Abstract
Chronic inflammatory conditions, such as in autoimmune disease, can disturb immune cell homeostasis and induce the expansion of normally rare cell populations. In our analysis of various murine models of lupus, we detect increased frequency of an uncommon subset identified as NK1.1+CD11c+CD122+MHC-II+. These cells share characteristics with the NK cell lineage and with cells previously described as IKDCs: (1) they depend on IL15 and express E4BP4; (2) they are cytotoxic and produce type I and type II interferon upon activation; (3) they are efficient antigen presenting cells both through MHC-II expression and in cross-presentation to CD8s. These atypical NK cells are responsive to TLR stimulation and thus are most abundant in mice with high copy number of the Tlr7 gene. They are highly proliferative as assessed by in vivo BrdU incorporation. In adoptive transfer experiments they persist in high numbers for months and maintain their surface marker profile, indicating that this population is developmentally stable. Gene expression analyses on both mRNA and microRNAs show a modified cell cycle program in which various miR15/16 family members are upregulated, presumably as a consequence of the proliferative signal mediated by the increased level of growth factors, Ras and E2F activity. On the other hand, low expression of miR150, miR181 and miR744 in these cells implies a reduction in their differentiation capacity. These results suggest that cells of the NK lineage that undergo TLR stimulation might turn on a proliferative program in detriment of their full differentiation into mature NK cells.
1. Introduction
The program of cell differentiation and maturation in the immune system is designed for great plasticity, especially in immature populations (1). In certain cases, an effective immunity requires rapid innate activation and thus cell lineages linked to innate responses are preferentially expanded. For example, TLR engagement of hematopoietic progenitor cells was reported to stimulate innate immune system replacement: TLR signaling drove differentiation of myeloid progenitors, bypassing some normal growth and differentiation requirements, and also drove lymphoid progenitors to become dendritic cells (2). As chronic and spontaneous TLR activation has been linked to autoimmunity (3), it is possible to assume that immune cell lineage differentiation is disrupted in autoimmune conditions.
In the context of autoimmune disease, and shown in various models of Systemic Lupus Erythematosus (SLE), there is accumulating data linking TLRs and the activation of both autoreactive B cells and dendritic cells (4-6). Elevated copy number of Tlr7 leads to spontaneous activation of this innate pathway and consequent pathology, as illustrated by the aggravation of disease in lupus-prone mice with the Yaa mutation where Tlr7 is duplicated (7-9), or the pathology developed in transgenic mice containing multiple copies of the endogenous Tlr7 gene (TLR7tg) (10). While genetic and mouse model studies show a clear link between spontaneous TLR7 activation and lupus-like pathologies, there is less certainty as to which cells are most sensitive to TLR7’s endogenous ligands and thus mediate this effect in vivo. TLR7 preferentially and intrinsically activates germinal center B cells and plasmablasts, but it does so in a T cell-dependent manner (11). Within the innate cell lineage populations, plasmacytoid dendritic cells (pDCs) have an important role in the initiation of SLE (12) and are very sensitive to TLR7 stimulation. In the context of tumor immunology and chronic stimulation, TLR7 stimulation in pDCs was reported to transform them into an efficient cell killing population. Application of a TLR7 ligand was shown to convert DCs into cells with cytotoxic capacity in patients with basal cell carcinoma (13). In mice, repeated injections of a TLR7 ligand leads to CCL2-dependent recruitment of pDCs and their transformation into a subset of killer DCs able to directly eliminate tumor cells through TRAIL expression and granzyme B secretion (14) Initial studies described the newly identified TLR7-sensitive killer cell subset as interferon-producing killer dendritic cells or IKDCs. These cells were originally categorized as DCs because of their potential to produce type I and type II interferon and their antigen-presenting activity (15, 16). Subsequent studies indicated that IKDCs belong to the NK-cell lineage because they are absent in Rag2 −/− Il2rg −/− mice and require IL-15 just as cells of the NK lineage do (17-19). A more recent assessment redefines IKDCs as intermediate precursors to mature NK cells (20, 21).
As IKDCs were reported to arise in the context of chronic TLR7 stimulation, and because TLR7 plays a critical role in a variety of models of SLE (22-25), it is possible that this type of cells appear in autoimmune conditions and might be a factor in the development of disease. In this manuscript we thoroughly characterize a cell population of atypical NK cells that expand in murine models of SLE and share functional characteristics with IKDCs. These cells lack some mature NK cell surface markers but they express NK1.1, CD122 and several NK-restricted transcription factors, therefore they most likely arise from activation of an immature precursor. Here we show that TLR activation of NK cells precursors induces considerable alterations of both microRNA and messenger RNA expression patterns that could explain the expansion of immature populations under chronic innate activation conditions.
Material and Methods
Mice and experimental protocols
The generation of TLR7tg and B6.FcγRIIB−/− mice has been described earlier (8, 10). All experiments in this manuscript use the transgenic line 7.1 that harbors 8-16 copies of the TLR7gene. All other C57BL/6, IL15−/− and B6.ptpa were obtained from the NIAID mouse colony at Taconic Farms. B6.FcγRIIB−/− were crossed to B6yaa mice to obtain B6.FcγRIIB−/−yaa. Housing at the NIH facility met the instructional animal care and use committee (IACUC) and NIH guidelines.
Mice were used at 8-12 weeks of age, and were age and sex matched within experiments. 12-wk-old C57BL/6 received interperitoneal injection every other day for 2 weeks as follow 50µg, 100µg and 200µg of imiquimod or R848 (Invivogen, CA).
In vivo assays for BrdU (Bromodeoxyuridine or 5-bromo-2’-deoxyuridine) incorporation by proliferating cells used the BrdU Staining Kit from eBioscience (San Diego, CA), used according to the manufacture’s protocol. In brief, mice were fed with water containing 0.8 mg ml−1 BrdU for 14 days. Incorporated BrdU was detected by flow cytometric analysis, after permeabilization and staining with anti-BrdU FITC antibody.
