Abstract
Shq1 is an essential protein involved in the early steps of biogenesis and assembly of H/ACA RNPs. Shq1 binds to dyskerin (Cbf5 in yeast) at an early step of H/ACA RNP assembly and is subsequently displaced by the H/ACA RNA. Shq1 contains an N-terminal CS and a C-terminal Shq1-specific domain (SSD). Dyskerin harbors many dyskeratosis congenita (DC) associated mutations. Structures of yeast Shq1 SSD bound to Cbf5 revealed that only a subset of these mutations is in the SSD binding site, implicating another subset in the putative CS binding site. Here we present the crystal structure of human Shq1 CS (hCS) and the NMR and crystal structure of hCS containing a serine substitution for proline 22 that is associated with some prostate cancers. The structure of hCS is similar to yeast Shq1 CS domain (yCS) and consists of two β-sheets that form an immunoglobulin-like β-sandwich fold. The N-terminal affinity tag sequence AHHHHHH associates with a neighboring protein in the crystal lattice to form an extra β-strand. Deletion of this tag was required to get spectra suitable for NMR structure determination, while the tag was required for crystallization. NMR chemical shift perturbation (CSP) experiments with dyskerin and Cbf5 derived peptides from putative CS binding sites revealed a conserved surface on CS important for Cbf5/dyskerin binding. A HADDOCK docking model of a Shq1-Cbf5 complex that defines the position of CS domain in the pre-H/ACA RNP was calculated using the CSP data.
Keywords: NMR, chemical shift perturbation, X-ray crystal structure, dyskerin, Cbf5, telomerase
Introduction
Box H/ACA ribonucleoparticles (RNPs) are responsible for conversion of uridine (U) to pseudouridine (ψ) at specific sites in ribosomal RNAs (rRNAs) and some spliceosomal RNAs (snRNAs) [1–5]. Depending on the site of localization they are classified as small nucleolar H/ACA RNPs (snoRNPs) or small Cajal body H/ACA RNPs (scaRNPs). Mature eukaryotic H/ACA RNPs contain a specific guide snoRNA and two sets of four proteins: dyskerin (also called NAP57, or Cbf5 in yeast), Nop10, Gar1, and Nhp2 (L7ae in archaea). Dyskerin/Cbf5 is the catalytic subunit that provides the pseudouridylase activity, and the snoRNA or scaRNA acts as a ‘guide’ RNA for the correct positioning of one or two specific target Us for modification to ψ. A few H/ACA RNPs have no known function in pseudouridylation. Notable among these is vertebrate telomerase, which contains a telomerase RNA (TR) that includes an H/ACA RNA domain that binds the H/ACA proteins [6–8]. Assembly of H/ACA RNPs in vivo involves multiple steps and other protein factors, including the conserved and essential specific assembly proteins Shq1 and Naf1 [4,9–13]. These proteins are nucleocytoplasmic shuttle proteins but accumulate mainly in the nucleoplasm [10–12] and are not components of the final RNPs.
Mutations in the H/ACA proteins have been linked to a genetic disorder, dyskeratosis congenita (DC) [14,15]. It is characterized by three classical conditions, abnormal skin pigmentation, nail dystrophy and oral leukoplakia, and results in premature aging, bone marrow failure, pulmonary fibrosis, and predisposition to cancers [14,16]. In addition to mutations in dyskerin, Nop10, and Nhp2, DC mutations in telomerase reverse transcriptase (TERT), telomerase RNA (TR), TIN2 (telomere protecting protein), and WDR79 (also known as TCAB1) which localizes scaRNPs and telomerase to Cajal bodies have also been reported [14,17,18]. DC is a telomere deficiency disorder as DC patients show short telomeres in proliferating tissues [17]. The best characterized and a predominant form of DC is the result of one or more mutations in the dyskerin gene (DKC) located on the X-chromosome (hence called X-DC) [15– 17]. X-DC is primarily characterized by bone marrow failure, which is the cause of early mortality in > 80% cases.
Shq1 interacts with the free dyskerin/Cbf5, independent of other H/ACA proteins, indicating that it acts at an early step of H/ACA RNP biogenesis in the cytoplasm [10,11]. The Shq1-dyskerin complex is then shuttled into the nucleus, presumably using the nuclear localization sequence in dyskerin/Cbf5. Nop10 and Nhp2 then associate with dyskerin to form a Shq1-dyskerin-Nop10-Nhp2 complex, followed by binding of assembly factor Naf1 to dyskerin using its ‘Gar1-like’ domain [19]. The H/ACA RNA then replaces Shq1 in the nuleoplasm to form a pre H/ACA RNP [10,11]. Release of Shq1 from dyskerin is at least partially dependent on its interaction with two evolutionarily conserved AAA+ ATPases, pontin and reptin, of the R2TP complex [20]. Reptin and pontin together with two other proteins RPAP3 and PIH1D1 constitute the R2TP complex. These ATPases have previously been implicated in H/ACA RNP and vertebrate telomerase biogenesis [21]. The final step in the assembly involves replacement of Naf1 by Gar1 [10,11].
Shq1 contains two well-defined, conserved domains, an N-terminal HSP90 co-chaperone p23-like CS (CHORD containing proteins and Sgt1) domain and a C-terminal Shq1 specific domain (SSD) (Fig. 1a) [12,22,23]. Structures of the yeast CS domain [22,23] and the yeast SSD domain have been reported [24,25]. However, no structures of any human H/ACA proteins or domains have been reported, including human Shq1. Structural studies of Shq1 are important for understanding its role in H/ACA RNP assembly and the effects of disease associated mutations on protein-protein interactions.
Fig. 1.
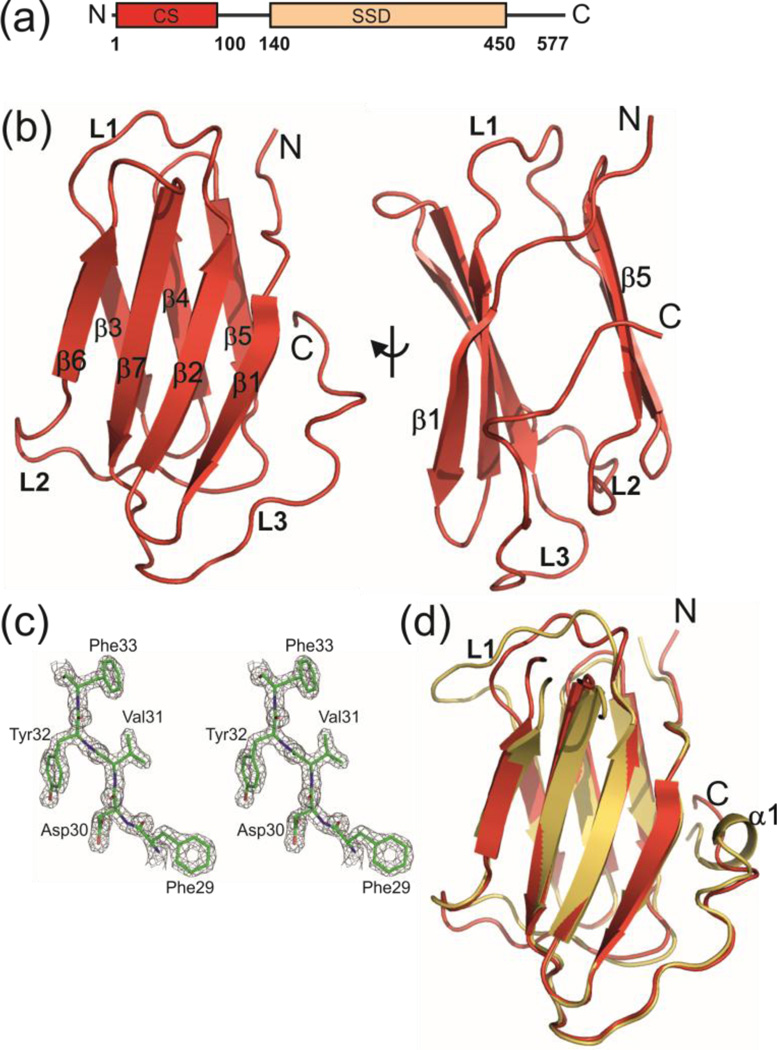
Crystal structure of N-terminal CS domain of human Shq1 with 6XHis-tag (his-hCS). (a) Domain organization of human Shq1 with CS domain and SSD indicated. (b) Ribbon representation of his-hCS structure. All the secondary structure elements and loops are marked. Front and side views are shown. (c) Stereo view of representative (residues F29-F33 of β3 strand) 2Fo-Fc electron density map of his-hCS contoured at 1.5σ and superimposed with the final model of the structure (d) Comparison of human Shq1 CS domain (his-hCS, red) and yeast Shq1 CS (yCS, gold) structures in ribbon representation.
Dyskerin/Cbf5 is a modular protein with an N-terminal extension (NTE), catalytic domain (CAT), RNA binding PUA domain, C-terminal extension (CTE), and a KKE/D repeat containing C-terminal tail (Fig. S1a,b) [9,26]. Structural studies have shown that the NTE forms a new structural layer extending the PUA domain beyond the canonical boundaries [9]. Crystal structures of Shq1 SSD bound to Cbf5 showed that the SSD binds to a region of the PUA domain and the CTE that overlaps considerably with the H/ACA RNA binding site in the mature RNP [24,25,27,28]. The electrostatic surface charge distribution and binding site of Shq1 SSD revealed that it acts as RNA mimic. Competition experiments with different snoRNAs showed that the RNA could, at least partially, compete with SSD for binding to Cbf5 [24,25]. The PUA and CTE domains harbor almost all of the X-DC mutations, however the SSD binding surface contains only a subset of these mutations. There is a cluster of X-DC mutations on the region of the PUA domain that does not interact with Shq1 SSD or other H/ACA proteins.
A previous study showed that neither CS nor SSD alone could restore growth in shq1 null strain of yeast in a complementation assay; however, when CS and SSD were provided together growth was fully restored in Shq1p depleted strain [20]. In pull down assays dyskerin was shown to retain the CS domain only when SSD is present (in trans or cis). X-DC mutations T66A and M350T in dyskerin were shown to modulate the interaction of dyskerin with Shq1 even though they are not part of SSD binding surface. Based on these biochemical and mutational studies, it has been proposed that Shq1 CS domain binds to the X-DC mutation-containing region of the PUA domain and that this interaction is SSD dependent [20]. However, the molecular details of the CS interaction with dyskerin remain elusive. In addition to binding to dyskerin, the CS domain along with dyskerin interacts with the R2TP complex that helps disrupt the Shq1/dyskerin complex required for dyskerin assembly with the H/ACA RNA [20].
