Abstract
In the past several years, a wealth of evidence has emerged illustrating how metabolism supports many aspects of T cell biology, as well as how metabolic changes drive T cell differentiation and fate. Here we outline developing principles in the regulation of T cell metabolism, and discuss how these processes are impacted in settings of inflammation and cancer. In this context we discuss how metabolic pathways might be manipulated for the treatment of human disease, including how metabolism may be targeted to prevent T cell dysfunction in inhospitable microenvironments, to generate more effective adoptive cellular immunotherapies in cancer, and to direct T cell differentiation and function towards non-pathogenic phenotypes in settings of autoimmunity.
Introduction
During the course of an immune response, naïve T cells recognize foreign antigen (Ag) in the form of peptide complexed to major histocompatibility complex (MHC) molecules, and with proper co-stimulation, become activated, rapidly proliferate, and produce a variety of effector molecules that lead to control of a pathogen. T cell activation, clonal expansion, and acquisition of effector functions are energetically demanding processes that are accompanied by and dependent upon marked changes in nutrient uptake and cellular metabolism [1, 2]. Once the antigen burden is diminished, the majority of antigen specific effector T cells die, leaving behind only a small number of stable memory T cells that persist and can rapidly respond to future Ag-challenge. Memory T cells must also reprogram cellular metabolic pathways in order to support their development, longevity, and ‘rapid recall’ ability [3, 4]. Thus, proper metabolic programming in T cells is required for a productive immune response.
The cellular activation, differentiation, and extensive proliferation that happens during a T cell response is unusual for cells in a healthy adult organism, where most cells have differentiated to a terminal phenotype [5]. This aspect of T cell biology, combined with the modern tools for assaying these cells and highly tractable in vivo systems, make them uniquely suitable for studying how metabolic pathways support vigorous changes in cellular activity. In addition, and perhaps more importantly from a human health standpoint, each of these metabolic changes that occur as part of the normal development of a T cell are intimately linked to cell fate and function, and as such, represent points for clinical intervention. Since many infections, cancers, and autoimmune diseases might be controlled, or at least mitigated, by eliciting a desired response from T cells, novel approaches to therapeutically target these cells have clinical potential. Many comprehensive and up to date reviews on T cell metabolism are available [1, 2, 6–9]. Here we focus on recent advances in the mechanisms that link metabolic changes with T cell fate and function and consider novel approaches in which T cells might be manipulated by blocking, or potentiating, metabolic pathways.
The basics of T cell metabolism
Naive T cells have a metabolically quiescent phenotype and generate energy by breaking down glucose, fatty acids, and amino acids to fuel oxidative phosphorylation (OXPHOS) [10–12]. The transition from a resting naïve T cell into activated and highly proliferative effector T cells requires substantial metabolic reprogramming. While mitochondrial OXPHOS and reactive oxygen species (ROS) production increase, and are critical for T cell activation and the development of effector T cells, a rapid induction of aerobic glycolysis also occurs during this time [4, 13, 14]. Aerobic glycolysis involves the mitochondrion-independent metabolism of glucose into pyruvate and its subsequent conversion into lactate. ATP can be generated through this pathway, in what is believed to be a rapid but inefficient fashion. Specifically, only 2 molecules of ATP are gained per molecule of glucose via aerobic glycolysis, whereas up to 36 ATP molecules per molecule of glucose are produced by OXPHOS [5]. Aerobic glycolysis may however afford a metabolic advantage to effector cells by not only allowing the rapid production of ATP in glucose replete environments, but also by supplying metabolic intermediates for the synthesis of lipids, protein, carbohydrates, and nucleic acids, as well as providing a means for maintaining redox balance [5, 15–17]. Additionally, it has been found that although T cells can use OXPHOS or aerobic glycolysis interchangeably depending on their environment, engagement of aerobic glycolysis is needed for the acquisition of full effector functions [18–20]. Glutamine metabolism is also required for proper effector T cell development and utilization of this amino acid is augmented following activation [21, 22]. Glutamine can be used as a carbon source for the tricarboxylic acid (TCA) cycle in the form of α-ketoglutarate through the process of glutaminolysis, or can contribute to the citrate pool, via reductive carboxylation [23, 24]. Deletion of glutamine or glucose transporters impairs T cell activation and function [15, 22, 25, 26].
Metabolic reprograming in activated T cells is driven by a number of signaling pathways and transcription factors. A key regulator of T cell metabolism is mechanistic target of rapamycin (mTOR), which functions as two distinct complexes, mTORC1 and mTORC2, that differ in their regulation and downstream targets [27]. mTOR integrates signaling pathways associated with nutrient levels, energy status, cell stress responses, T cell receptor and growth factor signaling, and can induce multiple pathways associated with cell growth, proliferation, and metabolism [28–30].
The metabolic transition towards increased glycolysis and glutaminolysis is associated with mTOR induction as well as the expression of the transcription factors Myc and hypoxia inducible factor 1-alpha (HIF-1α) [21, 31]. HIF-1α is a transcription factor that when induced by hypoxia or mTORC1 activity leads to increased glucose uptake and diverts glucose away from OXPHOS towards aerobic glycolysis [31–33]. Myc also promotes aerobic glycolysis and glutaminolysis through enzyme expression and enhances anabolic processes such as lipid, amino acid, and nucleic acid synthesis [21]. Collectively these factors enforce metabolic phenotypes in effector T cells appropriate for their function, and alterations in these pathways can be used to manipulate effector T cell differentiation.
AMP-activated protein kinase (AMPK) is another key metabolic regulator in T cells that acts as a metabolic stress sensor, becoming activated when the ratio of AMP to ATP increases. Despite being transiently activated upon T cell activation, it can work in opposition to mTOR-mediated anabolism by promoting catabolic pathways and energy conservation during metabolic stress [34, 35]. AMPK is important for the development of memory T cells [36], and more recently is was shown that it is also critical for effector T cell development and metabolic flexibility in response to changing nutrient environments [37]. These studies highlight the importance of this kinase in T cells during nutrient stress, and also help to sediment the idea that T cells are under metabolic constraints in vivo.
In contrast to effector T cells, memory T cells do not engage aerobic glycolysis highly, and instead preferentially rely on OXPHOS, a process that is fueled, at least in part, by the catabolism of intracellular fatty acids in the mitochondria of these cells [3, 38]. Memory T cells also maintain substantial spare respiratory capacity (SRC) and have increased mitochondrial mass, both of which confer a metabolic advantage for survival and recall following antigen challenge [3, 4]. Augmenting catabolic pathways in activated CD8+ T cells with rapamycin or the AMPK activator metformin reduces the differentiation of CD8+ effector T cells, and instead enhances CD8+ memory T cell development [39]. Attenuation of aerobic glycolysis or the enhancement of OXPHOS in activated CD4+ T cells also alters their phenotype dramatically. For example, in CD4+ T cells the inhibition of mTOR by rapamycin, or the inhibition of glycolysis by the hexokinase inhibitor 2-deoxyglucose (2-DG), blocks the differentiation of T helper 17 (Th17) cells while promoting regulatory T (Treg) cell development [14, 40, 41]. Likewise AMPK activation, which enhances FAO and energy conservation by antagonizing anabolic pathways, also alters this balance in favor of Treg cells [14, 42].
