Abstract
Contemporary Jews retain a genetic imprint from their Near Eastern ancestry, but obtained substantial genetic components from their neighboring populations during their history. Whether they received any genetic contribution from the Far East remains unknown, but frequent communication with the Chinese has been observed since the Silk Road period. To address this issue, mitochondrial DNA (mtDNA) variation from 55,595 Eurasians are analyzed. The existence of some eastern Eurasian haplotypes in eastern Ashkenazi Jews supports an East Asian genetic contribution, likely from Chinese. Further evidence indicates that this connection can be attributed to a gene flow event that occurred less than 1.4 kilo-years ago (kya), which falls within the time frame of the Silk Road scenario and fits well with historical records and archaeological discoveries. This observed genetic contribution from Chinese to Ashkenazi Jews demonstrates that the historical exchange between Ashkenazim and the Far East was not confined to the cultural sphere but also extended to an exchange of genes.
Consistent with their displaced ethno-history since the ancient Northern Kingdom of Israel was invaded and occupied by the Neo-Assyrian Empire1, contemporary Jews, including Ashkenazi Jews, Sephardic Jews, North African Jews and Middle Eastern Jews2, retain a genetic imprint of their Near Eastern ancestry, but have received a substantial contribution, to a variable extent, from their neighboring populations such as Europeans, Near Easterners, and North Africans2,3,4,5,6,7,8. However, it has hitherto remained unclear whether Jews received any genetic contribution from populations outside western Eurasia. Intriguingly, frequent communication has been observed between Jews and Chinese since the early centuries of the Common Era, plausibly initiated by the Silk Road. For instance, Hebrew letters and prayers in the 8th century from ancient Jewish merchants were found in the northwestern region of China9. Some unearthed pottery figurines from the Tang Dynasty (618–907AD) have Semitic characteristics9, and synagogues were recorded in the epigraphy from the Ming (1368–1644AD) and Qing Dynasties (1644–1912AD)9,10. Nonetheless, such connections, as revealed by the archaeological discoveries and historical records, have been confined to economic and cultural exchanges; so far, no direct evidence of a genetic contribution from Chinese into Jews has been reported.
To address the issue of whether Jews received any genetic contribution from the Far East, and thus shed more light on their ethno-origins, mitochondrial DNA (mtDNA) variation (mainly from the control region of the molecule, plus some coding-region variants) of 1,930 Jews and 21,191 East Asians, retrieved from previous studies as well as our unpublished data, were considered and analyzed, with especial attention to pinpointing eastern Eurasian haplogroups in Jews (Supplementary Table S1). Then, mtDNA control region variants of an additional 32,474 Eurasian individuals were analyzed to gain further insights into the phylogeographic distribution of M33c (Supplementary Table S1), so that the total number of Eurasian mtDNAs considered here was 55,595. Our results do reveal a direct genetic connection, as manifested by the sharing of some Eastern Eurasian haplogroups e.g. N9a, A, and M33c, between Jews and Chinese. Further analyses, including phylogeny reconstruction with the aid of new mtDNA genomes, confirm that this connection was established at least by a founder lineage M33c2. The differentiation time of this lineage is estimated to ~1.4 kilo-years ago (kya), which fits well with the historical records and, most importantly, indicates that the exchange between Jews and the Far East was not confined to culture but also extended to the demic.
Results and Discussion
Our analysis of the mtDNA variation in a total of 23,121 individuals from East Asian populations and Jews reveals that mtDNAs of four Ashkenazi Jewish individuals can be allocated into eastern Eurasian haplogroups A and N9a, suggesting that Ashkenazi Jews received a genetic contribution from East Asia (Table 1). Intriguingly, our results also disclose that 14 eastern Ashkenazi Jews belong to haplogroup M33c (Table 1), for which sister clusters, M33a, M33b and M33d, are prevalent in the Indian Subcontinent and thus most plausibly trace their origins there11,12.
