Abstract
Horizontal gene transfer is a major contributor to bacterial evolution and diversity. For a bacterial cell to utilize newly-acquired traits such as virulence and antibiotic resistance, new genes must be integrated into the existing regulatory circuitry to allow appropriate expression. Xenogeneic silencing of horizontally-acquired genes by H-NS or other nucleoid-associated proteins avoids adventitious expression and can be relieved by other DNA-binding counter-silencing proteins in an environmentally- and physiologically-responsive manner. Biochemical and genetic analyses have recently demonstrated that counter-silencing can occur at a variety of promoter architectures, in contrast to classical transcriptional activation. Disruption of H-NS nucleoprotein filaments by DNA bending is a suggested mechanism by which silencing can be relieved. This review discusses recent advances in our understanding of the mechanisms and importance of xenogeneic silencing and counter-silencing in the successful integration of horizontally-acquired genes into regulatory networks.
Introduction
A brief comparison of the genomes of a bacterial pathogen and its close non-pathogenic relatives will typically reveal that these strains differ primarily by the presence of horizontally-acquired virulence-associated genomic islets and islands. The model enteric pathogen Salmonella enterica serovar Typhimurium is a well-studied example, possessing five separate Salmonella pathogenicity islands (SPIs) that are not present in its close relative Escherichia coli [1]. This demonstrates the ability of bacteria to evolve in quantum leaps made possible by horizontal gene transfer [2]. Horizontal gene transfer can provide a bacterial cell with new traits or phenotypes in a single genetic event, rather than via the gradual accumulation of beneficial point mutations over time, allowing recipient cells to rapidly take advantage of and colonize new environmental niches. This has made horizontal gene transfer a primary driver of bacterial evolution [3].
To produce an adaptive advantage, newly-acquired genes must be integrated into existing regulatory circuits so that they are expressed in an environmentally and physiologically appropriate manner. If a new gene is too highly expressed, it risks imposing a significant fitness cost upon the host and placing it at a competitive disadvantage. Conversely, if a new gene is only weakly expressed, it may not provide the host with a selective advantage and hence will not be maintained. Thus, the acquisition or evolution of an appropriate regulatory circuit represents an evolutionary threshold. This review focuses on the mechanisms used by S. Typhimurium to integrate new genes into existing transcriptional regulatory networks. Recent findings to suggest the existence of analogous mechanisms in distantly related bacterial species will also be discussed.
Global repression by xenogeneic silencing
Horizontally-acquired DNA in bacteria is readily distinguished from ancestral DNA on the basis of its sequence characteristics, in particular, AT-content higher than that of the ancestral genome [3, 4]. Findings over the past decade have revealed that many species of bacteria possess DNA-binding proteins that recognize and silence the expression of AT-rich DNA. First described in S. Typhimurium, [5, 6], “xenogeneic silencing” of AT-rich DNA by nucleoid-associated proteins [5, 7] effectively allows a cell to discriminate between “self” and “non-self” sequences, repressing foreign gene expression to avoid potential fitness costs. This suggests that horizontally-acquired DNA that is relatively AT-rich is more likely to be retained by recipient cells because it is recognized by xenogeneic silencing proteins and thus better tolerated. Xenogeneic silencing proteins fall into at least three different classes based on structural similarity: the H-NS-like proteins of Salmonella and other species of proteobacteria [8, 9], the MvaT-like proteins of Pseudomonas spp. [10, 11], and the Lsr2-like proteins of the Actinomycetes [12–14]. These classes of proteins share an ability to selectively bind AT-rich DNA and prevent gene expression by forming higher order oligomers that further polymerize to comprise extensive nucleoprotein structures.
A remarkable feature common to all three classes of silencing proteins is their ability to target AT-rich DNA without strict sequence specificity. Studies of the DNA-binding domains of H-NS and Lsr2 have determined that both proteins recognize structural features unique to the minor groove of AT-rich DNA [15], which is typically narrower and deeper than that of GC-rich DNA [16]. AT-rich DNA sequences vary considerably in their affinity for H-NS, and several studies have shown that a critical determinant for high-affinity binding by H-NS and Lsr2 is the “TpA step”, a thymine base immediately followed by an adenine, which distorts the shape of the minor groove more than other dinucleotide steps and imparts a high degree of flexibility to DNA. Both H-NS and Lsr2 utilize a “prokaryotic AT-hook” motif (Q/RGR) that inserts into the minor groove and forms extensive interactions along the groove floor [15]. The flexibility and distortions provided by TpA steps facilitate the insertion of the AT-hook motif [15, 17]. It remains unclear how MvaT-like proteins, which lack an AT-hook motif, selectively bind AT-rich DNA.
