Abstract
Specific CD8+ T cells (CTLs) play an important role in resolving protracted infection with hepatitis B and C virus in humans and lymphocytic choriomeningitis virus (LCMV) in mice. The contribution of individual CTL specificities to chronic virus control, as well as epitope-specific patterns in timing and persistence of antiviral selection pressure remain, however, incompletely defined.
To monitor and characterize the antiviral efficacy of individual CTL specificities throughout the course of chronic infection, we co-inoculated mice with a mixture of wildtype LCMV and genetically engineered CTL epitope-deficient mutant virus. A quantitative longitudinal assessment of viral competition revealed that mice continuously exerted CTL selection pressure on the persisting virus population. The timing of selection pressure characterized individual epitope specificities, and its magnitude varied considerably between individual mice.
This longitudinal assessment of “antiviral efficacy” provides a novel parameter to characterize CTL responses in chronic viral infection. It demonstrates remarkable perseverance of all antiviral CTL specificities studied, thus raising hope for therapeutic vaccination in the treatment of persistent viral diseases.
Introduction
Estimated 350 and 170 million people worldwide are chronically infected with hepatitis B (HBV) and C virus (HCV), respectively, and thirty million people live with HIV (1). The resulting morbidity and mortality render these diseases major global health challenges, while strategies for therapeutic viral clearance remain unsatisfactory. In accordance with initial observations in LCMV-infected mice (2, 3), cytotoxic CD8+ T lymphocytes (CTL) have been shown to play a key role in containing and resolving persistent viral infections in humans. HIV and HCV viremia decline when the antiviral CD8+ T cell response emerges (4-6). HIV long-term non-progression as well as spontaneous clearance of HCV is associated with so-called "protective" HLA molecules (7, 8), corroborating the role of MHC class I-restricted T cells in viral control. Moreover, CD8+ T cell depletion experiments in simian immunodeficiency virus-infected macaques and HBV- or HCV-infected chimpanzees underscore the importance of CTLs in the acute phase of infection, as well as in long-term control and acquired immunity (9, 10).
Infection of mice with the Clone 13 (Cl13) strain of lymphocytic choriomeningitis virus (LCMV) has demonstrated governing principles of CTL responses in chronic viral infection (11-13), finding subsequent confirmation in the aforementioned human diseases (1). As a common finding, persisting viral infections with continuous exposure to a high antigen burden tend to subvert specific CTL responses (1). Fig. 1A,B and Figs. S1A-C provide an illustration of reduced CTL functionality in Cl13 chronically infected mice, as compared to CTLs originating from acute infection with the Armstrong strain of LCMV. If not physically deleted (11), such CTLs gradually lose IL-2 and TNF-α production but tend to retain IFN-γ secretion and CD107a expression (12-14). “Exhaustion” therefore refers to mostly mono- to bifunctional CTL populations with a relative paucity in polyfunctional cells, and is associated with the upregulation of inhibitory receptors such as programmed cell death 1 (PD-1, Fig. 1B, (15)).
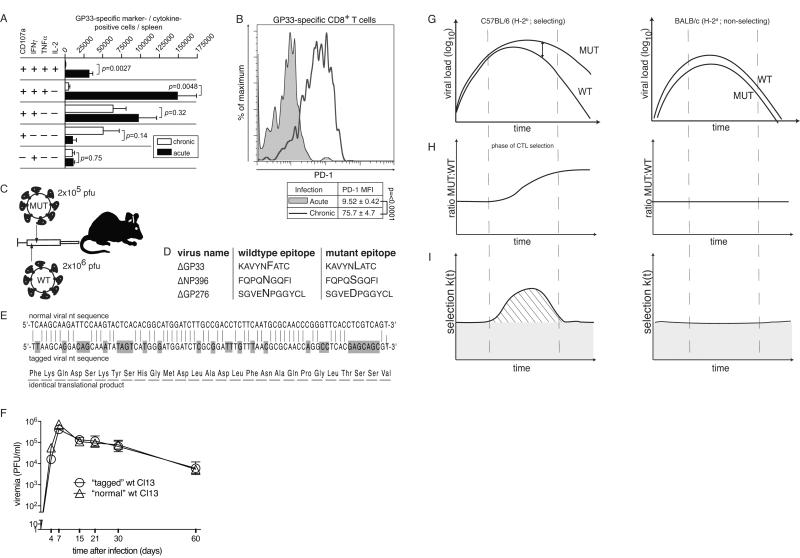
Figure 1. Characterization of exhausted CTLs and experimental approach for assessing their antiviral efficacy in protracted infection.
A-B: We infected C57BL/6 mice with LCMV Armstrong (acute) or LCMV strain Clone 13 (chronic). On day 20 we assessed GP33-specific CTLs by intracellular cytokine assay (A) and surface PD-1 expression (B). Bars in (A) represent the mean±SEM of three mice per group. Frequent cytokine-/marker-combinations are displayed (complete data sets in Fig. S1A-C). MFI in (B) is displayed as mean±SD of three mice. C: Experimental approach: Mice were co-infected with CTL epitope-mutant (MUT) and wildtype (WT) virus at the indicated dose. D: Amino acid sequences of the wildtype and mutant CTL epitopes. E: target sequences of the TaqMan RT-PCR assays utilized to individually quantify epitope-mutant virus (normal viral nt sequence) and wildtype virus (tagged viral nt sequence) resulting in the same translation product. F : We infected C57BL/ mice with 2×105 PFU of either « tagged » wt Cl13 or « normal » wt Cl13 and monitored viremia over time. G-I: Schematic of viral load curves (F) with epitope-mutant:wildtype virus ratios (G) and time dependent selection co-efficients k(t) of prototypic selecting (C57BL/6, H-2b) or non-selecting (BALB/c, H-2d) hosts. The shaded grey area in (I) denotes the 95% confidence interval of background k(t) determined in BALB/c mice.