Table 1. The shared eastern Eurasian haplotypes between Ashkenazi Jews and Chinese.
| Sample ID | Haplogroup | HVS-I (16000+) | HVS-II | Population | Region/Country | Reference |
|---|---|---|---|---|---|---|
| AS2C5 | A | 182C 183C 189 193.1C 223 290 319 362 | 73 150 152 204 235 263 | Ashkenazi | Hungary | 32 |
| 472 | N9a | 129 223 257A 261 | 73 146 150 263 | Ashkenazi | Russia | 7 |
| 75916 | N9a | 129 223 257A 261 | 73 146 150 263 309.1C 309.2C 315.1C | Ashkenazi | Belarus | Family Tree DNA |
| 110612 | N9a | 129 223 257A 261 | 73 146 150 263 309.1C 309.2C 315.1C | Ashkenazi | Belarus | Family Tree DNA |
| Bel 1a | M33c | 111 223 235 362 (519) | 73 263 315.1C (489) | Ashkenazi | Belarus | This study |
| EU148486a | M33c | 111 223 235 362 (519) | 73 263 315.1C (489) | Ashkenazi | Belarus | Family Tree DNA |
| 4130b | M33c | 111 223 235 362 (519) | 73 263 315.1C (489) | Ashkenazi | Belarus | Family Tree DNA |
| 135479b | M33c | 111 223 235 362 (519) | 73 263 315.1C (489) | Ashkenazi | Western Ukraine | Family Tree DNA |
| 266609b | M33c | 111 223 235 362 (519) | 73 263 315.1C (489) | Ashkenazi | Western Ukraine | Family Tree DNA |
| N50366b | M33c | 111 223 235 362 (519) | 73 263 315.1C (489) | Ashkenazi | Western Ukraine | Family Tree DNA |
| N105293b | M33c | 111 223 235 362 (519) | 73 263 315.1C (489) | Ashkenazi | Russia | Family Tree DNA |
| 50126b | M33c | 111 223 235 362 (519) | 73 263 315.1C (489) | Ashkenazi | Russia | Family Tree DNA |
| 289247 | M33c | 111 223 235 362 (519) | Ashkenazi | Western Ukraine | Family Tree DNA | |
| 403 | M33c | 111 223 235 362 (519) | 73 263 | Ashkenazi | Belarus | 7 |
| 404 | M33c | 111 223 235 362 (519) | 73 263 | Ashkenazi | Poland | 7 |
| 405 | M33c | 111 223 235 362 (519) | 73 263 | Ashkenazi | Poland | 7 |
| 406 | M33c | 111 223 235 362 (519) | 73 263 | Ashkenazi | Romania | 7 |
| Forum 1a,c | M33c | 111 223 235 362 (519) | 73 263 315.1C (489) | Jew | Belarus/Russia/Lithuania | A Genetic Genealogy Community |
| JQ702003a | M33c | 111 223 235 362 (519) | 73 263 315.1C (489) | N.A. | N.A. | 14 |
| HN-SZ420a | M33c | 111 192 223 362 (519) | 73 263 315.1C (489) | Han | Hunan, China | 33, This study |
| HN-SZ167a | M33c | 111 223 235 356 362 (519) | 73 263 309.1C 315.1C (489) | Han | Hunan, China | 33, This study |
| HN-SZ104a | M33c | 111 223 235 362 (519) | 73 263 308-310d 315.1C (489) | Han | Hunan, China | 33, This study |
| HN-SZ527a | M33c | 111 223 235 362 (519) | 73 263 309.1C 315.1C (489) | Han | Hunan, China | 33, This study |
| JSH08058 | M33c | 111 129 223 235 284 300 362 (519) | 73 150 263 315.1C | Han | Jiangsu, China | This study |
| K9534a | M33c | 111 129 140 223 235 300 362 (519) | 73 150 263 309.1C 315.1C (489) | Han | Jilin, China | This study |
| K9511a | M33c | 111 192 223 235 362 (519) | 73 263 315.1C 471 (489) | Han | Jilin, China | This study |
| H11694a | M33c | 104 111 223 362 (519) | 73 263 | Han | Shaanxi, China | This study |
| H11281a | M33c | 111 223 235 362 (519) | 73 263 309.1C 315.1C | Han | Shaanxi, China | This study |
