Abstract
In spite of substantial advances in defining the immunobiology and function of structural cells in lung diseases there is still insufficient knowledge to develop fundamentally new classes of drugs to treat many lung diseases. For example, there is compelling need for new therapeutic approaches to address severe persistent asthma that is insensitive to inhaled corticosteroids. Although the prevalence of steroid-resistant asthma is 5–10%, severe asthmatics require a disproportionate level of health care spending and constitute a majority of fatal asthma episodes. None of the established drug therapies including long-acting beta agonists or inhaled corticosteroids reverse established airway remodeling. Obstructive airways remodeling in patients with chronic obstructive pulmonary disease (COPD), restrictive remodeling in idiopathic pulmonary fibrosis (IPF) and occlusive vascular remodeling in pulmonary hypertension are similarly unresponsive to current drug therapy. Therefore, drugs are needed to achieve long-acting suppression and reversal of pathological airway and vascular remodeling. Novel drug classes are emerging from advances in epigenetics. Novel mechanisms are emerging by which cells adapt to environmental cues, which include changes in DNA methylation, histone modifications and regulation of transcription and translation by noncoding RNAs. In this review we will summarize current epigenetic approaches being applied to preclinical drug development addressing important therapeutic challenges in lung diseases. These challenges are being addressed by advances in lung delivery of oligonucleotides and small molecules that modify the histone code, DNA methylation patterns and miRNA function.
Keywords: Asthma, COPD, DNA methylation, fibrosis, histone code, noncoding RNA
1. Introduction
Epigenetic regulation of gene expression and protein abundance is carried out by a set of highly conserved processes that contribute to normal development and adaptation to changes in cellular and organ homeostasis. Any disease that alters the cellular milieu to the point of triggering adaptive changes in phenotype will likely involve one or more epigenetic mechanisms. Transcription is regulated by DNA methylation, often at CpG islands in promoters of protein-coding genes, and by posttranslational modification of histones. Noncoding RNAs also modify transcription, mRNA processing and mRNA stability to modulate protein abundance. Because DNA methylation, the histone code and microRNA-mediated gene silencing are highly conserved processes many clinically diverse conditions should respond to drugs that modify these epigenetic regulatory systems. It is also clear that epigenetic modifications can be cell and tissue specific, which suggests epigenetic therapies might be designed to be directed to particular diseases. It is also very important to note that changes in epigenetic modifications of DNA and histones and changes in miRNA expression may be a consequence of a disease as well as cause the disease. Substantial effort is now being spent defining the timing and the necessity of epigenetic adaptations in lung diseases. Many of those studies have identified potential new drug targets which are summarized in this review. The working hypothesis is that antagonizing causative epigenetic features of disease or antagonizing adaptive epigenetic responses might favor a return to more normal structure and function of the lung. There are two processes of particular interest in this regard - lung inflammation and subsequent lung remodeling. Lung inflammation is often well controlled by corticosteroid therapy, but in severe asthmatics may require additional approaches including novel antibody-based therapies targeting IgE, thymic stromal lymphopoietin or interleukin-5. For more information on these important developments in asthma therapy the reader is referred to recent advances in antibody-based antiinflammatory therapies (Gauvreau et al., 2014; Humbert et al., 2014; Bel et al., 2014; Ortega et al., 2014). In this review we will cite studies of epigenetic modifiers in lung disease as well as other diseases when they provide proof of key principles, but the focus will be on novel therapy of pulmonary hypertension, restrictive lung diseases and obstructive lung diseases. The reader should consult other recent reviews for a general appreciation of advances in epigenetic therapy in other organ systems (Dhanak & Jackson, 2014; Haldar & McKinsey, 2014; Natarajan, 2011; Tao et al., 2014).
In all cases the appeal of targeting epigenetic mechanisms is that they are reversible biochemical processes amenable to manipulation with small molecule drugs, and in some cases small oligonucleotides. This contrasts with mutations in genomic and mitochondrial DNA most of which are uncorrectable in a clinical setting. Identifying the best epigenetic targets and developing the best therapeutic strategies is a topic of great interest and high significance for developing novel drugs for lung diseases. In addition to targeting inflammation we suggest targeting tissue remodeling will be a fruitful strategy. Tissue remodeling is a common feature of pulmonary vascular disorders as well as obstructive and restrictive diseases of the airways. The key aspects of pathology of each disease will be described along with some emerging epigenetic targets. Recent progress in preclinical studies of small molecule and oligonucleotide modifiers will be summarized with the goal of focusing attention on promising therapeutic approaches that may add to the current standards of care.
2. Tissue remodeling in lung diseases
2.1 Pulmonary hypertension
Patients with pulmonary hypertension (PH) suffer from abnormally high pulmonary artery blood pressure that leads to right ventricular dysfunction.. In severe pulmonary arterial hypertension (Group I PH, PAH), the normally thin-walled, highly compliant pulmonary arteries become thickened, stiffer and hypercontractile (Chan & Loscalzo, 2008; Humbert et al., 2004; Rabinovitch, 2008). Peripheral pulmonary arteries become occluded by neointimal and plexiform lesions which are hypothesized to contribute to disease severity (Pietra et al., 2004; Tuder et al., 2007; Yi et al., 2000). Pathological vascular remodeling is due to increased proliferation and decreased apoptosis of endothelial cells, smooth muscle cells and adventitial fibroblasts. The resulting wall hypertrophy may also be due to disrupted autophagy, enhanced progenitor cell migration and differentiation as well as immune cell migration and differentiation. The biochemical processes that regulate these diverse processes are the subject of intense study, and are targets of numerous drugs in preclinical development (Morrell et al., 2013; Tuder et al., 2013). In addition to small molecule inhibitors of cell signaling pathways, microRNAs (miRNA) have become intensely studied molecular targets for novel anti-remodeling therapy (White et al., 2012). There is also emerging evidence for altered DNA methylation (Archer et al., 2010) and histone posttranslational modifications (Xu et al., 2010) in pulmonary hypertension, both of which are targets for new drug development (Saco et al., 2014).
2.2 Obstructive lung diseases – Asthma and COPD
Remodeling of the airways occurs in asthma and COPD resulting in obstruction of airflow. The pathophysiology of the two diseases is quite different, but there are some common features relevant to identifying novel targets for anti-remodeling drug therapy. Asthma is a multifactorial syndrome triggered by allergens, infections, aspirin, exercise or cold air that results in symptoms of obstruction. The wheezing, and shortness of breath is frequently, but not always, reversible with bronchodilators. Treatment with inhaled corticosteroids usually controls allergic inflammation and prevents asthma episodes. Acute therapy with beta agonists relaxes airway smooth muscle and relieves the symptoms in most subjects. However, in a significant minority of asthmatics (5–10%) airway obstruction is not reversible (Chipps et al., 2012; Sullivan et al., 2007). In severe cases steroid resistance and lack of response to beta agonists can result in potentially fatal attacks (Wenzel, 2005). The lack of adequate treatment of severe asthma makes a compelling case for developing new therapies. Recent progress in novel anti-inflammatory therapy is described elsewhere (Bel et al., 2014; Humbert et al., 2014; Gauvreau et al., 2014; Ortega et al., 2014). The goal of our prospective analysis is to describe potential targets for anti-remodeling therapy.
Aside from bronchial thermoplasty there are no clinically proven therapies to reverse airway obstruction in severe asthmatics, whether they are steroid-resistant or not. What is needed is an effective way to prevent or reverse airway smooth muscle hyperplasia and hypertrophy, mucosal metaplasia, submucosal and parenchymal fibrosis and persistent inflammation. This is a challenging task because multiple cell types contribute to airway and vascular remodeling and no single agent is likely to reverse all features of pathological remodeling. Thus, it is imperative that we identify drug targets that affect those cells most responsive to treatment.
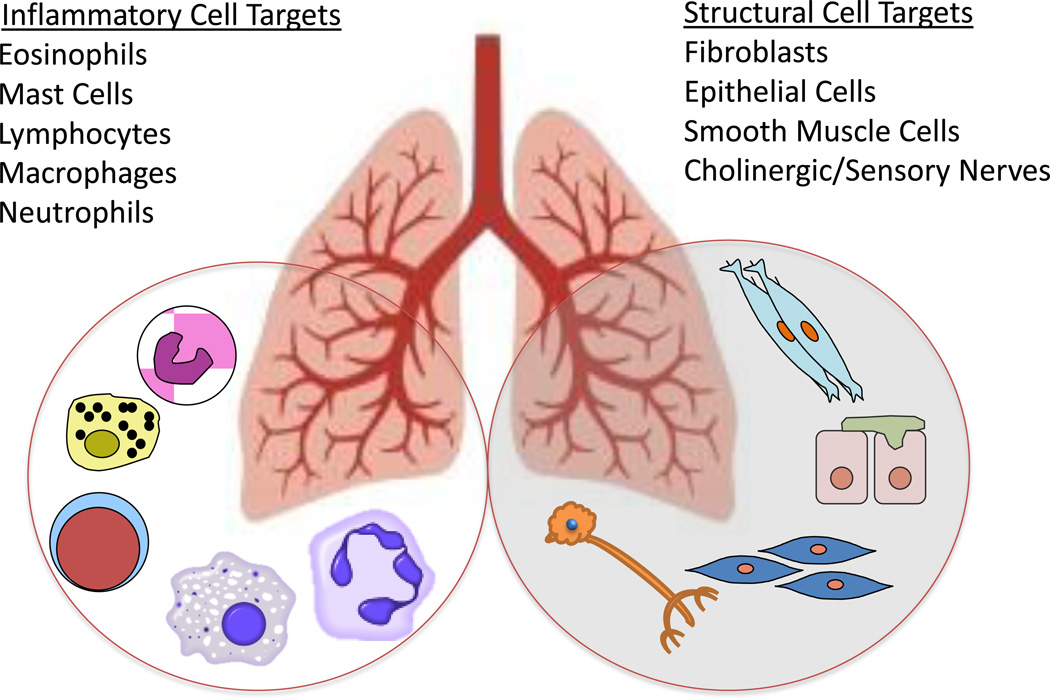
Therapy of COPD overlaps with asthma therapy in that corticosteroids are used for their anti-inflammatory effects and beta agonists for bronchodilation. A key difference between asthma and COPD is that obstruction is not fully reversible in COPD. Patients with COPD often present with emphysema, bronchitis or both. Smoking is a common insult that elicits mucosal hyperplasia and excess mucous production as well as smooth muscle hypertrophy in the small airways. In both asthma and COPD inflammation is a proximal event leading to symptoms, but there are key differences in the immune cell infiltrates; eosinophilia in many asthmatics versus neutrophils in COPD. Another key difference is destruction of elastin in the lung parenchyma in COPD by neutrophil elastases coincident with increased collagen deposition. This suggests that optimal therapy of COPD would include drugs to enhance parenchymal repair. In both COPD and asthma significant structural changes occur in the airways secondary to cell proliferation, cellular hypertrophy and fibrosis. The initiating insults are different, but there are common cellular targets (epithelial cells, smooth muscle, fibroblasts and immune cells) (Figure 1). Common cell types suggests there might be common molecular events required for remodeling and repair that will be suitable targets for novel anti-remodeling drugs.
Figure 1.
Lung cells involved in airway, vascular and parenchymal remodeling are potential targets for novel epigenetic therapies designed to prevent or repair remodeling. Both inflammatory and structural cells contribute to remodeling in asthma, COPD, IPF and pulmonary hypertension. The contributions of epigenetic processes to the function of each cell type in the lung is a topic of intense interest and rapid progress in lung cell biology.
2.3 Idiopathic pulmonary fibrosis (IPF)
IPF is a syndrome characterized by diffuse damage to alveoli of unknown etiology. It is thought to involve interstitial pneumonia and repeated bouts of alveolitis. Inflammation, oxidative stress and proteases of the coagulation cascade are thought to increase proliferation and reduce apoptosis of fibroblasts which synthesize inappropriate amounts of extracellular matrix proteins. There are several recent reviews of the mechanisms and novel targets of drug therapy for IPF that should be consulted for a broader perspective (Barkauskas & Noble, 2014; Baroke et al., 2013; Blackwell et al., 2014; Lota & Wells, 2013).
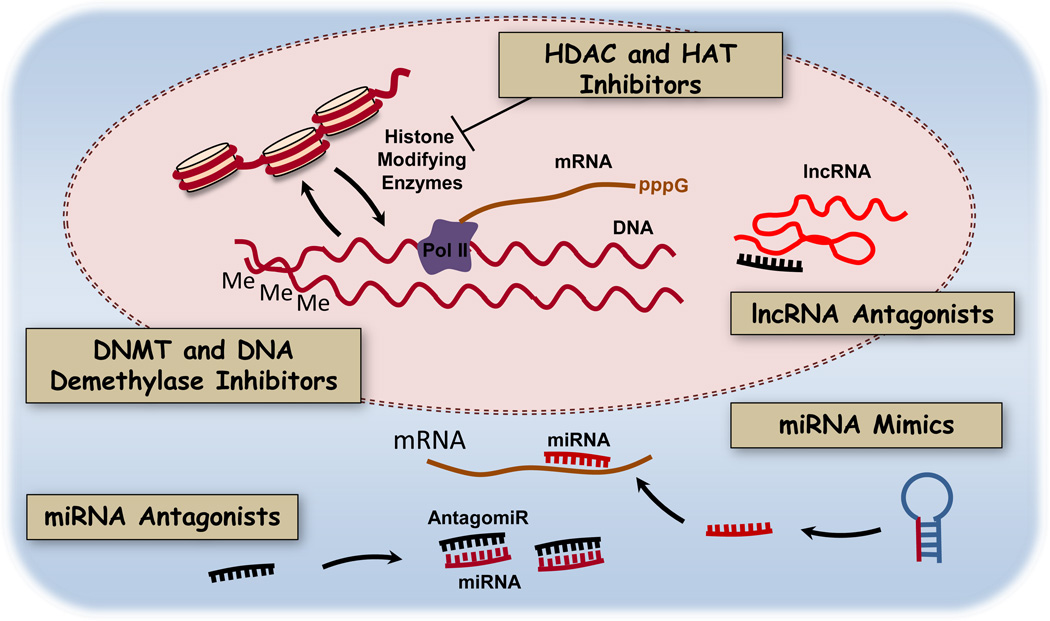
Several fundamental questions arise about targeting tissue remodeling in lung diseases. What are the cell types that should be targeted in epigenetic therapy (Figure 1)? What are the epigenetic processes contributing to lung remodeling (Figure 2)? Are there drug targets common to all remodeling lung tissues – eg. proteins promoting immune cell differentiation? Are there unique targets that could be hit to elicit disease-specific effects, such as regulation of bone morphogenetic protein receptor 2 (BMPR2) for therapy of pulmonary vascular remodeling, smooth muscle contractile proteins in asthma and COPD, or matrix remodeling enzymes in IPF? Our view is that druggable targets involved in lung remodeling and repair include enzymes that catalyze DNA methylation and demethylation, enzymes that catalyze posttranslational modifications of histones, and the noncoding RNAs - both microRNAs and long noncoding RNAs. We describe below potential targets as well as existing drugs that target each class of epigenetic regulator. Most studies are early stage preclinical trials or in many cases extrapolation of studies of lung cancer. In spite of the daunting complexities of multigenic, multifactorial chronic lung diseases there are opportunities presented by advances in epigenetics combined with advances in lung-directed delivery of small molecules and oligonucleotides. Examples of epigenetic targets are organized by biochemical and molecular classes and current progress in drug development is described for each clinical syndrome.
Figure 2.
Epigenetic processes in lung cells that mediate cell adaptation and tissue remodeling. Conserved biochemical processes in lung cells may be valid therapeutic targets for small molecule inhibitors of DNA methylation and histone modifications or oligonucleotide antagonists of short and long noncoding RNAs (miRNAs, lncRNAs). MiRNA mimics could be delivered to rescue miRNA function when it is dysregulated in disease.
3. Epigenetic control of gene expression in lung tissue remodeling
3.1 DNA methylation and inhibitors
DNA methylation normally regulates access of the transcriptional machinery to promoter loci and thus determines the set of genes expressed in a given tissue at any particular time (Figure 2). There has been rapid progress in studies of DNA methytransferase (DNMT) inhibitors and demethylase inhibitors, particularly in cancer chemotherapy (Gros et al., 2012). The data on DNA methylation patterns in lung diseases other than lung cancer are less extensive, but some interesting patterns have emerged that are relevant to new molecular interventions in asthma, COPD and IPF.
3.1.1 Asthma
In patients with asthma hypermethylation and hypomethylation of DNA have been reported in a number of genes in lung biopsies and postmortem samples. In some cases, differences in methylation correlated with functionally important differences in gene expression. For example, increased methylation of the STAT5A gene correlated with decreased expression of STAT5A in the asthmatic samples (Stefanowicz et al., 2012). Differences in DNA methylation might be intrinsic to asthma phenotypes, but they also might have been induced by allergen exposure. The latter possibility is supported by results from mouse models of asthma showing dynamic changes in DNA methylation from allergen challenge (Cheng et al., 2014; Shang et al., 2013). In support of allergen-induced changes in DNA methylation, ovalbumin sensitization of mice decreased DNA methyltransferase 1 (DNMT1) mRNA abundance which correlated with increased methylation of the DNMT1 gene (Verma et al., 2013). HDM sensitization of mice reduced global methylation and increased global hydroxymethylation which correlated with decreased DNA methyltransferase 3A (DMT3A) and increased ten-eleven translocation methylcytosine dioxygenase 1 respectively (Cheng et al., 2014). Furthermore, mice deficient in DMT3A in T-cells exhibit a higher degree of eosinophilia, more severe inflammation, and increased IL-13 secretion from splenocytes following ovalbumin challenge (Yu et al., 2012). These results indicate that DNMT activity limits asthma severity. However, acute inhibition of DNMT activity with 5-azacytidine, a nonselective inhibitor of DNA methyltransferases reduced airway hyperreactivity and inflammation in ovalabumin (OVA)-challenged mice, possibly due to increased numbers of regulatory T cells (Wu et al., 2013). Experiments using 5-azacytidine are informative but is possible some effects might be unrelated to DNMT inhibition. Additional studies utilizing more selective drugs are warranted. Taken together these studies demonstrate that DNA methylation is altered in asthma, possibly due to differential expression of DNMTs. The role of DNA methylation in asthma pathogenesis is complex and requires further investigation to identify appropriate drug targets.
3.1.2 COPD
In patients with COPD DNA methylation is also altered in a complex pattern similar to that seen in asthma in which there is hypermethylation of some loci and hypomethylation of others (Guzmán et al., 2012; Qiu et al., 2012; Vucic et al., 2014). Genes that have been identified as differentially regulated in COPD include components of the PI3K/Akt and anti-oxidant NFE2-related factor-2 (Nrf2) pathways (Vucic et al., 2014). Both pathways have been identified as treatment targets for COPD (Boutten et al., 2011; Bozinovski et al., 2006; Hosgood et al., 2009). CpG hypermethylation was observed in the PTEN and NFE2L2 (Nrf2) genes which correlated with reduced expression of their respective gene products. The Nrf2 pathway is anti-inflammatory and protects against oxidative stress in COPD (Boutten et al., 2011). PTEN is a negative regulator of PI3K/AKT signaling. PI3K/AKT signaling is thought to contribute to inflammation, remodeling, and proteolysis in COPD (Bozinovski et al., 2006). Thus, CpG hypermethylation appears to antagonize protective pathways and promote pathogenic pathways in COPD. Conversely, hypomethylation of the HDAC6 promoter has been linked to elevated expression of HDAC6 in COPD (Lam et al., 2013). HDAC6 is thought to contribute to cigarette smoke-mediated epithelial dysfunction in COPD by promoting autophagy (Lam et al., 2013). In addition, cigarette smoke has been linked to altered expression of DNMTs and alterations in CpG methylation highlighting its role in DNA structure in COPD (Liu et al., 2010). When DNA is hypermethylated therapies targeting DNMT may be beneficial, but further work is needed to determine whether altered DNA methylation is a cause or consequence of COPD.
3.1.3 IPF
Studies of DNMTs in IPF have reported either increased expression or enhanced promoter binding of DNMT-1 and DNMT-3a. Both DNA hypermethylation and hypomethylation have been observed in IPF (Cisneros et al., 2012; Coward et al., 2014; Dakhlallah et al., 2013; Huang et al., 2010; Rabinovich et al., 2012; Sanders et al., 2012). Many of these alterations in DNA methylation correlate with differences in gene expression. Some examples include hypermethylation of the cyclooxygenase-2 (COX-2) and PTGER2 promoters correlating with reduced gene expression (Coward et al., 2014; Huang et al., 2010). Interestingly, the miR-17~92 cluster promoter was hypermethylated in IPF correlating with reduced expression of miR-17~92 but also increased expression of DNMT-1, a target of miRNAs in the miR-17~92 cluster (Dakhlallah et al., 2013). This example highlights the multi-layered complexity of epigenetic regulation and begs the question of whether an alteration in miR-17~92 cluster expression or DNMT-1 expression was the initiating event in this imbalanced regulatory loop. The key question for epigenetic therapy of IPF is how to intervene to reestablish a normal regulatory circuit.
Studies of nucleoside inhibitors of DNMT suggest that molecular intervention is a tractable problem. Bleomycin-treated mice exhibited reduced expression of miR-17~92, elevated DNMT-1 expression and DNA hypermethylation (Dakhlallah et al., 2013). Inhibition of DNMT by 5-aza-2’-deoxycytidine (decitabine) enhanced miR 17~92 expression and reduced expression of profibrotic genes including collagen 1A1 and connective tissue growth factor (Dakhlallah et al., 2013). In a separate study, treatment of IPF fibroblasts with decitabine abrogated prostaglandin E2 resistance in these cells by increasing expression of the E prostanoid 2 receptor (encoded by PTGER2) which has a hypermethylated promoter in IPF (Huang et al., 2010). Furthermore, decitabine treatment of IPF fibroblasts reduced collagen I expression and reduced proliferation in response to prostaglandin E2 (Huang et al., 2010). These initial cell based studies and preclinical animal studies indicate that IPF may respond to treatment with DNMT inhibitors. However, it should be noted that even though decitabine is an inhibitor of DNMT, it also has effects independent of DNA methylation that enhance apoptosis. Thus, further investigations are warranted using more selective inhibitors of DNA methyltransferases to determine whether altered DNA methylation is a cause or consequence of IPF pathogenesis.
