIG 2.
Examples of domestication of ERV sequences by mouse and human embryonic cells. (A) MERV-L elements and their remnant “solo” long terminal repeats (LTRs) have been coopted to participate in gene regulatory networks by serving as primary or alternative promoters of nearby genes (31). A subset of mouse embryonic stem cells expresses MERV-L LTR-driven chimeric transcripts, which correlates with increased potency (46). (B) HERV-H interacts with Oct4 to promote the enhancer activities of LTR7 and nearby regions and to drive the expression of neighboring lncRNAs and protein-coding genes essential to hES cell identity (59, 61, 63). (C) ERV1 elements in the human and mouse genomes carry transcription factor-binding sites for Oct4 and Nanog, which can regulate genes that form the pluripotency network near insertion sites, leading to novel regulatory patterns in evolving mammals (9, 52).
One of the first mammalian genes shown to have a major ERV involvement was the mouse sex-limited protein (Slp) gene. An ancient ERV has integrated adjacent to the transcription start site (TSS) and dictates androgen-dependent regulation of this gene (38). Retrotransposons have also been shown to have had a complex role in the evolution of the regulation of the primate amylase gene family (39). Another intriguing example of how ERVs can facilitate the evolution of complex physiological processes involving gene expression on a global scale is their multifaceted involvement in the evolution of pregnancy and of the placenta (40). ERV sequences have been shown to be part of many functional promoters in mouse and human, often functioning as alternative, tissue-specific promoters in addition to the ancestral promoters (41, 42). Their contribution to the overall transcription may sometimes be small (43), but the number of genes affected may be very large. In a comprehensive survey of the repetitive element transcriptome, up to 30% of 5′cap-selected mouse and human RNA transcripts were found to initiate within repetitive elements (41). More than 25% of coding genes have ERV elements in their 3′ untranslated region (3′UTR), which negatively regulate their expression. Many of the ERV-initiated transcripts show high tissue specificity (44). Thirty percent of all transcripts in human embryonic tissues were associated with repetitive elements, pointing to a clear pattern of embryonic cell specificity for some viral promoters (9).
ERV EXPRESSION AND ES CELL PLURIPOTENCY
ERVs may play their most significant role in embryonic cells. The pluripotency of ES cells tracks closely with the expression levels of these elements (45, 46). The regulators of ERV expression, such as Trim28, are also key players in development (47, 48). The ERV regulatory elements are not only correlates of host gene elements but also used as host gene regulatory elements (49). ERV-dependent networks can be created when many copies of the same ERV element are used as promoters for different cellular genes. One remarkable example of such a network can be found in the early stages of embryogenesis, when particular class III ERVs regulate gene expression at the very early two-cell stage (50). At this stage, the murine endogenous retroviral element MERV-L is transiently and significantly derepressed, represents 3% of the total transcriptional output, and is very sharply regulated in time. Importantly, MERV-L DNAs represent a source of regulatory elements that are coopted by cellular genes to regulate their cell stage-specific expression. In the course of embryogenesis, more than 25% of MERV-L copies are activated and 307 protein-coding genes generate 626 different chimeric transcripts with MERV-L elements. MERV-L expression is regulated by histone modifications, such as H3K4me3, and in the absence of the H3K4me3 demethylase LSD1/KDM1A, MERV-L becomes overexpressed and embryonic development arrests at gastrulation (31). The pluripotency factor Rex1 can also bind MERV-L sequences and potentially provides sequence specificity to this silencing mechanism by LSD1 recruitment to the MERV-L DNA (32). In the rare subpopulation (∼1%) of ES cells that show high MERV-L expression, the same gene expression pattern typical for two-cell stage embryos can be observed. Hence, MERV-L regulatory sequences are necessary and sufficient to drive expression in rare ES cells that also display enhanced totipotent developmental potential. These studies highlight a critical role for ERVs in contributing to host cell fate decisions by activating a transcriptional network. This is mediated by epigenetic marks that are established and removed by the endogenous cellular machinery.
The major mechanism of action of ERV regulation of cellular genes is by providing binding sites for transcriptional regulatory factors (both positively and negatively acting). ERVs control or affect the regulation of a wide range of genes. These include genes in the human tumor suppressor protein p53 network, where over one-third of the protein binding sites harbor an ERV1 element (51). In ES cells, a striking example is the DNA binding network of two highly specific and very important pluripotency transcription factors, namely, Oct4 and Nanog. Comparison of the binding sites of these proteins in humans and mice has revealed a very low conservation, but about one-fourth of the sites harbor ERV1-like elements of the ERV class I family (52). That study identified a group of human-specific target genes that were recruited to the core regulatory network via the insertion of transposable elements. The expression of the majority of the genes with ERV1 sequences was affected by depletion of Oct4, suggesting that these genes are functional members of the pluripotency network. Using the ENCODE database, those researchers showed that the majority of the open chromatin regions in the human genome are driven from transposable element (TE) sequences, mainly ERVs (53). At least one-third of the open regions that harbor a TE sequence indeed serve as a regulatory element, as they were bound by at least one transcription factor (as shown by chromatin immunoprecipitation). These studies reveal the striking plasticity of the core regulatory network of mammalian ES cells and the importance that ERVs have had in facilitating this functional evolution.