Multiple studies have shown that the higher-order structure of the nucleoprotein complex plays a critical role in silencing, and that DNA binding alone is insufficient to block gene expression at most loci [18–20]. Structural and mutational studies of H-NS reveal that the molecule contains two separate dimerization domains. Oligomers consist of chains of H-NS molecules linked “head-to-head/tail-to-tail” [21]. Two dimerization interfaces have also been experimentally observed in MvaT and Lsr2 [22, 23], suggesting that these molecules oligomerize in a similar chain-like fashion. The manner in which xenogeneic silencing protein chains interact with DNA has recently been a subject of controversy. Atomic force microscopy studies of H-NS [24, 25], MvaT [26] and Lsr2 [20] have demonstrated that these molecules can bridge adjacent DNA duplexes, suggesting that silencing may occur when RNA polymerase is trapped in the loops formed between such bridges [25, 27, 28]. However other studies have shown that the bridging effect is strongly dependent on the concentration of intracellular magnesium [29, 30]. At ~1 mM magnesium concentrations thought to more closely approximate the intracellular environment [31], all three classes of xenogeneic silencing proteins form a stiffened filament when bound to DNA [19, 30, 32]. Furthermore, mutational analyses have demonstrated that mutant H-NS proteins that are incapable of forming stiffened filaments in vitro are also incapable of silencing gene expression in vivosuggesting that stiffening is the mode of DNA binding responsible for transcriptional repression [33].
Transcriptional activation is dependent on a conserved promoter architecture
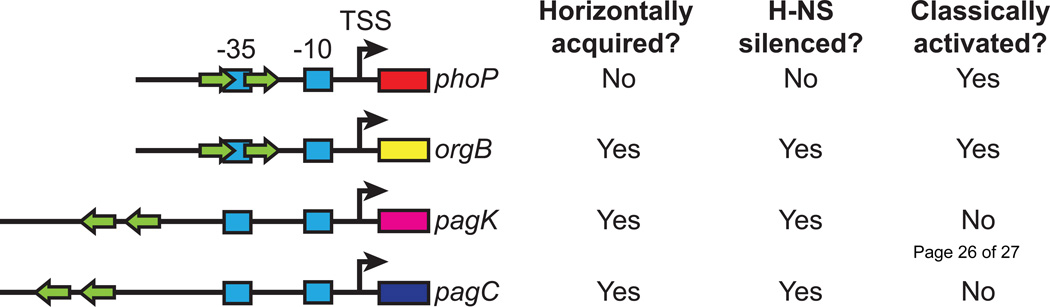
As H-NS is constitutively associated with the bacterial chromosome [34], xenogeneic silencing renders most horizontally-acquired genes transcriptionally inactive by default. This places H-NS in a central role within the regulatory network of many of virulence genes. Expression requires the regulated relief of H-NS-mediated repression, otherwise known as counter-silencing [7, 35]. The regulation of bacterial gene expression in response to environmental or physiological cues is often achieved by two-component systems, exemplified by the PhoPQ system, which is essential for Salmonella virulence [36, 37]. The PhoPQ response regulator, PhoP, is a prototypical transcriptional activator, regulating gene expression in response to low extracellular Mg2+ [38], acidic pH [39], and cationic antimicrobial peptides [40]. Recent studies have demonstrated that PhoP acts via different mechanisms at ancestral and horizontally-acquired promoters [41]. Bioinformatic analysis of the PhoP regulon has demonstrated that horizontally-acquired genes exhibit variable promoter architectures, with PhoP-binding sites at a variety of positions and orientations relative to the transcription start site [42, 43] (Figure 1). In contrast, ancestral genes exhibit a conserved promoter architecture, with a single PhoP binding site overlapping the −35 box. In vitro reconstitution of these regulatory circuits using supercoiled templates has revealed that PhoP is only capable of activating promoters with ancestral architectures, at which it presumably interacts directly with the RNA polymerase (RNAP) holoenzyme [41]. PhoP is unable to up-regulate promoters exhibiting alternative architecture unless they are silenced by H-NS, indicating that PhoP acts at these promoters by counter-silencing rather than by classical activation. However, H-NS-mediated silencing does not prevent activation, as horizontally-acquired promoters possessing an ancestral promoter architecture, such as orgBare capable of being activated by PhoP.
Figure 1. Architectural and regulatory logic of the PhoP regulon.
The PhoP regulon contains both ancestral and horizontally-acquired genes, a few of which are depicted here. Promoter architecture, i.e. the position and orientation of a transcription factor binding site (green arrows) relative to the transcription start site (TSS; bent arrow) of horizontally-acquired genes, is variable. Horizontally-acquired genes lacking conserved promoter architecture are not directly activated, as they do not permit an appropriate interaction between PhoP and RNAP. Genes exhibiting ancestral architecture are directly activated by PhoP, including those that have been horizontally-acquired (orgB). Blue boxes indicate the approximate positions of the −10 and −35 boxes. The figure is based on results presented elsewhere [5, 41, 42].