Apart from implicating the Shq1/dyskerin interaction in dyskerin-associated genetic diseases, several recent studies implicated Shq1 deletions or mutations in other human diseases [29]. In a prostate cancer genome profiling study, deletion and missense mutations in SHQ1 were identified that included a P22S somatic point mutation [30,31]. Genetic loss of chromosome locations 3p23 and 3p12-p14 found in many prostate and cervical cancers, respectively, frequently includes loss of SHQ1 [32,33]. In a microarray and RT-PCR based analysis of retinal gene expression in an imposed myopic defocus in chickens, SHQ1 was found to be differentially expressed, suggesting it has a role in generation of a stop signal for axial eye growth [34]. SHQ1 was also identified as a transcriptional target in the Notch signaling pathway in T-cell leukemia [35]. Together, these reports suggest that Shq1 functions as a tumor suppressor possibly via stabilization of dyskerin and assembly of H/ACA RNPs (including telomerase RNP in vertebrates).
Here, we report the crystal structure of human Shq1 CS domain (hCS) and crystal and NMR structures of hCS containing a serine to proline mutation at position 22 [hCS(P22S)]. The structure consists of immunoglobulin like β-sandwich fold similar to yeast Shq1 CS domain. We find that a peptide sequence from dyskerin and the equivalent sequence from Cbf5 specifically interact with hCS and with yeast S. cerevisiae Shq1 CS domain (yCS), respectively. Using NMR chemical shift perturbation experiments we mapped the binding site of the Cbf5 peptide on to the surface of yCS and generated a HADDOCK (High Ambiguity Driven protein-protein DOCKing) model of yCS bound to Cbf5 in an H/ACA pre-RNP complex. Our results provide insight into the Shq1/dyskerin(Cbf5) interaction that plays a critical role in H/ACA RNP biogenesis and assembly in eukaryotes.
Results and Discussion
X-ray crystal structure of human Shq1 CS domain
Human and yeast Shq1 CS domain share ~28% sequence identity. Using the sequence alignment and structure of the yeast CS domain (yCS) [22] as a guide, we designed and cloned human Shq1 CS domain (residues 1–96) into the E. coli expression vector pET46 Ek/LIC. The protein was expressed and purified using standard protocols (Material and Methods). The protein crystallized in its recombinant form with an N-terminal 6XHis-tag and enterokinase digestion sequence (sequence MAHHHHHHVDDDDK) (hereafter called his-hCS) in space group P41212. The structure was solved by the single anomalous dispersion (SAD) method using a Se-Met labeled protein to 1.68 Å resolution (Fig 1b,c). The X-ray data collection and refinement statistics are in Table 1. The his-hCS structure is an immunoglobulin like β-sandwich fold consisting of two β-sheets, and it is highly similar to yeast Shq1 CS domain structure (Fig 1d) [22,23]. The ‘front side’ of the β-sandwich consists of an antiparallel β-sheet comprising β1 (residues 6–11), β2 (residues 14–20), β6 (residues 62–66), β7 (residues 70–77) strands, and the ‘back side’ consists of another antiparallel β-sheet comprising β3 (residues 30–34), β4 (residues 37–42), and β5 (residues 45–50) strands. There are three major well structured loops: L1 between β2–β3 (residues 21–29), L2 between β5-β6 (residues 51–61), and C-terminal loop, L3 (residues 78–96) (Fig 1b). The most notable difference between hCS and yCS is in L1. L1 is two residues longer in yCS than hCS, resulting in a difference in the backbone conformation for this loop.
TABLE 1.
Data collection and refinement statistics for his-hCS and his-hCS(P22S)
| Data collection | hCS (Se-Met: peak) |
hCS(P22S) |
|---|---|---|
| Space group | P41212 | P3221 |
| Cell constants (Å) | a=41.61 | a=58.69 |
| b =41.61 | b=58.69 | |
| c =132.18 | c=146.83 | |
| (°) | α=β=γ=90 | α=β=90, γ=120 |
| Resolution range (Å) (last shell) | 7.51–1.68 (1.72–1.68) | 10.74–2.4 (2.46–2.4) |
| Wavelength (Å) | 0.97918 | 0.97920 |
| Observed reflections | 112617 | 57944 |
| Unique reflections | 24666 | 11757 |
| CC(1/2) | 99.7 (75.8) | 99.9 (76.3) |
| Completeness (%) (last shell) | 97.7 (98.8) | 97.5 (98.3) |
| Rcryst (last shell) | 7.7 (63.5) | 5.9 (68) |
| I/σ(I) (last shell) | 13.14 (2.4) | 19.07 (2.53) |
| SAD Phasing | ||
| Number of Se sites found | 2 | |
| FOM (after density modification) | 0.332(0.594) | |
| Refinement | ||
| No. of reflections | 13859 | 11744 |
| Resolution (Å) | 25.88-1.68 | 50.827-2.4 |
| R-factor (%)/Rfree (%) | 20.36/24.34 | 22.45/28.36 |
| Average B (Å2) | 19.83 | 51.63 |
| R.m.s.d bond length (Å)/ angles (°) | 0.007/1.142 | 0.007/1.144 |
| Content of asymmetric unit | ||
| No. of protein molecules | 1 | 2 |
| No. atoms in the model | 991 | 1709 |
| No. of solvent atoms | 165 | 6 |
| Ramachandran statistics | ||
| Allowed/generous/disallowed (%) | 94.9/5.1/0 | 92.61/6.9/0.49 |
| PDB accession number | 4PBD | 4PCK |
Rwork = ∑|Fo−Fc|/∑Fo
Rfree = ∑|Fo−Fc|/∑Fo, calculated using a random set containing 5% reflections that were not included throughout refinement.
Numbers in parentheses are for the last resolution shell.
Although his-hCS purified as a monomer, in the crystal the 6XHis-tag containing N-terminal residues interact with a neighboring molecule, in a manner analogous to the donor-strand complementation mechanism observed in the PapD-PapK chaperone-subunit complex that is involved in pilus formation in some Gram-negative bacteria (Fig. S2a–c) [36]. Residues AHHHHHH from the N-terminal His-tag extend the ‘back side’ β-sheet by forming a β3’ strand on a neighboring protein (Fig. S2c). This results in a continuous arrangement of protein molecules linked together by 6XHis-tag/domain interactions. This artificial interaction was apparently essential for crystallization of the protein as we failed to obtain crystals of a protein construct with the N-terminal tag cleaved off. In contrast, we note that the 6XHis-tag present in the yeast Shq1 CS construct used for crystallization did not form these contacts and is not visible in the electron density in the crystal structure [22].
NMR characterization of human CS domain
NMR 1H-15N HSQC spectra of his-hCS show good chemical shift dispersion indicative of a folded protein; however, only about 70% of the expected crosspeaks are observed (Fig. S3a). This suggests that in solution there may be a transient interaction between the 6XHis-tag and the CS domain similar to the stable interaction present in the crystal structure, leading to line broadening due to chemical exchange, or alternatively that only part of the domain was stably folded in solution. Surprisingly, removal of the 6XHis-tag using enterokinase was very inefficient (data not shown). Therefore, we re-cloned the hCS domain into pET41 EK/LIC vector, which contains a thrombin cleavage site between the hCS domain and N-terminal GST and a 6XHis-tag. The 1H-15N HSQC spectra of the purified and thrombin digested hCS without a 6XHis-tag (hereafter called hCS) shows almost all the expected amide crosspeaks for the sequence (Fig. S3a). Comparison of 1H-15N HSQC spectra of the hCS with and without the 6XHis-tag shows that most of the crosspeaks from his-hCS superimpose on those from hCS (Fig. S3a. Furthermore, the linewidths of the hCS crosspeaks are about two-fold narrower than his-hCS (data not shown).
The backbone resonances of hCS were assigned using standard CBCACONH/HNCACB and HNCACO/HNCO sets of triple resonance NMR experiments as well as by comparison with hCS(P22S) assignment (see later section) [37]. The missing residues in his-hCS compared to hCS spectra correspond to residues 21–32 (loop L1 and part of β3), 36–42 (β4 and part of β5), 55–58 (loop L2), 61–63 (β6), and 75–80 (β7) (Fig. S4). Overall the missing peaks are on the ‘β3– β6 face’ of the protein, where the 6XHis-tag interacts in the crystal structure. These results and the difference in linewidths are consistent with a transient intermolecular interaction of the N-terminal 6XHis-tag in solution, similar to what was observed in the crystal. Therefore, we used hCS for the dyskerin peptide chemical shift perturbation (CSP) experiments described below.
Binding of dyskerin peptides to the human Shq1 CS domain
We have previously shown that the CS domain is essential for Shq1p essential functions [22]. Subsequent in vitro pull down and in vivo complementation studies showed that the isolated Shq1 CS does not interact with dyskerin in the absence of the SSD. The CS domain was proposed to interact with dyskerin PUA domain, and it was further proposed that CS and SSD together constitute a high affinity-binding site for dyskerin [20]. Since the dyskerin PUA domain harbors most of the X-DC mutations (Fig. S5), it has been proposed that these mutations may affect Shq1 binding thereby affecting H/ACA RNP assembly [20]. Isolation and purification of free human dyskerin in quantities sufficient for NMR and other biochemical assays has not been successful as the protein expresses poorly and tends to aggregate and precipitate [24,25]. Therefore, to gain insights into the Shq1 CS domain binding to dyskerin, we investigated its interaction with peptides with sequences corresponding to the putative CS domain binding sites on dyskerin using NMR chemical shift mapping. Based on sequence alignment of dyskerin with Cbf5 and the structure of yeast Cbf5 in complex with other H/ACA proteins (yeast Shq1 SSD-Cbf5-Nop10-Gar1 (PDB ID 3UA1) and SSD-Cbf5 (PDB ID 3ZVO) [24,25], we designed peptide sequences corresponding to the regions of dyskerin harboring X-DC mutations, almost all of which are in loop regions (Fig. S1a and S5). We investigated 7 chemically synthesized peptides corresponding to regions in dyskerin or Cbf5, as well as 4 additional peptides containing mutations found in X-DC (Table 2). The locations of the peptides in the primary sequence of Cbf5 and dyskerin are shown in Figure S1 and their locations in the structure of yeast Cbf5 are shown in Figure S5.
TABLE 2.
