Abstract
Hepatitis B virus (HBV) remains a major human pathogen, with over 240 million individuals suffering from chronic HBV infections. These can persist for decades due to the lack of therapies that can effectively target the stable viral covalently closed circular (ccc) DNA molecules present in infected hepatocytes. Using lentiviral transduction of a bacterial Cas9 gene and single guide RNAs (sgRNAs) specific for HBV, we observed effective inhibition of HBV DNA production in in vitro models of both chronic and de novo HBV infection. Cas9/sgRNA combinations specific for HBV reduced total viral DNA levels by up to ~1000-fold and HBV cccDNA levels by up to ~10-fold and also mutationally inactivated the majority of the residual viral DNA. Together, these data provide proof of principle for the hypothesis that CRISPR/Cas systems have the potential to serve as effective tools for the depletion of the cccDNA pool in chronically HBV infected individuals.
Keywords: HBV, cccDNA, CRISPR/Cas, antiviral, DNA editing, lentiviral vector
Introduction
Hepatitis B virus (HBV) remains an important human pathogen despite the existence of a highly effective vaccine. Particularly in resource-limited contexts, vaccination rates remain well under 100% and vaccination at birth of infants born to HBV positive mothers is not fully protective against vertical transmission (Komatsu, 2014). Moreover, it is estimated that over 240 million individuals, ~4% of the world population, remain chronically infected with HBV and are therefore at high risk for cirrhosis and hepatocellular carcinoma (HCC), leading to ~800,000 deaths per year that are directly linked to HBV infection (Komatsu, 2014).
Treatment of HBV primarily relies on the use of nucleoside analog chain terminators, such as lamivudine (3TC), tenofovir disoproxil fumarate (TDF) and entecavir (ETV), which act as potent inhibitors of the HBV reverse transcriptase (RT). However, while these drugs can clearly slow the progression of HBV-induced disease, including both cirrhosis and HCC, they do not lead to a full elimination of this viral infection. This is due to the very high stability of an episomal DNA intermediate in the HBV replication cycle, called covalently closed circular DNA (cccDNA), that persists in the nuclei of infected hepatocytes and serves as the template for viral mRNA and pre-genomic RNA synthesis (Werle-Lapostolle et al., 2004). While RT inhibitors can prevent the de novo HBV infection of additional hepatocytes, the cells infected at the time of treatment initiation remain infected and only gradually decrease in number due to cell turnover. Therefore, in order to effectively clear an HBV infection and produce full remission, the viral cccDNA intermediates need to be destroyed. This goal has now become feasible due to the development of programmable RNA-guided DNA endonucleases derived from the Type II bacterial CRISPR (clustered regularly interspaced short palindromic repeats)/Cas mechanism of bacterial adaptive innate antiviral immunity (Hsu et al., 2014).
Type II CRISPR/Cas systems rely on a single effector protein, called CRISPR-associated protein 9 (Cas9), that is normally guided to a specific DNA sequence by two small RNA molecules called the tracrRNA and the crRNA (Barrangou and Marraffini, 2014; Hsu et al., 2014). The demonstration that the tracrRNA and crRNA could be combined into a single guide RNA (sgRNA) (Cong et al., 2013; Mali et al., 2013) enhanced the feasibility of using CRISPR/Cas systems as effective gene editing tools in eukaryotic cells. Previously, most research has focused on using the Cas9 protein from Streptococcus pyogenes (Spy), which uses a 20-nt guide sequence to target complimentary DNA sequences that are flanked 3′ by a so-called protospacer adjacent motif (PAM), with the sequence 5′-NGG-3′. The PAM, which forms an invariant part of every Spy Cas9-specific DNA target, is directly recognized by the Cas9 protein and initiates DNA targeting guided by the sgRNA (Barrangou and Marraffini, 2014; Cong et al., 2013; Hsu et al., 2014; Mali et al., 2013). Using transfection or lentiviral vector transduction, it is possible to induce cleavage of specific DNA sequences present in a mammalian genome by expression of Spy Cas9 and an sgRNA molecule transcribed by an RNA polymerase III promoter (Cong et al., 2013; Mali et al., 2013). Gene inactivation using this system is both efficient and specific, and Spy Cas9 has in fact been used for not only high throughput genetic screens in human cells in culture (Shalem et al., 2014; Wang et al., 2014; Zhou et al., 2014) but also for the generation of gene knockouts in several species, including mice and even monkeys (Yang et al., 2013). We therefore reasoned that the Spy Cas9 protein, in combination with sgRNAs designed to select a target present on the HBV cccDNA, might represent a powerful method to cleave and eradicate HBV cccDNAs. In the present study, we demonstrate that lentiviral vectors encoding Spy Cas9/sgRNA combinations targeted to the HBV reverse transcriptase (RT), surface antigen (Ag) or core protein genes are all able to effectively repress viral DNA production, including cccDNA accumulation, in chronically HBV infected cells and also inhibit de novo HBV infection. Moreover, these Spy Cas9/sgRNA combinations showed additive inhibition of HBV DNA accumulation when used in combination with known pharmacological inhibitors of the HBV RT enzyme. These experiments provide evidence in support of the hypothesis that HBV-specific Spy Cas9/sgRNA combinations, when delivered to HBV-infected hepatocytes in vivo by viral transduction, might provide a novel and effective approach to facilitate the elimination of HBV in chronically infected individuals.