Diversity in promoter architecture [44] suggests flexibility in the interaction of PhoP with horizontally-acquired promoters, whereas the interaction of PhoP with ancestral promoters appears to be highly constrained. Structural analyses of a related OmpR-family regulator, PhoB, suggests that a response regulator must be precisely positioned at the −35 box for interaction with RNAP holoenzyme and transcription activation to occur. The interaction results in remodeling of the linker between domains 3 and 4 of the σ-subunit, which allows passage of the nascent transcript through the RNA exit channel to result in promoter escape [45]. Such activation by RNAP remodeling is inconceivable at many horizontally-acquired promoter architectures, at which the PhoP binding site is situated at a variable distance and orientation upstream of the −35 box. It is unknown whether this mechanism applies to other response regulators with variable promoter architectures, such as ArcA [46] and CpxR [47] in E. coli. However, a distributional analysis of defined operator sites in E. coli in RegulonDB shows that although positive regulators bind a variety of promoter architectures, there is a strong selection for binding sites positioned near the −35 box [48], suggesting that such structural constraints are not unique to PhoP.
Disruptive counter-silencing
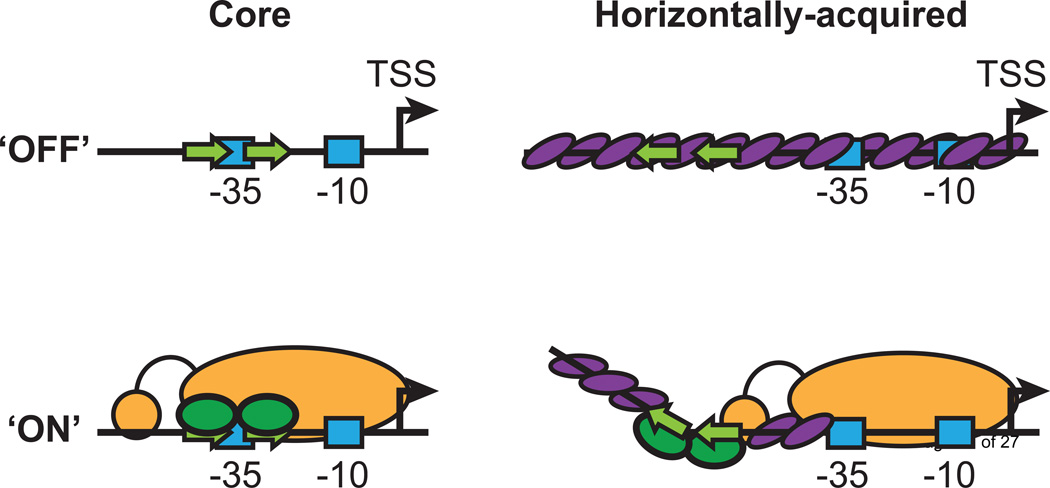
Although there are some observations to suggest that counter-silencing can occur via a supportive mechanism, wherein a counter-silencer stabilizes RNAP binding in the presence of H-NS [41], most recent studies suggest a disruptive mechanism, in which the H-NS-DNA complex is remodeled to allow RNAP binding or procession (Figure 2). This was first demonstrated by the construction of synthetic counter-silencing circuits, in which binding sites for the Lac and λ repressors were inserted within an H-NS-silenced domain upstream of the silenced bgl operon in E. coli [49]. Analogous experiments have been performed using the Shigella flexneri virulence regulator, VirB, to counter-silence the proU promoter [50]. In both studies, counter-silencing was observed at a variety of synthetic promoter architectures, likely due to the fact that the length of the H-NS-DNA filament provides multiple sites of potential interaction with a counter-silencer. VirB is thought to counter H-NS silencing by binding to DNA, oligomerizing, and bending DNA around itself, thereby disrupting the H-NS-DNA filament [51]. A similar model may be invoked to explain the activity of LeuO, a LysR-type transcriptional regulator that has been proposed to act as an H-NS counter-silencer in S. Typhimurium and E. coli [52, 53]. LeuO has been suggested to bind DNA between the H-NS nucleation site and promoter, thereby creating a barrier to H-NS polymerization [54, 55]. LeuO binds DNA as a tetramer [56] and, like VirB, is capable of bending DNA and wrapping it around its oligomerized form. Disruptive counter-silencing does not appear to require ejection of H-NS from the promoter region. In a recent analysis of PhoP-mediated counter-silencing, a quantitative comparative method of DNA footprint analysis designated Differential DNA Footprint Analysis (DDFA) was used to detect subtle structural changes in the H-NS-DNA nucleoprotein complex. DDFA demonstrated that H-NS remains bound to a promoter region even under counter-silencing conditions [41]. Rather than displacing H-NS, PhoP induces a bend in the H-NS-DNA filament to restore open complex formation. Bending of the H-NS-DNA filament may be essential for counter-silencing to occur, as mutational analyses have correlated stiffened filament formation with silencing in vivo [33].
Figure 2. Mechanisms of activation and counter-silencing.
Ancestral core genes are transcriptionally inactive (OFF) until a transcription activator (green ovals) binds at a conserved position (green arrows) relative to the transcription start site (TSS; bent arrow) and interacts directly with RNAP (yellow) to promote transcription (ON). In contrast, horizontally-acquired genes are repressed by oligomerized H-NS (purple ovals), which binds a large region of promoter DNA and occludes RNAP binding. A counter-silencing transcription factor can bind at an alternative site relative to the TSS to re-model the H-NS-DNA complex, thereby restoring RNAP binding and transcription. Blue boxes indicate the approximate positions of the −10 and −35 boxes at each promoter.