Cbf5 and dyskerin peptides sequences used in this study
| Peptide | Sequence | Protein and residue range | CS domain binding1 |
|---|---|---|---|
| Dys1 | AEIQHAEEFLIKPESKVA | Dyskerin, residues: 28–45 | No |
| Dys3 | GAKIMLPGVLRYEDG | Dyskerin, residues: 312–326 | No |
| Dys4 | LMTTAVISTCDHG | Dyskerin, residues: 349–361 | No |
| Dys2S | LNVRTTHYTPLA | Dyskerin, residues: 62–73 | Yes |
| Dys2L | LNVRTTHYTPLACGSNPLKR | Dyskerin, residues: 62–81 | Yes |
| Cbf5-2S | LLVRSGHYTPIP | Cbf5p, residues: 32–43 | Yes (yCS) |
| Cbf5-2L | LLVRSGHYTPIPAGSSPLKR | Cbf5p, residues: 32–51 | Yes (yCS) |
| Dys2S-R65T | LNVTTTHYTPLA | Dyskerin, residues: 62–73 | Yes |
| Dys2S-T66A | LNVRATHYTPLA | Dyskerin, residues: 62–73 | Yes (reduced) |
| Dys2S-H68Q | LNVRTTQYTPLA | Dyskerin, residues: 62–73 | Yes |
| Dys2-L72Y | LNVRTTHYTPYA | Dyskerin, residues: 62–73 | Yes |
Binding to human and yeast Shq1 CS domain monitored by NMR
In bold are the residues that are found mutated in X-DC. In red are the residues after point mutations. ND; not determined
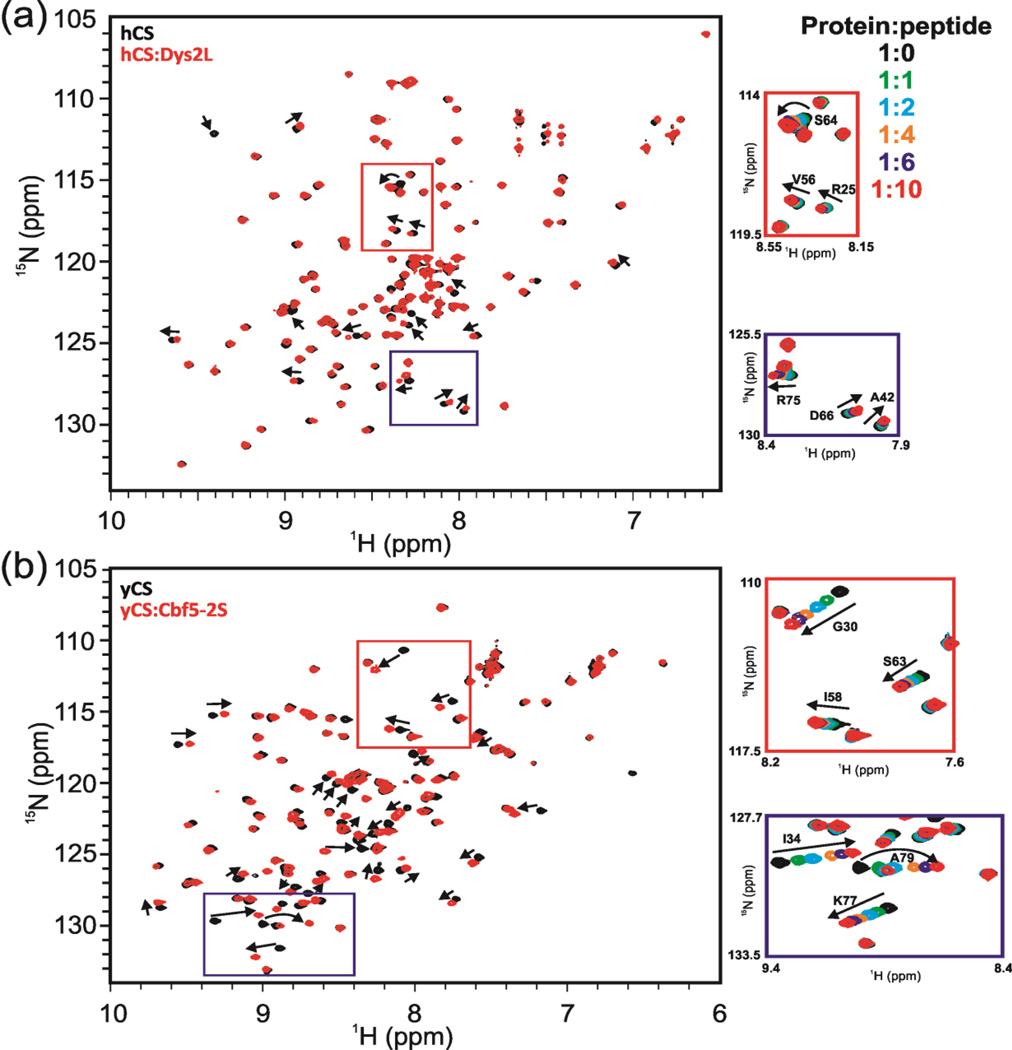
To investigate the interaction between hCS and dyskerin peptides, peptides were titrated into uniformly 15N labeled hCS, and a 1H-15N HQSC or SFHMQC [38] spectrum was recorded at each step of titration. NMR chemical shift perturbations (CSP) is a useful method to probe weak interactions between proteins and ligands, as NMR chemical shifts are sensitive to changes in the electronic environment of a nucleus that occur upon ligand binding. No chemical shift changes were observed for the titrations between hCS and peptides Dys1, Dys3, and Dys4, up to 1:10 protein to peptide ratio (Fig. S6a–c). Peptide Dys2S, in contrast, induced chemical shifts changes of several resonances in hCS as a function of added peptide, indicative of a specific interaction with hCS (Fig. 2a and Fig. S7a). The Dys2S peptide is in fast exchange with hCS on the NMR timescale, which indicates that it is a weak interaction (>500 µM) (Fig. 2a), consistent with previous observations that high affinity binding of the CS domain to dyskerin requires the SSD domain in cis or trans [20,26]. Dys2S was extended from 11 residues to 20 by adding C-terminal residues to include the entire loop region in the Cbf5 structure (Dys2L) (Table 2) to see if this improved the binding. The results of the NMR titration experiments with hCS show that the Dys2L peptide recapitulates the interaction of the shorter Dys2S interaction, with no new peaks getting significantly perturbed (Fig. 2a). This indicates that the binding site for the hCS domain on Dys2L is within Dys2S.
Fig. 2.
1H-15N HSQC spectra show specific binding between Shq1 CS domains (human and yeast) and dyskerin/Cbf5 peptides. (a) Interaction of Dys2L peptide with hCS. Overlay of 1H-15N HSQC spectra of hCS (black) and hCS:Dys2L peptide at 1:10 molar ratio (red). A few peaks showing chemical shifts are marked with arrows as examples. Red and blue boxed regions are shown expanded at right with overlay of spectra for multiple steps of the titration. Crosspeaks at each step in the titration are shown in different colors and the color code is shown. Crosspeaks that shift during the titration are labeled. (b) Titration of Cbf5-2S peptide with yCS. Overlay of 1H-15N HSQC spectra of yCS (black) and yCS:DCbf5-2L peptide at 1:10 molar ratio (red). A few peaks showing chemical shifts are marked with arrows as examples. Red and blue boxed regions are shown expanded at right with overlay of spectra for multiple steps of the titration. Protein:peptide ratios and color code are the same as in (a).
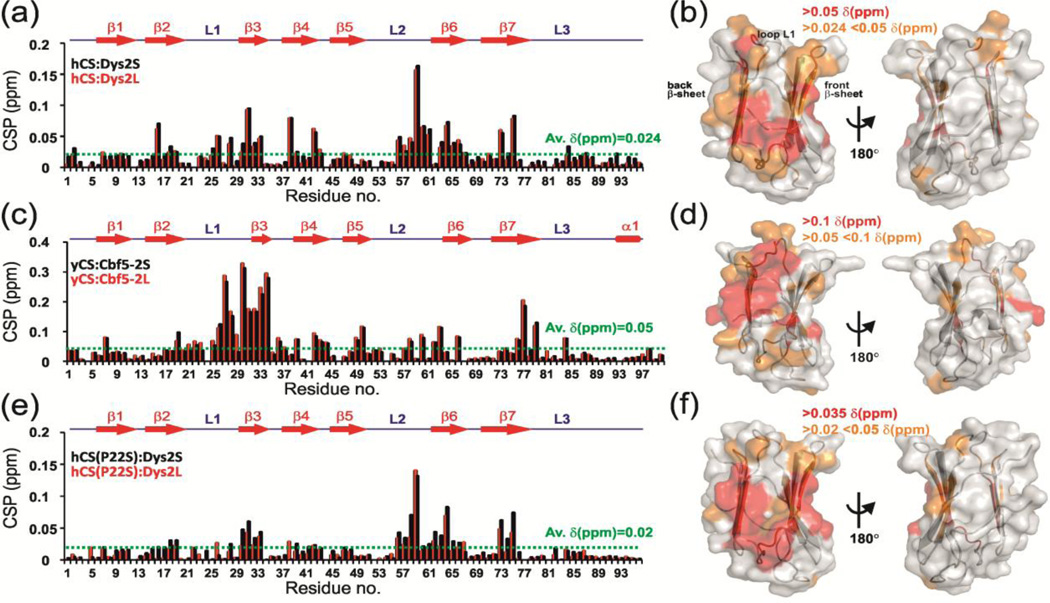
We calculated the magnitude of the induced chemical shift perturbations (CSP) at saturating 1:10 hCS to Dys2S or Dys2L ratio as described in Materials and Methods (Fig. 3a). Significant CSPs were classified as the CSPs above the average CSP of 0.024 ppm. CSPs between 0.024 – 0.05 ppm were considered weak and above 0.05 ppm were considered strong. Estimation of the dissociation constant (KD) in the case of weak interactions such as this can be erroneous due to significant dilution of the sample to reach saturation upon ligand addition and the small CSP observed. In such a situation however, qualitative analysis of CSP can still identify the binding interface. The CSPs were mapped on the crystal structure of his-hCS (Fig. 3b). The mapping shows that all induced shifts are located on the left side of the β-sandwich (β3-β6 face) of the structure, while there are no shifts on the opposite side (Fig. 3b). This suggests that the observed interaction is specific. All the residues having significant CSPs are located on β3 (residues D30, V31, Y32, and F33) and β4 (residues F38, K39, and A42) strands of the ‘back’ β-sheet and β6 (residues G53, S64, Y65, D66, and A67), β7 (residues T73 and R75), and β2 (residues T16, A18, and I19) strands of the ‘front’ β-sheet (Fig. 3b). Residue R25, V26, and E28 in loop L1 and residues V56, E57, N58, G59, S60, and E61 in loop L2 were also perturbed (Fig. 3b).
