Introduction
Hexameric helicases are ring-shaped motor proteins that encircle and move along nucleic acid strands to unwind duplex DNA and RNA substrates (Singleton et al., 2007). Enzymes of this type broadly fall into two evolutionarily related, but distinct families – RecA and AAA+ – that participate in a myriad number of vital DNA- and RNA-dependent transactions in the cell (Iyer et al., 2004; Lyubimov et al., 2011). Because of their complex action and essential functions, hexameric helicases have garnered significant interest both as archetypal molecular machines, and as prospective targets for therapeutic intervention.
Alongside biochemistry, structural investigations have helped generate key insights into hexameric helicase mechanism. However, while numerous members of disparate hexameric helicase families have been crystallized over the past decade, it has proven exceedingly difficult to obtain structures of these enzymes bound to both target nucleic acid substrates and the nucleotide co-factors that fuel motor movement. Studies of hexameric helicases have been complicated by many factors, including their large size and inherent flexibility, the presence of multiple ligand binding sites, and a tendency for hexamers to form mixtures of closely-related, symmetric and quasi-symmetric states of heterogeneous nucleotide occupancy.
This latter problem can be one of the most insidious. Hexameric helicases contain six ATPase sites (positioned between each subunit interface), along with six nucleic-acid binding sites housed in the center of the ring. These sites frequently display some level of positive and negative cooperativity between each other, giving rise to both strong and weak binding sites within a single hexamer (Bujalowski and Klonowska, 1993; Geiselmann and von Hippel, 1992; Hingorani and Patel, 1996; Kim and Patel, 1999; Kim et al., 1999; Seifried et al., 1992; Stitt, 1988; Xu et al., 2003). The presence of multiple classes of ATP binding sites can lead to difficulties in determining which ATP analogs and/or nucleotide mixtures might be optimal for use in structural studies. Even if this hurdle is surmounted, the high degree of conformational similarity between subunits in the hexamer can favor crystal packing arrangements that blur out essential details, such as when a nucleic acid segment bound in the center of a helicase ring lies on a crystallographic symmetry axis (Skordalakes and Berger, 2006). At its worst, pseudo-symmetry can produce crystals that display merohedral or non-merohedral twinning – a situation where single crystals are actually composed of blocks of overlapping microcrystals in similar but differing orientations – preventing structure solution and refinement (Yeates and Fam, 1999).
An approach we recently used in determining the structure of a fully-liganded hexameric helicase – the full-length E. coli Rho transcription termination factor – took these problems into account. Using prior biochemical knowledge of Rho’s catalytic mechanism, as well as our earlier experience with structural studies of this system, we implemented a crystallization strategy that screened a highly-focused set of chemical conditions against a panel of RNA substrates and different ATP mimetics. The result was three distinct, but related crystals forms, one of which contained the desired protein-ligand complex. We next conducted a time-dependent crystallization screen to overcome an otherwise intractable, interleaved form of coincident non-merohedral twinning that was present in our most promising crystals. Finally, by collecting data on specific regions of the crystal, we avoided contributions from the unwanted twin domain resulting in a complete 2.8 Å dataset that allowed structure determination (Thomsen and Berger, 2009). Here we present considerations for the crystallization of hexameric helicases bound to nucleic-acid and nucleotide cofactors, as well as a crystal screening method that can be used to mitigate the effects of non-merohedral twinning that can occur with pseudo-symmetric protein samples.
Biochemical Foundation
In designing a crystal screening strategy for enzymes that make use of multiple substrates, there is an overwhelmingly large combination of chemical and ligand space to explore (McPherson, 1999). Consideration of the activities and features unique to the system of interest is therefore critical to the design of a successful screen. Discovered in 1969, the Rho transcription termination factor has since been subject to more than forty years of study (Roberts, 1969). Rho is a 47 kDa protein consisting of an N-terminal oligonucleotide binding (OB) fold and a C-terminal RecA-like domain (Allison et al., 1998; Briercheck et al., 1998; Burgess and Richardson, 2001; Dombroski and Platt, 1988; Miwa et al., 1995; Opperman and Richardson, 1994; Skordalakes and Berger, 2003). The N-terminal region houses an RNA binding locus, termed the “primary site,” that preferentially associates with cytosine-rich nucleic acid segments, and is responsible for recruiting Rho to target mRNAs (Bear et al., 1985; Bogden et al., 1999; Martinez et al., 1996a; Martinez et al., 1996b; Modrak and Richardson, 1994). The C-terminal part of Rho also binds RNA, forming the so-called “secondary site,” and serves as the ATP-dependent motor element of the protein (Dolan et al., 1990; Wei and Richardson, 2001). Rho is capable of translocating 5′-3′ along RNA, and can both unwind RNA/DNA duplexes and release RNA from ternary transcription elongation complexes (Brennan et al., 1987; Park and Roberts, 2006; Richardson and Conaway, 1980; Roberts, 1969; Shigesada and Wu, 1980). At the time we began this project, it was known that RNA bound to Rho’s secondary site with maximal affinity only when the protein’s ATP binding sites were saturated. It was also established that Rho possessed a mixture of strong and weak ATP binding sites, whose relative strengths and number changed depending on the type of nucleotide being assayed. Hence, there was strong evidence that some type of ATP analog would be required to crystalize an intact Rho hexamer with RNA productively bound to the motor domains, and that the choice of analog would play a significant role in the type of conformational state we obtained.
Studies of Rho/RNA interactions provided boundary points for the length and sequence of RNA substrates used in our crystal screening strategy. Early experiments revealed that Rho could engage and wrap long stretches of cytosine-rich RNA through its primary RNA binding site (Lowery and Richardson, 1977; Lowery-Goldhammer and Richardson, 1974; Wang and von Hippel, 1993). By contrast, the secondary site had been shown to bind only ~8 bases of RNA (Richardson, 1982). Furthermore, although cytosine-rich pyrimidine RNAs >22 nucleotides in length were seen to maximally stimulate Rho’s ATPase activity at roughly stoichiometric concentrations, pyrimidine-based RNA substrates as short as nine nucleotides also strongly activated ATP turnover when present at saturating levels (Lowery and Richardson, 1977; Richardson, 1982). These data suggested that poly-r(CU)n or -r(U)n RNA polymers between eight and thirty nucleotides in length could target the secondary RNA binding site in the center of the Rho ring; poly-r(C)n oligos were not chosen for study, as we had seen earlier that these substrates can have solubility problems when used at crystallographic concentrations (Bogden et al., 1999).