Results
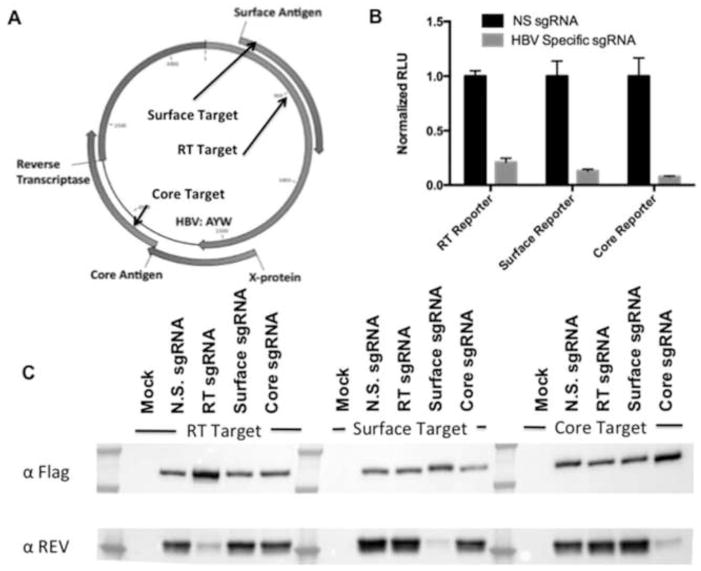
To target the HBV genome (subtype AYW) for elimination in cell culture, we first designed three sgRNAs specific for Spy Cas9 targeting the viral open reading frames (ORFs) encoding HBV surface Ag, core, and/or RT. For the HBV structural proteins surface Ag and core, N-terminal targets were selected to induce frame-shift mutagenesis. The surface Ag specific sgRNA was also predicted to cleave the RT gene (Fig. 1A). RT was also targeted in the highly conserved “YMDD” motif required for enzyme catalysis (Fig. 1A) using an sgRNA predicted to also cleave near the end of the surface Ag ORF. To verify effective sgRNA function, we employed a previously described (Kennedy et al., 2014) indicator assay based on co-transfection of Spy Cas9 and sgRNA expression plasmids, along with a cognate indicator plasmid, into human 293T cells. This assay quantitatively measures sgRNA efficacy by monitoring the firefly luciferase (FLuc) activity or steady state expression level of an HIV-1 Rev–sgRNA target–FLuc fusion protein; high cleavage activity results in a substantial knockdown of both. As shown in Figure 1B and 1C, all three sgRNAs induced an effective and specific knockdown of Fluc expression, as measured by dual luciferase assay, and Rev fusion protein expression, as measured by Western blot analysis, respectively. To directly test these three sgRNA constructs in relevant HBV cell-culture models, we next shuttled them into a lentiviral Cas9/sgRNA expression vector (Shalem et al., 2014) capable of efficient transduction of the cell lines HepAD38 and HepaRG (Fig. S1).
Figure 1. HBV targeting strategy and sgRNA optimization.
To target HBV DNA intermediates in infected cells, we designed sgRNAs specific for the HBV surface Ag, core, and RT ORFs, as depicted in panel A. To assess efficacy, fusion protein-based indicator plasmids, which encode an amino-terminal HIV-1 Rev-derived epitope tag, an in-frame HBV-derived target, and a carboxy-terminal FLuc indicator gene, were employed. As shown in panel B, all three sgRNA candidates effectively inhibited FLuc expression from their cognate reporter plasmid in co-transfected 293T cells when compared to a control, non-specific (N.S.) sgRNA. In panel C, expression of the same fusion protein was probed by Western blot using a previously described α-Rev rabbit polyclonal antiserum (Kennedy et al., 2014). Co-expression of Cas9 was confirmed using an antibody specific for the FLAG epitope tag present on this protein. When the reporter was cognate for the sgRNA, a marked reduction of the expression level of the fusion protein target could be observed, confirming the specificity and efficacy of each sgRNA tested.
Suppression of HBV replication by Cas9/sgRNA combinations specific for the HBV RT, surface Ag and core genes
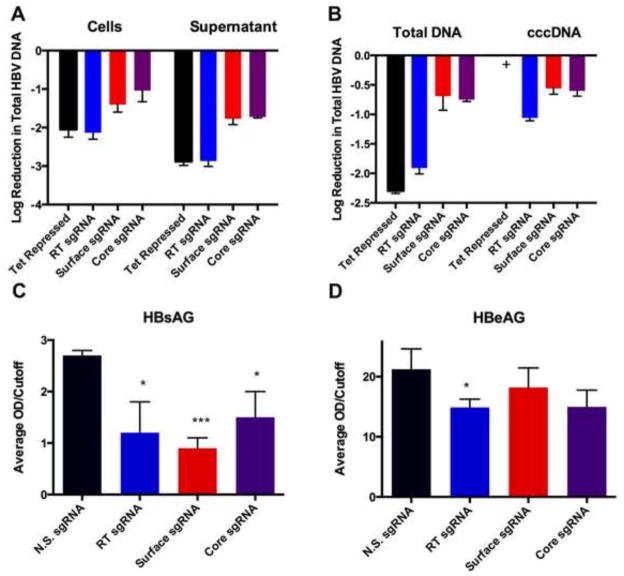
In the HepAD38 cell line, transcription initiation of an integrated HBV linear DNA genome is tightly regulated by a Tet repressed promoter (Ladner et al., 1997). In the absence of Tet in the culture medium, the system mimics HBV replication by transcribing HBV mRNAs and pre-genomic RNA, leading to the initiation of a robust HBV replication cycle, including the synthesis of substantial levels of HBV cccDNA, and the release of DNA-containing viral particles into the supernatant medium. Consequently, this cell line represents an excellent model to test suppression of HBV replication and cccDNA synthesis by Spy Cas9 loaded with sgRNAs specific for HBV DNA targets. Therefore, we tested the ability of this system to suppress HBV replication by transducing HepAD38 cells with lentiviral vectors encoding HBV-specific Cas9/sgRNA combinations. HBV transcription was activated 48 h prior to transduction by removal of Tet from the medium to ensure that an active HBV replication cycle was in progress prior to Cas9/sgRNA expression. Subsequently, Tet was added back to the medium again, repressing the chromosomal HBV locus, to ensure that cccDNA was the primary source of viral transcription. The lentiviral vectors used also encode a puromycin resistance gene (Shalem et al., 2014), thus permitting selection of the transduced cells. As a positive control for HBV gene expression, HepAD38 cells were also transduced with a non-specific (N.S.) sgRNA construct that does not target any HBV sequence. As a control for maximally repressed HBV expression, HepAD38 cells were also tested in the continuous presence of Tet, which in the HepAD38 cells blocks HBV transcription (Ladner et al., 1997). After transduction and selection, all three sgRNAs reduced HBV DNA levels inside cells by ~10 to ~125-fold and in the supernatant media by ~100-fold to ~800-fold, as measured by real-time PCR (Fig. 2A). Even more impressively, the RT sgRNA suppressed HBV DNA replication to the same extent as Tet addition, which essentially entirely blocks virus replication.
Figure 2. Suppression HBV replication by HBV-specific Cas9/sgRNA combinations.
HBV replication was first induced in the HepAD38 cells by culture in the absence of Tet for 48 hrs. Then, the cells were transduced with lentiviral vectors encoding Spy Cas9/sgRNA combinations specific for the HBV RT, surface Ag or core ORFs, puromycin selected, and cultured in the continued absence of Tet. HepAD38 cells were also maintained in Tet+ media as a negative control. The HepAD38 cells and supernatant media were harvested and total HBV DNA (A) and intracellular HBV DNA or cccDNA (B) quantified by qPCR after 10 or 14 days in culture, respectively. (+) indicates the sample was below the detection limit (> 45 cycles). All results of qPCR assays were normalized to the N.S. sgRNA control cell line. Levels of HBsAg (C) and HBeAg (D) secreted into the culture media were measured by ELISA after 10 days in culture. Data are displayed as the mean ± SD of replicates. Statistical analyses were performed using Student’s t-test for comparison between two groups using JMP pro 10 software. A value of P < 0.05 (*) was considered statistically significant. (***) represents P < 0.001.