In settings such as chronic infection and cancer, T cells can become anergic, or exhausted, losing the capacity to properly respond to stimulation. This hyporesponsiveness may be in large part due to an inability of the cells to optimally utilize appropriate metabolic pathways [2]. Gene expression analysis of exhausted T cells indicates that a number of genes involved in energy metabolism are transcriptionally downregulated [43], and inhibiting leucine or glucose metabolism during T cell activation can lead to an anergic phenotype [44]. Furthermore, ligation of inhibitory receptors that are highly expressed on exhausted T cells, such as cytotoxic Tlymphocyte associated protein-4 (CTLA-4) or programmed cell death protein-1 (PD-1), inhibits the upregulation of glucose and glutamine metabolism following TCR engagement and co-stimulation [45, 46]. Expression of these inhibitory receptors may restrain T cells from correctly remodeling their metabolism and hence, dampen their function. Targeting exhausted T cells with the aim of enhancing glycolysis may be a way to reactivate these cells. Consistent with this idea, T cells that lacked von Hippel-Lindau (VHL) tumor suppressor, a negative regulator of HIF-1α, had enhanced glycolysis and were resistant to exhaustion following persistent viral infection [31].
Metabolites within cells can also act as signaling molecules that influence diverse, and sometimes non-metabolic, processes [7, 18, 47, 48], thus the availability of particular metabolites can vastly impact both cellular metabolism and cell signaling. For example, the key metabolic intermediate acetyl-CoA is not only oxidized in the TCA cycle for energy production, but it is also needed for the acetylation of histones [47] and other proteins, including transcription factors and metabolic enzymes [49]. Levels of histone acetylation correlate to the activity of ATP citrate lysase (which converts citrate into acetyl-CoA and oxaloacetate), and availability of glucose (a major source of acetyl CoA) can alter histone acetylation in a ACL-dependent process [50]. The TCA cycle intermediate succinate also acts as an inflammatory signal in macrophages by inducing IL-1β through HIF-1α stabilization [48]. The accumulation of fumarate, due to fumarate hydratase deficiency, leads to a number of changes within cancer cells, including hypermethylation and HIF-1α stabilization [51]. Leucine transport into the cell is also required for T cell metabolic reprogramming [25]. Leucine can activate mTOR via leucyl-tRNA synthetase [52], thus low concentrations of intracellular leucine can impair mTOR activation. Accordingly it was shown that expression of cytosolic branched chain aminotransferase (BCATc), which transaminates leucine, regulates mTOR activity following T cell activation in a process that limits mTOR over activation [53]. Interestingly, it was found that BCATc is increased in anergic cells. This raises the intriguing possibility that leucine depletion by BCATc could contribute to T cell anergy through suppression of mTOR activity, and that inhibition of BCATc may be an interesting target within this context. Metabolites can also directly act as endogenious ligands for nuclear receptors, such as the liver X receptor (LXR) or the Aryl hydrocarbon receptor (AhR), both of which have been shown to regulate T cell differentiation and proliferation, [54, 55]. It is clear that more work needs to be done to understand how signaling from metabolites influences cell function.
Enhancing T cell function in tissue microenvironments
T cells are influenced by nutrients and other supportive signals, such as those provided by growth factor cytokines, available in their environment. We speculate that lymphoid organs are nutrient replete, but that other sites infiltrated by T cells may be much less nutrient rich. Manipulating the metabolism of tissues in which they reside, or substrates within a tissue, may provide a therapeutic approach to enhance T cell function. An example of this is provided by a consideration of a commonly expressed melanoma oncogenic mutation BRAF V600E, which generates a tumor that has a strongly immunosuppressive microenvironment [56]. This mutation induces constitutive activation of the MEK-MAPK pathway leading to enhanced tumor cell proliferation, suppression of OXPHOS, and a highly glycolytic phenotype [57, 58]. As glucose and glutamine are critical for T cell differentiation and function, and depletion of glucose impairs cytolytic activity as well as interferon-gamma (IFN-γ) production [2, 18, 19], it is likely that the highly glycolytic phenotype of BRAF V600E melanoma contributes to the immunosuppressive environment it imposes. This would be consistent with findings that effective therapeutic treatment using small molecule inhibitors of BRAF restricts glycolysis and glutaminolysis in BRAF V600E tumors, and that these inhibitors can reverse some of the immunosuppressive features within the tumor microenvironment [56–60]. Further supporting this concept, it was shown that T cells isolated from tumors had increased IFN-γ and CD40L expression after BRAF V600E inhibition and blockade of IFN-γ or CD40L compromised the tumor suppressive effects of BRAF V600E inhibition [56]. These data indicate that inhibition of BRAF V600E works, at least in part, through an immune cell dependent mechanism, and suggest that directly altering tumor metabolism allows anti-tumor T cells to work more effectively. The immunosuppressive metabolic environment induced by the BRAF V600E mutation could be further enhanced by tumor expression of inhibitory ligands for PD-1 and CTLA-4, which when bound to their cognate receptors on T cells, limit T cell-intrinsic glutaminolysis, glucose uptake, and glycolysis [45, 46]. It has also been shown that inhibition of such interactions (i.e. checkpoint blockade therapy) enhances tumor immune therapy [61]. Preliminary observations from our laboratory also indicate that checkpoint blockade therapy alters the metabolic balance between tumors and their infiltrating T cells. We postulate that immunosuppression in the tumor microenvironment is at least in part driven by the inability of T cells to acquire the nutrients to support their metabolism. Establishing that this is an important mechanism of immunosuppression may lead to new ways to manipulate the tumor microenvironment to better suit the metabolic needs of infiltrating T cells [62].
While it is relatively easy to envisage how T cells in a solid tumor could be at a competitive disadvantage for nutrients and that this would negatively effect their function, this paradigm can additionally be extended to other, perhaps less obvious, settings. For example, gut microbiota produce a number of metabolites that can interact with host tissues and the immune system, and these can have profound effects on T cell development and function [63, 64]. The bacterial production of short chain fatty acids, such as butyrate, within the gut has been shown to alter the balance of Th17 and Treg cells as well as altering T cell mTOR signaling [65–68]. It is also likely that microbially-derived amino acids and other fatty acids could regulate T cell responses [64]. Recently, it has been shown that de novo fatty acid synthesis controls the fate between Treg cells and Th17 cells [69]. Specifically, when acetyl-coA carboxylase, an important enzyme in fatty acid synthesis, is blocked either genetically or by the bacterial metabolite Soraphen A, Th17 cells fail to develop and instead, naïve T cells polarize to a Treg cell fate. Altering metabolic pathways to bias T cell differentiation away from Th17 cell development could be exploited in diseases such as multiple sclerosis or Crohn’s disease where Th17 mediated pathology has been implicated [70]. Perhaps other metabolites similar to Soraphen A are produced from gut bacteria that modulate cell metabolism and influence immune responses, possibly regulating autoimmune susceptibility in humans. Understanding how commensal organisms influence the intestinal microenvironment and how this environment then dictates metabolic pathway engagement by intestinal T cells and thus, their differentiation and function, is an important subject for future study.
