Abstract
Polyomaviruses (PyVs) are associated with tumors including Merkel cell carcinoma (MCC). Several PyVs encode microRNAs (miRNAs) but to date no abundant PyV miRNAs have been reported in tumors. To better understand the function of the Merkel cell PyV (MCPyV) miRNA, we examined phylogenetically-related viruses for miRNA expression. We show that two primate PyVs and the more distantly-related raccoon PyV (RacPyV) encode miRNAs that share genomic position and partial sequence identity with MCPyV miRNAs. Unlike MCPyV miRNA in MCC, RacPyV miRNA is highly abundant in raccoon tumors. RacPyV miRNA negatively regulates reporters of early viral (T antigen) transcripts, yet robust viral miRNA expression is tolerated in tumors. We also identify raccoon miRNAs expressed in RacPyV-associated neuroglial brain tumors, including several likely oncogenic miRNAs (oncomiRs). This work describes the first PyV miRNA abundantly expressed in tumors and is consistent with a possible role for both host and viral miRNAs in RacPyV-associated tumors.
Keywords: Polyomavirus, microRNA, miRNA, Merkel cell carcinoma, MCV, Raccoon Polyomavirus
INTRODUCTION
PyVs are small DNA viruses with ~5 kilobase genomes that likely infect most vertebrate species (1). There are at least 13 different human PyVs known and numerous PyVs have been identified that infect other animals (2-5). In a majority of cases, infection with these viruses is asymptomatic. However, in rare instances, typically involving immune suppression, infection with some PyVs can result in serious disease (6, 7). Under laboratory conditions, infection with PyVs can lead to a high incidence of tumors (8). In addition, two PyVs, McPyV and RacPyV, have been strongly associated with tumors arising in natural settings. In humans, MCPyV is associated with at least 80% of Merkel cell carcinomas (MCCs) through a mechanism that typically involves integration of the viral genome into tumor cells (3, 9-11). Integration of the MCPyV genome leads to expression of Tumor antigen (TAg) proteins derived from the early viral genomic region in the absence of late gene expression, consistent with the known transforming activities of the laboratory model of PyV TAgs (12-14). The fact that RacPyV-associated tumors harbor predominantly episomal viral genomes (3, 10) suggests that the modes of RacPyV and MCPyV-associated tumorigenesis are distinct.
We have previously shown that diverse PyVs encode miRNAs (15-20). miRNAs are small regulatory RNAs that are typically processed through a conserved set of machinery, including the Drosha and Dicer nucleases (21). Once generated, miRNAs enter the RNA induced silencing complex (RISC) where they bind to specific mRNAs and direct translation inhibition and/or transcript turnover. Although PyV miRNAs can vary in sequence and genomic position, all share the ability to negatively regulate early viral gene expression (22-24). The conserved nature of this activity implies importance, yet the functional role of the PyV miRNAs during natural infection and in PyV-associated disease remains obscure.
To better understand the function of the MCPyV miRNAs, we sought to apply a comparative evolutionary approach. Here we uncover that primate PyVs closely related to MCPyV, and a more distantly related RacPyV, each encode miRNAs of comparable genomic location. Each of these miRNAs possesses the ability to negatively regulate complementary regions of the early viral transcripts. These results are consistent with miRNA-mediated regulation of early gene expression being an important activity shared between all known PyV miRNAs. We demonstrate that the RacPyV miRNA is among the most abundant miRNAs detectable in RacPyV-associated tumors. This is in sharp contrast to the MCPyV-associated tumors where our meta-analysis of published reports shows that the viral miRNA is of extremely low abundance and unlikely to be bioactive. Therefore, despite the likely ability of RacPyV miRNAs to downregulate early gene expression levels, RacPyV tumors (known to express early T Antigen transcripts) readily tolerate high levels of the viral miRNA. We also identify for the first time numerous raccoon miRNAs, including several that are known oncogenic miRNAs (oncomiRs). Combined with previous work (3, 10), these observations further support a different mode of tumorigenesis for the MCPyV and RacPyV-associated tumors, and are consistent with a possible role of viral miRNAs in RacPyV-associated tumors.
MATERIALS AND METHODS
Cell culture, tissue samples and RNA isolation
Human embryonic kidney 293T cells (293T) were obtained from the American Type Culture Collection (Manassa, VA), wild type 293T and Dicer-deficient 293T cells were kind gifts from Bryan Cullen (Duke University). All cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Life Technologies, Carlsbad, CA), 100IU/mL penicillin, and 100μg/mL streptomycin (Corning Cellgro, Manassas, VA). Total RNA was harvested using an in-house PIG-B solution as previously described (15, 25-27). Animals were submitted for routine necropsy through the Veterinary Medical Teaching Hospital at UC Davis. Cells obtained from Rac14 tumor tissue was cultured as previously described (10). Briefly, cells were maintained in Neurobasal-A medium supplemented with N2 and B27 supplements (Life Technologies) and 25ng/mL of rhEGF and bFGF (R&D Systems Inc, Minneapolis, MN). Raccoon brain tissue samples were homogenized by placing the sample into Lysing Matrix A tubes (MP Biomedicals, Solon, OH) along with one milliliter of TRIzol RNA isolation reagents (Life Technologies) and subjected to shaking in a Mini-Beadbeater-24 (Bartlesville, OK) at the fastest setting for one minute followed by cooling on ice for one minute. The process of homogenization and cooling was repeated three times. The lysate was then centrifuged at 1000G for one minute to pellet. The supernatant was then transferred into four milliliters of TRIzol and subjected to RNA isolation according to the manufacturer's protocol.
Computational prediction and selection of viral pre-miRNA candidates
The complete genome sequences for Pan troglodytes verus polyomavirus 2a isolate 6512 (PtvPyV2a, accession number: HQ385748.1), Gorilla gorilla gorilla polyomavirus 1 isolate 5766 (GggPyVl, accession number: HQ385752.1) and Raccoon polyomavirus strain R45 (RacPyV, accession number: JQ178241.1) were subjected to miRNA prediction using VMir (18, 28, 29). A minimum cutoff score of 150 was applied to the pre-miRNA prediction for PtvPyV2a, GggPyV1 and RacPyV. Candidate pre-miRNAs were selected for verification if they met the following two criteria: 1) found in the late orientation and 2) the predicted genomic location of the pre-miRNA was either positioned similar to SV40-like (18) or muPyV-like (19) pre-miRNAs.. The pre-miRNA candidate for each PyV was predicted using the mfold RNA folding prediction web server (30).
Construction of PyV phylogenetic tree
The phylogenetic tree was constructed based on the complete genome sequences from the following PyVs: PtvPyV2a, GggPyV1, RacPyV, SV40 (Accession number: J02400.1), JCV (Accession number: NC_001699.1), BKV (Accession number: NC_001538.1), Murine Polyomavirus (Accession number: NC_001515.1), SA12 (Accession number: AY614708.1), Merkel Cell Carcinoma virus, 350 (Accession number: EU375803.1). The maximum likelihood phylogenetic tree was generated as previously described (31). Briefly, the phylogenetic tree was constructed using the PhyML version 3.0 aLRT program from the web-based Phylogeny.fr software, under the HKY85 substitution model. The SH-like approximate likelihood-ratio test (aLRT) was applied to test for branch support (32-37). The resulting phylogenetic tree was viewed using the FigTree software version 1.4.2 (tree.bio.ed.ac.uk/software/figtree/).
MiRNA expression vector construction, transfection, and Northern blot analysis
All DNA vector constructs were sequence verified through sequence analysis at the Institute of Cellular and Molecular Biology Sequencing Facility at the University of Texas at Austin. The primers used in the construction of the pre-miRNA candidates expression vectors have been listed in Table S1. Briefly, the primers are annealed and filled-in using Phusion High-Fidelity DNA polymerase (New England BioLabs, Ipswich, MA) according to the manufacturer's protocol. The PCR products were then cloned into the pcDNA3.1neo expression vector using the restriction sites listed in Table S1. 293T cells were plated in 6-wells plates and transfected with expression vector using the Lipofectamine 2000 transfection reagent (Life Technologies) according to the manufacturer's instruction. As a negative control, cells were transfected with empty pcDNA3.1neo vector. Total RNA was harvested at 48 hours post transfection. The total RNA was subjected Northern blot analysis as described previously (18). Briefly, 10 micrograms of total RNA was separated on a Tris-borate-EDTA-urea-15% denaturing polyacrylamide gel. The RNA was transferred onto a Hybond N+ membrane (GE Healthcare, Pittsburgh, PA). The probe sequences used are listed in Table S2.