ERVs ARE IMPORTANT FOR SUCCESSFUL REPROGRAMMING DURING THE FORMATION OF iPS CELLS
Differentiated cells can be reprogrammed by ectopic expression of specific transcription factors to become so-called induced pluripotent stem (iPS) cells. The ability of these cells to silence retroviral DNAs is a very effective marker for the “quality,” i.e., the pluripotency, of these cells (54). During mouse and human iPS cell reprograming, ERVs undergo erasure of gene-silencing marks, and their expression is thereby reactivated. However, in the resulting iPS clones, as in ES cells, most ERVs are silenced by Trim28-induced epigenetic silencing (55). Not all endogenous retroelements are silenced in reprogramming, however. In human iPS cells, the endogenous LINE-1 retroelements seem to be activated, rather than repressed, during the reprogramming of iPS cells (56). Curiously, a heterogeneous pattern of expression of some human ERVs (HERVs) was observed in human iPS clones. This heterogeneity seems to affect not only the HERVs themselves but also the nearby genes, some of them related to known human disease genes implicated in schizophrenia and cancer. In a different study, it was shown that iPS clones that retain high HERV expression levels after reprogramming are differentiation defective (57). Together, these studies suggest again that ERV silencing is generally a fundamental trait of pluripotency and that variability in ERV expression might contribute to the heterogeneity of iPS clones.
ERV RNAs AS lncRNAs
Many ERVs directly control expression of cellular, coding mRNAs, but others drive expression of long noncoding RNAs (lncRNAs), which act on genes at a distance (58, 59). A frequent association of the LTRs with lncRNA transcription start sites (TSSs) suggests a role for ERVs in the evolutionary origin of lncRNAs (60). Indeed, ERV-containing lncRNAs show increased expression in ES cells relative to non-ERV lncRNA expression. Deep profiling of the nuclear and cytoplasmic transcriptomes of human and mouse stem cells led to the identification of a class of stem cell-specific transcripts, mostly long terminal repeat (LTR)-derived transcripts (9). Some of these ERV-induced lncRNAs are associated with the maintenance of pluripotency, and many are associated with distal regulatory elements, which suggests that they make an extensive contribution to the complexity of the stem cell nuclear transcriptome. In humans, HERV-H and HERV-K families (class I and II ERVs, respectively) have a particularly strong effect in ES cells, as their transcription is regulated by Oct4 (61, 62). Moreover, HERV-H RNAs were recently shown to serve as cellular lncRNAs associated with Oct4 and Oct4-regulated genes and to be required for the maintenance of human ES (hES) cells.
Functional studies of the role of the HERV-H RNA in hES cells showed that the HERV-H LTR region first serves as a binding site for pluripotency factors such as Oct4 and thus that transcription of this element is ES cell specific (63). Subsequently, the transcribed RNA binds and recruits active chromatin modifiers and then acts to control an array of critical downstream genes. The new comprehension of ERVs' functions and their importance for pluripotency has recently led to an appreciation of their potential as markers for a holy grail of the field: naive hES cells (64).
OVERVIEW AND SPECULATIONS: WHY DO RETROVIRUSES OFTEN SERVE AS GENE REGULATORS IN ES CELLS?
A major aspect of the evolution of complex multicellular organisms has been the generation of a large repertoire of master regulators operating on an even larger repertoire of regulatory elements. The retroviral LTRs include many specific transcription factor binding sites, which, in different combinations, serve to create various patterns of gene expression. Retroviral genomes provide a rich source of such regulatory elements, which are available for exploitation during the evolution of the host genome. How did these viral promoters arise? Retroviruses have coevolved with their hosts throughout the history of life on earth (65). They replicate to their own benefit to spread as widely and effectively as possible, both by horizontal spread as infectious virus and by vertical transmission as inherited ERVs, but they do so under constant attack from the host to limit or minimize their potentially pathogenic effects. They are thus continually involved in an “arms race” with the host; the viruses are evolving to replicate as efficiently as possible, and the cell is evolving to repress their replication.