Metabolism and adoptive cellular immunotherapy
Substrate availability in the tissue microenvironment has a major impact on T cell function in vivoand clearly the environment will also impact T cells that are cultured in vitro. There has been intense interest in developing adoptive cellular immunotherapy (ACI) for cancer and chronic viral infections, whereby naturally occurring or engineered T cells are stimulated and expanded in vitrothen transferred to the patient [71]. Although some success has been achieved using this approach, many ACI strategies have failed or have had less than optimal therapeutic outcomes [72, 73]. While there has been a substantial amount of research focused on optimizing T cell activation and the use of appropriate adjuvants for ACI, relatively little research has been directed at manipulating metabolic pathways, which could potentially enhance therapeutic efficacy. Altering T cell metabolism can positively impact cell function and longevity [18, 39, 74], perhaps placing more consideration on metabolic parameters when designing and implementing ACI would lead to better patient outcomes.
In the context of cancer research, two of the most common forms of ACI involve either the in vitro expansion of tumor infiltrating lymphocytes (TILs) isolated from resected tumors, or the engineering of chimeric antigen receptor T cells (CARs) derived from patients’ peripheral T cell populations [75]. In both methods, large populations of Ag-specific T cells are reintroduced into the patient. It has been observed that therapeutic efficacy is enhanced when transferred T cells maintain both replicative capacity and the ability to persist for long periods [76] and restraining differentiation to a terminal phenotype in vitro can improve the efficacy of in vivo treatment [74, 77].
Altering culture conditions for ACI to take into account metabolism is one way in which T cells could be restrained from terminal differentiation while being optimized for persistence in vivo. For example, although some culture media contain levels of glucose that approximate blood glucose concentrations (around 5.5mM), many commonly used media (including media used for ACI) have glucose concentrations in the range of 10–35mM, which is substantially higher than normal physiological levels. This could potentially program proliferating T cells to become overly dependent on glycolysis [78], which would be expected to result in impaired function and survival when T cells for ACI are transferred back into patients and thus exposed to lower physiological glucose levels. It has been shown that augmentation of glycolysis in CD8+ T cells limits long term survival [74]. Often T cells expanded in vitro are larger in size compared to in vivo proliferating T cells, which may be a function of increased glucose availability [28, 79, 80]. Exaggerated glycolysis or cell size can negatively impact T cell survival in vivo [74, 81]. It has been demonstrated that independent from proliferation, increases in aerobic glycolysis in hepatocytes correlates to increased cell size, and conversely, cell size is inversely proportional to mitochondrial gene expression [82]. Likewise in T cells, constitutive activation of Akt, which enhances glycolysis [83], or increased cell surface expression of the glucose transporter Glut 1, results in increased basal T cell size [20]; and limiting glycolysis using low dose 2-DG (which inhibits hexokinase and thus glycolysis) in cultured T cells can reduce cell size and also increase longevity, without impairing proliferative capacity [74]. Collectively, these data suggest that limiting glycolysis and cell size, either through direct modulation of metabolism, or through careful consideration of culture conditions, could improve ACI. Furthermore, cell size and/or high glycolic rates could potentially be used as proxy indicators of poor in vivo survival and function of in vitro activated T cells. Monitoring these factors while optimizing in vitro metabolic conditions may provide an efficient and effective read out of T cell fitness.
Another way to enhance the replicative capacity and long-term persistence of ACI cells may be to promote OXPHOS or mitochondrial biogenesis. A recent study has demonstrated that inhibition of Akt in in vitro expanded TILs resulted in an altered metabolic profile with increased rates of OXPHOS and FAO and these cells exhibited enhanced in vivo persistence and improved anti-tumor immunity [84]. Consistent with this idea, in vitro activation of T cells in the presence of cytokines that signal via receptors containing the common gamma chain, such as interleukin-15 (IL-15) or IL-7 allows for substantial population expansion and improved in vivo T cell survival and anti-tumor efficacy [77, 81, 85]. These beneficial effects are likely to be related, at least in part, to metabolic changes induced by these cytokines; IL-15 reduces glycolysis while enhancing OXPHOS and SRC in activated CD8+ T cells, as well as increasing mitochondrial mass [3, 38]. In other types of tissue, cell longevity is often associated with a reliance on mitochondrial metabolism [86–88]. Unpublished observations from our laboratory indicate that memory T cells, in addition to gaining more mitochondrial mass, have mitochondria that are morphologically distinct and appear to be networked, as compared to those in effector T cells. Mitochondria are dynamic organelles that constantly fuse and divide, and these fission and fusion events can regulate metabolism, longevity, and cell fitness [89–91]. There also might be potential for enhancing T cell mitochondrial function though the use of Szeto-Schiller (SS) peptides, which target cardiolipin within mitochondria and optimize the efficiency of the ETC [92]. One of these peptides, SS-31 (or Bendavia), which is currently in phase II clinical trials as a treatment for ischemic reperfusion injury, could conceivably be used to enhance T cell mitochondrial health and integrity [93]. Pharmacologically modulating mitochondria to enhance their function [94, 95], for example by targeting fission/fusion events or enhancing ETC efficiency, may provide an effective way to improve the fitness and longevity of ACI T cells.
Optimizing the culture conditions for T cells used in ACI could also be combined with strategies that modify the tumor microenvironment to make it more metabolically favorable for T cells. Tumors are not composed of malignant cells alone, but also contain stromal and epithelial cells, as well as other immune cells, which often make up a substantial proportion of total tumor mass [96]. The presence of these various cell types, in addition to the tumor cells, can result in an unfavorable metabolic environment for effector T cells. For example, the tumor microenvironment can impact T cell metabolism through the depletion of amino acids such as arginine and tryptophan [97, 98], the competitive consumption of other nutrients [18, 99], and the production of metabolites like lactate [100]. A recent study showed that tumor-derived lactate drives the expression of Arginase 1 in tumor-associated macrophages, driving polarization into an M2 phenotype [101]. M2 macrophages can suppress T cell function and therefore, may support tumor growth [102]. It seems probable that not only tumor cells, but also infiltrating immune cells, contribute to the overall suppressive nature of the tumor microenvironment and could thus collectively contribute to impaired T cell function within the tumor. In fact, production of metabolites like lactate or kynurenine may also favor development of immunosuppressive cell populations such as myeloid derived suppressor cells and Treg cells [55, 103].
There is substantial interest in modifying tumor metabolism, and multiple metabolic pathways can be exploited in this regard [104]. However, as activated T cells and cancer cells often share similar metabolic traits [5], targeting tumor cell metabolism has the potential to also negatively impact infiltrating effector T cells. Some therapeutic targets are clearly tumor-specific [104], such as the aforementioned BRAF V600E mutation in melanoma; however, many therapeutic strategies in clinical trials target metabolic pathways that are active not only in tumor cells, but also in effector T cells, such as aerobic glycolysis [105]. Therefore, it is likely that such treatments could have a beneficial impact in terms of inhibition of metabolic pathways in neoplastic cells, but could also detrimentally effect tumor-infiltrating T cell populations, which could in turn limit the effectiveness of the therapy.