Extensive studies of ATP and ATP analog binding and hydrolysis by Rho further helped refine our crystallization strategy. As with ATP, Rho had been found to engage adenosine 5′-(γ –thio)triphosphate (ATPγS) using a mixture of three strong and three weak binding sites; however, these studies further showed that this cofactor could be readily hydrolyzed (Stitt and Webb, 1986; Stitt and Xu, 1998; Xu et al., 2003), indicating that it would not be suitable for stabilizing or trapping a translocation intermediate. The ATP analogs adenosine 5′-(β,γ-imido)triphosphate (AMP-PNP) and adenosine 5′-(β,γ-methylene)triphosphate (AMP-PCP) were found to bind much more weakly to the helicase, and tightly occupy only one of Rho’s six possible sites (Xu et al., 2003), properties shared with ADP rather than ATP. During pre-steady-state ATP hydrolysis assays, in which a single active site was filled with ATP, the addition of AMP-PCP or AMP-PNP produced burst hydrolysis rates significantly lower than seen from the addition of ATP (Browne et al., 2005), further suggesting that they would be poor substrate mimetics. In the end, only one non-hydrolyzable ATP analog, ADP•BeF3, was reported to bind Rho in the presence of RNA with an affinity similar to that of ATP (Adelman et al., 2006; Xu et al., 2003). This observation suggested that ADP•BeF3 might be the most viable candidate for a non-hydrolyzable ATP analog in co-crystallization trials.
Protein Expression, Purification and Storage
Although each type of hexameric helicase will necessitate a particular purification strategy, and will further exhibit a unique set of solution properties once pure, a number of considerations proved important for the crystallization of Rho that may be applicable to other systems. One such factor was the level of soluble protein that could be obtained during expression. Over the course of a crystallization project, expression levels correlate with the number of crystallization conditions that ultimately can be explored, a factor that can have a marked outcome on the success of a project. Expression levels further define a “signal to noise” ratio that influences the purity of the final preparation. Particularly high expression levels also provide more flexibility in the purification protocol, such as eliminating a need to use and remove affinity tags (see below), which can speed up purifications to produce protein of higher quality.
In looking for ways to boost expression, we found that full-length E. coli Rho was best overproduced from a pET-based vector in BL21 pLysS cells by inducing at an A600 of 0.6 at 37 °C for 3 h. In this instance, a range of IPTG concentrations produced fairly comparable results. BL21 pLysS cells provided approximately two-fold greater yields of soluble Rho protein compared to BL21 Codon (+) or Rosetta cells. With other Rho constructs (such as mutants), C41 cells have proven even more effective for expression, underlying the importance of screening a large number of cell lines at the earliest stages of a project. Once optimized, we found that Rho accounted for ~80% of the total soluble cellular protein content, obviating a need for affinity purification tags. Lysis was performed by resuspending induced cells into buffer A (25 mM Tris pH 7.5, 50 mM KCl, 10% glycerol, 1 mM TCEP), and sonicating on ice for a total of two minutes (30 seconds on, 1 min off, repeated four times) on a power setting of 5.5 using a (Sonicator 3000, Misonix), after which cells were centrifuged at 16,000 rpm (4°C) in an SS34 rotor (Sorvall) for 30 minutes. Cells were not frozen between harvesting and lysis steps.
Un-tagged Rho was purified to homogeneity by applying the clarified lysate to a cation-exchange column in buffer A (Poros HS, Life Technologies). Protein was eluted using a 100mL gradient into buffer B (25 mM Tris pH 7.5, 1 M KCl, 10% glycerol, 1 mM DTT), the dominant center peak fractions were pooled, the salt concentration was reduced to 75 mM using diluent buffer (25mM Tris pH 7.5, 10% glycerol, 1mM DTT), and the sample was reapplied to the same Poros HS column for a second round of enrichment. This procedure is fast, typically requiring ~2 hours. Rho hexamers were next concentrated by ultrafiltration (Amicon Ultra 10,000 MWCO, Millipore) and purified away from aggregated material using size exclusion chromatography. Two S-300 columns (GE Healthcare) were run in succession using buffer C (50 mM Tris pH 7.5, 500 mM KCl, 10% glycerol, 1 mM DTT), re-concentrating the peak hexamer fractions of protein between runs to ensure that all traces of aggregated protein or Rho subassemblies were removed. The purified protein was re-concentrated and stored in buffer C at 4°C.
The Rho protein produced from this procedure is of very high purity and homogeneity, and readily gives crystals in a number of conditions. Some of the steps, such as avoiding freezing and running two gel-filtration columns were empirically found to improve crystal quality and diffraction data resolution in our early work on this protein (Skordalakes and Berger, 2003, 2006). Similarly, we found it important to use only freshly prepared protein (<1 week old) for crystallization. Although these types of treatments are clearly not necessary for crystallization success with all proteins, their use may be good practice for increasing the chances of obtaining a hit in difficult cases.
Ligand Preparation and Storage
RNA oligonucleotides were purchased from IDT, dissolved in Ambion RNAse-free water to 4 mM final concentration, and stored at −80 °C in single-use aliquots. Nucleotides such as ADP, ATP, AMP•PNP and ATPγS were all dissolved in deionized H2O to 100 mM and stored at −80 °C, again in single-use aliquots. For our initial experiments, we prepared ADP•BeF3 and ADP•AlFx using ratios taken from the literature, with special attention paid to the work on the Bovine F1-ATPase by Walker, Leslie and colleagues (Braig et al., 2000; Kagawa et al., 2004; Menz et al., 2001). We used a ratio of 1 ADP:3 Be:15 F for ADP•BeF3 and a ratio of 1 ADP:4 Al:20 F for ADP•AlFx. To prepare the analogs, either 1 M AlCl3 or 1 M BeCl2 were mixed with 1 M NaF in the appropriate volumes to generate 1:5 ratios of Be:F and Al:F. These mixtures were then added to 100 mM stocks of ADP in order to produce a final concentration of 25mM ADP. Finally, all nucleotides were adjusted to an approximate pH of 7.5 using pH paper and drop wise addition of 1N HCl or NaOH.
