Abstract
Purpose
We previously found that Kcnj10, an inwardly-rectifying potassium channel, is a gene expressed in c-kit-positive retinal progenitor cells on P1. The shRNA-mediated screening of the functions of the genes for retinal development in retinal explant culture suggested a role for Kcnj10 in the differentiation of 23Müller glia. In the present study, we extended the work and focused on analyzing the role of Kcnj10 in retinal development.
Methods
shRNA-mediated downregulation of Kcnj10 in retinal explants and the in vivo mouse retina at the P1 stage was performed. Differentiation and proliferation of the retina were examined with immunohistochemistry. The effect of barium (Ba2+) treatment, which inhibits potassium currents by blocking potassium channels, on retinal development was examined.
Results
When Kcnj10 was downregulated at E18, cellular proliferation and morphological differentiation were perturbed; in particular, a decreased number of Müller glial cells with abnormal morphological maturation was observed. The overexpression of Kcnj10 in retinal progenitors did not result in gross abnormality during retinal development, but rescued the abnormal differentiation induced with sh-Kcnj10. The presence of Ba2+ in the retinal explant medium led to a phenotype similar to that seen with sh-Kcnj10. Ba2+ exerts an effect mainly during late retinal development, and sh-Kcnj10 in the P1 retina affected Müller glia maturation, suggesting that Kcnj10 plays a pivotal role in the maturation of retinal cell subsets. A previous study of Kcnj10-knockout mice showed no obvious abnormality in retinal differentiation, especially of Müller glia. We examined the effects of the downregulation of Kcnj10 with in vivo electroporation of sh-Kcnj10 in the P1 retina. Retinal differentiation was perturbed, as seen following the in vitro downregulation of Kcnj10, suggesting that compensatory gene expression and/or signaling occurred in the Kcnj10-knockout mice in the retina, leading to normal eye development.
Conclusion
Kcnj10 plays a role in Müller glia maturation during retinal development probably through ionic channel activities.
Introduction
The neural retina comprises various neurons and glial cells that are generated in a specific temporal order from a common pool of retinal progenitor cells [1]. There is diversity within each type of neuron [2,3], giving rise to complex neuronal networks and information processing [4]. Various molecules, such as transcription factors and neurotrophic factors, have been reported to play important roles in retinal cell differentiation [5].
We are interested in the transition of retinal progenitors during retinal development, and cell surface antigens gave us an opportunity to purify retinal cells at different retinal developmental stages. We found that SSEA-1 labels early embryonic retinal progenitor cells [6], and the c-kit labels retinal progenitor cells at around birth [7]. We then examined the gene expression signature during retinal development, focused on genes specifically expressed in the retinal progenitor at P1, and found that a number of genes specifically expressed in this retinal fraction were related to Müller glia development [8]. Kcnj10 (Kir4.1) is one of these genes, and we found that downregulation of Kcnj10 with shRNA in retinal explant resulted in the failure of proper differentiation of the retina, and expression of Kcnj10 was suppressed by the presence of Notch signaling inhibitor DAPT in the culture [8].
Kcnj10 is an inwardly rectifying potassium channel subunit and well-known since it plays essential roles in the function of the inner ear [9,10] and causes SeSAME/EAST (OMIM 612780) syndrome, which is characterized by sensorineural deafness and many neurologic symptoms [11,12]. Many genes have been identified as members of the Kcnj family, and the genes were classified into seven subfamilies (Kir1–Kir7) [13]. They participate in renal epithelial cells [14] and are related to various diseases of the central nervous system [15]. Kcnj10 was initially isolated in the brain by several groups [13], and is predominantly expressed in brain glial cells [16]. In the retina, Kcnj10 is expressed in Müller glia of the rat and rabbit eye [17]. However, previous work showed that retinal cells including Müller glia were morphologically normal in the eye of Kcnj10-knockout mice [18]. In situ whole-cell patch-clamp recordings showed a tenfold increase in the input resistance and large depolarization of Kir4.1-knockout Müller cells [18]. The normal morphological development of the retina of Kcnj10-knockout mice was different from our data that sh-RNA-mediated downregulation of Kcnj10 in retinal progenitor cells resulted in perturbation of morphological differentiation of the retina [8]. In this work, we extended of analysis of the function of Kcnj10 during retinal development using retinal explant culture in more detail. We found that Kcnj10 plays pivotal roles in the proliferation of retinal progenitors and maturation of subsets of retinal cells.
Methods
Animals, reagent, and plasmids
All animal experiments were approved by the Animal Care Committee of the Institute of Medical Science, University of Tokyo and conducted in accordance with the guidelines laid down by the U.S. National Institutes of Health regarding the care and use of animals for experimental procedures and the Association for Research in Vision and Ophthalmology (ARVO) Statement for the Use of Animals in Ophthalmic and Vision Research. ICR mice were obtained from Japan SLC Co (Shizuoka, Japan), and we confirmed that the mice were free of the Rd1 mutation. The day a vaginal plug was observed was embryonic day 0 (E0), and the day of birth was termed postnatal day 0 (P0). Barium chloride dihydrate (Wako, Osaka, Japan) was dissolved in Hank’s Balance Salt Solution (HBSS) at 100 mM. An expression plasmid of sh-Kcnj10 (pU6-shRNA expression vector: mouse U6 promoter in pBluescript-KS [19]) was described in a previous work [8]. Target sequences of sh-Kcnj10 are the first 5′-AAG GAA GTA TTC AGA CTT TAA CA-3′ and second 5′-AAG AGC CAG CTT ATG TGC TTT CA-3′ (Appendix 1). As a control, an empty pU6 vector was used. The efficiency of sh-Kcnj10 was examined using the mouse retina. The isolated retina at E18 was electroporated with 100 μg of sh-Kcnj10 or control vector, and after 48 h, the retinas were harvested. Kcnj10 levels were examined with RT–PCR using the whole retina. The results were obtained by using the first sh-Kcnj10, and essentially the same main data were obtained with the second sh-Kcnj10. As an enhanced green fluorescent protein (EGFP)-expressing vector, pCAG-EGFP, which is effective for expressing exogenous genes in most retinal subtypes, was used. Quantitative PCR (qPCR) for the Kcnj family members was performed using a Roche LightCycler 1.5 apparatus (Roshe, Basel, Switzerland) and was analyzed with the second derivative maximum method for quantification (Roche Diagnostics). The sequence of primers are as follows: Kcnj10 forward 5′-AGT CTT GGC CCT GCC T-3′, reverse 5′-AGC GAC CGA CGT CAT C-3′, Kcnj13 forward 5′-TGG ACT TCC ACC TTG A-3′, reverse 5′-TGG AGC AGG GTA GCT A-3′, Kcnj15 forward 5′-TTG CTG CCA CCT TTG T-3′, 5′-TAA TGC AGG GTG TGT G-3′. β-actin was used as the control. Full-length mouse Kcnj10 (Appendix 1), which covers from the first ATG to termination codon, was cloned with RT–PCR, and the sequence of an isolated Kcnj10 clone was confirmed with a DNA sequence. The primers used for cloning were forward 5′-ATG ACG TCG GTC GCT AAG GT -3′ and reverse 5′-TCA GAC GTT GCT GAT GCG CA-3′. Full-length Kcnj10 was subcloned into downstream of the CAG promoter. Total RNA was purified from the retinas using RNeasy Plus Micro (Qiagen, Venlo, Netherlands), and cDNA was synthesized using SuperScriptII (Life Technologies, Carlsbad, CA).