One way to avoid this problem is through the development of therapies that target metabolic pathways in a tumor-specific manner. An example of this are the experimental compounds AGI-5198 or AGI6780, which specifically inhibit the mutant forms of the enzymes isocitrate dehydrogenase (IDH) 1 and 2, respectively, and have shown anti-cancer potential against glioma and leukemia cell lines [106, 107]. Alternatively, antineoplastic therapies could be beneficially used prior to ACI treatments. Using this sequential treatment approach, initial administration of a therapeutic could be used to reduce tumor size and alter metabolism of the tumor, then T cells could be adoptively transferred after tumor metabolism has been altered. An example of this could be to use the Glut 1 inhibitor WZB117 [108] or the alkalizing agent 3-bromopyruvate that is transported into cells through MCT-1 [109], prior to ACI. Treatment should be sustained for a sufficient amount of time to reduce the tumor mass and to alter the metabolism of the remaining tumor cells, then drug treatment could cease and ACI T cells could be transferred into the patient. This method would theoretically provide a tumor microenvironment that is more metabolically favorable (i.e. nutrient replete) for the infiltrating ACI T cells and allow them to optimally function. It is possible that drug companies have developed compounds that modulate tumor cell metabolism for the specific purpose of killing tumor cells, but it is unlikely that these types of compounds have been tested for their potential ability to concomitantly augment the efficacy of TILs, through the creation of more nutrient rich tumor microenvironment. Considering how potential anti-cancer compounds could positively or negatively impact TIL metabolism, may lead to improved treatment options.
Targeting metabolism therapeutically
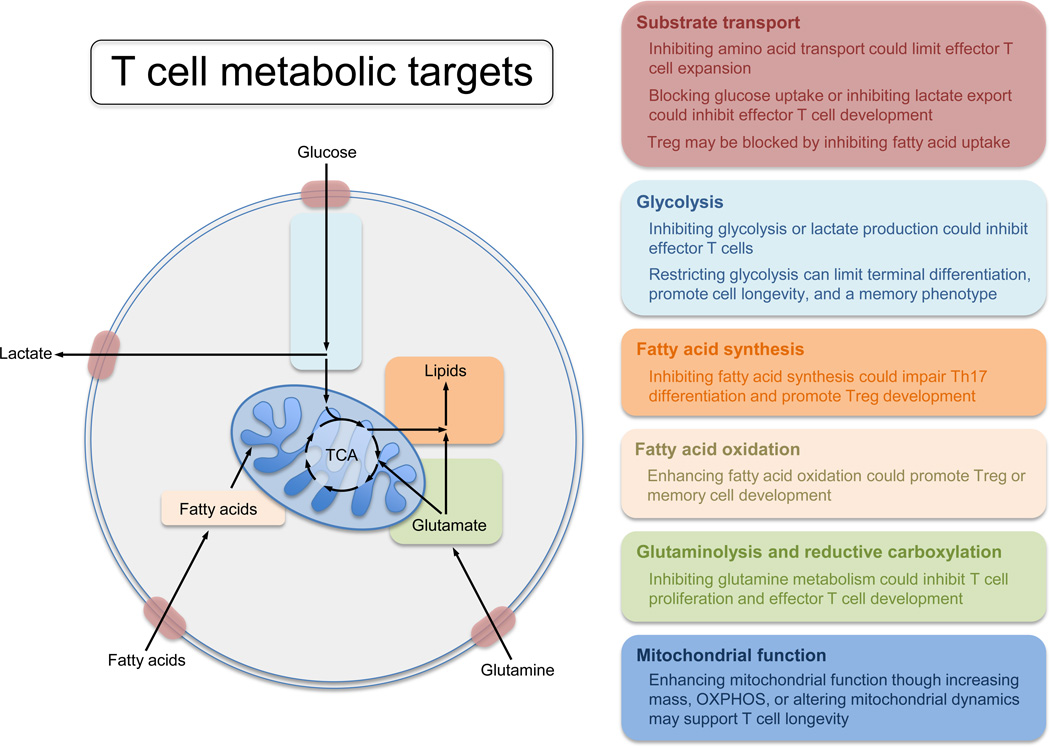
Metabolic reprogramming is needed to support T cell activation and function. Therefore modulating metabolism of T cells may be a way to target T cell function therapeutically (Figure 1). Depending on the setting, T cells can have distinct metabolic phenotypes. With this in mind, treatments with broad metabolic effects could potentially be used to target specific subsets of T cells. For example, as alloreactive T cells from graft versus host disease (GvHD) appear to be dependent on OXPHOS, targeting mitochondrial ATP production could be used to specifically inhibit these cells [110]. Since metformin has been recently shown to inhibit complex 1 of the ETC, it is possible that metformin may be useful in GvHD, particularly given that alloreactive T cells exhibit hyperpolarized mitochondria and metformin is predicated to concentrate in the mitochondria as a function of membrane potential [110, 111]. Pharmacological agents that inhibit the oxidation of long chain fatty acids in the mitochondria, such as the carnitine palmitoyl transferase (CPT)-1a inhibitors etomoxir or perhexiline could likewise be used to selectively target GvHD T cells, which have higher rates of FAO compared to other effector T cell populations [112, 113].
Figure 1.
Compounds such rapamycin can also have differing effects on T cells depending on the context, for example, although traditionally thought of as an immunosuppressant, when administered after acute viral or bacterial infection, this compound can promote memory CD8+ T cell formation [39, 114, 115]. As mitochondrial health and respiration is fundamental to effective memory T cell development it is conceivable that an enhancement of memory T cell development could also be achieved by the aforementioned SS peptides, which promote efficient ETC function [3, 92]. Although these examples provide possible therapeutic options that could be administered systemically, a limitation in systemic administration of many drugs that could modify T cell metabolic processes is the potential for off-target side effects, due to the alterations in metabolic pathways within other tissues. Therefore, future therapeutics may need to be directed more specifically to particular T cell types or subsets.
One potential way to target T cell populations might be through transporter-facilitated drug uptake, whereby a metabolite is conjugated to a drug to enhance delivery into a target cell population [116]. An example of this approach is glufosfamide, a cancer chemotherapeutic that reached phase III trails [117], which is a conjugate of d-glucose and the antineoplastic drug ifosfamide. This construct utilizes the d-glucose portion of the compound as a way to gain access to cells via glucose transporters, resulting in the preferential accumulation of ifosfamide in cells that have high glucose uptake [118]. A similar construct with direct relevance to metabolism is an experimental compound consisting of a conjugate of d-glucose and a N-hydroxyindole-based lactate dehydrogenase inhibitor; this compound accumulates at higher concentrations in glycolytic cells and results in the targeted inhibition of aerobic glycolysis within these cells by blocking lactate production [119]. Although these systems have used glucose to facilitate drug transport, it is conceivable that other metabolites, such as amino acids, could be conjugated to drugs in a similar manner, allowing for semi-selective targeting of T cell populations.