Retrovirus genomes in general are capable of rapid evolution. Their generation time as replicating viruses is much shorter than that of their hosts, and the typical mutation rate, determined by the relatively error-prone RNA polymerase II and reverse transcriptase enzymes responsible for their replication, is much higher than those of the hosts, determined by their more faithful DNA polymerase and repair systems. Retroviruses can also undergo sudden mutational changes induced by the cytidine deaminase activity of the APOBEC (apolipoprotein B mRNA-editing enzyme and catalytic) enzymes. The APOBECs can introduce multiple cytosine-to-uracil changes by deamination of the retroviral negative-strand DNA during the first steps of reverse transcription (66). Such deaminated DNA is subject to degradation by uracil removal and abasic nuclease cleavage, but genomes that escape that degradation and survive can be heavily mutated or edited. Editing modifies a large number of nucleotides simultaneously, so it can change a given element to such an extent that subsequent random mutagenesis may lead to a different evolutionary trajectory from that of the original element, without having to cross valleys of low fitness. Therefore, DNA editing can help explain how retroviruses have acquired such a diverse collection of functions (67). Active ERVs can also be edited (68), and such DNA editing has been a frequent event in the evolution of various ERV families (69).
One of the most rapidly evolving regions of the viral genome is that of the transcriptional promoter and enhancers, encoded in the U3 region of the LTR. This region determines the tissue tropism and cell type specificity of the virus; i.e., it identifies the cells in which the virus will be most strongly expressed to produce the most progeny virus. There is strong selective pressure for the virus to evolve and acquire good binding sites for host transcription factors that will drive viral transcription. Different viruses will evolve to grow in particular tissue niches (T cells, B cells, erythroid cells, or steroid-responsive cells) by virtue of the binding sites present in their LTRs. In some settings, over evolutionary times, it may also be an advantage for a virus to be able to lie quiet as an integrated provirus and so be transcriptionally silent. The virus can thereby evade host defenses for some period of time and then reactivate later to initiate a new period of replication. In this situation, the virus may be selected to acquire sites that mediate transcriptional silencing by the host. Thus, we can think of the retroviruses as evolving regulatory elements that will allow for their selective growth and survival in a wide range of different tissues and under different circumstances. Examination of the many exogenous retroviruses in mice and the ERVs in virtually all branches of life reveals a highly varied set of binding sites for transcription factors, both positive and negative, in the viral LTRs that affect expression and cell tropism (see, e.g., references 70 to 74).
Because retroviruses insert proviral DNAs throughout the genome during infection and are able to introduce DNAs into the germ line by infecting preimplantation embryos (3, 4), the ERVs provide a rich reservoir of regulatory elements that are scattered widely throughout the genome. Thus, it is not surprising that these viral elements can be coopted by the host over evolutionary times to serve useful roles in the regulation of developmentally important host genes. Genome rearrangements can quickly conjoin a viral LTR with a target gene and thus create a novel pattern of expression for that gene. These rearrangements can include retrovirus-mediated transposition or nonviral translocations.
The pool of regulatory elements in retroviral promoters is not only diverse and rapidly mutating but may have other useful features. One potentially important aspect of retroviral promoters is the metastable nature of ERV-containing sequences (75). Metastable alleles display the unusual characteristic of variable expressivity in the absence of genetic heterogeneity, which is dependent on their epigenetic state. Interestingly, the epigenetic state of ERVs is somewhat labile, resulting in phenotypic mosaicism between cells (variegation) and also between individuals (variable expressivity). Many studies suggest that retroviral expression is especially prone to variegation, due to its epigenetic regulation (76–80). Grown under different culture conditions, mouse ES cells were shown to display the transcriptional heterogeneity of many of their pluripotency markers, including Nanog (81) and REX1 (82). The establishment of the epigenetic state occurs during early embryogenesis and is a probabilistic and somewhat stochastic event that can produce major consequences (83). We now know that ES cells are not a uniform cell population but rather exist in a metastable state and shift between ICM- and epiblast-like states while retaining pluripotency. This equilibrium can shift in either direction in response to a variety of factors, including epigenetic regulators. Specifically, it is the interplay between variable transcriptional activation signals and the repressive influence of the nucleosome remodeling and deacetylation (NuRD) complex that results in transcriptional heterogeneity at pluripotency-associated genes in ES cell cultures (84). This model suggests regulation in which the NuRD complex regulates gene expression, not only by straightforward silencing but also by restricting the dynamic range of transcription. Remarkably, it was also shown that heterogeneous expression of class II and III ERVs can affect mouse ES cell pluripotency and differentiation potential (85). As mentioned above, the NuRD complex is recruited to the proviral LTR and plays an important role in exogenous and endogenous retroviral silencing in ES cells. These observations led to the suggestion that fluctuations in gene expression levels may help coordinate differentiation in a fraction of otherwise-identical cells. On a longer time scale, reequilibration would maintain a fully responsive stem cell population that might again be fractionally induced (86). Integrating these data, we suggest that retroviral sequences are especially fit to regulate pluripotency networks due to the heterogeneous and somewhat stochastic nature of their transcriptional regulation.