Disruptive counter-silencing also allows the evolutionary exaptation of DNA-binding proteins not otherwise involved in the up-regulation of transcription, as it does not require a productive interaction between a DNA-binding protein and RNAP. For example, VirB is not homologous to known transcription regulators, but rather is related to the ParB-family of plasmid partitioning proteins [57, 58]. The PhoP co-regulator SlyA and homologs such as RovA from Yersinia spp. also appear to have been co-opted from their original functions as transcriptional repressors to serve as counter-silencers [59–62].
H-NS-like proteins can also contribute to counter-silencing. This has been most clearly demonstrated in studies of Ler, an H-NS-like protein that paradoxically acts as a counter-silencer in pathogenic strains of E. coli. Structure-function and domain-swapping analyses have revealed that the ability of Ler to antagonize H-NS results from differences in their oligomerization domains, as the DNA-binding domains of these proteins are functionally interchangeable [63, 64]. This reinforces the notion that the structure of the DNA-bound H-NS oligomer is a critical determinant of its activity. Ler appears to disrupt the H-NS nucleoprotein structure by wrapping AT-rich DNA in non-cooperative toroidal complexes [64, 65]. Along similar lines, the H-NST family of proteins in E. coli are truncated derivatives of H-NS that interact with endogenous H-NS via its oligomerization domain to disrupt silencing [66]. H-NST proteins are themselves horizontally-acquired, and modulate the expression of genes on the LEE (locus of enterocyte effacement) pathogenicity island [67].
Architectural conservation of horizontally-acquired regulatory circuits
Horizontally-acquired genes are not regulated solely by counter-silencing, as regulation by classical activation or counter-silencing is determined by the evolutionary relationship between the regulator and the target promoter. Activation is more likely when an entire regulatory circuit is transferred. For example, the Salmonella SsrAB two-component system, which is horizontally-acquired, activates genes both within and outside of SPI-2 [68]. Because SsrAB is encoded by SPI-2, its SPI-2 targets are effectively ancestral, as opposed to horizontally-acquired targets outside of SPI-2. As a result, the response regulator SsrB binds many SPI-2 promoters at a conserved position located immediately upstream of the −35 box, where it appears to both disrupt the H-NS-DNA complex and activate transcription [69, 70]. Similarly, the SPI-1 regulatory genes hilC and hilD are situated within the pathogenicity island and are responsible for up-regulating the SPI-1 master regulator, hilA [71]. Notably, this activity is at least partly dependent on a direct interaction with the carboxyl-terminal domain of the RNAP α subunit (α-CTD) which is sensitive to the position of the HilC/D binding site [72, 73]. Whether this is true for HilC/D targets outside of SPI-1, such as ssrABwhere HilD is thought to oppose H-NS [74, 75], remains to be determined.
Concluding Remarks
The nucleoid-associated protein H-NS and related proteins are able to recognize and silence horizontally-acquired genes exhibiting a variety of promoter architectures. Architectural flexibility can explain why horizontally-acquired genes are more likely to be integrated into existing regulatory networks via silencing and counter-silencing, rather than by the de novo evolution of classical activation circuitry. The versatility of counter-silencing facilitates the evolution of complex regulatory circuits that allow signal integration and regulatory checkpoints [76]. Three classes of xenogeneic silencing proteins have been described, and it is likely that more remain to be discovered. High resolution genome-wide technologies such as ChIP-Seq and ChIP-exo, along with bioinformatic analyses, can help to further contrast the architectural and evolutionary logic of classical activation and counter-silencing mechanisms in other systems. Meanwhile, the reconstitution of regulatory circuits in vitro will allow the further characterization of the molecular events taking place during activation and counter-silencing. The ability of counter-silencing to accommodate multiple promoter architectures and the large number of counter-silenced genes already identified suggest that counter-silencing is a predominant mechanism of transcription regulation in bacteria that has played a crucial role in facilitating evolution by horizontal gene transfer.
Highlights.
H-NS and other nucleoid-associated proteins can silence expression of horizontally-acquired genes.
Transcriptional silencing is countered by other DNA-binding proteins in response to environmental and physiological cues.
DNA bending with disruption of the H-NS-DNA filament is one mechanism by which counter-silencing may occur.
Counter-silencing accommodates a variety of promoter architectures which facilitates the integration of horizontally-acquired genes into existing regulatory networks.