RNA-sequencing and microarray
RNA-sequencing (RNA-seq) data were obtained as follows. Total RNA was extracted from the retinas of wild-type C57/BL/6J mice at P7 or P14 using the RNeasy Plus Micro Kit (Qiagen), and the RNA quality was confirmed using a 2100 Bioanalyzer (Agilent Technologies). The DNA microarray was performed using cDNA prepared from the retinal explant from E15 (culture days 0, 5, and 14) using the Affymetrix mouse genome 430 2.0 array (2004).
Retinal explant culture, BrdU incorporation assay, and in vivo electroporation into the retina
Retinal explant cultures were prepared as described previously [20]. For the cell proliferation assay, bromodeoxyuridine (BrdU; Sigma-Aldrich, MO; 1.5 μg/ml at final concentration) was added to the explant media, and the retinas were then fixed after 24 h of culture. BrdU was visualized with immunostaining as described previously [21]. For transfection, the U6-Kcnj10 or U6-empty vector was transfected with pCAG-EGFP, which was used as a marker of transfected samples. In vitro plasmid transfection to the isolated mouse retina was performed using electroporation with an Electroporator CUY21 (Nepa Gene, Chiba, Japan) and electrode CYU520P5 (Nepa Gene). Briefly, retinas were transferred to a microelectroporation chamber filled with plasmid solution (25 μg of pCAG-EGFP and 75 μg of sh-Kcnj10 or U6-vector, plasmids were dissolved 1 mg/ml in HBSS), and four square pulses (25 V) of 50 μs duration with 950 μs intervals were applied using the CUY21 pulse generator. In vivo electroporation was performed as previously described [22] with slight modifications. Approximately 0.5 μl of 5 μg/μl DNA solution (mixture of pCAG-EGFP and sh-Kcnj10 or U6-vector, 1:3) was injected into the subretinal space of the P1 mouse retina. Square electric pulses (100 V, six 50-ms pulses at 950-ms intervals) by using a pulse generator, CUY21 SC (Nepa Gene, Japan) were delivered with the CUY650-P5 Platinum Plate Tweezers Electrode (Nepa Gene, Chiba, Japan). The electroporated retinas were harvested at P12.
Immunohistochemistry
Immunostaining of frozen sections was done as described previously [21]. Briefly, retinal explants were fixed with 4% paraformaldehyde for 10 min on ice, treated with 25% sucrose for 30 min, embedded in optimum cutting temperature (OCT) compound (Miles), and sectioned (10 μm thickness) using the cryostat (CM3050S, Leica Microsystems, Wetzlar, Germany). The primary antibodies used were mouse monoclonal antibodies anti-glutamine synthetase (Chemicon), PNR (ppmx), HuC/D (Molecular Probes), Ki67 (BD Bioscience), BrdU (Roche Diagnostics), rabbit polyclonal antibodies anti-Calbindin 28k (Chemicon), PNR, Ccnd3 active Caspase-3 (Promega), GFP (BD Biosciences), sheep polyclonal antibody anti-Chx10 (Exalpha Biologicals), and rat polyclonal antibody anti-GFP (Nakarai). Primary antibody signals were visualized using appropriate secondary antibodies conjugated with Alexa 488 or Alexa 594 (Molecular Probes).
Results
Examination of expression of members of the Kcnj family in the mouse retina with RNA sequence
Previously, we found that Kcnj10 was expressed in c-kit-positive retinal progenitor cells, and RT-qPCR analysis indicated that the expression level of Kcnj10 increased in the later stages of retinal development [8]. We first examined the retinal expression of Kcnj family genes with RNA-seq data from whole mouse retinas at P7 and P14. At P7, the expression level of Kcnj10 was the highest among all Kcnj family members considered (Table 1). At P14, the highest expression level was observed for Kcnj14, although its expression level at P7 was relatively low (Table 1). The downregulation of Kcnj14 in the photoreceptor-specific nuclear receptor (PNR)-knockout retina was reported previously [23], suggesting Kcnj14 is expressed in photoreceptors. Kcnj9 also showed a high level of expression at P14. Members of the Kcnj family are classified into four sub-families according to their structure; of these, Kcnj10 belongs to the potassium inwardly-rectifying channel family [13]. Kcnj1, -13, -15, and -16 belong to the same family, but these members showed negligible retinal expression at P7 and P14 based on our RNA-seq data (Table 1). These results suggest that Kcnj10 is one of the main potassium transport channels of the Kcnj family in the retina. Since we used retinal explant culture in the following experiments, we also examined the expression profiles of members of the Kcnj family in retinal explants using the gene expression profile of retinal explant cultures. DNA microarray data of retinal explant culture prepared from the mouse retina at E15 (0-day, 5-day, and 14-day cultured retina) was examined (Table 1). The Kcnj10 level increased during explant culture as in vivo, and relatively high expression levels of Kcnj3, -9, and -10 were observed in the RNA-seq and microarray data (Table 1).