Recent studies addressing the question of developmental noise, e.g., phenotypic variability in isogenic animals reared in controlled environments, show an involvement of epigenetic regulators in reducing the noise (87). Remarkably, two main factors that were shown to be involved are the NuRD complex and Trim28 (88), both important regulators of ERV silencing in ES cells. Thus, it might be speculated that downregulating ERVs is directly and causally linked to the observed reduction in the transcriptional noise. Although not much is known about the regulation of lentiviral ERVs, it is interesting to note that the Tat gene's origins can be traced back for 12 to 14 million years through the ERV lineage (89). Recently, a mechanistic model has been suggested for the involvement of Tat stochastic fluctuations in determining proviral latency (90, 91). These findings illustrate the importance of stochastic fluctuations in gene expression and suggest yet another mechanism through which ERVs' tendency toward heterogeneous expression may have contributed to mammalian evolution. We suggest that the regulatory elements of the ERVs are used by the genomes of mammals as “noise-promoting agents” to enhance the population variability and thus also long-term evolutionary fitness. Epigenetic mechanisms not only are one of the cell's weapons to fight the infecting retrovirus in the arms race but also are used as a valve to control and regulate phenotypic and genotypic variability.
CONCLUDING REMARKS
We have here speculated (i) on the evolution of the diversity of transcriptional regulatory elements in retroviral genomes as providing advantages to viruses replicating in varied cell types and under different circumstances and (ii) on the domestication of these elements as useful tools in the evolution of regulatory networks in host developmental pathways. The intimate relationships between retroviruses and their hosts are reflected in the way that their transcriptional regulatory pathways are so intertwined. Thus, retroviral transcription is often a close proxy for the transcription pattern of many host genes important for development, but more importantly, retroviral transcription in many cases actually determines the host patterns of gene expression because viral regulatory sequences were exploited in the formation of the cellular determinants of those patterns. We expect that the future will uncover many more examples of how the ERVs are not passive junk sequences nested within larger genomes but are important players in both health and disease.
Biographies

Sharon Elizur-Schlesinger received her Ph.D. from the Hebrew University Medical School under the cosupervision of Profs. Howard Cedar and Yehudit Bergman for work revealing the epigenetic regulation of ribosomal genes during early embryogenesis. She recently completed postdoctoral training in the laboratory of Stephen Goff at Columbia University. Her work defined the chromatin state of silenced proviruses in embryonic stem (ES) cells, revealed the importance of the cellular factor YY1 in silencing, and documented the asynchronous silencing of independent proviruses in a single ES cell. Currently Dr. Schlesinger is collaborating with Prof. Eran Meshorer from the life science faculty of the Hebrew University. The collaboration aims to study epigenetic regulation of genes in pluripotent cells and during early differentiation. Schlesinger's long-term goal is to broaden our understanding of known silencing mechanisms that function in stem cells, as well as to reveal novel ones.

Stephen P. Goff is presently Higgins Professor of Biochemistry and a Howard Hughes Investigator at Columbia University Medical Center in New York City. He received his A.B. from Amherst College and his Ph.D. from Stanford University, working in the laboratory of Paul Berg, and then trained as a postdoctoral fellow with David Baltimore at MIT before starting up his own lab at Columbia. He has been studying retrovirus replication and oncogenesis for more than 30 years. He is particularly interested in the roles of specific host factors in restricting or promoting virus replication. His laboratory continues to study cellular proteins affecting intracellular trafficking of virus, integration of viral DNA, expression of viral RNAs, and assembly of virions.