| DJY576a | M33c | 111 223 235 243 362 (519) | 73 263 309.1C 315.1C | Han | Sichuan, China | This study |
| Zhuang21a | M33c | 093 104 111 223 235 362 (519) | 73 263 309.1C 315.1C (489) | Zhuang | Guangxi, China | This study |
| Zhuang74 | M33c | 093 104 111 223 235 362 (519) | 73 263 309.1C 315.1C (489) | Zhuang | Guangxi, China | This study |
| Zhuang9 | M33c | 093 104 111 223 235 362 (519) | 73 263 315.1C (489) | Zhuang | Guangxi, China | This study |
| HN-SZ416 | M33c | 093 104 111 223 362 (519) | 73 263 309.1C 315.1C (489) | Han | Hunan, China | 33 |
| Dongguan-65 | M33c | 093 104 111 223 235 311 362 | 73 263 210 309.1C 310 315.1C | Han | Guangdong, China | 34 |
| 539 | M33c | 093 104 111 223 235 362 | Han | Guangxi, China | 35 | |
| 129 | M33c | 093 104 111 223 235 362 | Han | Guangxi, China | 35 | |
| 136 | M33c | 093 104 111 223 235 362 | Han | Guangxi, China | 35 | |
| 499 | M33c | 093 104 111 223 235 362 | Han | Guangxi, China | 35 | |
| 370 | M33c | 104 111 223 235 362 | Han | Guangxi, China | 35 | |
| 149 | M33c | 111 223 235 362 | Han | Guangxi, China | 35 | |
| Viet0121 | M33c | 111 223 362 (519) | 73 203 263 309.1C 309.2C 315.1C (489) | Vietnamese | Vietnam | 36 |
| GD7815a | M33c | 093 104 111 223 362 | 73 146 263 309.1C 315.1C | Han | Guangdong, China | 13 |
| Yao171a | M33c | 111 129 223 235 362 (519) | 73 263 309.1C 309.2C 315.1C | Yao | Hunan, China | 13 |
| MK13 | M33c | 111 129 223 235 300 362 | Kam-Tai | Guizhou, China | 37 | |
| Kinh117 | M33c | 111 223 362 (519) | Kinh | Vietnam | 38 | |
| (03B)045 | M33c | 093 104 111 223 235 362 (519) | 73 263 309.1C 315.1C | Han | Guangdong, China | 39 |
| YZ-Tib05-23 | M33c | 092 104 111 223 362 | Tibetan | Yunnan, China | 40 | |
| YZ-Tib05-8 | M33c | 092 104 111 223 362 | Tibetan | Yunnan, China | 40 | |
| MHN43 | M33c | 111 223 235 362 | Miao | Hunan, China | 41 | |
| YBP28 | M33c | 093 104 111 223 235 362 | Yao | Guangdong, China | 41 | |
| YLO22 | M33c | 093 104 111 223 235 305C 362 | Yao | Guangxi, China | 41 | |
| YLO03 | M33c | 111 223 235 362 | Yao | Guangxi, China | 41 | |
| YLO23 | M33c | 111 223 235 362 | Yao | Guangxi, China | 41 | |
| GD7821 | M33c | 093 104 111 223 235 362 | 73 263 309.1C 315.1C | Han | Guangdong, China | 42 |
| HG00457a | M33c | 111 223 362 (519) | Han | Hunan/Fujian, China | 15 | |
| TL397 | M33c | 093 101 111 223 235 362 (519) | Thai | Thailand | 43 |
aThe whole-mtDNA genome information is displayed on the phylogenetic tree (Figure 2).
bThe individual has coding region information confirming its haplogroup assignment from the Family Tree DNA (www.familytreedna.com).
cThis sequence is from A Genetic Genealogy Community (http://eng.molgen.org/viewtopic.php?f=41&t=141).
N.A. = not available.
All the samples in this study have confirmed G2361A mutation when assigning their haplogroup. The mutations (e.g. 489 and 16519) outside HVS-I and HVS-II are listed in the parentheses. The haplogroups A and N9a mtDNAs in the Far East are not shown here.