Drosha dependence assay
293T cells were seeded in 6-wells plates. Cells were transfected with either the Silencer Select negative control siRNA (scrambled) or a pool of 10 siRNAs against Drosha using the RNAiMAX transfection reagent (Life Technologies) according to the manufacturer's instructions. At 12 hours post siRNA transfection, the miRNA expression vectors were co-transfected with Drosha siRNA or negative control siRNA using the Lipofectamine 2000 transfection reagent (Life Technologies) according to the manufacturer's instructions. Transfection of the SV40 miRNA expression vector was included as a positive control. Five hundred ng of pIDT-MHV68 miR-M1-7, a plasmid that expresses the Murid herpesvirus 68 (MHV68) M1-7 miRNA, was included in every transfection to serve as an internal control. Total RNA was harvested at 48 hours post transfection and subjected to Northern blot analysis as described above. For these assays, probes that recognized the corresponding 5p derivative miRNA of each virus were utilized. For the internal control MHV68 M1-7 miRNAs, the 3p derivative miRNA was probed for. The probe sequences used are listed in Table S2.
Dicer dependence assay
Wild type 293T cells or the Dicer-deficient 293T cells (38, 39) were seeded in 6-well plates and were transfected at approximately 50% confluency with the polyomaviral miRNA expression vectors, using the TurboFect transfection reagent (Thermo Scientific). Total RNA was harvested at 48 hours post transfection using the PIG-B solution (27) followed by Northern blot analysis as described above. The SV40 miRNA expression vector and an empty pcDNA3.1neo vector were used as the positive and negative controls, respectively. The northern blot probes used are listed in Table S2.
Luciferase assays
The miRNA reporter plasmids were constructed by cloning the genomic region corresponding to the pre-miRNA of each PyV in the early (T antigen) orientation into the 3’ UTR of dsRLuc (primers listed in Table S1). To construct the RacPyV late region reporter plasmids, the RacPyV genomic region that encodes the late mRNA was divided into four overlapping fragments, each 500bp in length, as listed in Table S1. The four fragments were synthesized as gBlock gene fragments (Integrated DNA Technologies). PCR products were generated using Phusion High-Fidelity DNA polymerase (New England BioLabs) and cloned into the pcDNA3.1dsRluc vector, which expresses a destabilized version of the Renilla luciferase (using the restriction sites listed in Table S1). 293T cells were plated in 24-wells plates and transfected using the TurboFect transfection reagent. The SV40 miRNA expression vector (17) and the empty Renilla luciferase reporter construct were included as negative controls. The pcDNA3.1luc2CP vector, which expresses a destabilized version of the firefly luciferase, was included in the transfection reactions to normalize for transfection efficiency. Transfected cells were collected at 24 hours post transfection and analyzed with the Dual-luciferase reporter assay system (Promega, Fitchburg, WI) according to the manufacturer's instruction. The luciferase readings were collected using a Luminoskan Ascent microplate luminometer (Thermo Scientific). The readings from the Renilla luciferase were normalized to the readings from the firefly luciferase, with the ratios plotted as a bar graph relative to the empty Renilla luciferase vector control.
Small RNA library and computational analysis of sequencing reads
293T cells were seeded in 10cm tissue culture dishes and transfected with the PtvPyV2a, GggPyV1 or the RacPyV miRNA expression vector using the Lipofectamine 2000 transfection reagent (Life Technologies). Total RNA was harvested at 48 hours post transfection. Two raccoon brain tumor tissue samples (Rac 12 and Rac 14) were homogenized and total RNA was harvested as described above. Small RNAs from the transfected 293T cells and the brain tumor tissue samples were gel fractionated as previously described (17, 26). The small RNA cDNA library was generated using the NEBNext Multiplex Small RNA library prep set for Illumina kit (New England BioLabs) according to the manufacturer's instructions. The resulting cDNA library was then subjected to paired-end sequencing on the Illumina HiSeq (Illumina, San Diego, CA) as previously described (17). Briefly, the PtvPyV2a, GggPyV1 and RacPyV small RNA reads were mapped onto the respective virus genomes. The raccoon miRNAs from the tumor tissue samples were mapped to the mouse mature miRNA sequences build on miRBase (40-44), without allowing for any mismatches. To annotate each raccoon miRNA as an oncogene or a tumor suppressor, each miRNA was referenced to two independent databases, the OncomiRDB (45) and miRCancer (46).
RESULTS
PyVs closely related to MCPyV also encode miRNAs
We have previously demonstrated that multiple members of the PyV family encode miRNAs (15-20). To better understand the functions of the MCPyV miRNAs, we applied a comparative evolutionary approach to search for miRNAs in viruses related to MCPyV. To this end, we turned our attention to three recently discovered, non-human PyVs that are related to MCPyV (3, 4). Pan troglodytes verus polyomavirus 2a (PtvPyV2a) and Gorilla gorilla gorilla polyomavirus 1 (GggPyV1) are two of the closest known relatives of MCPyV, whereas the raccoon polyomavirus (RacPyV) is more distantly related to MCPyV (Fig. 1A). Using the vMir pre-miRNA prediction algorithm (18, 28, 29), we identified pre-miRNA candidates from all three PyVs. In all three viruses, the pre-miRNA candidates were found at similar genomic locations to the known pre-miRNA encoded by MCPyV (data not shown, (16,19)).
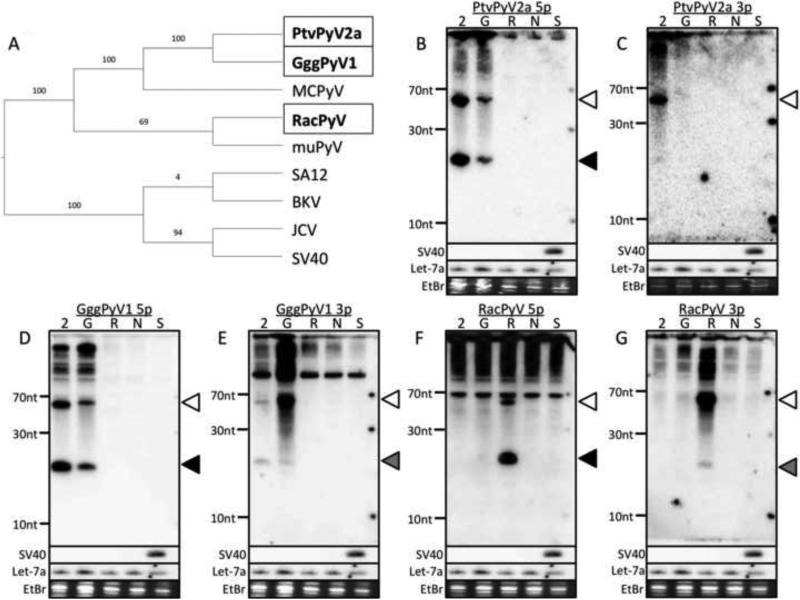
Fig. 1. PtvPyV2a, GggPyV1 and RacPyV encode miRNAs.
(A) Phylogenetic analysis of all PyVs that are known to encode miRNAs. A Phylogenetic tree was constructed based on the complete genome sequence of all known miRNA-expressing PyVs. The phylogeny was determined using a maximum likelihood analysis and represented as a cladogram. The three PyVs studied in this report are boxed and shown in bold. (B - G) Northern blot confirms miRNA expression. The pre-miRNA candidate from each PyV was cloned into a heterologous expression vector and transfected into 293T cells. Total RNA was subjected to Northern blot analysis. Probes designed to recognize the (B) PtvPyV2a 5p, (C) PtvPyV2a 3p, (D) GggPyV1 5p, (F) GggPyV1 3p, (G) RacPyV 5p and (H) RacPyV 3p were used. As controls, cells were transfected with either the empty pcDNA3.1neo vector (lane “N”) or an SV40 miRNA expression vector (lane “S”). As positive controls, the blots were probed for either the SV40 3p miRNA or the cellular miRNA, let-7a. The bands corresponding to the pre-miRNAs (white arrowhead), the 5p derivative miRNAs (black arrowhead) and the 3p derivative miRNAs (grey arrowhead) are indicated. Ethidium bromide-stained low-molecular-weight RNA served as a loading control. The identity of each lane is indicated at the top of each blot as follows: 2 = PtvPyV2a, G = GggPyV1 and R = RacPyV.
To test the validity of these candidate pre-miRNAs, an approximately 140 base pair (bp) portion of each PyV sequence (encompassing the pre-miRNA candidate and flanking genomic regions) was synthesized and cloned into individual expression vectors. We then transfected the expression vectors into 293T cells, harvested total RNA, and conducted Northern blot analysis. The results showed that all three candidates give rise to abundant bands consistent with the size and processing pattern of known miRNAs (Fig. 1B to 1G). Similar to the MCPyV miRNA, the 5p derivative miRNAs (Fig. 1B, 1D and 1F) were detected at a greater abundance than their 3p derivative counterparts (Fig. 1C, 1E and 1G) for PtvPyV2a, GggPyV1 and RacPyV. We note that this Northern blot analysis showed extensive cross-reactivity between the probes for the PtvPyV2a and GggPyV1 candidates (Fig. 1B and 1D), likely due to the high sequence similarity (86% identity) between the pre-miRNA genomic regions of these two viruses. These data are consistent with these viruses encoding miRNAs in the same genomic region as MCPyV.