REFERENCES
- 1.Payne GS, Bishop JM, Varmus HE. 1982. Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature 295:209–214. doi: 10.1038/295209a0. [DOI] [PubMed] [Google Scholar]
- 2.Neel BG, Gasic GP, Rogler CE, Skalka AM, Ju G, Hishinuma F, Papas T, Astrin SM, Hayward WS. 1982. Molecular analysis of the c-myc locus in normal tissue and in avian leukosis virus-induced lymphomas. J Virol 44:158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaenisch R. 1976. Germ line integration and Mendelian transmission of the exogenous Moloney leukemia virus. Proc Natl Acad Sci U S A 73:1260–1264. doi: 10.1073/pnas.73.4.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaenisch R. 1977. Germ line integration of Moloney leukemia virus: effect of homozygosity at the m-mulV locus. Cell 12:691–696. doi: 10.1016/0092-8674(77)90269-0. [DOI] [PubMed] [Google Scholar]
- 5.Hatziioannou T, Bieniasz PD. 2011. Antiretroviral restriction factors. Curr Opin Virol 1:526–532. doi: 10.1016/j.coviro.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng YH, Jeang KT, Tokunaga K. 2012. Host restriction factors in retroviral infection: promises in virus-host interaction. Retrovirology 9:112. doi: 10.1186/1742-4690-9-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teich NM, Weiss RA, Martin GR, Lowy DR. 1977. Virus infection of murine teratocarcinoma stem cell lines. Cell 12:973–982. doi: 10.1016/0092-8674(77)90162-3. [DOI] [PubMed] [Google Scholar]
- 8.Brulet P, Condamine H, Jacob F. 1985. Spatial distribution of transcripts of the long repeated ETn sequence during early mouse embryogenesis. Proc Natl Acad Sci U S A 82:2054–2058. doi: 10.1073/pnas.82.7.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fort A, Hashimoto K, Yamada D, Salimullah M, Keya CA, Saxena A, Bonetti A, Voineagu I, Bertin N, Kratz A, Noro Y, Wong CH, de Hoon M, Andersson R, Sandelin A, Suzuki H, Wei CL, Koseki H, Consortium F, Hasegawa Y, Forrest AR, Carninci P. 2014. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat Genet 46:558–566. doi: 10.1038/ng.2965. [DOI] [PubMed] [Google Scholar]
- 10.Evans MJ, Kaufman MH. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 11.Barklis E, Mulligan RC, Jaenisch R. 1986. Chromosomal position or virus mutation permits retrovirus expression in embryonal carcinoma cells. Cell 47:391–399. doi: 10.1016/0092-8674(86)90596-9. [DOI] [PubMed] [Google Scholar]
- 12.Harada F, Peters GG, Dahlberg JE. 1979. The primer tRNA for Moloney murine leukemia virus DNA synthesis. Nucleotide sequence and aminoacylation of tRNAPro. J Biol Chem 254:10979–10985. [PubMed] [Google Scholar]
- 13.Petersen R, Kempler G, Barklis E. 1991. A stem cell-specific silencer in the primer-binding site of a retrovirus. Mol Cell Biol 11:1214–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolf D, Goff SP. 2009. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sripathy SP, Stevens J, Schultz DC. 2006. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol Cell Biol 26:8623–8638. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolf D, Goff S. 2007. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 17.Schlesinger S, Lee AH, Wang GZ, Green L, Goff SP. 2013. Proviral silencing in embryonic cells is regulated by yin yang 1. Cell Rep 4:50–58. doi: 10.1016/j.celrep.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cherry SR, Biniszkiewicz D, van Parijs L, Baltimore D, Jaenisch R. 2000. Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol Cell Biol 20:7419–7426. doi: 10.1128/MCB.20.20.7419-7426.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pannell D, Ellis J. 2001. Silencing of gene expression: implications for design of retrovirus vectors. Rev Med Virol 11:205–217. doi: 10.1002/rmv.316. [DOI] [PubMed] [Google Scholar]
- 20.Wolf D, Cammas F, Losson R, Goff SP. 2008. Primer binding site-dependent restriction of murine leukemia virus requires HP1 binding by TRIM28. J Virol 82:4675–4679. doi: 10.1128/JVI.02445-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang GZ, Wolf D, Goff SP. 2014. EBP1, a novel host factor involved in primer binding site-dependent restriction of Moloney murine leukemia virus in embryonic cells. J Virol 88:1825–1829. doi: 10.1128/JVI.02578-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsui T, Leung D, Miyashita H, Maksakova I, Miyachi H, Kimura H, Tachibana M, Lorincz M, Shinkai Y. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 23.Schlesinger S, Goff S. 2013. Silencing of proviruses in embryonic cells: efficiency, stability and chromatin modifications. EMBO Rep 14:73–79. doi: 10.1038/embor.2012.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leeb M, Pasini D, Novatchkova M, Jaritz M, Helin K, Wutz A. 2010. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev 24:265–276. doi: 10.1101/gad.544410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gifford R, Tristem M. 2003. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes 26:291–315. doi: 10.1023/A:1024455415443. [DOI] [PubMed] [Google Scholar]