Acknowledgements
This work was supported by the National Institutes of Health (AI101084) and the Canadian Institutes of Health Research (MOP-86683).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Recommended Reading
Papers of particular interest, published within the period of review have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Groisman EA, Ochman H. How Salmonella became a pathogen. Trends Microbiol. 1997;5:343–349. doi: 10.1016/S0966-842X(97)01099-8. [DOI] [PubMed] [Google Scholar]
- 2.Groisman EA, Ochman H. Pathogenicity islands: bacterial evolution in quantum leaps. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 3.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 4.Groisman EA, Saier MHJ, Ochman H. Horizontal transfer of a phosphatase gene as evidence for mosaic structure of the Salmonella genome. EMBO J. 1992;11:1309–1316. doi: 10.1002/j.1460-2075.1992.tb05175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. A striking correlation between H-NS binding and AT-content was observed.
- 6. Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. H-NS does not co-localize with RNA polymerase.
- 7.Navarre WW, McClelland M, Libby SJ, Fang FC. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 2007;21:1456–1471. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]
- 8.Bertin P, Benhabiles N, Krin E, Laurent-Winter C, Tendeng C, Turlin E, Thomas A, Danchin A, Brasseur R. The structural and functional organization of H-NS-like proteins is evolutionarily conserved in gram-negative bacteria. Mol Microbiol. 1999;31:319–329. doi: 10.1046/j.1365-2958.1999.01176.x. [DOI] [PubMed] [Google Scholar]
- 9.Bertin P, Hommais F, Krin E, Soutourina O, Tendeng C, Derzelle S, Danchin A. H-NS and H-NS-like proteins in Gram-negative bacteria and their multiple role in the regulation of bacterial metabolism. Biochimie. 2001;83:235–241. doi: 10.1016/s0300-9084(01)01247-0. [DOI] [PubMed] [Google Scholar]
- 10.Tendeng C, Soutourina OA, Danchin A, Bertin PN. MvaT proteins in Pseudomonas spp.: a novel class of H-NS-like proteins. Microbiology. 2003;149:3047–3050. doi: 10.1099/mic.0.C0125-0. [DOI] [PubMed] [Google Scholar]
- 11.Castang S, McManus HR, Turner KH, Dove SL. H-NS family members function coordinately in an opportunistic pathogen. Proc Natl Acad Sci U S A. 2008;105:18947–18952. doi: 10.1073/pnas.0808215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon BR, Imperial R, Wang L, Navarre WW, Liu J. Lsr2 of Mycobacterium represents a novel class of H-NS-like proteins. J Bacteriol. 2008;190:7052–7059. doi: 10.1128/JB.00733-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon BR, Li Y, Wang L, Sintsova A, van Bakel H, Tian S, Navarre WW, Xia B, Liu J. Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2010;107:5154–5159. doi: 10.1073/pnas.0913551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colangeli R, Helb D, Vilcheze C, Hazbon MH, Lee CG, Safi H, Sayers B, Sardone I, Jones MB, Fleischmann RD, et al. Transcriptional regulation of multi-drug tolerance and antibiotic-induced responses by the histone-like protein Lsr2 in M. tuberculosis. PLoS Pathog. 2007;3:e87. doi: 10.1371/journal.ppat.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gordon BR, Li Y, Cote A, Weirauch MT, Ding P, Hughes TR, Navarre WW, Xia B, Liu J. Structural basis for recognition of AT-rich DNA by unrelated xenogeneic silencing proteins. Proc Natl Acad Sci U S A. 2011;108:10690–10695. doi: 10.1073/pnas.1102544108. Identification of an H-NS structural motif associated with recognition of AT-rich DNA, which has convergently evolved in distantly-related bacterial species.
- 16.Johnson RC, Stella S, Heiss JK. Bending and compaction of DNA by proteins. In: Rice PA, Correll CC, editors. Protein-Nucleic Acid Interactions. RSC Press; 2008. pp. 176–220. [Google Scholar]
- 17.Sette M, Spurio R, Trotta E, Brandizi C, Brandi A, Pon CL, Barbato G, Boelens R, Gualerzi CO. Sequence-specific recognition of DNA by the C-terminal domain of nucleoid-associated protein H-NS. J Biol Chem. 2009;284:30453–30462. doi: 10.1074/jbc.M109.044313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spurio R, Falconi M, Brandi A, Pon CL, Gualerzi CO. The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. EMBO J. 1997;16:1795–1805. doi: 10.1093/emboj/16.7.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winardhi RS, Fu W, Castang S, Li Y, Dove SL, Yan J. Higher order oligomerization is required for H-NS family member MvaT to form gene-silencing nucleoprotein filament. Nucleic Acids Res. 2012;40:8942–8952. doi: 10.1093/nar/gks669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen JM, Ren H, Shaw JE, Wang YJ, Li M, Leung AS, Tran V, Berbenetz NM, Kocincova D, Yip CM, et al. Lsr2 of Mycobacterium tuberculosis is a DNA-bridging protein. Nucleic Acids Res. 2008;36:2123–2135. doi: 10.1093/nar/gkm1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Arold ST, Leonard PG, Parkinson GN, Ladbury JE. H-NS forms a superhelical protein scaffold for DNA condensation. Proc Natl Acad Sci U S A. 2010;107:15728–15732. doi: 10.1073/pnas.1006966107. A structural model for H-NS oligomerization, which is capable of accommodating both H-NS binding modes.