3.2 Histone modifications and inhibitors
Histone post-translation modifications such as acetylation and methylation are profoundly important regulators of transcription comprising key components of the “histone code” initially described in 1964 by Vincent Allfrey (Allfrey et al., 1964; Jenuwein & Allis, 2001). These post-translational modifications regulate chromatin structure directly, presumably by charge effects, or by recruiting chromatin remodeling enzymes (Bannister & Kouzarides, 2011). Typically, histone lysine acetylation is permissive for transcription. Histone methylation can be permissive or restrictive depending upon the methylated residue. As an example, H3K4 di/tri-methylation is permissive while H3K9 di/tri-methylation is restrictive. These modifications are regulated by enzymes that are targets of small molecule inhibitors (Rodríguez-Paredes & Esteller, 2011). Histone acetylation is regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs). Histone methylation is regulated by histone lysine or arginine methyltransferases and histone demethylases. Serine residues on histones can also be phosphorylated but this area is less well studied than histone methylation and acetylation. Disease-specific differences in the histone code have been assigned to changes in activity or expression of histone modifying enzymes. Many studies suggest these differences are significant contributors to asthma, COPD, and IPF. Typically differences in histone acetylation and methylation have been linked to either a difference in enzyme activity or chromatin binding in a disease state. Essentially, an imbalance of the enzymes responsible for maintaining the “on” and “off” chromatin states exists in many disease states including lung diseases (see Figure 2).
3.2.1 Asthma
In asthmatic humans, HDAC activity is reduced and HAT activity is elevated in alveolar macrophages and bronchial biopsies (Cosío et al., 2004; Ito et al., 2002). Peripheral blood mononuclear cells from patients with severe asthma have even less histone deacetylase activity than non-severe asthmatic samples which correlated with steroid insensitivity (Hew et al., 2006). The reduction in histone deacetylase activity correlated with decreases in HDAC1 and HDAC2 protein abundance, but no difference in histone acetyltransferase protein abundance was observed. Also, elevated HATs (CBP, PCAF, p300) bound to chromatin at the CXCL8 promoter correlated with elevated histone H3 pan-acetylation and enhanced expression of CXCL8 in airway smooth muscle cells from patients with asthma (John et al., 2009). Reduced G9A methyltransferase bound to chromatin at the VEGF promoter correlated with reduced H3K9 trimethylation and enhanced expression of VEGF in airway smooth muscle cells from patients with asthma (Clifford et al., 2012). Taken together, these studies indicate that histone modifying enzyme expression, binding, and activity are altered in asthma and correlate with altered post-translational modifications. Reduced HDAC activity in asthma may promote enhanced expression of pro-inflammatory and pro-remodeling genes which presumably worsens symptoms. The correlations of histone modifications and asthma pathogenesis are provocative, but further work is needed to establish whether alterations in histone acetylation are a cause or consequence of asthma phenotypes.
In either case there is preclinical evidence that altering HDAC activity alters airway hyperreactivity, a hallmark of asthma in humans and in mouse models of asthma. Treatment of mice with trichostatin A, a non-selective HDAC inhibitor, reduced airway hyperresponsiveness and remodeling, but conflicting results exist regarding effects on inflammation (Banerjee et al., 2012; Choi et al., 2005; Royce et al., 2012). The effects of trichostatin A on experimental asthma in mice appeared to be due to alterations in calcium signaling thus discounting the role of epigenetic effects (Banerjee et al., 2012). Recent studies support the notion that HDAC inhibitors may prove to be effective bronchodilators by enhancing acetylation of substrates other than histones such as HSP20 and cortactin (Li et al. AJP Cell 2014; Chen et al. 2013 ; Karolczak-Byatti et al. 2011). Beneficial effects of HDAC inhibitors is paradoxical in asthma because histone acetylation is increased in disease. The evidence to date suggests the benefit of HDAC inhibition in asthma may be primarily due to acetylation of non-histone substrates. This raises interesting questions about the future role of HDAC inhibition as acute bronchodilators versus long term epigenetic modifiers.
Higher histone acetylation in asthma suggests a better approach for chronic epigenetic therapy will be drugs targeting HATs. However, development of clinically useful HAT inhibitors has lagged that of HDAC inhibitors due in part to the poor selectivity of natural product inhibitors such as curcumin and the low druggability of the catalytic domain of histone acetyltransferases (reviewed by Arrowsmith et al., 2012). One of the most promising candidates is C646, a competitive inhibitor of p300 that reduces histone acetylation, radiosensitizes lung cancer cells and causes cell cycle arrest in leukemia cells (Bowers et al., 2010; Gao et al., 2013; Oike et al., 2014). The effects of HAT inhibition with siRNA or C646 need to be tested in cells from asthmatics and in animal models of asthma.
3.2.2 COPD
As seen in patients with asthma, samples from patients with COPD also exhibit alterations in histone modifying enzymes that correlate with alterations in histone post-translational modifications. Reductions in mRNA abundance for HDACs 2, 5 and 8 as well as a reduction in HDAC2 protein abundance and HDAC activity were observed in lung tissue and macrophages (Ito et al., 2005). Reduced HDAC expression correlated with elevated IL-8 expression and greater H4 pan-acetylation at the IL-8 promoter (Ito et al., 2005). Conversely elevated HDAC1 expression was observed in epithelial biopsies from patients with COPD and positively correlated with the expression of beta-defensin 1 (Andresen et al., 2011). Interestingly, smoking alone was sufficient to elevate H4 pan-acetylation and reduce the expression of some HDAC isoforms (Ito et al., 2005). Cigarette smoke alone is a powerful modulator of histone post-translational modifications because it reduces HDAC activity and reduces HDAC expression, highlighting the importance for indicating whether samples were obtained from smokers or non-smokers (Adenuga et al., 2009; Lakshmi et al., 2014; Szulakowski et al., 2006; Yang et al., 2006). Elevated H3 pan-acetylation and H4 pan-acetylation have also been observed in lung tissue and airway smooth muscle cells (H4 pan-acetylation only) from patients with COPD. (Raidl et al., 2007; Szulakowski et al., 2006). Elevated H4 pan-acetylation in the VEGF promoter, typically considered a “permissive” transcription modification, correlated with reduced VEGF expression in airway smooth muscle from patients with COPD, illustrating the complex nature of the histone code. Sirtuin-1 (SIRT1) expression and deacetylase activity were reduced in peripheral lung tissues from patients with COPD (Nakamaru et al., 2009; Rajendrasozhan et al., 2008). Reduced SIRT1 activity correlated with increased expression of MMP9 and H4 pan-acetylation (Nakamaru et al., 2009). It should be noted that non-histone substrates for SIRT1 and HDAC2 have also been linked to the pathophysiology of COPD. FOXO3, TIMP-1, and RelA are substrates for SIRT1 (Rajendrasozhan et al., 2008; Yao et al., 2012; Yao et al., 2013). Nrf2 and the glucocorticoid receptor are substrates for HDAC2 (Ito et al., 2006; Mercado et al., 2011). Taken together these studies indicate that a deficiency in histone deacetylase activity may exist in COPD and that an imbalance between histone acetylases and deacetylases contributes to disease specific alterations in gene expression. Additional studies are warranted to determine whether these alterations in enzyme activity are a cause or consequence of COPD. In addition, non-histone protein modifications catalyzed by HDACs may also contribute to inflammation and remodeling in COPD. Epigenetic studies of asthma and COPD indicate a trend towards transcriptionally “permissive” histone modifications due to differences in enzymatic activity or binding to chromatin. If these epigenetic changes contribute to disease (either as a cause or consequence) then reversing or preventing the epigenetic changes might have therapeutic benefit. From this hypothesis one would predict drugs that inhibit HATs, histone methyltransferases or DNMTs may be appropriate candidates for further preclinical studies (Table 1). Conversely, compounds that selectively upregulate the activity and/or expression of HDACs or histone demethylases might also be therapeutically useful and should also be investigated in preclinical studies. Evidence from the COPD literature highlights the potential of natural products to upregulate histone deacetylase expression. In a mouse model of COPD, treatment with the plant flavonoid quercetin increased SIRT1 expression, reduced MMP9 and MMP12 expression, and reduced acetylation at their respective promoters in vitro (Ganesan et al., 2010). In addition, theophylline activates HDAC2 by inhibiting PI3K-δ and in combination with corticosteroids abrogates corticosteroid insensitivity in COPD (Cosio et al., 2004; Ford et al., 2010; Ito et al., 2002; To et al., 2010). Whether other small molecule drugs are able to upregulate HDAC activity or expression is an open question with high translational significance.
Table 1.
Epigenetic modifiers of lung disease in preclinical studies
| Mechanism | Disease/model | Drug | References |
|---|---|---|---|
| DNMT inhibitors | Asthma, OVA mouse | 5-azacytidine | Wu et al., 2013 |
| IPF, bleomycin mouse model | Decitabine | Dakhlallah et al., 2013 | |
| IPF fibroblasts | Decitabine | Huang et al., 2010 | |
| Histone acetyltransferase inhibitors | Lung cancer | C646 | Gao et al., 2013 |
| HDAC inhibitors | Asthma, OVA mouse | Trichostatin A | Banerjee et al., 2012 |
| Airway smooth muscle | OSU-HDAC-44 | Li et al., 2014 | |
| IPF fibroblasts | LBH589 and SAHA | Coward et al., 2009 | |
| IPF fibroblasts | Trichostatin A | Huang et al., 2013 | |
| IPF, bleomycin mouse model | SAHA | Sanders et al., 2014 | |
| IPF fibroblasts | SAHA | Zhang et al., 2013 | |
| HDAC upregulation | COPD, elastase mouse model | Quercetin | Ganesan et al., 2010 |
| COPD | Theophylline | Cosio et al., 2004 | |
| Histone methyltransferase inhibitors | IPF fibroblasts | BIX-01294 and 3-Deazaneplanocin |
Coward et al., 2010 Coward et al., 2014 |
| Bromodomain protein inhibitor | IPF fibroblasts | JQ1 | Filippakopoulos, et al., 2010 |
3.2.3 IPF
In contrast to asthma and COPD, which are predominantly characterized by increases in “permissive” histone post-translational modifications, IPF is characterized by increases in “restrictive” histone post-translational modifications that result in reduced anti-fibrotic and reduced pro-apoptotic gene expression. Reduced expression of COX-2 in IPF may be the result of elevated H3K9 and H3K27 trimethylation as well as reduced H3 and H4 pan-acetylation at the COX-2 promoter (Coward et al., 2009; Coward et al., 2014). Elevated H3K9 and H3K27 trimethylation correlated with elevated binding of G9A and EZH2, respectively, at the COX-2 promoter. Reduced H3 and H4 pan-acetylation correlated with reduced binding of HATs (CBP, p300 and PCAF), and increased binding of transcription-inhibitory complexes CoREST, NCoR and mSin3a. There was also reduced binding of heterochromatin-associated proteins (HP1, PRC1, MECP2) at the COX-2 promoter (Coward et al., 2009). Expression of the antiinflammatory cytokine CXCL10 is reduced in IPF, possibly due to elevated H3K9 trimethylation and reduced H3 and H4 pan-acetylation (Coward et al., 2010). Fas expression is also reduced in IPF and has been linked to elevated H3K9 trimethylation and reduced H3 pan-acetylation at the Fas promoter (Huang et al., 2013). Altogether these studies indicate that an imbalance in both histone acetylation and methylation exists in IPF which prevents the transcriptional activation of anti-fibrotic and pro-apoptotic genes. Further studies are needed to determine if these alterations in epigenetic modifications are cause or consequence of IPF. Nonetheless, current studies indicate that epigenetic dysregulation of the expression of these genes permits an exaggerated repair response leading to fibrosis.
Inhibitors targeting HDACs or histone methyltransferases responsible for “restrictive” histone acetylation and methylation patterns should correct the “restrictive” imbalance in IPF. This prediction was borne out by pre-clinical evidence demonstrating efficacy of HDAC inhibitors. In fibroblasts from patients with IPF, treatment with HDAC inhibitors LBH589 and suberoylanilide hydroxamic acid (SAHA) elevated the expression of COX-2 to levels similar to those observed in control cells (Coward et al., 2009). Trichostatin-A treatment of human IPF fibroblasts restored Fas expression to levels similar to those observed in control cells thereby increasing their ability to undergo apoptosis (Huang et al., 2013). Trichostatin A is not clinically useful but is a very useful broad-spectrum HDAC inhibitor for in vitro studies (Rodríguez-Paredes & Esteller, 2011). SAHA (Vorinostat) is a more promising candidate because it is currently approved for treatment of cutaneous T-cell lymphoma (Slingerland et al., 2014). SAHA treatment of bleomycin-treated mice reduced lung fibrosis, collagen III abundance, and improved lung function (Zhang et al., 2013; Sanders et al., 2014). LBH589 (Panobinostat) is another promising HDAC inhibitor in phase 3 clinical trials for therapy of multiple myeloma (San-Miguel et al., 2014). Vorinostat and Panobinostat along with other HDAC inhibitors in earlier stages of development may become effective anti-fibrotic therapy in IPF. In contrast to asthma and COPD, targeting HDACs appears to be the most promising therapeutic approach for IPF.
In addition to promising results with HDAC inhibitors there is some preclinical evidence for beneficial effects of histone methyltransferase inhibitors in IPF. Two inhibitors of the G9a and EZH2 histone methyltransferases (BIX-01294 and 3-Deazaneplanocin) increased COX-2 expression in IPF fibroblasts, which may enhance production of the anti-fibrotic prostanoid, PGE2 (Coward et al., 2014). BIX-01294 treatment of fibroblasts from patients with IPF restored CXCL10 expression to levels similar to those observed in control cells (Coward et al., 2010). Restoration of normal CXCL10 levels may have a beneficial anti-angiogenic effect in IPF. Additional EZH2 inhibitors have been developed including GSK126, EPZ005687, and UNC1999 that may also prove useful for preclinical testing for IPF treatment (Campbell & Tummino, 2014). Whether these newer agents upregulate COX-2 and CXCL10 expression in IPF cell and animal studies remains to be established.
Bromodomain-containing proteins of the histone code “reader” machinery bind acetylated lysine residues on histones and function as transcriptional coactivators (Arrowsmith et al., 2012; Zhang et al., 2012b). Inhibition of Bromodomain-containing protein 4 (BRD4) with JQ1 in bleomycin-treated mice reduced lung fibrosis in vivo (Filippakopoulos et al., 2010). JQ1 also reduced proliferation and migration of IPF fibroblasts (Tang et al., 2013). The effect of blocking BRD4 binding to acetylated histones is consistent with the observations that reduced histone acetylation correlates with reduced COX-2, CXCL10, and Fas expression in IPF fibroblasts (Coward et al., 2010; Coward et al., 2009; Huang et al., 2013). It seems likely that inhibiting BRD4 and other bromodomain proteins will be a useful strategy to rescue the expression of repressed genes in IPF. Additional inhibitors of the BET family bromodomain proteins are currently being developed as epigenetic modifiers for use in cancer chemotherapy (GSK-I-BET151 and PFI-1, GSK I-BET762). These agents might also have efficacy as anti-fibrotic therapy of IPF. Further preclinical trials in the bleomycin mouse model and in human IPF fibroblasts are required to test this idea.
3.3 MicroRNAs
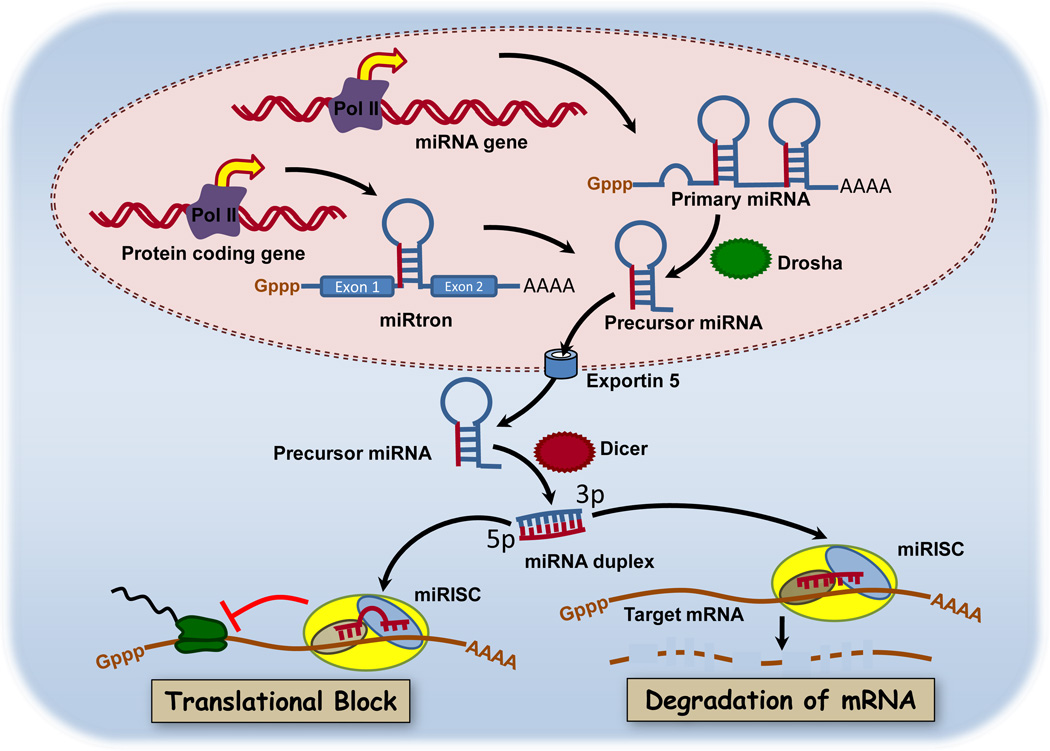
MicroRNAs (miRNAs) are 21–23 nucleotide, non-coding single stranded RNAs that negatively regulate post-transcriptional gene expression via mRNA destabilization or/and degradation. miRNA were first described in Caenorhabditis elegans, where lin-4 and let-7 were identified as novel regulators of developmental timing whose loss results in reiteration of cell lineages due to post-transcriptional silencing of lin-14 and lin-28 (Reinhart et al., 2000; Wightman et al., 1993). Both cloning and bioinformatics approaches have been used to identify individual miRNA or polycistronic clusters in introns, exons or intragenic regions (Lagos-Quintana et al., 2001; Rodriguez et al., 2004). Computational analysis estimates that miRNAs regulate >60% of the protein-coding genes in the human genome (Friedman et al., 2009). MiRNAs are generated by transcription of primary RNA transcripts by RNA polymerases II and III. The primary transcripts are processed into stem-loop structures of 70–100 nucleotides by the double-strand-RNA specific RNase Drosha (Figure 3). This pre-miRNA is transported into the cytoplasm via exportin-5, where it is digested by a second RNase Dicer (Chendrimada et al., 2005). This mature miRNA is then bound by a miRNA-associated RNA-induced silencing complex (RISC) (Chendrimada et al., 2005). Within this complex, the miRNA can bind to the 3’-untranslated region (3’-UTR) or in some cases the 5’UTR of target mRNA to either repress translation or induce cleavage (Lytle et al., 2007). For more information, the interested reader is directed to a recent detailed review of miRNA biogenesis and activity (Hausser & Zavolan, 2014).
Figure 3.
MicroRNA biogenesis and mechanisms of gene silencing. The primary RNA transcript of miRNA genes and intronic primary miRNAs are transcribed by RNA polymerase II and processed by Drosha to pre-miRNA. Pre-miRNAs are transported from the nucleus via exportin-5 where they are further processed by Dicer. The mature miRNA is then bound to miRNA-associated RNA-induced silencing complexes (miRISC). Within miRISC, the miRNA can bind to the 3’-untranslated region (3’-UTR) of target mRNA to either repress translation or induce cleavage. This simplified schematic does not show other known interactions of miRNAs with 5’UTRs or with long noncoding RNAs, both of which are known to regulate expression of some proteins. Reprinted under terms of the Creative Commons license from Joshi et al., 2011.
3.3.1 Normal functions in development and organogenesis of the lung
Normal lung development requires the temporal and spatial regulation of a complex set of genes, including transforming growth factor β1 (TGF-β1), fibroblast growth factors, hedgehogs and Wnts (Ramasamy et al., 2007; Wang et al., 2005; White et al., 2006; Zhang et al., 2012). Lung epithelial cells are initially differentiated from the endoderm to form an epithelial tube surrounded by mesenchymal cells that branch to form bronchioles. The epithelial cells subsequently differentiate into the respiratory epithelium. The first evidence that miRNA played a role in lung development came from the conditional knockout of Dicer in cells that express sonic hedgehog, which include lung epithelial cells (Harris et al., 2006). The loss of Dicer in these cells caused defective airway branching and continued distal epithelial cell proliferation. These changes correlated with abnormal distribution and increased expression of fibroblast growth factor 10 in the mesenchyme, which is thought to act as chemoattractant for epithelial branching.
Indeed, subsequent experiments have highlighted the function of miRNA in epithelial cell differentiation and branching during lung development. Microarray profiling experiments identified 21 miRNA with varying expression patterns during the different phases of rat lung development (Bhaskaran et al., 2009). Of these miRNA, miR-127 expression was highest at the later stages of development, having gradually shifted from mesenchymal cells to epithelial cells. Overexpression of miR-127 in early stages of fetal lung organ culture led to improper bud development. A semi-quantitative PCR-based approach was used to compare differential miRNA expression during mouse and human lung development (Williams et al., 2007). The overall expression profile was similar between mouse and human tissue, which suggested conservation of miRNA expression during lung development, with differences in expression patterns of miR-29a, -29b and -154. MiR-154 is of particular interest because it is found in a maternally imprinted miRNA cluster upregulated in IPF (Milosevic et al., 2012), while miR-29 directly targets the de novo DNMTs, DNMT3A and 3B (Fabbri et al., 2007). Moreover, the expression of miR-29 is decreased by TGF-β1 in human lung fibroblasts and blocks activation of phosphoinositide-3 kinase/Akt signaling (Yang et al., 2013b).
The miR-17~92 cluster was identified as a potential oncogene that is markedly overexpressed in several cancers and occasionally undergoes gene amplification in non-small cell lung carcinomas (Hayashita et al., 2005). This cluster contains miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92-1, which are highly expressed in embryonic stem cells and early stages of lung development, gradually decreasing as development progresses with little expression seen in the adult lung (Lu et al., 2007). Transgenic overexpression of the miR-17~92 cluster in lung epithelial cells caused delayed differentiation and hyperplasia in distal epithelial cells, while targeted deletion resulted in early post-natal mortality due to lung hypoplasia but no specific branching defects (Ventura et al., 2008).
Studies of miRNA expression and function have progressed rapidly in many areas of lung biology. Initial efforts concentrated on profiling differential miRNA expression between healthy and diseased tissues or isolated cells, generating lists of miRNA with limited utility. This was due to the need to identify target mRNAs and to understand the complexity of networks of gene expression that interact in different tissues during disease pathogenesis. Each disease is associated with changes in expression and function of particular miRNAs that have become important therapeutic targets. A summary of the miRNA studied in lung diseases described below is shown in Table 2.
Table 2.