- 26.Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, Spitz F, Constam DB, Trono D. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 27.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T-KK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Day DS, Luquette LJ, Park PJ, Kharchenko PV. 2010. Estimating enrichment of repetitive elements from high-throughput sequence data. Genome biology 11:R69. doi: 10.1186/gb-2010-11-6-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guallar D, Pérez-Palacios R, Climent M, Martínez-Abadía I, Larraga A, Fernández-Juan M, Vallejo C, Muniesa P, Schoorlemmer J. 2012. Expression of endogenous retroviruses is negatively regulated by the pluripotency marker Rex1/Zfp42. Nucleic Acids Res 40:8993–9007. doi: 10.1093/nar/gks686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan X, Xu X, Elkenani M, Smorag L, Zechner U, Nolte J, Engel W, Pantakani DV. 2013. Zfp819, a novel KRAB-zinc finger protein, interacts with KAP1 and functions in genomic integrity maintenance of mouse embryonic stem cells. Stem Cell Res 11:1045–1059. doi: 10.1016/j.scr.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 31.Macfarlan TS, Gifford WD, Agarwal S, Driscoll S, Lettieri K, Wang J, Andrews SE, Franco L, Rosenfeld MG, Ren B, Pfaff SL. 2011. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev 25:594–607. doi: 10.1101/gad.2008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoorlemmer J, Pérez-Palacios R, Climent M, Guallar D, Muniesa P. 2014. Regulation of mouse retroelement MuERV-L/MERVL expression by REX1 and epigenetic control of stem cell potency. Front Oncol 4:14. doi: 10.3389/fonc.2014.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hisada K, Sanchez C, Endo TA, Endoh M, Roman-Trufero M, Sharif J, Koseki H, Vidal M. 2012. RYBP represses endogenous retroviruses and preimplantation- and germ line-specific genes in mouse embryonic stem cells. Mol Cell Biol 32:1139–1149. doi: 10.1128/MCB.06441-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leung DC, Dong KB, Maksakova IA, Goyal P, Appanah R, Lee S, Tachibana M, Shinkai Y, Lehnertz B, Mager DL, Rossi F, Lorincz MC. 2011. Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proc Natl Acad Sci U S A 108:5718–5723. doi: 10.1073/pnas.1014660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casa V, Gabellini D. 2012. A repetitive elements perspective in Polycomb epigenetics. Front Genet 3:199. doi: 10.3389/fgene.2012.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AAHA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens JH, Jenuwein T. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12:1577–1589. doi: 10.1016/S1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 37.McClintock B. 1956. Controlling elements and the gene. Cold Spring Harb Symp Quant Biol 21:197–216. doi: 10.1101/SQB.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- 38.Stavenhagen JB, Robins DM. 1988. An ancient provirus has imposed androgen regulation on the adjacent mouse sex-limited protein gene. Cell 55:247–254. doi: 10.1016/0092-8674(88)90047-5. [DOI] [PubMed] [Google Scholar]
- 39.Robins DM, Samuelson LC. 1992. Retrotransposons and the evolution of mammalian gene expression. Genetica 86:191–201. doi: 10.1007/BF00133720. [DOI] [PubMed] [Google Scholar]
- 40.Lynch VJ, Leclerc RD, May G, Wagner GP. 2011. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat Genet 43:1154–1159. doi: 10.1038/ng.917. [DOI] [PubMed] [Google Scholar]
- 41.Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, Waki K, Hornig N, Arakawa T, Takahashi H, Kawai J, Forrest ARR, Suzuki H, Hayashizaki Y, Hume DA, Orlando V, Grimmond SM, Carninci P. 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet 41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 42.Jern P, Coffin JM. 2008. Effects of retroviruses on host genome function. Annu Rev Genet 42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- 43.Cohen CJ, Lock WM, Mager DL. 2009. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene 448:105–114. doi: 10.1016/j.gene.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 44.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y, FANTOM Consortium, RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group). 2005. The transcriptional landscape of the mammalian genome. Science 309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 45.Gifford WD, Pfaff SL, Macfarlan TS. 2013. Transposable elements as genetic regulatory substrates in early development. Trends Cell Biol 23:218–226. doi: 10.1016/j.tcb.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macfarlan T, Gifford W, Driscoll S, Lettieri K, Rowe H, Bonanomi D, Firth A, Singer O, Trono D, Pfaff S. 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB. 2012. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science 335:1499–1502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- 48.Rowe HM, Kapopoulou A, Corsinotti A, Fasching L, Macfarlan TS, Tarabay Y, Viville S, Jakobsson J, Pfaff SL, Trono D. 2013. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res 23:452–461. doi: 10.1101/gr.147678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mey A, Acloque H, Lerat E, Gounel S, Tribollet V, Blanc S, Curton D, Birot AM, Nieto MA, Samarut J. 2012. The endogenous retrovirus ENS-1 provides active binding sites for transcription factors in embryonic stem cells that specify extra embryonic tissue. Retrovirology 9:21. doi: 10.1186/1742-4690-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kigami D, Minami N, Takayama H, Imai H. 2003. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol Reprod 68:651–654. doi: 10.1095/biolreprod.102.007906. [DOI] [PubMed] [Google Scholar]