Table 1. Expression of members of Kcnj family in retina and retinal explants.
| Gene name | RNaseq |
microarray |
||||
|---|---|---|---|---|---|---|
| P7 | P14 | probe | E15 | DIV5 (P0) | DIV14 (P9) | |
| Kcnj1 |
0.01 |
0.01 |
1418614_at
1418613_at |
7.2
3.6 |
10.8
7.6 |
11.7
3.9 |
| Kcnj2 |
1.02 |
2.83 |
1422871_at
1450503_at |
4.4
8.6 |
1.5
7.9 |
26.7
1.8 |
| Kcnj3 |
3.22 |
6.74 |
1421468_at |
48.1 |
25.5 |
64.6 |
| Kcnj4 |
0.10 |
0.37 |
1451808_at |
5.1 |
4.8 |
6.5 |
| Kcnj5 |
1.34 |
6.50 |
421762_at |
1.9 |
8.6 |
5.8 |
| Kcnj6 |
2.23 |
2.63 |
11425707_a_at
1421581_at
1425044_at
1450294_a_at |
1.7
3.9
24.1
1.3 |
1.5
7.3
16
1.6 |
4.2
13.5
16.5
1 |
| Kcnj8 |
2.31 |
3.70 |
1418142_at |
3.9 |
1.5 |
3.1 |
| Kcnj9 |
3.70 |
30.0 |
1426115_a_at
1450712_at
1433029_at |
10.7
10.2
9.6 |
3.7
31.5
2.7 |
12.3
62.1
10.1 |
| Kcnj10 |
6.15 |
20.34 |
1419601_at |
20.3 |
34.8 |
50.7 |
| Kcnj11 |
3.90 |
4.25 |
1450515_at |
14.3 |
8.1 |
6.2 |
| Kcnj12 |
2.20 |
6.65 |
||||
| Kcnj13 |
0.72 |
2.18 |
1456418_at |
10.6 |
2.7 |
12.1 |
| Kcnj14 |
2.63 |
240.57 |
||||
| Kcnj15 |
0.03 |
0.03 |
1450185_at |
8.1 |
2.1 |
1.8 |
| Kcnj16 | 0.04 | 0.17 | ||||
RNA sequence was done using RNA extracted from whole retinas of wild type ICR mice. Values of expression levels are FPKM value. wt; wild type. Retinal explant was prepared from E15 retina, and gene expression pattern of culture day 0, 5 and 14 was examined by DNA microarray (Affymetrix). DIV, days in vitro.
Sh-RNA mediated downregulation of Kcnj10 during retinal development
We previously showed that the downregulation of Kcnj10 expression by sh-RNA in retinal explants resulted in malformed retinas [8]. Thus, here we examined the detailed development of retinas in which Kcnj10 was downregulated with sh-RNA. We transfected an expression plasmid carrying sh-RNA against Kcnj10 (sh-Kcnj10) into isolated retinas at E18, and the retinas were cultured as explants. We first examined the efficiency of the suppression of the expression level of Kcnj10 by the expression of sh-Kcnj10 in the retina. The efficiency of electroporation using the mouse retina at around birth is more than 50% (Appendix 1). sh-Kcnj10 was electroporated into the retina at E18, and after 48 h of the culture, the retinas were harvested and subjected to RT-qPCR. The level of Kcnj10 in sh-Kcnj10 was less than 50% of that in the control (Appendix 1). At the same time, we examined the expression level of Kcnj13 and -15, which are closely related to Kcnj10 in their structure [13]. The expression level of Kcnj13 was not changed by the expression of sh-Kcnj10, and that of Kcnj15 was not detectable in the control and sh-Kcnj10-expressing retina (Appendix 1). We first examined the sub-retinal localization of sh-Kcnj10-expressing cells in the retina (Figure 1A,B). On day 3 of culture, strong EGFP signals were observed the inside the neuroblastic layer; most cells on the outer half of the layer showed weak EGFP signals in the control and sh-Kcnj10 plasmid-transfected groups (Figure 1A). On day 7 of culture, the intensity of the EGFP signal in the cells on the outer side of the neuroblastic layer became stronger than on day 3, and the distribution of EGFP expression between the control and sh-Kcnj10-transfected retinas was similar (Figure 1A,B). On day 14, cells showing strong EGFP positivity were observed in the outer nuclear layer (ONL), but not in the inner nuclear layer (INL) in the control group; in contrast, in the sh-Kcnj10-expressing retina, strong EGFP positivity was observed mostly in the INL (Figure 1A,B). These cells were large compared to the small cells in the ONL, which showed weak EGFP signals (Figure 1A). In addition, a rosette-like structure was observed in the ONL of the sh-Kcnj10-expressing retinas.
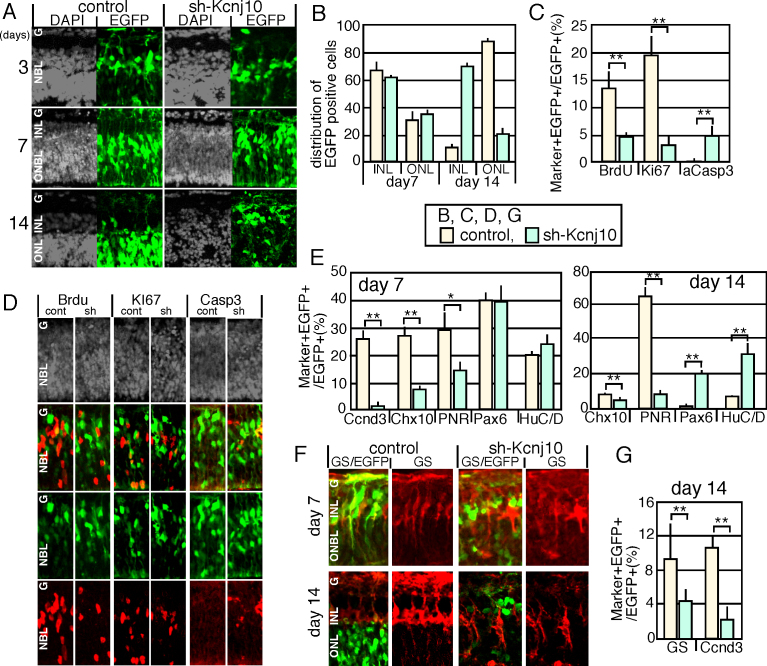
Figure 1.
Expression of sh-Kcnj10 in mouse retinal explants. A: Control-enhanced green fluorescent protein (EGFP) or sh-Kcnj10/EGFP expression plasmids were electroporated into isolated retinas at E18, and cultured as explants on the indicated days. B: Sub-retinal distribution of EGFP-positive cells was calculated on days 7 and 14. C, D: Proliferated and apoptotic cells were analyzed after 3 days of culture with immunostaining. The population of the immunostaining signal-positive cells (C) and immunostained pattern (D) are shown. E–H: Differentiation was examined with immunostaining of markers for retinal subpopulation. Calculation of the population of marker and EGFP double-positive cells in the total EGFP positive cells (E, G). The number of marker-positive cells in each layer was counted in 150-μm-wide retina (F). Staining patterns of GS and EGFP (H) are shown. Nuclei were visualized with 4',6-diamidino-2-phenylindole (DAPI) staining (gray) in A, D. P value; * <0.05, **<0.01 (the Student t test from at least three independent samples). cont, control; sh, sh-Kcnj10; ONL, outer nuclear layer; INL, inner nuclear layer; ONBL, outer neuroblastic layer; GCL, ganglion cell layer; aCasp3, active caspase 3; GS, glutamine synthetase.