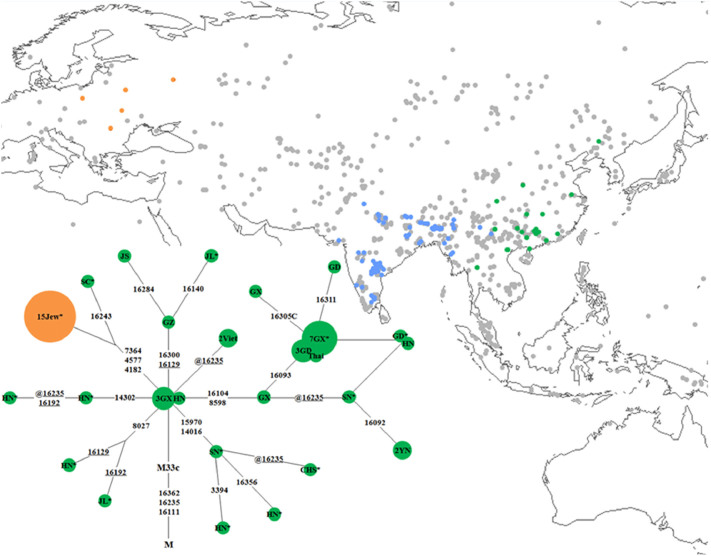
To achieve further insight into the phylogeographic distribution of M33c, mtDNA variants (mainly from the control region) of an additional 32,474 Eurasian individuals were analyzed, so that the total number of Eurasian mtDNAs considered here was 55,595. As shown in Table 1, besides the 14 Ashkenazi Jewish M33c lineages, an additional 38 M33c mtDNAs (with the specific control-region motif showing transitions at positions 16111, 16223, 16235, and 16362) were pinpointed, among which 34 are from China, 2 from Vietnam, and 1 from Thailand, with the remaining individual most likely from Europe but with ambiguous ancestry. Thus, despite the restricted distribution of M33a, M33b and M33d in South Asia, it is most likely that M33c originated, or at least differentiated, in eastern Asia. This notion receives clear support from the median network, in which virtually all of the diversity of this haplogroup is observed in China (Figure 1).
Figure 1. Median-joining network of haplogroup M33c.
The median-joining network is reconstructed on the basis of mtDNA hypervariable segment I (HVS-I) variation. The sampling locations are shown by different colors in the map. Transversions are highlighted by adding suffixes “A”, “C”, “G”, and “T”. The prefix @ designates back mutation, whereas recurrent variants are underlined. * denotes that this individual's whole-mtDNA genome information is shown on the phylogenetic tree. The size of the circle is in proportion to the number of individuals. The geographic locations are abbreviated as follows: CHS (Hunan or Fujian), GD (Guangdong), GX (Guangxi), GZ (Guizhou), HN (Hunan), JL (Jilin), JS (Jiangsu), SC (Sichuan), SN (Shaanxi), Thai (Thailand), Viet (Vietnam), and YN (Yunnan). Note:  M33c individuals in Europe.
M33c individuals in Europe.  M33c individuals in Asia.
M33c individuals in Asia.  M33a, M33b or M33d individuals.
M33a, M33b or M33d individuals.  Sampling locations of all the other samples considered in this study. The map was created by the Kriging algorithm of the Surfer 8.0 package. More details regarding the populations are displayed in Supplementary Table S1.
Sampling locations of all the other samples considered in this study. The map was created by the Kriging algorithm of the Surfer 8.0 package. More details regarding the populations are displayed in Supplementary Table S1.
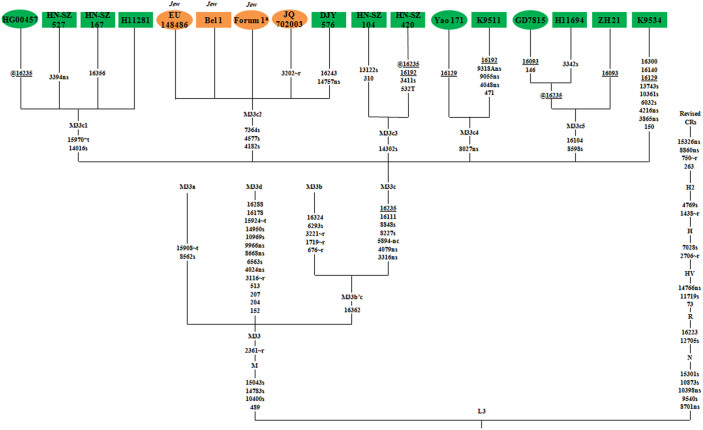
To shed light on the phylogeny within haplogroup M33c, 11 mtDNAs, covering the widest range of internal variation within the haplogroup, were chosen for whole-mtDNA genome sequencing. In good agreement with the previous result13, the resulting phylogenetic tree (Figure 2), incorporating five previously reported mtDNA genomes13,14,15 as well as one whose information was released online (A Genetic Genealogy Community; http://eng.molgen.org), confirms that M33c is defined by mutations at positions 3316, 4079, 5894, 8227, 8848, 16111, and 16235. Of note is that five clades within M33c appear respectively characterized by diagnostic coding-region variant(s), and these are named M33c1 to M33c5 here. With the exception of M33c2, all the samples in these clades are from China. The likely origin of M33 in South Asia and the restriction to China of M33c, dating to 10 kya according to the estimation based on whole-mtDNA genome, implies some dispersal from South to East Asia in the immediate postglacial.