Suppression of cccDNA formation by HBV specific Cas9/sgRNA combinations
To test the capacity of HBV-specific Cas9/sgRNA combinations to eliminate cccDNA accumulation, transduced HepAD38 cells were also analyzed for the level of cccDNA accumulation by real-time PCR after 14 days in culture using previously described cccDNA-specific primers (Chen et al., 2004). Remarkably, the HBV RT-specific sgRNA suppressed cccDNA formation by ~10-fold, while the surface Ag and core-specific sgRNAs repressed cccDNA levels by ~4-fold (Fig. 2B). Total intracellular HBV DNA accumulation was reduced by ~8-fold by the HBV surface Ag and core-specific sgRNAs, and were repressed by ~80-fold by the RT specific sgRNA (Fig. 2B). Impressively, the HBV RT specific sgRNA was therefore able to reduce HBV cccDNA formation by ~90% and total HBV intracellular DNA accumulation by ~99%.
HBV-specific Cas9/sgRNA combinations reduce HBV surface Ag secretion
In HepAD38 cells, analysis of the level of HBV surface Ag (HBsAg) and “e” Ag (HBeAg) secretion represents a useful assay for screening for antivirals that inhibit HBV replication and/or viral gene expression. To determine if HBV-specific Cas9/sgRNA combinations can suppress secretion of these viral antigens, we performed an ELISA assay using the supernatant media of transduced HepAD38 cells harvested after 12 days in culture. Low levels of secreted HBsAg were observed in the supernatant of HepAD38 cells transduced with HBV-specific sgRNAs targeting the RT, surface and core genes but not the N.S. sgRNA. Indeed, the surface Ag-specific sgRNA suppressed HBsAg production to nearly undetectable levels (Fig. 2C), possibly consistent with the mutational inactivation of this viral gene due to editing by the cognate Cas9/sgRNA combination. We also observed a statistically significant reduction in HBeAg secretion in the cells transduced with the RT specific sgRNA, indicating perhaps a global reduction in cccDNA in this case (Fig. 2D). In contrast, we did not observe a significant reduction in HBeAg for the Core and Surface sgRNAs, and this likely results from this protein’s secretion and exceptional stability. In contrast to the HBsAg ORF, it should be noted that none of the sgRNAs employed directly target this viral ORF for mutagenic inactivation.
Mutational inactivation of HBV by Cas9/sgRNA combinations
While the observed depletion of total HBV DNA and cccDNA accumulation was extensive (Figs. 2A and 2B), we were also curious as to the mutational status of the residual viral DNA as Cas9 cleavage of chromosomal targets usually results in the introduction of small sequence insertions or deletions (indels) (Shalem et al., 2014). For this purpose, we focused on HepAD38 cells transduced with the sgRNA specific for the active site YMDD motif of the HBV RT gene (Fig. 1A). We harvested total DNA from the HepAD38 cells transduced with the RT-specific Cas9/sgRNA combination that we had previously shown effectively knocked down viral DNA levels (Figs. 2A and 2B) and subjected the DNA to PCR amplification using HBV RT specific primers followed by cloning and sequencing of the resultant HBV DNA fragments. Seventy-three deletion mutations located at or immediately adjacent to the predicted Cas9 cleavage site present 3 bp 5′ to the target DNA PAM sequence were recovered, and we also detected five insertion mutations at this same location in the HBV genome (Fig. 3). Of the 103 recovered viral sequences, 78 (76%) were found to be mutated and, interestingly, the bulk of the amplicons recovered were predicted to be lethally mutagenized; even an in-frame insertion or deletion would be highly detrimental as these residues are required for HBV RT function and therefore highly conserved. In conclusion, expression of a Cas9/sgRNA combination specific for the HBV RT gene not only dramatically reduced viral DNA levels, as expected (Figs. 2A and 2B), but also mutationally inactivated the majority of the low level of residual viral DNA.
Figure 3. An HBV RT-specific Cas9/sgRNA combination induces extensive mutagenesis of the residual HBV DNA.
Total cellular DNA was used as a template to amplify viral DNA sequences flanking the predicted HBV RT sgRNA target site (Fig. 1A). The resultant PCR fragment was then cloned and sequenced. Shown above the multiple alignments of the recovered clones are the key RT active site residues “YMDD”, which includes the two aspartate residues required to chelate the Mg2+ ion required for catalysis. The cognate sgRNA sequence is underlined and shown in red, while the Spy Cas9 PAM sequence (5′-NGG-3′) is shown in green. Cleavage by Spy Cas9 is expected to occur 3 base pairs 5′ of this sequence. To the right of each HBV sequence variant we list the number of times each sequence was recovered. Of the 103 sequences analyzed, 78 bore deletions while 5 contained insertions adjacent to the predicted cleavage site.