Nanotechnology is now being used to target antigens or immune-modulatory compounds to specific cell types [120]. This involves encapsulating drugs in biodegradable nanospheres that are then conjugated to antibodies, a technique that is already being explored for the delivery of cytokines and other therapeutics [121]. As this method allows for the controlled and sustained release of a compound in a cell-specific manner, it could be an extremely useful tool for delivering metabolism-modifying compounds to T cells. An example of this type of drug encapsulation and delivery system is the use of biodegradable poly(lactide-co-glyceride) (PLGA) nanoparticles, which can be manipulated in a number of ways to optimize biocompatibility/biodegradability to specific applications and allows for the controlled release of drug ranging from days to months [122]. The use of PLGA nanoparticles conjugated to anti-CD4 antibodies has proven to be an effective delivery system of leukemia inhibitory factor, a cytokine used to oppose Th17 cell differentiation and enhance Treg cell development in a mouse model of allograft rejection [123]. It is conceivable that this technology could be used to manipulate the metabolic microenvironment of T cells. For example, encapsulation of the glycolysis inhibitors 2-DG [14] or dichloroacetate [124], the L-type amino acid transport inhibitor JPH203 [125], or lactate transporter inhibitors like AR-C141990 [126], could be targeted to T cells as a way to block metabolic pathways in these cells, which could be useful in settings where T cells are hyperactive, as in autoimmunity. The de novo fatty acid synthesis inhibitor Soraphen A [69] could be targeted in a similar way to CD4 T cells to modify the Treg to Th17 cell balance in autoimmune conditions. Finally, the AMPK agonist AICAR or 6-Diazo-5-oxy-L-norleucine (DON), an inhibitor of glutaminase (enzyme that converts glutamine to glutamate), could be targeted to effector T cells to limit their inflammatory responses [21, 127–129].
Another potential approach to specifically target T cell metabolism for therapy would be to use antibodies to block nutrient transporters. The SLC family of membrane transporters includes over 300 genes that code for solute carrier proteins [130]. Proteins in the SLC family transport various molecules across the membranes surrounding the cell and its component parts [130]. There is often competition for nutrients between cells in a given microenvironment, for example, as is the case between tumor cells and T cells. During cancer progression, tumor cells can outcompete T cells for nutrients in a solid tumor [18, 99]. This differential usage of nutrients, as well as the fact that many transporter families have multiple isoforms with different substrate specificities, transport kinetics, and expression levels between different cell populations, suggests targeting distinct transport proteins with antibodies might render only one cell type susceptible to therapy, while leaving the other unperturbed.
In terms of gaining a higher level of specificity, bi-specific antibodies could be applied to this approach. Bi-specific antibodies are engineered proteins composed of two independently targeted Fab regions within an antibody, or two separate antibodies, targeted against distinct epitopes, connected by a linker protein [131, 132]. These proteins have been primarily explored as cancer therapies where the bi-specific antibody is used to simultaneously bind both a tumor cell and cytotoxic T cell [133, 134], or in the case of tri-functional antibodies, the protein can additionally bind a cell with a Fc receptor, such as a macrophage or dendritic cell [135]. The result of this association is to bring these cells into close proximity, so that the T cells can kill the tumor cells in a targeted fashion [131, 132]. In the context of T cell metabolism, the use of a strong-binding antibody against a cell surface marker, such as CD8 on T cells, could be used in conjunction with a weak-binding antagonistic antibody against a specific substrate transporter. This system could allow for the preferential binding to the target cell population by the dominant antibody followed by specific inhibition of cell surface transporter by the auxiliary antibody. The advantage of this approach would be that the lower affinity antibody against the nutrient transport protein would only bind if held in direct proximity by the high affinity antibody, resulting in nutrient transporter inhibition only on cells that express CD8.
It is fairly obvious how blocking Glut1 on T cells would dampen their ability to use glucose and thus, inhibit their activation and function. However, there are many other transporters, which could be targeted to alter T cell metabolism. For example, blocking amino acid transporters like Slc1a5, Slc7a5, and Slc3a2, which are required for effector T cell metabolic reprogramming and differentiation following stimulation [25, 125, 136] could effectively inhibit T cell effector function or activation in vivo. This approach could be particularly useful in autoimmune disease, where dampening effector T cell function would be advantageous. Recent data suggest that Treg cells acquire exogenous fatty acids, while Th17 cells synthesize fatty acids intracellularly [69]. Perhaps targeting fatty acid transporters on T cells would be a way to inhibit Treg cell development. A strategy such as this might be useful in a tumor setting where infiltration with Treg cells is associated with poor prognosis.
Concluding remarks
Cells in the immune system undergo dynamic changes in metabolism during an immune response. In the past several years a wealth of exciting data has emerged illustrating how metabolism supports many aspects of T cell biology, as well as how metabolic changes drive T cell differentiation and fate. Since cellular metabolism is linked to immune cell function, understanding more about metabolic pathways in T cells will likely illuminate new ways to exploit these pathways to harness immunity in vivo. In addition to exploring the mechanisms involved in T cell metabolism, designing ways to exploit cellular metabolism for therapy is another challenge for this field. Approaching this task with an appreciation for the role of metabolism in dictating immune cell function could likely result in revolutionary new treatments, as well as prove to be a fruitful area of exciting future research.
Highlights.
T cells undergo metabolic remodeling to support their function
Metabolic pathways impact T cell differentiation decisions and function in the periphery
Manipulating metabolic microenvironments may enhance T cell function in cancer
Metabolic pathways could be targeted for the treatment of human disease
Acknowledgements
We thank Edward Pearce and Michael Buck for critical reading of the manuscript, and Chih-Hao Chang, Jing Qiu, and Michael Buck for discussing unpublished data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pollizzi KN, et al. Integrating canonical and metabolic signalling programmes in the regulation of T cell responses. Nature Reviews Immunology. 2014;14:435–446. doi: 10.1038/nri3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacIver NJ, et al. Metabolic Regulation of T Lymphocytes. Annual Review of Immunology. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Windt Gerritje JW, et al. Mitochondrial Respiratory Capacity Is a Critical Regulator of CD8+ T Cell Memory Development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Windt GJW, et al. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proceedings of the National Academy of Sciences. 2013;110:14336–14341. doi: 10.1073/pnas.1221740110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macintyre A, et al. Activated lymphocytes as a metabolic model for carcinogenesis. Cancer & Metabolism. 2013;1:5. doi: 10.1186/2049-3002-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearce EL, et al. Fueling Immunity: Insights into Metabolism and Lymphocyte Function. Science. 2013;342 doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang R, et al. Metabolic checkpoints in activated T cells. Nat Immunol. 2012;13:907–915. doi: 10.1038/ni.2386. [DOI] [PubMed] [Google Scholar]