Flow cytometry
Spleen, liver and lymph nodes prepared to single-cells suspension were stained with various fluorochrome-conjugated antibodies purchased from eBioscience (San Diego, CA), and BD Biosciences (San Jose, CA). Fc block (CD16/32) was added to prevent unspecific binding. Fluorescence-activated cell sorting (FACS) analysis was done on FACS Calibur and LSRII (BD Bioscience) machine and data were analyzed using Flowjo software.
Bone marrow chimeras
Bone marrow cells were isolated from femurs and tibias of TLR7.1tg or B6.ptpa mice aged 6-7 weeks and then mixed in equal proportions for the transfer. 8-10 week old C57BL/6 recipient mice were irradiated with 940 k-rads 24h prior the transfer. 1×107 Bone marrow cells were injected retro-orbitally into each recipient. All mice, donors and recipients, were female. Blood was drawn one month after transfer, and cellular fractions were stained with antibodies to CD3 and B220 (BD Biosciences, San Jose CA) to confirm reconstitution. Reconstituted mice were followed up for a total of 3 months.
Antigen presentation assays
The antigen presenting cells, ie. splenocytes or NK1.1+ purified from spleens of WT /TLR7tg, were irradiated with 4000 rads to prevent their proliferation and 2×105 were incubated for 72h with OVA-specific T cells (CD4+ selection of OTII splenocytes by beads from Stemcell Technologies) in presence of different concentration of OVA(323-339) peptide (AnaSpec, Inc. CA). CD4+ were labeled with CFSE (Molecular probe Inc. Invitrogen, V12883) before incubation so that T cell proliferation could be measured by CFSE dilution in flow cytometry. The procedure for cross-presentation of cell associated antigen was as described earlier (26). Briefly, OVA-specific CD8 T cells were isolated form spleens of OT-I mice using a RoboSep CD8 positive selection kit (Stemcell Technologies) and were labeled with CFSE (Molecular probe Inc. Invitrogen, V12883). They were then cultured with 2×105 OVA-albumin-coated irradiated H-2bm1 splenocytes or NK1.1+ cells purified from either WT or TLR7tg spleens. Proliferation was quantified 60-65 hours later by calculating the number of CD8 cells with reduced green fluorescence by flow cytometry.
Cytotoxic responses
YAC-1 cells (susceptible to NK cytotoxic activity) and reference cell line EL4 were labeled with 1 or 5 CFSE µm respectively. NK1.1+ isolated from WT or TLR7tg spleens were incubated for 20h with these two cell lines at different ratios between effector and target cells. The change is ratio between CFSE hi and CFSE lo cells was determined by flow cytometry and interpreted as cytotoxic activity relative to background apoptosis of cells. Additionally, cytotoxic activity was measured by caspase activity in live cells by using CyToxiLux PLUS (OncoImmunin, Inc.) according to the manufacturer’s instructions.
Adoptive transfer
Transferred NK1.1+CD11c+ cells were sorted by FACSAria (BD Bioscience) or RoboSep (Stemcell Technology). Transferred NK1.1+ cells were purified by a combination of CD4-negative / NK1.1 and CD11c-positive bead selection (RoboSep, Stemcell Technologies) from cell suspension depleted of CD4 cells by CD4-positive-selection kit (Stemcell Technologies). 3-5×106 cells were injected i.v. per mouse. Recipients were untouched WT mice.
Genotyping and real-time PCR
For genotyping IL15−/− mice we used following primers: neo GAA TGG GCT GAC CGC TTC CTC G, downstream TCA TAT CCT CTG CAC CTT GAC TG, upstream GAG GGC TAA ATC TGA TGC GTG TG, exon 3’ GAG CTG GCT ATG GCG ATG GGC. Quantitative PCR on genomic DNA and cDNA were used to measure levels of following genes: TLR7 as described previously (6), mRNA level of samples were normalize to actin. For isolation of mRNA, purified cells were resuspended in Trizol (Invitrogen, Carlsbad CA) and incubated for 5 min. RNA was then extracted using chloroform and precipitated with ethanol. RNA pellets were washed and resuspended in nuclease-free water, and integrity was tested by using Experion RNA StdSens Electrophoresis machine. cDNA were synthesized using iScript cDNA synthesis kit according to the manufacturer's instructions (Bio-Rad Laboratories, Hercules, CA).
Transmission electron microscopy
Cells were sorted using NK1.1-PE, CD11c-APC and PDCA-1-FITC antibodies and samples were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate/0.05 M sucrose buffer. The samples were post-fixed with 0.5% osmium tetroxide-0.8% potassium ferricyanide followed by 1% tannic acid and stained overnight at 4 °C en bloc in 1% uranyl acetate. Samples were dehydrated in a graded ethanol series and embedded in Spurr’s resin. Thin sections were cut with an RMC MT-7000 ultramicrotome (Ventana, Tucson, AZ), stained with 1% uranyl acetate and Reynold’s lead citrate prior to being observed at 120 kV on a Tecnai BT Spirit transmission electron microscope (FEI, Hillsboro, OR). Digital images were acquired with a Hammamatsu XR-100 side mount digital camera system (Advanced Microscopy Techniques, Chazy, NY).
Immunohistochemistry
Spleens were obtained from WT and TLR7tg mice, frozen in Tissue Freezing Medium (Triangle Biomedical Sciences, Durham, NC) on dry ice, and stored at −80 ºC until sectioning. Organs were sectioned at 5–10 µm using a Leica 1850 cryostat (Leica, Wetzlar, Germany) and stored at −20 °C until used. Before staining, sections were allowed to equilibrate to room temperature, fixed in cold acetone, and rehydrated in PBS. To reduce nonspecific staining 10% horse serum were applied for 20 min to all sections before addition of Abs. Slides were stained with primary antibodies for 30 min. and examined using Zeizz LSM 710 microscope and data were acquired using Zen2009 software.