Next, using Illumina high throughput sequencing, we characterized the small RNAs derived from the PtvPyV2a, GggPyV1, RacPyV, and MCPyV candidate pre-miRNA expression vectors. 293T cells were transfected with individual expression vectors and total RNA was size fractionated to enrich for small RNAs. We generated cDNA libraries from these small RNAs and conducted deep sequencing using the Illumina platform (Fig. 2). The predominant derivative RNAs displayed all the features expected of true miRNAs including: appropriate size (~22 nucleotides), ability to ligate to library linkers (demonstrating correct end structure), and mapped position within the putative hairpin pre-miRNA structure. We conclude that PtvPyV2a, GggPyV1 and RacPyV viruses encode bona fide miRNAs.
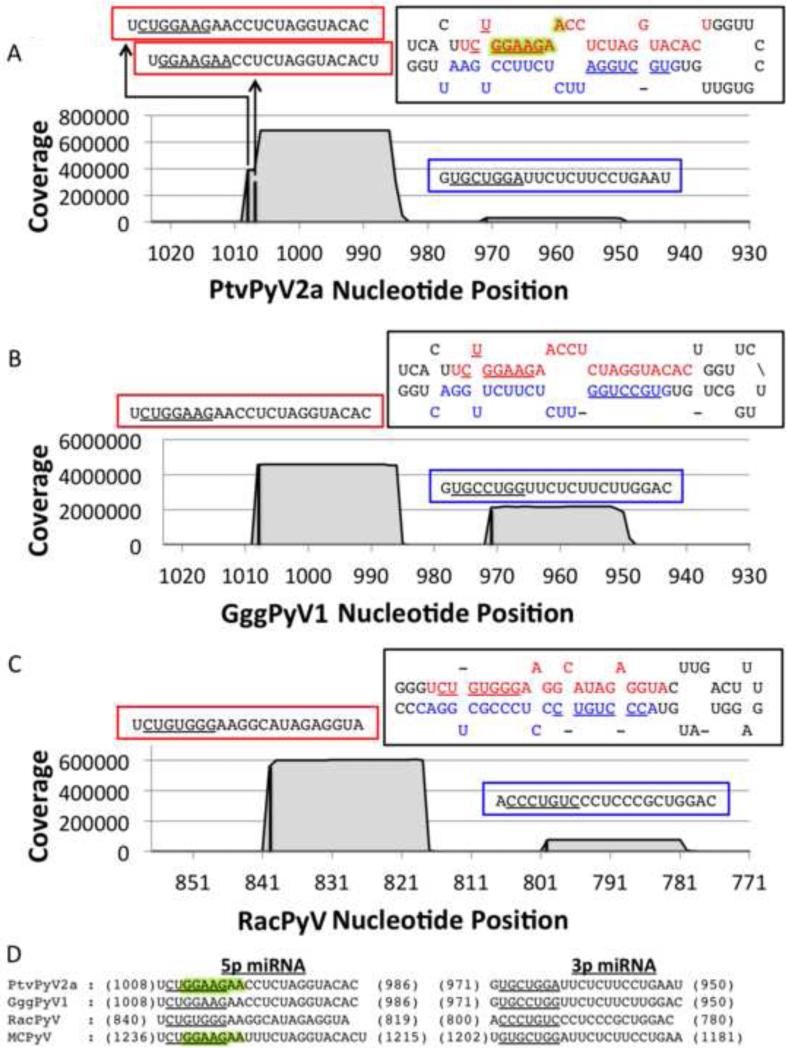
Fig. 2. The PtvPyV2a, GggPyV1 and RacPyV miRNAs are partially conserved.
(A - C) Coverage plot of the small RNA deep sequencing reads from (A) PtvPyV2a, (B) GggPyV1 and (C) RacPyV miRNA expression-vector-transfected 293T cells. The number of reads (y axis) was mapped onto the corresponding PyV genome (x axis). The black bars indicate the start position of each miRNA and the coverage is represented by the grey filled area. All three PyVs produce dominant 5p derivative miRNAs (indicated in the red boxes) and less abundant 3p derivative miRNAs (indicated in the blue boxes). Each miRNA sequence is indicated above its corresponding peak, with the seed sequences (nucleotide positions 2 to 8) underlined. The 5p (red) and 3p (blue) derivative miRNAs are indicated in each hairpin structure, with their seed sequence underlined. PtvPyV2a expresses two different species of the 5p derivative miRNAs. The seed sequences of the major and minor species are underlined and highlighted in green, respectively, in the hairpin structure. (D) The PtvPyV2a, GggPyV1 and RacPyV miRNAs share partial sequence identity with the MCPyV miRNAs. Sequence alignment of the 5p and 3p derivative miRNAs from PtvPyV2a, GggPyV1a and RacPyV and MCPyV miRNA sequences (16, 47). The 5’ and 3’ coordinates of each derivative miRNA are indicated and the seed sequence for each miRNA is underlined. The seed sequences from the major and minor form of the PtvPyV2a 5p derivative miRNAs are underlined and highlighted in green respectively. The seed sequence from the MCPyV 5p derivative miRNA reported by Lee et al. and Seo et al. (16, 47) are underlined and highlighted in green respectively.
This deep sequencing analysis revealed that, consistent with the Northern blot results, the 5p derivative miRNAs for all three pre-miRNA expression vectors (PtvPyV2a, GggPyV1 and RacPyV) are dominant (Fig. 2A, B and C). We also detected 5’ end heterogeneity for most of these 5p miRNAs, which likely explains earlier apparently contradictory reports (16, 47) identifying MCPyV 5p miRNA start sites that differ by 2 nucleotides. These sequences reveal extensive identity between the PtvPyV2a, GggPyV1, and MCPyV miRNAs including shared seed sequences (Fig. 2D). In contrast, the RacPyV miRNA shared only limited sequence identity with the MCPyV miRNA. We conclude that the miRNAs from PtvPyV2a, GggPyV1 and MCPyV, and possibly RacPyV, are orthologous.
The PtvPyV2a, GggPyVl and RacPyV miRNAs derive from canonical mechanisms
Although most host miRNAs are produced from canonical Drosha/Dicer-dependent processing, there are several viral miRNAs that arise via non-conventional mechanisms (48-55). To determine if canonical miRNA biogenesis machinery contributes to the abundance of the PtvPyV2a, GggPyV1 and RacPyV miRNAs, we transfected 293T cells with an siRNA to knockdown Drosha and then individually transfected cells with expression vectors for either the PtvPyV2a, GggPyV1 or RacPyV miRNAs. We then performed northern blot analysis on total RNA harvested from these cells. This showed that knockdown of Drosha led to a substantial decrease in the pre-miRNA signals for all the miRNAs we tested (Fig. 3A). As expected, these results mirrored the results for cells transfected with our positive control SV40 miRNA expression vector, but not the negative control MHV68 miR-M1-7 whose biogenesis is independent of Drosha processing (55, 56). Thus, the PtvPyV2a, GggPyV1 and RacPyV genomes encode miRNAs whose biogenesis is dependent on Drosha processing.
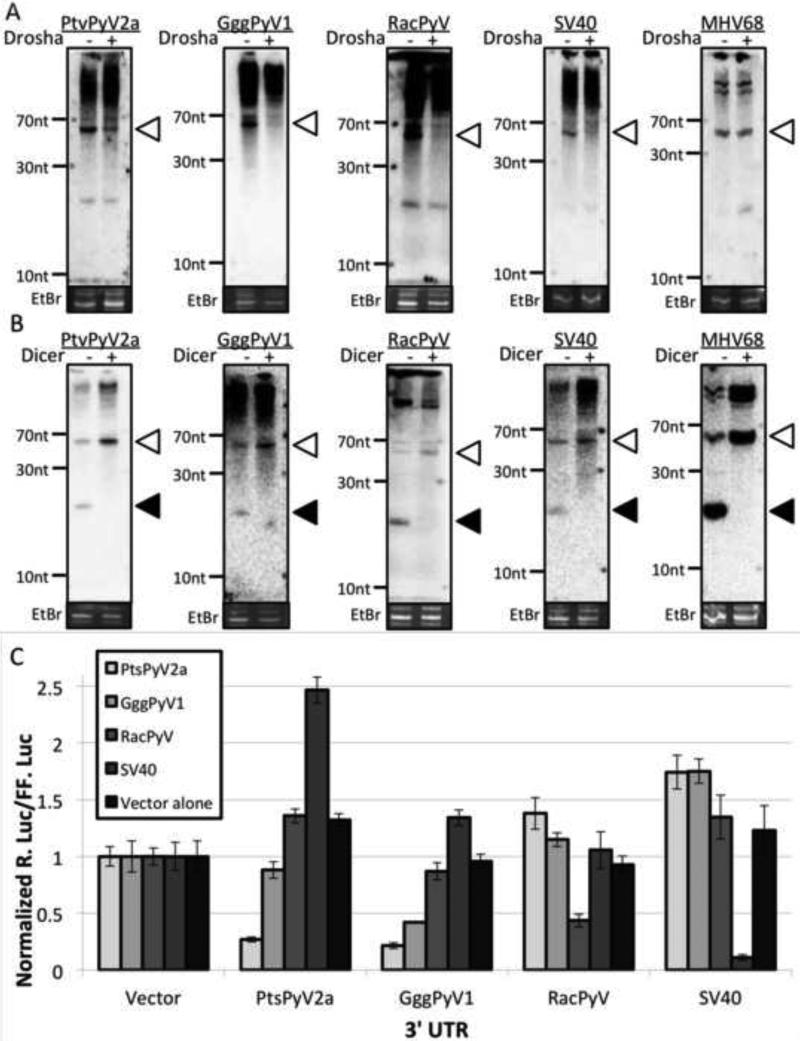
Fig. 3. The PtvPyV2a, GggPyV1 and RacPyV miRNAs can negatively regulate early transcripts.