MicroRNAs involved in obstructive and restrictive lung diseases
3.3.2 Pulmonary hypertension
The role of miRNA in pulmonary hypertension has been studied largely in rat models of chronic hypoxia and monocrotaline treatment with correlations in human PAH samples. Initial studies identified up- and down-regulation of miRNA expression in both rat models, although significant differences in down-regulation of miR-21 that correlated with human PAH samples were observed only in the monocrotaline-treated animals. (Caruso et al., 2010). Expression profiling experiments in pulmonary artery smooth muscle cells from PAH patients and monocrotaline-treated rats identified down-regulation of miR-204 (Courboulin et al., 2011), whose expression was confined to pulmonary arteries with little expression seen in pulmonary endothelial cells, veins or surrounding bronchi. Expression of miR-204 correlated with PAH severity and miR-204 inhibition increased proliferation and resistance to apoptosis. Targets of miR-204 include, among others, SHP2, an upstream activator of Src tyrosine kinase and nuclear factor of activated T cells (Courboulin et al., 2011) and TGF-β1 receptor 2 (Wang et al., 2010). The role of miR-204 in PAH and vascular remodeling events may be mediated by effects of proliferative signaling pathways since loss of miR-204 activates the small GTPase Rac1 to stimulate actin reorganization via Akt/mTOR in cancer cells (Imam et al., 2012). Several miRNA clusters have also been suggested to play a role in PAH. BMPR2 protein expression is reduced in PAH animal models via STAT3-mediated expression of the miR-17~92 cluster, which directly targets BMPR2 (Brock et al., 2009). The miR-143~145 cluster, which plays a key role in smooth muscle cell phenotype, has also been implicated in PAH. MiR-145-5p is elevated in PAH patients, which correlates with increased miR-145-5p expression in the hypoxic mouse model of PH (Caruso et al., 2010). In contrast, inhibition or knockout of miR-145 protects animals from hypoxia-induced PH (Caruso et al., 2012). A thorough analysis of miRNA expression in PAH lesions in human lungs found miR-143 and miR-145 expression was reduced (Bockmeyer et al., 2012), underscoring the contribution of smooth muscle cell phenotype and remodeling in PAH progression.
3.3.3 Asthma
MiRNAs have been implicated in all key features of asthma including proliferation, migration, inflammation and hyper-contractility in animal models and isolated human cells. Evidence linking miRNAs to the development and pathogenesis of asthma are mainly derived from mouse models of asthma. Garbacki and colleagues identified a large panel of miRNAs that were dynamically regulated in acute, intermediate and chronic murine models of asthma (Garbacki et al., 2011). Additional studies by other research groups have highlighted individual miRNAs for their contributions to asthma pathogenesis (see Table 2). It should be noted there are contradictory results concerning contributions of let-7 and miR-21 to asthma pathogenesis (Collison et al., 2011;Kumar et al., 2011;Lu et al., 2009; Polikepahad et al., 2010). Animal studies are supported by profiling studies identifying differentially expressed miRNAs in tissues, cells and exosomes from human subjects with asthma (Jardim et al., 2012; Jude et al., 2012; Levanen et al., 2013; Liu et al., 2012; Nakano et al., 2013; Perry et al., 2014; Qin et al., 2012; Solberg et al., 2012; Suojalehto et al., 2014). The evidence clearly shows that miRNA expression changes in asthma which may cause disease-related alterations in cell phenotypes that contribute to tissue remodeling and inflammation.
Altered expression in microRNAs in samples from patients with asthma have been linked to numerous features of asthma pathogenesis. Reduced expression of miR-140-3p in asthmatic airway smooth muscle was linked to elevated expression of CD38 which may contribute to altered calcium dynamics in asthma (Jude et al., 2010; Jude et al., 2012). MiR-221 expression was observed to be elevated in airway smooth muscle from severe asthmatics and was linked to hyperproliferation and hypersecretion of IL-6 in these cells (Perry et al., 2014). CD4+ T-Cells from patients with asthma exhibit reduced expression of miR-15a and elevated expression of its validated target VEGFA highlighting the possible role for microRNA dysregulation to the increased vascularization observed in asthma (Nakano et al., 2013). Epithelial cells from patients with asthma exhibit an altered miRNA expression profile including reduced expression of miR-203 that correlated with elevated expression of aquaporin-4, a predicted target of miR-203 (Jardim et al., 2012; Solberg et al., 2012). The pro-inflammatory miRNA, miR-155, was also observed to be upregulated in asthmatic human epithelial and airway smooth muscle cells (Jardim et al., 2012; Comer et al. 2014a). MiR-155−/− mice subjected to ovalbumin sensitization and challenge had reduced eosinophilia and mucus hypersecretion along with a reduced Th2 cell numbers and cytokines (Malmhall et al., 2014). In vitro studies of miR-155 in human airway smooth muscle support its role in promoting pro-inflammatory gene expression (Comer et al. 2014a) These examples demonstrate the mechanism by which miRNAs contribute to the pathogenesis of asthma. Potential pathogenic mechanisms include dysregulation of inflammatory protein expression, altered cell signaling proteins and changes in T-cell differentiation patterns. The evidence suggests either rescue or inhibition of select miRNAs may serve as a beneficial treatment for patients with severe asthma who do not respond to current therapies.
A growing body of cell-based evidence and preclinical animal studies focuses interest on therapies that reduce inflammation, airway hyperreactivity, mucus hypersecretion, and airway remodeling. Inhibition of miR-106a, miR-126, miR-145, miR-221, and Let-7 in mouse models of asthma have successfully reduced several of these pathological features of asthma. Inhibition of miR-106a in a mouse model of asthma reduced Th2 cytokine production, reduced airway hyperreactivity, reduced mucin content, and reduced subepithelial collagen (Sharma et al., 2012). Inhibition of miR-126 in a mouse model of asthma reduced airway inflammation, airway hyperresponsiveness, and mucus hypersecretion (Collison et al., 2011). Inhibition of miR-145 alleviated the hallmark features of allergic inflammation such as eosinophilic inflammation, mucus hyper-secretion, Th2 differentiation and airway hyperreactivity (Collison et al., 2011; Mattes et al., 2009). Inhibition of miR-221 reduced airway inflammation in a mouse model (Qin et al., 2012). MiR-221 regulates mast cell activation, which is consistent with reduced inflammation in mice treated with a miR-221 inhibitor (Mayoral et al., 2011).
MiR-25, miR-133a, and miR-146a/b are additional miRNAs that are promising targets for asthma therapy. These miRNAs target contractile proteins and inflammatory proteins, RhoA and immune signaling respectively. Study of miR-25 in human airway smooth muscle cells demonstrated its broad role in regulating contractile, proliferative and inflammatory phenotype through negative regulation of Kruppel like factor-4 (Kuhn et al., 2010; Petty & Singer, 2012). Transgenic smooth-muscle targeted overexpression of miR-25 in mice appears to ameliorate asthmatic changes in lung function in preliminary studies (Copley-Salem et al., 2013). Moreover, miR-25 expression was significantly down-regulated following chronic sensitization of mice with ovalbumin (Garbacki et al., 2011). In the ovalbumin mouse model of asthma miR-133a was also down-regulated resulting in increased expression of RhoA, a target of miR-133a (Chiba et al., 2009). Transfection of human bronchial smooth muscle cells with a miR-133a antagonist upregulated Rho A suggesting endogenous miR-133a normally restrains RhoA expression. The study by Chiba and colleagues highlights the potential for a miR-133a rescue therapy of asthma using miRNA mimics.
In contrast to miR-25 and miR-133a, which are downregulated, miR miR-146a and miR-146b are upregulated in the ovalbumin mouse model of asthma and miR-146a expression in elevated in human airway smooth muscle from patients with asthma (Garbacki et al., 2011; Comer et al. 2014b). MiR-146a/b inhibits innate immune and IL-1 receptor-mediated signaling and the expression of HuR, an RNA binding protein that typically stabilizes pro-inflammatory transcripts (Cheng et al., 2013; Larner-Svensson et al., 2010; Perry et al., 2009; Taganov et al., 2006; Comer et al., 2014b). Upregulation of miR146a/b in the ovalbumin mouse model may reflect a compensatory mechanism to blunt airway inflammation. As a therapeutic approach miR-146a/b mimics would be expected to antagonize the development of allergic airway inflammation. In cultured human airway smooth muscle cells miR-146a was induced by a mixture of IL1-β, TNFα and IFNγ (Comer et al., 2014b) consistent with induction by ovalbumin in the mouse asthma model. A mir-146a mimic reduced expression of HuR, which was associated with downregulation of COX-2 and downregulation of IL1-β expression. Therefore, a miR-146a mimic or strategies to upregulate miR-146a expression should antagonize allergic airway inflammation and provide a novel therapeutic approach based on RNA interference.
3.3.4 COPD
While there are a variety of environmental, genetic and epigenetic factors that contribute to COPD, cigarette smoke is the most important risk factor that correlates with dysregulation of several miRNAs even though only 20% of smokers will eventually develop COPD (Pauwels & Rabe, 2004). The expression of miRNAs are generally downregulated in smokers compared to non-smokers in airway epithelium (De Flora et al., 2012), alveolar macrophages (Graff et al., 2012) and in lung tissue (Ezzie et al., 2012). This down regulation of miRNA expression in smokers may be due to reduced Dicer activity following sumoylation of Dicer (Gross et al., 2014). Thus, as with histone modifications and DNA methylation, cigarette smoke has profound effects on miRNA expression by directly affecting the expression of enzymes that regulate the levels of the epigenetic modifiers. Microarray-based studies have been performed by a number of labs and have identified a large number of differentially expressed miRNAs in primary fibroblasts and lung tissue from patients with COPD (Chatila et al., 2014; Ezzie et al., 2012; Gross et al., 2014; Molina-Pinelo et al., 2014; Sato et al., 2010). In addition studies of BALF cells, sputum, serum, and plasma samples from patients with COPD have identified numerous differentially expressed miRNAs (Akbas et al., 2012; Soeda et al., 2013; Van Pottelberge et al., 2011). Many of the miRNAs implicated in COPD have also been associated with various cancers, and COPD is associated with increased risk of squamous cell non-small cell lung carcinoma (Papi et al., 2004). To determine whether miRNA expression was differentially expressed between lung cancer and COPD patients, microarray experiments were performed from whole blood RNA (Leidinger et al., 2011) and plasma (Sanfiorenzo et al., 2013). Both studies identified a subset of miRNA that discriminated COPD from lung cancer, including miRNAs 26a, 641, 383, 940, 662, 92a, 369-5p, and 636 (Leidinger et al., 2011) and miRNAs 20a, 24, 25, 152, 145, 199a (Sanfiorenzo et al., 2013). These results suggest patterns of miRNA expression might be developed as clinical biomarkers to assess cancer risk and progression.
The miRNAs dysregulated in COPD modulate expression of genes that contribute to pathogenesis of the disease. Decreased expression of let-7c and miR-126a was observed in sputum of COPD patients who continued to smoke compared to smokers without COPD. Reduced let-7c correlated with increased tumor necrosis factor receptor 2 (TNFR2) (Van Pottelberge et al., 2011), which is increased in the sputum of COPD patients. Increased TNFR2 indicates continued inflammation even when the patients stop smoking (Vernooy et al., 2002). The expression of miR-101 is elevated in the lungs of patients with COPD and in mice treated with cigarette smoke. Elevated miR-101 expression correlates with reduced expression of CFTR in COPD which may contribute to mucus accumulation, chronic infection, and inflammation (Hassan et al., 2014). A microarray analysis of cytokine-stimulated fibroblasts from COPD patients identified downregulation of miR146a. Decreased miR-146a was shown to upregulate COX-2 expression which increases prostaglandin E2 and increases disease severity (Sato et al., 2010).
Cellular heterogeneity in expression patterns is a complicating factor in establishing the role of a particular miRNA in COPD pathology. Expression of miR-199a-5p has been observed to be elevated in some studies and reduced in different cell types in patients with COPD (Chatila et al., 2014; Hassan et al., 2014; Mizuno et al., 2012). Previously, Lindsay and colleagues observed cell specific miRNA profiles in human lung samples indicating that in disease different cell types can exhibit differential expression of a particular miRNA (Williams et al., 2007). Reduced expression of miR-199a-5p in COPD was linked to miR-199a-2 promoter CpG hypermethylation in monocytes from patients with COPD and may contribute to enhancement of the unfolded protein response in patients with α1-antitrypsin deficiency (Chatila et al., 2014). Furthermore, reduced expression of miR-199a-5p was observed in regulatory T cells from patients with COPD and transcriptome analysis revealed that a number of miR-199a-5p targets are expressed in regulatory T cells including members of the TGFβ superfamily (Chatila et al., 2014). Reduced miR-199a-5p expression in regulatory T cells in COPD may enhance regulatory T cell function resulting in abnormal bias towards Th1 immune responses (Kalathil et al., 2014). In contrast, elevated miR-199a-5p expression was observed in lung tissue from patients with COPD that correlated with reduced expression of hypoxia inducible factor-1α (HIF1α). HIF1α is a validated target of miR-199a-5p that is reduced in COPD (Mizuno et al., 2012; Yasuo et al., 2011). Reduced HIF1α is hypothesized to impair the normal “lung structure maintenance program” (Yasuo et al., 2011). Mizuno and colleagues later observed elevated miR-34a expression in COPD, and in vitro experiments demonstrated that miR-34a controls expression of miR-199a-5p (Mizuno et al., 2012). Thus, in COPD, altered expression of miRNAs contributes to changes in both protein coding and miRNA coding genes that each contribute to pathogenesis. These dysregulated miRNAs appear to be ideal treatment targets in COPD although additional studies are warranted to help determine whether observed differences in miRNAs are a cause or consequence of COPD.
3.3.5 IPF
Studies of miRNA expression in IPF have estimated that 10% of all miRNA are dysregulated in IPF lungs (Pandit et al., 2010), including let-7d, miR-15b, miR-26a/b and miR-29, as well as miR-17~92 cluster, miR-29a/b/c, and miR-30c. An additional study identified a large number of differential expressed miRNAs including 43 upregulated miRNAs in samples from patients with IPF (Milosevic et al., 2012). MicroRNA profiling in bleomycin-treated mice (Xie et al., 2011) support human profiling studies and suggest temporal differences in miRNA expression may exist during the development and progression of IPF. Furthermore, differential miRNA expression was observed in lung biopsies from patients with rapidly progressing IPF when compared to slow progressing IPF (Oak et al., 2011). Interestingly, the expression of argonaute 1 mRNA was reduced in both slow and rapidly progressing IPF lung biopsies. The expression of arogonaute 2 mRNA was elevated in rapidly progressing IPF lung biopsies but argonaute 2 immunoreactivity was lowest in rapidly progressing IPF. Oak and colleagues were unable to detect argonaute 1 immunoreactivity reliably. In cultured IPF fibroblasts, cells from rapidly progressing IPF had reduced expression of DICER1 and argonaute 1 compared to slow progressing IPF and control fibroblasts. Reduced DICER1 expression is hypothesized to reduce miRNA biogenesis while reduced Argonaute expression is hypothesized to reduce both miRNA biogenesis and miRNA-mediated silencing of gene expression (Diederichs et al. 2007) Taken together, these studies indicate that differential miRNA expression occurs in IPF in human subjects and alterations in miRNA biogenesis and function may also contribute to rapidly progressing IPF.
The function of numerous miRNAs that are elevated in IPF have been investigated to establish relevance to disease pathogenesis. Many miRNAs associated with IPF regulate TGF-β1 expression or function, inflammation, actin expression or cell signaling pathways, all of which contribute to fibrosis. MiR-155 expression is also elevated in IPF in human subjects and in the lungs of bleomycin-treated mice (Pandit et al., 2010; Pottier et al., 2009). Interestingly, miR-155 is a negative regulator of TGF-β1 signaling in macrophages by silencing Smad2 (Louafi et al., 2010), but it promotes epithelial-mesenchymal transition (EMT) in response to TGF-β1 by silencing RhoA in epithelial cells (Kong et al., 2008). MiR-155 also repressed the expression of keratinocyte growth factor in lung fibroblasts (Pottier et al., 2009) which may promote fibrosis. These results highlight the potential of a miR-155 inhibitor as an anti-fibrotic therapy in IPF.
The functions of several miRNA that are elevated in IPF converge on TGF-β1 signaling. MiR-154 is an example of an miRNA upregulated in IPF and induced by TGF-β1 in human lung fibroblasts (Milosevic et al., 2012). MiR-154 induced proliferation and migration of fibroblasts which was mediated by the Wnt/β-catenin pathway. The Wnt/β-catenin pathway was shown previously to be upregulated in lungs biopsies obtained from patients with IPF (Königshoff et al., 2008). MiR-21 is another well-studied miRNA that is upregulated in lung fibroblasts of IPF patients (Liu et al., 2010), and is induced by TGF-β1 in primary fibroblasts. Upregulation of alpha smooth muscle actin by miR-145 is another example of a miRNA regulating profibrotic gene expression. MiR-145 is elevated in the lungs of patients with IPF (Yang et al., 2013a), and it promotes fibroblast to myofibroblast transition in response to TGF-β1 treatment. MiR-145 expression is necessary for bleomycin-induced fibrosis because miR-145 −/− mice do not develop pulmonary fibrosis in response to bleomycin (Yang et al., 2013a). The profibrotic mechanism of miR-145 is due in part to upregulation of smooth muscle actin expression by repression of Kruppel-like factor 4, which is a negative regulator of serum response factor-dependent gene expression.
Elements of TGF-β signaling pathways are also influenced by reduced expression of miRNAs, which results in upregulation of target transcripts that drive fibrosis. For example, Let-7d expression is decreased in alveolar epithelium of IPF lungs, correlating with increased high mobility group AT-hook 2 (HMGA2), which is a mediator of TGF-β1 induction of EMT (Pandit et al., 2010; Thuault et al., 2006). Inhibition of Let-7d in mice yielded a lung pathology similar to IPF including alveolar septal thickening, increased collagen 1 expression, increased HMGA2 expression, increased lung collagen deposition, decreased epithelial cells markers (CDH1 and TJP1), and increased numbers of smooth muscle α-actin positive cells. Promotion of fibrosis by let-7d repression suggests a rescue therapy might be beneficial in IPF. Similar arguments can be made for a number of other profibrotic miRNAs downregulated in IPF. MiR-26a expression is reduced in lung tissue from patients with IPF correlating with increased expression of connective tissue growth factor and enhanced collagen production (Liang et al., 2014). MiR-29 expression is lower in patients with IPF and in bleomycin-treated mice (Cushing et al., 2011; Pandit et al., 2010). Reduced miR-29 correlated with increased collagen and extracellular matrix gene expression (Cushing et al., 2011). The expression of miR-200a and -200c are also reduced in IPF lungs (Yang et al., 2012). Members of the miR-200 family inhibit TGF-β1-mediated EMT transitions and reduce smooth muscle α-actin and fibronectin expression (Yang et al., 2012). There is also evidence that miR-326 negatively regulates TGF-β1 expression in IPF and the bleomycin mouse model (Das et al., 2014). Normalizing TGF-β1 signaling in IPF may be possible using a rescue strategy where one or more the downregulated miRNAs are delivered to the lung.
In addition to miRNA regulation of fibrous structural proteins there is evidence for miRNA control of signaling proteins in fibrosis. MiR-199a is elevated in IPF patients and in bleomycin-treated mice (Lino Cardenas et al., 2013), and it is correlated with reduced caveolin-1 expression. MiR-199a promoted human MRC-5 lung fibroblast proliferation, migration, invasion, and differentiation into myofibroblasts in vitro (Lino Cardenas et al., 2013). Caveolin-1 is a validated target of miR-199a that has been was implicated previously in IPF pathogenesis (Wang et al., 2006).
Correlations of miRNA expression and mechanistic validation studies in IPF and the bleomycin mouse model suggest a profibrotic miRNA gene expression program that involves both upregulation (miRNAs 21, 145, 155, 199a) and downregulation of miRNAs (miRNAs Let7d, 17~92, 26a, 29, 200 family and 326). In all cases target proteins regulated by these miRNAs either directly or indirectly promote fibrosis. In many of the preclinical animal studies rescue and inhibition strategies were effective antifibrotic therapies. Translation of these exciting findings to first in human safety and efficacy trials is the next important step.
3.3.6 Extracellular miRNA and miRNA SNPs: New avenues of exploration
The discovery of extracellular miRNA in biological fluids, including serum, plasma, urine and saliva (Weber et al., 2010) has emerged as a growing area of study in the search for non-invasive diagnostic and prognostic biomarkers. These extracellular miRNAs are surprisingly resistant to nuclease digestion (Szafranska et al., 2008) and concentrated in circulating exosomes (Gallo et al., 2012). Both up- and down-regulated circulating miRNA were isolated from serum of PAH patients (Rhodes et al., 2013; Schlosser et al., 2013). Five circulating miRNA have been identified in the serum of COPD patients with downregulation of miRNAs 20, 28-3p, 34c-5p and 100 and upreglation of miR-7 compared to healthy controls (Akbas et al., 2012). In the serum of IPF patients, levels of miR-21, miR-101-3p and miR-155 correlates with disease progression, suggesting that these miRNA could be promising biomarkers for IPF prognosis (Li et al., 2014). A report of circulating miRNA relevant to an asthmatic phenotype identified 100 exosomal miRNAs in the serum of mice exposed to HDM extract (Shuichiro et al., 2014). While serum is a convenient, accessible source of miRNA-containing exosomes, the identification of circulating miRNA in bronchoalveolar lavage fluid (BALF), saliva or sputum could also provide useful information about localized expression of miRNA more relevant to lung diseases. Significant differences in exosomal miRNA isolated from BAL fluid of asthmatic patients identified let-7 and miR-200 family members that could be used to predict asthma severity (Levanen et al., 2013). These intriguing early discovery studies need to be confirmed in new validation cohorts in prospective longitudinal studies. Significant new data are needed to determine specificity and sensitivity of patterns of miRNAs in blood, BALF or sputum before they can be used as prognostic indicators.
Single nucleotide polymorphisms (SNPs) are emerging as another important area of study in lung disease. SNPs occur at a frequency of 1200–1500 base pairs in the human genome (Sherry et al., 1999). When they occur in miRNA transcripts, they could alter expression, processing or binding to target mRNA. SNPs in the 3’UTRs of target genes could affect mRNA stability, polyadenylation or regulatory interactions with RNA-binding proteins. SNPs in miRNA target sites, known as miR-SNPs, could lead to heritable defects in miRNA targeting that modifies gene expression. A systemic meta-analysis of genetic association studies determined that polymorphisms of miRNA-196a2 rs11614913 and the basic-loop-helix transcription factor MYCL1 rs3134615 could be potential biomarkers of lung cancer (Chen et al., 2013). Moreover, a SNP identified in the 3’UTR of KRAS altered targeting by let-7 and was significantly associated with increased lung cancer risk (Chin et al., 2008). In asthma, a SNP in the 3’UTR of the asthma susceptibility gene HLA-G affects targeting by the miR-148/152 family (Tan et al., 2007). MiR-SNPs identified in the pre-miRNA of miR-146a and miR-149 were significantly associated with a lower risk of asthma in a Chinese population and among a group of Mexican pediatric patients (Jimenez-Morales et al., 2012). In COPD patients, SNPs in alleles for miR-196a and miR-499 were associated with decreased risk of the disease (Li et al., 2011). Understanding polymorphisms in miRNAs and their target transcripts will become highly relevant as miRNA-based therapies evolve. Designing optimal miRNA antagonists and mimics may eventually require a deep understanding of SNPs in an individual’s protein coding genes as well as SNPs in the noncoding RNA loci being targeted.