Trapping Rho-RNA-nucleotide complexes
Protein/nucleic acid complexes can in principle be prepared either by mixing the constituents at the desired ratios/concentrations and setting trays directly, or by mixing at lower concentrations, purifying the complex using column chromatography, and then reconcentrating prior to setting trays. Given the high cost of synthesized RNA and the toxicity of the ADP•BeF3 and ADP•AlFx nucleotide preparations, along with a concern that low-affinity RNA or nucleotide binding sites might be stripped during the course of a column purification strategy, we chose to directly mix purified protein and ligands immediately prior to crystallization screening.
To prevent precipitation and produce the best protein crystals, a specific procedure for ligand addition was developed. Purified Rho at ~60 mg/mL in buffer C (50 mM Tris 7.5, 500 mM KCl, 10% glycerol, 1 mM DTT) was first exchanged into dialysis buffer (10 mM Tris pH 7.5, 100 mM NaCl, 1 mM DTT) for approximately three hours using a 10,000 MWCO Slide-A-Lyzer dialysis cup (Thermo). We empirically found that lower quality crystals resulted if Rho was dialyzed into a low salt buffer for longer than approximately six hours, suggesting that dialysis might not have gone to completion. Following dialysis, the concentration of Rho typically approached ~ 40 mg/mL due to sample dilution. As trays were set at 20 mg/mL, this procedure provided a 2x protein stock with ample room for ligand addition. More importantly, it allowed us to pre-dilute various ligands into dialysis buffer, thus buffering them and reducing their local concentration upon addition to the protein solution. Reasoning that flooding Rho with high concentrations of a non-hydrolyzable ATP analog alone might trap a fraction of the helicase population in non-productive conformations incompatible with RNA binding, we first added individual RNA substrates, and let the sample sit on ice for 15 minutes. We next added non-hydrolyzable nucleotide complexes at 2.5 mM (and spiked with 5–10 mM MgCl2), mixed the solution and incubated the reagents together on ice for another 15 minutes prior to setting trays. Early crystal trials suggested that incubation of Rho-ligand complexes for up to 30 minutes produced better crystals than setting trays immediately after mixing.
Design of a Substrate-Centric Crystal Screening Strategy
We next used the biochemical data to inform the design of a substrate-centered screening strategy. Because our early work with Rho had found that the protein crystallized preferentially under low-salt conditions with organic (e.g., PEG- or MPD-based) precipitants, we focused on diversifying the number and types substrates and cofactors screened, rather than on thoroughly searching chemical space. To maximize our exploration of ligand space, we conducted screens using r(CU)n and rUn nucleic acid polymers between 8 and 30 nucleotides in length, alongside a variety of adenosine nucleotides (Table 1). We initially focused on longer r(CU)n RNA’s between 20 and 30 nucleotides in length, reasoning that they might bind first to the high affinity primary RNA binding site and then feed down into the central channel in the presence of the proper adenosine nucleotide. For the nucleotides themselves, we individually tested ADP, ATPγS, AMP-PNP, ADP•BeFx, and ADP•AlFx. After hits were obtained, we screened various combinations of nucleotides as part of our optimization efforts. Mixtures of ADP and AMPPNP have proved important for the crystallization of Rho’s closest homolog, the F1-ATPase (Abrahams et al., 1994), but such mixtures did not improve the quality of our crystals. For each substrate combination, we selected a relatively limited set of commercial crystallization screens, typically including Index HT (Qiagen), The MPD’s (Qiagen), PEG-Ion (Hampton Research), and Natrix (Hampton Research). Ultimately, the screening strategy yielded three different crystal-forms (Figure 1).
Table 1.
Substrate matrix used for Rho crystal screening
| ANP | ATPγS | ADP | ADP•AlF4 | ADP•BeF3 | |
|---|---|---|---|---|---|
| r(CU)4 | X | ||||
| r(U)12 | 2, 3 | ||||
| r(CU)6 | 2, 3 | ||||
| r(CU)8 | 2, 3 | ||||
| r(CU)10 | X | 1 | 1 | 1 | 1, 2 |
| r(CU)12C | X | 1 | 1 | 1 | 1, 2 |
| r(CU)15 | X | 1 | 1 | 1 | 1, 2 |
Key: (X) – no crystal, (1–3) – crystal-forms, (blank) – not screened
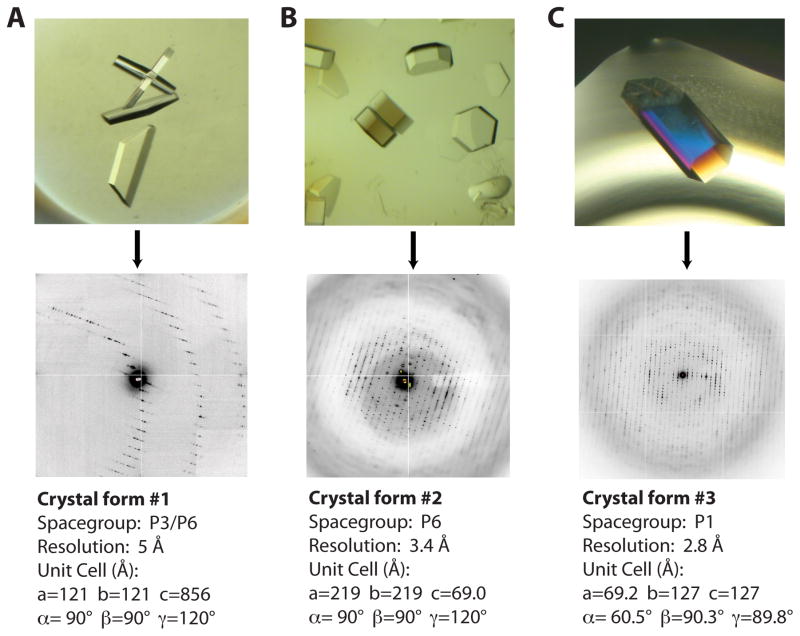
Figure 1. Three Rho crystal-forms obtained from substrate matrix screening.
A) Crystal-form I (top) grew as thin hexagonal plates that diffract to relatively low resolution. A close-up view of the diffraction pattern (bottom) reveals the long unit cell edge that caused severe overlaps and prevented collection of a complete dataset.
B) Crystal-form II grew as thick hexagonal “lug-nuts” (top). Diffraction patterns revealed a P6 packing arrangement. Usable diffraction maxima extended to 3.4 Å resolution.
C) Crystal-form III grew as large hexagonal rods free from any obvious morphological defects (top). While the diffraction pattern is highly mosaic, these crystals diffracted to the highest resolution of any of the forms.