(A) The biogenesis of PtvPyV2a, GggPyV1 and RacPyV miRNAs are Drosha dependent. 293T cells were transfected with an siRNA against Drosha or an irrelevant siRNA followed by co-transfection of the miRNA expression vectors with the siRNAs. Northern blot analysis was performed on the total RNA at 48 hours post transfection. The SV40 miRNA expression vector and the MHV68 miR-M1-7 expression vector were transfected as positive and negative control, respectively. The bands corresponding to the pre-miRNAs (white arrowheads) are indicated. Ethidium bromide-stained low-molecular-weight RNA served as a loading control. Irrelevant siRNA transfections are indicated by the “−” lanes and the Drosha siRNA transfections are indicated by the “+” lanes. (B) The biogenesis of PtvPyV2a, GggPyV1 and RacPyV miRNAs are Dicer dependent. Wild type or Dicer-deficient 293T cells are transfected with the corresponding miRNA expression vector. Northern blot analysis was performed on the total RNA at 48 hours post transfection. The SV40 miRNA and MHV68 miR-M1-7 expression vectors were transfected as positive controls. The bands corresponding to the pre-miRNAs (white arrowheads) and the mature miRNAs (black arrowheads) are indicated. As a loading control, ethidium bromide_stained low-molecular-weight RNA is shown. (C) The PtvPyV2a, GggPyV1 and RacPyV miRNAs can autoregulate early mRNA expression. The antisense Renilla luciferase construct for each of the three PyV was co-transfected with the firefly luciferase and various miRNA expression vectors into 293T cells. Normalized R. Luc for the different reporter constructs is shown. As a negative control, the empty pcDNA3.1neo vector was utilized. To show specificity of the reporter assay, an SV40 miRNA expression vector was co-transfected with a reporter construct containing the SV40 miRNA binding sites in the 3’ UTR.
Almost all known miRNAs depend on the Dicer nuclease for maturation of the pre-miRNA to final effector miRNA. To determine if the biogenesis of PtvPyV2a, GggPtV1 and RacPyV miRNAs is dependent on Dicer, the PtvPyV2a, GggPyV1 or RacPyV miRNA expression vectors were transfected into either wild type 293T or Dicer-deficient 293T cells (38, 39), followed by Northern blot analysis of the total RNA. For all three miRNAs tested, there was less detectable mature miRNA signal and a corresponding build-up of the pre-miRNA signals in the Dicer-deficient cell line (Fig. 3B). The increase in the ratio of pre-miRNA-to-miRNA bands for PtvPyV2a, GggPyV1 and RacPyV was also observed for the control SV40 miRNA and MHV68 M1-7 miRNAs, which were previously demonstrated to be Dicer-dependent (56). These results demonstrate that the PtvPyV2a, GggPyV1 and RacPyV genomes encode miRNAs whose biogenesis is dependent on Dicer processing.
The PtvPyV2a, GggPyV1 and RacPyV miRNAs negatively regulate early viral transcripts
Similar to all other known polyomaviral miRNAs, the PtvPyV2a, GggPyV1, and RacPyV miRNAs are located antisense to the viral early transcripts. Therefore, we hypothesized that, similar to other polyomaviral miRNAs (15-19), these miRNAs can regulate early transcripts. To test this, we generated luciferase reporters containing the corresponding genomic region from the early transcripts. As expected, co-transfection of the PtvPyV2a, the GggPyV1 or the RacPyV miRNA expression vectors with the negative control reporter plasmid had no effect on luciferase activity (Fig. 3C). However, co-transfection of any of the three individual miRNA expression vectors, with their appropriate corresponding reporter, demonstrated a significant reduction in luciferase activity (Fig. 2C). Interestingly, we observed extensive cross reactivity of the PtvPyV2a miRNA on the GggPyV1 reporter, likely due to the extensive sequence similarity between these viruses. Combined, these results demonstrate that the PtvPyV2a, GggPyV1 and RacPyV miRNAs are fully active in RISC, and suggest that these miRNAs can regulate early transcript expression.
The RacPyV miRNA is highly expressed in tumors
RacPyV was first discovered in neuroglial frontal/olfactory lobe tumors in free-ranging raccoons in the Western United States, and 100% of these tumors (19/19) contain RacPyV DNA (3). To determine if the RacPyV miRNAs are expressed in tumors, we conducted northern blot analysis on RNA harvested from four different RacPyV-associated tumors (Rac 10, 12, 14 and 16). The RacPyV 5p derivative miRNA was readily detectable in all four tumor tissue samples, but not in the RacPyV-negative non-tumor raccoon tissue (Fig. 4A). To rule out that the miRNA signal we detected was due to non-specific RNA degradation, we stripped and reprobed our blots with control probes designed to recognize the terminal loop and flanking regions of the RacPyV pre-miRNA (which should not be processed into stable ~22 nt RNAs). These probes failed to detect specific bands thereby demonstrating the specificity of the miRNA probes (data not shown). These results demonstrate that the RacPyV miRNA is readily detectable in tumors.
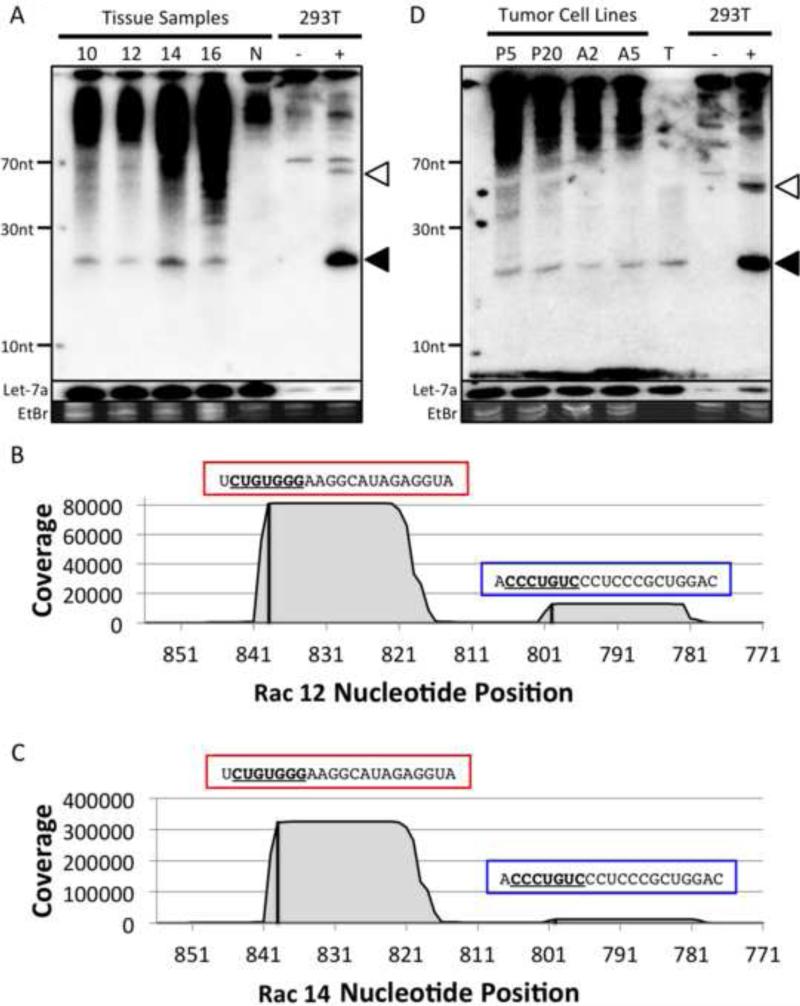
Fig. 4. The RacPyV miRNA is detected in RacPyV-associated tumors and cell lines.