Microarray and microRNA analysis
For microarrays, RNA was purified from splenic NK1.1+ cells isolated from WT and TLR7tg mice as described above. The Ambion® Illumina® TotalPrep™ RNA Amplification Kit (Ambion, Inc, Austin, TX) was used according to manufacturer instructions and as previously described (27) to prepare biotinylated cRNA from isolated total RNA. Input was normalized to 500 ng per sample. Labelled cRNA is then used for hybridization to Illumina’s MouseWG-6 per manufacturer’s instructions. BeadChips were imaged on an Illumina HiScan-SQ scanner. Microarray target preparation and BeadChip hybridization was performed by the Genomic Technologies Section, Research Technologies Branch, NIAID. Microarray data was submitted to the GEO repository (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63829). Initially, a splenic NK sample from a single B6 WT mouse and splenic NK1.1+ sample from a sample TLR7tg mouse were each probed in duplicate on a single MouseWG-6 array (number 7011). Later, a second MouseWG-6 array (number 4020) was probed with samples from two B6 WT mice and two TLR7tg mice. The two technical replicates from the first array were averaged together to allow the two unique biological samples from the first array to be analyzed with the four biological replicates from the second array.
For the miRNA expression analysis, total RNA was isolated from cell sorted NK1.1+CD11c+PDCA-1− and conventional NKs (NK1.1+CD11c−) using MiRNeasy Mini Kit (Qiagen). The quality of RNA was assessed by the Experion system (Bio-Rad), and only RNAs with RNA quality Indicator (RQI) over 9 were used. cDNA was generated using the miScript II RT Kit (Qiagen) and reverse-transcription reaction with SYBR Green (Qiagen). MiRNAs expression profiles were detected using the mouse miFinder and Inflammatory Response & Autoimmunity miScript miRNA PCR Arrays (Qiagen). Data was analyzed further with SA Bioscience microarray data analysis software.
Statistical analysis
Illumina microarray data were log2-transformed and normalized for each array per the instructions from Illumina by NIAID Research Technologies Branch. All subsequent analyses were computed using the R Project for Statistical Computing (http://www.R-project.org) (28). Microarray data were inspected for possible failed chips using density plots, boxplots and principal components analysis (PCA) plots. Even after initial platform-specific normalization at NIAID Research Technologies Branch, there were still systematic differences in the distribution of expression values between the samples from the first array (7011) with two technical replicates (Ave 7011A+7011C B6 and Ave 7011D+7011E Tg) and the samples from the second array (4020) with no technical replicates (4020A, 4020B, 4020D and 4020F). Quantile normalization was applied to the six samples to eliminate these differences. Gene probe sets with log2 expression standard deviation ≤ 0.2 among all samples were removed to eliminate low information genes prior to statistical testing. Differential expression was tested using a pairwise empirical Bayes modified T-test from the limma R package library (29). A probe set was considered to be differentially expressed if its false discovery rate (FDR) adjusted p-value was less than 0.05 and its absolute fold change was greater than 2. Any statistically significant differentially expressed genes were imported into Ingenuity Pathways Analysis (IPA) to identify any enriched pathway groups among the significant genes.
Micro-RNA analyses were computed using Qiagen software and p-values are reported from unpaired Student’s T-tests. All other statistical tests were computed using GraphPad Prism, with p-values reported from one-way ANOVA with Dunnett’s Multiple Comparisons against Control (MCC) adjustments for multiple comparisons.
Results
NK1.1+ CD11c+ cells expand in autoimmune conditions
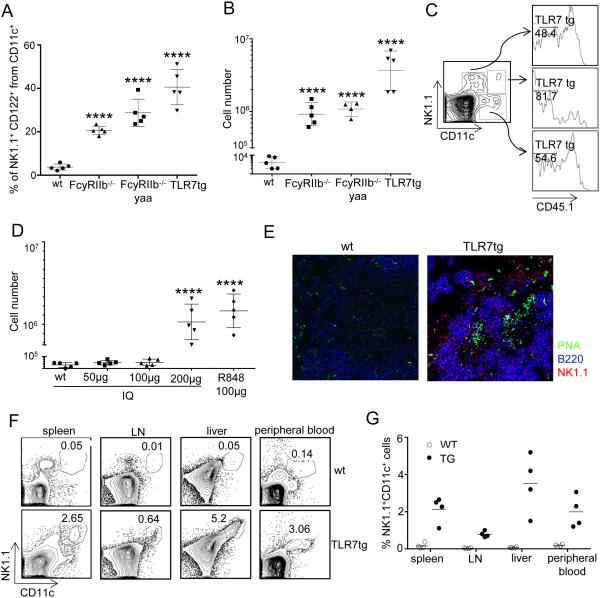
During our characterization of dendritic cell populations associated with disease in various mouse models of SLE, we found that a sizable number of CD11c+ cells in the spleen also expressed NK1.1 and CD122, a combination of surface markers normally expressed by NK cells (Fig 1A). The mouse strains we selected represent a varied range of autoimmune pathology: (1) FcγRIIB−/− mice develop moderate lupus-like disease with 8 months median survival (8), (2) FcγRIIB−/−.Yaa mice succumb to severe disease by 5 months of age (8, 30) and (3) TLR7tg mice, containing about 10 copies of the endogenous Tlr7 gene (maintaining the expression profile), suffer from autoimmune and inflammatory pathology that is lethal by 4 months of age (10). We found that within the WT CD11c+ splenocytes, less than 5% are also NK1.1+, making the entire splenic population of CD11c+NK1.1+ cells less than 2×104 cells in WT mice (Figure 1B). In contrast, in SLE-prone mice the percentage of NK1.1+ within the CD11c population increases considerably to 20% in FcγRIIB−/−, 30% in FcγRIIB−/−.Yaa and 40% in TLR7tg mice (Figure 1A). As a whole, spleens from 6 month-old FcγRIIB−/− mice or 3 month-old FcγRIIB−/−.Yaa mice contained on average 1×106 of these cells (Figure 1B). TLR7tg mice of 2-3 months of age had the highest number, with an average of 4×106 NK1.1+CD11c+ cells in the spleen. In fact, over 90% of NK1.1+ cells originated in TLR7tg were also CD11c+CD122+ (not shown).