Targeting HBV with Cas9/sgRNA combinations reduced antigen secretion and viral replication and leads to cccDNA degradation in infected HepaRG cells
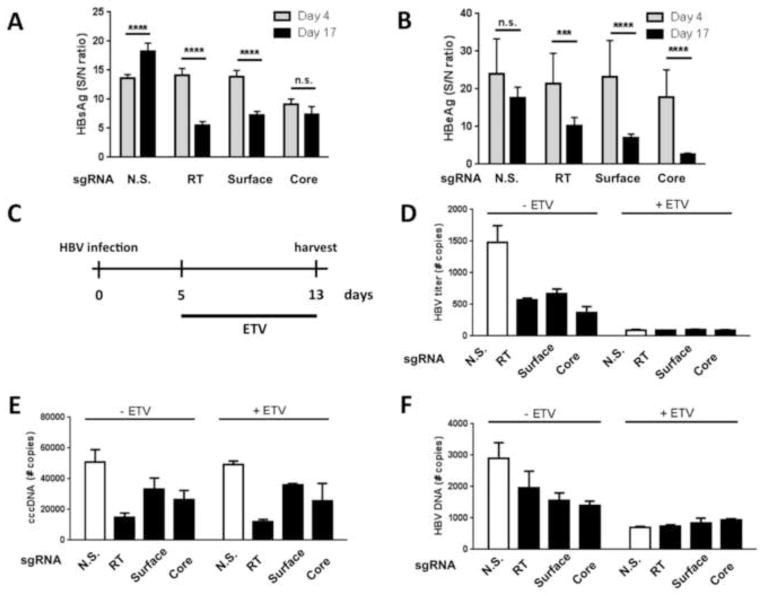
HepAD38 cells are an excellent model of post-entry chronic infection, but to test whether Cas9/sgRNA combinations specific for HBV are also capable of inhibiting de novo infection we employed a second cell culture model, HepaRG cells. These cells are permissive for infection with live HBV virus subsequent to differentiation (Gripon et al., 2002) and are an excellent complement to the HepAD38 system, which models the late stage of the HBV lifecycle (Ladner et al., 1997). To assess the effect of HBV-specific sgRNAs on HBV replication in HepaRG cells, we transduced these cells with the lentiviral constructs described above, expressing HBV RT, surface Ag, core, or control N.S. sgRNAs. After selection for puromycin resistance and differentiation, these cell lines were infected with HBV at a high MOI of ~30 and the antiviral effects of the sgRNAs evaluated (Fig. 4). Both HBsAg and HBeAg) secretion was reduced by up to ~80% by 17 days after infection in the presence of HBV-specific sgRNAs (Fig. 4A and 4B) and this reduction was highly significant. This effect could be due either to a direct effect of the sgRNA on the expression of the specific viral gene or, perhaps more likely, a global reduction in cccDNA production and HBV transcription. To further investigate the impact and specificity of the panel of HBV-specific Cas9/sgRNA combinations, we isolated DNA from the supernatant media and cell lysates of the HepaRG RT, surface Ag, core, and N.S. sgRNA-expressing cells following infection with HBV in the presence or absence of ETV treatment (see schematic in Fig. 4C). ETV, a nucleoside analogue, has been shown to reduce HBV DNA levels by inhibiting reverse transcription, but does not directly affect cccDNA stability in HepaRG cells (Lucifora et al., 2014). Moreover, in the context ETV treatment and viral inhibition, Cas9/sgRNAs might represent an effective means to eliminate cccDNA from the treated and suppressed cells. We therefore isolated total DNA from ETV-treated and untreated HepaRG cells and used these for qPCR analysis of total HBV DNA levels present either in viral particles released into the supernatant media (Fig. 4D) or present intracellularly (Fig. 4F). As expected, in the presence of ETV, HBV DNA was greatly reduced both in the supernatant (by ~96%) and intracellularly (by ~80%). In addition, all three sgRNAs targeting the HBV genome also induced a reduction in HBV total DNA levels in the supernatant media and inside infected cells, arguing that not only are HBe- and HBs-antigen gene expression specifically inhibited, but that HBV replication is also globally reduced by the HBV specific Cas9/sgRNA combinations tested. To further investigate if this reduction is due to degradation of the viral genome in the infected cells, intracellular DNA was subjected to qPCR using primers that specifically amplify cccDNA. All three sgRNAs led to reductions in cccDNA levels in infected HepaRG cells (up to 67%) (Fig. 4E), providing evidence that the Cas9/sgRNA combinations directly act on viral cccDNA intermediates. As expected, ETV treatment had no effect on cccDNA levels, and the observed almost complete inhibition of overall HBV DNA production by ETV in the supernatant media and intracellularly (Figs. 4D and 4F) further confirms the high specificity of the primers used for cccDNA detection. Overall, these data therefore suggest that HBV DNA cleavage by Cas9/sgRNA combinations has the potential to not only disrupt HBV replication but also viral cccDNA stability.
Figure 4. HBV-specific Cas9/sgRNA combinations can reduce both viral production and cccDNA accumulation.
HepaRG cells transduced with lentiviral Cas9/sgRNA expression vectors were infected with HBV (MOI=30) and HBsAg- (A) and HBeAg- (B) secretion measured 4 and 17 days post-infection. Results are presented relative to non-infected control cultures. Data are displayed as mean ± SD of replicates and statistics were performed using unpaired Student’s t-test (p>0.05 (n.s.); p<0.001(***); p<0.0001(****)). HBV-infected, transduced HepaRG cells were treated with 1 μM ETV 5 dpi for 8 days (shown schematically in (C). On day 13, HBV DNA secretion (D), intracellular cccDNA expression (E) and total intracellular HBV DNA (G) were measured by specific qPCR. HBV secreted DNA levels and total intracellular HBV DNA accumulation were quantified by using an HBV-containing plasmid as the standard. Levels of cccDNA are given relative to the non-infected control culture. Data are displayed as mean ± SD of replicates. N.S., non-specific sgRNA.
