Abstract
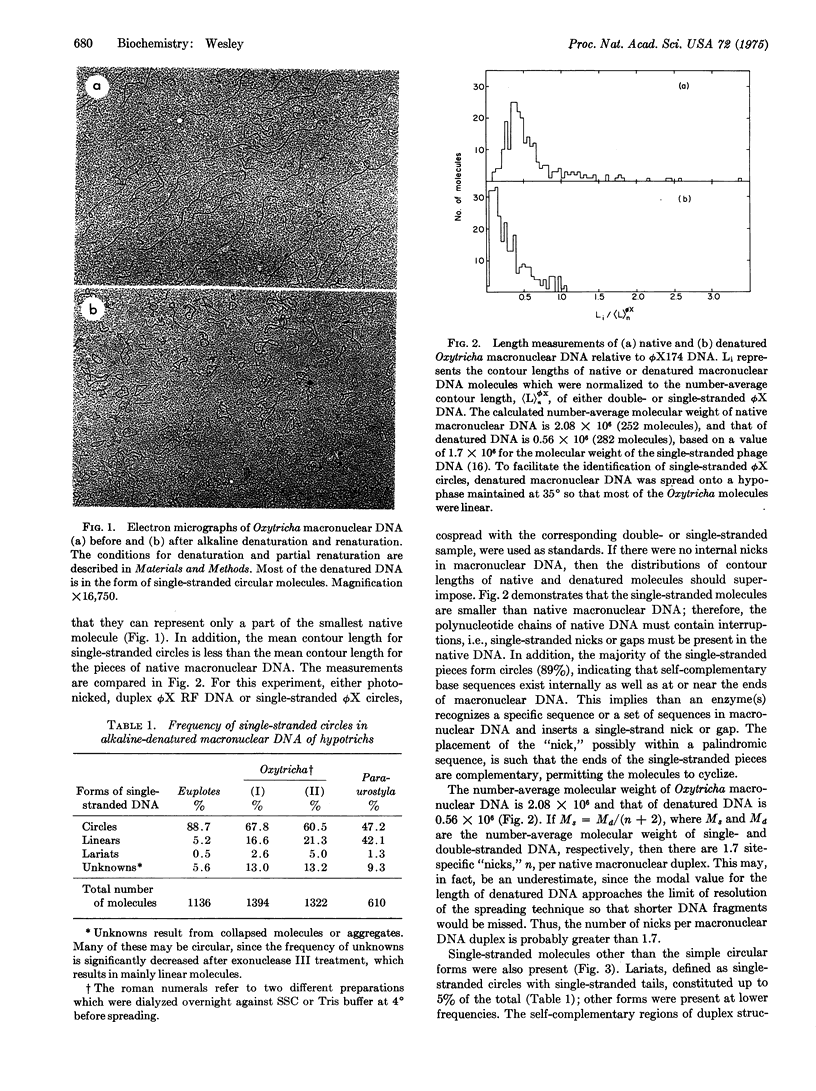
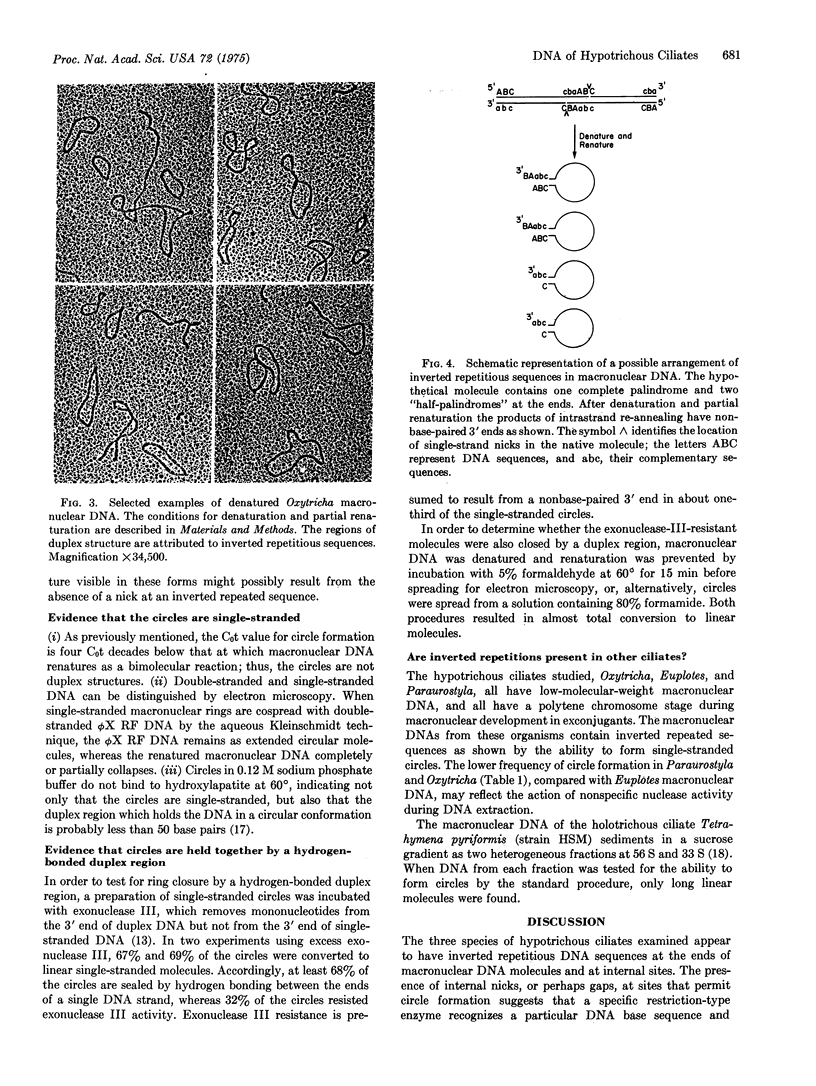
The low-molecular-weight macronuclear DNA of the hypotrichous ciliates Oxytricha, Euplotes, and Paraurostyla contains inverted repetitious sequences. Up to 89% of the denatured macronuclear DNA molecules form single-stranded circles due to intramolecular renaturation of complementary sequences at or near the ends of the same polynucleotide chain. Other ciliated protozoans, such as Tetrahymena, with high-molecular-weight macronuclear DNA and an alternative mode of macronuclear development, appear to lack these selfcomplementary sequences. The denatured macronuclear molecules of hypotrichs are held in the circular conformation by a hydrogen-bonded duplex region, which is probably less than 50 base pairs in length, since the duplex regions are not visible by electron microscopy and since the circles in 0.12 M phosphate buffer are not retained during hydroxylapatite chromatography at 60 degrees. The existence of extremely small circles, with contour lengths shorter than the smallest pieces of native DNA, suggests that inverted repetitions containing nicks (broken phosphodiester bonds in duplex DNA) or gaps (interruptions with missing nucleotides) are present at internal positions as well as at the ends of native molecules. Estimates from length measurements of native and denatured Oxytricha macronuclear DNA indicate an average of 1.7 nicks per duplex molecule. Thus, in order to account for the high frequency of circle formation, a restriction-type enzyme(s) must exist which inserts single-strand, site-specific nicks or gaps at internal positions in the macronuclear DNA of Oxytricha.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bostock C. J., Prescott D. M. Evidence of gene diminution during the formation of the macronucleus in the protozoan, Stylonychia (DNA density-melting curves-micronuclear DNA-polytene chromosomes). Proc Natl Acad Sci U S A. 1972 Jan;69(1):139–142. doi: 10.1073/pnas.69.