Fig. 3.
NMR chemical shift perturbation (CSP) studies reveal a conserved surface on Shq1 CS domain for dyskerin/Cbf5 binding. (a, c, e) Plot of CSP vs. residue number and (b, d, f) CSP mapped on CS structure for interaction of (a, b) hCS with Dys2S/L, (c, d) yCS with Cbf5-2S/L and (e, f) hCS(P22S) with Dys2S/L. In (a, c, e) the CS secondary structure is shown on top. CSP are for 1:10 protein:peptide ratio and are shown in black and red, respectively, for (a) hCS:Dys2S and hCS:Dys2L, (c) yCS:Cbf5-2S and yCS:Cbf5-2L, and (d) hCS(P22S):Dys2S and hCS(P22S):Dys2L. Dotted green line shows the average chemical shift differences upon peptide addition. In (b,d,f) different limits to classify shifts as strong (red) and weak (orange) are noted on each panel. Two views are represented, showing surface that is perturbed and 180° rotation showing no significant perturbation. ‘Back’ and ‘front’ β-sheets as well as L1 loop are marked.
NMR chemical shift mapping of Cbf5 peptides interaction on yeast Shq1 CS domain
We next investigated the interaction of yeast Shq1 CS domain (yCS) with Cbf5-2S and Cbf5-2L peptides from Cbf5, equivalent to Dys2S and Dys2L (Table 2). NMR titration of Cbf5-2S and Cbf5-2L with yCS also showed chemical shift perturbations in the fast exchange timescale by NMR (Fig. 2b and Fig. S7b). As with hCS, a set of peaks was perturbed for both Cbf5-2S and Cbf5-2L (Fig. 2b and Fig. 3c). The residues that were perturbed more than the average CSP of 0.05 ppm were mapped onto the surface of the yCS crystal structure [22] (for Cbf5-2S peptide titration). Chemical shift mapping showed that most of the residues that showed chemical shift perturbations are on the left side of the ‘front” face of the β-sandwich (β3-β6 face) (Fig. 3d). Significant CSPs were located on the β3 (residues E32, I33, and I34), β4 (residues F42, H43, and L44), and β5 (residues L49, and R50) strands of ‘back’ β-sheet, and β6 (residue Q66), β7 (residues V76, K77, and A79) strands of ‘front’ β-sheet (Fig. 3c,d). Almost all the residues in loop L1 (residues I21-E32) and residues I58, D60, R62, and S63 loop L2 were also significantly perturbed. The most notable difference between Cbf5 peptides binding to yCS and dyskerin peptides binding to hCS was the perturbation of the entire L1 loop residues in Cbf5-2S/L peptide titrations in comparison to only a few L1 loop residues for the Dys2-S/L and hCS interaction (Fig. 3b,d). As mentioned previously, yCS L1 is two residues longer than in hCS, resulting in a difference in the backbone conformation in comparison to hCS that may be the reason for the observed CSP on the entire L1 in yCS. This may have functional consequences given that a disease associated P22S mutation is found in L1. It is however, also possible that we simply did not capture the entire binding surface due to weak binding with the peptide. Overall, the CSP analysis showed that generally Cbf5-2S/L peptides are binding to the same surface on yCS where Dys2S/L peptides bind to hCS.
NMR data driven HADDOCK docking model of Shq1 yCS on Shq1 ySSD-Cbf5-Nop10-Gar1 complex
Since crystal structures of a complex of yeast Shq1 SSD with Cbf5-Nop10-Gar1 (PDB 3UAI) and the structure of yeast Shq1 CS (PDB 3EUD) have been determined [22,25], we modeled the interaction of the yCS to this complex using the modeling program HADDOCK [39,40] with incorporation of the CSP data for the interaction of Cbf5-2S and Cbf5-2L. All of the proteins in Cbf5-SSD-Nop10-Gar1 were used in the HADDOCK run. This was especially important as the position of SSD could act as a restraint for docking the CS domain. NMR CSP information and solvent accessibility of residues were used to classify ‘active’ residues in yCS (see Materials and Methods). For Cbf5, the ‘active’ residues were classified as residues found mutated in X-DC (in the Cbf5-2S/L peptide region) and postulated to be important for Shq1 interaction. ‘Passive’ residues were defined automatically by HADDOCK as the surface exposed neighbors of ‘active’ residues.
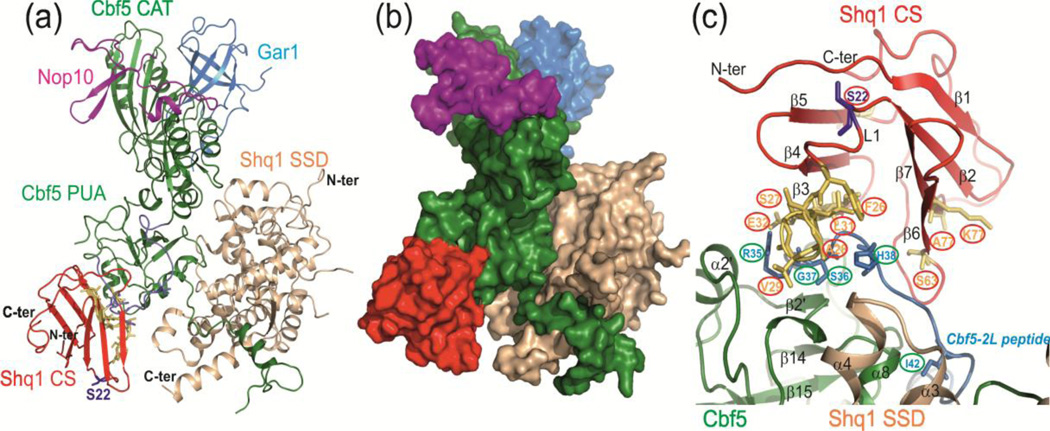
Analysis of the final 200 HADDOCK model structures of the CS-SSD-Cbf5-Nop10-Gar1 complex resulted in only two clusters. The top cluster (cluster 1) contained 163 structures with HADDOCK score of −17.7 and a Z-score of −1.0. The HADDOCK score is an arbitrary score that is used to group solutions into clusters, and the Z-score is a standard score that indicates the statistical significance of the HADDOCK scores. A lower Z-score and a larger gap with the next cluster indicate a better cluster in HADDOCK docking. The next cluster (cluster 2) had only 19 structures with Z-score of 1.0 (HADDOCK score 34.4). This suggests that cluster 1 can be taken as a representative ensemble of the CS-SSD-Cbf5-Nop10-Gar1 complex. The results of the HADDOCK model are shown in Figure 4. The backbone RMSD from the overall lowest-energy structure in this cluster was 0.7±0.4 Å with the total buried surface area of 2000.2 Å2 between the CS domain and Cbf5.
Fig. 4.
NMR CSP data-driven HADDOCK model structure of yCS bound to yeast Cbf5-Nop10-Gar1-Shq1 SSD complex. (a) Model with lowest HADDOCK score with proteins shown as ribbons. The model is based on the structures of yeast Cbf5-Nop10-Gar1-Shq1 SSD complex (PDB 3UAI) and Shq1 yCS (PDB 3EUD). Cbf5 is green, Nop10 is magenta, Gar1 is cyan, Shq1 SSD is wheat, and Shq1 CS is red. The yCS surface identified to interact with Cbf5 peptide using NMR CSP is shown in yellow, and the Cbf5-2S/L peptide position is shown in blue. (b) Surface view of the model, showing the ‘capture’ of Cbf5 by Shq1 CS and SSD. (c) Close up view of interactions of yCS with the Cbf5 PUA domain. Positions of key residues used to drive modeling are shown.
In the HADDOCK model, yCS is positioned on the PUA domain opposite to the SSD and the H/ACA RNA binding site (Fig. 4a,b). In crystal structures of yeast Shq1 SSD-Cbf5 or Shq1-Cbf5-Nop10-Gar1 complexes [24,25], this side of the PUA domain of Cbf5 has a large free area that is also the site of several DC mutations, and has been proposed to be the region where the CS domain binds [20]. In the HADDOCK model, the region of Cbf5 containing the peptide sequence in Cbf5-2S/L binds to the CS domain between the β-sandwich on the left side of the structure (β3-β6 face). The conformations of the side chains of several residues of Cbf5 in the HADDOCK model are changed in comparison to the free Cbf5 structure. Most notably, the histidine 68 side-chain inserts into the β-sandwich in a small hydrophobic cavity that involves loop L2 residues (Fig. 4c).
We mapped the designed dyskerin peptides (Dys1–4) and corresponding Cbf5 peptides (Cbf5 1–4) on the CS-Cbf5-SSD complex (Fig. S5). Dys1 peptide sequence falls in the region that is not present in the crystal structure of Cbf5 (which lacks N-terminal residues 1 to 17) and hence it is not placed in the CS-Cbf5-SSD structure. Dys3 and Dys4 (and the corresponding Cbf5-3 and Cbf5-4) peptides are not contributing to the CS-Cbf5 interaction except R292 from (R322 in dyskerin) Dys4 (Fig. S5). Interestingly, R322Q is one of the X-DC mutations found in dyskerin. Almost all the CS/dyskerin (Cbf5) interaction is due to Dys2 (Cbf2-2S) in our HADDOCK model. Finally, our HADDOCK model (CS-Cbf5-SSD) supports the previously proposed model that suggested that CS and SSD together create a viselike high affinity-binding grip for Cbf5/dyskerin [20].