The course of Cl13 infection in C57BL/6 mice has resemblance to spontaneous HBV or HCV control in humans in that the immune system eventually prevails and clears the virus from blood after 2-3 months (16). The underlying mechanisms remain, however, incompletely understood. Specific antibody responses and T follicular helper cell-driven germinal center reactions are necessary but not sufficient to resolve protracted infection (17, 18). C57BL/6 (H-2b) mice also mount a very potent and broad antiviral CTL response, comprising 28 known epitope specificities (19). MHC class I deficiency results in life-long viral persistence, documenting the key contribution of CTLs to control of chronic LCMV infection (3). However, several factors have rendered it difficult to address the contribution of individual CTL specificities to clearance: I) Virus strain and dose as well as host parameters influence LCMV persistence. In settings of life-long viremia, LCMV-specific CTLs may eventually be deleted (11, 20). Conversely, CTL function recovers when cognate antigen levels decline in Cl13-infected C57BL/6 mice, irrespective of the mechanism for viral clearance (21, 22). Thus, resurgence of CTL functionality might be a cause or a consequence of declining viremia. II) CTL epitope-mutant viruses evoke compensatory alterations in CTL immunodominance hierarchies (23). Therefore, a comparison of clearance kinetics of epitope-mutant and wild type viruses, when individually administered to animals, cannot assess the efficacy of the respective CTL population. III) Immunodominance hierarchies change significantly as chronic infection progresses (13). Hence the undisputed role of CTLs for chronic virus control, as established by CD8+ T cell depletion and gene knockout in experimental animals (2, 24), might have reflected the efficacy of subdominant CTL specificities (25), which surface during protracted infection and tend to be more functional than early immunodominant ones (23). The combined evidence of several recent reports supported the idea that the phenotypically “exhausted” CTL response is of functional relevance (14, 26-28). Still, the protective efficacy of individual antiviral CTL specificities and their contribution to resolution of chronic virus infection remained to be defined.
Here we exploited viral reverse genetics for a novel methodological approach to assess the antiviral selection pressure exerted by individual CTL specificities of chronically infected mice. This quantitative analysis of the extent, timing and inter-individual variability of protective CTL efficacy lends a novel perspective on CTL-mediated control of chronic viral infection.
Materials and Methods
Ethics statement
Animal experiments were performed at the Universities of Geneva and Zurich, and have been approved by the Direction générale de la santé (permissions 1005/3312/2 and 1005/3312/2-R) of the Canton of Geneva, and the Cantonal Veterinary Office of the Canton of Zurich (permission 176/2005), respectively. All animal experiments were performed in accordance with the Swiss law for animal protection.
Mice
C57BL/6, H-2Db−/− and H-2Kb−/− (29) mice were bred at the Institute for Laboratory Animal Sciences of the University of Zurich, Switzerland and BALB/c mice were obtained from Harlan laboratories. All mice were maintained in specific pathogen-free conditions.
Generation and titration of viruses, antibody neutralization and infection of mice
LCMV Clone 13 (Cl13) and variants thereof were generated from cDNA by reverse genetic techniques (30). To generate a « tagged » virus for discrimination from a competitor virus by TaqMan RT-PCR (Fig. 1C,E) and to introduce single amino acid changes resulting in deletion of select CTL epitopes (Fig. 1D, Fig. S1E-I) we used standard two-way site-directed mutagenesis on the LCMV S segment expression plasmid pI-S-Cl13(−)* (30) that was used for the generation of LCMV Cl13 from cDNA (primer sequences avilable from the authors upon request). The codons of the mutated amino acids were chosen to be as different from the wildtype amino acid’s codons as possible, to avoid viral reversion by a single nucleotide change. All viruses were grown on BHK-21 cells. Infectious titers and the potency of antiviral antibodies were assessed by immunofocus assay and focus reduction neutralization tests, respectively, as previously described (30, 31). Mice were infected with a 10:1 ratio of wildtype : epitope-mutant virus, i.e.2×106 PFU and 2×105 PFU, respectively, mixed for intravenous (i.v.) co-administration in one single syringe. When given as individual infection, Cl13 was given at a dose of 200 PFU (Fig. S1E-I) or 2×106 PFU i.v. (Fig. 1, Fig. S1A-C), and 105 PFU LCMV Armstrong were administered intraperitoneally (i.p.).
T cell assays
Intracellular cytokine assays were performed on splenocytes as previously described (30, 32). For the assessment of degranulation, anti-CD107a antibody was added to the restimulation culture at a concentration of 1 μg/ml. Peptides (90% pure) of the sequences displayed in Fig. 1D and Fig. S1E-I were purchased from ProImmune. MHC class I tetramers (GP33: H-2Db/KAVYNFATC; GP276: H-2Db/ SGVENPGGYCL) were supplied by Beckman Coulter. Antibodies were from BD biosciences and Biolegend. Flow cytometry was performed on BD FACSCalibur and Beckman Coulter Gallios flow cytometers.
Assessment of viral loads by TaqMan RT-PCR
Mouse serum was collected into microtainer tubes (BD Biosciences) and viral RNA was extracted using the Qiagen 96 DNA blood kit. TaqMan RT-PCR was performed using Invitrogen Superscript III One Step Platinum Taq. Primers and probes were ordered through Sigma Aldrich. To detect the natural Cl13 NP sequence (present in epitope-escape variants) we utilized fwd primer 5`-ACTGACGAGGTCAACCCGG-3’, reverse primer 5`-CAAGTACTCACACGGCATGGA-3’ and the probe 5`-FAM- CTTGCCGACCTCTTCAATGCGCAA-BHQ1-3’ (33). For detection of the “tagged” sequence (in epitope-sufficient wildtype virus, utilized in competition experiments), we designed fwd primer 5'-TGCTCGTGAGGCCTGGTT-3', reverse primer 5'-TTAAGCAGGACAGCAAATATAGTCATG-3' and the probe 5'-HEX-caaTttGtTTaggCgc-BHQ1-3' (small letters indicating locked nucleotides). Measurements were performed on an ABI TaqMan device. Absolute viral RNA copies per 3.3 μl of serum were calculated by comparison to in vitro transcript RNAs (compare Fig. S1J-L), which served as reference in all TaqMan assays. Further details on cycling conditions and the generation of in vitro transcripts are available from the authors upon request.