- 8.Pearce Erika L, et al. Metabolic Pathways in Immune Cell Activation and Quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finlay D, et al. Metabolism, migration and memory in cytotoxic T cells. Nat Rev Immunol. 2011;11:109–117. doi: 10.1038/nri2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox CJ, et al. Fuel feeds function: energy metabolism and the T-cell response. Nat Rev Immunol. 2005;5:844–852. doi: 10.1038/nri1710. [DOI] [PubMed] [Google Scholar]
- 11.Yusuf I, et al. Regulation of quiescence in lymphocytes. Trends in Immunology. 2003;24:380–386. doi: 10.1016/s1471-4906(03)00141-8. [DOI] [PubMed] [Google Scholar]
- 12.van der Windt GJW, et al. Metabolic switching and fuel choice during T-cell differentiation and memory development. Immunological Reviews. 2012;249:27–42. doi: 10.1111/j.1600-065X.2012.01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sena Laura A, et al. Mitochondria Are Required for Antigen-Specific T Cell Activation through Reactive Oxygen Species Signaling. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michalek RD, et al. Cutting Edge: Distinct Glycolytic and Lipid Oxidative Metabolic Programs Are Essential for Effector and Regulatory CD4+ T Cell Subsets. The Journal of Immunology. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anastasiou D, et al. Inhibition of Pyruvate Kinase M2 by Reactive Oxygen Species Contributes to Cellular Antioxidant Responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kidani Y, et al. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat Immunol. 2013;14:489–499. doi: 10.1038/ni.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vander Heiden MG, et al. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang C-H, et al. Posttranscriptional Control of T Cell Effector Function by Aerobic Glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cham CM, et al. Glucose deprivation inhibits multiple key gene expression events and effector functions in CD8+ T cells. European Journal of Immunology. 2008;38:2438–2450. doi: 10.1002/eji.200838289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobs SR, et al. Glucose Uptake Is Limiting in T Cell Activation and Requires CD28-Mediated Akt-Dependent and Independent Pathways. The Journal of Immunology. 2008;180:4476–4486. doi: 10.4049/jimmunol.180.7.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang R, et al. The Transcription Factor Myc Controls Metabolic Reprogramming upon T Lymphocyte Activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carr EL, et al. Glutamine Uptake and Metabolism Are Coordinately Regulated by ERK/MAPK during T Lymphocyte Activation. The Journal of Immunology. 2010;185:1037–1044. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullen AR, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sinclair LV, et al. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nature Immunology. 2013;14:500–508. doi: 10.1038/ni.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macintyre Andrew N, et al. The Glucose Transporter Glut1 Is Selectively Essential for CD4 T Cell Activation and Effector Function. Cell Metabolism. 2014;20:61–72. doi: 10.1016/j.cmet.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laplante M, et al. Regulation of mTORC1 and its impact on gene expression at a glance. Journal of Cell Science. 2013;126:1713–1719. doi: 10.1242/jcs.125773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laplante M, et al. mTOR Signaling in Growth Control and Disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powell JD, et al. Regulation of immune responses by mTOR. In. Annual Review of Immunology. 2012:39–68. doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Düvel K, et al. Activation of a Metabolic Gene Regulatory Network Downstream of mTOR Complex 1. Molecular Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doedens AL, et al. Hypoxia-inducible factors enhance the effector responses of CD8+ T cells to persistent antigen. Nat Immunol. 2013;14:1173–1182. doi: 10.1038/ni.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J-w, et al. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metabolism. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Finlay DK, et al. PDK1 regulation of mTOR and hypoxia-inducible factor-1 integrate metabolism and migration of CD8+ T cells. The Journal of Experimental Medicine. 2012;209:2441–2453. doi: 10.1084/jem.20112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamás P, et al. Regulation of the energy sensor AMP-activated protein kinase by antigen receptor and Ca2+ in T lymphocytes. The Journal of Experimental Medicine. 2006;203:1665–1670. doi: 10.1084/jem.20052469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacIver NJ, et al. The Liver Kinase B1 Is a Central Regulator of T Cell Development, Activation, and Metabolism. The Journal of Immunology. 2011;187:4187–4198. doi: 10.4049/jimmunol.1100367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rolf J, et al. AMPKα1: A glucose sensor that controls CD8 T-cell memory. European Journal of Immunology. 2013;43:889–896. doi: 10.1002/eji.201243008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blagih J, et al. The energy sensor AMPK regulates T cell metabolic adaptation and effector responses in vivo. Immunity. 2015 doi: 10.1016/j.immuni.2014.12.030. In Press. [DOI] [PubMed] [Google Scholar]
- 38.O’Sullivan D, et al. Memory CD8+ T Cells Use Cell-Intrinsic Lipolysis to Support the Metabolic Programming Necessary for Development. Immunity. 2014;41:75–88. doi: 10.1016/j.immuni.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pearce EL, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kopf H, et al. Rapamycin inhibits differentiation of Th17 cells and promotes generation of FoxP3+ T regulatory cells. International Immunopharmacology. 2007;7:1819–1824. doi: 10.1016/j.intimp.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi LZ, et al. HIF1α–dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. The Journal of Experimental Medicine. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hardie DG, et al. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wherry EJ, et al. Molecular Signature of CD8+ T Cell Exhaustion during Chronic Viral Infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 44.Zheng Y, et al. Anergic T Cells Are Metabolically Anergic. The Journal of Immunology. 2009;183:6095–6101. doi: 10.4049/jimmunol.0803510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parry RV, et al. CTLA-4 and PD-1 Receptors Inhibit T-Cell Activation by Distinct Mechanisms. Molecular and Cellular Biology. 2005;25:9543–9553. doi: 10.1128/MCB.25.21.9543-9553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boussiotis VA, et al. Biochemical signaling of PD-1 on T cells and its functional implications. Cancer Journal (United States) 2014;20:265–271. doi: 10.1097/PPO.0000000000000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wellen KE, et al. A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol. 2012;13:270–276. doi: 10.1038/nrm3305. [DOI] [PubMed] [Google Scholar]