We next examined the proliferative activity in the retinas with BrdU incorporation and Ki67 staining. After 3 days of culture, both analyses showed that the proliferative activity of retinal progenitor cells expressing sh-Kcnj10 was severely suppressed (Figure 1C,D). In addition, the number of apoptotic cells, identified based on the expression of active caspase-3, was slightly increased (Figure 1C,D). We then examined the differentiation of retinal cells with immunostaining for markers of various retinal subtypes. On day 7 in the sh-Kcnj10-expressing retina, few EGFP-positive and Ccnd3-positive Müller glia (Figure 1E,H and Appendix 1) were observed, as expected based on our previous data [8]. The number of Chx10-positive bipolar cells and PNR-positive rods was also significantly decreased (Figure 1E and Appendix 1). On day 14, the number of Müller glia was still low, as shown by counting the number of GS- and Ccnd3-positive cells (Figure 1F,H). In addition, GS staining indicated that the morphology of the Müller glia was severely perturbed (Figure 1H).
The number of PNR-positive rods was still low on day 14 in the sh-Kcnj10-expressing retinas (Figure 1E and Appendix 1), but the number of cells positive for Pax6 and HuC/D, which label amacrine cells, was increased on day 14 in sh-Kcnj10-expressing retina (Figure 1E and Appendix 1). We then counted the total number of HuC/D or Pax6 positive cells in each layer (Figure 1G). We found the cell number of HuC/D or Pax6 in INL was indistinguishable between the control and sh-Kcnj10-expressing samples, but the Pax6 positive cells in the ganglion cell layer (GCL) were slightly increased in the sh-Kcnj10-expressing samples (Figure 1G). Therefore, these results suggest that the increased percentage of amacrine cells in Figure 1E may be caused by the decreased rod photoreceptors rather than the increased number of amacrine cells. We found ectopically localized HuC/D or Pax6-positive cells in the ONL in sh-Kcnj10-expressing samples, but these cells were EGFP negative. We surmise that the cells were miss-localized probably because the Müller glia cells failed to form properly.
Overexpression of Kcnj10 rescued phenotype induced by sh-Kcnj10
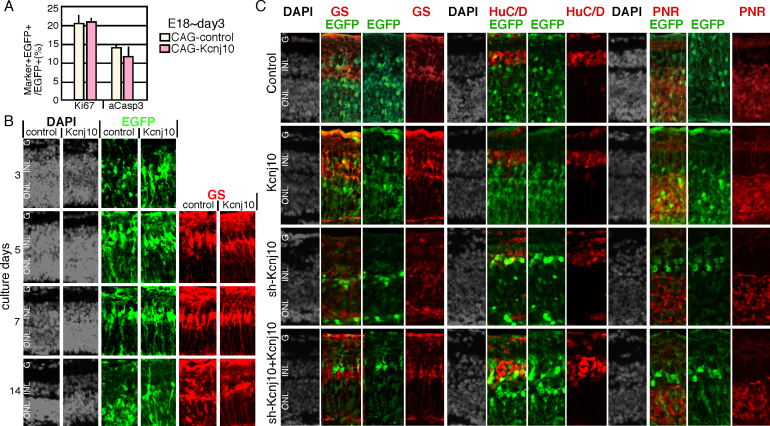
We then examined the effects of Kcnj10 overexpression during retinal development. A Kcnj10 expression plasmid or empty vector was electroporated into isolated retinas at E18 and cultured for the indicated number of days. On day 3, proliferation and apoptosis were examined based on Ki67 or active caspase-3 expression, respectively, and was not perturbed (Figure 2A). We then examined Müller glial cell differentiation. We hypothesized that Müller glial cell differentiation would occur earlier in the Kcnj10-overexpressing retina, but a time-course analysis did not support this possibility (Figure 2B).
Figure 2.
Overexpression of Kcnj10 in retinal explant culture and rescue of phenotype induced by sh-Kcnj10. A, B: Expression plasmid of Kcnj10 or empty vector and enhanced green fluorescent protein (EGFP)-expressing plasmid was electroporated into the isolated retina at E18, and cultured for the indicated days. A: Proliferation and apoptotic cells were examined with immunostaining of Ki67 or active caspase 3 (aCasp3), respectively. B: Immunostaining patterns of 4',6-diamidino-2-phenylindole (DAPI), EGFP, and glutamine synthetase (GS) are shown. C: Control, CAG-Kcnj10, sh-Kcnj10, or CAG-Kcnj10 + sh-Kcnj10 was introduced into the retina at E18, and cultured for 2 weeks. Then, morphology and differentiation were examined with immunostaining of frozen sections. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer.
To confirm that the effect of sh-Kcnj10 was through specific suppression of Kcnj10 expression, we performed a rescue experiment. A plasmid encoding wild-type Kcnj10 was transfected with sh-Kcnj10, and the transfected retina was cultured as the explant for 14 days. Differentiation of the retina was examined with immunostaining of frozen sections (Figure 2C). We examined the distribution of the EGFP-positive cells, sub-layer structure, and expression of GS, HuC/D, and PNR, and all criteria indicated the expression of the Kcnj10 rescued phenotype was induced with sh-Kcnj10.