Figure 2. Phylogenetic tree of haplogroup M33c.
The nucleotide positions in the sequences are scored relative to the rCRS27. Transversions are annotated by adding suffixes “A” and “T”. The recurrent variants are underlined and prefix @ designates a back mutational event; “s” means synonymous and “ns” means nonsynonymous mutation; “nc” refers to mutations at the intergenic noncoding regions in segments 577–16023; and “r” and “t” denote mutations in rRNA genes and tRNA genes, respectively. Length polymorphisms (e.g., 309.1C, 309.2C and 315.1C) are disregarded from the analysis. The newly sequenced samples in this study are marked in rectangles, while mtDNAs from the published literature are displayed in ellipses. Note: a This individual is from A Genetic Genealogy Community (http://eng.molgen.org/viewtopic.php?f=41&t=141).
Intriguingly, sub-haplogroup M33c2 (defined by three additional coding-region variants at positions 4182, 4577, and 7364) consists of three different haplotypes (one seen in three Ashkenazi Jews, another in a single Chinese individual and the third in the likely European with unknown ethnicity). Although there is no control-region variant in the defining motif of M33c2, multiple lines of evidence suggest that the pinpointed 14 Ashkenazi Jewish M33c mtDNAs most likely all belong to this clade: (1) all of the 14 mtDNAs share an identical control-region motif (Table 1); (2) the three completely sequenced Ashkenazi Jewish mtDNAs with this motif (EU148486, Bel 1 and Forum 1) belong to M33c2 (Figure 2); (3) M33c shows a virtually exclusive distribution in Ashkenazi Jews in western Eurasia, even though 55,595 mtDNAs have been checked (Table 1 and Supplementary Table S1). Thus, it is plausible that the unknown European individual (JQ702003) was in fact from a Jewish population or had Ashkenazi Jewish ancestry.
Age estimates for M33c2 are similar whether based either on the whole genome or on the control region alone (Table 2), and the age of ~1.4 kya fits well with the medieval operation of the Silk Road. We note that this is an upper bound for the gene flow event during which the lineage was assimilated into the Ashkenazim; it is the age of the subclade overall, which most likely arose within China, and indeed there is no variation at all within the Jewish lineages, suggesting a very recent event. If we assume that the unidentified European lineage belongs within the Ashkenazi diversity, we can date the Ashkenazi subclade itself more specifically to about 640 years ago – around 1350AD. This in turn would then provide a minimum point estimate for the age of the gene flow event (although the range taking account of errors in the estimates is of course much wider).
Table 2. Ages of the major clades of haplogroup M33c estimated from control-region and whole-mtDNA genome data with 95% confidence intervals.
| Control region | Whole-mtDNA genome | ||||||
|---|---|---|---|---|---|---|---|
| Rho | Rho | Maximum Likelihood | |||||
| Haplogroup | n | ρ ± σ | Age (kya) | n | ρ ± σ | Age (kya) | Age (kya) |
| M33c | 52 | 1.13 ± 0.47 | 21.38 [4.1;38.7] | 17 | 3.88 ± 0.76 | 10.29 [6.2;14.4] | 8.95 [5.2;12.8] |
| M33c2 | 16 | 0.06 ± 0.06 | 1.18 [0;3.5] | 5 | 0.60 ± 0.35 | 1.55 [0;3.3] | 1.55 [0;3.3] |
| M33c2* | – | – | – | 4 | 0.25 ± 0.25 | 0.64 [0;1.9] | 0.64 [0;1.9] |
M33c2*: the Chinese individual is not considered for the estimation.