Figure 1. TLR7-induced expansion of NK1.1+CD11c+ cells in lupus-prone mice.
(A) Representative percentages of NK1.1+ CD122+within CD11c+ splenocytes in mice of the indicated genotype. (B) Number of NK1.1+CD11c+CD122+ cells per spleen. Data are representative of tree independent experiments. (C) Flow cytometry of splenotyes from WT mice that were irradiated and reconstituted with an equal mix of BM from B6.ptpa/b (CD45.1+/2+) and TLR7tg (CD45.2+/2+) mice. Analysis was performed two months after the BM transfer and it is representative of 3 experiments. Numbers represent the percentage of CD45.1− cells. (D) Number of NK1.1+CD11c+CD122+ splenocytes in WT mice injected every other day for 2 weeks with Imiquimod or R848. One-way ANOVA with Dunnett’s Multiple Comparisons against Control (MCC) adjustments for multiple comparisons, ****p<0.0001. (E) Immunostaining of spleen sections from WT and TLR7tg mice showing the distribution of NK1.1+ cells using the antibodies anti-NK1.1-PE (red), anti-B220-APC (blue) and anti-PNA-FITC (green). Data are representative of four independent experiments. (F) Representative flow cytometric analysis and (G) percentage of splenic NK1.1+CD11c+ cells in different organs.
Given that NK1.1+CD11c+ cells were found to be most abundant in mice with increased expression of TLR7, we considered that this cell population might be especially sensitive to TLR7 stimulation. To determine whether the NK1.1+CD11c+ cells were intrinsically responding to increased TLR7 activity, we generated bone marrow chimeras using an equal mixture of WT (CD45.1+/CD45.2+) and TLR7tg (CD45.2+) bone marrow (BM) to reconstitute an irradiated WT host. Two months after reconstitution, NK1.1+CD11c+ cells were four times more likely to be derived from TLR7tg BM than from WT cells (81.7% CD45.1− vs 19.3% CD45.1+). In contrast, conventional dendritic cells (CD11c+NK1.1−) or NK cells (NK1.1+CD11c−) were equally likely to originate from TLR7tg or from WT BM (Figure 1C). Thus, the CD11c+NK1.1+ population expands as a consequence of the higher expression of TLR7 within these cells, which presumably leads to continued activation of this innate pathway. To test whether chronic activation of TLR7 was enough for the expansion of NK1.1+CD11c+ cells in the spleen, we repeatedly injected WT mice with a TLR7 ligand: we used Imiquimod (IQ) at doses ranging from 50 to 200 µg or Resimiquimod (R848) at 100 µg. We observed that twice weekly IP injections of a TLR7 ligand for three weeks significantly increased the number of NK1.1+CD11c+ cells up to 1.5 million per spleen in WT mice (Fig 1D).
Next we studied the in situ distribution of NK1.1+CD11c+ cells in the spleen by confocal microscopy. We used fluorescent antibodies against the NK1.1 cell surface marker, in addition to B220 antibodies to detect splenic B cells, and PNA for germinal center identification. As shown in Figure 1E, NK1.1 staining is greatly amplified in TLR7tg spleens compared to WT spleens. Conventional NK cells in WT spleens are scarce and located in the red pulp; NK1.1 staining on TLR7tg spleen shows cells primarily in the red pulp but also in T cell areas surrounding germinal centers. Since the majority of NK1.1+ cells in TLR7tg are NK1.1+CD11c+ CD122+, we infer that this particular cell population resides primarily in the red pulp but also spreads into the T cell areas. Moreover, CD11c+NK1.1+ cells were found in organs beyond the spleen: they represented 2-4% of splenocytes, 1% of lymph node cells, 2-3% of peripheral blood and 5-6% of liver-associated immune cells in TLR7tg mice of 2-3 months of age (Figure 1F and G).
NK1.1+CD11c+ cells from TLR7tg mice share features of the NK lineage
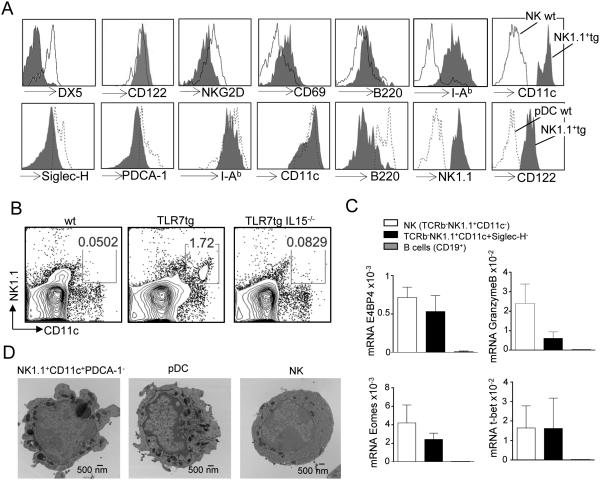
A flow cytometric analysis showed that, compared to conventional NK cells, TLR7tg-derived NK1.1+CD11c+ cells expressed no DX5, very low B220, low NKG2D and high MHC-II surface expression, but comparable levels of CD122 (Figure 2A top). Compared to WT pDCs, TLR7tg-derived NK1.1+CD11c+ cells expressed similar levels of MHC-II and CD11c, very low B220, and lacked Siglec-H and PDCA-1, two hallmark markers for pDCs (Figure 2A bottom). The lack of DX5 in the CD11c+NK1.1+ cells rules out a mature NK cell population, however expression of NK1.1 and CD122 is usually restricted to the NK lineage (31, 32). We then determined whether these cells required IL15 for development, as this cytokine is essential for cells of the NK lineage but dispensable for pDCs (33). We found that TLR7tg mice that lacked IL15 had negligible levels of CD11c+NK1.1+ cells by flow cytometry (Figure 2B). We further confirmed the NK cell lineage of CD11c+NK1.1+ cells by detecting the NK-cell specific transcription factor E4BP4, as well as high levels of Tbet and intermediate levels of Eomes and Granzyme B (Figure 2C). Overall our analysis shows that these cells belong to the NK cell lineage and that their surface expression pattern is reminiscent to the one reported for IKDCs (21), although these cells seem to express higher levels of MHC-II and lower levels of B220 compared to IKDCs.