- 22.Castang S, Dove SL. High-order oligomerization is required for the function of the H-NS family member MvaT in Pseudomonas aeruginosa. Mol Microbiol. 2010;78:916–931. doi: 10.1111/j.1365-2958.2010.07378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Summers EL, Meindl K, Uson I, Mitra AK, Radjainia M, Colangeli R, Alland D, Arcus VL. The structure of the oligomerization domain of Lsr2 from Mycobacterium tuberculosis eveals a mechanism for chromosome organization and protection. PLoS One. 2012;7:e38542. doi: 10.1371/journal.pone.0038542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dame RT, Wyman C, Goosen N. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acids Res. 2000;28:3504–3510. doi: 10.1093/nar/28.18.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dame RT, Wyman C, Wurm R, Wagner R, Goosen N. Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1. J Biol Chem. 2002;277:2146–2150. doi: 10.1074/jbc.C100603200. [DOI] [PubMed] [Google Scholar]
- 26.Dame RT, Luijsterburg MS, Krin E, Bertin PN, Wagner R, Wuite GJ. DNA bridging: a property shared among H-NS-like proteins. J Bacteriol. 2005;187:1845–1848. doi: 10.1128/JB.187.5.1845-1848.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schroder O, Wagner R. The bacterial DNA-binding protein H-NS represses ribosomal RNA transcription by trapping RNA polymerase in the initiation complex. J Mol Biol. 2000;298:737–748. doi: 10.1006/jmbi.2000.3708. [DOI] [PubMed] [Google Scholar]
- 28.Shin M, Song M, Rhee JH, Hong Y, Kim YJ, Seok YJ, Ha KS, Jung SH, Choy HE. DNA looping-mediated repression by histone-like protein H-NS: specific requirement of Esigma70 as a cofactor for looping. Genes Dev. 2005;19:2388–2398. doi: 10.1101/gad.1316305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amit R, Oppenheim AB, Stavans J. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys J. 2003;84:2467–2473. doi: 10.1016/S0006-3495(03)75051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Y, Chen H, Kenney LJ, Yan J. A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev. 2010;24:339–344. doi: 10.1101/gad.1883510. Biophysical analysis helps to resolve the controversy concerning H-NS binding by showing alternative modes of binding in response to divalent cation concentrations.
- 31.Martin-Orozco N, Touret N, Zaharik ML, Park E, Kopelman R, Miller S, Finlay BB, Gros P, Grinstein S. Visualization of vacuolar acidification-induced transcription of genes of pathogens inside macrophages. Mol Biol Cell. 2006;17:498–510. doi: 10.1091/mbc.E04-12-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qu Y, Lim CJ, Whang YR, Liu J, Yan J. Mechanism of DNA organization by Mycobacterium tuberculosis protein Lsr2. Nucleic Acids Res. 2013;41:5263–5272. doi: 10.1093/nar/gkt249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim CJ, Lee SY, Kenney LJ, Yan J. Nucleoprotein filament formation is the structural basis for bacterial protein H-NS gene silencing. Sci Rep. 2012;2:509. doi: 10.1038/srep00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kahramanoglou C, Seshasayee AS, Prieto AI, Ibberson D, Schmidt S, Zimmermann J, Benes V, Fraser GM, Luscombe NM. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res. 2011;39:2073–2091. doi: 10.1093/nar/gkq934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoebel DM, Free A, Dorman CJ. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology. 2008;154:2533–2545. doi: 10.1099/mic.0.2008/020693-0. [DOI] [PubMed] [Google Scholar]
- 36.Fields PI, Groisman EA, Heffron F. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science. 1989;243:1059–1062. doi: 10.1126/science.2646710. [DOI] [PubMed] [Google Scholar]
- 37.Miller SI, Kukral AM, Mekalanos JJ. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci U S A. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia Vescovi E, Soncini FC, Groisman EA. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 39.Prost LR, Daley ME, Le Sage V, Bader MW, Le Moual H, Klevit RE, Miller SI. Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol Cell. 2007;26:165–174. doi: 10.1016/j.molcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 40.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 41. Will WR, Bale DH, Reid PJ, Libby SJ, Fang FC. Evolutionary expansion of a regulatory network by counter-silencing. Nat Commun. 2014 doi: 10.1038/ncomms6270. in press. Reconstitution of PhoP regulatory circuits in vitro shows that activation only occurs at ancestral promoters with conserved architecture, whereas the architecture of horizontally-acquired counter-silenced promoters is less constrained, thereby facilitating regulatory integration.
- 42. Zwir I, Latifi T, Perez JC, Huang H, Groisman EA. The promoter architectural landscape of the Salmonella PhoP regulon. Mol Microbiol. 2012;84:463–485. doi: 10.1111/j.1365-2958.2012.08036.x. Comprehensive analysis of PhoP regulon demonstrates greater variability of promoter architecture in horizontally-acquired genes.