(A) The RacPyV miRNA are detectable in RacPyV-associated raccoon brain tumors. Total RNA was harvested from four different RacPyV-associated raccoon brain tumor samples (labeled as 10, 12, 14 and 16) and one normal raccoon brain sample (labeled N) and subjected to Northern blot analysis. A probe designed to recognize the RacPyV 5p derivative miRNA was used. (B and C) The RacPyV miRNAs expressed in tumor tissues are identical to the transfection samples. The number of small RNA deep sequencing reads from two RacPyV-associated tumor tissues, Rac 12 and 14, were mapped onto the RacPyV genome. The 5p and 3p derivative miRNAs are indicated on top of each peak and indicated by a red box and a blue box, respectively, with the seed sequences (nucleotide position 2 to 8) underlined. (D) The RacPyV miRNA is preserved throughout passage in cultured tumor cells and detectable in the mouse xenograft system. Total RNA from an early and late passage of tumor cultures derived from the Rac 14 tumor, and two different Rac 14 derived mouse xenograft tumors, were subjected to Northern blot detection of the RacPyV 5p derivative miRNA. The tumor cell lines are indicated at the top of the blot as follows: tumor cultures passages 5 and 20 (lanes “P5” and “P20”), mouse xenograft tumor (lanes “A2” and “A5”). Small RNAs isolated from the Rac14 tumor were included as a control (lane “T”). Also, 293T cells were transfected with either the empty pcDNA3.1neo vector (indicated as “-”) or the RacPyV miRNA expression vector (indicated as “+”). As a control, the blot was stripped and re-probed for the cellular miRNA, let-7a. The bands corresponding to the pre-miRNAs (white arrowhead) and 5p derivative miRNA (black arrowhead) are indicated. As a loading control, ethidium bromide-stained low-molecular-weight RNA is shown.
The above northern blot results suggested that the RacPyV miRNA is abundant in tumors. To better characterize the miRNAs in RacPyV tumors we deep sequenced small RNAs from two different tumors. This analysis showed that both in sequence and 5p dominance of the derivative miRNAs, the RacPyV miRNAs in tumors are identical to those we characterized in our transfection studies (Fig. 2C) (Fig. 4B and 4C). This analysis also identified numerous host (raccoon) miRNAs that share sequence identity with known miRNAs from other mammals, including some that have been reported as having oncogenic properties (oncomiRs) (Table 1). Importantly, in both tumor samples analyzed, the RacPyV miRNA was among the most abundant miRNAs detectable in our libraries (6th and 12th most abundant, Table 1, Fig. S1 and S2). These data uncover for the first time abundant viral miRNAs in a PyV-associated tumor, and suggest the possibility that viral and host miRNAs are active in the progression or stability of the tumor.
Table 1. Small RNA deep sequencing mapping of RacPyV and raccoon miRNAs.
Small RNA deep sequencing reads from the Rac 12 and Rac 14 tumor tissue samples were mapped onto the RacPyV genome or the miRBase miRNA database. The top fifty most abundant raccoon miRNAs were calculated for each sample and listed according to their abundance. The RacPyV 5p derivative miRNA is indicated in bold, underlined and highlighted in yellow. Each raccoon miRNA was designated as an oncogene (“O”), a tumor suppressor (“TS”) or both, according to the annotation from two different databases, OncomiRDB (45) and miRCancer (46). A miRNA is designated “NA” when it failed to match any annotation in both of the databases. Raccoon miRNAs that were sequenced from both tumor specimens are shaded in gray.
| miRNAs | Rac 12 | miRNAs | |||
|---|---|---|---|---|---|
| Counts | OncomiRDB | miRCancer | Counts | ||
| mmu-miR-423-5p | 240090 | na | O | mmu-miR-23b | 800100 |
| mmu-miR-191-5p | 230969 | Both | Both | mmu-let-7a | 781650 |
| mmu-miR-23b | 221848 | Both | Both | mmu-miR-15b | 684100 |
| mmu-miR-320a | 220030 | TS | TS | mmu-miR-184 | 564300 |
| mmu-miR-99a | 203606 | TS | Both | mmu-miR-191 | 385800 |
| mmu-miR-184 | 176451 | Both | Both | RacPyV | 220456 |
| mmu-let-7a | 167357 | TS | TS | mmu-miR-101 | 177600 |
| mmu-miR-335-5p | 166738 | Both | Both | mmu-miR-99a | 176800 |
| mmu-miR-342-3p | 158181 | TS | TS | mmu-mir-320a | 176500 |
| mmu-miR-204-3p | 121788 | na | Both | mmu-miR-342 | 176300 |
| mmu-miR-24-3p | 94515 | Both | Both | mmu-mir-423-5p | 165600 |
| RacPyV | 77568 | mmu-miR-185-5p | 162200 | ||
| mmu-miR-718 | 73838 | na | na | mmu-miR-1a | 144500 |
| mmu-miR-140-3p | 73835 | TS | Both | mmu-miR-24-3p | 144400 |
| mmu-miR-3195 | 73693 | na | na | mmu-miR-204 | 110800 |
| mmu-miR-320b | 73440 | na | na | mmu-miR-503-5p | 107700 |
| mmu-miR-4634 | 73115 | na | na | mmu-miR-335 | 103600 |
| mmu-miR-188 | 69885 | na | na | mmu-miR-195-5p | 103300 |
| mmu-miR-6087 | 63857 | na | na | mmu-miR-140-3p | 100900 |
| mmu-miR-3613-3p | 58992 | na | na | mmu-miR-9-3p | 90727 |
| mmu-miR-330-3p | 51746 | na | TS | mmu-miR-214-3p | 90454 |
| mmu-miR-503-5p | 33063 | TS | Both | mmu-miR-92b-5p | 90000 |
| mmu-miR-669 | 33054 | na | na | mmu-miR-140-5p | 86818 |
| mmu-miR-6724-5p | 31560 | na | na | mmu-miR-320b | 86454 |
| mmu-miR-4306 | 30960 | na | na | mmu-miR-6087 | 83181 |
| mmu-miR-4484 | 30960 | na | na | mmu-miR-4484 | 78090 |
| mmu-miR-4306+T | 30600 | na | na | mmu-miR-30d-5p | 74030 |
| mmu-miR-129-2-3p | 28800 | na | TS | mmu-miR-188 | 72820 |
| mmu-miR-30d-5p | 22320 | Both | Both | mmu-miR-671-5p | 72490 |
| mmu-miR-195-5p | 21600 | TS | Both | mmu-miR-208b-3p | 71060 |
| mmu-miR-214-3p | 18360 | Both | Both | mmu-miR-129-1-3p | 68200 |
| mmu-miR-3178 | 18360 | na | na | mmu-miR-874 | 68181 |
| mmu-miR-4442 | 16200 | na | na | mmu-miR-501-3p | 68090 |
| mmu-miR-7847-3p | 16200 | na | na | mmu-miR-378a-3p | 67980 |
| mmu-miR-4459 | 15120 | na | na | mmu-miR-486-3p | 66440 |
| mmu-miR-92b-5p | 12960 | na | na | mmu-miR-494-3p | 64790 |
| mmu-miR-185-5p | 12600 | TS | Both | mmu-miR-125a | 60060 |
| mmu-miR-3960 | 11160 | na | na | mmu-miR-3960 | 55770 |
It has been previously demonstrated that in cultured primary tumor cells, RacPyV genome is continually maintained throughout multiple passages (10). To determine if the expression of the RacPyV miRNA is also maintained, total RNA was harvested from cultured cells from the Rac 14 tumor at passage 5 and passage 20. Northern blot analysis indicated that the RacPyV 5p derivative miRNA is detectable at comparable intensities in both the early and the late passage cells (Fig. 4D,). This suggests that the RacPyV miRNA is stably maintained throughout passage. We also tested whether the RacPyV miRNA is detectable in RacPyV-associated mouse xenograft tumors (10). These tumors arise from implantation of the RacPyV tumor cell lines into NOD scid gamma mice (http://jaxmice.jax.org/nod-scid-gamma/). Similar to the naturally arising tumors in the wild, northern blot analysis confirmed the RacPyV 5p miRNA was readily detectable in the xenograft tumors. Combined, these data demonstrate that expression of the RacPyV miRNA is not prohibitive to tumor cell growth and are consistent with a possible pro-tumorigenic activity of this miRNA.