The phenotype induced by sh-Kcnj10 was mimicked by the presence of Ba2+ in retinal explants
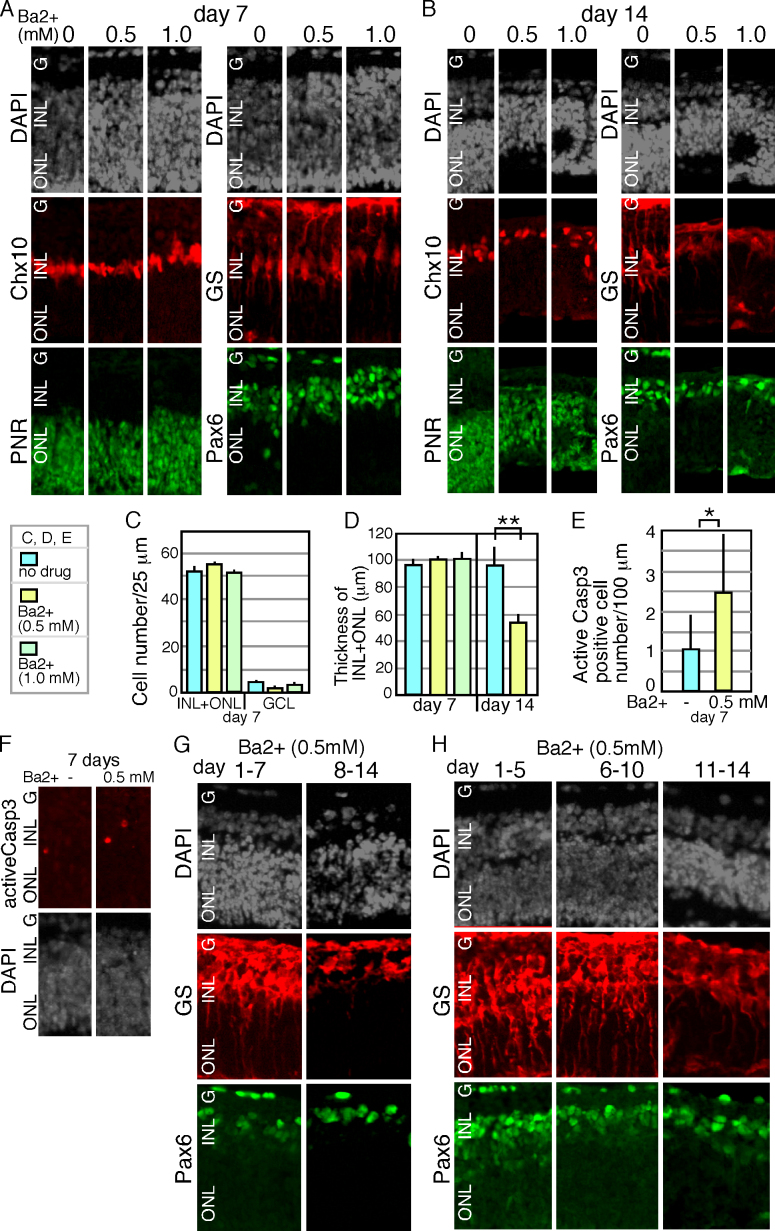
Since Kcnj10 encodes a potassium inwardly-rectifying channel [9], we next examined whether the phenotype observed in the absence of Kcnj10 was related to potassium channel activity using a potassium channel blocker. Almost all known Kir channel subunits are highly sensitive to extracellular Ba2+ [24,25], and Ba2+ is often used to test the physiologic roles of potassium channels in various tissues, including retinal glial cells [26]. We started retinal explant cultures using retinas at E18 with (0.5 and 10 mM, final concentration) or without Ba2+ in the culture medium. After 7 days of culture, we examined the gross morphology and differentiation of the retinal cells with immunostaining (Figure 3A). DAPI staining showed no significant difference between the control and Ba2+-treated samples (Figure 3A). The thickness and the cell number of the control and Ba2+-treated retinas were indistinguishable (Figure 3C,D). On day 14, the morphology of the 1.0 mM Ba2+-treated samples was severely perturbed; a rosette-like structure formed in the ONL, while the INL looked relatively normal (Figure 3B). In addition, the 0.5 mM Ba2+-treated samples were much thinner than the control samples on day 14 (Figure 3D). Immunostaining for active caspase-3 revealed a slightly higher number of apoptotic cells in the Ba2+-treated (0.5 mM) samples (Figure 3E,F). Immunostaining on days 7 and 14 for retinal markers showed well-developed amacrine cells (Pax6), bipolar cells (Chx10), and photoreceptors (PNR; Figure 3A,B). GS staining of the Ba2+-treated retina was relatively normal on day 7 (Figure 3A), but the signal became faint by day 14 (Figure 3B). Since GS was expressed on day 7, we surmise that the commitment of retinal progenitor cells to become Müller glia took place, at least partially, but morphological differentiation did not proceed. We analyzed the effects of Ba2+ on Müller glia maturation in greater detail using 0.5 mM Ba2+.
Figure 3.
Effects of barium chloride for retinal explants. Retinal explants were prepared from isolated mouse retinas at E18, and cultured as indicated days in the presence or absence of barium chloride (Ba2+) in the culture medium. Explants were frozen sectioned, and immunostaining was done with indicated antibodies. C: The number of retinal cells, which was counted with 4',6-diamidino-2-phenylindole (DAPI) signals, in each layer. Numbers of cells of the GCL, and the sum of the INL and the ONL are shown. D: The thickness of the INL and the ONL. E: Number of active caspase 3 (Casp3) positive cells is shown. Nuclei were visualized with DAPI staining (gray) in A, F–H. P value; * <0.05, **<0.01 (the Student t test from at least three independent samples).
Ba2+ exerts effects on retinal development mainly in the late stage of differentiation
We next set up retinal explant cultures in the presence of Ba2+ during either the first or second half of the culture period to examine when the inward potassium current affected Müller glial cell development. Retinal differentiation was examined after 14 days of culture in both cases. When Ba2+ was present during the first week of culture, the Müller glia differentiated normally (Figure 3G), suggesting that potassium channel function was dispensable during this period. However, when Ba2+ was present during the second week of culture, Müller glia development was severely perturbed. We next divided the culture period into three phases, and Ba2+ was added during one of these time periods as follows: 1 to 5, 6 to 10, and 11 to 14 days of culture. The morphology of the Müller glia, which was judged based on the number and location of the cell body and process extension, was quite similar to that of the control when Ba2+ was present during the second phase, but it was severely perturbed in the presence of Ba2+ during the third phase (Figure 3H). Thus, functional potassium channels may be required during the later stages of retinal differentiation.