- 43.Zwir I, Shin D, Kato A, Nishino K, Latifi T, Solomon F, Hare JM, Huang H, Groisman EA. Dissecting the PhoP regulatory network of Escherichia coli and Salmonella enterica. Proc Natl Acad Sci U S A. 2005;102:2862–2867. doi: 10.1073/pnas.0408238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez JC, Groisman EA. Evolution of transcriptional regulatory circuits in bacteria. Cell. 2009;138:233–244. doi: 10.1016/j.cell.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blanco AG, Canals A, Bernues J, Sola M, Coll M. The structure of a transcription activation subcomplex reveals how sigma(70) is recruited to PhoB promoters. EMBO J. 2011;30:3776–3785. doi: 10.1038/emboj.2011.271. Structural analysis of an OmpR-family regulator helps to explain the architectural requirements of ancestral promoters.
- 46.Park DM, Akhtar MS, Ansari AZ, Landick R, Kiley PJ. The bacterial response regulator ArcA uses a diverse binding site architecture to regulate carbon oxidation globally. PLoS Genet. 2013;9:e1003839. doi: 10.1371/journal.pgen.1003839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Price NL, Raivio TL. Characterization of the Cpx regulon in Escherichia colistrain MC4100. J Bacteriol. 2009;191:1798–1815. doi: 10.1128/JB.00798-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cox RS, Surette MG, Elowitz MB. Programming gene expression with combinatorial promoters. Mol Syst Biol. 2007;3:145. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caramel A, Schnetz K. Lac and lambda repressors relieve silencing of the Escherichia coli bgl promoter. Activation by alteration of a repressing nucleoprotein complex. J Mol Biol. 1998;284:875–883. doi: 10.1006/jmbi.1998.2191. [DOI] [PubMed] [Google Scholar]
- 50. Kane KA, Dorman CJ. Rational design of an artificial genetic switch: Co-option of the H-NS-repressed proU operon by the VirB virulence master regulator. J Bacteriol. 2011;193:5950–5960. doi: 10.1128/JB.05557-11. Construction of a synthetic counter-silencing circuit using unrelated components demonstrates the inherent flexibility of counter-silencing.
- 51.Gao X, Zou T, Mu Z, Qin B, Yang J, Waltersperger S, Wang M, Cui S, Jin Q. Structural insights into VirB-DNA complexes reveal mechanism of transcriptional activation of virulence genes. Nucleic Acids Res. 2013;41:10529–10541. doi: 10.1093/nar/gkt748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dillon SC, Espinosa E, Hokamp K, Ussery DW, Casadesus J, Dorman CJ. LeuO is a global regulator of gene expression in Salmonella enterica serovar Typhimurium. Mol Microbiol. 2012;85:1072–1089. doi: 10.1111/j.1365-2958.2012.08162.x. [DOI] [PubMed] [Google Scholar]
- 53.Shimada T, Bridier A, Briandet R, Ishihama A. Novel roles of LeuO in transcription regulation of E. coli genome: antagonistic interplay with the universal silencer H-NS. Mol Microbiol. 2011;82:378–397. doi: 10.1111/j.1365-2958.2011.07818.x. [DOI] [PubMed] [Google Scholar]
- 54.Chen CC, Ghole M, Majumder A, Wang Z, Chandana S, Wu HY. LeuO-mediated transcriptional derepression. J Biol Chem. 2003;278:38094–38103. doi: 10.1074/jbc.M300461200. [DOI] [PubMed] [Google Scholar]
- 55.Chen CC, Wu HY. LeuO protein delimits the transcriptionally active and repressive domains on the bacterial chromosome. J Biol Chem. 2005;280:15111–15121. doi: 10.1074/jbc.M414544200. [DOI] [PubMed] [Google Scholar]
- 56.Guadarrama C, Medrano-Lopez A, Oropeza R, Hernandez-Lucas I, Calva E. The Salmonella enterica serovar Typhi LeuO global regulator forms tetramers: residues involved in oligomerization, DNA binding, and transcriptional regulation. J Bacteriol. 2014;196:2143–2154. doi: 10.1128/JB.01484-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beloin C, McKenna S, Dorman CJ. Molecular dissection of VirB, a key regulator of the virulence cascade of Shigella flexneri. J Biol Chem. 2002;277:15333–15344. doi: 10.1074/jbc.M111429200. [DOI] [PubMed] [Google Scholar]
- 58.Watanabe H, Arakawa E, Ito K, Kato J, Nakamura A. Genetic analysis of an invasion region by use of a Tn3-lactransposon and identification of a second positive regulator gene invE, for cell invasion of Shigella sonnei: significant homology of invEwith ParB of plasmid P1. J Bacteriol. 1990;172:619–629. doi: 10.1128/jb.172.2.619-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heroven AK, Nagel G, Tran HJ, Parr S, Dersch P. RovA is autoregulated and antagonizes H-NS-mediated silencing of invasin and rovA expression in Yersinia pseudotuberculosis. Mol Microbiol. 2004;53:871–888. doi: 10.1111/j.1365-2958.2004.04162.x. [DOI] [PubMed] [Google Scholar]