Quantification of epitope-specific CTL selection pressure
(a) The mathematical model to determine selection
To infer the selection pressure exerted by CTLs specific for individual epitopes, we used the following mathematical model published by Ganusov and De Boer (34). Wildtype, w(t), and epitope-mutant virus, m(t), are assumed to grow at a time-dependent rate r(t), and die at a common time-dependent death rate d(t). In addition, the wildtype virus can be killed at a time-dependent killing rate k(t) denoting the epitope-specific CTL response. The change of wildtype and epitope-mutant virus over time was then described by the following system of ordinary differential equations.
| (1) |
| (2) |
By considering the ratio of wildtype and epitope-mutant virus we could directly estimate the time-dependent CTL selection pressure (TDSP), k(t), exerted on the epitope in question. Defining z(t) = w(t)/m(t) we obtained
| (3) |
Thus, the selection pressure could be obtained from the change over time of the log ratio of wildtype and epitope-mutant virus. Since we were only interested in the change of ln(z(t)) over time, the initial ratio between wildtype and epitope-mutant was irrelevant. The parameter k(t) denotes the net-selection pressure, i.e., the difference between the selective advantage of the epitope-mutant compared to the wildtype, due to the escaped CTL response, and the disadvantage due, for example, to fitness costs. Hence, negative values of k(t) could indicate that the fitness cost of the epitope-mutant outweighs its selective advantage over the wildtype. In most instances, however, negative k(t) values are due to viral loads falling below detection limits, thus preventing an accurate determination of wildtype : mutant virus ratios at the respective time points (outlined in more detail in section (c) below). It is equally conceivable that mechanisms such as transient and compartmentalized CTL selection pressure result in periods of negative k(t).
(b) Application to the data
The time-dependent selection curve k(t) for each individual mouse were determined by using Eq. (3). A smooth cubic spline function was fitted to the individual data points given by −ln(w(ti)/m(ti)), where w(ti) and m(ti), i=1,…,n denote the measured viral load for wildtype and epitope-mutant virus, respectively, at time point ti. The time-dependent selection curves k(t) were then estimated by the first derivative of the obtained spline function. Measurements of wildtype and epitope-mutant virus below the detection limit (100 copies / 3.3 μl) were set to half of the detection limit (50 copies/ 3.3 μl). Using other values for the replacement, e.g. setting measurements below the detection limit to 99 copies / 3.3 μl did not change the results overall.
(c) Calculating the average selection coefficient κ
We assumed that the observed variation in the log-ratio between wildtype and epitope-mutant virus in BALB/c mice represented effects unrelated to the epitope specific CTL response. Therefore, for each epitope, we used the BALB/c mice to calculate a function, kBG(t), representing background noise in k(t) due to fluctuation of ratios of wildtype and epitope-mutant virus in the absence of selection and also reflecting potential fitness costs for the epitope-mutant viruses. To this end, a smooth cubic spline function was fitted to the pooled data of all BALB/c mice tested for a given epitope, and the first derivative was estimated to obtain kBALB/c(t). The “background-selection” function, kBG(t), was then determined by performing a bootstrap analysis with 10,000 replicates re-sampling from the data of the BALB/c mice and taking the time-wise 97.5% upper limit of the obtained values of kBALB/c(t).
To calculate the average selection coefficient over time for each individual mouse we considered only those parts of the individual selection curves where the individual selection was larger than the estimated background noise, k(t)> kBG(t). Hence, the average selection coefficient κ until time point T was calculated by
| (4) |
with k+(t)= k(t)−kBG(t) if k(t)> kBG(t) and 0 otherwise. Hereby, t+ denoted the total length of the time intervals in [0,T] where k(t)> kBG(t). By only considering the time intervals where the individual selection was larger than the background selection, k(t)>kBG(t), we accounted for the fact that negative selection can be due to viral loads falling below detection limits (see e.g. wt virus in strongly selecting C57BL/6 mice; top panels of Fig. 2). This prevented an accurate determination of wildtype : mutant virus ratios at the respective time points, with TDSP artificially turning negative. By considering only k(t)>kBG(t) values, our method to calculate the average selection coefficient κ focused on unambiguous signals of selection.
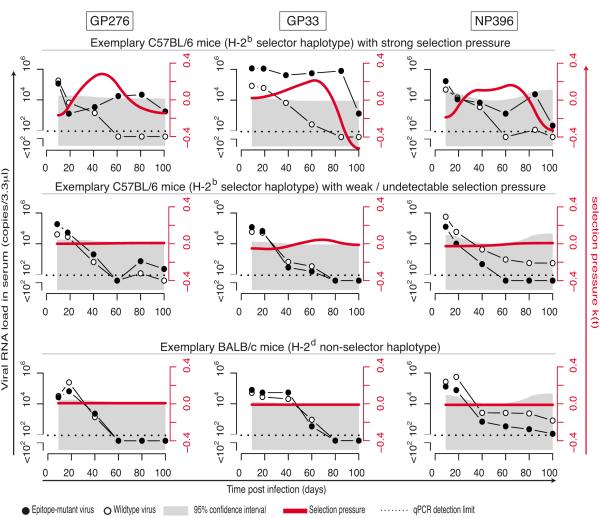
Figure 2. Epitope-specific CTL selection pressure in protracted LCMV infection.
We infected C57BL/6 (top and center) and BALB/c mice (bottom) with 2×05 PFU of either ΔGP276, ΔGP33 or ΔNP396 virus, in combination with 2×106 PFU of wildtype LCMV (see schematic in Fig. 1C). Each virus was individually quantified in serum by TaqMan RT-PCR. Time-dependent selection coefficients k(t) (red lines) were calculated (see Materials & Methods and schematic in Fig. 1G-I). The grey shaded area denotes the 95% confidence interval of background k(t) in the absence of epitope-specific selection pressure. Representative C57BL/6 mice with either strong (top) or weak/absent selection pressure (center) are shown in comparison to non-selecing BALB/c mice (bottom). Analogous plots for all mice tested are displayed in Figs. S2-S4.
(d) Time-dependent selection bands for C57BL/6 mice
Analogously to the function representing background noise in k(t) (due to fluctuation in ratios of wildtype and epitope-mutant virus in the absence of selection), time-dependent selection bands were calculated for the C57BL/6 mice, individually for each epitope. A smooth cubic spline function was fitted to the pooled data of all C57BL/6 mice for one epitope, and the first derivative was estimated to obtain kC57BL/6(t). A 95% confidence band for kC57BL/6(t) was calculated by performing a bootstrap analysis with 10,000 replicates re-sampling from the data of the C57BL/6 mice and taking the time-wise 97.5% upper limit and 2.75% lower limits of the obtained values of k C57BL/6 (t). For these latter analyses, only those C57BL/6 mice were considered, where the average selection pressure, κ, was larger than the maximal selection pressure determined for BALB/c mice.