Crystallization of Rho bound to RNA and Adenosine Nucleotides
Crystal-form I
Crystal-form one was obtained with r(CU)n polymers between 20 and 30 bases in length and either ADP, ATPγS, or ADP•AlF4 (Table 1). These crystals were grown by mixing 1 μL of the Rho-substrate complex in dialysis buffer (see above) with 1 μL of a crystallization solution containing 100 mM HEPES pH 7.5, 8% PEG 6000, 5% MPD, and incubating under paraffin oil at 18° C. Crystals grew within 24 hours and reached full size in approximately one week. Since the crystals that grew in three unique nucleotide conditions were all indistinguishable morphologically, we reasoned that Rho likely associated with ADP in each instance. Crystal-form I diffracted to approximately 6Å in a hexagonal space group and possessed an unusually long unit cell edge (>850Å) (Figure 1A). Matthews analysis suggested that as many as 12 Rho hexamers were present per unit cell, while the space group (P3 or P6) implied that the hexamers were likely centered around crystallographic symmetry axes (Kantardjieff and Rupp, 2003; Matthews, 1968). Numerous approaches were explored in order to improve the resolution of these crystals and/or alter the symmetry or unit cell dimensions, including dehydration, freeze/thaw annealing, and chemical crosslinking using glutaraldehyde; however, none of these techniques improved diffraction (Newman, 2006). Data processing was further hindered by the long unit cell, which caused severe overlaps in the diffraction patterns and ultimately prevented us from solving the structure. Given these results, we continued to explore substrate space with the goal of obtaining crystals that diffracted to higher resolution and possessed more manageable unit cell dimensions.
Crystal-form II
Crystal-form II was obtained with r(CU)n polymers between 12 and 30 bases in length and the ATP mimic ADP•BeF3 (Table 1). Crystals were grown by mixing 1 μL of the Rho-substrate complex in dialysis buffer with 1 μL of a crystallization solution containing 100 mM HEPES pH 7.0, and 4% MPD and incubating under paraffin oil at 18° C. The initial hit took approximately one month to appear, although later optimization produced crystals that would appear within a 3–7 days. Crystals were cryoprotected by the addition of mother liquor supplemented with 20% MPD and flash frozen in liquid nitrogen. The hexagonal crystals resembled “lug-nuts” and proved capable of diffracting to approximately 3.4 Å after refinement of the initial growth conditions (Table 2). Molecular replacement using a Rho monomer model (Skordalakes and Berger, 2003) was used to phase the data (which processed cleanly as P6), and indicated that three copies were present per asymmetric unit (Figure 2A). Application of crystal symmetry operators to these solutions revealed that the monomers belonged to two different Rho hexamers, one exhibiting three-fold symmetry and the other six-fold (Figure 2B).
Table 2.
Data Collection and Partial Refinement for Crystal-form II
| Data | |
| Space Group | P6 |
| Unit Cell | a=218.9 b=218.9 c=68.95 α=90 β=90γ=120 |
| Wavelength (Å) | 0.9796 |
| Resolution (Å) | 50 – 3.4 |
| Unique Reflections | 50,668 |
| Redundancy | 2.3 (2.3) |
| Completeness (%) | 99.7 (99.8) |
| I/σ | 8.3 (2.5) |
| Rmerge | 0.104 (0.348) |
| Refinement | |
| Rwork (%) | 33.5a |
| Rfree (%) | 35.7b |
| RMSD bonds (Å) | 0.010 |
| RMSD angles (°) | 1.174 |
| Ramachandran | |
| Preferred (%) | 99.0 |
| Allowed (%) | 1.0 |
| Outliers (%) | 0 |
| Number of Atoms | 9,936 |
| Protein | 9,840 |
| Ligands | 96 |
Values in parenthesis correspond to the highest resolution bin
Rwork = ΣΣ|Fo−Fc|/ΣFo
Rfree is calculated using 5% of the data omitted from refinement
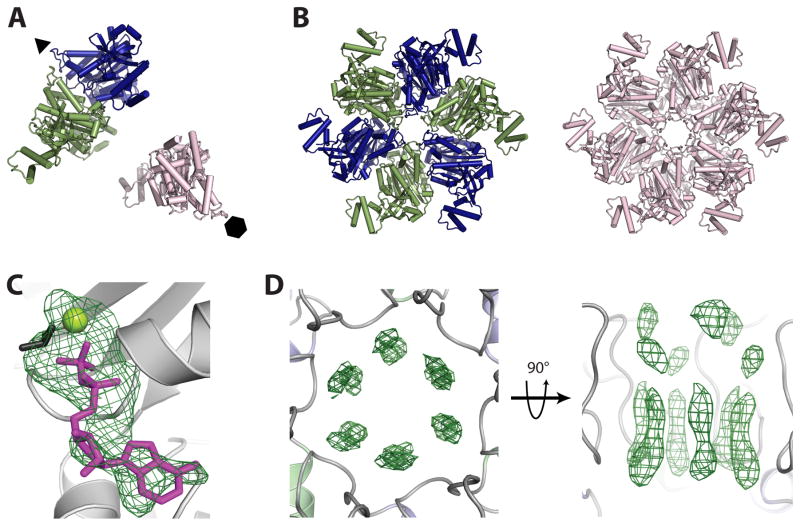
Figure 2. Structure solution and ligand binding in crystal-form II.
A) Molecular replacement reveals one Rho dimer (blue/dark grey and green/grey) and one monomer (pink/light grey) per asymmetric unit. Crystallographic six- and three-fold symmetry axes are indicated by a hexagon and triangle, respectively.
B) Crystallographic symmetry generates one three-fold symmetric trimer-of-dimers (blue/dark grey and green/grey) and one six-fold symmetric hexamer (pink/light grey). These structures represented our first crystallographic views of a full-length, closed-ring Rho hexamer.
C) Initial refinement of molecular replacement solutions produced Fo−Fc difference electron density maps revealing the presence of nucleotide (modeled as ADP•BeF3) at all three ATP binding sites per asymmetric unit. ADP is colored magenta/grey, BeF3 is colored black, and the Mg2+ ion is colored yellow-green/light grey. Electron density (green mesh) is contoured at 3σ.
D) Initial refinement of molecular replacement solutions produced Fo−Fc difference electron density maps revealing the presence of RNA in the central channel of each Rho hexamer. The RNA density appeared to be averaged around the crystallographic symmetry axis, producing un-interpretable maps. Electron density (green mesh) is contoured at 2.5σ.