- 48.Tannahill GM, et al. Succinate is an inflammatory signal that induces IL-1[bgr] through HIF-1[agr] Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choudhary C, et al. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 50.Wellen KE, et al. ATP-Citrate Lyase Links Cellular Metabolism to Histone Acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sullivan Lucas B, et al. The Proto-oncometabolite Fumarate Binds Glutathione to Amplify ROS-Dependent Signaling. Molecular Cell. 51:236–248. doi: 10.1016/j.molcel.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han Jung M, et al. Leucyl-tRNA Synthetase Is an Intracellular Leucine Sensor for the mTORC1-Signaling Pathway. Cell. 149:410–424. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 53.Ananieva EA, et al. Cytosolic Branched Chain Aminotransferase (BCATc) Regulates mTORC1 Signaling and Glycolytic Metabolism in CD4+ T cells. Journal of Biological Chemistry. 2014 doi: 10.1074/jbc.M114.554113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bensinger SJ, et al. LXR Signaling Couples Sterol Metabolism to Proliferation in the Acquired Immune Response. Cell. 2008;134:97–111. doi: 10.1016/j.cell.2008.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mezrich JD, et al. An Interaction between Kynurenine and the Aryl Hydrocarbon Receptor Can Generate Regulatory T Cells. The Journal of Immunology. 2010;185:3190–3198. doi: 10.4049/jimmunol.0903670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ho PC, et al. Immune-based antitumor effects of BRAF inhibitors rely on signaling by CD40L and IFNγ. Cancer Research. 2014;74:3205–3217. doi: 10.1158/0008-5472.CAN-13-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hall A, et al. Dysfunctional oxidative phosphorylation makes malignant melanoma cells addicted to glycolysis driven by the V600EBRAF oncogene. Oncotarget. 2013;4:584–599. doi: 10.18632/oncotarget.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haq R, et al. Oncogenic BRAF Regulates Oxidative Metabolism via PGC1α and MITF. Cancer Cell. 2013;23:302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knight DA, et al. Host immunity contributes to the anti-melanoma activity of BRAF inhibitors. The Journal of Clinical Investigation. 2013;123:1371–1381. doi: 10.1172/JCI66236. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Baudy A, et al. FDG-PET is a good biomarker of both early response and acquired resistance in BRAFV600 mutant melanomas treated with vemurafenib and the MEK inhibitor GDC-0973. EJNMMI Research. 2012;2:22. doi: 10.1186/2191-219X-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mockler MB, et al. Targeting T cell immunometabolism for cancer immunotherapy; understanding the impact of the tumor microenvironment. Frontiers in Oncology. 2014;4 doi: 10.3389/fonc.2014.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nicholson JK, et al. Host-Gut Microbiota Metabolic Interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 64.Brestoff JR, et al. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 67.Park J, et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 2014 doi: 10.1038/mi.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith PM, et al. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berod L, et al. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat Med. 2014 doi: 10.1038/nm.3704. advance online publication. [DOI] [PubMed] [Google Scholar]
- 70.Wilke CM, et al. Deciphering the role of Th17 cells in human disease. Trends in Immunology. 2011;32:603–611. doi: 10.1016/j.it.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maus MV, et al. Adoptive Immunotherapy for Cancer or Viruses. Annual Review of Immunology. 2014;32:189–225. doi: 10.1146/annurev-immunol-032713-120136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Drake CG, et al. Breathing new life into immunotherapy: review of melanoma, lung and kidney cancer. Nat Rev Clin Oncol. 2014;11:24–37. doi: 10.1038/nrclinonc.2013.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Melero I, et al. Therapeutic vaccines for cancer: An overview of clinical trials. Nature Reviews Clinical Oncology. 2014;11:509–524. doi: 10.1038/nrclinonc.2014.111. [DOI] [PubMed] [Google Scholar]
- 74.Sukumar M, et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. The Journal of Clinical Investigation. 2013;123:4479–4488. doi: 10.1172/JCI69589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Restifo NP, et al. Adoptive immunotherapy for cancer: harnessing the T cell response. Nat Rev Immunol. 2012;12:269–281. doi: 10.1038/nri3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crompton JG, et al. Uncoupling T-cell expansion from effector differentiation in cell-based immunotherapy. Immunological Reviews. 2014;257:264–276. doi: 10.1111/imr.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gattinoni L, et al. Acquisition of full effector function in vitro paradoxically impairs the in vivo antitumor efficacy of adoptively transferred CD8+ T cells. Journal of Clinical Investigation. 2005;115:1616–1626. doi: 10.1172/JCI24480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Glick GD, et al. Anaplerotic Metabolism of Alloreactive T Cells Provides a Metabolic Approach To Treat Graft-Versus-Host Disease. Journal of Pharmacology and Experimental Therapeutics. 2014;351:298–307. doi: 10.1124/jpet.114.218099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim D-H, et al. mTOR Interacts with Raptor to Form a Nutrient-Sensitive Complex that Signals to the Cell Growth Machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 80.MacIver NJ, et al. Glucose metabolism in lymphocytes is a regulated process with significant effects on immune cell function and survival. Journal of Leukocyte Biology. 2008;84:949–957. doi: 10.1189/jlb.0108024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Manjunath N, et al. Effector differentiation is not prerequisite for generation of memory cytotoxic T lymphocytes. The Journal of Clinical Investigation. 2001;108:871–878. doi: 10.1172/JCI13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miettinen Teemu P, et al. Identification of Transcriptional and Metabolic Programs Related to Mammalian Cell Size. Current Biology. 2014;24:598–608. doi: 10.1016/j.cub.2014.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rathmell JC, et al. Activated Akt promotes increased resting T cell size, CD28-independent T cell growth, and development of autoimmunity and lymphoma. European Journal of Immunology. 2003;33:2223–2232. doi: 10.1002/eji.200324048. [DOI] [PubMed] [Google Scholar]
- 84.Crompton JG, et al. Akt inhibition enhances expansion of potent tumorspecific lymphocytes with memory cell characteristics. Cancer Res. 2014 doi: 10.1158/0008-5472.CAN-14-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fewkes NM, et al. Novel gamma-chain cytokines as candidate immune modulators in immune therapies for cancer. Cancer Journal. 2010;16:392–398. doi: 10.1097/PPO.0b013e3181eacbc4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shen E-Z, et al. Mitoflash frequency in early adulthood predicts lifespan in Caenorhabditis elegans. Nature. 2014;508:128–132. doi: 10.1038/nature13012. [DOI] [PubMed] [Google Scholar]
- 87.Amiel E, et al. Mechanistic Target of Rapamycin Inhibition Extends Cellular Lifespan in Dendritic Cells by Preserving Mitochondrial Function. The Journal of Immunology. 2014;193:2821–2830. doi: 10.4049/jimmunol.1302498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee SS, et al. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 89.Westermann B. Bioenergetic role of mitochondrial fusion and fission. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2012;1817:1833–1838. doi: 10.1016/j.bbabio.2012.02.033. [DOI] [PubMed] [Google Scholar]
- 90.Chan DC. Fusion and Fission: Interlinked Processes Critical for Mitochondrial Health. Annual Review of Genetics. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 91.Youle RJ, et al. Mitochondrial Fission, Fusion, and Stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Szeto HH, et al. Serendipity and the Discovery of Novel Compounds That Restore Mitochondrial Plasticity. Clinical Pharmacology and Therapeutics. 2014;96:672–683. doi: 10.1038/clpt.2014.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chakrabarti AK, et al. Rationale and design of the EMBRACE STEMI Study: A phase 2a, randomized, double-blind, placebo-controlled trial to evaluate the safety, tolerability and efficacy of intravenous Bendavia on reperfusion injury in patients treated with standard therapy including primary percutaneous coronary intervention and stenting for ST-segment elevation myocardial infarction. American Heart Journal. 2013;165:509.e507–514.e507. doi: 10.1016/j.ahj.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 94.Andreux PA, et al. Pharmacological approaches to restore mitochondrial function. Nat Rev Drug Discov. 2013;12:465–483. doi: 10.1038/nrd4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smith RAJ, et al. Mitochondrial pharmacology. Trends in Pharmacological Sciences. 2012;33:341–352. doi: 10.1016/j.tips.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 96.Li H, et al. Tumor microenvironment: The role of the tumor stroma in cancer. Journal of Cellular Biochemistry. 2007;101:805–815. doi: 10.1002/jcb.21159. [DOI] [PubMed] [Google Scholar]