Results
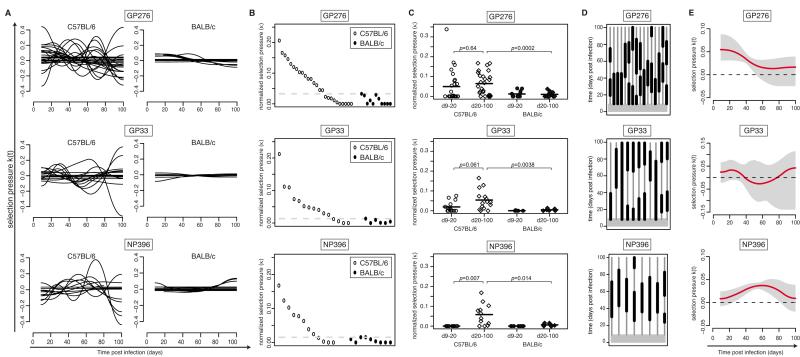
To determine the protective efficacy of individual CTL specificities in protracted LCMV infection we designed viral in vivo competition experiments (Fig. 1C). We infected C57BL/6 mice with 2×106 PFU of wildtype cDNA-derived Cl13 virus, in combination with 2×105 PFU of genetically engineered CTL epitope-mutant virus (see below). In this setting, a relative enrichment of the epitope-mutant virus over time would be evidence of CTL selection pressure on the epitope in question. We engineered CTL epitope-mutant viruses, differing from the wildtype virus by only one amino acid substitution in the GP276, GP33 or NP396 CTL epitope, respectively (Fig. 1D). Each of these mutations were known and/or predicted to prevent MHC presentation to CD8+ T cells (H-2Db for GP276 and NP396; H-2Db and H-2Kb for GP33 (35)). Conversely, these mutations did not affect their neutralization by monoclonal antibodies or virus-specific antiserum (Fig. S1D). This was expected since neutralizing antibodies are uniformely targeted against the outer globular GP-1 domain of the viral glycoprotein complex whereas the mutations introduced were situated in the membrane-proximal glycoprotein stalk (GP276), signal peptide (GP33) and virion-internal nucleoprotein (NP396). We validated the epitope-mutant viruses by verifying their inability to induce CTL responses against the wildtype and mutated epitope sequences and we also tested the lack of recognition of the mutant epitopes by wildtype virus-induced CTLs (Fig. S1E-I). To individually quantify wildtype and epitope-mutant viruses in co-infected animals, we « tagged » the wildtype virus by introducing non-coding changes in the target sequence of our TaqMan RT-PCR assay (Fig. 1E). As expected, this « tagged » wt virus reached normal titers and persisted in the blood of mice analogously to non-tagged wt virus (Fig. 1F). The respective TaqMan assays were validated for accuracy and discrimination of the two viral target sequences (Fig. S1J-L; Materials and Methods), and we utilized them to individually follow serum loads of epitope-mutant and wildtype virus in co-infected animals over time (Fig. 1G). From these values, we calculated ratios of epitope-mutant:wildtype virus (Fig. 1H) and derived a time-dependent selection coefficient k(t), which quantifies the time-dependent selection pressure (TDSP) CTLs exert on the epitope under study (Fig. 1I; see also Materials and Methods). Given the MHC molecules presenting these epitopes, CTL-driven selection of epitope-mutant virus was only expected in C57BL/6 (H-2b) but not in BALB/c mice of the H-2d haplotype (control). Cohorts of 26, 15 and 12 inbred C57BL/6 mice were subject to coinfection experiments with GP276-, GP33- or NP396-deficient epitope-mutant and wildtype competitor virus, respectively. We collected serum samples over time to monitor viral loads and calculate TDSP by GP276-, GP33- and NP396-specific CTLs. Viral load curves and deducted selection coefficients are shown in Figure 2 (representative animals) and Figs. S2-S4. 6-10 BALB/c mice per epitope were included as controls. As expected, wt and epitope-mutant viruses showed largely parallel clearance kinetics in these H-2d non-selecting mice. There were minor residual fluctuations in epitope-mutant:wildtype virus ratios. These were used to calculate a time-dependent background noise in the selection coefficient k(t) for each epitope, which we defined as being unrelated to specific CTL selection pressure (gray area in Fig. 2 and Figs. S2-S4). Unlike in BALB/c mice, C57BL/6 mice tended to clear the wt virus more readily than GP276-, GP33- or NP396-deleted viruses, indicating selection pressure on the respective epitopes. The timing and magnitude of selection pressure (red lines in Fig. 2 and Figs. S2-S4) exhibited, however, extensive diversity between individual animals. Exemplary C57BL/6 mice with either strong or weak to undetectable selection pressure as shown in Fig. 2 represented the extremes of a spectrum (Figs. S2-S4). This finding matched earlier observations that Cl13 control in the late phase of infection varies considerably within cohorts of genetically identical mice (17, 20). The resulting diversity of individual TDSP curves is illustrated in Fig. 3A. Conversely, very minor fluctuations of TDSP in BALB/c mice attest to the technical accuracy and overall reliability of the measurements and experimental set-up. Note that periods of negative TDSP in C57BL/6 mice were mostly due to viral loads falling below detection limits (see e.g. Fig. 2 top panels and Materials & Methods). The LCMV polymerase is error-prone, and viral variants with increased fitness can be selected for in chronic infection. Such random mutation and selection occurs, however, irrespective the MHC haplotype of the host. Consistently low TDSP values in BALB/c mice indicate therefore that the selection of epitope-mutant viruses, as observed in C57BL/6 mice, was not due to random mutations but resulted from MHC-restricted CTL pressure on the very epitope, which had been deliberately mutated. Next we calculated the area under each mouse’ individual selection curve (striped in Fig. 1I; see Materials & Methods). Thereby we obtained an average value for epitope-specific CTL pressure (κ) during time intervals the TDSP curve was above background. The resulting values showed that about 60-75% of C57BL/6 mice exerted CTL selection pressure on GP276, GP33 and NP396 