The ancient Silk Road was an important transportation hub connecting China and the Mediterranean region from the Han Dynasty (206BC–220AD) onwards, and there are likely to have been Jewish merchants at the eastern end of the Silk Road from the early centuries AD. Moreover, Jewish merchants in Europe, referred to as Radhanites, were involved in trade between west and east as early as the ninth century16. It has been suggested, on the basis of contrasts between patterns of mtDNA and Y-chromosome variation17, that such merchants may have formed the nucleus for a number of extant Jewish communities.
Ashkenazi origins are controversial18. According to recent archaeological evidence, the Jewish community of Cologne, mentioned by Emperor Constantine in 321AD, existed in the city continuously until they had to leave in 1423–1424AD19. This suggests that Ashkenazi Jewry may date to Roman times, possibly originating in Italy, which is also suggested by analysis of mtDNA8 and autosomal data20. An early eastern European Ashkenazi origin from Italy (first millennium and earlier) would also agree with the finding that an origin mainly from Germany21 or another central or western European country18 during the late Middle Ages, is demographically not possible. Recent work also suggests a sizable Jewish presence in eastern Germany (the Danube region, rather than the Rhineland) prior to the expansion in Poland between 1500 and 1650AD22. The M33c2 mtDNAs are confined to eastern European Ashkenazim in the present database (the single unknown example is of likely East European ancestry14), suggesting that these groups had contacts to the east to the extent that they mediated female gene flow.
Extensive genetic admixture has been observed in populations residing around the ancient Silk Road region23,24. Our currently observed genetic imprint echoes the previously observed ancient communications between Jews and Chinese and, most significantly, implies that such historical exchanges were not confined to the cultural realm but involved gene flow. This unexpected ancient genetic connection between Ashkenazi Jews and the Far East, as witnessed at least by mtDNA haplogroup M33c2, provides the first evidence for a significant genetic contribution from Chinese to eastern European Ashkenazi Jews that was most likely mediated by the Silk Road between around 640 and 1400 years ago. Although the involvement of male Jewish traders has been suggested before17, our results, focusing on the female line of descent, specifically point to the involvement also of women. Well-resolved evidence from the male-specific part of the Y chromosome and from the autosomes would help to further illustrate the rather complex, pan-Eurasian ethno-history of Jews.
Methods
mtDNA Data collection and mining
mtDNA variation (mainly from control region) of 23,121 East Asians and Jews, retrieved from previous studies as well as our unpublished data, were considered and analyzed, with especial attention to pinpointing the eastern Eurasian haplogroups in Jews. Then, additional 32,474 individuals were analyzed to gain further insights into the phylogeographic distribution of M33c, leading the total number of Eurasian mtDNAs considered here to 55,595. The study project was approved by the Ethics Committee at Kunming Institute of Zoology, Chinese Academy of Sciences. Each participant was informed about the study and provided informed consent. All mtDNAs collected and considered in the present study were first allocated to haplogroups, based mainly on their control-region motifs, which were then further confirmed by typing specific coding-region variation according to the PhyloTree (mtDNA tree Build 1625; http://www.phylotree.org/).
DNA amplification and sequencing
For haplogroups of interest, special attention was paid to the intrinsic phylogeny reconstructed on entire mitogenome information. In this way, entire mitogenomes for 11 selected representatives from haplogroup M33c were amplified, sequenced, and dealt with as described elsewhere13,26. The sequencing outputs were edited and aligned by Lasergene (DNAStar Inc., Madison, Wisconsin, USA) and compared with the revised Cambridge Reference Sequence (rCRS)27.
Data analysis
The median-joining network of M33c was constructed manually28 and then confirmed using Network 4.612 (http://www.fluxus-engineering.com/sharenet.htm). The most parsimonious phylogenetic tree (Figure 2) was reconstructed by hand as carried out previously13,26. The coalescence ages were estimated by the ρ ± σ method29,30 and maximum likelihood (ML) analysis. Recently corrected calibrated mutation rates31 were adopted in the ρ statistic and the ML analysis.