Figure 2. Characterization of NK1.1+CD11c+ cells in comparison to NKs and pDCs.
(A) Surface marker analysis of TLR7tg NK1.1+CD11c+ cells (grey histogram above and below) compared to WT NK cells (open histogram above) and compared to WT pDCs (doted histogram below). (B) Flow cytometric determination of the number of splenic NK1.1+CD11c+ cells in mice of the genotype shown, at least three mice per group. (C) Expression of E4BP4, Granzyme B, T-bet and Eomes determined by qPCR in cell sorted WT NK cells (cell sorted TCRb−NK1.1+CD11c−), TLR7tg NK (TCRb−NK1.1+CD11c+Siglec-H−) and WT B cells (CD19+). Expression was calculated relative to actin. (D) Representative morphology of sorted splenic pDCs (NK1.1−CD11c+PDCA-1+), TLR7tg NK (NK1.1+CD11c+PDCA-1−) and conventional NKs (NK1.1+CD11c−) using transmission electron microscopy (Tecnai BT Spirit transmission electron microscope FEI, Hillsboro, OR); Bar 500nm; Direct mag. 13000x.
The overall morphology of CD11c+NK1.1+ cells, determined by electron microscopy, was distinct from conventional pDCs or conventional NK cells: they presented small size, high nuclear/cytoplasmic ratio, round shape, smooth plasma membrane with short pseudopodia, and abundant and large electron-dense granules in the cytoplasm (Figure 2D). Overall, these cells had a general lymphoid morphology and a vesicular-rich cytoplasm.
Multiple functionalities of lupus-associated atypical NKs
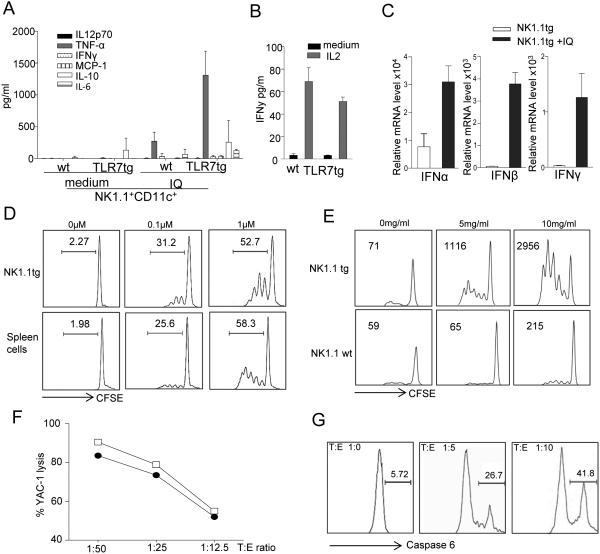
Given that we found atypical NK cells in high numbers associated to lupus models, we then investigated whether their functionalities might make these cells potentially pathogenic in autoimmune conditions. We first evaluated the ability of a highly purified NK1.1+CD11c+ population to produce cytokines upon stimulation. Single-cell sorted (FACSAria) NK1.1+CD11c+ cells from TLR7tg spleens produced TNFα, IL10 and IL6 when stimulated with the TLR7 agonist imiquimod (Fig 3A). Additionally, purified TLR7tg NK1.1+ cells produced IFNγ in response to IL2 (Fig. 3B) as well as IFNα/β and IFNγ upon TLR7 stimulation (Fig. 3C).
Figure 3. Functional properties of NK1.1+CD11c+ cells from TLR7tg.
(A) Secretion of inflammatory cytokines from cell sorted NK1.1+CD11c+PDCA− splenocytes isolated from wild type or TLR7 tg mice. Cells sorted to 99% purity were cultured with Imiquimod for 24h and supernatants were tested for the presence of inflammatory cytokines using the BD Biosciences CBA kit or (B) cultured with IL2 for 48h and supernatants were tested for secretion of IFNγ using the CBA kit; (C) IFNα, IFNβ or IFNγ mRNA was measured by RT-PCR in cell extracts of samples treated for 24h with Imiquimod. Actin mRNA levels were used for normalization.
(D) MHC-II dependent antigen presentation by WT or TLR7tg NK1.1+ cells. Purified splenic NK1.1+ cells from the indicated genotype were loaded with OVA(323-339) peptide and cultured with CFSE-labeled CD4+ OT-II cells as specified in Methods. Numbers in each graph represent the percentage of divided cells. Data shown representative of three separate experiments. (E) Cross-presentation of cell-associated antigen by WT or TLR7tg NK1.1+ cells. CFSE-labeled OT-I CD8 cells were cultured with irradiated OVA-coated B6.H2bm1splenocytes, in the presence of splenic NK1.1+ cells from WT or TLR7tg mice. CD8+ proliferation was quantified 60 hours later by calculating the number of cells with CFSE-reduced levels after division, shown as numbers within each graph. Data shown representative of three separate experiments; (F) Cytotoxic activity of IL2-pretreated NK1.1+ cells isolated from TLR7tg mice (open square) or WT (closed circle) was determined by incubating them with a mixture of CFSEhi YAC-1 cells (susceptible to NK killing) and CFSElo EL-4 cells (resistant to natural killing) at the various target: effector ratios and cytotoxic activity was measured either by the ratio of CFSEhi versus CFSElo or (G) caspase activation through the CyToxiLux PLUS method (OncoImmunin, Inc.). Numbers in each gate represent percentage of apoptotic target cells.