Enhanced inhibition of HBV DNA accumulation by antivirals in combination with Cas9/sgRNAs
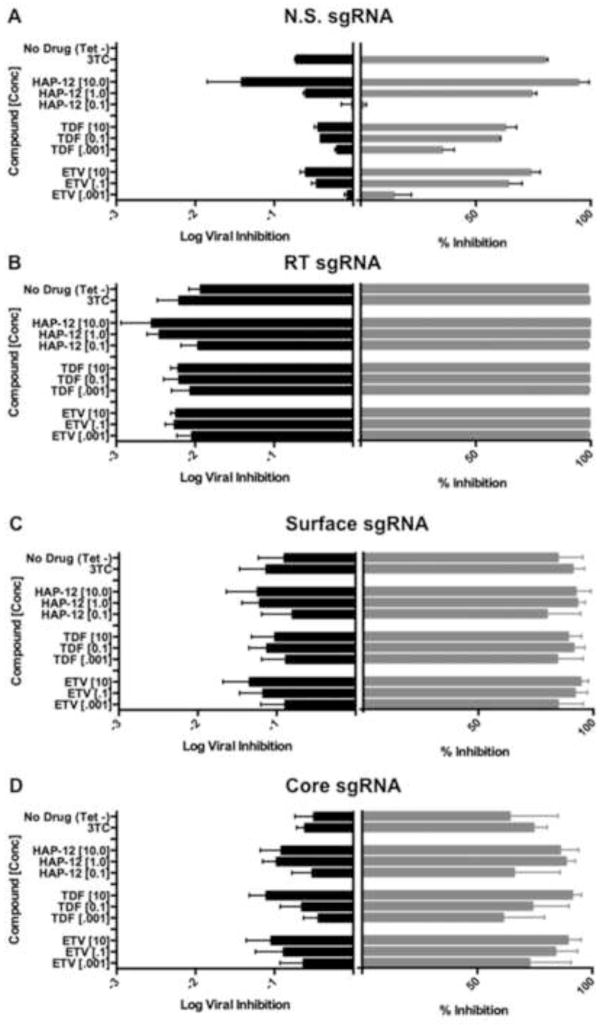
To further assess the possibility that other potent RT inhibitors (TDF, ETV or 3TC) or a viral assembly inhibitor (HAP12) could enhance elimination of residual virus from HBV-specific Cas/sgRNA expressing cells, we treated HepAD38 cells at concentrations that only partly inhibit HBV DNA replication in this system, as measured by real-time PCR of secreted HBV DNA. First, we assessed the level of viral inhibition in cells expressing Cas9 and the control N.S. sgRNA and all inhibitors tested exhibited the expected dose response (Fig. 5A). Remarkably, and as previously observed (Fig. 2A), HepAD38 cells expressing the HBV RT-specific sgRNA showed essentially complete suppression of secreted HBV DNA accumulation and gained no added benefit from the viral replication inhibitors employed (Fig. 5B). However, with the less effective surface Ag and core-specific sgRNAs, there was a modest but readily detectable enhancement of the level of induced inhibition of secreted HBV DNA in the presence of the antivirals, as shown in Fig. 5C and 5D. To further confirm these results, we used a constitutively HBV-expressing cell system, HBV2.2.15 cells (Liu et al., 2004; Sells et al., 1987). After transduction with Cas9/sgRNA expressing lentiviral constructs and selection, we observed that extracellular levels of HBV DNA were suppressed by ~98%, ~80%, and ~90%, respectively, by the RT, surface Ag, and core specific Cas9/sgRNA combinations (Table 1). Importantly, treatment of these cells with 1 μM of TDF, ETV, HAP12 or 3TC demonstrated a clear at least additive effect, leading to more efficient elimination of residual HBV DNA replication, for the less effective surface Ag- and core-specific sgRNA in 2.2.15 cells (Table 1). In the case of the RT-specific sgRNA, the detection of a possibly synergistic level of inhibition was again largely prevented by the extremely high efficacy of this Cas9/sgRNA combination.
Figure 5. Antiviral activity of TDF, ETV, HAP12, and 3TC in HepAD38 cells.
HepAD38 cells transduced with lentiviral vectors expressing various Cas9/sgRNA combinations were treated with 0.1 μM 3TC or with several concentrations (μM) of HAP12, TDF or ETV, as indicated in the y-axes. On day 7, total levels of secreted HBV DNA were measured by real-time PCR, and are here shown as both log viral inhibition and percent inhibition of the level of HBV DNA replication seen in the positive control. All data were normalized to untreated cells expressing Cas9 and the N.S. sgRNA. Panels A, B, C and D show results obtained using HepAD38 cells transduced with lentivectors expressing the N.S., RT, surface Ag and core sgRNAs, respectively. Log total viral inhibition is shown in the left panel, and percent inhibition is shown in the right panel in each case. Data are displayed as mean ± SD of replicates. –Tet indicates absence of Tet in culture media; TDF, tenofovir disoproxil fumarate; ETV, entecavir; HAP12, capsid assembly effector 12; 3TC, lamivudine.
Table 1. Antiviral activity of TDF, ETV, HAP12, and 3TC in HBV2.2.15 cells.
Percent inhibition of HBV DNA release into the supernatant medium relative to untreated, HBV2.2.15 cells expressing the non-specific control sgRNA.
| Compound | N.S. sgRNA | HBV RT sgRNA | HBV surface sgRNA | HBV core sgRNA |
|---|---|---|---|---|
| Untreated | 0.00 | 97.6 ± 0.2 | 79.6 ± 3.9 | 90.3 ± 1.4 |
| TDF, 1 μM | 68.1 ± 1.5 | 98.9 ± 0.2 | 95.9 ± 0.7 | 97.0 ± 0.3 |
| ETV, 1 μM | 78.3 ± 7.9 | 98.5 ± 1.0 | 97.6 ± 1.1 | 97.9 ± 0.6 |
| HAP12, 1 μM | 77.5 ± 5.2 | 97.5 ± 0.7 | 98.0 ± 0.3 | 97.6 ± 0.1 |
| 3TC, 1 μM | 61.4 ± 6.2 | 99.1 ± 0.03 | 96.3 ± 0.1 | 97.1 ± 0.5 |
N.S., non-specific, RT, reverse transcriptase.
All values represent the average ± SD of an experiment performed in triplicate.
Cytotoxicity analysis
For both HepAD38 and HepaRG cells transduced with lentiviral sgRNA/Cas9 expression vectors cytotoxicity assays were performed, and no cellular toxicity was observed using two distinct assays at late timepoints (see Materials and Methods and Table S1).
Discussion
Despite an effective vaccine, HBV remains a serious human pathogen, with over 240 million infected individuals worldwide leading to ~800,000 deaths annually, primarily from cirrhosis and HCC (Komatsu, 2014). Nucleoside inhibitors such as ETV and TDF are effective inhibitors of the HBV RT enzyme and can prevent the release of infectious virions from HBV infected cells, but they are unable to cure HBV due to the remarkable stability of the viral cccDNA intermediate, which is not a direct target for these drugs (Lucifora et al., 2014). Recently, RNA-guided DNA endonucleases belonging to the CRISPR/Cas family (Hsu et al., 2014) were shown to effectively cleave and inactivate viral DNA in both HIV-1 and HPV-infected human cells (Ebina et al., 2013; Hu et al., 2014; Kennedy et al., 2014), and we therefore wished to test whether Cas9/sgRNA combinations specific for the HBV DNA genome might be able to also cleave and reduce HBV DNA levels, including levels of cccDNA, in infected human cells in culture.