- 51.Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, Burgess SM, Brachmann RK, Haussler D. 2007. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci U S A 104:18613–18618. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kunarso G, Chia N-YY, Jeyakani J, Hwang C, Lu X, Chan Y-SS, Ng H-HH, Bourque G. 2010. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet 42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 53.Jacques P-É, Jeyakani J, Bourque G. 2013. The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet 9:e1003504. doi: 10.1371/journal.pgen.1003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okada M, Yoneda Y. 2011. The timing of retroviral silencing correlates with the quality of induced pluripotent stem cell lines. Biochim Biophys Acta 1810:226–235. doi: 10.1016/j.bbagen.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 55.Friedli M, Turelli P, Kapopoulou A, Rauwel B, Castro-Diaz N, Rowe HM, Ecco G, Unzu C, Planet E, Lombardo A, Mangeat B, Wildhaber BE, Naldini L, Trono D. 2014. Loss of transcriptional control over endogenous retroelements during reprogramming to pluripotency. Genome Res 24:1251–1259. doi: 10.1101/gr.172809.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wissing S, Munoz-Lopez M, Macia A, Yang Z, Montano M, Collins W, Garcia-Perez JL, Moran JV, Greene WC. 2012. Reprogramming somatic cells into iPS cells activates LINE-1 retroelement mobility. Hum Mol Genet 21:208–218. doi: 10.1093/hmg/ddr455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koyanagi-Aoi M, Ohnuki M, Takahashi K, Okita K, Noma H, Sawamura Y, Teramoto I, Narita M, Sato Y, Ichisaka T, Amano N, Watanabe A, Morizane A, Yamada Y, Sato T, Takahashi J, Yamanaka S. 2013. Differentiation-defective phenotypes revealed by large-scale analyses of human pluripotent stem cells. Proc Natl Acad Sci U S A 110:20569–20574. doi: 10.1073/pnas.1319061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kelley D, Rinn J. 2012. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol 13:R107. doi: 10.1186/gb-2012-13-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. 2013. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 9:e1003470. doi: 10.1371/journal.pgen.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson R, Guigo R. 2014. The RIDL hypothesis: transposable elements as functional domains of long noncoding RNAs. RNA 20:959–976. doi: 10.1261/rna.044560.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santoni FA, Guerra J, Luban J. 2012. HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology 9:111. doi: 10.1186/1742-4690-9-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fuchs NV, Loewer S, Daley GQ, Izsvák Z, Löwer J, Löwer R. 2013. Human endogenous retrovirus K (HML-2) RNA and protein expression is a marker for human embryonic and induced pluripotent stem cells. Retrovirology 10:115. doi: 10.1186/1742-4690-10-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu X, Sachs F, Ramsay L, Jacques PE, Goke J, Bourque G, Ng HH. 2014. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol 21:423–425. doi: 10.1038/nsmb.2799. [DOI] [PubMed] [Google Scholar]
- 64.Wang J, Xie G, Singh M, Ghanbarian AT, Rasko T, Szvetnik A, Cai H, Besser D, Prigione A, Fuchs NV, Schumann GG, Chen W, Lorincz MC, Ivics Z, Hurst LD, Izsvak Z. 2014. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature 516:405–409. doi: 10.1038/nature13804. [DOI] [PubMed] [Google Scholar]
- 65.Stoye JP. 2012. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol 10:395–406. doi: 10.1038/nrmicro2783. [DOI] [PubMed] [Google Scholar]
- 66.Sheehy AM, Gaddis NC, Malim MH. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med 9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 67.Carmi S, Church GM, Levanon EY. 2011. Large-scale DNA editing of retrotransposons accelerates mammalian genome evolution. Nat Commun 2:519. doi: 10.1038/ncomms1525. [DOI] [PubMed] [Google Scholar]