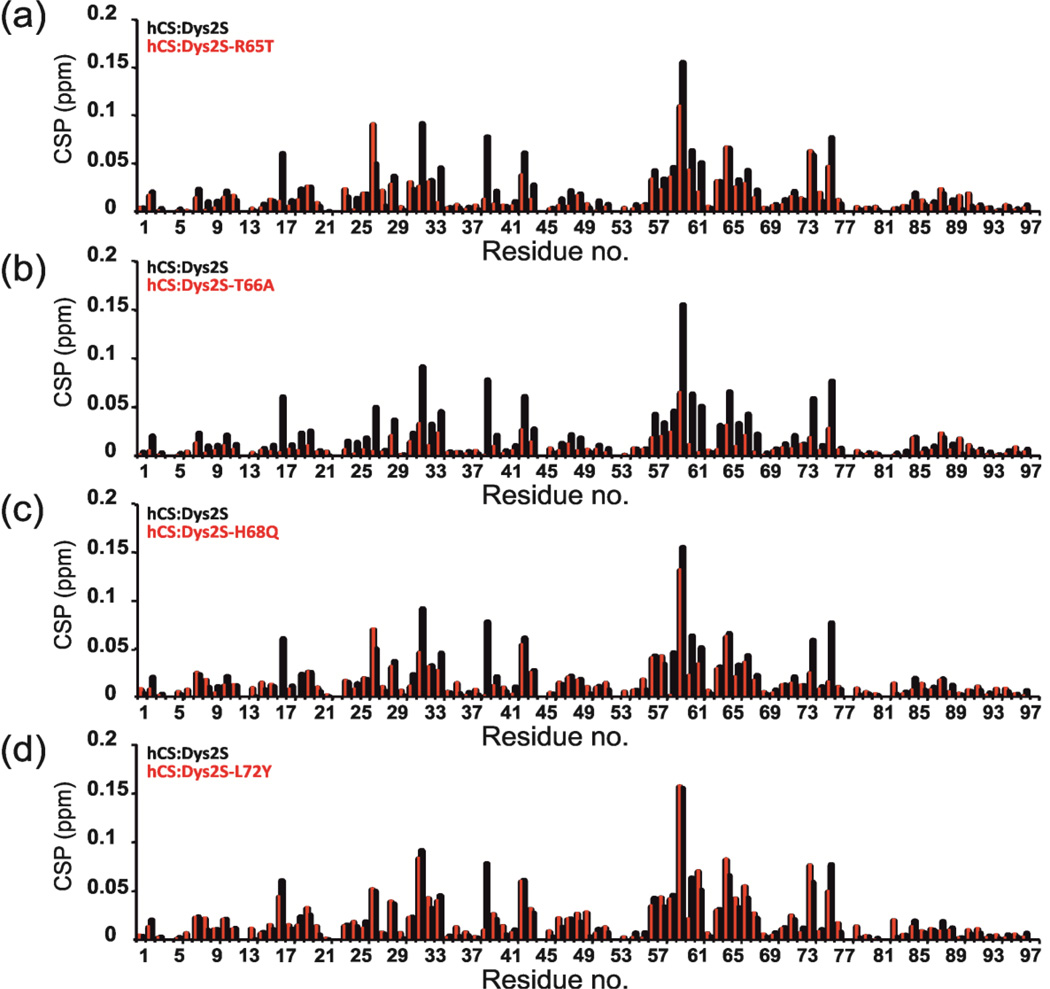
Binding of X-DC mutation-containing dyskerin/Cbf5 peptides to hCS
To gain the further insight into the binding specificity of the Dys2S peptide to hCS and effects of DC mutations, we designed single amino acid substitutions in Dys2S corresponding to mutations found in X-DC patients: Dys2S–R65T, Dys2S–T66A, Dys2S–H68Q, Dys2S–L72Y (Table 2). NMR titrations were carried out under the same conditions as for Dys2S and hCS, and the relative magnitudes of the CSP with respect to WT were plotted at saturating protein/RNA ratio (Fig. 5 and Fig. S8). Dys2S–R65T, Dys2S–H68Q, and Dys2S–L72Y did not show much reduction of CSP with hCS compared to Dys2S. (Fig. 5a,c,d and Fig. S8a,c,d) However, Dys2S-T66A showed a significant reduction in binding (Fig. 5b and Fig. S8b). We note that the dyskerin T66A mutation has been shown to reduce its association with Shq1 [26]. In our NMR CSP driven HADDOCK model of yeast Shq1/Cbf5 model S36 (equivalent to T66 in dyskerin) is located at the binding interface (Fig. 4c). Taken together, these results support our HADDOCK model and putative CS domain-binding surface onto the dyskerin/Cbf5.
Fig. 5.
Effect of X-DC mutations in Dys2S peptide on hCS binding. Plots of CSP for interaction of Dys2S and Dys2S mutant peptides with hCS are shown vs. hCS residue number for (a) Dys2S-R65T, (b) Dys2S-T66A, (c) Dys2S-H68Q, and (d) Dys2S-L72Y (shown in red). For comparison, the CSP of Dys2S with hCS is shown in black for each plot. hCS to peptide ratio is 1:10 in all cases.
Structure of human Shq1 CS P22S
In a prostate cancer profiling study a specific deletion at chromosome 3p14 was found that corresponds to the SHQ1 locus. A somatic point mutation in the conserved CS domain of Shq1, proline 22 to serine (P22S), was also confirmed in prostate cancer [30]. In S. cerevisae, the equivalent position to residue 22 based on sequence alignment is a serine, located in the β2-β3 loop (loop 1); however, this loop is two residues longer in yCS than hCS. We therefore investigated the effect of the hCS P22S mutation on the structure, stability, and function of the CS domain.
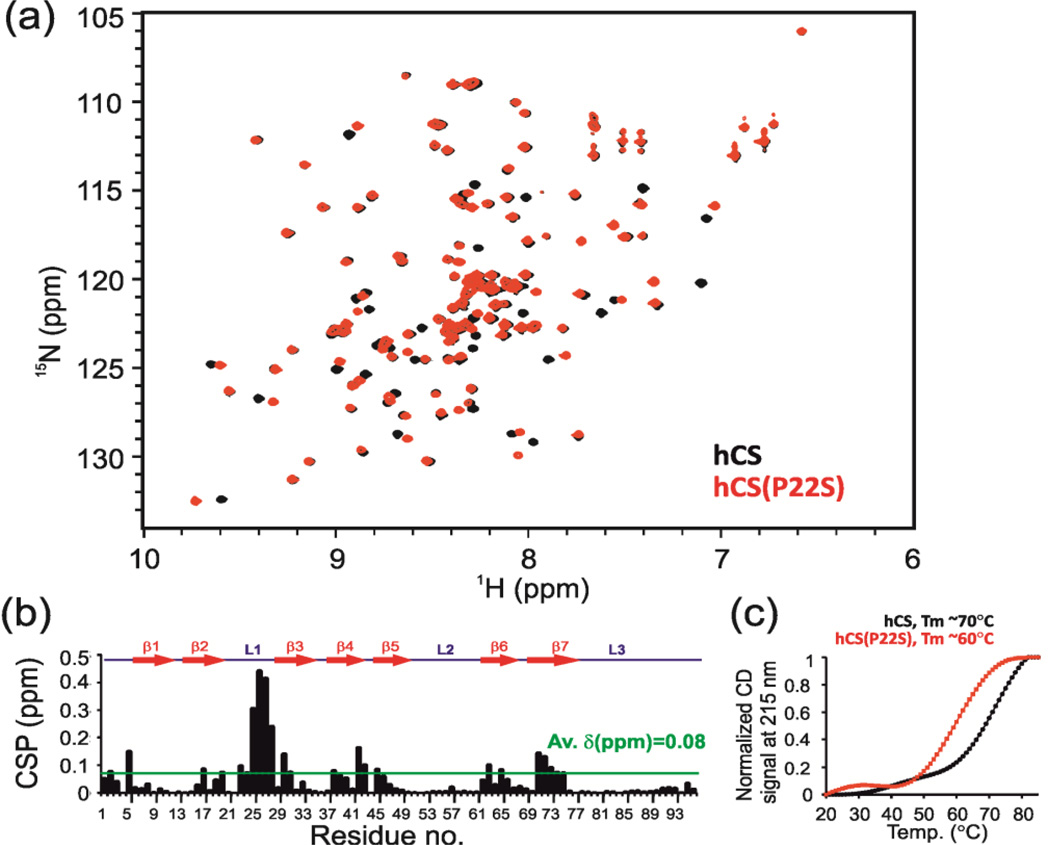
We cloned, expressed in E. coli, and purified hCS(P22S) (using a GST tag that was cleaved off by thrombin digestion, see Materials and Methods). NMR 1H-15N HSQC spectra showed that hCS(P22S) is folded with well-dispersed peaks of uniform intensities and a crosspeak pattern similar to the hCS spectra (Fig. 6a). The CSP between hCS and hCS(P22S) showed that the majority of peaks that were shifted in hCS(P22S) were in loop L1, around the P22 position (Fig 6a,b and Fig. S9).
Fig. 6.
Comparison of hCS and hCS(P22S). (a) An overlay of 1H-15N HSQC spectra of hCS (black) and hCS(P22S) (red). (b) A plot of the chemical shift difference in backbone amides of hCS and hCS(P22S) vs. residue number. Secondary structure is drawn on top. Dotted green line depicts the average chemical shift differences between the two proteins. (c) Comparison of thermal melting profile of hCS and hCS(P22S) using CD spectroscopy. The estimated melting temperatures of hCS and hCS(P22S) are ~70°C and 60°C respectively.