Refinement of the models from these data could not proceed beyond an Rwork/Rfree of 33.6/35.7% (Table 2). However, Fo−Fc difference electron density maps obtained after rigid body fitting of the MR solution revealed clear density for a nucleotide bound at the active site of each Rho monomer (Figure 2C). This density was modeled as a complex of ADP•BeF3•Mg2+, although the presence of the BeF3 group and the Mg2+ ion could not be confirmed at this resolution and level of overall model accuracy. Six strong bands of Fo−Fc difference density also were observed around the three- and six-fold crystal symmetry axes at the center of each hexamer (Figure 2D), suggesting that RNA was bound in the center of each ring; however, the electron density was not of sufficient quality for modeling, likely due to rotational averaging of the RNA about the crystallographic symmetry axes. Unfortunately, these issues obscured any details that might have allowed us to identify the ATP and RNA binding status of individual subunits, preventing us from gaining useful insights into Rho’s translocation mechanism. Nonetheless, these data revealed that short RNA substrates and the ATP analog ADP•BeF3 could be used to capture a fully liganded complex with RNA bound in the center of the ring. We therefore expanded our search of chemical space around these substrate conditions to identify a crystal-form that would contain at least one complete hexamer whose conformational state was not influenced by crystal symmetry.
Crystal-form III
Our inability to fully refine crystal-form II suggested either that certain pathologies were present in the diffraction data, or that some insurmountable form of model bias was carrying over from molecular replacement. Reasoning that any bias might be due to significant conformational differences between our search model (which was from an RNA-free, open-ring Rho state) and the subunits in a closed-ring, fully-liganded Rho complex, we purified selenomethionine-labeled Rho for use with phasing. Crystal-form III was obtained with this protein, using short r(CU)6 (and eventually rU12) RNA polymers in the presence of ADP•BeF3 (Table 1). Crystals (100 × 500 μm hexagonal rods) were grown by mixing 2 μL of protein-ligand solution at 20 mg/mL in modified dialysis buffer (10 mM Tris 7.5, 300 mM NaCl, and 1 mM TCEP) with 2μL of a solution of 5% MPD, 100 mM HEPES pH 7.9, 20 mM NaCl, and 10 mM spermidine-HCl and incubating under paraffin oil at 18 °C (Figure 1C). After three days, the crystals were cryoprotected by adding mother liquor supplemented with 25% MPD directly to the 4 uL drop in 1 uL increments separated by 1 – 5 minute intervals. Once 4 uL of cryoprotectant was added in this way, the drop was fully exchanged into cryoprotectant by: 1) removal of ½ drop volume; 2) addition of ½ drop volume of cryoprotectant, and; 3) incubation for 1 minute intervals. The entire cryoprotection process was conducted under paraffin oil in the original crystallization plates to mimimize crystal manipulations. Upon complete exchange, the crystals were looped flash frozen in liquid nitrogen (Thomsen and Berger, 2009).
Attempts to collect data from these crystals were not initially promising. Diffraction images obtained from the central region of the rods showed a mixture of lattices. However, exposures taken from different points along the length of the rods revealed that one end (the tip that nucleated from the drop or crystallization plate surface) diffracted to approximately 3.5 Å resolution and indexed as spacegroup P6, whereas the distal end diffracted to approximately 3.0 Å and appeared to be P1. Interestingly, the P6 end possessed the same unit cell dimensions as seen in crystal-form II. Molecular replacement solutions obtained with data from the P1 end of the crystals revealed that a single Rho hexamer was present in the asymmetric unit, and revealed clear difference density for both RNA and nucleotide (Thomsen and Berger, 2009). However, the MR-phased maps were of low quality and refinement stalled at an early stage. Close inspection of the diffraction patterns revealed that the P1 lattice was conflated with the overlapping hexagonal lattice at low resolution (Figure 3A), suggesting that these crystals were non-merohedrally twinned.
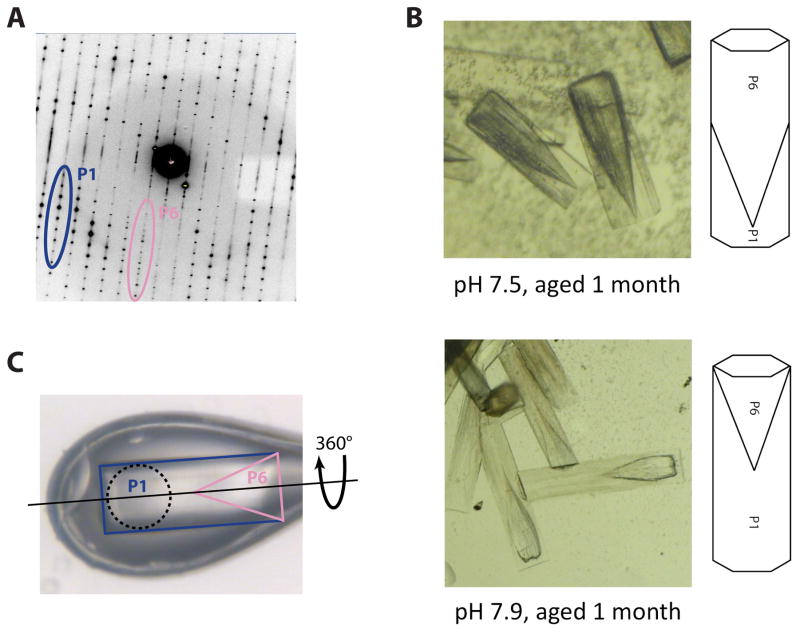
Figure 3. Overcoming a vexing form of coincident non-merohedral twinning.
A) Initial diffraction patterns obtained from crystal-form III (Figure 1C), revealed two distinct lattices. While the P1 diffraction dominated the pattern (blue/dark grey), a P6 diffraction pattern corresponding to crystal-form II was present at low resolution (pink/light grey). Since crystal-forms I and II shared a coincident ~69 Å unit cell edge, the diffraction patterns almost perfectly overlapped and prevented accurate measurement of low resolution P1 data.
B) Aging caused the P1 portion of each crystal to degrade at a faster rate than the P6 portion. When viewed under bright-field illumination, the two twin domains are clearly distinguishable. The P6 twin domain forms at the nucleating end of the crystals and has a “cone-shaped” protrusion that interleaved with the P1 twin domain (top). An X-ray beam aimed at the P1 tip of the crystal thus passes through a small fraction of the P6 twin domain, producing P6 diffraction only at low resolution as seen in panel A. Increasing the pH of crystal growth increased the relative size of the P1 twin domain (bottom).