Author Contributions
Q.-P.K. designed the research; Y.-C.L., W.Z. and Y.-G.Y. collected the samples; J.-Y.T., H.-W.W. and Y.-C.L. collected the data; J.-Y.T. and H.-W.W. performed the experiments; J.-Y.T., H.-W.W., Y.-C.L. and Q.-P.K. analyzed data; J.-Y.T., Y.-C.L., J.v.S., M.B.R. and Q.-P.K. wrote the paper.
Supplementary Material
Sample information of populations analyzed in present study
Acknowledgments
We thank Dr. Pedro Soares for his help in time estimation. This work was supported by grants from the National Natural Science Foundation of China (81272309, 31123005, 31322029) and the Chinese Academy of Sciences.
References
- Josephus F. The Antiquities of the Jews (Echo Library, 2006). [Google Scholar]
- Ostrer H. A genetic profile of contemporary Jewish populations. Nat. Rev. Genet. 2, 891–898 (2001). [DOI] [PubMed] [Google Scholar]
- Behar D. M. et al. The genome-wide structure of the Jewish people. Nature 466, 238–242 (2010). [DOI] [PubMed] [Google Scholar]
- Atzmon G. et al. Abraham's children in the genome era: major Jewish diaspora populations comprise distinct genetic clusters with shared Middle Eastern Ancestry. Am. J. Hum. Genet. 86, 850–859 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopelman N. et al. Genomic microsatellites identify shared Jewish ancestry intermediate between Middle Eastern and European populations. BMC Genet. 10, 80 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need A. C., Kasperavičiūtė D., Cirulli E. T. & Goldstein D. B. A genome-wide genetic signature of Jewish ancestry perfectly separates individuals with and without full Jewish ancestry in a large random sample of European Americans. Genome Biol. 10, R7 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar D. M. et al. The matrilineal ancestry of Ashkenazi Jewry: portrait of a recent founder event. Am. J. Hum. Genet. 78, 487–497 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M. D. et al. A substantial prehistoric European ancestry amongst Ashkenazi maternal lineages. Nat. Commun. 4, 2543 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G. The Jews in China (China Intercontinental Press, 2007). [Google Scholar]
- Shapiro S. Jews in old China: Studies by Chinese Scholars (Hippocrene Books, 2001). [Google Scholar]
- Sun C. et al. The dazzling array of basal branches in the mtDNA macrohaplogroup M from India as inferred from complete genomes. Mol. Biol. Evol. 23, 683–690 (2006). [DOI] [PubMed] [Google Scholar]
- Chandrasekar A. et al. Updating phylogeny of mitochondrial DNA macrohaplogroup m in India: dispersal of modern human in South Asian corridor. PLoS ONE 4, e7447 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q. P. et al. Large-scale mtDNA screening reveals a surprising matrilineal complexity in East Asia and its implications to the peopling of the region. Mol. Biol. Evol. 28, 513–522 (2011). [DOI] [PubMed] [Google Scholar]
- Behar D. M. et al. A “Copernican” reassessment of the human mitochondrial DNA tree from its root. Am. J. Hum. Genet. 90, 675–684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H. X. et al. Major population expansion of East Asians began before Neolithic Time: Evidence of mtDNA genomes. PLoS ONE 6, e25835 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick M. Origins of the European Economy: Communications and Commerce AD 300–900 688–693 (Cambridge University Press, 2002). [Google Scholar]
- Goldstein D. B. Jacob's Legacy: A Genetic View of Jewish History. (Yale University Press, 2008). [Google Scholar]
- van Straten J. The Origin of Ashkenazi Jewry: The Controversy Unraveled. (New York: Walter de Gruyter & Co, 2011). [Google Scholar]
- Schütte S. & Gechter M. Köln: archäologische zone Jüdisches Museum: von der Ausgrabung zum Museum-Kölner Archäologie zwischen Rathaus und Praetorium: Ergebnisse und Materialien 2006–2011. (Jüdisches Museum, 2011). [Google Scholar]
- Behar D. M. et al. No evidence fom genome-wide data of a Khazar origin for the Ashekanzi Jews. (Hum. Biol., in press) (2013). [DOI] [PubMed] [Google Scholar]
- van Straten J. Early modern Polish Jewry: The Rhineland hypothesis revisited. Hist. Method. 40, 39–50 (2007). [Google Scholar]