- 68.Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, Heidmann T, Schwartz O. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- 69.Jern P, Stoye JP, Coffin JM. 2007. Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS Genet 3:2014–2022. doi: 10.1371/journal.pgen.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Speck NA, Baltimore D. 1987. Six distinct nuclear factors interact with the 75-base-pair repeat of the Moloney murine leukemia virus enhancer. Mol Cell Biol 7:1101–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Flanagan JR, Krieg AM, Max EE, Khan AS. 1989. Negative control region at the 5′ end of murine leukemia virus long terminal repeats. Mol Cell Biol 9:739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Golemis EA, Speck NA, Hopkins N. 1990. Alignment of U3 region sequences of mammalian type C viruses: identification of highly conserved motifs and implications for enhancer design. J Virol 64:534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Granger SW, Fan H. 1998. In vivo footprinting of the enhancer sequences in the upstream long terminal repeat of Moloney murine leukemia virus: differential binding of nuclear factors in different cell types. J Virol 72:8961–8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lewis AF, Stacy T, Green WR, Taddesse-Heath L, Hartley JW, Speck NA. 1999. Core-binding factor influences the disease specificity of Moloney murine leukemia virus. J Virol 73:5535–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ekram MB, Kang K, Kim H, Kim J. 2012. Retrotransposons as a major source of epigenetic variations in the mammalian genome. Epigenetics 7:370–382. doi: 10.4161/epi.19462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ellis J. 2005. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum Gene Ther 16:1241–1246. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- 77.Katz R, Jack-Scott E, Narezkina A, Palagin I, Boimel P, Kulkosky J, Nicolas E, Greger J, Skalka A. 2007. High-frequency epigenetic repression and silencing of retroviruses can be antagonized by histone deacetylase inhibitors and transcriptional activators, but uniform reactivation in cell clones is restricted by additional mechanisms. J Virol 81:2592–2604. doi: 10.1128/JVI.01643-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Minoguchi S, Iba H. 2008. Instability of retroviral DNA methylation in embryonic stem cells. Stem Cells 26:1166–1173. doi: 10.1634/stemcells.2007-1106. [DOI] [PubMed] [Google Scholar]
- 79.Lo MY, Rival-Gervier S, Pasceri P, Ellis J. 2012. Rapid transcriptional pulsing dynamics of high expressing retroviral transgenes in embryonic stem cells. PLoS One 7:e37130. doi: 10.1371/journal.pone.0037130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schlesinger S, Meshorer E, Goff SP. 2014. Asynchronous transcriptional silencing of individual retroviral genomes in embryonic cells. Retrovirology 11:31. doi: 10.1186/1742-4690-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. 2007. Nanog safeguards pluripotency and mediates germline development. Nature 450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 82.Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. 2008. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development 135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- 83.Hayashi K, Lopes S, Tang F, Surani M. 2008. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell 3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reynolds N, Latos P, Hynes-Allen A, Loos R, Leaford D, O'Shaughnessy A, Mosaku O, Signolet J, Brennecke P, Kalkan T, Costello I, Humphreys P, Mansfield W, Nakagawa K, Strouboulis J, Behrens A, Bertone P, Hendrich B. 2012. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell 10:583–594. doi: 10.1016/j.stem.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ramírez MA, Pericuesta E, Fernandez-Gonzalez R, Moreira P, Pintado B, Gutierrez-Adan A. 2006. Transcriptional and post-transcriptional regulation of retrotransposons IAP and MuERV-L affect pluripotency of mice ES cells. Reprod Biol Endocrinol 4:55. doi: 10.1186/1477-7827-4-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Eldar A, Elowitz MB. 2010. Functional roles for noise in genetic circuits. Nature 467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Whitelaw NC, Chong S, Whitelaw E. 2010. Tuning in to noise: epigenetics and intangible variation. Dev Cell 19:649–650. doi: 10.1016/j.devcel.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 88.Whitelaw NC, Chong S, Morgan DK, Nestor C, Bruxner TJ, Ashe A, Lambley E, Meehan R, Whitelaw E. 2010. Reduced levels of two modifiers of epigenetic gene silencing, Dnmt3a and Trim28, cause increased phenotypic noise. Genome Biol 11:R111. doi: 10.1186/gb-2010-11-11-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gifford RJ, Katzourakis A, Tristem M, Pybus OG, Winters M, Shafer RW. 2008. A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc Natl Acad Sci U S A 105:20362–20367. doi: 10.1073/pnas.0807873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV. 2005. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 122:169–182. doi: 10.1016/j.cell.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 91.Dar RD, Hosmane NN, Arkin MR, Siliciano RF, Weinberger LS. 2014. Screening for noise in gene expression identifies drug synergies. Science 344:1392–1396. doi: 10.1126/science.1250220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schultz DC, Friedman JR, Rauscher FJ III. 2001. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev 15:428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rowe HM, Friedli M, Offner S, Verp S, Mesnard D, Marquis J, Aktas T, Trono D. 2013. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development 140:519–529. doi: 10.1242/dev.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]