Since we observed that these atypical NK cells from TLR7tg spleens expressed high levels of MHC-II, we tested their ability to process and present antigen to CD4 cells. TLR7tg NK1.1+ cells were as efficient as whole splenocytes at presenting OVA peptide at two differect concentrations (Fig. 3D) to OT-II CD4+ cells. We also tested these cells’ ability to cross-pressent antigen to CD8s, as this has been shown to be one important aspect described for IKDCs (33). We confirmed this function in TLR7tg-derived NK1.1+ cells using cell-associated antigen on class I-mismatched APCs (Figure 3E). NK1.1+ cells purified from WT or TLR7tg spleens were incubated with CFSC-labeled OT-I CD8+ cells as well as with 2×105 OVA-albumin-coated irradiated H-2bm1 splenocytes. H-2bm1 APCs cannot present OVA to OT-I cells due to the bm1 mutation. Thus, any proliferation of CD8 cells in this experiment comes as a result of capture of antigen by the APC harboring the unmutated H-2b and its re-routing through the MHC-I pathway. As shown in Figure 3E, NK1.1+ cells from TLR7tg very efficiently cross-present antigen to CD8 cells.
Finally, we determined that TLR7tg-derived NK1.1+ CD11c+ cells pretreated with IL2 have quite efficient cytotoxic activity compared to conventional NK cells. This test was performed in two separate systems: using the YAC-1 lysis assay (Fig. 3F) and by the Caspase 6 assay (Fig. 3G).
NK1.1+CD11c+ cells from TLR7tg are highly replicative and developmentally stable
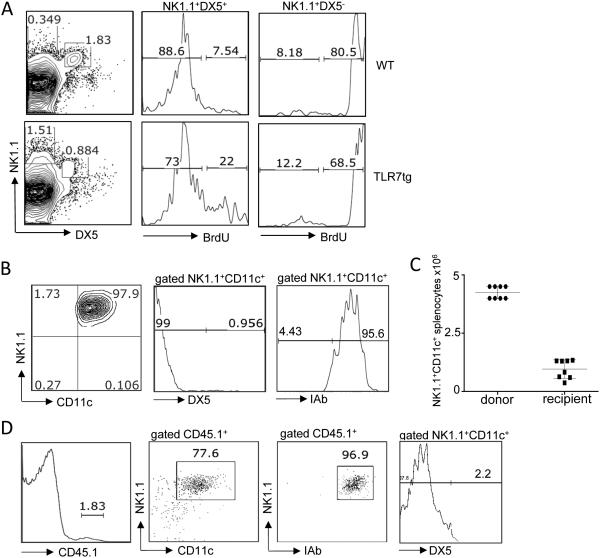
As NK1.1+CD11c+ cells appear in great numbers in TLR7tg spleens, we sought to determine whether they are expanding and actively dividing. We fed BrdU to TLR7tg mice for 14 days and tested BrdU incorporation in various splenic populations. As shown in Figure 4A, the majority of CD11c+NK1.1+ cells, also DX5−, were positive for BrdU compared to less than one fourth BrdU+ cells within the DX5+NK1.1+ conventional NK cells, independent of their transgenic or WT origin. To determine their survival rate after adoptive transfer, we sorted CD11c+NK1.1+ cells from TLR7tg spleens and injected 3-5 million IP into WT mice. Sorted cells were of 98% purity and were almost exclusively DX5− and MHC-II+ (Figure 4B). After the transfer, donor cells could be identified as CD45.1+ while cells from recipient mice were CD45.1−. Four weeks later, donor CD11c+NK1.1+ cells represented on average 2% of the spleen, equivalent to one million cells, in reconstituted mice (Figure 4C). This is a remarkable 30% recovery rate of the total number of cells injected. The surface marker expression in donor cells was maintained as NK1.1+DX5−MHC-II+, although a few of the donor cells had lost their CD11c expression (Figure 4D). Overall, the CD11c+NK1.1+ cells that developed in TLR7tg demonstrated an elevated in vivo proliferation rate and an exceptional capacity for survival.
Figure 4. Proliferative nature and immature phenotype of NK1.1+ cells isolated from TLR7tg mice.
(A) Flow cytometric analysis of BrdU incorporation after 2 weeks of BrdU drinking water in conventional NK cells (NK1.1+DX5+) and atypical/immature NK cells (NK1.1+DX5−) from WT or TLR7tg spleens. Data were from two experiments with three mice per group. (B) Flow cytometric analysis of splenic cells purified from recipient mice 1 month after injection NK1.1+ cells purified from TLR7tg. ptpa/b spleens. Splenocytes were gated for CD45.1 positive expression and analyzed for MHC II (I-Ab), NK1.1, and CD11c and DX5 expression as shown. Data are from three experiments with three mice per group.