above BALB/c backgrounds (16/26, 11/15 and 9/12 mice tested for GP276, GP33 and NP396, respectively; Fig. 3B). When broken down in early and late phases of chronic infection (day 9-20 vs. day 20-100), the average epitope-specific CTL selection pressure (κ) on GP276 and GP33 did not differ significantly between the two time windows, yet for NP396 was higher in the late phase of infection (Fig. 3C). Furthermore, the average CTL selection pressure in the late phase of infection (day 20-100) was significantly above BALB/c backgrounds for all three epitopes tested. These results documented that antiviral CTL selection pressure was not lost during the chronic phase of infection, which is in line with earlier reports on at least partially retained CTL killing capacity during chronic Cl13 infection (20, 27). Next we analyzed the timing of CTL selection pressure at the level of individual C57BL/6 mice. We confirmed the trend for early onset CTL pressure on the GP276 and GP33 epitopes, whereas NP396-specific CTL selection tended to occur later (Fig. 3D). A bootstrap analysis was performed to investigate overarching patterns in TDSP, which characterize its timing across all mice despite considerable inter-individual variability. The TDSP 95% confidence interval (gray in Fig. 3E) confirmed that C57BL/6 mice on average exerted GP276-specific CTL pressure during the first period of persistent infection up until day 50. The average GP33-specific selection pressure was biphasic, with early and late peaks. This pattern may be reflective of two distinct GP33-specific CTL populations restricted by H2-Kb- and H2-Db, respectively, yet prevented a meaningful statistical assessment of CTL timing. In concordance with the results of Fig. 3C, NP396-specific CTL efficacy was significant at the cohort level in a later phase between around day 30-80 of infection (Fig. 3E). Measurements of GP276-specific CTL frequencies in peripheral blood have failed to reveal a correlation with normalized epitope-specific CTL pressure (κ, Fig. 4). This finding was compatible with earlier observations that the quantitative assessment of virus-specific CTLs in peripheral blood of HIV patients showed little correlation to viral loads (36 , 37).
Figure 3. Interindividual diversity, magnitude and timing of epitope-specific CTL selection pressure.
A: Superimposition of k(t) curves from individual C57BL/6 (left) and BALB/c (right) mice, assessing selection pressure on the respective epitopes. B: Symbols represent the average selection pressure, κ, for individual C57BL/6 (white circles) and BALB/c mice (black circles) on the indicated epitopes over 100 days. Horizontal dashed lines denote the respective highest values recorded in BALB/c mice. C: Symbols represent the average epitope-specific selection pressure, κ, for individual C57BL/6 (white symbols) and BALB/c mice (black synbols) calculated for the period from day 9-20 or from day 20-100, as indicated. For pair-wise comparisons paired t tests (C57BL/6 d9-20 vs. C57BL/6 d20-100) and unpaired t tests (C57BL/6 d20-d100 vs. BALB/c d20-d100) were used. The resulting p-values were subject to Bonferroni correction for multiple comparisons. D: Time windows during which the TDSP k(t) curves of individual mice (Fig. 2 and Figs. S2-S4) are above the 95% confidence interval of background noise (see Methods). Individual mice are displayed as vertical lines, with thick segments denoting detectable selection pressure. Only C57BL/6 mice with average selection coefficients above BALB/c backgrounds (dashed lines in Fig. 3B) were included in the analysis. E: Derived TDSP based on all mice shown in (D). The mean (red line) and 95% confidence interval (grey shaded) of 10,000 bootstrap replicates are shown. The dashed line indicates the zero reference line, above which selection is recorded.
Figure 4. Lack of correlation between normalized GP276-specific selection pressure and GP276-specific CD8+ T cell frequencies in peripheral blood.
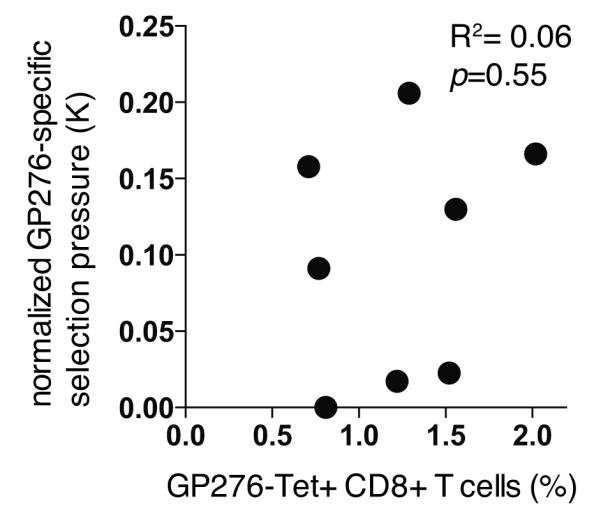
We infected C57BL/6 with 2×105 PFU of ΔGP276 virus in combination with 2×106 PFU of wildtype LCMV as outlined in Fig. 2 and Fig. S2. In one out of 3 analogous experiments summarized in this manuscript, we determined GP276-specific CD8+ T cell frequencies in peripheral blood on day 40 after infection by using MHC class I tetramers. Symbols represent individual mice. Normalized GP276-specific selection pressure as displayed in Fig. 3B is plotted against the percentage Tet+ cells amongst CD8+B220- lymphocytes. Pearson correlation coefficient and p-value are indicated.
Discussion
The present quantification and characterization of antiviral CTL efficacy adds to our understanding how the immune system can prevail in chronic infection. Immunotherapeutic interventions bear considerable potential for treating persistent viral diseases, notably in HBV (38), and should aim for mimicking successful immune responses in spontaneous controllers.
The mechanisms underlying differential epitope-specific timing of selection pressure remain to be investigated. Several contributing factors can be envisioned including i) viral protein abundance (39) and resulting peptide ligand availability on target cells, ii) peptide – MHC binding affinity (23) and iii) the relative susceptibility of responding CTL populations to clonal deletion (12, 13). A better understanding thereof should help in the choice of target antigens and epitopes for antiviral immunotherapy.