The system we have used relies on the delivery of the Spy Cas9 protein and an HBV-specific sgRNA to HBV-infected hepatoma cells using a lentiviral vector system (Shalem et al., 2014). We selected three distinct target sites in the HBV genome, focused on the viral surface Ag, core and RT ORFs (Fig. 1A) and were able to confirm that all three sgRNAS were active using indicator constructs containing cognate HBV DNA segments (Fig. 1). We next introduced each Cas9/sgRNA combination, or a control non-specific sgRNA, into the cell line HepaRG (Ladner et al., 1997) by lentiviral transduction, selected transduced cells using puromycin and then quantified the ability of the HBV DNA genome chromosomally integrated in these cells to support a complete viral replication cycle. To ensure that any observed inhibition did not reflect simply the mutational activation of the single integrated HBV copy, we removed Tet, which blocks HBV transcription in HepAD38 cells, 48 h prior to transduction resulting in the production of readily detectable levels of unintegrated HBV DNA, including cccDNA. Nevertheless, we observed a strong inhibition of viral DNA accumulation upon introduction of any of the three HBV-specific Cas9/sgRNA combinations, with the RT-specific sgRNA, which targets the essential “YMDD” motif in the HBV RT ORF for cleavage, being particularly effective. In fact, the RT-specific sgRNA reduced total viral DNA released into the supernatant media, by ~1000-fold and total intracellular HBV DNA levels by ~100-fold, which is comparable to the inhibitory effect seen upon retention of Tet in the culture media (Fig. 2A). The Cas9/sgRNA combinations used also inhibited the accumulation of cccDNA in this system, with the RT-specific sgRNA again most effective, though the maximal reduction in this case was ~10-fold (Fig. 2B). Measurement of viral surface Ag production also revealed a clear inhibition caused by all three Cas9/sgRNA combinations, though in this case the surface Ag-specific sgRNA was most effective, possibly due to mutational inactivation of the surface Ag ORF due to frameshifting (Fig. 2C). In contrast, only a modest inhibitory effect on HBeAg secretion was observed, likely in part because no sgRNA specific for HBV ORF was tested.
To examine whether HBV-specific Cas9/sgRNA combinations were indeed inducing inactivating indels at their predicted cleavage site in the HBV genome, as expected (Shalem et al., 2014), we cloned and sequenced PCR-generated DNA fragments that traversed the predicted cleavage site in the HepAD38 cells expressing the highly effective RT-specific sgRNA and observed deletions or insertions in 78 out of 103 sequenced clones. The RT-specific sgRNA already reduced HBV DNA levels by 100–1000-fold, and it is therefore clear that not only is very little HBV DNA present in the cells transduced with the RT-specific Cas9/sgRNA combination but also what little viral DNA is left has almost all been mutationally inactivated by error-prone repair of the Cas9-induced cleavage site.
To extend these studies to a system that supports de novo HBV infection, we also analyzed the effect of these Cas9/sgRNA combinations on HBV replication in the permissive HepaRG cell line (Gripon et al., 2002). As may be observed in Fig. 4, we again were able to detect a substantial inhibition in the accumulation of HBV DNA in these cells, including cccDNA (Fig. 4E) as well as a clear, statistically significant reduction in both HBgAg and HBeAg production at later time points (Fig. 4A and B). Finally, we also tested whether expression of Cas9/sgRNA combinations would be able to inhibit HBV replication more effectively when used together with known inhibitors of the HBV life cycle, including 3TC, TDF, ETV and HAP-12. As shown in Fig. 5 and Table 1, we were indeed able to observe enhanced inhibition of HBV replication by HBV-specific Cas9/sgRNA combinations in both HepAD38 cells and in the chronically HBV-infected cell line HBV2.2.15 (Sells et al., 1987) in the presence of various antiviral drugs. We therefore believe that the use of CRISPR/Cas system to directly target HBV cccDNA in the infected liver clearly merits further testing to assess whether there is real potential for a possible cure for chronic HBV infections when effective Cas9/sgRNA combinations are used together with current antiviral drugs.
While this current study used the Spy Cas9 protein, and cognate sgRNAs, delivered by lentiviral vector transduction (Shalem et al., 2014) we believe that future efforts should focus on the use of adeno-associated virus (AAV) vectors, as these can potentially be generated at very high titers, up to 1014 virion particles per ml, and have a high degree of tropism for hepatocytes. However, the Spy Cas9 gene, at ~4.2 kb, is too large for incorporation into AAV vectors, as these can only accommodate ~4.6 kb of DNA, which would need to include not only the Cas9 ORF but also the sgRNA, as well as the transcriptional regulatory regions required for Cas9 mRNA and sgRNA expression and, finally, the AAV inverted terminal repeats. However, there are several smaller (~3.2 kb) Cas9 genes known in other bacterial species of which at least one, expressed by Neisseria meningitidis (Nme) has been characterized (Esvelt et al., 2013). Our initial data indeed suggest that Nme Cas9 is as effective as Spy Cas9 in inducing target DNA cleavage and inactivation (data not shown), thus raising the clear possibility of generating single AAV vectors able to deliver complete, fully functional Cas9/sgRNA combinations specific for HBV to the liver in vivo at high efficiency. Whether Cas9/sgRNA combinations are entirely safe, and whether their efficiency can be high enough to effectively clear the HBV cccDNA pool, are issues that will next need to be carefully addressed in animal models of HBV infection (Dandri and Lutgehetmann, 2014).
While this article was in preparation, Lin et al. (2014) reported that HBV-specific Spy Cas9/sgRNA combinations were able to cleave HBV DNA when Cas9 and HBV expression plasmids were co-transfected into Huh7 cells. Neither HBV-infected human cells nor HBV cccDNA levels were examined in this previous work. Here, we substantially extend these recent findings by demonstrating HBV DNA cleavage and suppression, including repression of cccDNA accumulation, in both a model of chronic HBV infection (HepAD38 cells) and during de novo infection (HepaRG cells) in culture. These novel observations therefore provide substantial support for the idea that bacterial CRISPR/Cas systems have the potential to function as an effective treatment for DNA virus infections.
Material and methods
Single guide RNA design and lentiviral vector production
Multiple sgRNAs for each HBV DNA target were screened using a dual luciferase indicator assay, essentially as described previously (Kennedy et al., 2014). Briefly, Spy Cas9/sgRNA coexpression constructs based upon pX330 (Cong et al., 2013) were co-transfected into 293T cells at an 8:1 ratio relative to an indicator plasmid expressing a fusion protein consisting of an amino-terminal HIV-1 Rev derived epitope tag, a central target region derived from an HBV open reading frame and lastly a carboxy-terminal firefly luciferase (FLuc) indicator gene. A Renilla luciferase expression plasmid was also co-transfected as an internal control. Transfections were analyzed at 72 h post-transfection by Promega dual luciferase assay and Western blot for the expression of the encoded Rev-target-Fluc fusion protein to confirm the specific knockdown of the DNA target. The HBV DNA targets for the sgRNAs are depicted in Fig. 1A. These candidate sgRNAs were shuttled into the LentiCRISPR lentiviral expression vector (Shalem et al., 2014), which was produced at high titer in 293T cells by co-transfection, as previously described (Kennedy et al., 2014).