C) Crystals grown at a pH of 7.9 were mounted in bendable cryo-loop and oriented such that the axis of crystal rotation (dashed line) was parallel with the long axis of the hexagonal rods. A 100 μM collimated beam (dashed circle) was directed into the P1 tip (blue/dark grey) of the crystal. Data collection in this orientation avoided contributions from the P6 twin domain (pink/light grey).
Although one end of the crystal was primarily P1, the nearly perfect crystal morphology prevented us from identifying the nature of the P6 contribution. Without this knowledge, isolation of the two twin domains or improvement of the crystals proved exceedingly difficult. However, one element of the two lattice arrangements that proved fortuitously variable was their stability over time. While some crystals remained intact for several months, others exhibited age dependent defects and eventually degraded or disappeared. Reasoning that the two different crystal-forms in the non-merohedral twins might degrade over time at different relative rates, we conducted a series of crystallization screens around 10 mM Spermidine-HCl, 20 mM NaCl, 100 mM HEPES (pH values between 7.5 and 8.2) and MPD (1 to 12 % MPD). After reaching full size, crystals were left to slowly degrade over a period of two months as the drops slowly dehydrated. By one month, we observed that one portion of the rod was degrading, leaving behind a cone-shaped crystal at its nucleating end (Figure 3B). This finding indicated that the P1 portion of the rods was being corrupted by the invasion of a highly related P6 packing arrangement, explaining why twinning was present in our diffraction experiments.
Significantly, this “aging” screen also revealed that the size of the P1 twin domain was dependent on pH. Crystallization at pH 7.5 produced a nearly complete overlap of the two twin domains. By contrast, crystallization at pH 7.9 caused the P1 twin domain to increase in size relative to the P6 domain, and dominate the length of the crystal. Using this information, we grew fresh crystals at pH 7.9 for data collection. Data collection was carried out using bendable crystal mounting loops (Hampton Research), which together with our knowledge of the twin domain structure, allowed us to properly orient the crystals in the X-ray beam to avoid contributions from the P6 portion (Figure 3C). Although the P1 datasets still displayed a relatively high degree of disorder (1.5–2° mosaicity) that caused some difficulties in data processing, and also likely contained some residual amount of pseudo-merohedral twinning, the data were of sufficiently high quality to successfully solve and refine a Rho model bound to both RNA and the ATP mimic ADP•BeF3. The resultant structure shows how a homohexameric helicase adopts an asymmetric ring-shaped structure around a single-stranded RNA coil, and provides a model for how ATP turnover is coupled to translocation. Together with a DNA- and ADP-bound structure of a distantly-related helicase – the papillomavirus E1 protein – the Rho model further provides an explanation for how helicases can translocate with opposite polarities along nucleic acid substrates (Enemark and Joshua-Tor, 2006; Thomsen and Berger, 2009).
Conclusion
Our efforts with Rho yielded many useful lessons for crystallizing a substrate-bound hexameric helicase complex. First and foremost is the importance of varying the sample itself during crystal screening. Although we could have applied this principle by looking at Rho homologs from different bacterial species, we chose instead to focus on the well-characterized E. coli Rho and screen different ligand combinations. Extensive efforts to improve crystal-form I using only our starting panel of RNA and nucleotide substrates failed: while thousands of crystallization experiments were performed, only a single relatively low quality crystal-form was obtained. However, immediately upon switching from ADP to ADP•BeF3, new higher quality crystals (form II) were obtained. Efforts to improve upon these crystals also proved extremely difficult, and only after shortening the RNA oligonucleotides did crystal-form III arise. The most promising strategy with Rho, which may be useful when working with other multi-substrate enzymes, was thus to screen a relatively limited region of chemical space (200–300 conditions), but a wide range of substrate conditions (Table 1). This recommendation is in line with meta-analyses of protein crystallization experiments, which have suggested that most proteins crystallize from a relatively small subset of conditions (Kimber et al., 2003; Page et al., 2003). In the same vein, biochemical data is essential to help focus early screens around the most useful substrate combinations. This is particularly important when looking at closely-related cofactors like ATP analogs, which can have unexpected effects on the target system. For example, AMP-PNP behaves more like ADP (and ADP•BeF3 like ATP) in how it interacts with Rho; whereas, in other systems we have studied, the opposite has been true. Finally, it may be worth taking additional care in how protein samples are treated during purification, for instance, avoiding sample freezing or doubly ensuring that no aggregated material is present during crystallization.
Rho also exemplified many of the challenges that can arise when crystallizing pseudo-symmetric protein-ligand complexes, such as those formed by many homo-hexameric protein systems. These problems include the “averaging-out” of interpretable ligand density due to disadvantageous crystal packing, high mosaicity, and twinning. Nonetheless, even non-ideal crystals can provide indispensible clues as to how to move forward. For instance, the solution and partial refinement of models from crystal-form II, while not useful for directly understanding Rho mechanism per se, did reveal that we had succeeded in capturing a fully liganded complex. This observation in turn allowed us to confidently focus on a set of substrate conditions that were likely to trap the desired complex, allowing us to expand our search of chemical space while screening for new crystals. Moreover, although non-merohedral twinning with coincident cell orientations can cause severe difficulties during structure solution and refinement, this work showed that crystal-trays are the experiment that keeps on giving: just as it is worth running out old crystallization drops on gels from time to time to ascertain sample integrity, visually inspecting crystals over time enabled us to identify an insidious twin domain morphology and uncover the conditions that best mitigated its effects. Although each macromolecular system presents its own set of unique challenges insofar as purification and crystallization, these experiences have shaped our thinking and approach to future projects, and may prove useful to those working on similarly complex systems.
Additional Methods
Structure solution and refinement
Data on all crystals were collected at Beamline 8.3.1 at the Advanced Light Source (ALS) (MacDowell et al., 2004), and processed with HKL-2000 (Otwinowski and Minor, 1997). Crystal-form II was solved using molecular replacement (Phaser) (McCoy et al., 2007), by searching for three copies of a Rho monomer from the open ring Rho structure (Skordalakes and Berger, 2003). Initial rigid body and translation libration screw (TLS) refinement was conducted using Phenix (Adams et al., 2010). Crystal-form III was solved using molecular replacement and refined as described (Thomsen and Berger, 2009).