3.4 Long noncoding RNAs
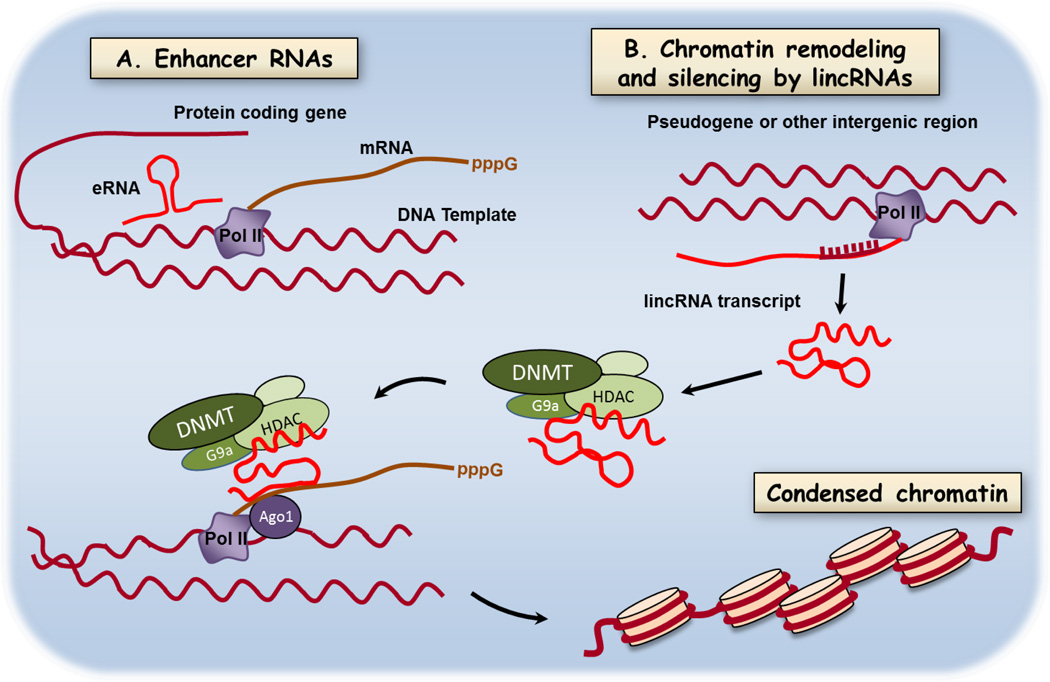
Long noncoding RNAs (lnc RNA) are RNAs >200nt transcribed from genomic sequences that are not translated into proteins. They are transcribed from enhancer sequences of protein coding genes (eRNAs, Natoli & Andrau, 2012), from intergenic, multiexomic regions (lincRNAs, Ulitsky & Bartel, 2013), and from noncoding strands of protein-coding genes (naturally occurring antisense transcripts, NAT, Cabili et al., 2011; Rinn & Chang, 2012) (see Figure 4). We will use the term long noncoding RNAs, abbreviated lncRNA to refer to the set of all noncoding RNAs > 200 nt. Long noncoding RNAs are emerging as versatile adapter molecules that bind to proteins, to DNA, and in some cases other RNAs. Intense investigation into lncRNA function has revealed multiple effects on gene expression that are often tissue-restricted. Functions of lncRNAs include regulation of transcription by controlling gene looping (Natoli & Andrau, 2012; Rinn et al., 2007), regulation of mRNA splicing (Zong et al., 2011), regulation of mRNA decay (Gong & Maquat, 2011; Kretz et al., 2013) and organizing chromosomes into gene neighborhoods (Hacisuleyman et al., 2014) (Figure 4). HOTAIR is a well-studied example of a lncRNA that binds components of the chromatin remodeling machinery. HOTAIR is a lncRNA in the HOXC locus (Rinn et al., 2007) that silences expression of HOXD genes during differentiation of myogenic progenitors (Tsumagari et al., 2013). It acts as a scaffold for assembly of the histone modifying enzyme complexes described above in section 3.2 and shown schematically in Figure 4. Altered histone modifications influence transcription of HOXD genes. In the lung HOXA and HOXB family members are important during development and in disease (Golpon et al., 2001). In non-small cell lung cancer HOTAIR regulates expression of HOXA5 and high expression of HOTAIR is a negative prognostic indicator (Liu et al., 2013). We describe below the rapid progress being made in studies of other lncRNAs in lung development and in nonneoplastic lung diseases.
Figure 4.
Mechanisms of regulation of gene expression by long noncoding RNAs. Long nonprotein coding RNAs are transcribed from a variety of genomic loci including enhancer sequences (eRNAs), intergenic, multiexomic sequences (lincRNAs), and noncoding strands of protein-coding genes (naturally occurring antisense transcripts. Shown here are a few of the rapidly emerging mechanisms by which lncRNAs modify gene expression and protein abundance. A. Binding of RNAs coded in enhancer regions to effect DNA looping and B. lncRNA serving as an adapter molecule in chromatin remodeling by DNA methylation and histone modifications. A salient feature of lncRNAs is the potential bind chromatin remodeling proteins, DNA, and in some cases other RNAs. Two important functions not shown are regulation of mRNA splicing and regulating mRNA decay. Targeting new oligonucleotide drugs to lncRNA function will require identification of key lncRNAs in lung diseases and defining the contribution these mechanisms to pathology.
3.4.1 Normal functions in development and organogenesis of the lung
Several hundred lncRNAs were identified in the developing mouse lung by sequencing polyadenylated RNAs in embryonic (E12.5) and adult lung tissue (Herriges et al., 2014). Analysis of the genomic structure of lncRNA loci in mouse lung revealed many loci contain binding sites for serum response factor, Fox and SP1 transcription factors, which collectively regulate early stages of lung mesoderm and endoderm development. A loss of function strategy was used to validate the role of several lncRNAs at loci close to transcription factors that regulate embryonic lung development. Two of these candidates, NANCI and LL34, were found to regulate expression of hundreds of genes in mouse airway epithelial cell culture. Some of the regulated genes control key steps in differentiation and development of airway epithelial cells. Similar studies in lncRNAs in lung diseases are underway and new data relevant to disease mechanisms and novel targets of therapy are eagerly anticipated.
The mechanistic studies of lncRNAs in mouse lung are highly relevant to the role of lncRNA deletions in the rare lethal neonatal lung disorder, capillary dysplasia and misalignment of pulmonary veins (ACD/MPV) (Szafranski et al., 2013). Lung tissues from subjects with ACD/MPV contain a “shared deletion region” which includes loci for several lncRNAs. The authors speculated that loss of chromatin looping function mediated by lncRNAs in the shared deletion region might underlie loss of forkhead box F1 expression leading to some aspects of the vascular pathology of ACD/MPV. The study illustrates the principle that loss of function of lncRNA can have profound effects on lung structure which includes pathological effects on neovascularization in this example. It remains to be seen whether similar changes in lncRNA expression and function occur in pulmonary vascular remodeling in PAH or bronchial vascular remodeling in asthma (Avdalovic, 2014).
Some of the nonvascular remodeling events in asthma, COPD and IPF might also be due to changes in lncRNA expression or function. Important lncRNA targets in lung diseases will emerge rapidly in the next few years, and much work remains to define relevant lncRNAs in various lung cell types. A good example of early stage lncRNA discovery is a recent survey of noncoding RNAs in human airway smooth muscle (Perry et al., 2014). Several lncRNAs were described that the authors suggest might act as sponges for microRNAs that regulate smooth muscle differentiation (Perry et al., 2014). They also observed expression of a noncoding RNA from the PVT-1 locus. PVT-1 lncRNA is upregulated in many cancers where it is known to interact with c-myc and contribute to dysregulated cell proliferation through an unknown mechanism. Further studies on the lncRNAs expressed in lung cells and the functions of those lncRNAs must be defined to identify valid targets for novel therapy of lung diseases.
3.4.2 Experimental and therapeutic potential of lncRNA inhibitors
The earliest information on lncRNAs in lung cancers suggested they might be drug targets. Changes in expression of lncRNAs is associated with several lung cancers (Enfield et al., 2012; Ji et al., 2003) and lncRNAs have been nominated as diagnostic and prognostic biomarkers in several cancers. However, the functions of lncRNAs in disease pathogenesis are still being defined. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1) was one of the first lncRNAs described in non-small cell lung cancer (Ji et al., 2003). It is one of the few lncRNAs for which there is mechanistic information. MALAT-1 is associated with greater potential for metastasis, and elevated MALAT-1 was suggested to be a prognostic indicator of lower survival rate (Schmidt et al., 2011). MALAT-1 is necessary for metastasis of lung cancer cells (Gutschner et al., 2013; Ji et al., 2003; Tano et al., 2010). It regulates more than 20 genes required for cancer cell metastasis (Gutschner et al., 2013). Using both in vitro and in vivo antisense knockdown strategies and zinc finger nuclease-mediated gene silencing Gutschner et al. (2013) conducted a preclinical efficacy trial in A549 cells and a xenograft lung cancer model. This important study showed that reducing MALAT-1 expression could prevent metastasis of lung cancer.
In a prior study of HUVECs and Hela cells MALAT-1 was shown to regulate RNA splicing (Lin et al., 2011). Although this activity was not associated with metastatic potential (Gutschner et al., 2013), it suggests the function of MALAT-1 may be relevant to pulmonary hypertension, which is a disease of endothelial dysfunction. The notion that MALAT-1 is relevant to pulmonary hypertension was later explored in a study of lncRNAs in vascular endothelial cells (Michalik et al., 2014). MALAT-1 was increased by hypoxia and inhibited proliferation while promoting endothelial cell migration (Michalik et al., 2014). These findings strongly support the role of MALAT-1 in regulating genes important in signaling, cell communication and cell adhesion, which are processes dysregulated in pulmonary hypertension. Whether MALAT-1 influences other structural cells in the lung, such as airway smooth muscle and lung fibroblasts is unknown. If so, MALAT-1 might be a suitable target for reducing or reversing airway and parenchymal remodeling in both pulmonary vascular and obstructive lung diseases.
The BIC gene transcript is another noncoding RNA that has high potential for influencing lung remodeling and inflammation in asthma, COPD and PAH. After posttranscriptional processing the BIC transcript yields miR-155. MiR-155 is a widely expressed multifunctional miRNA that influences T cell differentiation by modulating development of Th2, Th17 and regulatory T cell subtypes (Seddiki et al., 2013). The effects are complex and can vary with the disease model, but there is evidence miR-155 is necessary for chronic inflammatory responses (Malmhall et al., 2014; Seddiki et al., 2013). Many protein targets of miR-155 have been identified in immune cells including Pu.1, SHIP1, SOCS1 and c-Maf (Lu et al., 2009; Rodriguez et al., 2007). In cultured human airway smooth muscle cells miR-155 expression is enhanced in cells from asthmatics, and a miR-155 mimic enhances COX-2 expression and PGE2 synthesis (Comer et al., 2014a). In an important animal study miR-155 knockout reduced airway inflammation, reduced airway hyperreactivity and reduced Th2 cytokine expression in ovalbumin-sensitized mice (Malmhall et al., 2014). These intriguing results strongly suggest the BIC lncRNA and its miRNA end product, miR-155, are potential targets for novel anti-inflammatory and antiremodeling therapy.
Although recent studies reveal association between lncRNAs, lung development and lung cell biology (Herriges et al., 2014; Perry, 2014), there are few details of how lncRNAs modify lung structure and function in lung diseases. Important new information is needed to define how lncRNAs function in tissue remodeling, in inflammation and in tissue repair. There is also a question of what strategies can be applied to modify lncRNA expression or function in the lungs. These strategies are evolving, but some clear examples include using antisense oligonucleotides (also called “antagoNATs”) to inactivate lncRNAs in cells and in animal models of disease (Wahlestedt, 2013). The response to a lncRNA antagonist at the protein level might be positive if the lncRNA is a repressor of gene expression, or negative if the lncRNA is an enhancer. For example, inhibiting the function of MALAT-1 reduces endothelial cell differentiation in vitro (Michalik et al., 2014). A MALAT-1 antagonist active in vivo might reduce occlusive lesions in pulmonary arterial hypertension, or inhibit airway epithelial cell proliferation and reduce obstructive remodeling of the airways in asthma and COPD. Proliferation of airway smooth muscle cells might also be inhibited by antagonists of lncRNA in the PVT-1 locus, which is expressed in human airway smooth muscle cells (Perry, 2014). Inhibiting lncRNAs that act as “sponges” for miRNAs might enhance expression of proteins normally repressed by those miRNAs. In addition to oligonucleotide antagonists lncRNAs might be amenable to manipulation with small molecules that bind to secondary structure or disrupt tertiary structure of ribonucleoprotein complexes (Li & Chen, 2013). Whether the outcome of these various molecular interventions is therapeutic or an adverse reaction will depend on the role the target protein(s) play in a given disease.
Antisense oligonucleotides are promising tools to modify lncRNA function in vivo (see Figure 2 and section 4 below). However, there are two major limitations to this approach shared by miRNA antagonists and mimics: effective tissue-directed delivery and minimizing off-target effects in the lung and other uninvolved organs. There have been exciting advances in lung delivery of oligonucleotides that are described below in section 4. The tissue-restricted nature of lncRNAs suggests that off target effects might be tolerable when using antisense oligonucleotides (Guttman et al., 2010). The set of lncRNAs in the mature lung is different than the set of lncRNAs in the developing lung, which may provide another degree of selectivity for carefully designed antagonists (Herriges et al., 2014). Critical testing of this idea awaits further studies of in vivo delivery of oligonucleotide antagonists and discovery of small molecules that disrupt binding of lncRNAs to binding partners.
4. Delivery and therapeutic potential of noncoding RNA mimics and antagonists
The two therapeutic approaches developed to modulate miRNA expression and function are miRNA antagonists and miRNA mimics. MiRNA antagonists are used to treat disease that arises from miRNA gain of function by using chemically modified small interfering RNA (siRNA) oligonucleotides complementary to the endogenous miRNA mature strand. These antagonists bind with high affinity to the target miRNAs preventing them from being processed by RISC, thereby inhibiting target mRNA degradation (Bader et al., 2010). In contrast, miRNA mimics are duplex oligonucleotides composed of a guide strand and a passenger strand used in replacement therapy to restore loss of miRNA function in diseased cells or tissues. The guide strand contains the same sequence as the mature endogenous miRNA, while the passenger strand sequence is complementary to the mature miRNA. These miRNAs are processed via similar pathways as the endogenous miRNA of interest and, therefore, are thought to have fewer adverse affects compared to miRNA antagonists (Bader et al., 2010). The success of miRNAs-based therapy requires specific, safe, efficient and stable delivery of siRNA/miRNA into the target cell, tissue organ or organism. Thus, several delivery systems have been developed to overcome challenges in administration, stability, extracellular and intracellular barriers and intracellular uptake to the cytoplasm associated with miRNA-based treatment. Here, we will review delivery of unmodified, chemically modified or vector-mediated expression of siRNAs/miRNAs in different systems necessary for future therapeutic development, concentrating on lung delivery, when data are available (summarized in Table 3).
Table 3.
Selected miRNA targets in lung diseases
| Delivery system | Targeted miRNA |
Disease model |
Delivery route |
Therapeutic strategy |
References |
|---|---|---|---|---|---|
| Uncomplexed 2′-O-methyl-modied phosphorothioate |
let-7 | Lung cancer | Intratumoral | Replacement | Trang et al., 2010 |
| Locked nucleic acid (LNA) | miR-21 | Lung fibrosis | Intraperitoneal Intratracheal |
Inhibition | Liu et al., 2010 |
| Cationic lipoplexes | miR-29b | Lung cancer | Intravenous | Replacement | Wu et al., 2013 |
| Lipid- based | miR-34a | Lung cancer | Intravenous Intratumoral |
Replacement | Wiggins et al., 2010 |
| DOTMA:Clolesterol:TPGS lipoplexes | miR-133b | Lung cancer | Intravenous | Replacement | Wu et al., 2011 |
| Cationic polyurethanes-short branch PEI | miR-145 | Lung adenocarcinoma | Subcutaneous Intrabronchial Intravenous |
Replacement | Chiou et al., 2012 |
| PR-ASO-150 plasmids | miR-150 | Lung cancer | Intratumoral | Inhibition | Li et al., 2012 |
4.1 Unmodified siRNA
Naked siRNAs are highly susceptible to degradation by RNases. Thus, they have a short half-life in vivo and are unstable in biological systems (Akhtar & Benter, 2007). Despite the challenges encountered with this method of delivery, naked siRNA has been administered successfully (Dorn et al., 2004; Filleur et al., 2003; Thakker et al., 2004; Tolentino et al., 2004; Zhang et al., 2004). For instance, systemic intraperitoneal administration of siRNA targeting vascular endothelial growth factor (VEGF) led to the efficient and specific inhibition of subcutaneous fibrosarcoma tumor growth in mice (Filleur et al., 2003). In the lung, naked siRNA targeting heme oxygenase-1 was specifically detected in the airway and lung parenchyma 4–16 hours post-intranasal administration and improved ischemia-reperfusion-induced lung apoptosis (Zhang et al., 2004). Several subsequent studies confirmed that intranasal delivery of naked siRNA can be applied to the treatment of lung diseases (reviewed by Bitko and Barik, 2008). More recent work on delivery of oligonucleotides to the lung show that the stability and the specificity of naked siRNA in vivo can be improved with one of several chemical modification strategies.
4.2 Chemically modified siRNA
The most commonly used chemical modifications include incorporation of a phosphorothioate group and 2’-fluoro, 2’-O- methyl or 2’-O- methoxyethyl group, peptide nucleic acids (PNA), as well as locked and unlocked nucleic acid (LNA and UNA) oligonucleotides, (Guzman-Villanueva et al., 2012; Zhang et al., 2013). Phosphorothioate modification of the phosphodiester backbone improves siRNA biostability and RNase H activation to facilitate target RNA degradation (Crooke et al., 1996; Eckstein, 2000). However, this modification did not improve low binding affinities to complementary sequences or reduce off-target and cytotoxic effects (Brown et al., 1994; Levin, 1999). PNAs are uncharged oligonucleotide derivatives containing a pseudo-peptide backbone composed of N-(2-aminoethyl) glycine units (Nielsen et al., 1991). They display enhance stability and specificity, but are not readily taken up by eukaryotic cells (Brognara et al., 2014; Nielsen, 2010). This limitation hinders the use of PNA conjugated siRNA to treat human diseases, and resulted in the discovery of other modifications with improved properties including unlocked nucleic acids (UNA) and locked nucleic acids (LNA). UNAs are a helix destabilizing derivative of RNA in which the C2′–C3′ bond of the ribose ring is omitted. This method consists of inserting the UNA at the 5′ end of the sense strand to inhibit its loading into RISC, which leads to enhancement of the silencing potency of the antisense strand (Vaish et al., 2011). For instance, 5′-UNA modification of an siRNA targeting a conserved region of the HIV transcript improved silencing activity to potentially attenuate HIV (Snead et al., 2013). This suggests that UNA is a promising tool for the treatment of viral diseases and can further be explored in the treatment of lung diseases, although to date nothing has been reported.
LNAs are RNA analogs containing a methylene bridge that links the 2′-oxygen of ribose with the 4′-carbon (Braasch & Corey, 2001). LNAs are very stable oligonucleotides with low off-target effects and improved siRNA functionality (Elmen et al., 2005; Puri et al., 2008). The administration of 1 mg/kg/day of LNA oligonucleotide targeting the gene of the large subunit of RNA polymerase II (POLR2A) significantly inhibited tumor growth in a xenograft mouse model (Fluiter et al., 2003). Further, in vivo and in vitro administrations of LNA anti-miR-21 led to substantial down-regulation of miR-21 as a biomarker of chemotherapeutic response in lung cancer in A549 cells, accompanied by enhanced chemosensitivity of these cells to cisplatin (Xu et al., 2014). These findings indicate that LNAs are a promising tool for siRNA-based treatment of proliferative lung diseases.
4.3 Viral and lipid-based delivery systems
A safe, efficient and specific delivery of siRNA into cytoplasm requires the stability of siRNA in the blood stream, transport across the vascular endothelium, diffusion through the extracellular matrix and endosomal escape. Thus, while chemical modifications of siRNA led to the significant enhancement of naked siRNA stability the problem of efficient intracellular uptake and adequate RISC loading of siRNA required additional approaches. To overcome these limitations, various viral and non-viral vector systems have been developed (Lin et al., 2014; Martin & Caplen, 2007). Both methods have been shown to effectively induce targeted over-expression of miR-145 in vascular smooth muscle and miR-34a in the lungs (Lovren et al., 2012; Trang et al., 2011). Lentiviruses possess favorable features that promote more efficient, stable and long-term expression of target miRNAs in the host cell or organism, including the presence of accessory genes that facilitate RNA transport, gene expression, and assembly. However, the clinical applications of viral vectors are limited because of their high immunogenicity, potential induction of carcinogenesis and random integration into the host genome (reviewed by Yin et al., 2014). Thus, viral vectors are only useful in experimental animal models. The benefit of synthetic nonviral delivery systems is that they can be easily manipulated, and possess lower immunogenicity compared to the viral vectors. Thus, lipid-based systems are the most advanced and commonly used vectors for siRNA delivery in vivo with therapeutic potential.
Lipid-based siRNA delivery includes, neutral, cationic and anionic liposomes and lipoplexes that are biocompatible, biodegradable. Despite some unwanted effects associated with neutral lipid systems, including the lack of electrostatic interactions with anionic siRNA, some successes have been obtained with this method of delivery. Neutral lipid emulsion (NLE) is a lipid-based system that specifically accumulates in the lungs. Thus, systemic in vivo replacement therapy of miR-34a and let-7 using NLE led to a significant decrease in lung tumor size, indicating that NLE may be a promising tool for lung-specific miRNAs delivery (Trang et al., 2011).
The advantages of cationic systems is the positively-charged amines that facilitate their interaction with negatively charged cell membranes, and confer the ability to bind and form complexes, known as lipoplexes, with polyanionic siRNA (Akhtar & Benter, 2007). DOPE (1,2-dioleoyl-sn-glycerol-3-phosphoethanolamine) is an advanced cationic lipid-based delivery system designed to improve the in vivo delivery of existing cationic lipid systems including DOTAP (1,2-dioleoyl-3-trimethylammonium-propane). Recently, DOPE cationic liposomes harboring miR-7 have been used efficiently in vitro and in vivo to inhibited lung cancer cell growth (Rai et al., 2011). Transfection of a miR-7–expressing plasmid using cationic liposomes into cell lines resistant to epidermal growth factor receptor tyrosine kinase inhibitors significantly reduced proliferation. Further, systemic injection of DOPE cationic liposome complexes containing miR-7 into a lung cancer xenograft mouse model caused a dramatic inhibition in tumor growth. Additionally, intratumoral and systemic administration of miR-34a in mouse model of lung tumor showed a significant increase of miR-34a expression accompanied by down-regulation of its target genes (c-Met, Bcl-2 and CDK4) and inhibition of tumor growth (Wiggins et al., 2010). Systemic delivery of these liposomes containing miR-34a did not cause any toxicity, and did not elicit any immune response, indicating safe and efficient in vivo lipid-based delivery of miR-34a to the lung (Wiggins et al., 2010). Further modifications to enhance cationic liposomes loading efficiency of siRNA led to the development of AtuPLEX technology. AtuPLEX is AtuFECT–liposome–siRNA complex containing AtuFECT01, neutral/helper lipid, 1,2-diphytanoylsn-glycero-3-phosphoethanolamine, and N-(carbonylmethoxy-polyethyleneglycol)-1,2-distearoyl-sn-glycero-3-phosphoethanolamine sodium salt (Guzman-Villanueva et al., 2012). Atu027, an AtuPLEX based formulation targeting the expression of the protein kinase N3 (PKN3) was shown potent to treat advance solid cancer and prevent lung cancer metastasis (Aleku et al., 2008; Santel et al., 2010). The success of Atu027 initiated several clinical trials for the treatment of advanced solid cancers (Lin et al., 2014).