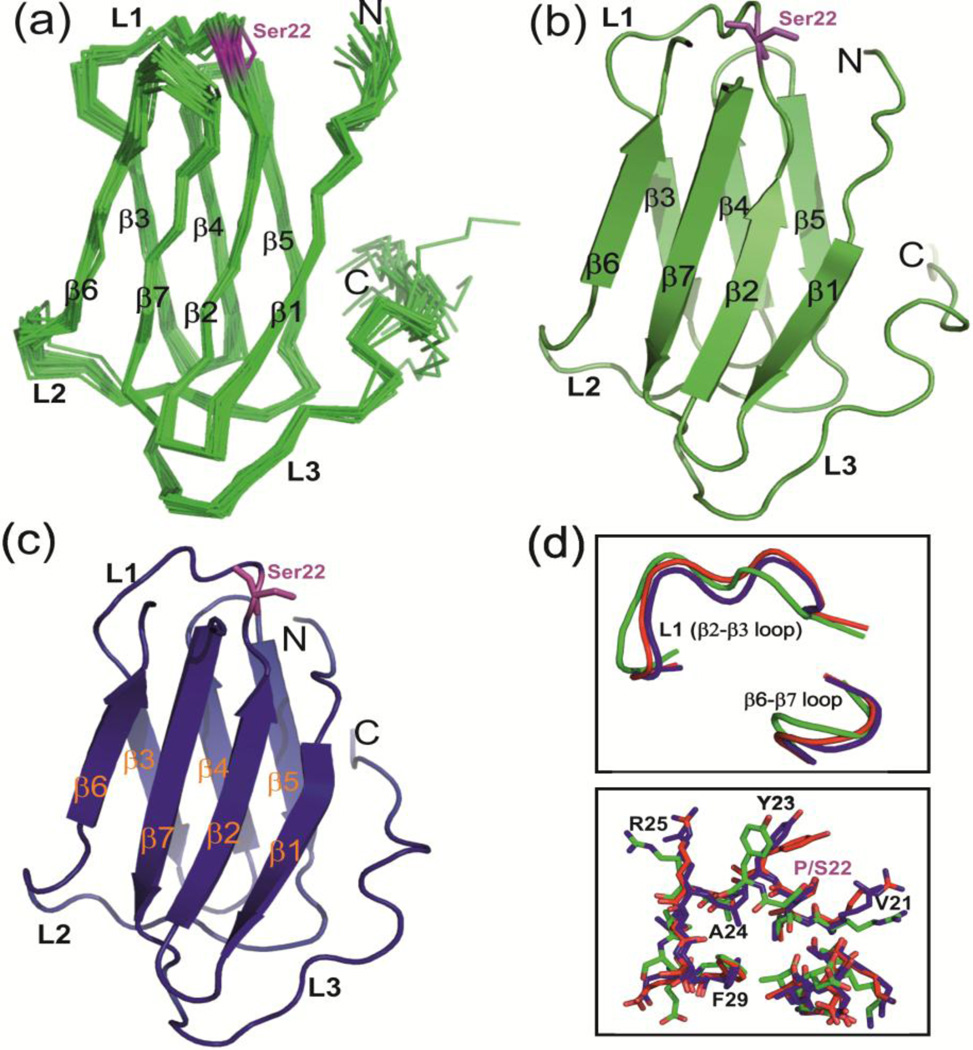
To gain further insights into the effect of P22S mutation on CS domain structure, we determined the solution NMR and X-ray crystal structures of hCS(P22S) and his-hCS(P22S), respectively. For the NMR structure, resonance assignment was achieved using standard sets of triple resonance experiments at 25 °C. The NMR structure was determined using NOEs, dihedral angle, hydrogen bonds, and RDC restraints [41,42]. The 20 lowest energy structures have an rmsd to the mean of 0.51 Å for residues 6–90 (Fig. 7a,b). The NMR structural statistics are in Table 3. The overall structure consisting of an immunoglobulin like β-sandwich fold is very similar to the wild type hCS structure as described below.
Fig. 7.
NMR and crystal structures of hCS(P22S). (a) Ensemble of 20 lowest energy solution NMR structures of hCS(P22S). (b) The lowest energy structure of hCS(P22S) in cartoon representation. (c) Crystal structure of hCS(P22S). All the secondary structure elements are marked. Ser22 is shown in magenta in stick representation. (d) Comparison of L1 loops in hCS (crystal, red), hCS(P22S) (NMR, green), and hCS(P22S) (crystal, blue) in backbone tube (top) and stick (bottom) representation. Residues in L1 loop are marked.
TABLE 3.
NMR structure statistics for hCS(P22S)
| Distance and dihedral restraints | ||
| Total NOE restraints | 2852 | |
| Intraresidue | 674 | |
| Sequential | 843 | |
| Medium (i+2 to i+4) | 329 | |
| Long range (> i+4) | 1006 | |
| Hydrogen bond restraints | 60 | |
| RDC restraints | 87 | |
| Dihedral angle restraints | 156 | |
|
Structure statistics (20 lowest energy structures) |
||
| No. of NOE violations>0.2Å | 0 | |
| No. of NOE violations>0.5Å | 0 | |
| No. of Dihedral violations>5° | 0 | |
| No. of RDC violations>2(Hz) | 0 | |
| RMSD of RDC, Hz | 0.17±0.02 | |
| RMSD from ideal covalent geometry | ||
| Bond lengths (Å) | 0.0013±0.00005 | |
| Bond angles (°) | 0.378±0.006 | |
| Impropers (°) | 0.267±0.012 | |
| RMSD from the mean structure (Å) | Backbone | Heavy atoms |
| Residues 6–90 | 0.51±0.10 | 1.04±0.09 |
| Ramachandran statistics | ||
| Most favored regions | 81.4% | |
| Additional allowed regions | 15.3% | |
| Generously allowed regions | 2.6% | |
| Disallowed regions | 0.7% | |
| PDB/BMRB accession numbers | 2MNW/19910 |
His-hCS(P22S) crystallized at 4 °C in space group P3221 and diffracted to 2.4 Å. The crystal structure was solved by molecular replacement using the his-hCS structure as a search model. There are two nearly identical copies of his-hCS(P22S) in the asymmetric unit, with minor differences between the two proteins located in loop L1. The N-terminal 6XHis-tag residues form a β3’ strand with neighboring molecules in the crystal similar to what was observed for his-hCS (Fig. S10). Overall the crystal structure is similar to the NMR structure with some differences in the conformations of several side chain residues (with global backbone RMSD of 1.2 Å) (Fig. 7b,c). We note that the NMR data were collected at 25°C whereas crystals for this construct only appeared at 4°C, therefore some of the observed differences may reflect this temperature difference.
The NMR and crystal structures of hCS(P22S) are overall similar to hCS with only small differences near the P22S mutation site in loop L1 (Fig. S11). The conformation of L1 in his-hCS(P22S) (crystal structure) is similar to his-hCS (crystal structure) (Fig. 7d, upper inset). There is a difference in the conformations of the side chains of residues V21, Y23, R25, and F29 in loop L1 near P22 (Fig. 7d, lower inset). The differences in side chain conformations of these residues are more significant in the NMR structure of hCS(P22S) where the difference in L1 position is more pronounced (Fig. 7d). The change in side chain position between hCS and hCS(P22S) may have consequences for dyskerin binding, as our NMR CSP results as well as HADDOCK model show that loop L1 residues are perturbed by Dys2S/L and Cbf5-2S/L peptides.
Thermal stability and dyskerin peptide interactions of hShq1(P22S)
We investigated the relative stabilities of hCS and hCS(P22S) using CD spectroscopy. We monitored the CD signal as a function of increasing temperature at 215 nm wavelength, which is the characteristic minimum for the β-sheet containing proteins. The calculated melting transition temperature (Tm) using CD spectroscopy for hCS and hCS(P22S) are 70°C and 60°C, respectively (Fig. 6c). In addition, the CD melting curve shows that hCS(P22S) has an early melting transition in the 20–40°C range, suggesting that at least some part of the protein is undergoing a structural transition even at room temperature (Fig. 6c). In NMR studies, the majority (> 70% as judged by SDS-PAGE) of hCS(P22S) precipitates out of solution at 25°C (especially at concentration >0.1 mM in < 2h), while hCS is stable for weeks at 25°C. These results indicate that the P22S mutation makes the CS domain thermally less stable.
In our previous study we showed that yeast Shq1 CS domain had a Tm of ~60°C, which is close to the Tm of hCS(P22S) [22], but the CD spectrum does not show an early melting transition. The higher stability of hCS vs yCS may be due to the difference in optimal growth temperatures in the two organisms. Two mutations, yCS(F6A) and yCS(Q10A) that were found to be thermally destabilized (Tm decreased by 15°C and 20°C respectively) had a temperature sensitive phenotype, consistent with the hypothesis that thermal destabilization of hCS(P22S) could affect its function in vivo.
We also investigated the binding of the Dys1, Dys2, Dys3, and Dys4 peptides to hCS(P22S). As with hCS, no binding was observed for Dys1, Dys3, and Dys4 (data not shown). Residue P22 is in a part of L1 that does not interact directly with dyskerin based on the HADDOCK model of yCS with Cbf5. The Dys2S and Dys2L peptides interact with hCS(P22S) in the same region as observed with hCS and with similar affinity (Fig. S12a,b). It is therefore possible that in addition to destabilizing the protein the P22S mutation affects binding of the CS domain to pontin/reptin or other unknown factors rather than binding to dyskerin.
Conclusions
Shq1 is essential for H/ACA RNP biogenesis and interacts with dyskerin at an early step in H/ACA RNP assembly. Previous studies reported the structures of the yeast Shq1 N-terminal CS domain and the C-terminal SSD domain in complex with Cbf5. The structures and functional analysis of human Shq1 CS domain and its point mutant P22S, presented here, are the first WT and mutant structures of a protein domain of a vertebrate H/ACA RNP protein. The human Shq1 CS structure consists of an immunoglobulin like β-sandwich fold similar to yeast Shq1 CS domain. However, in the crystal structures of his-hCS and his-hCS(P22S), the 6XHis tag forms an extra β3’ strand with a neighboring protein, while the 6XHis tag in yCS is disordered [22]. This interaction also seems to be at least transiently present in solution, and cleavage of the tag was required to get suitable spectra for structure determination in solution by NMR.
Using NMR spectroscopy we found a peptide from dyskerin/Cbf5 that showed weak but specific interaction with the CS domain of human and yeast Shq1. Most of the X-DC mutations in dyskerin are clustered on the Shq1 binding surface (both CS and SSD binding sites), thereby implicating a defect in Shq1-dyskerin interactions in the etiology of X-DC in these cases [20,26]. The NMR CSP data-driven HADDOCK model of full length Shq1 (both CS and SSD) bound to Cbf5 in an H/ACA RNP complex indicated an interaction surface that encompasses known DC mutations sites separate from the SSD binding surface and is consistent with previous studies [20,26].
In S. cerevisiae, mutations that destabilize the N-terminal CS domain (F6A, Q10A, and K80A) in vitro result in temperature sensitive mutants and show growth defects, H/ACA snoRNA depletion, and defects in rRNA processing, even in conditions where Shq1p protein levels are normal (Q10A and K80A) [22]. In a human genome profiling study, deletion of the SHQ1 locus corresponding to the CS domain and a point mutation (P22S) in the CS domain were found in prostrate cancers, indicating that the CS domain is equally important for Shq1 function in humans. Comparison of the NMR spectra and structures of hCS and hCS(P22S) revealed that the point mutation, which is located in the β2–β3 loop (loop 1), has little effect on the overall structure although there are changes in positions of some sidechains. However, CD melting studies show that hCS(P22S) is thermally less stable than hCS, with the major melting transition about 10°C lower than hCS and an early pre-melting transition at around 25°C not present in hCS. Based on the hCS-Cbf5 HADDOCK model, P22 does not appear to be part of the interaction surface of hCS with dyskerin but does affect the position of some residues that are in the interaction surface. However, hCS and hCS(P22S) have similar affinities for Dys peptides as judged by NMR CSP results. Therefore, the P22S mutation may affect Shq1 function either by lowering its overall stability as previously observed in S. cerevisiae where it may affect assembly and/or by affecting the interaction with pontin/reptin, thereby affecting removal of Shq1 for H/ACA RNA binding. Our structural study combined with recent results in the literature [20,26] provides insights into the molecular basis of CS domain interaction with dyskerin/Cbf5 and its importance in H/ACA RNP assembly.
