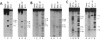Abstract
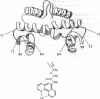
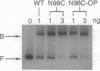
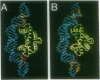
The Escherichia coli Fis (factor for inversion stimulation) protein functions in many diverse biological systems including recombination, transcription, and DNA replication. Although Fis is a site-specific DNA-binding protein, it lacks a well-defined consensus recognition sequence. The electrophoretic mobility of Fis-DNA complexes, along with considerations of the Fis crystal structure, indicates that significant deformation of DNA occurs upon Fis binding. To investigate the structure of Fis-DNA complexes, the chemical nuclease 1,10-phenanthroline-copper complex (OP-Cu) has been linked to four specific sites within the Fis DNA-binding domain. Two of these Fis-OP derivatives were active in cleaving DNA. The scission patterns obtained on four different Fis binding sites indicate that Fis positions itself on these highly divergent DNA sequences in a very similar fashion. The patterns of cleavage of a derivative at Asn-98 generally support a model of a Fis-DNA complex that contains specific bends within the core-recognition sequence. Data from a second Fis-OP derivative at Asn-73 provides evidence for greater wrapping of flanking DNA around the sides of the Fis protein than was previously postulated. The cleavage efficiency of flanking segments varies, suggesting that the extent of DNA wrapping is sequence dependent. Specific amino acids on Fis are implicated in promoting this DNA wrapping.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chen C. H., Sigman D. S. Chemical conversion of a DNA-binding protein into a site-specific nuclease. Science. 1987 Sep 4;237(4819):1197–1201. doi: 10.1126/science.2820056. [DOI] [PubMed] [Google Scholar]
- Ebright R. H., Ebright Y. W., Pendergrast P. S., Gunasekera A. Conversion of a helix-turn-helix motif sequence-specific DNA binding protein into a site-specific DNA cleavage agent. Proc Natl Acad Sci U S A. 1990 Apr;87(8):2882–2886. doi: 10.1073/pnas.87.8.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel S. E., Johnson R. C. The Fis protein: it's not just for DNA inversion anymore. Mol Microbiol. 1992 Nov;6(22):3257–3265. doi: 10.1111/j.1365-2958.1992.tb02193.x. [DOI] [PubMed] [Google Scholar]
- Gosink K. K., Ross W., Leirmo S., Osuna R., Finkel S. E., Johnson R. C., Gourse R. L. DNA binding and bending are necessary but not sufficient for Fis-dependent activation of rrnB P1. J Bacteriol. 1993 Mar;175(6):1580–1589. doi: 10.1128/jb.175.6.1580-1589.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübner P., Arber W. Mutational analysis of a prokaryotic recombinational enhancer element with two functions. EMBO J. 1989 Feb;8(2):577–585. doi: 10.1002/j.1460-2075.1989.tb03412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Simon M. I. Hin-mediated site-specific recombination requires two 26 bp recombination sites and a 60 bp recombinational enhancer. Cell. 1985 Jul;41(3):781–791. doi: 10.1016/s0092-8674(85)80059-3. [DOI] [PubMed] [Google Scholar]
- Kostrewa D., Granzin J., Koch C., Choe H. W., Raghunathan S., Wolf W., Labahn J., Kahmann R., Saenger W. Three-dimensional structure of the E. coli DNA-binding protein FIS. Nature. 1991 Jan 10;349(6305):178–180. doi: 10.1038/349178a0. [DOI] [PubMed] [Google Scholar]
- Landt O., Grunert H. P., Hahn U. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene. 1990 Nov 30;96(1):125–128. doi: 10.1016/0378-1119(90)90351-q. [DOI] [PubMed] [Google Scholar]
- Osuna R., Finkel S. E., Johnson R. C. Identification of two functional regions in Fis: the N-terminus is required to promote Hin-mediated DNA inversion but not lambda excision. EMBO J. 1991 Jun;10(6):1593–1603. doi: 10.1002/j.1460-2075.1991.tb07680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddles P. W., Blakeley R. L., Zerner B. Reassessment of Ellman's reagent. Methods Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
- Sigman D. S., Chen C. H. Chemical nucleases: new reagents in molecular biology. Annu Rev Biochem. 1990;59:207–236. doi: 10.1146/annurev.bi.59.070190.001231. [DOI] [PubMed] [Google Scholar]
- Sluka J. P., Horvath S. J., Bruist M. F., Simon M. I., Dervan P. B. Synthesis of a sequence-specific DNA-cleaving peptide. Science. 1987 Nov 20;238(4830):1129–1132. doi: 10.1126/science.3120311. [DOI] [PubMed] [Google Scholar]
- Sutton C. L., Mazumder A., Chen C. H., Sigman D. S. Transforming the Escherichia coli Trp repressor into a site-specific nuclease. Biochemistry. 1993 Apr 27;32(16):4225–4230. doi: 10.1021/bi00067a009. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Moitoso de Vargas L., Koch C., Kahmann R., Landy A. Cellular factors couple recombination with growth phase: characterization of a new component in the lambda site-specific recombination pathway. Cell. 1987 Sep 11;50(6):901–908. doi: 10.1016/0092-8674(87)90516-2. [DOI] [PubMed] [Google Scholar]
- Yuan H. S., Finkel S. E., Feng J. A., Kaczor-Grzeskowiak M., Johnson R. C., Dickerson R. E. The molecular structure of wild-type and a mutant Fis protein: relationship between mutational changes and recombinational enhancer function or DNA binding. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9558–9562. doi: 10.1073/pnas.88.21.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]