The therapeutic potential of cationic lipid carriers is also significantly enhanced by PEGylation, which improves lung uptake and retention of siRNAs (Polach et al., 2012; Sparks et al., 2012). A cationic lipopolyamine formulation (Staramine-mPEG) delivered relatively high levels of caveolin-1 silencing siRNA to the lung in mice with a longer long half-life in the lung compared to the liver and other tissues sufficient to reduce caveolin-1 expression by ~60% after a single dose. The major advantages of Staramine-mPEG are that intravenous delivery achieves effective target repression with no detectable toxicity (Polach et al., 2012). Delivery of oligonucleotides from the blood compartment may prove to be a significant advantage over inhaled oligonucleotides when airways are obstructed. Effective delivery to blood vessels in the microcirculation is also the therapeutic goal in pulmonary hypertension. Recent advances in oligonucleotide delivery methods confirm the efficacy of cationic lipid-based carriers for therapeutic delivery of siRNAs into lung and suggests that this approach is highly promising for a variety of lung diseases.
Solid nucleic acid lipid particles (SNALPs) are cationic liposomes that have been coated by PEGylation to improve siRNA serum stability and decrease immune responses. The efficacy of siRNA delivery into lung using these particles has been demonstrated. For instance, in vivo delivery of miRNA-34a containing SNALPs into lung tissues of cancer stem cell-bearing mice resulted in enhanced miR-34a stability in serum and led to the increased survival (Shi et al., 2013). There are several ongoing clinical trials targeting other forms of cancers using SNALPs derivatives (Guzman-Villanueva et al., 2012), demonstrating that SNALPs are promising delivery systems for novel epigenetic therapies of lung diseases.
Anionic liposomes have a limited entrapment capacity for negatively charged oligonucleotides. However, this problem was quickly solved by incorporating siRNA into anionic liposomes using stable calcium ion bridges (Kapoor & Burgess, 2012). Anionic liposomes can also be used to enhance the efficiency of cationic lipoplexes. Recently, Schlegel et al. (2011) developed an efficient lipoplex system composed of an anionic polymer of polyglutamate and cationic siRNA lipoplexes. Polyglutamate is a biodegradable, non-toxic and non- immunogenic water soluble anionic polymer that has been used previously in a biomedical setting (Shih et al., 2004), and the cationic liposomes contained 2-(3-[Bis-(3-amino-propyl)-amino]-propylamino)-N-ditetradecyl carbamoyl methyl-acetamide and DOPE (1,2-dioleoyl-sn-glycerol-3-phosphoethanolamine). Further, an external polyethyleneglycol (PEG) layer was added to the lipoplexes to enhance in vivo administration. The intravenous administration of fluorescent PEG-siRNA lipoplexes prepared with polyglutamate showed a substantial enhancement of siRNA recovery in the liver and lung, increased gene silencing potency and decreased cellular toxicity compared with cationic lipoplexes alone (Schlegel et al., 2013). These data indicate that combination of anionic polymers with cationic liposome is a viable tool to improve cationic-based delivery of siRNA.
In conclusion, miRNA-based therapies are promising tools for targeting miRNA expression in the treatment of various lung diseases. However, these therapies have not been fully explored for the treatment of obstructive and restrictive lung diseases. Most miRNA-based therapy targeting the lung relies on local intranasal and intratracheal administration. The advantages of these routes of delivery include a simple formulation that does not require delivery vectors, localized effects with minimal off- target effects, and cost efficiency. Even though there are multiple reports of successful local delivery of naked siRNA/miRNA into lung, this method remains ineffective and challenging because of the complexity of lung anatomy, the presence of mucous plugs in asthma and the existence of small airway obstruction in COPD. Therefore, the success of miRNA-based treatment of inflammatory lung diseases such as COPD and asthma will require the development of lipid- based vectors with more effective systemic delivery features.
5. Summary and Future Directions
The epigenetic processes that regulate cell phenotype and determine tissue architecture are reversible and amenable to modification with drugs. Significant progress has been made in chemotherapy of cancers by employing drugs that modify each of the epigenetic processes described above. Monotherapy and combination therapies with epigenetic modifiers have efficacy, but optimum therapeutic regimens are still being defined. The success of epigenetic reprogramming in therapy of lung cancer is encouraging with minimal reported side effects. These results are highly relevant to other lung diseases where inhaled epigenetic modifiers might be expected to be tolerated equally well or perhaps even better. One view of airway and vascular remodeling in asthma, IPF and pulmonary hypertension is that increased tissue mass is analogous to benign tumor growth (Tuder et al., 2007). Normal control of cell proliferation, cell survival and tissue boundaries are compromised. Therefore, as in cancer chemotherapy, inappropriate airway and vascular tissue growth might be reversible with agents that reestablish the proper balance of epigenetic processes controlling tissue architecture. However, treating nonneoplastic lung diseases with epigenetic modifying drugs is in its infancy with most studies in early proof of principle stages and initial preclinical animal models (Table 1). The current gaps in knowledge that limit development of new epigenetic modifying drugs include descriptions of which miRNAs, lncRNAs, histone modifications, and DNA methylation sites are present in a given cell type in various lung diseases. Studies are needed to define which epigenetic events are causes and which are consequences of disease. The relevant pathways modified by epigenetic regulation of gene expression must be defined in vitro and the function of those pathways validated in vivo in clinically relevant animal models. Other important areas for future research are to establish efficacy, lung-directed delivery, and off-target effects of epigenetic modifiers in vivo. If effective and safe agents can be delivered preferentially to the lungs they could become important adjuncts to bronchodilators, corticosteroids and other established agents used in obstructive and restrictive lung diseases.
Acknowledgements
Preparation of the manuscript was support in part by NIH grant HL077726 to WTG.
Abbreviations
- ACD/MPV
alveolar capillary dysplasia and misalignment of pulmonary veins
- BMPR2
bone morphogenetic protein receptor 2
- COPD
chronic obstructive pulmonary disease
- COX-2
cyclooxygenase-2
- DNMT
DNA methyltransferase
- EMT
epithelial-mesenchymal transition
- eRNA
enhancer RNA
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HDM
house dust mite
- HIF1α
hypoxia inducible factor-1α
- HOTAIR
HOX antisense intergenic RNA
- IL-β
interleukin -1β
- IFN-γ
interferon-γ
- IPF
idiopathic pulmonary fibrosis
- lncRNA
long noncoding RNA
- lincRNA
long intergenic noncoding RNA
- LNA
locked nucleic acid
- MALAT-1
metastasis associated lung adenocarcinoma transcript 1
- miRNA
microRNA
- Nrf2
NF-E2-related factor 2
- OVA
ovalbumin
- PAH
pulmonary arterial hypertension
- PH
pulmonary hypertension
- PVT-1
Pvt1 non-protein coding RNA
- SIRT1
Sirtuin-1
- SNPs
single nucleotide polymorphisms
- SAHA
suberoylanilide hydroxamic acid
- TGF-β1
transforming growth factor β1
- TNF-α
tumor necrosis factor-α
- UNA
unlocked nucleic acid
Footnotes
Conflict of Interest
BSC, MB, CAS and WTG declare they have no conflicts of interest.
References
- Adenuga D, Yao H, March TH, Seagrave J, Rahman I. Histone deacetylase 2 is phosphorylated, ubiquitinated, and degraded by cigarette smoke. American Journal of Respiratory Cell and Molecular Biology. 2009;40:464–473. doi: 10.1165/rcmb.2008-0255OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbas F, Coskunpinar E, Aynaci E, Oltulu YM, Yildiz P. Analysis of serum micro-RNAs as potential biomarker in chronic obstructive pulmonary disease. Exp Lung Res. 2012;38:286–294. doi: 10.3109/01902148.2012.689088. [DOI] [PubMed] [Google Scholar]
- Akhtar S, Benter IF. Nonviral delivery of synthetic siRNAs in vivo. J Clin Invest. 2007;117:3623–3632. doi: 10.1172/JCI33494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleku M, Schulz P, Keil O, Santel A, Schaeper U, Dieckhoff B, Janke O, Endruschat J, Durieux B, Roder N, Loffler K, Lange C, Fechtner M, Mopert K, Fisch G, Dames S, Arnold W, Jochims K, Giese K, Wiedenmann B, Scholz A, Kaufmann J. Atu027, a liposomal small interfering RNA formulation targeting protein kinase N3, inhibits cancer progression. Cancer Res. 2008;68:9788–9798. doi: 10.1158/0008-5472.CAN-08-2428. [DOI] [PubMed] [Google Scholar]
- Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulaton of RNA synthesis. Proceedings of the National Academy of Sciences of the United States of America. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andresen E, Günther G, Bullwinkel J, Lange C, Heine H. Increased expression of beta-defensin 1 (DEFB1) in chronic obstructive pulmonary disease. PloS One. 2011;6 doi: 10.1371/journal.pone.0021898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SL, Marsboom G, Kim GH, Zhang HJ, Toth PT, Svensson EC, Dyck JR, Gomberg-Maitland M, Thebaud B, Husain AN, Cipriani N, Rehman J. Epigenetic attenuation of mitochondrial superoxide dismutase 2 in pulmonary arterial hypertension: a basis for excessive cell proliferation and a new therapeutic target. Circulation. 2010;121:2661–2671. doi: 10.1161/CIRCULATIONAHA.109.916098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nature Reviews. Drug Discovery. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- Avdalovic M. Pulmonary Vasculature and Critical Asthma Syndromes: a Comprehensive Review. Clin Rev Allergy Immunol. 2014 doi: 10.1007/s12016-014-8420-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70:7027–7030. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Trivedi CM, Damera G, Jiang M, Jester W, Hoshi T, Epstein JA, Panettieri RA. Trichostatin A abrogates airway constriction, but not inflammation, in murine and human asthma models. American Journal of Respiratory Cell and Molecular Biology. 2012;46:132–138. doi: 10.1165/rcmb.2010-0276OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Research. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas CE, Noble PW. Cellular mechanisms of tissue fibrosis. 7. New insights into the cellular mechanisms of pulmonary fibrosis. Am J Physiol Cell Physiol. 2014;306:C987–C996. doi: 10.1152/ajpcell.00321.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroke E, Gauldie J, Kolb M. New treatment and markers of prognosis for idiopathic pulmonary fibrosis: lessons learned from translational research. Expert Rev Respir Med. 2013;7:465–478. doi: 10.1586/17476348.2013.838015. [DOI] [PubMed] [Google Scholar]
- Bel EH, Wenzel SE, Thompson PJ, Prazma CM, Keene ON, Yancey SW, Ortega HG, Pavord ID, Investigators S. Oral glucocorticoid-sparing effect of mepolizumab in eosinophilic asthma. N Engl J Med. 2014;371:1189–1197. doi: 10.1056/NEJMoa1403291. [DOI] [PubMed] [Google Scholar]
- Berschneider B, Ellwanger DC, Baarsma HA, Thiel C, Shimbori C, White ES, Kolb M, Neth P, Konigshoff M. miR-92a regulates TGF-beta1-induced WISP1 expression in pulmonary fibrosis. Int J Biochem Cell Biol. 2014;53:432–441. doi: 10.1016/j.biocel.2014.06.011. [DOI] [PubMed] [Google Scholar]
- Bhaskaran M, Wang Y, Zhang H, Weng T, Baviskar P, Guo Y, Gou D, Liu L. MicroRNA-127 modulates fetal lung development. Physiol Genomics. 2009;37:268–278. doi: 10.1152/physiolgenomics.90268.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitko V, Barik S. Nasal delivery of siRNA. Methods Mol Biol. 2008;442:75–82. doi: 10.1007/978-1-59745-191-8_6. [DOI] [PubMed] [Google Scholar]
- Blackwell TS, Tager AM, Borok Z, Moore BB, Schwartz DA, Anstrom KJ, Bar-Joseph Z, Bitterman P, Blackburn MR, Bradford W, Brown KK, Chapman HA, Collard HR, Cosgrove GP, Deterding R, Doyle R, Flaherty KR, Garcia CK, Hagood JS, Henke CA, Herzog E, Hogaboam CM, Horowitz JC, King TE, Jr, Loyd JE, Lawson WE, Marsh CB, Noble PW, Noth I, Sheppard D, Olsson J, Ortiz LA, O'Riordan TG, Oury TD, Raghu G, Roman J, Sime PJ, Sisson TH, Tschumperlin D, Violette SM, Weaver TE, Wells RG, White ES, Kaminski N, Martinez FJ, Wynn TA, Thannickal VJ, Eu JP. Future directions in idiopathic pulmonary fibrosis research. An NHLBI workshop report. Am J Respir Crit Care Med. 2014;189:214–222. doi: 10.1164/rccm.201306-1141WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockmeyer CL, Maegel L, Janciauskiene S, Rische J, Lehmann U, Maus UA, Nickel N, Haverich A, Hoeper MM, Golpon HA, Kreipe H, Laenger F, Jonigk D. Plexiform vasculopathy of severe pulmonary arterial hypertension and microRNA expression. J Heart Lung Transplant. 2012;31:764–772. doi: 10.1016/j.healun.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Boutten A, Goven D, Artaud-Macari E, Boczkowski J, Bonay M. NRF2 targeting: a promising therapeutic strategy in chronic obstructive pulmonary disease. Trends in Molecular Medicine. 2011;17:363–371. doi: 10.1016/j.molmed.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, Crump NT, Hazzalin CA, Liszczak G, Yuan H, Larocca C, Saldanha SA, Abagyan R, Sun Y, Meyers DJ, Marmorstein R, Mahadevan LC, Alani RM, Cole PA. Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem Biol. 2010;17:471–482. doi: 10.1016/j.chembiol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozinovski S, Vlahos R, Hansen M, Liu K, Anderson GP. Akt in the pathogenesis of COPD. International Journal of Chronic Obstructive Pulmonary Disease. 2006;1:31–38. doi: 10.2147/copd.2006.1.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch DA, Corey DR. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem Biol. 2001;8:1–7. doi: 10.1016/s1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- Brock M, Trenkmann M, Gay RE, Michel BA, Gay S, Fischler M, Ulrich S, Speich R, Huber LC. Interleukin-6 modulates the expression of the bone morphogenic protein receptor type II through a novel STAT3-microRNA cluster 17/92 pathway. Circ Res. 2009;104:1184–1191. doi: 10.1161/CIRCRESAHA.109.197491. [DOI] [PubMed] [Google Scholar]
- Brognara E, Fabbri E, Bianchi N, Finotti A, Corradini R, Gambari R. Molecular methods for validation of the biological activity of peptide nucleic acids targeting microRNAs. Methods Mol Biol. 2014;1095:165–176. doi: 10.1007/978-1-62703-703-7_14. [DOI] [PubMed] [Google Scholar]
- Brown DA, Kang SH, Gryaznov SM, DeDionisio L, Heidenreich O, Sullivan S, Xu X, Nerenberg MI. Effect of phosphorothioate modification of oligodeoxynucleotides on specific protein binding. J Biol Chem. 1994;269:26801–26805. [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RM, Tummino PJ. Cancer epigenetics drug discovery and development: the challenge of hitting the mark. The Journal of Clinical Investigation. 2014;124:64–69. doi: 10.1172/JCI71605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso P, Dempsie Y, Stevens HC, McDonald RA, Long L, Lu R, White K, Mair KM, McClure JD, Southwood M, Upton P, Xin M, van Rooij E, Olson EN, Morrell NW, MacLean MR, Baker AH. A role for miR-145 in pulmonary arterial hypertension: evidence from mouse models and patient samples. Circ Res. 2012;111:290–300. doi: 10.1161/CIRCRESAHA.112.267591. [DOI] [PubMed] [Google Scholar]
- Caruso P, MacLean MR, Khanin R, McClure J, Soon E, Southgate M, MacDonald RA, Greig JA, Robertson KE, Masson R, Denby L, Dempsie Y, Long L, Morrell NW, Baker AH. Dynamic changes in lung microRNA profiles during the development of pulmonary hypertension due to chronic hypoxia and monocrotaline. Arterioscler Thromb Vasc Biol. 2010;30:716–723. doi: 10.1161/ATVBAHA.109.202028. [DOI] [PubMed] [Google Scholar]
- Chan SY, Loscalzo J. Pathogenic mechanisms of pulmonary arterial hypertension. J. Mol. Cell Cardiol. 2008;44:14–30. doi: 10.1016/j.yjmcc.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatila WM, Criner GJ, Hancock WW, Akimova T, Moldover B, Chang JK, Cornwell W, Santerre M, Rogers TJ. Blunted expression of miR-199a-5p in regulatory T cells of patients with chronic obstructive pulmonary disease compared to unaffected smokers. Clinical and Experimental Immunology. 2014;177:341–352. doi: 10.1111/cei.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Xu L, Ye X, Shen S, Li Z, Niu X, Lu S. Polymorphisms of microRNA sequences or binding sites and lung cancer: a meta-analysis and systematic review. PloS One. 2013;8:e61008. doi: 10.1371/journal.pone.0061008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Karolczak-Bayatti M, Sweeney M, Treumann A, Morrissey K, Ulrich SM, Europe-Finner GN, Taggart MJ. Lysine deacetylase inhibition promotes relaxation of arterial tone and C-terminal acetylation of HSPB6 (Hsp20) in vascular smooth muscle cells. Physiol Rep. 2013;1:e00127. doi: 10.1002/phy2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HS, Sivachandran N, Lau A, Boudreau E, Zhao JL, Baltimore D, Delgado-Olguin P, Cybulsky MI, Fish JE. MicroRNA-146 represses endothelial activation by inhibiting pro-inflammatory pathways. EMBO molecular medicine. 2013;5:949–966. doi: 10.1002/emmm.201202318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng RY, Shang Y, Limjunyawong N, Dao T, Das S, Rabold R, Sham JS, Mitzner W, Tang W-Y. Alterations of the lung methylome in allergic airway hyper-responsiveness. Environmental and Molecular Mutagenesis. 2014;55:244–255. doi: 10.1002/em.21851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba Y, Tanabe M, Goto K, Sakai H, Misawa M. Down-regulation of miR-133a contributes to up-regulation of Rhoa in bronchial smooth muscle cells. Am J Respir Crit Care Med. 2009;180:713–719. doi: 10.1164/rccm.200903-0325OC. [DOI] [PubMed] [Google Scholar]
- Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, Muller RU, Straka E, Su L, Burki EA, Crowell RE, Patel R, Kulkarni T, Homer R, Zelterman D, Kidd KK, Zhu Y, Christiani DC, Belinsky SA, Slack FJ, Weidhaas JB. A SNP in a let-7 microRNA complementary site in the KRAS 3' untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou GY, Cherng JY, Hsu HS, Wang ML, Tsai CM, Lu KH, Chien Y, Hung SC, Chen YW, Wong CI, Tseng LM, Huang PI, Yu CC, Hsu WH, Chiou SH. Cationic polyurethanes-short branch PEI-mediated delivery of mir145 inhibited epithelial-mesenchymal transdifferentiation and cancer stem-like properties and in lung adenocarcinoma. J Control Release. 2012;159:240–250. doi: 10.1016/j.jconrel.2012.01.014. [DOI] [PubMed] [Google Scholar]
- Chipps BE, Zeiger RS, Borish L, Wenzel SE, Yegin A, Hayden ML, Miller DP, Bleecker ER, Simons FE, Szefler SJ, Weiss ST, Haselkorn T. Key findings and clinical implications from The Epidemiology and Natural History of Asthma: Outcomes and Treatment Regimens (TENOR) study. J Allergy Clin. Immunol. 2012;130:332–342. doi: 10.1016/j.jaci.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Oh SW, Kang MS, Kwon HJ, Oh GT, Kim DY. Trichostatin A attenuates airway inflammation in mouse asthma model. Clinical and Experimental Allergy: Journal of the British Society for Allergy and Clinical Immunology. 2005;35:89–96. doi: 10.1111/j.1365-2222.2004.02006.x. [DOI] [PubMed] [Google Scholar]
- Cisneros J, Hagood J, Checa M, Ortiz-Quintero B, Negreros M, Herrera I, Ramos C, Pardo A, Selman M. Hypermethylation-mediated silencing of p14(ARF) in fibroblasts from idiopathic pulmonary fibrosis. American Journal of Physiology. Lung Cellular and Molecular Physiology. 2012;303:L295–L303. doi: 10.1152/ajplung.00332.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford RL, John AE, Brightling CE, Knox AJ. Abnormal histone methylation is responsible for increased vascular endothelial growth factor 165a secretion from airway smooth muscle cells in asthma. Journal of Immunology (Baltimore, Md.: 1950) 2012;189:819–831. doi: 10.4049/jimmunol.1103641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collison A, Herbert C, Siegle JS, Mattes J, Foster PS, Kumar RK. Altered expression of microRNA in the airway wall in chronic asthma: miR-126 as a potential therapeutic target. BMC Pulm Med. 2011;11:29. doi: 10.1186/1471-2466-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collison A, Mattes J, Plank M, Foster PS. Inhibition of house dust mite-induced allergic airways disease by antagonism of microRNA-145 is comparable to glucocorticoid treatment. J Allergy Clin Immunol. 2011;128:160–167. e164. doi: 10.1016/j.jaci.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Comer BS, Camoretti-Mercado B, Kogut PC, Halayko AJ, Solway J, Gerthoffer WT. Cyclooxygenase-2 and MicroRNA-155 Expression are elevated in asthmatic airway smooth muscle cells. Am J Respir Cell Mol Biol. 2014a doi: 10.1165/rcmb.2014-0129OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer BS, Camoretti-Mercado B, Kogut PC, Halayko AJ, Solway J, Gerthoffer WT. MicroRNA-146a and microRNA-146b expression and anti-inflammatory function in human airway smooth muscle. Am J Physiol Lung Cell Mol Physiol. 2014b doi: 10.1152/ajplung.00174.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley-Salem C, Thompson SE, Kaminski M, Evans R, Burkin HR, Singer CA. Airway smooth muscle-specific expression of miR-25 in ovalbumin-sensitized mice. Am J Respir Crit Care Med. 2013;187:A5262. [Google Scholar]