Material and Methods
Protein expression and purification
Human Shq1 CS domain (residues 1–96) was sub-cloned into two E. coli expression vectors, pET46 Ek/LIC and pET41 Ek/LIC. pET46 Ek/LIC vector imparts a short N-terminal 6XHis-tag and enterokinase cleavage site preceding the protein coding sequence (sequence, MAHHHHHHVDDDDK). pET41 Ek/LIC vector incorporates an N-terminal GST tag (~26 KDa) followed by a 6XHis-tag, thrombin cleavage site, S-tag, and an enterokinase cleavage site. Construct with point mutant, P22S, of hCS domain hCS(P22S) was made by site-directed mutagenesis (Stratagene) in both vectors, pET46 Ek/LIC and pET41 Ek/LIC. For selenomethionine labeling, hCS was expressed in E. coli B834 (DE3) cells (Novagen). For all other purposes, plasmids were transformed into E. coli BL21 (DE3) cells for protein expression. Cultures were grown at 37°C until an OD600 of 0.6 was reached, then the temperature was lowered to 28°C and the cells were induced with 1 mM IPTG for over-night (~16h).
hCS and hCS(P22S) from pET46 Ek/LIC vector were expressed and purified for the crystallization. Cells were harvested by centrifugation and the pellets were resuspended in the binding buffer (buffer A: 50 mM Tris, pH 8, 500 mM NaCl, 5% Glycerol v/v, 10 mM β-mercaptoethanol). Cells were lysed by sonication on ice and centrifuged to remove cell debris. The supernatant was applied to a Ni-NTA agarose column (5 ml), which had been pre-equilibrated with buffer A. The column was washed with buffer A, followed by buffer B (buffer A supplemented with 20 mM imidazole) and were eluted with buffer C (buffer A supplemented with 400 mM imidazole). Proteins were further purified by Superdex S75 preparative gel-filtration column.
hCS and hSC-P22S from pET41 Ek/LIC vector were expressed and purified for the NMR titration, CD spectroscopy and other experiments. Cell were suspended in phosphate buffer saline (PBS), sonicated and loaded onto a sepharose GSTrap FF column (5 ml) (GE Lifesciences). Column was washed with 10 column volumes of PBS. Thrombin protease (GE Healthcare) dissolved in 4.5 ml of PBS (250U) were loaded onto the column and column was incubated for ~16 h at 22°C for the cleavage of the GST-tag. GST-tag free hCS and hCS(P22S) GST-tag were eluted using PBS. Free GST-tag and uncut GST-fusion proteins were eluted using PBS supplemented with 10 mM reduced glutathione. Removal of GST-tag by thrombin leaves hCS and hCS(P22S) with following N-terminal vector specific residues preceding protein coding sequence: GSTAIGMKETAAAKFERQHMDSPDLGTGGGSGDDDDK.
Yeast Shq1 CS domain (yCS, 1–101) was purified as described previously [22]. All proteins were >95% purified as judged by SDS-PAGE. The buffer for NMR and CD spectroscopy was 20 mM NaH2PO4, pH 7, 100 mM NaCl, and 2 mM DTT, 0.02% NaN3 and for crystallization was 20 mM Tris base, pH 7.2, 100 mM NaCl, 2 mM DTT, and 0.02% NaN3.
Crystallization of hCS and hCS(P22S)
Crystallization of hCS and hCS(P22S) were carried out using a hanging drop, vapor-diffusion method by mixing equal volumes of protein and reservoir solutions. hCS was crystallized in following buffer: 0.2 M Ammonium tartrate dibasic, pH 6.6; 20% PEG 3350 at 18 °C. Crystals appeared under the same conditions for both unlabeled and selenomethionine derivatives within 24 h. Crystals grew to maximum size of 0.2×0.2×0.3 mm in two days and belonged to space group P41212. Unit cell dimensions and other parameters are given in Table 1.
hCS(P22S) was crystallized in following buffer: 0.1 M HEPES pH 7.5; 0.02 M MgCl2.6H2O; 22% w/v Poly(acrylic acid sodium salt) 5,100; 15% Glycerol v/v at 4 °C. Crystals appeared in 24 h and grew to maximum size of 0.1×0.1×0.2 mm in one week and belonged to space group P3221. Unit cell dimensions and other parameters are given in Table 1
X-ray structure determination of hCS and hCS(P22S)
Data for both hCS and hCS(P22S) were collected at the Advanced Photon Source at beamline 24-ID-C. Data were processed with the XDS package [43]. The selenomethionine derivative of hCS was measured at peak wavelength of 0.97949 Å. Datasets were of high quality and showed a strong anomalous signal. Macromolecular phasing was carried out by HKL2MAP using single anamolous dispersion (SAD) method [44]. The resulting phases were improved by using the DM program and used for an automated model building with the Arp/Warp software [45,46]. The resulting model of about 90% completeness was inspected and finished manually with the program Coot [47]. Restrained refinement enforced by the PHENIX software was then performed and was followed by the addition of solvent molecules [48]. Final iterative rounds of model building and refinement were carried out using Coot [47] and PHENIX [48]. Final data collection, phasing, and refinement statistics are presented in Table 1.
hCS(P22S) was measured at wavelength of 0.97949 Å. Macromolecular phasing was carried out using molecular replacement using structure of hCS and software Phaser [46,49]. Resulting model after Phaser was subjected to restraints refinement using program PHENIX [48]. Final iterative round of model building and refinement were carried out using Coot and PHENIX [47,48]. Final data collection, phasing, and refinement statistics are presented in Table 1. All the figures based on the structures of hCS and hCS(P22S) were prepared using Pymol (www.pymol.org).
NMR spectroscopy
Uniformly 13C,15N-labeled samples of hCS and hCS(P22S) were used for NMR assignments and 15N labeled samples were used for other NMR experiments. NMR spectra were recorded on Bruker DRX 500 MHz, 600 MHz, and 800 MHz spectrometers equipped with cryoprobes. NMR samples contained 0.1 to 0.8 mM protein. The backbone assignments for hCS, hCS(P22S), and yCS were obtained by standard methods using the CBCACONH/HNCACB and HNCACO/HNCO pairs of triple resonance experiments at 22°C [37].
NMR Chemical shift mapping of CS and dyskerin/Cbf5 peptides
The peptides used in this study were commercially synthesized and purified (GenScript). All the peptides dissolved easily dissolved in either NMR buffer or water. For NMR titrations, unlabeled peptides solution were added to 15N-labeled protein samples (hCS, hCS(P22S), and yCS) and 1H-15N HSQC or SFHMQC [50] spectra were recorded at each step of titration. The titrations were carried out at 22°C on 500 MHz, 600 MHz, or 800 MHz NMR spectrometers. Datasets were processed using the Bruker program XWIN-NMR version 3.5 and assigned using the SPARKY (T.D. Goddard & D.G. Kneller, SPARKY 3, University of California, San Francisco). The magnitude of the induced chemical shift perturbations (CSP) at saturating 1:10 protein to peptide ratio was calculated using a standard formula, . CSPs above the average CSP were considered significant and arbitrarily classified as strong or weak. The strong and weak CSPs were mapped on the structure of proteins.
NMR structure determination of hCS(P22S)
For hCS(P22S), side-chain and NOE assignments were performed using standard NMR 3D experiments: HBHACONH, CCCONH, HCCH-TOCSY, 15N–edited NOESY, and 13C–edited NOESY [37]. One-bond 1H-15N RDCs were measured from HSQC-IPAP experiments in the presence and absence of C12E5/hexanol on a 600 MHz spectrometer [51]. Slowly exchanging amides involved in hydrogen bonds were identified by re-dissolving the lyophilized protein in 100% 2H2O and collecting 1H-15N HSQC spectra at various time intervals to monitor the rate of disappearance of the amide resonances. Dihedral angle restraints were generated using TALOS+ [52]. The structures were calculated using the NIH-XPLOR package following standard protocols including NOE, dihedral angle restraints, and hydrogen bond restraints [41,42]. The 200 lowest energy structures were refined with incorporation of RDCs restraints using XPLOR-NIH [41,42] Structures were validated by PROCHECK-NMR [53] and visualized by and PyMol (www.pymol.org). Structural statistics are presented in Table 3.
CD spectroscopy
CD spectra of the proteins were obtained on a J-715 spectropolarimeter (Jasco J715 model). Spectra were recorded using 1 mm path length quartz cuvettes. For thermal denaturation experiments spectra were monitored at 215 nm and the sample temperature was increased at a rate of 30°C/h.
HADDOCK docking
HADDOCK docking was performed using the ‘easy’ web interface on the WeNMR website [39,40]. PBD 3UAI for the Cbf5 structure and PDB 3EUD for yCS were used as input [22,25]. All chains in 3UAI (Cbf5, Nop10, Shq1 SSD, and Gar1) were used. Docking CS domain to Cbf5 in SSD bound form would constraint orientation of CS domain relative to SSD. Residues that showed >average chemical shifts were first selected. For HADDOCK modeling only residues that are more than 30% solvent exposed [46] were choose as ‘active residues’. For yCS the active residues were: S7, S22, N23, R25, F26, S27, A28, L31, E32, I33, I34, Q36, M39, H43, R50, I58, D60, R62, Q66, K77, and N84 (CSP >0.024 (ppm) and > 30% solvent accessibility). For hCS the active residues were: L2, R25, V26, E28, D30, V31, Y32, F33, K39, V56, E57, N58, G59, S60, E61, G63, S64, Y65, D66, A67, T73, R75, and H84 (CSP >0.05 (ppm) and > 30% solvent accessibility). Passive residues were selected automatically as the exposed surface neighbors of the active residues. On Cbf5, the area corresponding to Cbf5-2/Dys2 peptide was chosen for docking (residue L32-P43 in Cbf5 and L62-A73 in dyskerin). The residues that are found mutated in X-DC were classified as active residues. These ‘active residues’ are: R35, S36, G37, H38, and I42 in Cbf5 and R65, T66, T67, H68, and L71 in dyskerin. Program was allowed to choose the neighboring exposed residues as ‘passive residues’. Clusters with the lowest HADDOCK scores were analyzed and presented in the paper.
Supplementary Material
Highlights.