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W. Restriction and modification of DNA: enzymes and substrates. Introductory remarks. Fed Proc. 1974 May;33(5):1125–1127. [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Palindromic base sequences and replication of eukaryote chromosome ends. Nature. 1974 Aug 9;250(5466):467–470. doi: 10.1038/250467a0. [DOI] [PubMed] [Google Scholar]
- GALL J. G. Macronuclear duplication in the ciliated protozoan Euplotes. J Biophys Biochem Cytol. 1959 Mar 25;5(2):295–308. doi: 10.1083/jcb.5.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey A. D., Burgi E., Ingraham L. COHESION OF DNA MOLECULES ISOLATED FROM PHAGE LAMBDA. Proc Natl Acad Sci U S A. 1963 May;49(5):748–755. doi: 10.1073/pnas.49.5.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou P. General model for the replication of double stranded DNA molecules. Nat New Biol. 1973 Aug 29;244(139):257–260. doi: 10.1038/newbio244257a0. [DOI] [PubMed] [Google Scholar]
- Lewin B. Interaction of regulator proteins with recognition sequences of DNA. Cell. 1974 May;2(1):1–7. doi: 10.1016/0092-8674(74)90002-6. [DOI] [PubMed] [Google Scholar]
- Mertz J. E., Davis R. W. Cleavage of DNA by R 1 restriction endonuclease generates cohesive ends. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3370–3374. doi: 10.1073/pnas.69.11.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishi A., Ando T. The DNA of Tetrahymena pyriformis GL strain. A mild method for preparation and its characterization. Biochim Biophys Acta. 1973 Apr 11;299(4):507–515. doi: 10.1016/0005-2787(73)90222-0. [DOI] [PubMed] [Google Scholar]
- Murti K. G., Prescott D. M., Pène J. J. DNA of ciliated protozoa. 3. Binding of RNA polymerase and denaturation at the ends of the low-molecular weight DNA in Stylonychia. J Mol Biol. 1972 Jul 28;68(3):413–416. doi: 10.1016/0022-2836(72)90095-2. [DOI] [PubMed] [Google Scholar]