Inter-individual variability of CTL selection pressure in genetically identical animals, both in terms of strength and timing, matches earlier reports from TCR spectratype analyses in Cl13-infected mice (40). Differences in T cell repertoire due to stochastic events in TCR rearrangement and subsequent thymic selection may account for this, and were reported to influence HIV control (41). Similarly, de novo recruitment of thymic emigrants into the ongoing CTL response may contribute to time-dependent fluctuations in CTL selection pressure of individual mice (42).
Our observations suggest that therapeutic induction of antiviral CTL responses in chronic infection, even if partially dysfunctional by commonly used ex vivo criteria, can represent a correlate of immune control and a valuable goal of such interventions. This interpretation is in line with earlier reports that viremic Cl13-infected mice reject GP33- or NP396-pulsed syngeneic splenocytes in so-called «in vivo CTL assays». Similarly, NP396-specific CTLs were found to resurge in the late phase of infection (20 , 27), matching the timing of selection pressure on this epitope found here. When adoptively transferred and rested in an antigen-free environement, phenotypically « exhausted » CTLs can afford protection against subsequent virus challenge (28). Our report adds to these earlier observations by establishing and characterizing the protective antiviral efficacy of CTL populations in the chronically infected host.
Exhaustion is a gradual process (1, 13) and invidual cells of exhausted CTL populations cover a wide range of differential functionality (compare Fig. 1A,B and Fig. S1A-C). Hence, it seems plausible that subsets of more functional cells contributed overproportionally to the epitope-specific CTL pressure. It also is possible that fluctuations in CTL functionality account for the observed « waves » of TDSP.
The novel approach outlined herein provides a means to directly determine the impact of defined CTL specificities on viral loads, thus lending itself to future studies aimed at validating immune correlates of CTL efficacy. Specific CTL numbers are likely of relevance but their abundance in peripheral blood seems insufficient a predictor for antiviral efficacy (Fig. 4, (36, 37, 43)). Potential qualitative correlates of CTL efficacy comprise, among others, functional avidity (44), proliferative capacity (45), polyfunctionality (46), lytic granule loading (47), perforin expression (48), resistance to immunoregulation (49-51) and TCR clonotype composition (41). Unfortunately, peripheral blood may not represent the most relevant compartment to monitor in this context, albeit clearly the only option in longitudinal studies. This possibility is suggested by a dramatic skewing of virus-specific CTLs to LCMV-infected tissues such as bone marrow, liver, lungs and brain, in combination with compartment-related differences in CTL functionality (13). A timely assessment of such tissue compartments would require the experimental animal to be euthanized. This conflicts with the assessment of CTL efficacy, relying on the longitudinal sampling of serum from the same animals until CTL-mediated control is evident in reduced viral loads. Conversely, an analysis of CTL populations at such a later stage of the experiment would be confounded by differential antigen levels, since high viral antigen burden per se is known to compromise the functionality of antiviral CTLs (52). One potential strategy to overcome these limitations may consist in the identification of gene expression profiles or clonotype composition as biomarkers of CTL efficacy, an endeavor for which the technological approach presented herein seems well suited.
Taken together, this longitudinal assessment of epitope-specific CTL selection pressure adds to our understanding how the immune system can prevail in chronic viral infection. Our findings are encouraging and should help in the rational design of CTL-based immunotherapies for persistent viral diseases.
Supplementary Material
Acknowledgements
We thank M. Fernandez, E. Horvath and M. Lu for excellent technical support, R. Sommerstein, R. Zinkernagel and J. Luban for discussions.
Non-standard abbreviations
- LCMV
lymphocytic choriomeningitis virus
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- Cl13
LCMV strain Clone 13
- PD-1
programmed cell death 1
Footnotes
S.J was an NHMRC fellow #628934. A.B. was an EMBO long-term fellow, held a Boehringer Ingelheim Fonds PhD scholarship, a Roche Research Foundation postdoctoral scholarship and was funded by the SSMBS, Swiss National Science Foundation (SNF) and Austrian Academy of Sciences. F.G. was supported by NIH grant AI028433 and the Center for Modeling and Simulation in the Biosciences. L.F. was an SSMBS fellow. RRR was funded by the SNF (grant number 315230-130855) and D.D.P was supported by an SNF professorship.
References
- 1.Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138:30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 2.Fung-Leung WP, Kundig TM, Zinkernagel RM, Mak TW. Immune response against lymphocytic choriomeningitis virus infection in mice without CD8 expression. J Exp Med. 1991;174:1425–1429. doi: 10.1084/jem.174.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matloubian M, Concepcion RJ, Ahmed R. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J Virol. 1994;68:8056–8063. doi: 10.1128/jvi.68.12.8056-8063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koup RA, Safrit JT, Cao Y, Andrews CA, McLeod G, Borkowsky W, Farthing C, Ho DD. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thimme R, Oldach D, Chang KM, Steiger C, Ray SC, Chisari FV. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med. 2001;194:1395–1406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goulder PJ, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2008;8:619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKiernan SM, Hagan R, Curry M, McDonald GS, Kelly A, Nolan N, Walsh A, Hegarty J, Lawlor E, Kelleher D. Distinct MHC class I and II alleles are associated with hepatitis C viral clearance, originating from a single source. Hepatology. 2004;40:108–114. doi: 10.1002/hep.20261. [DOI] [PubMed] [Google Scholar]