Structural analysis
All structural superpositions and figures were prepared using PyMol (Schrödinger) (DeLano, 2002).
References
- Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman JL, Jeong YJ, Liao JC, Patel G, Kim DE, Oster G, Patel SS. Mechanochemistry of transcription termination factor Rho. Mol Cell. 2006;22:611–621. doi: 10.1016/j.molcel.2006.04.022. [DOI] [PubMed] [Google Scholar]
- Allison TJ, Wood TC, Briercheck DM, Rastinejad F, Richardson JP, Rule GS. Crystal structure of the RNA-binding domain from transcription termination factor rho. Nature structural biology. 1998;5:352–356. doi: 10.1038/nsb0598-352. [DOI] [PubMed] [Google Scholar]
- Bear DG, Andrews CL, Singer JD, Morgan WD, Grant RA, von Hippel PH, Platt T. Escherichia coli transcription termination factor rho has a two-domain structure in its activated form. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:1911–1915. doi: 10.1073/pnas.82.7.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogden CE, Fass D, Bergman N, Nichols MD, Berger JM. The structural basis for terminator recognition by the Rho transcription termination factor. Molecular cell. 1999;3:487–493. doi: 10.1016/s1097-2765(00)80476-1. [DOI] [PubMed] [Google Scholar]
- Braig K, Menz RI, Montgomery MG, Leslie AG, Walker JE. Structure of bovine mitochondrial F(1)-ATPase inhibited by Mg(2+) ADP and aluminium fluoride. Structure. 2000;8:567–573. doi: 10.1016/s0969-2126(00)00145-3. [DOI] [PubMed] [Google Scholar]
- Brennan CA, Dombroski AJ, Platt T. Transcription termination factor rho is an RNA-DNA helicase. Cell. 1987;48:945–952. doi: 10.1016/0092-8674(87)90703-3. [DOI] [PubMed] [Google Scholar]
- Briercheck DM, Wood TC, Allison TJ, Richardson JP, Rule GS. The NMR structure of the RNA binding domain of E. coli rho factor suggests possible RNA-protein interactions. Nature structural biology. 1998;5:393–399. doi: 10.1038/nsb0598-393. [DOI] [PubMed] [Google Scholar]
- Browne RJ, Barr EW, Stitt BL. Catalytic cooperativity among subunits of Escherichia coli transcription termination factor Rho. Kinetics and substrate structural requirements. The Journal of biological chemistry. 2005;280:13292–13299. doi: 10.1074/jbc.M500221200. [DOI] [PubMed] [Google Scholar]
- Bujalowski W, Klonowska MM. Negative cooperativity in the binding of nucleotides to Escherichia coli replicative helicase DnaB protein. Interactions with fluorescent nucleotide analogs. Biochemistry. 1993;32:5888–5900. doi: 10.1021/bi00073a023. [DOI] [PubMed] [Google Scholar]
- Burgess BR, Richardson JP. RNA passes through the hole of the protein hexamer in the complex with the Escherichia coli Rho factor. The Journal of biological chemistry. 2001;276:4182–4189. doi: 10.1074/jbc.M007066200. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
- Dolan JW, Marshall NF, Richardson JP. Transcription termination factor rho has three distinct structural domains. J Biol Chem. 1990;265:5747–5754. [PubMed] [Google Scholar]
- Dombroski AJ, Platt T. Structure of rho factor: an RNA-binding domain and a separate region with strong similarity to proven ATP-binding domains. Proc Natl Acad Sci U S A. 1988;85:2538–2542. doi: 10.1073/pnas.85.8.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- Geiselmann J, von Hippel PH. Functional interactions of ligand cofactors with Escherichia coli transcription termination factor rho. I. Binding of ATP. Protein Sci. 1992;1:850–860. doi: 10.1002/pro.5560010703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani MM, Patel SS. Cooperative interactions of nucleotide ligands are linked to oligomerization and DNA binding in bacteriophage T7 gene 4 helicases. Biochemistry. 1996;35:2218–2228. doi: 10.1021/bi9521497. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Kagawa R, Montgomery MG, Braig K, Leslie AG, Walker JE. The structure of bovine F1-ATPase inhibited by ADP and beryllium fluoride. EMBO J. 2004;23:2734–2744. doi: 10.1038/sj.emboj.7600293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantardjieff KA, Rupp B. Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein science : a publication of the Protein Society. 2003;12:1865–1871. doi: 10.1110/ps.0350503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DE, Patel SS. The mechanism of ATP hydrolysis at the noncatalytic sites of the transcription termination factor Rho. J Biol Chem. 1999;274:32667–32671. doi: 10.1074/jbc.274.46.32667. [DOI] [PubMed] [Google Scholar]
- Kim DE, Shigesada K, Patel SS. Transcription termination factor Rho contains three noncatalytic nucleotide binding sites. J Biol Chem. 1999;274:11623–11628. doi: 10.1074/jbc.274.17.11623. [DOI] [PubMed] [Google Scholar]
- Kimber MS, Vallee F, Houston S, Necakov A, Skarina T, Evdokimova E, Beasley S, Christendat D, Savchenko A, Arrowsmith CH, Vedadi M, Gerstein M, Edwards AM. Data mining crystallization databases: knowledge-based approaches to optimize protein crystal screens. Proteins. 2003;51:562–568. doi: 10.1002/prot.10340. [DOI] [PubMed] [Google Scholar]
- Lowery C, Richardson JP. Characterization of the nucleoside triphosphate phosphohydrolase (ATPase) activity of RNA synthesis termination factor p. II. Influence of synthetic RNA homopolymers and random copolymers on the reaction. J Biol Chem. 1977;252:1381–1385. [PubMed] [Google Scholar]
- Lowery-Goldhammer C, Richardson JP. An RNA-dependent nucleoside triphosphate phosphohydrolase (ATPase) associated with rho termination factor. Proc Natl Acad Sci U S A. 1974;71:2003–2007. doi: 10.1073/pnas.71.5.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubimov AY, Strycharska M, Berger JM. The nuts and bolts of ring-translocase structure and mechanism. Current opinion in structural biology. 2011;21:240–248. doi: 10.1016/j.sbi.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDowell AA, Celestre RS, Howells M, McKinney W, Krupnick J, Cambie D, Domning EE, Duarte RM, Kelez N, Plate DW, Cork CW, Earnest TN, Dickert J, Meigs G, Ralston C, Holton JM, Alber T, Berger JM, Agard DA, Padmore HA. Suite of three protein crystallography beamlines with single superconducting bend magnet as the source. J Synchrotron Radiat. 2004;11:447–455. doi: 10.1107/S0909049504024835. [DOI] [PubMed] [Google Scholar]