Downregulation of Kcnj10 in later stage of retinal development
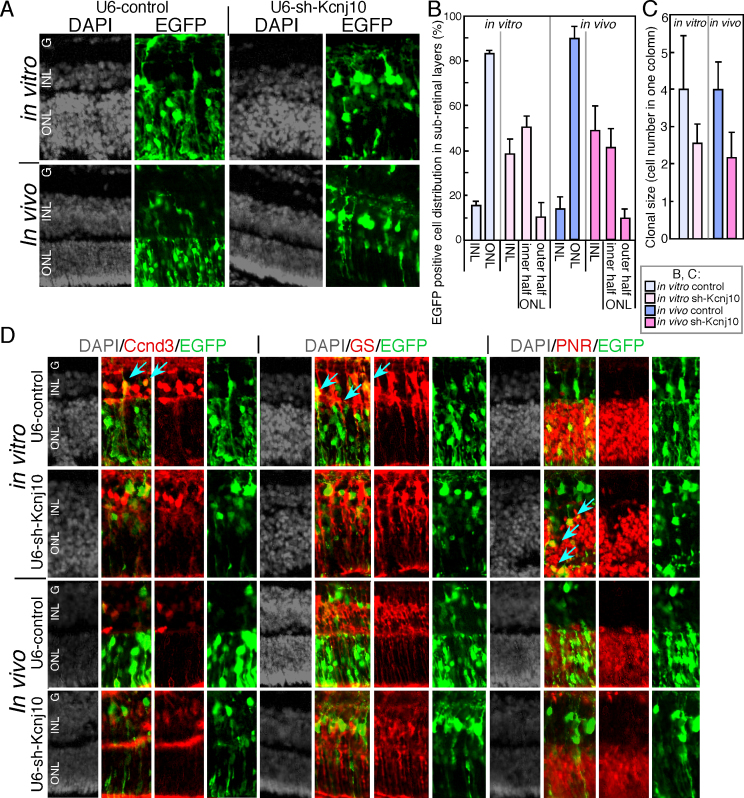
Our experiments with Ba2+ indicated that Kcnj10 plays a role in postnatal Müller glia differentiation; thus, we examined the effects of sh-Kcnj10 when it was expressed in the postnatal retina. Proliferation stops quickly in the postnatal retina; therefore, we focused on the effects of the downregulation of Kcnj10 on late-phase retinal differentiation without considering the effects on proliferation. Sh-Kcnj10 was electroporated into isolated retinas at P1, and the retinas were cultured as explants for 12 days. The sub-retinal distribution of EGFP-positive cells showed that, in the controls, most of the EGFP-positive cells were in the ONL (Figure 4A). In contrast, large cells that were strongly positive for EGFP were observed on the inner side of the INL in the sh-Kcnj10-expressing samples, while small cells that were weakly positive for EGFP were observed in the INL and (to a lesser extent) in the ONL (Figure 4A, upper panels). Counting of the cells in each layer showed a clear difference in distribution between the control and sh-Kcnj10-expressing cells (Figure 4B), and the EGFP-positive cells in the ONL of the sh-Kcnj10-expressing samples tended to localize to the inner half of the ONL (Figure 4B). We next examined retinal differentiation by immunostaining for retinal markers. None of the EGFP-positive cells in the sh-Kcnj10-expressing samples expressed Ccnd3 or GS, and few cells in the ONL expressed PNR (Figure 4D, blue arrows). The expression patterns of Chx10, HuC/D, and Pax6 suggest that some of the EGFP-positive cells differentiated into bipolar and amacrine cells; moreover, the staining patterns were similar between the control and sh-Kcnj10-expressing samples (Appendix 1).
Figure 4.
In vitro and in vivo electroporation of sh-Kcnj10 in postnatal retina. A-D: Control or Kcnj10 expressing plasmids with enhanced green fluorescent protein (EGFP) expression plasmid were electroporated into the isolated retina at P1 (in vitro electroporation) or the retina of live mice at P1 (in vivo electroporation). Then, the retinas were harvested (in vitro samples), or the mice were euthanized, and the retinas were isolated (in vivo samples), and the EGFP and retinal markers were immunostained. B: The subretinal distribution of EGFP-positive cells. In the sh-Kcnj10 sample, the outer nuclear layer (ONL) was horizontally divided equally, and the number of cells was counted in each area (the outer half and the inner half of the ONL). C: Clonal size was counted by the number of EGFP-positive cells in vertically the same column. A and C show immunostaining pattern, and nuclei were visualized with4',6-diamidino-2-phenylindole (DAPI) staining.
Downregulation of Kcnj10 by sh-RNA in the in vivo retina resulted in failed differentiation of Müller glia
Since the Müller glia of retinas from the Kcnj10-knockout mice showed no morphological abnormalities [18], we next asked whether sh-Kcnj10 affects Müller glia development when sh-Kcnj10 is expressed in vivo in the retina. In vivo electroporation was performed using retinas from P1 mouse pups, and the mice were euthanized by cervical dislocation after 12 days. The retinas were then isolated, and their differentiation examined with immunostaining in frozen sections. We first examined the sub-retinal localization of EGFP-positive cells. As seen with in vitro electroporation, after 12 days, the sh-Kcnj10-expressing cells were distributed mainly in the INL (Figure 4A, lower panels). Immunostaining for the Müller glia markers Ccnd3 and GS showed that the sh-Kcnj10-transfected EGFP-positive cells did not differentiate into Müller glia (Figure 4D). Staining for Chx10, HuC/D, and Pax6 showed that some EGFP-positive cells merged with these markers in the control and sh-Kcnj10-expressing retinas, and the staining patterns were similar in these samples (Appendix 1).
Discussion
In this study, we found that Kcnj10 is required for proper cellular proliferation and differentiation in the retina, especially in the late phase of Müller glia development. Our experiments with Ba2+ suggest that the functional effects of Kcnj10 on the retina are mediated through channel activity. Our initial query in this study involved the question of why sh-Kcnj10 affected retinal explants even though Müller glia from Kcnj10-knockout mice were reported to be morphologically normal [18]. We first surmised that differences between the retinas in vitro and in vivo (e.g., ionic strength) resulted in the difference in phenotype caused by the downregulation of Kcnj10. However, the in vivo downregulation of Kcnj10 at stage P1 via the expression of sh-Kcnj10 resulted in the failure of appropriate differentiation, suggesting that this explanation was insufficient to elucidate the difference. In terms of structure, Kcnj13 and Kcnj15 are closely related to Kcnj10 [13]. Therefore, other members of the Kcnj family, which have redundant functions with Kcnj10, might be expressed in a compensatory manner in Kcnj10-knockout cells. This situation was observed in the case of retinoblastoma protein (Rb). When Rb was genetically knocked out in mice, p107 was upregulated, and functional compensation by p107 or p130 was reported [27,28]. Although we did not observe induction of expression of Kcnj13 and Kcnj15 by sh-Kcnj10 in explants, similar examination in the Kcnj10-knockout retina may provide conclusive evidence.