- 9.Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, Ghrayeb J, Forman MA, Montefiori DC, Rieber EP, Letvin NL, Reimann KA. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 10.Thimme R, Wieland S, Steiger C, Ghrayeb J, Reimann KA, Purcell RH, Chisari FV. CD8(+) T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J Virol. 2003;77:68–76. doi: 10.1128/JVI.77.1.68-76.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moskophidis D, Lechner F, Pircher H, Zinkernagel RM. Virus persistence in acutely infected immunocompetent mice by exhaustion of antiviral cytotoxic effector T cells. Nature. 1993;362:758–761. doi: 10.1038/362758a0. [DOI] [PubMed] [Google Scholar]
- 12.Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, Ahmed R. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med. 1998;188:2205–2213. doi: 10.1084/jem.188.12.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol. 2003;77:4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agnellini P, Wolint P, Rehr M, Cahenzli J, Karrer U, Oxenius A. Impaired NFAT nuclear translocation results in split exhaustion of virus-specific CD8+ T cell functions during chronic viral infection. Proc Natl Acad Sci U S A. 2007;104:4565–4570. doi: 10.1073/pnas.0610335104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 16.Rehermann B, Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol. 2005;5:215–229. doi: 10.1038/nri1573. [DOI] [PubMed] [Google Scholar]
- 17.Bergthaler A, Flatz L, Verschoor A, Hegazy AN, Holdener M, Fink K, Eschli B, Merkler D, Sommerstein R, Horvath E, Fernandez M, Fitsche A, Senn BM, Verbeek JS, Odermatt B, Siegrist CA, Pinschewer DD. Impaired antibody response causes persistence of prototypic T cell-contained virus. PLoS Biol. 2009;7:e1000080. doi: 10.1371/journal.pbio.1000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harker JA, Lewis GM, Mack L, Zuniga EI. Late Interleukin-6 Escalates T Follicular Helper Cell Responses and Controls a Chronic Viral Infection. Science. 2011 doi: 10.1126/science.1208421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotturi MF, Peters B, Buendia-Laysa F, Jr., Sidney J, Oseroff C, Botten J, Grey H, Buchmeier MJ, Sette A. The CD8+ T-cell response to lymphocytic choriomeningitis virus involves the L antigen: uncovering new tricks for an old virus. J Virol. 2007;81:4928–4940. doi: 10.1128/JVI.02632-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuller MJ, Khanolkar A, Tebo AE, Zajac AJ. Maintenance, loss, and resurgence of T cell responses during acute, protracted, and chronic viral infections. J Immunol. 2004;172:4204–4214. doi: 10.4049/jimmunol.172.7.4204. [DOI] [PubMed] [Google Scholar]
- 21.Blattman JN, Wherry EJ, Ha SJ, van der Most RG, Ahmed R. Impact of epitope escape on PD-1 expression and CD8 T-cell exhaustion during chronic infection. J Virol. 2009;83:4386–4394. doi: 10.1128/JVI.02524-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brooks DG, McGavern DB, Oldstone MB. Reprogramming of antiviral T cells prevents inactivation and restores T cell activity during persistent viral infection. J Clin Invest. 2006;116:1675–1685. doi: 10.1172/JCI26856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Most RG, Murali-Krishna K, Lanier JG, Wherry EJ, Puglielli MT, Blattman JN, Sette A, Ahmed R. Changing immunodominance patterns in antiviral CD8 T-cell responses after loss of epitope presentation or chronic antigenic stimulation. Virology. 2003;315:93–102. doi: 10.1016/j.virol.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 24.Matano T, Shibata R, Siemon C, Connors M, Lane HC, Martin MA. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J Virol. 1998;72:164–169. doi: 10.1128/jvi.72.1.164-169.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frahm N, Kiepiela P, Adams S, Linde CH, Hewitt HS, Sango K, Feeney ME, Addo MM, Lichterfeld M, Lahaie MP, Pae E, Wurcel AG, Roach T, St John MA, Altfeld M, Marincola FM, Moore C, Mallal S, Carrington M, Heckerman D, Allen TM, Mullins JI, Korber BT, Goulder PJ, Walker BD, Brander C. Control of human immunodeficiency virus replication by cytotoxic T lymphocytes targeting subdominant epitopes. Nat Immunol. 2006;7:173–178. doi: 10.1038/ni1281. [DOI] [PubMed] [Google Scholar]
- 26.Paley MA, Kroy DC, Odorizzi PM, Johnnidis JB, Dolfi DV, Barnett BE, Bikoff EK, Robertson EJ, Lauer GM, Reiner SL, Wherry EJ. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science. 2012;338:1220–1225. doi: 10.1126/science.1229620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graw F, Richter K, Oxenius A, Regoes RR. Comparison of cytotoxic T lymphocyte efficacy in acute and persistent lymphocytic choriomeningitis virus infection. Proc Biol Sci. 2011;278:3395–3402. doi: 10.1098/rspb.2011.0453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Utzschneider DT, Legat A, Fuertes Marraco SA, Carrie L, Luescher I, Speiser DE, Zehn D. T cells maintain an exhausted phenotype after antigen withdrawal and population reexpansion. Nat Immunol. 2013;14:603–610. doi: 10.1038/ni.2606. [DOI] [PubMed] [Google Scholar]
- 29.Perarnau B, Saron MF, San Martin BR, Bervas N, Ong H, Soloski MJ, Smith AG, Ure JM, Gairin JE, Lemonnier FA. Single H2Kb, H2Db and double H2KbDb knockout mice: peripheral CD8+ T cell repertoire and anti-lymphocytic choriomeningitis virus cytolytic responses. Eur J Immunol. 1999;29:1243–1252. doi: 10.1002/(SICI)1521-4141(199904)29:04<1243::AID-IMMU1243>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 30.Flatz L, Bergthaler A, de la Torre JC, Pinschewer DD. Recovery of an arenavirus entirely from RNA polymerase I/II-driven cDNA. Proc Natl Acad Sci U S A. 2006;103:4663–4668. doi: 10.1073/pnas.0600652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Battegay M, Cooper S, Althage A, Banziger J, Hengartner H, Zinkernagel RM. Quantification of lymphocytic choriomeningitis virus with an immunological focus assay in 24- or 96-well plates. J Virol Methods. 1991;33:191–198. doi: 10.1016/0166-0934(91)90018-u. [DOI] [PubMed] [Google Scholar]