HBV strain AYW targets for the sgRNAs used in this work were as follows: HBV RT (GTTCAGTTATATGGATGATG), HBV surface antigen (Ag) (GCCTGTCCTCCAACTTGTCC), HBV core protein (GTACCGCCTCAGCTCTGTAT), and nonspecific control (N.S.) (GAAATCCTGCAGAAAGACCT). The initial G required for efficient RNA polymerase III transcription from the U6 promoter is underlined and is not complementary to the DNA target.
To assess the mutagenic spectrum generated by Spy Cas9/sgRNA cleavage, primers bearing unique restriction sites were designed to anneal to HBV sequences flanking the predicted Cas9 cleavage site in the RT gene. Total HBV genomic DNA was extracted from HepAD38 cells following transduction with the Cas9/sgRNA combination specific for the HBV RT gene described above, PCR amplified, cloned into pcDNA3 (Invitrogen), and Sanger sequenced. The recovered sequences were then aligned to the wild-type HBV strain AYW genome.
Cell culture
The human 293T cells employed in the reporter assays and for lentiviral vector production were cultured in Dulbecco’s modified Eagle medium (DMEM) including 10% fetal bovine serum (FBS) and antibiotics. The HepAD38 cell line regulates HBV replication through the presence or absence of Tet in the culture medium (King and Ladner, 2000; Ladner et al., 1997). Upon removal of Tet from the medium, HBV replicates and is secreted from these cells, while Tet addition completely represses HBV replication. HepAD38 cells were cultured in DMEM/F12 medium (Life Technologies) supplemented with 10% heat-inactivated FBS, 100 IU/ml penicillin, 100 μg/ml streptomycin, 100 μg/ml kanamycin, 400 μg/ml G418, and with 0.3 μg/ml Tet (for inhibition of HBV replication) or without any Tet (for induction of HBV replication). HepaRG (Biopredic) cells (Gripon et al., 2002) were cultured in Williams E medium containing HepaRG supplement medium. HBV 2.2.15 cells (Sells et al., 1987) were cultured in DMEM containing 10% FBS, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 2 mM L-glutamine
Lentiviral transduction of cells
HepAD38 or HBV2.2.15 cells were transduced with lentiviral vectors expressing a puromycin resistance gene as well as Spy Cas9 and an sgRNA, as described above, at 85–90% confluency after growth in medium lacking Tet for 48 h, to ensure expression of HBV mRNAs and DNA. Beginning at 48 h post-transduction, cells were selected in 4 μg/ml puromycin (Life Technologies) for 10 days to eliminate any non-transduced cells. Supernatant media and cells were then harvested for ELISA and real-time PCR assays. Control cells including non-transduced HepAD38 cells were included as positive controls for HBV replication (in medium lacking Tet) or virtual absence of HBV replication (in medium containing Tet). Non-transduced HBV 2.2.15 cells were included as a control in experiments where relevant.
HepaRG (Biopredic) cells were plated at a density of 8×105 cells per well in a 6-well cell tissue culture plate (NUNC). Cells were transduced 24 h later with the same Spy Cas9/sgRNA expressing lentiviral vectors used above and selected with 5 μg/ml puromycin. Cells were subsequently differentiated for 4 weeks in HepaRG culture medium containing 2% DMSO (Sigma-Aldrich) for HBV infection experiments.
HBV infection of Spy Cas9/sgRNA-expressing HepaRG stable cell line
HepaRG cells were infected with HBV particles produced from HBV-producing 2.2.15 cells at an MOI of ~30. HepaRG culture conditions, differentiation and HBV infection have been described previously (Hantz et al., 2009). One day post-infection, the cells were washed three times with phosphate-buffered saline and medium was replaced every two days during the experiment. To evaluate the impact on HBV antigen expression and secretion, supernatants were collected on day 4 and Day 17. HBV HBsAg and HBeAg levels were measured using CLIA ELISA Kits (Autobio Diagnostic), according to the manufacturer’s protocol. After the removal of supernatant media, cells were incubated with CellTiter-Glo One Solution (Promega) to measure cell viability.
To measure the effect of anti-HBV Cas9/sgRNA combinations on viral production and cccDNA accumulation, from day 5 post-infection onwards, some cultures were treated with entecavir (Ark Pharma) at 1 μM and retreated every two days till harvest on day 13.
On day 13 post-infection, HepaRG supernatant media were collected and HBV HBsAg and HBeAg levels measured using CLIA ELISA Kits (Autobio Diagnostic), according to the manufacturer’s protocol. In addition, DNA isolation from the supernatant media was performed using the MagNA Pure 96 DNA and Viral DNA Small Volume Kit (Roche). To quantify HBV DNA, supernatant and HBV plasmid (Genotype D) serial dilutions, as quantification standards, were used as templates for Taqman qPCR using an HBV core gene specific primer. After removal of the supernatants, the cells were lysed and total DNA was extracted (Macherey-Nagel, Tissue Kit). HBV DNA and cccDNA were detected using specific PCR primers and the LightCycler system (Roche), as described elsewhere (Lucifora et al., 2014).
Quantification of HBV DNA and cccDNA in HepAD38 cells
For HBV DNA quantification in HepAD38 cells, a set of primers for the HBV pre-S gene region was assessed according to a published real-time PCR protocol (Pas et al., 2000), which yielded an 89-bp product. For HBV cccDNA amplification, we used TaqMan primers previously shown (Chen et al., 2004) to specifically amplify cccDNA using an AB7900 HT sequence detection system (Applied Biosystems) or the Light Cycler 480 instrument (Roche). Closely similar data (not shown) were also obtained using a second set of previously described, HBV cccDNA-specific primers (Malmstrom et al., 2012)
Nuclear extraction for cccDNA analysis
Trypsinized HepAD38 cells were collected by centrifugation, washed with PBS and the supernatant medium discarded. We then added 500 μl of hypotonic buffer (10 mM HEPES, 10mM NaCl, 1.5 mM MgCl2, 0.5 mM DTT) to the cell pellets, along with zirconium oxide beads (ZROB05, Next Advance), and the cytoplasmic membranes were disrupted using a Bullet Blender (Next Advance), set at speed “4”, for 15 sec. Nuclear pellets were then collected by centrifugation at 2500 rpm for 5 min at 4°C and total nuclear DNA extracted following the protocol in the DNeasy Blood & Tissue Kit (Qiagen).
Antiviral activity in HBV-CRISPR and HepAD38 systems
Four compounds were tested against the HepAD38 cells, including lamivudine (3TC), tenofovir disoproxil fumarate (TDF), entecavir (ETV), and the nucleocapsid assembly inhibitor heteroaryldihydropyrimidine (HAP-12). All the antivirals were synthesized in our laboratories. HepAD38 cells were seeded at 50,000 cells/well in collagen-coated 96-well plates. Test compounds were added to cells to a final concentration ranging from 0.001 to 10 μM.