- 60.Navarre WW, Halsey TA, Walthers D, Frye J, McClelland M, Potter JL, Kenney LJ, Gunn JS, Fang FC, Libby SJ. Co-regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Mol Microbiol. 2005;56:492–508. doi: 10.1111/j.1365-2958.2005.04553.x. [DOI] [PubMed] [Google Scholar]
- 61.Ellison DW, Miller VL. H-NS represses inv transcription in Yersinia enterocolitica through competition with RovA and interaction with YmoA. J Bacteriol. 2006;188:5101–5112. doi: 10.1128/JB.00862-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cathelyn JS, Ellison DW, Hinchliffe SJ, Wren BW, Miller VL. The RovA regulons of Yersinia enterocolitica and Yersinia pestis are distinct: evidence that many RovA-regulated genes were acquired more recently than the core genome. Mol Microbiol. 2007;66:189–205. doi: 10.1111/j.1365-2958.2007.05907.x. [DOI] [PubMed] [Google Scholar]
- 63.Mellies JL, Larabee FJ, Zarr MA, Horback KL, Lorenzen E, Mavor D. Ler interdomain linker is essential for anti-silencing activity in enteropathogenic Escherichia coli. Microbiology. 2008;154:3624–3638. doi: 10.1099/mic.0.2008/023382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Winardhi RS, Gulvady R, Mellies JL, Yan J. Locus of Enterocyte Effacement-encoded Regulator (Ler) of Pathogenic Escherichia coli Competes Off Histone-like Nucleoid-structuring Protein (H-NS) through Noncooperative DNA Binding. J Biol Chem. 2014;289:13739–13750. doi: 10.1074/jbc.M113.545954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mellies JL, Benison G, McNitt W, Mavor D, Boniface C, Larabee FJ. Ler of pathogenic Escherichia coli forms toroidal protein-DNA complexes. Microbiology. 2011;157:1123–1133. doi: 10.1099/mic.0.046094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williamson HS, Free A. A truncated H-NS-like protein from enteropathogenic Escherichia coli acts as an H-NS antagonist. Mol Microbiol. 2005;55:808–827. doi: 10.1111/j.1365-2958.2004.04421.x. [DOI] [PubMed] [Google Scholar]
- 67.Levine JA, Hansen AM, Michalski JM, Hazen TH, Rasko DA, Kaper JB. H-NST induces LEE expression and the formation of attaching and effacing lesions in enterohemorrhagic Escherichia coli. PLoS One. 2014;9:e86618. doi: 10.1371/journal.pone.0086618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tomljenovic-Berube AM, Mulder DT, Whiteside MD, Brinkman FS, Coombes BK. Identification of the regulatory logic controlling Salmonella pathoadaptation by the SsrA-SsrB two-component system. PLoS Genet. 2010;6:e1000875. doi: 10.1371/journal.pgen.1000875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walthers D, Li Y, Liu Y, Anand G, Yan J, Kenney LJ. Salmonella enterica response regulator SsrB relieves H-NS silencing by displacing H-NS bound in polymerization mode and directly activates transcription. J Biol Chem. 2011;286:1895–1902. doi: 10.1074/jbc.M110.164962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walthers D, Carroll RK, Navarre WW, Libby SJ, Fang FC, Kenney LJ. The response regulator SsrB activates expression of diverse Salmonella pathogenicity island 2 promoters and counters silencing by the nucleoid-associated protein H-NS. Mol Microbiol. 2007;65:477–493. doi: 10.1111/j.1365-2958.2007.05800.x. [DOI] [PubMed] [Google Scholar]
- 71.Schechter LM, Lee CA. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol Microbiol. 2001;40:1289–1299. doi: 10.1046/j.1365-2958.2001.02462.x. [DOI] [PubMed] [Google Scholar]
- 72.Boddicker JD, Knosp BM, Jones BD. Transcription of the Salmonella invasion gene activator hilA, requires HilD activation in the absence of negative regulators. J Bacteriol. 2003;185:525–533. doi: 10.1128/JB.185.2.525-533.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Olekhnovich IN, Kadner RJ. Contribution of the RpoA C-terminal domain to stimulation of the Salmonella enterica hilA promoter by HilC and HilD. J Bacteriol. 2004;186:3249–3253. doi: 10.1128/JB.186.10.3249-3253.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martinez LC, Banda MM, Fernandez-Mora M, Santana FJ, Bustamante VH. HilD Induces Expression of SPI-2 Genes by Displacing the Global Negative Regulator H-NS from ssrAB. J Bacteriol. 2014 Aug 18; doi: 10.1128/JB.01799-14. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Petrone BL, Stringer AM, Wade JT. Identification of HilD-regulated genes in Salmonella enterica serovar Typhimurium. J Bacteriol. 2014;196:1094–1101. doi: 10.1128/JB.01449-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shen S, Fang FC. Integrated stress responses in Salmonella. Int J Food Microbiol. 2012;152:75–81. doi: 10.1016/j.ijfoodmicro.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]