We found that the downregulation of Kcnj10 and treatment with Ba2+ resulted in different phenotypes (sh-Kcnj10 suppressed retinal progenitor cell proliferation whereas Ba2+ did not). Sh-Kcnj10 expression also perturbed the differentiation of bipolar cells and rods, which are late-differentiating cell types; therefore, we surmise that the phenotype was secondary to the effects of suppression on proliferation rather than fate change of retinal progenitors. Kcnj16 modulates Kcnj10 channel conductance [29], and heteromeric Kcnj10/16 channels were reported to be expressed in Müller glia [13]. Perturbation of the stoichiometry of Kcnj10 with interacting molecules by sh-Kcnj10 may be one cause of the phenotypic differences that resulted from sh-Kcnj10 and Ba2+ treatment. Interestingly, perisynaptic processes and the end feet of Müller cells harbor homomeric or heteromeric Kcnj channels in opposite directions to the cell membrane [13]. Therefore, a lack of Kcnj10 in Müller glia may result in complex phenotypes compared with simple blockage of the channel from the outside. However, Kcnj10 overexpression had no effect on retinal development. The modification of Kcnj10 by pH is well-known. PtdIns(4,5)P2 also modifies Kcnj10 function, and binding proteins such as Mag1 regulate the localization of Kcnj10 [30]. We suspect that these modifiers did not function appropriately for exogenous Kcnj10, or that retinal Kcnj10 expression may have been saturated when we overexpressed Kcnj10.
A previous study indicated the negative involvement of Kcnj10 in proliferation during wound healing in human corneal epithelial cells through calcium signaling [31]. Ba2+ treatment also promoted the proliferation of neural precursor cells [32]. Therefore, our observations may not be explained by simple blockage of the inward potassium current. In the case of Kv potassium channels, the involvement of Kv channels in epithelial cell proliferation has been reported, and Kv4.1 is related to gastric cancer cell proliferation [33]. Kv and Kir channels are clearly different channels, but calcium mobilization occurs downstream of both channels. The positive involvement of calcium signaling in cellular proliferation under certain situations by Kv4.1 and Kcnj10 is one possible mechanism.
Changes in the Kcnj10 level in Müller glial cells of RGS rats during retinal degeneration has been reported [34]. In addition to changing the membrane potential of Müller glia during degeneration, changes in membrane potential have also been demonstrated during Müller glia development [35,36]. At an early stage of Müller glia differentiation, no inward potassium current was observed; however, the inward current increased during maturation of the cells. These results suggest important roles for potassium channels in Müller glia development. Taken together, in this study we uncovered a role for Kcnj10 in retinal development using an explant culture system and an in vivo electroporation system. Further molecular analysis using these systems may shed new light on the role and molecular mechanisms of Kcnj10 in retinal development.
Acknowledgments
This work was supported by a grant-in-aid from the Ministry of Education, Culture, Sports, Science (15118708).
Appendix 1. Immunostaining pattern of sh-Kcnj10 expressing retinal explant.
To access the data, click or select the words “Appendix 1.”
References
- 1.Marquardt T, Gruss P. Generating neuronal diversity in the retina: one for nearly all. Trends Neurosci. 2002;25:32–8. doi: 10.1016/s0166-2236(00)02028-2. [DOI] [PubMed] [Google Scholar]
- 2.Masland RH. The Neuronal Organization of the Retina. Neuron. 2012;76:266–80. doi: 10.1016/j.neuron.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masland RH. The fundamental plan of the retina. Nat Neurosci. 2001;4:877–86. doi: 10.1038/nn0901-877. [DOI] [PubMed] [Google Scholar]
- 4.Gollisch T, Meister M. Eye Smarter than Scientists Believed: Neural Computations in Circuits of the Retina. Neuron. 2010;65:150–64. doi: 10.1016/j.neuron.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jean D, Ewan K, Gruss P. Molecular regulators involved in vertebrate eye development. Mech Dev. 1998;76:3–18. doi: 10.1016/s0925-4773(98)00117-8. [DOI] [PubMed] [Google Scholar]
- 6.Koso H, Ouchi Y, Tabata Y, Aoki Y, Satoh S, Arai K, Watanabe S. SSEA-1 marks regionally restricted immature subpopulations of embryonic retinal progenitor cells. Dev Biol. 2006;292:265–76. doi: 10.1016/j.ydbio.2005.09.051. [DOI] [PubMed] [Google Scholar]
- 7.Koso H, Satoh S, Watanabe S. c-kit marks late retinal progenitor cells and regulates their differentiation in developing mouse retina. Dev Biol. 2007;301:141–54. doi: 10.1016/j.ydbio.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 8.Mochizuki Y, Iida A, Lyons E, Kageyama R, Nakauchi H, Murakami A, Watanabe S. Use of cell type-specific transcriptome to identify genes specifically involved in Müller glia differentiation during retinal development. Dev Neurobiol. 2014;74:426–37. doi: 10.1002/dneu.22131. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Zhao HB. The role of an inwardly rectifying K+ channel (Kir4.1) in the inner ear and hearing loss. Neuroscience. 2014;265:137–46. doi: 10.1016/j.neuroscience.2014.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rozengurt N, Lopez I, Chiu C-S, Kofuji P, Lester HA, Neusch C. Time course of inner ear degeneration and deafness in mice lacking the Kir4.1 potassium channel subunit. Hear Res. 2003;177:71–80. doi: 10.1016/s0378-5955(02)00799-2. [DOI] [PubMed] [Google Scholar]