Real-time PCR for HBV DNA species in HepAD38
On day 7, total DNA was purified from supernatant using a commercially available kit (DNeasy Blood & Tissue kit, Qiagen). The HBV DNA was amplified in a real-time PCR assay using the AB 7900HT sequence detection system (Applied Biosystems) or the LightCycler 480 (Roche) as described by Stuyver et al. (2002). All samples were tested in duplicate. The concentration of compound that inhibited HBV DNA replication by 50% (EC50) or 90% (EC90) was determined by linear regression. HBV log viral reduction was also determined.
Cytotoxiticy
Cell viability was checked using two assays at late time points. For HepaRG, cell viability was monitored on Day 17 post-infection using CellTiter-Glo One Solution (Promega), according to the manufacturer’s protocol. No differences were observed between controls and experimental samples (data not shown). For the HepAD38 cells, Real-Time PCR for mitochondrial DNA [cytochrome c oxidase subunit II (COXII)] and nuclear DNA (ribosomal DNA) was performed. The mitochondrial DNA (MtDNA) and nuclear DNA (ribosomal DNA, Applied Biosystems) were amplified in parallel in a real-time PCR assay, and the amount of target mtDNA was normalized to the amount of an endogenous control and was then calculated relative to the untreated control. Again, no evidence of cytotoxicity was observed (Table S1).
Supplementary Material
We inactivated the HBV genome by targeted DNA editing using CRISPR/Cas.
while no cytotoxicity was observed, HBV cccDNA was effectively downregulated.
This paper provides proof of principle for a novel approach to HBV treatment.
Acknowledgments
The authors thank Souphalone Luangsay for technical assistance. EMK was supported by a postdoctoral fellowship from Hofffmann-LaRoche. These studies were supported in part by funds from the Department of Veterans Affairs (RFS) and by NIH CFAR grant P30-AI050409 (RFS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barrangou R, Marraffini LA. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sze J, He ML. HBV cccDNA in patients’ sera as an indicator for HBV reactivation and an early signal of liver damage. World J Gastroenterol. 2004;10:82–85. doi: 10.3748/wjg.v10.i1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandri M, Lutgehetmann M. Mouse models of hepatitis B and delta virus infection. J Immunol Methods. 2014;410C:39–49. doi: 10.1016/j.jim.2014.03.002. [DOI] [PubMed] [Google Scholar]
- Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gripon P, Rumin S, Urban S, Le Seyec J, Glaise D, Cannie I, Guyomard C, Lucas J, Trepo C, Guguen-Guillouzo C. Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci U S A. 2002;99:15655–15660. doi: 10.1073/pnas.232137699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantz O, Parent R, Durantel D, Gripon P, Guguen-Guillouzo C, Zoulim F. Persistence of the hepatitis B virus covalently closed circular DNA in HepaRG human hepatocyte-like cells. J Gen Virol. 2009;90:127–135. doi: 10.1099/vir.0.004861-0. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Kaminski R, Yang F, Zhang Y, Cosentino L, Li F, Luo B, Alvarez-Carbonell D, Garcia-Mesa Y, Karn J, Mo X, Khalili K. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc Natl Acad Sci U S A. 2014;111:11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Kornepati AV, Goldstein M, Bogerd HP, Poling BC, Whisnant AW, Kastan MB, Cullen BR. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J Virol. 2014;88:11965–11972. doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Ladner SK. Hep AD38 Assay: A High-Throughput, Cell-Based Screen for the Evaluation of Compounds Against Hepatitis B Virus. Methods Mol Med. 2000;24:43–50. doi: 10.1385/1-59259-245-7:43. [DOI] [PubMed] [Google Scholar]
- Komatsu H. Hepatitis B virus: where do we stand and what is the next step for eradication? World J Gastroenterol. 2014;20:8998–9016. doi: 10.3748/wjg.v20.i27.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner SK, Otto MJ, Barker CS, Zaifert K, Wang GH, Guo JT, Seeger C, King RW. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother. 1997;41:1715–1720. doi: 10.1128/aac.41.8.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SR, Yang HC, Kuo YT, Liu CJ, Yang TY, Sung KC, Lin YY, Wang HY, Wang CC, Shen YC, Wu FY, Kao JH, Chen DS, Chen PJ. The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo. Mol Ther Nucleic Acids. 2014;3:e186. doi: 10.1038/mtna.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MC, Yu M, Zhang NL, Gong WB, Wang Y, Piao WH, Wang QH, Wang GQ. Dynamic analysis of hepatitis B virus DNA and its antigens in 2.2.15 cells. J Viral Hepat. 2004;11:124–129. doi: 10.1046/j.1365-2893.2003.00485.x. [DOI] [PubMed] [Google Scholar]
- Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, Remouchamps C, Chou WM, Thasler WE, Huser N, Durantel D, Liang TJ, Munk C, Heim MH, Browning JL, Dejardin E, Dandri M, Schindler M, Heikenwalder M, Protzer U. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343:1221–1228. doi: 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrom S, Larsson SB, Hannoun C, Lindh M. Hepatitis B viral DNA decline at loss of HBeAg is mainly explained by reduced cccDNA load--down-regulated transcription of PgRNA has limited impact. PLoS One. 2012;7:e36349. doi: 10.1371/journal.pone.0036349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pas SD, Fries E, De Man RA, Osterhaus AD, Niesters HG. Development of a quantitative real-time detection assay for hepatitis B virus DNA and comparison with two commercial assays. J Clin Microbiol. 2000;38:2897–2901. doi: 10.1128/jcm.38.8.2897-2901.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci U S A. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuyver LJ, Lostia S, Adams M, Mathew JS, Pai BS, Grier J, Tharnish PM, Choi Y, Chong Y, Choo H, Chu CK, Otto MJ, Schinazi RF. Antiviral activities and cellular toxicities of modified 2′,3′-dideoxy-2′,3′-didehydrocytidine analogues. Antimicrob Agents Chemother. 2002;46:3854–3860. doi: 10.1128/AAC.46.12.3854-3860.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werle-Lapostolle B, Bowden S, Locarnini S, Wursthorn K, Petersen J, Lau G, Trepo C, Marcellin P, Goodman Z, Delaney WEt, Xiong S, Brosgart CL, Chen SS, Gibbs CS, Zoulim F. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology. 2004;126:1750–1758. doi: 10.1053/j.gastro.2004.03.018. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.