- Martinez A, Burns CM, Richardson JP. Residues in the RNP1-like sequence motif of Rho protein are involved in RNA-binding affinity and discrimination. Journal of molecular biology. 1996a;257:909–918. doi: 10.1006/jmbi.1996.0211. [DOI] [PubMed] [Google Scholar]
- Martinez A, Opperman T, Richardson JP. Mutational analysis and secondary structure model of the RNP1-like sequence motif of transcription termination factor Rho. Journal of molecular biology. 1996b;257:895–908. doi: 10.1006/jmbi.1996.0210. [DOI] [PubMed] [Google Scholar]
- Matthews BW. Solvent content of protein crystals. Journal of molecular biology. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson A. Crystallization of biological macromolecules. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. [Google Scholar]
- Menz RI, Walker JE, Leslie AG. Structure of bovine mitochondrial F(1)-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis. Cell. 2001;106:331–341. doi: 10.1016/s0092-8674(01)00452-4. [DOI] [PubMed] [Google Scholar]
- Miwa Y, Horiguchi T, Shigesada K. Structural and functional dissections of transcription termination factor rho by random mutagenesis. J Mol Biol. 1995;254:815–837. doi: 10.1006/jmbi.1995.0658. [DOI] [PubMed] [Google Scholar]
- Modrak D, Richardson JP. The RNA-binding domain of transcription termination factor rho: isolation, characterization, and determination of sequence limits. Biochemistry. 1994;33:8292–8299. doi: 10.1021/bi00193a016. [DOI] [PubMed] [Google Scholar]
- Newman J. A review of techniques for maximizing diffraction from a protein crystal in stilla. Acta crystallographica Section D, Biological crystallography. 2006;62:27–31. doi: 10.1107/S0907444905032130. [DOI] [PubMed] [Google Scholar]
- Opperman T, Richardson JP. Phylogenetic analysis of sequences from diverse bacteria with homology to the Escherichia coli rho gene. Journal of bacteriology. 1994;176:5033–5043. doi: 10.1128/jb.176.16.5033-5043.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW Jr, Sweet RM, editors. Methods in Enzymology. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Page R, Grzechnik SK, Canaves JM, Spraggon G, Kreusch A, Kuhn P, Stevens RC, Lesley SA. Shotgun crystallization strategy for structural genomics: an optimized two-tiered crystallization screen against the Thermotoga maritima proteome. Acta crystallographica Section D, Biological crystallography. 2003;59:1028–1037. doi: 10.1107/s0907444903007790. [DOI] [PubMed] [Google Scholar]
- Park JS, Roberts JW. Role of DNA bubble rewinding in enzymatic transcription termination. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4870–4875. doi: 10.1073/pnas.0600145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JP. Activation of rho protein ATPase requires simultaneous interaction at two kinds of nucleic acid-binding sites. J Biol Chem. 1982;257:5760–5766. [PubMed] [Google Scholar]
- Richardson JP, Conaway R. Ribonucleic acid release activity of transcription termination protein rho is dependent on the hydrolysis of nucleoside triphosphates. Biochemistry. 1980;19:4293–4299. doi: 10.1021/bi00559a022. [DOI] [PubMed] [Google Scholar]
- Roberts JW. Termination factor for RNA synthesis. Nature. 1969;224:1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- Seifried SE, Easton JB, von Hippel PH. ATPase activity of transcription-termination factor rho: functional dimer model. Proc Natl Acad Sci U S A. 1992;89:10454–10458. doi: 10.1073/pnas.89.21.10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigesada K, Wu CW. Studies of RNA release reaction catalyzed by E. coli transcription termination factor rho using isolated ternary transcription complexes. Nucleic acids research. 1980;8:3355–3369. doi: 10.1093/nar/8.15.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- Skordalakes E, Berger JM. Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell. 2003;114:135–146. doi: 10.1016/s0092-8674(03)00512-9. [DOI] [PubMed] [Google Scholar]
- Skordalakes E, Berger JM. Structural insights into RNA-dependent ring closure and ATPase activation by the Rho termination factor. Cell. 2006;127:553–564. doi: 10.1016/j.cell.2006.08.051. [DOI] [PubMed] [Google Scholar]
- Stitt BL. Escherichia coli transcription termination protein rho has three hydrolytic sites for ATP. J Biol Chem. 1988;263:11130–11137. [PubMed] [Google Scholar]
- Stitt BL, Webb MR. Absence of a phosphorylated intermediate during ATP hydrolysis by Escherichia coli transcription termination protein rho. The Journal of biological chemistry. 1986;261:15906–15909. [PubMed] [Google Scholar]
- Stitt BL, Xu Y. Sequential hydrolysis of ATP molecules bound in interacting catalytic sites of Escherichia coli transcription termination protein Rho. The Journal of biological chemistry. 1998;273:26477–26486. doi: 10.1074/jbc.273.41.26477. [DOI] [PubMed] [Google Scholar]
- Thomsen ND, Berger JM. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–534. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, von Hippel PH. Escherichia coli transcription termination factor rho. I. ATPase activation by oligonucleotide cofactors. J Biol Chem. 1993;268:13940–13946. [PubMed] [Google Scholar]
- Wei RR, Richardson JP. Identification of an RNA-binding Site in the ATP binding domain of Escherichia coli Rho by H2O2/Fe-EDTA cleavage protection studies. J Biol Chem. 2001;276:28380–28387. doi: 10.1074/jbc.M102444200. [DOI] [PubMed] [Google Scholar]
- Xu Y, Johnson J, Kohn H, Widger WR. ATP binding to Rho transcription termination factor. Mutant F355W ATP-induced fluorescence quenching reveals dynamic ATP binding. J Biol Chem. 2003;278:13719–13727. doi: 10.1074/jbc.M212979200. [DOI] [PubMed] [Google Scholar]
- Yeates TO, Fam BC. Protein crystals and their evil twins. Structure. 1999;7:R25–29. doi: 10.1016/S0969-2126(99)80016-1. [DOI] [PubMed] [Google Scholar]