- King R. D. Migration and linguistics as illustrated by Yiddish. In Reconstructing languages and cultures, Polomé P. C., & Winter W., eds. ed. 419–439 (Berlin/New York: Mouton de Gruyter, 1992). [Google Scholar]
- Comas D. et al. Trading genes along the silk road: mtDNA sequences and the origin of central Asian populations. Am. J. Hum. Genet. 63, 1824–1838 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y. G., Kong Q. P., Wang C. Y., Zhu C. L. & Zhang Y. P. Different matrilineal contributions to genetic structure of ethnic groups in the Silk Road region in China. Mol. Biol. Evol. 21, 2265–2280 (2004). [DOI] [PubMed] [Google Scholar]
- van Oven M. & Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 30, E386–E394 (2009). [DOI] [PubMed] [Google Scholar]
- Kong Q. P. et al. Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations. Hum. Mol. Genet. 15, 2076–2086 (2006). [DOI] [PubMed] [Google Scholar]
- Andrews R. M. et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 23, 147–147 (1999). [DOI] [PubMed] [Google Scholar]
- Bandelt H.-J., Macaulay V. & Richards M. Median networks: speedy construction and greedy reduction, one simulation, and two case studies from human mtDNA. Mol. Phylogenet. Evol. 16, 8–28 (2000). [DOI] [PubMed] [Google Scholar]
- Forster P., Harding R., Torroni A. & Bandelt H.-J. Origin and evolution of Native American mtDNA variation: a reappraisal. Am. J. Hum. Genet. 59, 935 (1996). [PMC free article] [PubMed] [Google Scholar]
- Saillard J., Forster P., Lynnerup N., Bandelt H.-J. & Nørby S. mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am. J. Hum. Genet. 67, 718–726 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares P. et al. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am. J. Hum. Genet. 84, 740–759 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandstätter A. et al. Mitochondrial DNA control region variation in Ashkenazi Jews from Hungary. Forensic Sci Int Genet 2, e4–e6 (2008). [DOI] [PubMed] [Google Scholar]
- Zhang W. et al. A matrilineal genetic legacy from the Last Glacial Maximum confers susceptibility to schizophrenia in Han Chinese. J. Genet. Genomics 41, 397–407 (2014). [DOI] [PubMed] [Google Scholar]
- Chen F. et al. Analysis of mitochondrial DNA polymorphisms in Guangdong Han Chinese. Forensic Sci Int Genet 2, 150–153 (2008). [DOI] [PubMed] [Google Scholar]
- Gan R. J. et al. Pinghua population as an exception of Han Chinese's coherent genetic structure. J. Hum. Genet. 53, 303–313 (2008). [DOI] [PubMed] [Google Scholar]
- Irwin J. A. et al. Mitochondrial control region sequences from a Vietnamese population sample. Int. J. Legal Med. 122, 257–259 (2008). [DOI] [PubMed] [Google Scholar]
- Li H. et al. Mitochondrial DNA diversity and population differentiation in southern East Asia. Am. J. Phys. Anthropol. 134, 481–488 (2007). [DOI] [PubMed] [Google Scholar]
- Peng M. S. et al. Tracing the Austronesian footprint in Mainland Southeast Asia: a perspective from mitochondrial DNA. Mol. Biol. Evol. 27, 2417–2430 (2010). [DOI] [PubMed] [Google Scholar]
- Wang W. Z. et al. Tracing the origins of Hakka and Chaoshanese by mitochondrial DNA analysis. Am. J. Phys. Anthropol. 141, 124–130 (2010). [DOI] [PubMed] [Google Scholar]
- Wen B. et al. Analyses of genetic structure of Tibeto-Burman populations reveals sex-biased admixture in southern Tibeto-Burmans. Am. J. Hum. Genet. 74, 856–865 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B. et al. Genetic structure of Hmong-Mien speaking populations in East Asia as revealed by mtDNA lineages. Mol. Biol. Evol. 22, 725–734 (2005). [DOI] [PubMed] [Google Scholar]
- Yao Y. G., Kong Q. P., Bandelt H.-J., Kivisild T. & Zhang Y.-P. Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am. J. Hum. Genet. 70, 635–651 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampuansai J. et al. Mitochondrial DNA Variation of Tai Speaking Peoples in Northern Thailand. ScienceAsia 33, 443–448 (2007). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sample information of populations analyzed in present study