Shq1 binds to dyskerin/Cbf5 at the initial step of H/ACA RNP assembly
Crystal structure of human Shq1 CS domain is determined
Crystal and NMR structure of prostate cancer mutant CS-P22S are reported
NMR CSP experiments revealed a conserved surface on CS for Cbf5/dyskerin binding
HADDOCK model of Shq1-Cbf5 complex defines position of CS on pre-H/ACA RNP
Acknowledgements
This research was supported by NIH grant GM048123 and NSF MCB1022379 to J.F. The NMR core facility and X-ray core facility are supported in part by DOE grant DE-FC0302ER63421. We thank M. Capel, K. Rajashankar, N. Sukumar, F. Murphy and I. Kourinov of Northeastern Collaborative Access Team (NE-CAT) beamline ID-24 at the Advanced Photon Source (APS) of Argonne National Laboratory, which are supported by NIH grants P41 RR015301 and P41 GM103403. Use of the APS is supported by DOE under Contract DE-AC02-06CH11357. We also would like to thank R. Peterson for help with NMR data collection, M. Collazzo for help with crystallization screening, and M. Sawaya for helpful discussions.
Abbreviations
- RNP
ribonucleoprotein particle
- NMR
nuclear magnetic resonance
- CD
circular dichroism
- HADDOCK
High Ambiguity Driven protein-protein DOCKing
- DC
dyskeratosis congenita
- X-DC
X-linked dyskeratosis congenita
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest.
Accession numbers PDB coordinates and the structure factors for crystal structures of his-hCS and his-hCS(P22S) have been deposited to Protein Data Bank under accession numbers 4PBD and 4PCK respectively. NMR Coordinates and restraints for the 20 lowest energy structures of hCS(P22S) have been deposited in the Protein Data Bank, accession number 2MNW and chemical shifts in the BioMagResBank, accession number 19910. Backbone chemical shifts for hCS has been deposited to BioMagResBank under accession number 25167.
References
- 1.Kiss T, Fayet-Lebaron E, Jady BE. Box H/ACA small ribonucleoproteins. Mol Cell. 2010;37:597–606. doi: 10.1016/j.molcel.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 2.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 3.Meier UT. The many facets of H/ACA ribonucleoproteins. Chromosoma. 2005;114:1–14. doi: 10.1007/s00412-005-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye K. H/ACA guide RNAs, proteins and complexes. Curr Opin Struct Biol. 2007;17:287–292. doi: 10.1016/j.sbi.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 5.Ge J, Yu YT. RNA pseudouridylation: new insights into an old modification. Trends Biochem Sci. 2013;38:210–218. doi: 10.1016/j.tibs.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell JR, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3’ end. Mol Cell Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell JR, Collins K. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol Cell. 2000;6:361–371. doi: 10.1016/s1097-2765(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 8.Egan ED, Collins K. Biogenesis of telomerase ribonucleoproteins. RNA. 2012;18:1747–1759. doi: 10.1261/rna.034629.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S, Duan J, Li D, Yang B, Dong M, et al. Reconstitution and structural analysis of the yeast box H/ACA RNA-guided pseudouridine synthase. Genes Dev. 2011;25:2409–2421. doi: 10.1101/gad.175299.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grozdanov PN, Roy S, Kittur N, Meier UT. SHQ1 is required prior to NAF1 for assembly of H/ACA small nucleolar and telomerase RNPs. RNA. 2009;15:1188–1197. doi: 10.1261/rna.1532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darzacq X, Kittur N, Roy S, Shav-Tal Y, Singer RH, et al. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J Cell Biol. 2006;173:207–218. doi: 10.1083/jcb.200601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang PK, Rotondo G, Porras T, Legrain P, Chanfreau G. The Shq1p.Naf1p complex is required for box H/ACA small nucleolar ribonucleoprotein particle biogenesis. J Biol Chem. 2002;277:45235–45242. doi: 10.1074/jbc.M207669200. [DOI] [PubMed] [Google Scholar]
- 13.Li L, Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006;443:302–307. doi: 10.1038/nature05151. [DOI] [PubMed] [Google Scholar]
- 14.Dokal I, Vulliamy T. Dyskeratosis congenita: its link to telomerase and aplastic anaemia. Blood Rev. 2003;17:217–225. doi: 10.1016/s0268-960x(03)00020-1. [DOI] [PubMed] [Google Scholar]
- 15.Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, et al. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 16.Bessler M, Wilson DB, Mason PJ. Dyskeratosis congenita. FEBS Lett. 2010;584:3831–3838. doi: 10.1016/j.febslet.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walne AJ, Dokal I. Advances in the understanding of dyskeratosis congenita. Br J Haematol. 2009;145:164–172. doi: 10.1111/j.1365-2141.2009.07598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong F, Savage SA, Shkreli M, Giri N, Jessop L, et al. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Dev. 2011;25:11–16. doi: 10.1101/gad.2006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leulliot N, Godin KS, Hoareau-Aveilla C, Quevillon-Cheruel S, Varani G, et al. The box H/ACA RNP assembly factor Naf1p contains a domain homologous to Gar1p mediating its interaction with Cbf5p. J Mol Biol. 2007;371:1338–1353. doi: 10.1016/j.jmb.2007.06.031. [DOI] [PubMed] [Google Scholar]
- 20.Machado-Pinilla R, Liger D, Leulliot N, Meier UT. Mechanism of the AAA+ ATPases pontin and reptin in the biogenesis of H/ACA RNPs. RNA. 2012;18:1833–1845. doi: 10.1261/rna.034942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh M, Gonzales FA, Cascio D, Heckmann N, Chanfreau G, et al. Structure and functional studies of the CS domain of the essential H/ACA ribonucleoparticle assembly protein SHQ1. J Biol Chem. 2009;284:1906–1916. doi: 10.1074/jbc.M807337200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Godin KS, Walbott H, Leulliot N, van Tilbeurgh H, Varani G. The box H/ACA snoRNP assembly factor Shq1p is a chaperone protein homologous to Hsp90 cochaperones that binds to the Cbf5p enzyme. J Mol Biol. 2009;390:231–244. doi: 10.1016/j.jmb.2009.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walbott H, Machado-Pinilla R, Liger D, Blaud M, Rety S, et al. The H/ACA RNP assembly factor SHQ1 functions as an RNA mimic. Genes Dev. 2011;25:2398–2408. doi: 10.1101/gad.176834.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S, Duan J, Li D, Ma S, Ye K. Structure of the Shq1-Cbf5-Nop10-Gar1 complex and implications for H/ACA RNP biogenesis and dyskeratosis congenita. EMBO J. 2011;30:5010–5020. doi: 10.1038/emboj.2011.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grozdanov PN, Fernandez-Fuentes N, Fiser A, Meier UT. Pathogenic NAP57 mutations decrease ribonucleoprotein assembly in dyskeratosis congenita. Hum Mol Genet. 2009;18:4546–4551. doi: 10.1093/hmg/ddp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamma T, Ferre-D’Amare AR. The box H/ACA ribonucleoprotein complex: interplay of RNA and protein structures in post-transcriptional RNA modification. J Biol Chem. 2010;285:805–809. doi: 10.1074/jbc.R109.076893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meier UT. How a single protein complex accommodates many different H/ACA RNAs. Trends Biochem Sci. 2006;31:311–315. doi: 10.1016/j.tibs.2006.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nallar SC, Kalvakolanu DV. Regulation of snoRNAs in cancer: close encounters with interferon. J Interferon Cytokine Res. 2013;33:189–198. doi: 10.1089/jir.2012.0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trotman L, Powers S. New views into the prostate cancer genome. Cancer Cell. 2010;18:1–2. doi: 10.1016/j.ccr.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 32.Krohn A, Seidel A, Burkhardt L, Bachmann F, Mader M, et al. Recurrent deletion of 3p13 targets multiple tumour suppressor genes and defines a distinct subgroup of aggressive ERG fusion-positive prostate cancers. J Pathol. 2013;231:130–141. doi: 10.1002/path.4223. [DOI] [PubMed] [Google Scholar]
- 33.Lando M, Wilting SM, Snipstad K, Clancy T, Bierkens M, et al. Identification of eight candidate target genes of the recurrent 3p12-p14 loss in cervical cancer by integrative genomic profiling. J Pathol. 2013;230:59–69. doi: 10.1002/path.4168. [DOI] [PubMed] [Google Scholar]
- 34.Schippert R, Schaeffel F, Feldkaemper MP. Microarray analysis of retinal gene expression in chicks during imposed myopic defocus. Mol Vis. 2008;14:1589–1599. [PMC free article] [PubMed] [Google Scholar]
- 35.Chadwick N, Zeef L, Portillo V, Fennessy C, Warrander F, et al. Identification of novel Notch target genes in T cell leukaemia. Mol Cancer. 2009;8:35. doi: 10.1186/1476-4598-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waksman G, Hultgren SJ. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat Rev Microbiol. 2009;7:765–774. doi: 10.1038/nrmicro2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cavanagh J, Fairbrother WJ, Skelton NJ, Rance M, Palmer IAG. Protein NMR Spectroscopy: Principles and Practice. Elsevier Science. 2006 [Google Scholar]
- 38.Schanda P, Kupce E, Brutscher B. SOFAST-HMQC experiments for recording two- dimensional heteronuclear correlation spectra of proteins within a few seconds. J Biomol NMR. 2005;33:199–211. doi: 10.1007/s10858-005-4425-x. [DOI] [PubMed] [Google Scholar]
- 39.de Vries SJ, van Dijk M, Bonvin AM. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 40.Wassenaar T, Dijk M, Loureiro-Ferreira N, Schot G, Vries S, et al. WeNMR: Structural Biology on the Grid. Journal of Grid Computing. 2012;10:743–767. [Google Scholar]
- 41.Schwieters CD, Kuszewski JJ, Clore GM. Using Xplor-NIH for NMR Molecular Structure Determination. ChemInform. 2006;37 doi: 10.1016/s1090-7807(02)00014-9. no-no. [DOI] [PubMed] [Google Scholar]
- 42.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 43.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pape T, Schneider TR. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. Journal of Applied Crystallography. 2004;37:843–844. [Google Scholar]
- 45.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 46.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schanda P, Kupƒçe ƒ, Brutscher B. SOFAST-HMQC Experiments for Recording Two-dimensional Deteronuclear Correlation Spectra of Proteins within a Few Seconds. Journal of Biomolecular NMR. 2005;33:199–211. doi: 10.1007/s10858-005-4425-x. [DOI] [PubMed] [Google Scholar]
- 51.Ottiger M, Delaglio F, Marquardt JL, Tjandra N, Bax A. Measurement of dipolar couplings for methylene and methyl sites in weakly oriented macromolecules and their use in structure determination. J Magn Reson. 1998;134:365–369. doi: 10.1006/jmre.1998.1546. [DOI] [PubMed] [Google Scholar]
- 52.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.