The MCPyV miRNA is unlikely to be biologically relevant in tumors
Given that RacPyV and MCPyV are phylogentically related and both are associated with tumors, we wanted to determine if similar to RacPyV-associated tumors, the MCPyV miRNA was a likely regulator of gene expression in tumors. Previously, Lee et al. identified MCPyV miRNA expression in MCCs, and had suggested that host targets of this miRNA may be involved in tumor biology of MCCs (47). More recently, Renwick et al. collected a larger deep sequencing dataset on small RNAs from multiple independent MCC tumor samples (57). To determine the plausibility that the MCPyV miRNA is important in tumor biology, we analyzed the existing published MCC tumor small RNA-seq libraries (47, 57, 58) to determine the relative abundance of the MCPyV miRNAs. This meta-analysis showed that the MCPyV miRNA is only detectable in less than half of MCPyV-positive MCC tumors (Table 2). When present, the McPyV miRNA is not detectable at high levels, as it ranks among the least abundant miRNAs in the 6 MCC tumors and cell lines analyzed (Table 3). Given that miRNAs that are expressed at low levels (less than 0.1% of total miRNA reads or less than 100 copies/ cell) are unlikely to be of functional significance (59, 60), we conclude that MCPyV miRNAs are not relevant regulators of gene expression in tumor cells, at least by the current understanding of canonical miRNA function. Combined with previous publications (3, 10), these results further emphasize the differences in MCPyV and RacPyV-associated tumors.
Table 2. Meta-analysis of MCC tumor small RNA library data.
Small RNA deep sequencing reads were obtained from three independent studies on MCC tumors (47, 57, 58). The number of reads of mcv-miR-M1-5p miRNA was tabulated and represented as a percent of total miRNAs in the tumors.
| Publication | Lee 2011 | Ning 2014 | Renwick 2013 |
|---|---|---|---|
| Total MCC tumors studied | 52 | 2 | 14 |
| MCPyV(+) tumors | 38 | 0 | 7 |
| mcv-miR-M1-5p(+) tumors | 19 | 0 | 1 |
| % of total miRNAs in MCPyV(+) tumors | 0.005 | 0 | 0.025 |
Table 3. Summary of small RNA library data from MCV-positive tumors and cell lines.
The mcv-miR-M1-5p deep sequencing reads and their relative rank were obtained from a previously report study on an MCC tumor and cell lines (57).
| Sample name | MCC Tumor Sample | MCC Cell Line | ||
|---|---|---|---|---|
| MCC10 | MKL-1a | MKL-1b | MKL-1c | |
| Total miRNA reads | 138,293 | 361,783 | 434,595 | 197,841 |
| mcv-miR-M1-5p reads | 35 | 24 | 3 | 5 |
| % of total miRNA reads | 0.0253 | 0.0067 | 0.0007 | 0.0025 |
| mcv-miR-M1-5p rank (out of total miRNAs) | 196/702 | 273/707 | 544/763 | 395/666 |
DISCUSSION
The fraction of PyVs that encode miRNAs and their functional relevance remain poorly understood (61). In this work, we have applied a comparative evolutionary approach to MCPyV and related viruses to better understand PyV miRNA function. We demonstrate that closely related gorilla and chimpanzee PyVs, as well as the more distantly related RacPyV, encode miRNAs in similar genomic locations (Fig. 1). As predicted by the genomic location, each of these newly-discovered miRNAs are active at directing repression of PyV-early-gene-reporter chimeric transcripts (Fig. 1 and 3). This suggests a functional role for the MCPyV and related miRNAs in the negative regulation of early gene expression, consistent with all other known PyV miRNAs (15-20).
In addition to a role in regulating the early transcripts, the sequence similarity between MCPyV and related viruses is also consistent with other shared common targets. Our work demonstrates that PtvPyV2a and GggPyV1 share high sequence identity with the MCPyV miRNA (Fig. 2D). The 5p derivative miRNA encoded by RacPyV also shares partial sequence identity (10 out of 22 nucleotides on a shifted alignment, Fig. 2D) with the 5p derivative miRNA of MCPyV. This might be unexpected since RacPyV and MCPyV are presumably separated by over 94 million years of evolution when their respective raccoon and human hosts diverged from a common ancestor (62). Although the shared sequences do not encompass identical shared seed regions (Fig. 2D), the striking common sequence elements do imply the possibility that RacPyV and MCPyV diverged from a common miRNA locus precursor. If true, then this would represent one of the oldest examples known of viral miRNA divergence from a common ancestor.
Viral miRNAs have been established or suggested as playing a role in various herpesviral-associated tumors. These include chicken sarcomas associated with Marek's disease virus (MDV) and various human lymphocyte and solid tumors associated with Epstein Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV) (63-70). Additionally, we have identified a miRNA encoded by the bovine leukemia retrovirus (BLV) that mimics a known host oncogenic miRNA (56), and this miRNA was later identified in BLV-associated tumors (71). We have also previously shown that a PyV-Papillomavirus hybrid virus, BPCV1, encodes a miRNA that is readily detectable in tumors (25). However, to date, no PyV miRNAs have been implicated in tumorigenesis.
Our finding that the RacPyV miRNA is among the most abundant miRNAs in RacPyV-associated tumors opens up the possibility of a role for viral miRNAs in these tumors. Our transfection-based reporter studies suggest that the RacPyV miRNA can downregulate early T antigen transcripts (Fig. 3C), yet enigmatically, early transcripts are robust in tumors (10). Thus, despite the likely activity of the RacPyV miRNA in reducing levels of a transforming early viral protein(s) in some contexts (Fig. 3C), these tumors readily tolerate robust levels of this miRNA (Fig. 4A). Several non-mutually exclusive explanations could account for this. First, the relevant RacPyV transforming proteins might result from a transcript that escapes RacPyV miRNA regulation. For example, miRNA regulation could be blocked by alternative splicing that eliminates the miRNA docking site in the derivative mRNA or through an mRNA folding structure that is incompatible with RISC access. Second, an “optimal” amount of early protein might be required for these tumors such that too much early protein might have growth inhibitory properties. Third, any penalty of the RacPyV miRNA for negatively regulating viral gene expression might be outweighed by other positive activities of the miRNA such as targeting host tumor suppressors or even other viral proteins. We considered the possibility that the RacPyV miRNA might be additionally targeting the late transcripts, but results from luciferase reporter-based experiments do not support this idea (Fig. S3). Deciphering which, if any, of these models apply will require loss of function and target identification studies for the RacPyV miRNA in these tumors.
Given the similarities of RacPyV and MCPyV miRNAs, we explored the possibility that MCPyV miRNA could function similarly in the tumor biology of MCCs. Based on our understanding of canonical miRNA function, our findings do not support such a model. Meta-analysis of two different small RNA-seq studies of MCCs and derivative cell lines, showed that the MCPyV miRNA was often below the limit of detection. Furthermore, in the tumors where the MCPyV miRNA was detected, their levels never rose above 0.025% of total miRNAs present in these cells (Table 2). In addition, the analysis performed by Renwick et al. placed the MCPyV miRNA in the bottom 30% of all detectable miRNAs (Table 3). Despite the fact it has been argued that host targets of the MCPyV miRNA could contribute to the tumor phenotype (47), our findings strongly argue that MCPyV miRNAs are so low in MCCs as to be insignificant, at least based on the current understanding of canonical miRNA function. These findings suggest that the process of viral genome integration and/or the cell type of MCCs limits promoter activity or post-transcriptional activities required for robust MCPyV miRNA expression, lending further support that the MCPyV and RacPyV-associated tumors arise via separate mechanisms (3,10).
miRNAs are generally “fine-tuning regulators” that balance “transcriptional noise,” and in doing so, promote homeostasis for a variety of physiological processes (72-74). An emerging model for viral miRNA function is that diverse viruses utilize miRNAs to alter both host and viral gene expression to promote the “homeostasis” of persistent infection (22, 23, 75). In this scenario, viral miRNAs are expected to suppress non-beneficial host and viral gene expression to promote infected cell viability in the face of the immune response. Although the mechanisms of persistent infection are poorly understood for PyVs, the majority of PyVs that have been studied take up long-term infections. In combination with previous work (8,15-19, 25, 31, 76), our current findings are consistent with a model whereby all PyV miRNAs regulate early gene expression, with miRNAs from individual clades of viruses possessing unique host targets. As it is likely that most PyV-associated disease results from dysregulated control of persistent infection, continued deciphering of the biological activities of diverse PyV miRNAs may be useful for understanding the genesis of PyV-associated disease.
Supplementary Material
Fig. S1. The RacPyV miRNA is the 12th most abundant miRNA in the Rac 12 tumor tissue sample. The fifty most abundant raccoon miRNAs were plotted on the y axis and their corresponding abundance plotted on the x axis. The RacPyV miRNA was included in the plot and represented as a black bar.
Fig. S2. The RacPyV miRNA is the 6th most abundant miRNA in the Rac 14 tumor tissue sample. The fifty most abundant raccoon miRNAs were plotted on the y axis and their corresponding abundance plotted on the x axis. The RacPyV miRNA was included in the plot and represented as a black bar.