- Prescott D. M., Murti K. G., Bostock C. J. Genetic apparatus of Stylonychia sp. Nature. 1973 Apr 27;242(5400):576, 597-600. doi: 10.1038/242576a0. [DOI] [PubMed] [Google Scholar]
- RICHARDSON C. C., LEHMAN I. R., KORNBERG A. A DEOXYRIBONUCLEIC ACID PHOSPHATASE-EXONUCLEASE FROM ESCHERICHIA COLI. II. CHARACTERIZATION OF THE EXONUCLEASE ACTIVITY. J Biol Chem. 1964 Jan;239:251–258. [PubMed] [Google Scholar]
- Salser W., Fry K., Wesley R. D., Simpson L. Appendix to studies on kinetoplast DNA: use of nucleic acid fingerprints to estimate the complexity of minicircle DNA. Biochim Biophys Acta. 1973 Sep 7;319(3):277–280. doi: 10.1016/0005-2787(73)90166-4. [DOI] [PubMed] [Google Scholar]
- Smith C. A., Vinograd J. Small polydisperse circular DNA of HeLa cells. J Mol Biol. 1972 Aug 21;69(2):163–178. doi: 10.1016/0022-2836(72)90222-7. [DOI] [PubMed] [Google Scholar]
- Sobell H. M. Molecular mechanism for genetic recombination. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2483–2487. doi: 10.1073/pnas.69.9.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. A., Thomas C. A., Jr Hydroxyapatite chromatography of short double-helical DNA. Biochim Biophys Acta. 1973 Dec 21;331(3):333–340. doi: 10.1016/0005-2787(73)90019-1. [DOI] [PubMed] [Google Scholar]
- Wilson D. A., Thomas C. A., Jr Palindromes in chromosomes. J Mol Biol. 1974 Mar 25;84(1):115–138. doi: 10.1016/0022-2836(74)90216-2. [DOI] [PubMed] [Google Scholar]
- Wolfson J., Dressler D. Adenovirus-2 DNA contains an inverted terminal repetition. Proc Natl Acad Sci U S A. 1972 Oct;69(10):3054–3057. doi: 10.1073/pnas.69.10.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R., Taylor E. Nucleotide sequence analysis of DNA. II. Complete nucleotide sequence of the cohesive ends of bacteriophage lambda DNA. J Mol Biol. 1971 May 14;57(3):491–511. doi: 10.1016/0022-2836(71)90105-7. [DOI] [PubMed] [Google Scholar]