- 97.Rodriguez PC, et al. Arginase I Production in the Tumor Microenvironment by Mature Myeloid Cells Inhibits T-Cell Receptor Expression and Antigen-Specific T-Cell Responses. Cancer Research. 2004;64:5839–5849. doi: 10.1158/0008-5472.CAN-04-0465. [DOI] [PubMed] [Google Scholar]
- 98.Uyttenhove C, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nature Medicine. 2003;9:1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 99.Wang T, et al. The intercellular metabolic interplay between tumor and immune cells. Frontiers in Immunology. 2014;5 doi: 10.3389/fimmu.2014.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fischer K, et al. Inhibitory effect of tumor cell–derived lactic acid on human T cells. Blood. 2007;109:3812–3819. doi: 10.1182/blood-2006-07-035972. [DOI] [PubMed] [Google Scholar]
- 101.Colegio OR, et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513:559–563. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hao NB, et al. Macrophages in tumor microenvironments and the progression of tumors. Clinical and Developmental Immunology. 2012;2012 doi: 10.1155/2012/948098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Husain Z, et al. Tumor-Derived Lactate Modifies Antitumor Immune Response: Effect on Myeloid-Derived Suppressor Cells and NK Cells. The Journal of Immunology. 2013;191:1486–1495. doi: 10.4049/jimmunol.1202702. [DOI] [PubMed] [Google Scholar]
- 104.Galluzzi L, et al. Metabolic targets for cancer therapy. Nat Rev Drug Discov. 2013;12:829–846. doi: 10.1038/nrd4145. [DOI] [PubMed] [Google Scholar]
- 105.Tennant DA, et al. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10:267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- 106.Wang F, et al. Targeted Inhibition of Mutant IDH2 in Leukemia Cells Induces Cellular Differentiation. Science. 2013;340:622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 107.Rohle D, et al. An Inhibitor of Mutant IDH1 Delays Growth and Promotes Differentiation of Glioma Cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu Y, et al. A Small-Molecule Inhibitor of Glucose Transporter 1 Downregulates Glycolysis, Induces Cell-Cycle Arrest, and Inhibits Cancer Cell Growth In Vitro and In Vivo. Molecular Cancer Therapeutics. 2012;11:1672–1682. doi: 10.1158/1535-7163.MCT-12-0131. [DOI] [PubMed] [Google Scholar]
- 109.Birsoy K, et al. MCT1-mediated transport of a toxic molecule is an effective strategy for targeting glycolytic tumors. Nature Genetics. 2013;45:104–108. doi: 10.1038/ng.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gatza E, et al. Manipulating the Bioenergetics of Alloreactive T Cells Causes Their Selective Apoptosis and Arrests Graft-Versus-Host Disease. Science Translational Medicine. 2011;3:67ra68. doi: 10.1126/scitranslmed.3001975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wheaton WW, et al. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. eLife. 2014;2014 doi: 10.7554/eLife.02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Byersdorfer CA, et al. Effector T cells require fatty acid metabolism during murine graft-versus-host disease. Blood. 2013;122:3230–3237. doi: 10.1182/blood-2013-04-495515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ashrafian H, et al. Perhexiline. Cardiovascular Drug Reviews. 2007;25:76–97. doi: 10.1111/j.1527-3466.2007.00006.x. [DOI] [PubMed] [Google Scholar]
- 114.Wullschleger S, et al. TOR Signaling in Growth and Metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 115.Araki K, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Calvaresi EC, et al. Glucose conjugation for the specific targeting and treatment of cancer. Chemical Science. 2013;4:2319–2333. doi: 10.1039/C3SC22205E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ciuleanu TE, et al. A randomised Phase III trial of glufosfamide compared with best supportive care in metastatic pancreatic adenocarcinoma previously treated with gemcitabine. European Journal of Cancer. 2009;45:1589–1596. doi: 10.1016/j.ejca.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 118.Mazur L, et al. Glufosfamide as a new oxazaphosphorine anticancer agent. Anti-Cancer Drugs. 2011;22:488–493. doi: 10.1097/CAD.0b013e328345e1e0. [DOI] [PubMed] [Google Scholar]
- 119.Calvaresi EC, et al. Dual Targeting of the Warburg Effect with a Glucose-Conjugated Lactate Dehydrogenase Inhibitor. ChemBioChem. 2013;14:2263–2267. doi: 10.1002/cbic.201300562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Smith DM, et al. Applications of nanotechnology for immunology. Nat Rev Immunol. 2013;13:592–605. doi: 10.1038/nri3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Metcalfe SM, et al. Targeted nanotherapy for induction of therapeutic immune responses. Trends in Molecular Medicine. 2012;18:72–80. doi: 10.1016/j.molmed.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 122.Mundargi RC, et al. Nano/micro technologies for delivering macromolecular therapeutics using poly(d,l-lactide-co-glycolide) and its derivatives. Journal of Controlled Release. 2008;125:193–209. doi: 10.1016/j.jconrel.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 123.Park J, et al. Modulation of CD4+ T lymphocyte lineage outcomes with targeted, nanoparticle-mediated cytokine delivery. Molecular Pharmaceutics. 2011;8:143–152. doi: 10.1021/mp100203a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Caro-Maldonado A, et al. Metabolic Reprogramming Is Required for Antibody Production That Is Suppressed in Anergic but Exaggerated in Chronically BAFF-Exposed B Cells. The Journal of Immunology. 2014;192:3626–3636. doi: 10.4049/jimmunol.1302062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hayashi K, et al. LAT1 Is a Critical Transporter of Essential Amino Acids for Immune Reactions in Activated Human T Cells. The Journal of Immunology. 2013;191:4080–4085. doi: 10.4049/jimmunol.1300923. [DOI] [PubMed] [Google Scholar]
- 126.Påhlman C, et al. Immunosuppressive properties of a series of novel inhibitors of the monocarboxylate transporter MCT-1. Transplant International. 2013;26:22–29. doi: 10.1111/j.1432-2277.2012.01579.x. [DOI] [PubMed] [Google Scholar]
- 127.Nath N, et al. 5-Aminoimidazole-4-carboxamide ribonucleoside: A novel immunomodulator with therapeutic efficacy in experimental autoimmune encephalomyelitis. Journal of Immunology. 2005;175:566–574. doi: 10.4049/jimmunol.175.1.566. [DOI] [PubMed] [Google Scholar]
- 128.Bai A, et al. Novel anti-inflammatory action of 5-aminoimidazole-4-carboxamide ribonucleoside with protective effect in dextran sulfate sodium-induced acute and chronic colitis. Journal of Pharmacology and Experimental Therapeutics. 2010;333:717–725. doi: 10.1124/jpet.109.164954. [DOI] [PubMed] [Google Scholar]
- 129.Hensley CT, et al. Glutamine and cancer: cell biology, physiology, and clinical opportunities. The Journal of Clinical Investigation. 2013;123:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.He L, et al. Analysis and update of the human solute carrier (SLC) gene superfamily. Human Genomics. 2009;3:195–205. doi: 10.1186/1479-7364-3-2-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Byrne H, et al. A tale of two specificities: bispecific antibodies for therapeutic and diagnostic applications. Trends in Biotechnology. 2013;31:621–632. doi: 10.1016/j.tibtech.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Staerz UD, et al. Hybrid antibodies can target sites for attack by T cells. Nature. 1985;314:628–631. doi: 10.1038/314628a0. [DOI] [PubMed] [Google Scholar]
- 133.Baeuerle PA, et al. Bispecific T-Cell Engaging Antibodies for Cancer Therapy. Cancer Research. 2009;69:4941–4944. doi: 10.1158/0008-5472.CAN-09-0547. [DOI] [PubMed] [Google Scholar]
- 134.Lameris R, et al. Bispecific antibody platforms for cancer immunotherapy. Critical Reviews in Oncology/Hematology. 2014 doi: 10.1016/j.critrevonc.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 135.Müller D, et al. Bispecific antibodies for cancer immunotherapy: Current perspectives. BioDrugs. 2010;24:89–98. doi: 10.2165/11530960-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 136.Nakaya M, et al. Inflammatory T Cell Responses Rely on Amino Acid Transporter ASCT2 Facilitation of Glutamine Uptake and mTORC1 Kinase Activation. Immunity. 2014;40:692–705. doi: 10.1016/j.immuni.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]