- Cosío BG, Mann B, Ito K, Jazrawi E, Barnes PJ, Chung KF, Adcock IM. Histone acetylase and deacetylase activity in alveolar macrophages and blood mononocytes in asthma. Am J Respir Crit Care Med. 2004;170:141–147. doi: 10.1164/rccm.200305-659OC. [DOI] [PubMed] [Google Scholar]
- Cosio BG, Tsaprouni L, Ito K, Jazrawi E, Adcock IM, Barnes PJ. Theophylline restores histone deacetylase activity and steroid responses in COPD macrophages. The Journal of Experimental Medicine. 2004;200:689–695. doi: 10.1084/jem.20040416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courboulin A, Paulin R, Giguere NJ, Saksouk N, Perreault T, Meloche J, Paquet ER, Biardel S, Provencher S, Cote J, Simard MJ, Bonnet S. Role for miR-204 in human pulmonary arterial hypertension. J Exp Med. 2011;208:535–548. doi: 10.1084/jem.20101812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coward WR, Feghali-Bostwick CA, Jenkins G, Knox AJ, Pang L. A central role for G9a and EZH2 in the epigenetic silencing of cyclooxygenase-2 in idiopathic pulmonary fibrosis. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2014;28:3183–3196. doi: 10.1096/fj.13-241760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coward WR, Watts K, Feghali-Bostwick CA, Jenkins G, Pang L. Repression of IP-10 by interactions between histone deacetylation and hypermethylation in idiopathic pulmonary fibrosis. Molecular and Cellular Biology. 2010;30:2874–2886. doi: 10.1128/MCB.01527-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coward WR, Watts K, Feghali-Bostwick CA, Knox A, Pang L. Defective histone acetylation is responsible for the diminished expression of cyclooxygenase 2 in idiopathic pulmonary fibrosis. Molecular and Cellular Biology. 2009;29:4325–4339. doi: 10.1128/MCB.01776-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke ST, Graham MJ, Zuckerman JE, Brooks D, Conklin BS, Cummins LL, Greig MJ, Guinosso CJ, Kornbrust D, Manoharan M, Sasmor HM, Schleich T, Tivel KL, Griffey RH. Pharmacokinetic properties of several novel oligonucleotide analogs in mice. J Pharmacol Exp Ther. 1996;277:923–937. [PubMed] [Google Scholar]
- Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, Thannickal VJ, Cardoso WV, Lu J. miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol. 2011;45:287–294. doi: 10.1165/rcmb.2010-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakhlallah D, Batte K, Wang Y, Cantemir-Stone CZ, Yan P, Nuovo G, Mikhail A, Hitchcock CL, Wright VP, Nana-Sinkam SP, Piper MG, Marsh CB. Epigenetic regulation of miR-17~92 contributes to the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med. 2013;187:397–405. doi: 10.1164/rccm.201205-0888OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Kumar M, Negi V, Pattnaik B, Prakash YS, Agrawal A, Ghosh B. MicroRNA-326 regulates profibrotic functions of transforming growth factor-β in pulmonary fibrosis. American Journal of Respiratory Cell and Molecular Biology. 2014;50:882–892. doi: 10.1165/rcmb.2013-0195OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Flora S, Balansky R, D'Agostini F, Cartiglia C, Longobardi M, Steele VE, Izzotti A. Smoke-induced microRNA and related proteome alterations. Modulation by chemopreventive agents. Int J Cancer. 2012;131:2763–2773. doi: 10.1002/ijc.27814. [DOI] [PubMed] [Google Scholar]
- Dhanak D, Jackson P. Development and Classes of Epigenetic Drugs for Cancer. Biochem. Biophys. Res. Commun. 2014 doi: 10.1016/j.bbrc.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Donaldson A, Natanek SA, Lewis A, Man WD, Hopkinson NS, Polkey MI, Kemp PR. Increased skeletal muscle-specific microRNA in the blood of patients with COPD. Thorax. 2013;68:1140–1149. doi: 10.1136/thoraxjnl-2012-203129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn G, Patel S, Wotherspoon G, Hemmings-Mieszczak M, Barclay J, Natt FJ, Martin P, Bevan S, Fox A, Ganju P, Wishart W, Hall J. siRNA relieves chronic neuropathic pain. Nucleic Acids Res. 2004;32:e49. doi: 10.1093/nar/gnh044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein F. Phosphorothioate oligodeoxynucleotides: what is their origin and what is unique about them? Antisense Nucleic Acid Drug Dev. 2000;10:117–121. doi: 10.1089/oli.1.2000.10.117. [DOI] [PubMed] [Google Scholar]
- Elmen J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y, Wahren B, Liang Z, Orum H, Koch T, Wahlestedt C. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enfield KS, Pikor LA, Martinez VD, Lam WL. Mechanistic Roles of Noncoding RNAs in Lung Cancer Biology and Their Clinical Implications. Genet. Res. Int. 2012;2012:737416. doi: 10.1155/2012/737416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzie ME, Crawford M, Cho JH, Orellana R, Zhang S, Gelinas R, Batte K, Yu L, Nuovo G, Galas D, Diaz P, Wang K, Nana-Sinkam SP. Gene expression networks in COPD: microRNA and mRNA regulation. Thorax. 2012;67:122–131. doi: 10.1136/thoraxjnl-2011-200089. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filleur S, Courtin A, Ait-Si-Ali S, Guglielmi J, Merle C, Harel-Bellan A, Clezardin P, Cabon F. SiRNA-mediated inhibition of vascular endothelial growth factor severely limits tumor resistance to antiangiogenic thrombospondin-1 and slows tumor vascularization and growth. Cancer Res. 2003;63:3919–3922. [PubMed] [Google Scholar]
- Fluiter K, ten Asbroek AL, de Wissel MB, Jakobs ME, Wissenbach M, Olsson H, Olsen O, Oerum H, Baas F. In vivo tumor growth inhibition and biodistribution studies of locked nucleic acid (LNA) antisense oligonucleotides. Nucleic Acids Res. 2003;31:953–962. doi: 10.1093/nar/gkg185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford PA, Durham AL, Russell REK, Gordon F, Adcock IM, Barnes PJ. Treatment effects of low-dose theophylline combined with an inhaled corticosteroid in COPD. Chest. 2010;137:1338–1344. doi: 10.1378/chest.09-2363. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauvreau GM, O'Byrne PM, Boulet LP, Wang Y, Cockcroft D, Bigler J, FitzGerald JM, Boedigheimer M, Davis BE, Dias C, Gorski KS, Smith L, Bautista E, Comeau MR, Leigh R, Parnes JR. Effects of an anti-TSLP antibody on allergen-induced asthmatic responses. N Engl J Med. 2014;370:2102–2110. doi: 10.1056/NEJMoa1402895. [DOI] [PubMed] [Google Scholar]
- Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PloS One. 2012;7:e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan S, Faris AN, Comstock AT, Chattoraj SS, Chattoraj A, Burgess JR, Curtis JL, Martinez FJ, Zick S, Hershenson MB, Sajjan US. Quercetin prevents progression of disease in elastase/LPS-exposed mice by negatively regulating MMP expression. Respiratory Research. 2010;11 doi: 10.1186/1465-9921-11-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XN, Lin J, Ning QY, Gao L, Yao YS, Zhou JH, Li YH, Wang LL, Yu L. A histone acetyltransferase p300 inhibitor C646 induces cell cycle arrest and apoptosis selectively in AML1-ETO-positive AML cells. PloS One. 2013;8:e55481. doi: 10.1371/journal.pone.0055481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbacki N, Di Valentin E, Huynh-Thu VA, Geurts P, Irrthum A, Crahay C, Arnould T, Deroanne C, Piette J, Cataldo D, Colige A. MicroRNAs profiling in murine models of acute and chronic asthma: a relationship with mRNAs targets. PloS One. 2011;6:e16509. doi: 10.1371/journal.pone.0016509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauvreau GM, O'Byrne PM, Boulet LP, Wang Y, Cockcroft D, Bigler J, FitzGerald JM, Boedigheimer M, Davis BE, Dias C, Gorski KS, Smith L, Bautista E, Comeau MR, Leigh R, Parnes JR. Effects of an anti-TSLP antibody on allergen-induced asthmatic responses. N Engl J Med. 2014;370:2102–2110. doi: 10.1056/NEJMoa1402895. [DOI] [PubMed] [Google Scholar]
- Golpon HA, Geraci MW, Moore MD, Miller HL, Miller GJ, Tuder RM, Voelkel NF. HOX genes in human lung: altered expression in primary pulmonary hypertension and emphysema. Am J Pathol. 2001;158:955–966. doi: 10.1016/S0002-9440(10)64042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golpon HA, et al. HOX genes in human lung: altered expression in primary pulmonary hypertension and emphysema. Am J Pathol. 2001;158(3):955–966. doi: 10.1016/S0002-9440(10)64042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff JW, Powers LS, Dickson AM, Kim J, Reisetter AC, Hassan IH, Kremens K, Gross TJ, Wilson ME, Monick MM. Cigarette smoking decreases global microRNA expression in human alveolar macrophages. PloS One. 2012;7:e44066. doi: 10.1371/journal.pone.0044066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros C, Fahy J, Halby L, Dufau I, Erdmann A, Gregoire JM, Ausseil F, Vispe S, Arimondo PB. DNA methylation inhibitors in cancer: recent and future approaches. Biochimie. 2012;94:2280–2296. doi: 10.1016/j.biochi.2012.07.025. [DOI] [PubMed] [Google Scholar]
- Gross TJ, Powers LS, Boudreau RL, Brink B, Reisetter A, Goel K, Gerke AK, Hassan IH, Monick MM. A microRNA processing defect in smokers' macrophages is linked to SUMOylation of the endonuclease DICER. J Biol Chem. 2014;289:12823–12834. doi: 10.1074/jbc.M114.565473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, Zornig M, MacLeod AR, Spector DL, Diederichs S. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, Rinn JL, Lander ES, Regev A. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman-Villanueva D, El-Sherbiny IM, Herrera-Ruiz D, Vlassov AV, Smyth HD. Formulation approaches to short interfering RNA and MicroRNA: challenges and implications. J Pharm Sci. 2012;101:4046–4066. doi: 10.1002/jps.23300. [DOI] [PubMed] [Google Scholar]
- Guzmán L, Depix MS, Salinas AM, Roldán R, Aguayo F, Silva A, Vinet R. Analysis of aberrant methylation on promoter sequences of tumor suppressor genes and total DNA in sputum samples: a promising tool for early detection of COPD and lung cancer in smokers. Diagnostic Pathology. 2012;7 doi: 10.1186/1746-1596-7-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, Morse M, Engreitz J, Lander ES, Guttman M, Lodish HF, Flavell R, Raj A, Rinn JL. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar SM, McKinsey TA. BET-ting on chromatin-based therapeutics for heart failure. J Mol Cell Cardiol. 2014;74C:98–102. doi: 10.1016/j.yjmcc.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci U S A. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan T, Carroll TP, Buckley PG, Cummins R, O'Neill SJ, McElvaney NG, Greene CM. miR-199a-5p silencing regulates the unfolded protein response in chronic obstructive pulmonary disease and α1-antitrypsin deficiency. Am J Respir Crit Care Med. 2014;189:263–273. doi: 10.1164/rccm.201306-1151OC. [DOI] [PubMed] [Google Scholar]
- Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions - beyond repression of gene expression. Nat Rev Genet. 2014 doi: 10.1038/nrg3765. [DOI] [PubMed] [Google Scholar]
- Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- Herriges MJ, Swarr DT, Morley MP, Rathi KS, Peng T, Stewart KM, Morrisey EE. Long noncoding RNAs are spatially correlated with transcription factors and regulate lung development. Genes Dev. 2014;28:1363–1379. doi: 10.1101/gad.238782.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hew M, Bhavsar P, Torrego A, Meah S, Khorasani N, Barnes PJ, Adcock I, Chung KF. Relative corticosteroid insensitivity of peripheral blood mononuclear cells in severe asthma. Am J Respir Crit Care Med. 2006;174:134–141. doi: 10.1164/rccm.200512-1930OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honeyman L, Bazett M, Tomko TG, Haston CK. MicroRNA profiling implicates the insulin-like growth factor pathway in bleomycin-induced pulmonary fibrosis in mice. Fibrogenesis Tissue Repair. 2013;6:16. doi: 10.1186/1755-1536-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosgood HD, Menashe I, He X, Chanock S, Lan Q. PTEN identified as important risk factor of chronic obstructive pulmonary disease. Respiratory Medicine. 2009;103:1866–1870. doi: 10.1016/j.rmed.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SK, Fisher AS, Scruggs AM, White ES, Hogaboam CM, Richardson BC, Peters-Golden M. Hypermethylation of PTGER2 confers prostaglandin E2 resistance in fibrotic fibroblasts from humans and mice. The American Journal of Pathology. 2010;177:2245–2255. doi: 10.2353/ajpath.2010.100446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SK, Scruggs AM, Donaghy J, Horowitz JC, Zaslona Z, Przybranowski S, White ES, Peters-Golden M. Histone modifications are responsible for decreased Fas expression and apoptosis resistance in fibrotic lung fibroblasts. Cell Death & Disease. 2013;4 doi: 10.1038/cddis.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert M, Morrell NW, Archer SL, Stenmark KR, MacLean MR, Lang IM, Christman BW, Weir EK, Eickelberg O, Voelkel NF, Rabinovitch M. Cellular and molecular pathobiology of pulmonary arterial hypertension. J. Am. Coll. Cardiol. 2004;43:13S–24S. doi: 10.1016/j.jacc.2004.02.029. [DOI] [PubMed] [Google Scholar]
- Humbert M, Busse W, Hanania NA, Lowe PJ, Canvin J, Erpenbeck VJ, Holgate S. Omalizumab in asthma: an update on recent developments. J. Allergy Clin Immunol Pract. 2014;2:525–536. e521. doi: 10.1016/j.jaip.2014.03.010. [DOI] [PubMed] [Google Scholar]
- Imam JS, Plyler JR, Bansal H, Prajapati S, Bansal S, Rebeles J, Chen HI, Chang YF, Panneerdoss S, Zoghi B, Buddavarapu KC, Broaddus R, Hornsby P, Tomlinson G, Dome J, Vadlamudi RK, Pertsemlidis A, Chen Y, Rao MK. Genomic loss of tumor suppressor miRNA-204 promotes cancer cell migration and invasion by activating AKT/mTOR/Rac1 signaling and actin reorganization. PloS One. 2012;7:e52397. doi: 10.1371/journal.pone.0052397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam JS, et al. Genomic loss of tumor suppressor miRNA-204 promotes cancer cell migration and invasion by activating AKT/mTOR/Rac1 signaling and actin reorganization. PloS One. 2012;7(12):e52397. doi: 10.1371/journal.pone.0052397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Caramori G, Lim S, Oates T, Chung KF, Barnes PJ, Adcock IM. Expression and activity of histone deacetylases in human asthmatic airways. Am J Respir Crit Care Med. 2002;166:392–396. doi: 10.1164/rccm.2110060. [DOI] [PubMed] [Google Scholar]
- Ito K, Ito M, Elliott WM, Cosio B, Caramori G, Kon OM, Barczyk A, Hayashi S, Adcock IM, Hogg JC, Barnes PJ. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. The New England Journal of Medicine. 2005;352:1967–1976. doi: 10.1056/NEJMoa041892. [DOI] [PubMed] [Google Scholar]
- Ito K, Lim S, Caramori G, Cosio B, Chung KF, Adcock IM, Barnes PJ. A molecular mechanism of action of theophylline: Induction of histone deacetylase activity to decrease inflammatory gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:8921–8926. doi: 10.1073/pnas.132556899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Yamamura S, Essilfie-Quaye S, Cosio B, Ito M, Barnes PJ, Adcock IM. Histone deacetylase 2-mediated deacetylation of the glucocorticoid receptor enables NF-kappaB suppression. The Journal of Experimental Medicine. 2006;203:7–13. doi: 10.1084/jem.20050466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalali S, Ramanathan GK, Parthasarathy PT, Aljubran S, Galam L, Yunus A, Garcia S, Cox RR, Jr, Lockey RF, Kolliputi N. Mir-206 regulates pulmonary artery smooth muscle cell proliferation and differentiation. PLoS One. 2012;7:e46808. doi: 10.1371/journal.pone.0046808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardim MJ, Dailey L, Silbajoris R, Diaz-Sanchez D. Distinct microRNA expression in human airway cells of asthmatic donors identifies a novel asthma-associated gene. American Journal of Respiratory Cell and Molecular Biology. 2012;47:536–542. doi: 10.1165/rcmb.2011-0160OC. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science (New York, N.Y.) 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Muller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- Jimenez-Morales S, Gamboa-Becerra R, Baca V, Del Rio-Navarro BE, Lopez-Ley DY, Velazquez-Cruz R, Saldana-Alvarez Y, Salas-Martinez G, Orozco L. MiR-146a polymorphism is associated with asthma but not with systemic lupus erythematosus and juvenile rheumatoid arthritis in Mexican patients. Tissue Antigens. 2012;80:317–321. doi: 10.1111/j.1399-0039.2012.01929.x. [DOI] [PubMed] [Google Scholar]
- John AE, Zhu YM, Brightling CE, Pang L, Knox AJ. Human airway smooth muscle cells from asthmatic individuals have CXCL8 hypersecretion due to increased NF-kappa B p65, C/EBP beta, and RNA polymerase II binding to the CXCL8 promoter. Journal of Immunology (Baltimore, Md.: 1950) 2009;183:4682–4692. doi: 10.4049/jimmunol.0803832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jude JA, Dileepan M, Subramanian S, Solway J, Panettieri RA, Walseth TF, Kannan MS. miR-140-3p regulation of TNF-α-induced CD38 expression in human airway smooth muscle cells. American Journal of Physiology. Lung Cellular and Molecular Physiology. 2012;303:L460–L468. doi: 10.1152/ajplung.00041.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jude JA, Solway J, Panettieri RA, Walseth TF, Kannan MS. Differential induction of CD38 expression by TNF-{alpha} in asthmatic airway smooth muscle cells. American Journal of Physiology. Lung Cellular and Molecular Physiology. 2010;299:L879–L890. doi: 10.1152/ajplung.00021.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalathil SG, Lugade AA, Pradhan V, Miller A, Parameswaran GI, Sethi S, Thanavala Y. T-regulatory cells and programmed death 1(+) T cells contribute to effector T-cell dysfunction in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2014;190:40–50. doi: 10.1164/rccm.201312-2293OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K, Peng X, Zhang X, Wang Y, Zhang L, Gao L, Weng T, Zhang H, Ramchandran R, Raj JU, Gou D, Liu L. MicroRNA-124 suppresses the transactivation of nuclear factor of activated T cells by targeting multiple genes and inhibits the proliferation of pulmonary artery smooth muscle cells. J Biol Chem. 2013;288:25414–25427. doi: 10.1074/jbc.M113.460287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, Burgess DJ. Efficient and safe delivery of siRNA using anionic lipids: Formulation optimization studies. Int J Pharm. 2012;432:80–90. doi: 10.1016/j.ijpharm.2012.04.058. [DOI] [PubMed] [Google Scholar]
- Karolczak-Bayatti M, Sweeney M, Cheng J, Edey L, Robson SC, Ulrich SM, Treumann A, Taggart MJ, Europe-Finner GN. Acetylation of heat shock protein 20 (Hsp20) regulates human myometrial activity. J Biol Chem. 2011;286:34346–34355. doi: 10.1074/jbc.M111.278549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kang Y, Kojima Y, Lighthouse JK, Hu X, Aldred MA, McLean DL, Park H, Comhair SA, Greif DM, Erzurum SC, Chun HJ. An endothelial apelin-FGF link mediated by miR-424 and miR-503 is disrupted in pulmonary arterial hypertension. Nat Med. 2013;19:74–82. doi: 10.1038/nm.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W, Yang H, He L, Zhao J-j, Coppola D, Dalton WS, Cheng JQ. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Molecular and Cellular Biology. 2008;28:6773–6784. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Königshoff M, Balsara N, Pfaff E-M, Kramer M, Chrobak I, Seeger W, Eickelberg O. Functional Wnt signaling is increased in idiopathic pulmonary fibrosis. PloS One. 2008;3 doi: 10.1371/journal.pone.0002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, Johnston D, Kim GE, Spitale RC, Flynn RA, Zheng GX, Aiyer S, Raj A, Rinn JL, Chang HY, Khavari PA. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn AR, Schlauch K, Lao R, Halayko AJ, Gerthoffer WT, Singer CA. MicroRNA expression in human airway smooth muscle cells: Role of miR-25 in regulation of airway smooth muscle phenotype. Am J Respir Cell Mol Biol. 2010;42:506–513. doi: 10.1165/rcmb.2009-0123OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Ahmad T, Sharma A, Mabalirajan U, Kulshreshtha A, Agrawal A, Ghosh B. Let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. J Allergy Clin Immunol. 2011;128:1077–1085. e1071–e1010. doi: 10.1016/j.jaci.2011.04.034. [DOI] [PubMed] [Google Scholar]
- Kumar M, Ahmad T, Sharma A, Mabalirajan U, Kulshreshtha A, Agrawal A, Ghosh B. Let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. J Allergy Clin Immunol. 2011;128:1077–1085. e1071–e1010. doi: 10.1016/j.jaci.2011.04.034. [DOI] [PubMed] [Google Scholar]