Activated expression pattern in NK1.1+CD11c+ cells
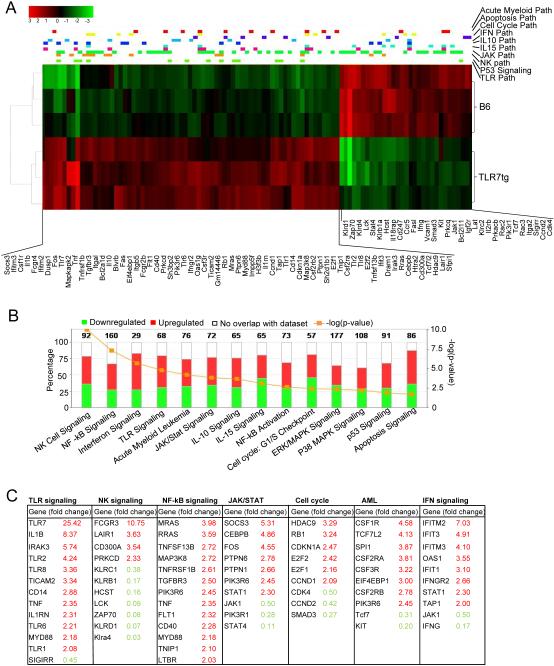
We characterized the gene expression profile of NK1.1+CD11c+ cells isolated from TLR7tg mice and compared them to conventional NK1.1+ cells. Data was acquired from an Illumina whole genome microarray using 3 samples per genotype. The complete array featured 45,281 probe sets representing 21,563 unique genes. After transformation, normalization and quality control assessments, we removed any probe set with log2-expression standard deviation ≤ 0.2 among all 6 samples, leaving 14,762 probes representing 9,581 unique genes. We used empirical Bayes pairwise T-tests to identify 1,490 probe sets with statistically significant differences among the two cell populations (FDR < 0.05 and absolute fold change > 2), the 97 with best scores shown in the heatmap of Figure 5A. We narrowed this list of probe sets down further to identify 964 probe sets with FDR ≤ 0.00003 and absolute fold change > 2) for use in Ingenuity Pathway Analysis (IPA) software. IPA identified major molecular pathways and group patterns differentially expressed between the two experimental groups. The graph in Figure 5B shows the number of genes in each of 14 significantly enriched pathways. The most statistically significant (p = 10e-10) enriched pathway was NK cell signaling, with 92 genes differentially expressed (those with more than two-fold difference shown in Figure 5C). Genes that are normally expressed in mature NK cells but not in NK cell precursors (KlLRC1, KlLRB1, KLRD1, KLRA4, HCST, LCK, ZAP70) were significantly down regulated in NK1.1+CD11c+ cells compared to conventional NK cells (Figure 5C). In contrast, the inhibitory receptors LAIR1 and CD300A were upregulated, as well as FcγRIII, perhaps as a sign of cell activation.
Figure 5. Gene expression profile of NK1.1+CD11c+ cells from TLR7tg.
(A) Heatmap analysis of the gene expression comparison between conventional NK cells isolated from WT mice and NK1.1+ cells isolated from TLR7tg mice. Using 3 samples per group, the identified genes that showed statistically significant differences (FDR<0.05) and absolute fold change >2. (B) genes identified in the heatmap analysis were filetered slightly further with FDR<0.005 yielding 959 genes for pathway analysis. (C) List of genes whose expression was at least 2-fold different in NK1.1+CD11c+ samples compared to conventional NK cells.
NK1.1+CD11c+ cells also differentially expressed a number of TLR signaling genes compared with conventional NK cells: they expressed high levels of TLR1, TLR6 plus the signaling adaptors MYD88, IRAK3, CD14 (Figures 5B and C). TLR7 was highly upregulated, even more than expected from the expression of the transgene. On the other hand, the inhibitory receptor SIGIRR, which has TLR regulatory function, was down regulated in the same cells. Overall this pattern of expression would make NK1.1+CD11c+ cells highly sensitive to innate stimulation. Moreover, signaling pathways downstream of TLR or interferon were also significantly different in NK1.1+CD11c+ cells compared to conventional NK cells, with a pattern of activation in NF-kB, and cytokine/Jak/Stat signaling genes. Interferon inducible genes (several IFIT- genes, OAS1, STAT1) are all highly expressed in NK1.1+CD11c+, as expected from cells that have chronic TLR stimulation. Uniquely among cytokine signaling genes, STAT4 was downregulated in NK1.1+CD11c+ cells (Figure 5C). STAT4 is an adaptor molecule important for NK cell activity and its expression is high in mature NK cells compared to precursor NK cells (35).
A recurrent pattern in the groups selected by the IPA software in this analysis is the differential expression of cell cycle related genes, and those expressed in oncogenic transformation. Cyclins and cell cycle regulators RB1, CDKN1A, E2F, and CCND1 were upregulated while CDK4, CCND2 were downregulated in NK1.1+CD11c+ cells compared to conventional NK cells (Figures 5A, B and C). Finally, a number of colony stimulating factor receptors (CSF-Rs) showed high expression in NK1.1+CD11c+ cells and might account for the rapid growth in vivo of these cells.