- 32.Bergthaler A, Flatz L, Hegazy AN, Johnson S, Horvath E, Lohning M, Pinschewer DD. Viral replicative capacity is the primary determinant of lymphocytic choriomeningitis virus persistence and immunosuppression. Proc Natl Acad Sci U S A. 2010;107:21641–21646. doi: 10.1073/pnas.1011998107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinschewer DD, Flatz L, Steinborn R, Horvath E, Fernandez M, Lutz H, Suter M, Bergthaler A. Innate and adaptive immune control of genetically engineered live-attenuated arenavirus vaccine prototypes. Int Immunol. 2010;22:749–756. doi: 10.1093/intimm/dxq061. [DOI] [PubMed] [Google Scholar]
- 34.Ganusov VV, De Boer RJ. Estimating Costs and Benefits of CTL Escape Mutations in SIV/HIV Infection. PLoS Comput Biol. 2006;2:e24. doi: 10.1371/journal.pcbi.0020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuler MM, Nastke MD, Stevanovikc S. SYFPEITHI: database for searching and T-cell epitope prediction. Methods Mol Biol. 2007;409:75–93. doi: 10.1007/978-1-60327-118-9_5. [DOI] [PubMed] [Google Scholar]
- 36.Betts MR, Ambrozak DR, Douek DC, Bonhoeffer S, Brenchley JM, Casazza JP, Koup RA, Picker LJ. Analysis of total human immunodeficiency virus (HIV)-specific CD4(+) and CD8(+) T-cell responses: relationship to viral load in untreated HIV infection. J Virol. 2001;75:11983–11991. doi: 10.1128/JVI.75.24.11983-11991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Addo MM, Yu XG, Rathod A, Cohen D, Eldridge RL, Strick D, Johnston MN, Corcoran C, Wurcel AG, Fitzpatrick CA, Feeney ME, Rodriguez WR, Basgoz N, Draenert R, Stone DR, Brander C, Goulder PJ, Rosenberg ES, Altfeld M, Walker BD. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J Virol. 2003;77:2081–2092. doi: 10.1128/JVI.77.3.2081-2092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horiike N, Fazle Akbar SM, Michitaka K, Joukou K, Yamamoto K, Kojima N, Hiasa Y, Abe M, Onji M. In vivo immunization by vaccine therapy following virus suppression by lamivudine: a novel approach for treating patients with chronic hepatitis B. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2005;32:156–161. doi: 10.1016/j.jcv.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 39.Oldstone MB, Buchmeier MJ. Restricted expression of viral glycoprotein in cells of persistently infected mice. Nature. 1982;300:360–362. doi: 10.1038/300360a0. [DOI] [PubMed] [Google Scholar]
- 40.Lin MY, Welsh RM. Stability and diversity of T cell receptor repertoire usage during lymphocytic choriomeningitis virus infection of mice. J Exp Med. 1998;188:1993–2005. doi: 10.1084/jem.188.11.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen H, Ndhlovu ZM, Liu D, Porter LC, Fang JW, Darko S, Brockman MA, Miura T, Brumme ZL, Schneidewind A, Piechocka-Trocha A, Cesa KT, Sela J, Cung TD, Toth I, Pereyra F, Yu XG, Douek DC, Kaufmann DE, Allen TM, Walker BD. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nat Immunol. 2012;13:691–700. doi: 10.1038/ni.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vezys V, Masopust D, Kemball CC, Barber DL, O'Mara LA, Larsen CP, Pearson TC, Ahmed R, Lukacher AE. Continuous recruitment of naive T cells contributes to heterogeneity of antiviral CD8 T cells during persistent infection. J Exp Med. 2006;203:2263–2269. doi: 10.1084/jem.20060995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goulder PJ, Watkins DI. HIV and SIV CTL escape: implications for vaccine design. Nat Rev Immunol. 2004;4:630–640. doi: 10.1038/nri1417. [DOI] [PubMed] [Google Scholar]
- 44.Almeida JR, Sauce D, Price DA, Papagno L, Shin SY, Moris A, Larsen M, Pancino G, Douek DC, Autran B, Saez-Cirion A, Appay V. Antigen sensitivity is a major determinant of CD8+ T-cell polyfunctionality and HIV-suppressive activity. Blood. 2009;113:6351–6360. doi: 10.1182/blood-2009-02-206557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Migueles SA, Laborico AC, Shupert WL, Sabbaghian MS, Rabin R, Hallahan CW, Van Baarle D, Kostense S, Miedema F, McLaughlin M, Ehler L, Metcalf J, Liu S, Connors M. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat Immunol. 2002;3:1061–1068. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- 46.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, Roederer M, Koup RA. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Migueles SA, Osborne CM, Royce C, Compton AA, Joshi RP, Weeks KA, Rood JE, Berkley AM, Sacha JB, Cogliano-Shutta NA, Lloyd M, Roby G, Kwan R, McLaughlin M, Stallings S, Rehm C, O'Shea MA, Mican J, Packard BZ, Komoriya A, Palmer S, Wiegand AP, Maldarelli F, Coffin JM, Mellors JW, Hallahan CW, Follman DA, Connors M. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity. 2008;29:1009–1021. doi: 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hersperger AR, Pereyra F, Nason M, Demers K, Sheth P, Shin LY, Kovacs CM, Rodriguez B, Sieg SF, Teixeira-Johnson L, Gudonis D, Goepfert PA, Lederman MM, Frank I, Makedonas G, Kaul R, Walker BD, Betts MR. Perforin expression directly ex vivo by HIV-specific CD8 T-cells is a correlate of HIV elite control. PLoS Pathog 6: e1000917. 2010 doi: 10.1371/journal.ppat.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elahi S, Dinges WL, Lejarcegui N, Laing KJ, Collier AC, Koelle DM, McElrath MJ, Horton H. Protective HIV-specific CD8+ T cells evade Treg cell suppression. Nat Med. 2011;17:989–995. doi: 10.1038/nm.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaufmann DE, Kavanagh DG, Pereyra F, Zaunders JJ, Mackey EW, Miura T, Palmer S, Brockman M, Rathod A, Piechocka-Trocha A, Baker B, Zhu B, Le Gall S, Waring MT, Ahern R, Moss K, Kelleher AD, Coffin JM, Freeman GJ, Rosenberg ES, Walker BD. Upregulation of CTLA-4 by HIV-specific CD4+ T cells correlates with disease progression and defines a reversible immune dysfunction. Nat Immunol. 2007;8:1246–1254. doi: 10.1038/ni1515. [DOI] [PubMed] [Google Scholar]
- 51.Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, DePierres C, Mncube Z, Duraiswamy J, Zhu B, Eichbaum Q, Altfeld M, Wherry EJ, Coovadia HM, Goulder PJ, Klenerman P, Ahmed R, Freeman GJ, Walker BD. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350–354. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 52.Mueller SN, Ahmed R. High antigen levels are the cause of T cell exhaustion during chronic viral infection. Proc Natl Acad Sci U S A. 2009;106:8623–8628. doi: 10.1073/pnas.0809818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.