- Kuo Y-C, Li Y-SJ, Zhou J, Shih Y-RV, Miller M, Broide D, Lee OK-S, Chien S. Human mesenchymal stem cells suppress the stretch-induced inflammatory miR-155 and cytokines in bronchial epithelial cells. PloS One. 2013;8 doi: 10.1371/journal.pone.0071342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lakshmi SP, Reddy AT, Zhang Y, Sciurba FC, Mallampalli RK, Duncan SR, Reddy RC. Down-regulated peroxisome proliferator-activated receptor γ (PPARγ) in lung epithelial cells promotes a PPARγ agonist-reversible proinflammatory phenotype in Chronic Obstructive Pulmonary Disease (COPD) J Biol Chem. 2014;289:6383–6393. doi: 10.1074/jbc.M113.536805. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lam HC, Cloonan SM, Bhashyam AR, Haspel JA, Singh A, Sathirapongsasuti JF, Cervo M, Yao H, Chung AL, Mizumura K, An CH, Shan B, Franks JM, Haley KJ, Owen CA, Tesfaigzi Y, Washko GR, Quackenbush J, Silverman EK, Rahman I, Kim HP, Mahmood A, Biswal SS, Ryter SW, Choi AMK. Histone deacetylase 6-mediated selective autophagy regulates COPD-associated cilia dysfunction. The Journal of Clinical Investigation. 2013;123:5212–5230. doi: 10.1172/JCI69636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larner-Svensson HM, Williams AE, Tsitsiou E, Perry MM, Jiang X, Chung KF, Lindsay MA. Pharmacological studies of the mechanism and function of interleukin-1beta-induced miRNA-146a expression in primary human airway smooth muscle. Respiratory Research. 2010;11 doi: 10.1186/1465-9921-11-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidinger P, Keller A, Borries A, Huwer H, Rohling M, Huebers J, Lenhof HP, Meese E. Specific peripheral miRNA profiles for distinguishing lung cancer from COPD. Lung Cancer. 2011;74:41–47. doi: 10.1016/j.lungcan.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Levanen B, Bhakta NR, Torregrosa Paredes P, Barbeau R, Hiltbrunner S, Pollack JL, Skold CM, Svartengren M, Grunewald J, Gabrielsson S, Eklund A, Larsson BM, Woodruff PG, Erle DJ, Wheelock AM. Altered microRNA profiles in bronchoalveolar lavage fluid exosomes in asthmatic patients. J Allergy Clin Immunol. 2013;131:894–903. doi: 10.1016/j.jaci.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin AA. A review of the issues in the pharmacokinetics and toxicology of phosphorothioate antisense oligonucleotides. Biochim Biophys Acta. 1999;1489:69–84. doi: 10.1016/s0167-4781(99)00140-2. [DOI] [PubMed] [Google Scholar]
- Lewis A, Riddoch-Contreras J, Natanek SA, Donaldson A, Man WD, Moxham J, Hopkinson NS, Polkey MI, Kemp PR. Downregulation of the serum response factor/miR-1 axis in the quadriceps of patients with COPD. Thorax. 2012;67:26–34. doi: 10.1136/thoraxjnl-2011-200309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int. J Biochem. Cell Biol. 2013;45:1895–1910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- Li J, Chen S, Cleary RA, Wang R, Gannon OJ, Seto E, Tang DD. Histone deacetylase 8 regulates cortactin deacetylation and contraction in smooth muscle tissues. Am J Physiol Cell Physiol. 2014;307:C288–C295. doi: 10.1152/ajpcell.00102.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LJ, Gao LB, Lv ML, Dong W, Su XW, Liang WB, Zhang L. Association between SNPs in pre-miRNA and risk of chronic obstructive pulmonary disease. Clin Biochem. 2011;44:813–816. doi: 10.1016/j.clinbiochem.2011.04.021. [DOI] [PubMed] [Google Scholar]
- Li P, Li J, Chen T, Wang H, Chu H, Chang J, Zang W, Wang Y, Ma Y, Du Y, Zhao G, Zhang G. Expression analysis of serum microRNAs in idiopathic pulmonary fibrosis. Int J Mol Med. 2014;33:1554–1562. doi: 10.3892/ijmm.2014.1712. [DOI] [PubMed] [Google Scholar]
- Li P, Zhao GQ, Chen TF, Chang JX, Wang HQ, Chen SS, Zhang GJ. Serum miR-21 and miR-155 expression in idiopathic pulmonary fibrosis. J Asthma. 2013;50:960–964. doi: 10.3109/02770903.2013.822080. [DOI] [PubMed] [Google Scholar]
- Li YJ, Zhang YX, Wang PY, Chi YL, Zhang C, Ma Y, Lv CJ, Xie SY. Regression of A549 lung cancer tumors by anti-miR-150 vector. Oncol Rep. 2012;27:129–134. doi: 10.3892/or.2011.1466. [DOI] [PubMed] [Google Scholar]
- Liang H, et al. The antifibrotic effects and mechanisms of microRNA-26a action in idiopathic pulmonary fibrosis. Mol Ther. 2014;22(6):1122–1133. doi: 10.1038/mt.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Chen J, Zhang Z, Zheng G. Lipid-based nanoparticles in the systemic delivery of siRNA. Nanomedicine (Lond) 2014;9:105–120. doi: 10.2217/nnm.13.192. [DOI] [PubMed] [Google Scholar]
- Lin R, Roychowdhury-Saha M, Black C, Watt AT, Marcusson EG, Freier SM, Edgington TS. Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Lett. 2011;585:671–676. doi: 10.1016/j.febslet.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lino Cardenas CL, Henaoui IS, Courcot E, Roderburg C, Cauffiez C, Aubert S, Copin M-C, Wallaert B, Glowacki F, Dewaeles E, Milosevic J, Maurizio J, Tedrow J, Marcet B, Lo-Guidice J-M, Kaminski N, Barbry P, Luedde T, Perrais M, Mari B, Pottier N. miR-199a-5p Is upregulated during fibrogenic response to tissue injury and mediates TGFbeta-induced lung fibroblast activation by targeting caveolin-1. PLoS genetics. 2013;9 doi: 10.1371/journal.pgen.1003291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Killian JK, Yang M, Walker RL, Hong JA, Zhang M, Davis S, Zhang Y, Hussain M, Xi S, Rao M, Meltzer PA, Schrump DS. Epigenomic alterations and gene expression profiles in respiratory epithelia exposed to cigarette smoke condensate. Oncogene. 2010;29:3650–3664. doi: 10.1038/onc.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Qin H-B, Xu B, Zhou H, Zhao D-Y. Profiling of miRNAs in pediatric asthma: upregulation of miRNA-221 and miRNA-485-3p. Molecular Medicine Reports. 2012;6:1178–1182. doi: 10.3892/mmr.2012.1030. [DOI] [PubMed] [Google Scholar]
- Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 2010;207:1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lota HK, Wells AU. The evolving pharmacotherapy of pulmonary fibrosis. Expert. Opin. Pharmacother. 2013;14:79–89. doi: 10.1517/14656566.2013.758250. [DOI] [PubMed] [Google Scholar]
- Louafi F, Martinez-Nunez RT, Sanchez-Elsner T. MicroRNA-155 targets SMAD2 and modulates the response of macrophages to transforming growth factor-{beta} J Biol Chem. 2010;285:41328–41336. doi: 10.1074/jbc.M110.146852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovren F, Pan Y, Quan A, Singh KK, Shukla PC, Gupta N, Steer BM, Ingram AJ, Gupta M, Al-Omran M, Teoh H, Marsden PA, Verma S. MicroRNA-145 targeted therapy reduces atherosclerosis. Circulation. 2012;126:S81–S90. doi: 10.1161/CIRCULATIONAHA.111.084186. [DOI] [PubMed] [Google Scholar]
- Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. Journal of Immunology (Baltimore, Md.: 1950) 2009;182:4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Xiao J, Lin H, Bai Y, Luo X, Wang Z, Yang B. A single anti-microRNA antisense oligodeoxyribonucleotide (AMO) targeting multiple microRNAs offers an improved approach for microRNA interference. Nucleic Acids Res. 2009;37:e24. doi: 10.1093/nar/gkn1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR. Proc Natl Acad Sci U S A. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmhall C, Alawieh S, Lu Y, Sjostrand M, Bossios A, Eldh M, Radinger M. MicroRNA-155 is essential for T(H)2-mediated allergen-induced eosinophilic inflammation in the lung. J Allergy Clin Immunol. 2014;133:1429–1438. 1438 e1421–1438 e1427. doi: 10.1016/j.jaci.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Martin SE, Caplen NJ. Applications of RNA interference in mammalian systems. Annu Rev Genomics Hum Genet. 2007;8:81–108. doi: 10.1146/annurev.genom.8.080706.092424. [DOI] [PubMed] [Google Scholar]
- Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc Natl Acad Sci U S A. 2009;106:18704–18709. doi: 10.1073/pnas.0905063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayoral RJ, Deho L, Rusca N, Bartonicek N, Saini HK, Enright AJ, Monticelli S. MiR-221 influences effector functions and actin cytoskeleton in mast cells. PloS One. 2011;6 doi: 10.1371/journal.pone.0026133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado N, Thimmulappa R, Thomas CMR, Fenwick PS, Chana KK, Donnelly LE, Biswal S, Ito K, Barnes PJ. Decreased histone deacetylase 2 impairs Nrf2 activation by oxidative stress. Biochemical and Biophysical Research Communications. 2011;406:292–298. doi: 10.1016/j.bbrc.2011.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, Boon RA, Dimmeler S. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ. Res. 2014;114:1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- Milosevic J, Pandit K, Magister M, Rabinovich E, Ellwanger DC, Yu G, Vuga LJ, Weksler B, Benos PV, Gibson KF, McMillan M, Kahn M, Kaminski N. Profibrotic role of miR-154 in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2012;47:879–887. doi: 10.1165/rcmb.2011-0377OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno S, Bogaard HJ, Gomez-Arroyo J, Alhussaini A, Kraskauskas D, Cool CD, Voelkel NF. MicroRNA-199a-5p is associated with hypoxia-inducible factor-1α expression in lungs from patients with COPD. Chest. 2012;142:663–672. doi: 10.1378/chest.11-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Pinelo S, Pastor MD, Suarez R, Romero-Romero B, Gonzalez De la Pena M, Salinas A, Garcia-Carbonero R, De Miguel MJ, Rodriguez-Panadero F, Carnero A, Paz-Ares L. MicroRNA clusters: dysregulation in lung adenocarcinoma and COPD. Eur Respir J. 2014;43:1740–1749. doi: 10.1183/09031936.00091513. [DOI] [PubMed] [Google Scholar]
- Morrell NW, Archer SL, Defelice A, Evans S, Fiszman M, Martin T, Saulnier M, Rabinovitch M, Schermuly R, Stewart D, Truebel H, Walker G, Stenmark KR. Anticipated classes of new medications and molecular targets for pulmonary arterial hypertension. Pulm. Circ. 2013;3:226–244. doi: 10.4103/2045-8932.109940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamaru Y, Vuppusetty C, Wada H, Milne JC, Ito M, Rossios C, Elliot M, Hogg J, Kharitonov S, Goto H, Bemis JE, Elliott P, Barnes PJ, Ito K. A protein deacetylase SIRT1 is a negative regulator of metalloproteinase-9. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2009;23:2810–2819. doi: 10.1096/fj.08-125468. [DOI] [PubMed] [Google Scholar]
- Nakano T, Inoue Y, Shimojo N, Yamaide F, Morita Y, Arima T, Tomiita M, Kohno Y. Lower levels of hsa-mir-15a, which decreases VEGFA, in the CD4+ T cells of pediatric patients with asthma. J Allergy Clin Immunol. 2013;132:1224.e1212–1227.e1212. doi: 10.1016/j.jaci.2013.06.041. [DOI] [PubMed] [Google Scholar]
- Natarajan R. Drugs targeting epigenetic histone acetylation in vascular smooth muscle cells for restenosis and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2011;31:725–727. doi: 10.1161/ATVBAHA.111.222976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G, Andrau JC. Noncoding transcription at enhancers: general principles and functional models. Annu. Rev. Genet. 2012;46:1–19. doi: 10.1146/annurev-genet-110711-155459. [DOI] [PubMed] [Google Scholar]
- Nielsen PE. Peptide nucleic acids (PNA) in chemical biology and drug discovery. Chem Biodivers. 2010;7:786–804. doi: 10.1002/cbdv.201000005. [DOI] [PubMed] [Google Scholar]
- Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- Oak SR, Murray L, Herath A, Sleeman M, Anderson I, Joshi AD, Coelho AL, Flaherty KR, Toews GB, Knight D, Martinez FJ, Hogaboam CM. A micro RNA processing defect in rapidly progressing idiopathic pulmonary fibrosis. PloS One. 2011;6 doi: 10.1371/journal.pone.0021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oike T, Komachi M, Ogiwara H, Amornwichet N, Saitoh Y, Torikai K, Kubo N, Nakano T, Kohno T. C646, a selective small molecule inhibitor of histone acetyltransferase p300, radiosensitizes lung cancer cells by enhancing mitotic catastrophe. Radiother Oncol. 2014;111:222–227. doi: 10.1016/j.radonc.2014.03.015. [DOI] [PubMed] [Google Scholar]
- Ortega HG, Liu MC, Pavord ID, Brusselle GG, FitzGerald JM, Chetta A, Humbert M, Katz LE, Keene ON, Yancey SW, Chanez P, Investigators M. Mepolizumab treatment in patients with severe eosinophilic asthma. N Engl J Med. 2014;371:1198–1207. doi: 10.1056/NEJMoa1403290. [DOI] [PubMed] [Google Scholar]
- Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, Gibson KF, Konishi K, Yousem SA, Singh M, Handley D, Richards T, Selman M, Watkins SC, Pardo A, Ben-Yehudah A, Bouros D, Eickelberg O, Ray P, Benos PV, Kaminski N. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2010;182:220–229. doi: 10.1164/rccm.200911-1698OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papi A, Casoni G, Caramori G, Guzzinati I, Boschetto P, Ravenna F, Calia N, Petruzzelli S, Corbetta L, Cavallesco G, Forini E, Saetta M, Ciaccia A, Fabbri LM. COPD increases the risk of squamous histological subtype in smokers who develop non-small cell lung carcinoma. Thorax. 2004;59:679–681. doi: 10.1136/thx.2003.018291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels RA, Rabe KF. Burden and clinical features of chronic obstructive pulmonary disease (COPD) Lancet. 2004;364:613–620. doi: 10.1016/S0140-6736(04)16855-4. [DOI] [PubMed] [Google Scholar]
- Perry MM, Baker JE, Gibeon DS, Adcock IM, Chung KF. Airway smooth muscle hyperproliferation is regulated by microRNA-221 in severe asthma. American Journal of Respiratory Cell and Molecular Biology. 2014;50:7–17. doi: 10.1165/rcmb.2013-0067OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MM, Tsitsiou E, Austin PJ, Lindsay MA, Gibeon DS, Adcock IM, Chung KF. Role of non-coding RNAs in maintaining primary airway smooth muscle cells. Respir Res. 2014;15:58. doi: 10.1186/1465-9921-15-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MM, Williams AE, Tsitsiou E, Larner-Svensson HM, Lindsay MA. Divergent intracellular pathways regulate interleukin-1beta-induced miR-146a and miR-146b expression and chemokine release in human alveolar epithelial cells. FEBS Lett. 2009;583:3349–3355. doi: 10.1016/j.febslet.2009.09.038. [DOI] [PubMed] [Google Scholar]
- Petty D, Singer CA. miR-25 mediates airway smooth muscle cell proliferation through inhibition of mitogenic signaling pathways. Am J Respir Crit Care Med. 2012;185:A4133. [Google Scholar]
- Pietra GG, Capron F, Stewart S, Leone O, Humbert M, Robbins IM, Reid LM, Tuder RM. Pathologic assessment of vasculopathies in pulmonary hypertension. J. Am. Coll. Cardiol. 2004;43:25S–32S. doi: 10.1016/j.jacc.2004.02.033. [DOI] [PubMed] [Google Scholar]
- Polach KJ, Matar M, Rice J, Slobodkin G, Sparks J, Congo R, Rea-Ramsey A, McClure D, Brunhoeber E, Krampert M, Schuster A, Jahn-Hofmann K, John M, Vornlocher HP, Fewell JG, Anwer K, Geick A. Delivery of siRNA to the mouse lung via a functionalized lipopolyamine. Mol Ther. 2012;20:91–100. doi: 10.1038/mt.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikepahad S, Knight JM, Naghavi AO, Oplt T, Creighton CJ, Shaw C, Benham AL, Kim J, Soibam B, Harris RA, Coarfa C, Zariff A, Milosavljevic A, Batts LM, Kheradmand F, Gunaratne PH, Corry DB. Proinflammatory role for let-7 microRNAS in experimental asthma. J Biol Chem. 2010;285:30139–30149. doi: 10.1074/jbc.M110.145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pottier N, Maurin T, Chevalier B, Puisségur M-P, Lebrigand K, Robbe-Sermesant K, Bertero T, Lino Cardenas CL, Courcot E, Rios G, Fourre S, Lo-Guidice J-M, Marcet B, Cardinaud B, Barbry P, Mari B. Identification of keratinocyte growth factor as a target of microRNA-155 in lung fibroblasts: implication in epithelial-mesenchymal interactions. PloS One. 2009;4 doi: 10.1371/journal.pone.0006718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri N, Wang X, Varma R, Burnett C, Beauchamp L, Batten DM, Young M, Sule V, Latham K, Sendera T, Echeverri C, Sachse C, Magdaleno S. LNA incorporated siRNAs exhibit lower off-target effects compared to 2'-OMethoxy in cell phenotypic assays and microarray analysis. Nucleic Acids Symp Ser (Oxf) 2008:25–26. doi: 10.1093/nass/nrn013. [DOI] [PubMed] [Google Scholar]
- Qin H, Xu B, Mei J, Li D, Liu J, Zhao D, Liu F. Inhibition of miRNA-221 suppresses the airway inflammation in asthma. Inflammation. 2012;35:1595–1599. doi: 10.1007/s10753-012-9474-1. [DOI] [PubMed] [Google Scholar]
- Qiu W, Baccarelli A, Carey VJ, Boutaoui N, Bacherman H, Klanderman B, Rennard S, Agusti A, Anderson W, Lomas DA, DeMeo DL. Variable DNA methylation is associated with chronic obstructive pulmonary disease and lung function. Am J Respir Crit Care Med. 2012;185:373–381. doi: 10.1164/rccm.201108-1382OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich EI, Kapetanaki MG, Steinfeld I, Gibson KF, Pandit KV, Yu G, Yakhini Z, Kaminski N. Global methylation patterns in idiopathic pulmonary fibrosis. PloS One. 2012;7 doi: 10.1371/journal.pone.0033770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovitch M. Molecular pathogenesis of pulmonary arterial hypertension. J. Clin. Invest. 2008;118:2372–2379. doi: 10.1172/JCI33452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Takigawa N, Ito S, Kashihara H, Ichihara E, Yasuda T, Shimizu K, Tanimoto M, Kiura K. Liposomal delivery of MicroRNA-7-expressing plasmid overcomes epidermal growth factor receptor tyrosine kinase inhibitor-resistance in lung cancer cells. Mol Cancer Ther. 2011;10:1720–1727. doi: 10.1158/1535-7163.MCT-11-0220. [DOI] [PubMed] [Google Scholar]
- Raidl M, Sibbing B, Strauch J, Müller K, Nemat A, Schneider PM, Hag H, Erdmann E, Koch A. Impaired TNFalpha-induced VEGF expression in human airway smooth muscle cells from smokers with COPD: role of MAPkinases and histone acetylation--effect of dexamethasone. Cell Biochemistry and Biophysics. 2007;49:98–110. doi: 10.1007/s12013-007-0043-4. [DOI] [PubMed] [Google Scholar]
- Rajendrasozhan S, Yang S-R, Kinnula VL, Rahman I. SIRT1, an antiinflammatory and antiaging protein, is decreased in lungs of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008;177:861–870. doi: 10.1164/rccm.200708-1269OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy SK, Mailleux AA, Gupte VV, Mata F, Sala FG, Veltmaat JM, Del Moral PM, De Langhe S, Parsa S, Kelly LK, Kelly R, Shia W, Keshet E, Minoo P, Warburton D, Bellusci S. Fgf10 dosage is critical for the amplification of epithelial cell progenitors and for the formation of multiple mesenchymal lineages during lung development. Dev Biol. 2007;307:237–247. doi: 10.1016/j.ydbio.2007.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rhodes CJ, et al. Reduced microRNA-150 is associated with poor survival in pulmonary arterial hypertension. Am J Respir Crit Care Med. 2013;187(3):294–302. doi: 10.1164/rccm.201205-0839OC. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nature Medicine. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royce SG, Dang W, Yuan G, Tran J, El-Osta A, Karagiannis TC, Tang MLK. Effects of the histone deacetylase inhibitor, trichostatin A, in a chronic allergic airways disease model in mice. Archivum Immunologiae Et Therapiae Experimentalis. 2012;60:295–306. doi: 10.1007/s00005-012-0180-3. [DOI] [PubMed] [Google Scholar]
- Saco TV, Parthasarathy PT, Cho Y, Lockey RF, Kolliputi N. Role of epigenetics in pulmonary hypertension. Am J Physiol Cell Physiol. 2014;306:C1101–C1105. doi: 10.1152/ajpcell.00314.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders YY, Ambalavanan N, Halloran B, Zhang X, Liu H, Crossman DK, Bray M, Zhang K, Thannickal VJ, Hagood JS. Altered DNA methylation profile in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2012;186:525–535. doi: 10.1164/rccm.201201-0077OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders YY, Hagood JS, Liu H, Zhang W, Ambalavanan N, Thannickal VJ. Histone deacetylase inhibition promotes fibroblast apoptosis and ameliorates pulmonary fibrosis in mice. The European Respiratory Journal. 2014;43:1448–1458. doi: 10.1183/09031936.00095113. [DOI] [PubMed] [Google Scholar]
- Sanfiorenzo C, Ilie MI, Belaid A, Barlesi F, Mouroux J, Marquette CH, Brest P, Hofman P. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PloS One. 2013;8:e54596. doi: 10.1371/journal.pone.0054596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Miguel JF, et al. Panobinostat plus bortezomib and dexamethasone versus placebo plus bortezomib and dexamethasone in patients with relapsed or relapsed and refractory multiple myeloma: a multicentre, randomised, double-blind phase 3 trial. Lancet Oncol. 2014;15(11):1195–1206. doi: 10.1016/S1470-2045(14)70440-1. [DOI] [PubMed] [Google Scholar]
- Santel A, Aleku M, Roder N, Mopert K, Durieux B, Janke O, Keil O, Endruschat J, Dames S, Lange C, Eisermann M, Loffler K, Fechtner M, Fisch G, Vank C, Schaeper U, Giese K, Kaufmann J. Atu027 prevents pulmonary metastasis in experimental and spontaneous mouse metastasis models. Clin Cancer Res. 2010;16:5469–5480. doi: 10.1158/1078-0432.CCR-10-1994. [DOI] [PubMed] [Google Scholar]
- Sarkar J, Gou D, Turaka P, Viktorova E, Ramchandran R, Raj JU. MicroRNA-21 plays a role in hypoxia-mediated pulmonary artery smooth muscle cell proliferation and migration. Am J Physiol Lung Cell Mol Physiol. 2010;299:L861–L871. doi: 10.1152/ajplung.00201.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Liu X, Nelson A, Nakanishi M, Kanaji N, Wang X, Kim M, Li Y, Sun J, Michalski J, Patil A, Basma H, Holz O, Magnussen H, Rennard SI. Reduced miR-146a increases prostaglandin E(2)in chronic obstructive pulmonary disease fibroblasts. Am J Respir Crit Care Med. 2010;182:1020–1029. doi: 10.1164/rccm.201001-0055OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel A, Largeau C, Bigey P, Bessodes M, Lebozec K, Scherman D, Escriou V. Anionic polymers for decreased toxicity and enhanced in vivo delivery of siRNA complexed with cationic liposomes. J Control Release. 2011;152:393–401. doi: 10.1016/j.jconrel.2011.03.031. [DOI] [PubMed] [Google Scholar]
- Schlegel A, et al. Reduced in vitro and in vivo toxicity of siRNA-lipoplexes with addition of polyglutamate. J Control Release. 2013;165(1):1–8. doi: 10.1016/j.jconrel.2012.10.018. [DOI] [PubMed] [Google Scholar]