- 11.Reichold M, Adebik AA, Lieerer E, Rapedium M, Schmidt K, Bandulik S, Sterner C, Tegtmeier I, Penton D, Baukrowitz T, Hulton S-A, Witzgall R, Ben-Zeev B, Howie AJ, Kleta R, Bockenhauer D, Warth R. KCNJ10 gene mutations causing EAST syndrome (epilepsy, ataxia, sensorineural deafness, and tubulopathy) disrupt channel function. Proc Natl Acad Sci USA. 2010;107:14490–5. doi: 10.1073/pnas.1003072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scholl UI, Choi M, Liu T, Ramaekers VT, Hausler MG, Grimmer J, Tobe SW, Farhi A, Nelson-Williams C, Lifton RP. Seizures, sensorineural deafness, ataxia, mental retardation, and electrolyte imbalance (SeSAME syndrome) caused by mutations in KCNJ10. Proc Natl Acad Sci USA. 2009;106:5842–7. doi: 10.1073/pnas.0901749106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hibino H, Inanobe A, Furutani K, Murakami S, Findlay I. KUrachi Y. Inwardly rectifying potassium channels; Their structure, function, and physiological roles. Physiol Rev. 2010;90:291–366. doi: 10.1152/physrev.00021.2009. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton KL, Devor DC. Basolateral membrane K+ channels in renal epithelial cells. Am J Physiol Renal Physiol. 2012;302:F1069–81. doi: 10.1152/ajprenal.00646.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Judge SI, Smith PJ, Stewart PE, Bever CTJ. Potassium channel blockers and openers as CNS neurologic therapeutic agents. Recent Patents CNS Drug Discov. 2007;2:200–28. doi: 10.2174/157488907782411765. [DOI] [PubMed] [Google Scholar]
- 16.Takumi T, Ishii T, Horio Y, Morishige K, Takahashi N, Yamada M, Yamashita T, Kiyama H, Sohmiya K, Nakanishi S, Kurachi Y. A novel ATP-dependent inward rectifier potassium channel expressed predominantly in glial cells. J Biol Chem. 1995;270:16339–46. doi: 10.1074/jbc.270.27.16339. [DOI] [PubMed] [Google Scholar]
- 17.Ishii M, Horio Y, Tada Y. HIbino H, Inanobe A, Ito M, Yamada M, Gotow T, Uchiyama Y, Kurachi Y. Expression and clustered distribution of an inwardly rectifying potassium channel, KAB-2/Kir4.1, on mammalian retinal Muller cell membrane: their regulation by insulin and laminin signals. J Neurosci. 1997;17:7725–35. doi: 10.1523/JNEUROSCI.17-20-07725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kofuji P, Ceelen P, Zahs KR, Surbeck LW, Lester HA, Newman EA. Genetic Inactivation of an Inwardly Rectifying Potassium Channel (Kir4.1 Subunit) in Mice: Phenotypic Impact in Retina. J Neurosci. 2000;20:5733–40. doi: 10.1523/JNEUROSCI.20-15-05733.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamichi N, Yamamichi-Nishina M, Mizutani T, Watanabe H, Minoguchi S, Kobayashi N, Kimura S, Ito T, Yahagi N, Ichinose M, Omata M, Iba H. The Brm gene suppressed at the post-transcriptional level in various human cell lines is inducible by transient HDAC inhibitor treatment, which exhibits anti-oncogenic potential. Oncogene. 2005;24:5471–81. doi: 10.1038/sj.onc.1208716. [DOI] [PubMed] [Google Scholar]
- 20.Tabata Y, Ouchi Y, Kamiya H, Manabe T, Arai K, Watanabe S. Retinal fate specification of mouse embryonic stem cells by ectopic expression of Rx/rax, a homeobox gene. Mol Cell Biol. 2004;24:4513–21. doi: 10.1128/MCB.24.10.4513-4521.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iida A, Shinoe T, Baba Y, Mano H, Watanabe S. Dicer plays essential roles for retinal development by regulation of survival and differentiation. Invest Ophthalmol Vis Sci. 2011;52:3008–17. doi: 10.1167/iovs.10-6428. [DOI] [PubMed] [Google Scholar]
- 22.Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci USA. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng H, Aleman TS, Cideciyan AV, Khanna R, Jacobson SG, Swaroop A. In vivo function of the orphan nuclear receptor NR2E3 in establishing photoreceptor identity during mammalian retinal development. Hum Mol Genet. 2006;15:2588–602. doi: 10.1093/hmg/ddl185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ransom CB, Sontheimer H. Biophysical and pharmacological characterization of inwardly rectifying K+ curernts in rat spinal cord astrocytes. J Neurophysiol. 1995;73:333–46. doi: 10.1152/jn.1995.73.1.333. [DOI] [PubMed] [Google Scholar]
- 25.Coetzee WA, Amarillo Y, Chiu J, Chow A, Lau D, McCormack T, Moreno H, Nadal MS, Ozaita A, Pountney D, Saganich M, De Miera EVS, Rudy B. Molecular diversity of K+ channels. Ann N Y Acad Sci. 1999;868:233–85. doi: 10.1111/j.1749-6632.1999.tb11293.x. [DOI] [PubMed] [Google Scholar]
- 26.Newman EA. Inward-rectifying potassium channels in retinal glial (Mueller) cells. J Neurosci. 1993;13:3333–45. doi: 10.1523/JNEUROSCI.13-08-03333.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wirt SE, Sage J. p107 in the public eye: and Rb understudy and more. Cell Div. 2010;5:9–13. doi: 10.1186/1747-1028-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dannenberg J-H, Schuijff L, Dekker M. vander Valk M, te Riele H. Tissue-specific tumor suppressor activity of retinoblastoma gene homologs p107 and p130. Genes Dev. 2004;18:2952–62. doi: 10.1101/gad.322004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pessia M, Tucker SJ, Lee K, Bond CT, Adelman JP. Subunit positional effects revealed by novel heteromeric inwardly rectifyin gK+ channels. EMBO J. 1996;15:2980–7. [PMC free article] [PubMed] [Google Scholar]
- 30.Tanemoto M, Toyohara T, Abe T, Ito S. Mag1-Ia functions as a scaffolding protein for the distal renal tubular basolateral K+ channels. J Biol Chem. 2008;283:12241–7. doi: 10.1074/jbc.M707738200. [DOI] [PubMed] [Google Scholar]
- 31.Lin D, Halilovic A, Yue P, Bellner L, Wang K, Wang L, Zhang C. Inhibition of miR-205 impairs the wound-healing process in human corneal epithelial cells by targeting Kir4.1 (Kcnj10). IOVS. 2013;54:6167–78. doi: 10.1167/iovs.12-11577. [DOI] [PubMed] [Google Scholar]
- 32.Yasuda T, Bartlett PF, Adams DJ. Kir and Kv channels regulate electrical properties and proliferation of adult neural precursor cells. Mol Cell Neurosci. 2008;37:284–97. doi: 10.1016/j.mcn.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 33.Kim HJ, Jang SH, Jeong YA, Ryu PD, Kim D-Y, Lee SY. Involvement of Kv4.1 K+ channels in gastric cancer cell proliferation. Biol Pharm Bull. 2010;33:1754–7. doi: 10.1248/bpb.33.1754. [DOI] [PubMed] [Google Scholar]
- 34.Zhao TT. C. LY, Weng CH, Yin ZQ. The changes of potassium currents in RGS rat Muller cell during retinal degeneration. Brain Res. 2012;1427:78–87. doi: 10.1016/j.brainres.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 35.Bringmann A, Francke M, Pannicke T, Biedermann B, Kodal H, Faude F, Reichelt W, Reichenbach A. Role of glial K+ channels in ontogeny and gliosis: a hypothesis based upon studies on Mueller cells. Glia. 2000;29:35–44. doi: 10.1002/(sici)1098-1136(20000101)29:1<35::aid-glia4>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 36.Felmy F, Pannicke T, Richt JA, Reichenbach A, Guenther E. Electrophysiological properties of rat retinal Mueller (glial) cells in postnatally developing and in pathologically altered retinae. Glia. 2001;34:190–9. doi: 10.1002/glia.1053. [DOI] [PubMed] [Google Scholar]