Fig. S3. The RacPyV miRNA does not regulate the late mRNA expression. (A) The late genomic regions of RacPyV was divided into four overlapping fragments (indicated as “Late 1” to “Late 4”), each 500bp in length, and cloned into the 3’ UTR of the Renilla luciferase reporter construct. As a positive control, the region antisense to the SV40 pre-miRNA region (blue) was cloned into the 3’ UTR of Renilla luciferase reporter construct (indicated as grey, dashed arrows). (B) The reporter constructs were co-transfected with the firefly luciferase and miRNA expression plasmids into 293T cells. The Renilla luciferase readings (R. Luc) were normalized to the readings from the firefly luciferase (FF. Luc). Empty pcDNA3.1neo vector was transfected as a negative control. As a control to show specificity of the reporter assay, an SV40 miRNA expression vector was co-transfected with a Renilla luciferase reporter construct containing the SV40 miRNA binding sites in the 3’ UTR.
HIGHLIGHTS.
MCPyV and related viruses (PtvPyV2a, GggPyVl, RacPyV) encode a miRNA in similar genomic positions
These miRNAs can negatively regulate reporters of early viral gene expression
Unlike MCPyV positive tumors, RacPyV miRNA is abundant in RacPyV-associated tumors
The RacPyV miRNA is the first polyomaviral miRNA shown to be abundant in tumors
Acknowledgements
We gratefully acknowledge Bryan Cullen, Duke University, for the 293T Dicer knockout cells. This work was supported by Grant R01AI077746 from the National Institutes of Health, a Burroughs Wellcome Investigators in Pathogenesis Award, Grant RP110098 from the Cancer Prevention and Research Institute of Texas, and a University of Texas at Austin Institute for Cellular and Molecular Biology fellowship to CSS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Cole CN. Fields Virology. Third Edition. Lippincott-Raven; 1996. Polyomaviridae: the viruses and their replication; pp. 1997–2043. [Google Scholar]
- 2.DeCaprio JA, Garcea RL. A cornucopia of human polyomaviruses. Nat. Rev. Microbiol. 2013;11:264–276. doi: 10.1038/nrmicro2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dela Cruz FN, Jr, Giannitti F, Li L, Woods LW, Del Valle L, Delwart E, Pesavento PA. Novel polyomavirus associated with brain tumors in free-ranging raccoons, western United States. Emerg. Infect. Dis. 2013;19:77–84. doi: 10.3201/eid1901.121078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leendertz FH, Scuda N, Cameron KN, Kidega T, Zuberbuhler K, Leendertz SAJ, Couacy-Hymann E, Boesch C, Calvignac S, Ehlers B. African Great Apes Are Naturally Infected with Polyomaviruses Closely Related to Merkel Cell Polyomavirus. J. Virol. 2011;85:916–924. doi: 10.1128/JVI.01585-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehlers B, Moens U. Genome analysis of non-human primate polyomaviruses. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2014;26:283–294. doi: 10.1016/j.meegid.2014.05.030. [DOI] [PubMed] [Google Scholar]
- 6.Dalianis T, Hirsch HH. Human polyomaviruses in disease and cancer. Virology. 2013;437:63–72. doi: 10.1016/j.virol.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 7.White MK, Gordon J, Khalili K. The rapidly expanding family ofhuman polyomaviruses: recent developments in understanding their life cycle and role in human pathology. PLoS Pathog. 2013;9:e1003206. doi: 10.1371/journal.ppat.1003206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang S, Sroller V, Zanwar P, Chen CJ, Halvorson SJ, Ajami NJ, Hecksel CW, Swain JL, Wong C, Sullivan CS, Butel JS. Viral MicroRNA Effects on Pathogenesis ofPolyomavirus SV40 Infections in Syrian Golden Hamsters. PLoS Pathog. 2014;10:e1003912. doi: 10.1371/journal.ppat.1003912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brostoff T, Dela Cruz FN, Church ME, Woolard KD, Pesavento PA. The Raccoon Polyomavirus Genome and Tumor Antigen Transcription are Stable and Abundant in Neuroglial Tumors. J. Virol. 2014 doi: 10.1128/JVI.01912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, Chang Y. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16272–16277. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.An P, Sáenz Robles MT, Pipas JM. Large T antigens of polyomaviruses: amazing molecular machines. Annu. Rev. Microbiol. 2012;66:213–236. doi: 10.1146/annurev-micro-092611-150154. [DOI] [PubMed] [Google Scholar]
- 13.Cheng J, DeCaprio JA, Fluck MM, Schaffhausen BS. Cellular transformation by Simian Virus 40 and Murine Polyoma Virus T antigens. Semin. Cancer Biol. 2009;19:218–228. doi: 10.1016/j.semcancer.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fluck MM, Schaffhausen BS. Lessons in Signaling and Tumorigenesis from Polyomavirus Middle T Antigen. Microbiol. Mol. Biol. Rev. MMBR. 2009;73:542563. doi: 10.1128/MMBR.00009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seo GJ, Fink LHL, O'Hara B, Atwood WJ, Sullivan CS. Evolutionarily Conserved Function of a Viral MicroRNA. J. Virol. 2008;82:9823–9828. doi: 10.1128/JVI.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seo GJ, Chen CJ, Sullivan CS. Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology. 2009;383:183–187. doi: 10.1016/j.virol.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Chen CJ, Cox JE, Kincaid RP, Martinez A, Sullivan CS. Divergent MicroRNA targetomes of closely related circulating strains of a polyomavirus. J. Virol. 2013;87:11135–11147. doi: 10.1128/JVI.01711-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 19.Sullivan CS, Sung CK, Pack CD, Grundhoff A, Lukacher AE, Benjamin TL, Ganem D. Murine Polyomavirus encodes a microRNA that cleaves early RNA transcripts but is not essential for experimental infection. Virology. 2009;387:157–167. doi: 10.1016/j.virol.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cantalupo P, Doering A, Sullivan CS, Pal A, Peden KWC, Lewis AM, Pipas JM. Complete Nucleotide Sequence ofPolyomavirus SA12. J. Virol. 2005;79:1309413104. doi: 10.1128/JVI.79.20.13094-13104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox JE, Sullivan CS. Balance and Stealth: The Role of Noncoding RNAs in the Regulation ofVirus Gene Expression. Annu. Rev. Virol. 2014;1 doi: 10.1146/annurev-virology-031413-085439. null. [DOI] [PubMed] [Google Scholar]
- 23.Kincaid RP, Sullivan CS. Virus-Encoded microRNAs: An Overview and a Look to the Future. PLoS Pathog. 2012;8:e1003018. doi: 10.1371/journal.ppat.1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagatie O, Tritsmans L, Stuyver LJ. The miRNA world of polyomaviruses. Virol. 2013;10:268. doi: 10.1186/1743-422X-10-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen CJ, Kincaid RP, Seo GJ, Bennett MD, Sullivan CS. Insights into Polyomaviridae MicroRNA Function Derived from Study ofthe Bandicoot Papillomatosis Carcinomatosis Viruses. J. Virol. 2011;85:4487–4500. doi: 10.1128/JVI.02557-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin Y- T, Kincaid RP, Arasappan D, Dowd SE, Hunicke-Smith SP, Sullivan CS. Small RNA profiling reveals antisense transcription throughout the KSHV genome and novel small RNAs. RNA. 2010;16:1540–1558. doi: 10.1261/rna.1967910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weber K, Bolander ME, Sarkar G. PIG-B: A homemade monophasic cocktail for the extraction ofRNA. Mol. Biotechnol. 1998;9:73–77. doi: 10.1007/BF02752699. [DOI] [PubMed] [Google Scholar]
- 28.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA N. Y. N. 2006;12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sullivan CS, Grundhoff A. Identification ofviral microRNAs. Methods Enzymol. 2007;427:3–23. doi: 10.1016/S0076-6879(07)27001-6. [DOI] [PubMed] [Google Scholar]
- 30.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. NucleicAcids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen CJ, Burke JM, Kincaid RP, Azarm KD, Mireles N, Butel JS, Sullivan CS. Naturally arising strains of polyomaviruses with severely attenuated microRNA expression. J. Virol. 2014 doi: 10.1128/JVI.01933-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard J- F, Guindon S, Lefort V, Lescot M, Claverie J-M, Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dereeper A, Audic S, Claverie J-M, Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 2010;10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 36.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 37.Chevenet F, Brun C, Banuls A-L, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bogerd HP, Skalsky RL, Kennedy EM, Furuse Y, Whisnant AW, Flores O, Schultz KLW, Putnam N, Barrows NJ, Sherry B, Scholle F, Garcia-Blanco MA, Griffin DE, Cullen BR. Replication of many human viruses is refractory to inhibition by endogenous cellular microRNAs. J. Virol. 2014;88:8065–8076. doi: 10.1128/JVI.00985-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bogerd HP, Whisnant AW, Kennedy EM, Flores O, Cullen BR. Derivation and characterization of Dicer- and microRNA-deficient human cells. RNAN.Y.N. 2014;20:923–937. doi: 10.1261/rna.044545.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griffiths - Jones S. The microRNA Registry. NucleicAcids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol. Biol. Clifton NJ. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 42.Griffiths-Jones S, Saini HK, Dongen S van, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. NucleicAcids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang D, Gu J, Wang T, Ding Z. OncomiRDB: a database for the experimentally verified oncogenic and tumor-suppressive microRNAs. Bioinformatics btu1. 2014;55 doi: 10.1093/bioinformatics/btu155. [DOI] [PubMed] [Google Scholar]