- Schlosser K, et al. miR-26a linked to pulmonary hypertension by global assessment of circulating extracellular microRNAs. Am J Respir Crit Care Med. 2013;188(12):1472–1475. doi: 10.1164/rccm.201308-1403LE. [DOI] [PubMed] [Google Scholar]
- Schmidt LH, Spieker T, Koschmieder S, Schaffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, Hillejan L, Wiebe K, Berdel WE, Wiewrodt R, Muller-Tidow C. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J. Thorac. Oncol. 2011;6:1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- Seddiki N, Phetsouphanh C, Swaminathan S, Xu Y, Rao S, Li J, Sutcliffe EL, Denyer G, Finlayson R, Gelgor L, Cooper DA, Zaunders J, Kelleher AD. The microRNA-9/B-lymphocyte-induced maturation protein-1/IL-2 axis is differentially regulated in progressive HIV infection. Eur J Immunol. 2013;43:510–520. doi: 10.1002/eji.201242695. [DOI] [PubMed] [Google Scholar]
- Shang Y, Das S, Rabold R, Sham JSK, Mitzner W, Tang W-y. Epigenetic alterations by DNA methylation in house dust mite-induced airway hyperresponsiveness. American Journal of Respiratory Cell and Molecular Biology. 2013;49:279–287. doi: 10.1165/rcmb.2012-0403OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Kumar M, Ahmad T, Mabalirajan U, Aich J, Agrawal A, Ghosh B. Antagonism of mmu-mir-106a attenuates asthma features in allergic murine model. J Appl Physiol (1985) 2012;113:459–464. doi: 10.1152/japplphysiol.00001.2012. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward M, Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. [PubMed] [Google Scholar]
- Shi S, Han L, Gong T, Zhang Z, Sun X. Systemic delivery of microRNA-34a for cancer stem cell therapy. Angew Chem Int Ed Engl. 2013;52:3901–3905. doi: 10.1002/anie.201208077. [DOI] [PubMed] [Google Scholar]
- Shih IL, Van YT, Shen MH. Biomedical applications of chemically and microbiologically synthesized poly(glutamic acid) and poly(lysine) Mini Rev Med Chem. 2004;4:179–188. doi: 10.2174/1389557043487420. [DOI] [PubMed] [Google Scholar]
- Shuichiro M, Yasuhiro G, Sotaro S, Yoshitaka S, Daisuke K, Tdataka S, Hisato H, Toshio I, Ikuko T, Eriko T, Kaori S, Shu H. C32. ASTHMA: PRE-CLINICAL STUDIES. American Thoracic Society; 2014. Exosomal MicroRNAs In The Serum Are Potential Real-Time Biomarkers For Allergic Inflammation In The Airway Of Mice; pp. A4211–A4211. [Google Scholar]
- Slingerland M, Guchelaar H-J, Gelderblom H. Histone deacetylase inhibitors: an overview of the clinical studies in solid tumors. Anti-Cancer Drugs. 2014;25:140–149. doi: 10.1097/CAD.0000000000000040. [DOI] [PubMed] [Google Scholar]
- Snead NM, Escamilla-Powers JR, Rossi JJ, McCaffrey AP. 5? Unlocked Nucleic Acid Modification Improves siRNA Targeting. Mol Ther Nucleic Acids. 2013;2:e103-. doi: 10.1038/mtna.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda S, Ohyashiki JH, Ohtsuki K, Umezu T, Setoguchi Y, Ohyashiki K. Clinical relevance of plasma miR-106b levels in patients with chronic obstructive pulmonary disease. Int J Mol Med. 2013;31:533–539. doi: 10.3892/ijmm.2013.1251. [DOI] [PubMed] [Google Scholar]
- Solberg OD, Ostrin EJ, Love MI, Peng JC, Bhakta NR, Hou L, Nguyen C, Solon M, Nguyen C, Barczak AJ, Zlock LT, Blagev DP, Finkbeiner WE, nsel KM, Arron JR, Erle DJ, Woodruff PG. Airway epithelial miRNA expression is altered in asthma. Am J Respir Crit Care Med. 2012;186:965–974. doi: 10.1164/rccm.201201-0027OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Ma H, Yao C, Tao X, Gan H. Alveolar macrophage-derived vascular endothelial growth factor contributes to allergic airway inflammation in a mouse asthma model. Scand J Immunol. 2012;75:599–605. doi: 10.1111/j.1365-3083.2012.02693.x. [DOI] [PubMed] [Google Scholar]
- Sparks J, Slobodkin G, Matar M, Congo R, Ulkoski D, Rea-Ramsey A, Pence C, Rice J, McClure D, Polach KJ, Brunhoeber E, Wilkinson L, Wallace K, Anwer K, Fewell JG. Versatile cationic lipids for siRNA delivery. J Control Release. 2012;158:269–276. doi: 10.1016/j.jconrel.2011.11.006. [DOI] [PubMed] [Google Scholar]
- Stefanowicz D, Hackett T-L, Garmaroudi FS, Günther OP, Neumann S, Sutanto EN, Ling K-M, Kobor MS, Kicic A, Stick SM, Paré PD, Knight DA. DNA methylation profiles of airway epithelial cells and PBMCs from healthy, atopic and asthmatic children. PloS One. 2012;7 doi: 10.1371/journal.pone.0044213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan SD, Rasouliyan L, Russo PA, Kamath T, Chipps BE. Extent, patterns, and burden of uncontrolled disease in severe or difficult-to-treat asthma. Allergy. 2007;62:126–133. doi: 10.1111/j.1398-9995.2006.01254.x. [DOI] [PubMed] [Google Scholar]
- Suojalehto H, Lindström I, Majuri M-L, Mitts C, Karjalainen J, Wolff H, Alenius H. Altered microRNA expression of nasal mucosa in long-term asthma and allergic rhinitis. International Archives of Allergy and Immunology. 2014;163:168–178. doi: 10.1159/000358486. [DOI] [PubMed] [Google Scholar]
- Szafranska AE, Davison TS, Shingara J, Doleshal M, Riggenbach JA, Morrison CD, Jewell S, Labourier E. Accurate molecular characterization of formalin-fixed, paraffin-embedded tissues by microRNA expression profiling. J Mol Diagn. 2008;10:415–423. doi: 10.2353/jmoldx.2008.080018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafranski P, Dharmadhikari AV, Brosens E, Gurha P, Kolodziejska KE, Zhishuo O, Dittwald P, Majewski T, Mohan KN, Chen B, Person RE, Tibboel D, de KA, Pinner J, Chopra M, Malcolm G, Peters G, Arbuckle S, Guiang SF, III, Hustead VA, Jessurun J, Hirsch R, Witte DP, Maystadt I, Sebire N, Fisher R, Langston C, Sen P, Stankiewicz P. Small noncoding differentially methylated copy-number variants, including lncRNA genes, cause a lethal lung developmental disorder. Genome Res. 2013;23:23–33. doi: 10.1101/gr.141887.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulakowski P, Crowther AJL, Jiménez LA, Donaldson K, Mayer R, Leonard TB, MacNee W, Drost EM. The effect of smoking on the transcriptional regulation of lung inflammation in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;174:41–50. doi: 10.1164/rccm.200505-725OC. [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang K-J, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z, Randall G, Fan J, Camoretti-Mercado B, Brockman-Schneider R, Pan L, Solway J, Gern JE, Lemanske RF, Nicolae D, Ober C. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet. 2007;81:829–834. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Peng R, Phillips JE, Deguzman J, Ren Y, Apparsundaram S, Luo Q, Bauer CM, Fuentes ME, DeMartino JA, Tyagi G, Garrido R, Hogaboam CM, Denton CP, Holmes AM, Kitson C, Stevenson CS, Budd DC. Assessment of Brd4 inhibition in idiopathic pulmonary fibrosis lung fibroblasts and in vivo models of lung fibrosis. The American Journal of Pathology. 2013;183:470–479. doi: 10.1016/j.ajpath.2013.04.020. [DOI] [PubMed] [Google Scholar]
- Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, Ijiri K, Akimitsu N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584:4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- Tao H, Shi KH, Yang JJ, Huang C, Zhan HY, Li J. Histone deacetylases in cardiac fibrosis: current perspectives for therapy. Cell Signal. 2014;26:521–527. doi: 10.1016/j.cellsig.2013.11.037. [DOI] [PubMed] [Google Scholar]
- Thakker DR, Natt F, Husken D, Maier R, Muller M, van der Putten H, Hoyer D, Cryan JF. Neurochemical and behavioral consequences of widespread gene knockdown in the adult mouse brain by using nonviral RNA interference. Proc Natl Acad Sci U S A. 2004;101:17270–17275. doi: 10.1073/pnas.0406214101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuault S, Valcourt U, Petersen M, Manfioletti G, Heldin C-H, Moustakas A. Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. The Journal of Cell Biology. 2006;174:175–183. doi: 10.1083/jcb.200512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To Y, Ito K, Kizawa Y, Failla M, Ito M, Kusama T, Elliott WM, Hogg JC, Adcock IM, Barnes PJ. Targeting phosphoinositide-3-kinase-delta with theophylline reverses corticosteroid insensitivity in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;182:897–904. doi: 10.1164/rccm.200906-0937OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolentino MJ, Brucker AJ, Fosnot J, Ying GS, Wu IH, Malik G, Wan S, Reich SJ. Intravitreal injection of vascular endothelial growth factor small interfering RNA inhibits growth and leakage in a nonhuman primate, laser-induced model of choroidal neovascularization. Retina. 2004;24:132–138. doi: 10.1097/00006982-200402000-00018. [DOI] [PubMed] [Google Scholar]
- Trang P, Medina PP, Wiggins JF, Ruffino L, Kelnar K, Omotola M, Homer R, Brown D, Bader AG, Weidhaas JB, Slack FJ. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29:1580–1587. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trang P, Wiggins JF, Daige CL, Cho C, Omotola M, Brown D, Weidhaas JB, Bader AG, Slack FJ. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19:1116–1122. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsiou E, Williams AE, Moschos SA, Patel K, Rossios C, Jiang X, Adams OD, Macedo P, Booton R, Gibeon D, Chung KF, Lindsay MA. Transcriptome analysis shows activation of circulating CD8+ T cells in patients with severe asthma. J Allergy Clin Immunol. 2012;129:95–103. doi: 10.1016/j.jaci.2011.08.011. [DOI] [PubMed] [Google Scholar]
- Tsumagari K, Baribault C, Terragni J, Chandra S, Renshaw C, Sun Z, Song L, Crawford GE, Pradhan S, Lacey M, Ehrlich M. DNA methylation and differentiation: HOX genes in muscle cells. Epigenetics. Chromatin. 2013;6:25. doi: 10.1186/1756-8935-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuder RM, Archer SL, Dorfmuller P, Erzurum SC, Guignabert C, Michelakis E, Rabinovitch M, Schermuly R, Stenmark KR, Morrell NW. Relevant issues in the pathology and pathobiology of pulmonary hypertension. J Am Coll. Cardiol. 2013;62:D4–D12. doi: 10.1016/j.jacc.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuder RM, Marecki JC, Richter A, Fijalkowska I, Flores S. Pathology of pulmonary hypertension. Clin. Chest Med. 2007;28:23–42. vii. doi: 10.1016/j.ccm.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaish N, Chen F, Seth S, Fosnaugh K, Liu Y, Adami R, Brown T, Chen Y, Harvie P, Johns R, Severson G, Granger B, Charmley P, Houston M, Templin MV, Polisky B. Improved specificity of gene silencing by siRNAs containing unlocked nucleobase analogs. Nucleic Acids Res. 2011;39:1823–1832. doi: 10.1093/nar/gkq961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Pottelberge GR, Mestdagh P, Bracke KR, Thas O, van Durme YM, Joos GF, Vandesompele J, Brusselle GG. MicroRNA expression in induced sputum of smokers and patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2011;183:898–906. doi: 10.1164/rccm.201002-0304OC. [DOI] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma M, Chattopadhyay BD, Paul BN. Epigenetic regulation of DNMT1 gene in mouse model of asthma disease. Molecular Biology Reports. 2013;40:2357–2368. doi: 10.1007/s11033-012-2317-1. [DOI] [PubMed] [Google Scholar]
- Vernooy JH, Kucukaycan M, Jacobs JA, Chavannes NH, Buurman WA, Dentener MA, Wouters EF. Local and systemic inflammation in patients with chronic obstructive pulmonary disease: soluble tumor necrosis factor receptors are increased in sputum. Am J Respir Crit Care Med. 2002;166:1218–1224. doi: 10.1164/rccm.2202023. [DOI] [PubMed] [Google Scholar]
- Vucic EA, Chari R, Thu KL, Wilson IM, Cotton AM, Kennett JY, Zhang M, Lonergan KM, Steiling K, Brown CJ, McWilliams A, Ohtani K, Lenburg ME, Sin DD, Spira A, Macaulay CE, Lam S, Lam WL. DNA methylation is globally disrupted and associated with expression changes in chronic obstructive pulmonary disease small airways. American Journal of Respiratory Cell and Molecular Biology. 2014;50:912–922. doi: 10.1165/rcmb.2013-0304OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat. Rev. Drug Discov. 2013;12:433–446. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- Wang FE, Zhang C, Maminishkis A, Dong L, Zhi C, Li R, Zhao J, Majerciak V, Gaur AB, Chen S, Miller SS. MicroRNA-204/211 alters epithelial physiology. FASEB J. 2010;24:1552–1571. doi: 10.1096/fj.08-125856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Inoue S, Gu J, Miyoshi E, Noda K, Li W, Mizuno-Horikawa Y, Nakano M, Asahi M, Takahashi M, Uozumi N, Ihara S, Lee SH, Ikeda Y, Yamaguchi Y, Aze Y, Tomiyama Y, Fujii J, Suzuki K, Kondo A, Shapiro SD, Lopez-Otin C, Kuwaki T, Okabe M, Honke K, Taniguchi N. Dysregulation of TGF-beta1 receptor activation leads to abnormal lung development and emphysema-like phenotype in core fucose-deficient mice. Proc Natl Acad Sci U S A. 2005;102:15791–15796. doi: 10.1073/pnas.0507375102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XM, Zhang Y, Kim HP, Zhou Z, Feghali-Bostwick CA, Liu F, Ifedigbo E, Xu X, Oury TD, Kaminski N, Choi AMK. Caveolin-1: a critical regulator of lung fibrosis in idiopathic pulmonary fibrosis. The Journal of Experimental Medicine. 2006;203:2895–2906. doi: 10.1084/jem.20061536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Henderson H, Spradley C, Li L, Kim IK, Kumar S, Hong N, Arroliga AC, Gupta S. Circulating miRNAs as potential marker for pulmonary hypertension. PloS One. 2013;8:e64396. doi: 10.1371/journal.pone.0064396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel S. Severe asthma in adults. Am J Respir Crit Care Med. 2005;172:149–160. doi: 10.1164/rccm.200409-1181PP. [DOI] [PubMed] [Google Scholar]
- White AC, Xu J, Yin Y, Smith C, Schmid G, Ornitz DM. FGF9 and SHH signaling coordinate lung growth and development through regulation of distinct mesenchymal domains. Development. 2006;133:1507–1517. doi: 10.1242/dev.02313. [DOI] [PubMed] [Google Scholar]
- White K, Loscalzo J, Chan SY. Holding our breath: The emerging and anticipated roles of microRNA in pulmonary hypertension. Pulm. Circ. 2012;2:278–290. doi: 10.4103/2045-8932.101395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, Bader AG. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70:5923–5930. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Williams AE, Moschos SA, Perry MM, Barnes PJ, Lindsay MA. Maternally imprinted microRNAs are differentially expressed during mouse and human lung development. Dev Dyn. 2007;236:572–580. doi: 10.1002/dvdy.21047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-J, Yang C-Y, Chen Y-H, Chen C-M, Chen L-C, Kuo M-L. The DNA methylation inhibitor 5-azacytidine increases regulatory T cells and alleviates airway inflammation in ovalbumin-sensitized mice. International Archives of Allergy and Immunology. 2013;160:356–364. doi: 10.1159/000343030. [DOI] [PubMed] [Google Scholar]
- Wu Y, Crawford M, Mao Y, Lee RJ, Davis IC, Elton TS, Lee LJ, Nana-Sinkam SP. Therapeutic Delivery of MicroRNA-29b by Cationic Lipoplexes for Lung Cancer. Mol Ther Nucleic Acids. 2013;2:e84. doi: 10.1038/mtna.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Crawford M, Yu B, Mao Y, Nana-Sinkam SP, Lee LJ. MicroRNA delivery by cationic lipoplexes for lung cancer therapy. Mol Pharm. 2011;8:1381–1389. doi: 10.1021/mp2002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Meng XM, Huang XR, Chung AC, Feng YL, Hui DS, Yu CM, Sung JJ, Lan HY. miR-29 inhibits bleomycin-induced pulmonary fibrosis in mice. Mol Ther. 2012;20:1251–1260. doi: 10.1038/mt.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Liang J, Guo R, Liu N, Noble PW, Jiang D. Comprehensive microRNA analysis in bleomycin-induced pulmonary fibrosis identifies multiple sites of molecular regulation. Physiol Genomics. 2011;43:479–487. doi: 10.1152/physiolgenomics.00222.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Huang Y, Chen D, He J, Zhu W, Zhang Y, Liu X. Downregulation of miR-21 increases cisplatin sensitivity of non-small-cell lung cancer. Cancer Genet. 2014 doi: 10.1016/j.cancergen.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Xu XF, Ma XL, Shen Z, Wu XL, Cheng F, Du LZ. Epigenetic regulation of the endothelial nitric oxide synthase gene in persistent pulmonary hypertension of the newborn rat. J Hypertens. 2010;28:2227–2235. doi: 10.1097/HJH.0b013e32833e08f1. [DOI] [PubMed] [Google Scholar]
- Yang S-R, Chida AS, Bauter MR, Shafiq N, Seweryniak K, Maggirwar SB, Kilty I, Rahman I. Cigarette smoke induces proinflammatory cytokine release by activation of NF-kappaB and posttranslational modifications of histone deacetylase in macrophages. American Journal of Physiology. Lung Cellular and Molecular Physiology. 2006;291:L46–L57. doi: 10.1152/ajplung.00241.2005. [DOI] [PubMed] [Google Scholar]
- Yang S, Banerjee S, de Freitas A, Sanders YY, Ding Q, Matalon S, Thannickal VJ, Abraham E, Liu G. Participation of miR-200 in pulmonary fibrosis. The American Journal of Pathology. 2012;180:484–493. doi: 10.1016/j.ajpath.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Banerjee S, Freitas A, Cui H, Xie N, Abraham E, Liu G. miR-21 regulates chronic hypoxia-induced pulmonary vascular remodeling. Am J Physiol Lung Cell Mol Physiol. 2012;302:L521–L529. doi: 10.1152/ajplung.00316.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Cui H, Xie N, Icyuz M, Banerjee S, Antony VB, Abraham E, Thannickal VJ, Liu G. miR-145 regulates myofibroblast differentiation and lung fibrosis. FASEB J. 2013a;27:2382–2391. doi: 10.1096/fj.12-219493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Liang Y, Lin Q, Liu J, Luo F, Li X, Zhou H, Zhuang S, Zhang H. miR-29 mediates TGFbeta1-induced extracellular matrix synthesis through activation of PI3K-AKT pathway in human lung fibroblasts. J Cell Biochem. 2013b;114:1336–1342. doi: 10.1002/jcb.24474. [DOI] [PubMed] [Google Scholar]
- Yao H, Chung S, Hwang J-w, Rajendrasozhan S, Sundar IK, Dean DA, McBurney MW, Guarente L, Gu W, Rönty M, Kinnula VL, Rahman I. SIRT1 protects against emphysema via FOXO3-mediated reduction of premature senescence in mice. The Journal of Clinical Investigation. 2012;122:2032–2045. doi: 10.1172/JCI60132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Hwang J-w, Sundar IK, Friedman AE, McBurney MW, Guarente L, Gu W, Kinnula VL, Rahman I. SIRT1 redresses the imbalance of tissue inhibitor of matrix metalloproteinase-1 and matrix metalloproteinase-9 in the development of mouse emphysema and human COPD. American Journal of Physiology. Lung Cellular and Molecular Physiology. 2013;305:L615–L624. doi: 10.1152/ajplung.00249.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuo M, Mizuno S, Kraskauskas D, Bogaard HJ, Natarajan R, Cool CD, Zamora M, Voelkel NF. Hypoxia inducible factor-1α in human emphysema lung tissue. The European Respiratory Journal. 2011;37:775–783. doi: 10.1183/09031936.00022910. [DOI] [PubMed] [Google Scholar]
- Yi ES, Kim H, Ahn H, Strother J, Morris T, Masliah E, Hansen LA, Park K, Friedman PJ. Distribution of obstructive intimal lesions and their cellular phenotypes in chronic pulmonary hypertension. A morphometric and immunohistochemical study. Am. J. Respir. Crit Care Med. 2000;162:1577–1586. doi: 10.1164/ajrccm.162.4.9912131. [DOI] [PubMed] [Google Scholar]
- Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014 doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- Yu Q, Zhou B, Zhang Y, Nguyen ET, Du J, Glosson NL, Kaplan MH. DNA methyltransferase 3a limits the expression of interleukin-13 in T helper 2 cells and allergic airway inflammation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:541–546. doi: 10.1073/pnas.1103803109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J, Guan J, Wang X, Zhang L, Yang Z, Ao Q, Deng Y, Zhu P, Wang G. MicroRNA-206 is involved in hypoxia-induced pulmonary hypertension through targeting of the HIF-1alpha/Fhl-1 pathway. Lab Invest. 2013;93:748–759. doi: 10.1038/labinvest.2013.63. [DOI] [PubMed] [Google Scholar]
- Zhang M, Shi J, Huang Y, Lai L. Expression of canonical WNT/beta-CATENIN signaling components in the developing human lung. BMC Dev Biol. 2012;12:21. doi: 10.1186/1471-213X-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Prakash C, Sum C, Gong Y, Li Y, Kwok JJT, Thiessen N, Pettersson S, Jones SJM, Knapp S, Yang H, Chin K-C. Bromodomain-containing protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. J Biol Chem. 2012b;287:43137–43155. doi: 10.1074/jbc.M112.413047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Liu H, Hock T, Thannickal VJ, Sanders YY. Histone deacetylase inhibition downregulates collagen 3A1 in fibrotic lung fibroblasts. International Journal of Molecular Sciences. 2013;14:19605–19617. doi: 10.3390/ijms141019605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shan P, Jiang D, Noble PW, Abraham NG, Kappas A, Lee PJ. Small interfering RNA targeting heme oxygenase-1 enhances ischemia-reperfusion-induced lung apoptosis. J Biol Chem. 2004;279:10677–10684. doi: 10.1074/jbc.M312941200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang Z, Gemeinhart RA. Progress in microRNA delivery. J Control Release. 2013;172:962–974. doi: 10.1016/j.jconrel.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong X, Tripathi V, Prasanth KV. RNA splicing control: yet another gene regulatory role for long nuclear noncoding RNAs. RNA. Biol. 2011;8:968–977. doi: 10.4161/rna.8.6.17606. [DOI] [PMC free article] [PubMed] [Google Scholar]