- 46.Xie B, Ding Q, Han H, Wu D. miRCancer: a microRNA-cancer association database constructed by text mining on literature. Bioinformatics. 2013;29:638–644. doi: 10.1093/bioinformatics/btt014. [DOI] [PubMed] [Google Scholar]
- 47.Lee S, Paulson KG, Murchison EP, Afanasiev OK, Alkan C, Leonard JH, Byrd DR, Hannon GJ, Nghiem P. Identification and validation of a novel mature microRNA encoded by the Merkel cell polyomavirus in human Merkel cell carcinomas. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2011;52:272–275. doi: 10.1016/j.jcv.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cazalla D, Xie M, Steitz JA. A primate herpesvirus uses the integrator complex to generate viral microRNAs. Mol. Cell. 2011;43:982–992. doi: 10.1016/j.molcel.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kincaid RP, Chen Y, Cox JE, Rethwilm A, Sullivan CS. Noncanonical microRNA (miRNA) biogenesis gives rise to retroviral mimics of lymphoproliferative and immunosuppressive host miRNAs. mBio. 2014;5:e00074. doi: 10.1128/mBio.00074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whisnant AW, Kehl T, Bao Q, Materniak M, Kuzmak J, Lochelt M, Cullen BR. Identification of novel, highly expressed retroviral microRNAs in cells infected by bovine foamy virus. J. Virol. 2014;88:4679–4686. doi: 10.1128/JVI.03587-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diebel KW, Claypool DJ, van Dyk LF. A conserved RNA polymerase III promoter required for gammaherpesvirus TMER transcription and microRNA processing. Gene. 2014;544:8–18. doi: 10.1016/j.gene.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diebel KW, Smith AL, van Dyk LF. Mature and functional viral miRNAs transcribed from novel RNA polymerase III promoters. RNA N. Y. N. 2010;16:170–185. doi: 10.1261/rna.1873910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feldman ER, Kara M, Coleman CB, Grau KR, Oko LM, Krueger BJ, Renne R, van Dyk LF, Tibbetts SA. Virus-encoded microRNAs facilitate gammaherpesvirus latency and pathogenesis in vivo. mBio. 2014;5:e00981–00914. doi: 10.1128/mBio.00981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. Identification of microRNAs ofthe herpesvirus family. Nat. Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 55.Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs. Mol. Cell. 2010;37:135–142. doi: 10.1016/j.molcel.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kincaid RP, Burke JM, Sullivan CS. From the Cover: RNA virus microRNA that mimics a B-cell oncomiR. Proc. Natl. Acad. Sci. 2012;109:3077–3082. doi: 10.1073/pnas.1116107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Renwick N, Cekan P, Masry PA, McGeary SE, Miller JB, Hafner M, Li Z, Mihailovic A, Morozov P, Brown M, Gogakos T, Mobin MB, Snorrason EL, Feilotter HE, Zhang X, Perlis CS, Wu H, Suarez-Farinas M, Feng H, Shuda M, Moore PS, Tron VA, Chang Y, Tuschl T. Multicolor microRNA FISH effectively differentiates tumor types. J. Clin. Invest. 2013;123:2694–2702. doi: 10.1172/JCI68760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ning MS, Kim AS, Prasad N, Levy SE, Zhang H, Andl T. Characterization of the Merkel Cell Carcinoma miRNome. J. Skin Cancer. 2014;2014:289548. doi: 10.1155/2014/289548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cullen BR. Viruses and microRNAs: RISCy interactions with serious consequences. Genes Dev. 2011;25:1881–1894. doi: 10.1101/gad.17352611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brown BD, Gentner B, Cantore A, Colleoni S, Amendola M, Zingale A, Baccarini A, Lazzari G, Galli C, Naldini L. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 61.Imperiale MJ. Polyomavirus miRNAs: the beginning. Curr. Opin. Virol. 2014;7:29–32. doi: 10.1016/j.coviro.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinforma. Oxf. Engl. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- 63.Yao Y, Nair V. Role ofvirus-encoded microRNAs in Avian viral diseases. Viruses. 2014;6:1379–1394. doi: 10.3390/v6031379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gottwein E, Corcoran DL, Mukherjee N, Skalsky RL, Hafner M, Nusbaum JD, Shamulailatpam P, Love CL, Dave SS, Tuschl T, Ohler U, Cullen BR. Viral MicroRNA Targetome ofKSHV-Infected Primary Effusion Lymphoma Cell Lines. Cell Host Microbe. 2011;10:515–526. doi: 10.1016/j.chom.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi J-TA, Braich R, Manoharan M, Soutschek J, Ohler U, Cullen BR. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Skalsky RL, Corcoran DL, Gottwein E, Frank CL, Kang D, Hafner M, Nusbaum JD, Feederle R, Delecluse H-J, Luftig MA, Tuschl T, Ohler U, Cullen BR. The Viral and Cellular MicroRNA Targetome in Lymphoblastoid Cell Lines. PLoS Pathog. 2012;8:e1002484. doi: 10.1371/journal.ppat.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boss IW, Nadeau PE, Abbott JR, Yang Y, Mergia A, Renne R. A Kaposi's sarcoma-associated herpesvirus-encoded ortholog of microRNA miR-155 induces human splenic B-cell expansion in NOD/LtSz-scid IL2Rynull mice. J. Virol. 2011;85:9877–9886. doi: 10.1128/JVI.05558-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seto E, Moosmann A, Gromminger S, Walz N, Grundhoff A, Hammerschmidt W. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 2010;6:e1001063. doi: 10.1371/journal.ppat.1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vereide DT, Seto E, Chiu Y-F, Hayes M, Tagawa T, Grundhoff A, Hammerschmidt W, Sugden B. Epstein-Barr virus maintains lymphomas via its miRNAs. Oncogene. 2014;33:1258–1264. doi: 10.1038/onc.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dahlke C, Maul K, Christalla T, Walz N, Schult P, Stocking C, Grundhoff A. A microRNA encoded by Kaposi sarcoma-associated herpesvirus promotes B-cell expansion in vivo. PloS One. 2012;7:e49435. doi: 10.1371/journal.pone.0049435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rosewick N, Momont M, Durkin K, Takeda H, Caiment F, Cleuter Y, Vernin C, Mortreux F, Wattel E, Burny A, Georges M, Van den Broeke A. Deep sequencing reveals abundant noncanonical retroviral microRNAs in B-cell leukemia/lymphoma. Proc. Natl. Acad. Sci. U. S. A. 2013;110:2306–2311. doi: 10.1073/pnas.1213842110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gurtan AM, Sharp PA. The role of miRNAs in regulating gene expression networks. J. Mol. Biol. 2013;425:3582–3600. doi: 10.1016/j.jmb.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virology. 2011;411:325343. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Broekema NM, Imperiale MJ. miRNA regulation of BK polyomavirus replication during early infection. Proc. Natl. Acad. Sci. 2013;110:8200–8205. doi: 10.1073/pnas.1301907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. The RacPyV miRNA is the 12th most abundant miRNA in the Rac 12 tumor tissue sample. The fifty most abundant raccoon miRNAs were plotted on the y axis and their corresponding abundance plotted on the x axis. The RacPyV miRNA was included in the plot and represented as a black bar.
Fig. S2. The RacPyV miRNA is the 6th most abundant miRNA in the Rac 14 tumor tissue sample. The fifty most abundant raccoon miRNAs were plotted on the y axis and their corresponding abundance plotted on the x axis. The RacPyV miRNA was included in the plot and represented as a black bar.
Fig. S3. The RacPyV miRNA does not regulate the late mRNA expression. (A) The late genomic regions of RacPyV was divided into four overlapping fragments (indicated as “Late 1” to “Late 4”), each 500bp in length, and cloned into the 3’ UTR of the Renilla luciferase reporter construct. As a positive control, the region antisense to the SV40 pre-miRNA region (blue) was cloned into the 3’ UTR of Renilla luciferase reporter construct (indicated as grey, dashed arrows). (B) The reporter constructs were co-transfected with the firefly luciferase and miRNA expression plasmids into 293T cells. The Renilla luciferase readings (R. Luc) were normalized to the readings from the firefly luciferase (FF. Luc). Empty pcDNA3.1neo vector was transfected as a negative control. As a control to show specificity of the reporter assay, an SV40 miRNA expression vector was co-transfected with a Renilla luciferase reporter construct containing the SV40 miRNA binding sites in the 3’ UTR.