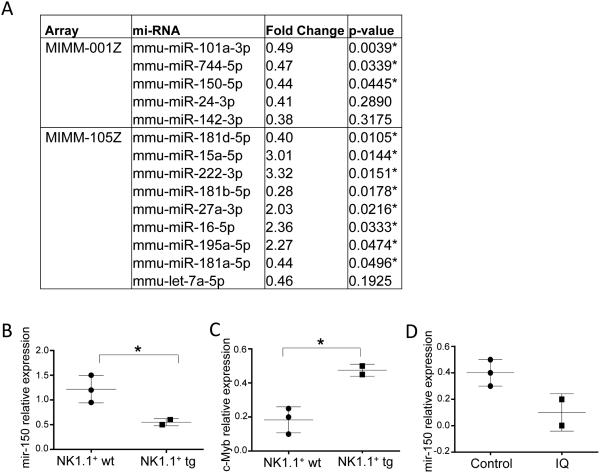
miRNA expression patterns in NK1.1+CD11c+ cells suggest a change in maturation potential and cell cycle programing
We characterized micro RNA (miRNA) expression differences in NK1.1+CD11c+ cells compared to conventional NK cells so that we could identify gene regulatory patterns in the newly characterized cell population. We tested our samples in two separate arrays: one miRNA PCR array that contained 84 miRNAs abundantly expressed in immune cells (Mouse miFinder miRNA PCR Array, MIMM-1001Z, Qiagen) and another array that contained 84 miRNAs linked to autoimmunity and inflammation (Mouse Inflammatory Response & Autoimmunity miRNA PCR Array, MIMM-105Z,Qiagen). We used these two arrays to compare miRNAs prepared from NK1.1+CD11c+ TLR7tg cells with conventional NK cells sorted as NK1.1+CD11c− cells. Figure 6A shows the list of microRNAs that were expressed at detectable levels in all samples. Among the miRNAs with significantly lower expression in NK1.1+CD11c+ cells compared to conventional NK cells, we found two that have been reported to be important for NK cell maturation in mice (miR-150) and in human (miR-181) (36, 37). We validated the differences on miR-150 expression by real time PCR (Figure 6B). A well-characterized target of miRNA150 is the Myb gene, which is highly expressed in lymphoid progenitors and downregulated upon maturation (38). We confirmed using RT-RCP that the c-Myb transcript was upregulated in TLR7tg NK1.1+CD11c+ cells compared to WT NK cells (Figure 6C). Importantly, miR-150 levels are directly regulated by TLR7, because the addition of Imiquimod to NK1.1+CD11c+ cells resulted in further downregulation of mi-150 expression (Figure 6D).
Figure 6. MiRNA expression in atypical NK cells from TLR7tg compared to conventional NK cells.
(A) WT NK cells sorted as TCRb−NK1.1+CD11c− and TLR7tg NK1.1+ sorted as TCRb−NK1.1+CD11c+SiglecH− were used to prepare miRNAs and differential expression was analyzed with Mouse miFinder (MIMM-001Z) or Inflammatory Response & Autoimmunity (MIMM-105Z) miRNA PCR Arrays (Qiagen). Shown is the list of genes that were different between the two samples. (B) mir-150 differential expression was confirmed by real time PCR on samples prepared as in (A). (C) c-Myb was one of the mir-150 target genes predicted using the TargetScan software, c-Myb mRNA was measured in WT NK cells or TLR7tg NK purified as in (A). (D) Purified NK-ILCs from TLR7tg were incubated with Imiquimod (IQ) or left unstimulated (Control) for 24h. mir-150 expression was measured by RT-PCR relative to actin levels.ntrol) for 24h. Mir-150 expression was measured by RT-PCR relative to U6 levels.
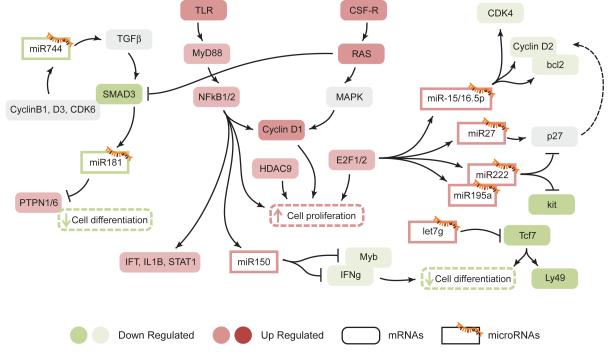
The IPA software provided information on the regulatory network between miRNAs and their targets from the combined analysis of the Illumina array data and the miRNA PCR assay. Figure 7 shows a number of identified connections among differentially regulated genes and miRNAs stemming from the comparison between NK1.1+CD11c+ cells and conventional NK cells. NK1.1+CD11c+ cells had very low expression of miR-744 compared to conventional NK cells (Figure 6A). miR-744 has been linked to tumor growth through regulation of Cyclin B1 expression (39). In the context of the immune response, miR-744 also regulates expression TGFα and its receptor (40). High levels of TGFβR can compensate low expression of the signaling molecule SMAD3 and directly impacts miR-181 expression and cell differentiation through increase expression of phosphatases PTPN1 and PTPN6 (Figure 7) (41).
Figure 7. The regulatory network of miRNAs and their target genes in atypical NK cells.
Representation of expression networks defined by the IPA analysis shown in Figure 5 in combination with the miRNA expression analysis shown in Figure 6. Genes with changes in mRNA expression in Figure 5 are shown as colored rounded boxes while miRNAs differentially expressed from data in Figure 6 are shown as rectangles marked with a miRNA symbol.
Among the microRNAs that showed higher expression in NK1.1+CD11c+ cells relative to conventional NK cells, we found the miR-15/16 family and the related miR27/195/222. All of them are induced by high expression of the transcription factors E2F1 and E2F2 and as a consequence of RAS and NFkB activation in the cells (42). Overall, NK1.1+CD11c+ cells activated by TLRs or by CSF-Rs turn on the transcription of genes involved in proliferation (eg. Cyclin D1, HDAC9 and E2F). Ultimately this program is regulated in feed-back fashion by the expression of miR-15 and miR-16, both of which target the cell cycle regulators CDK4, Cyclin D2 and the apoptotic factor Bcl2. Activation of NFkB also leads to upregulation of Bcl2-like genes such as BCL2A1D, with anti-apoptotic function (Figures 6A, 7) (43). miR222 and miR195a, both with high expression in NK1.1+CD11c+ cells, have been shown to affect the cell cycle by regulating p27 (44). Overall the data is consistent with an activated expression pattern in NK1.1+CD11c+ with concomitant induction of regulatory microRNAs (miR15/16/27/222/195) that set a specific cell cycle transcription program.
Discussion
Activation of innate immune pathways in inflammatory and autoimmune conditions can lead to preferential expansion of cell populations that are especially sensitive to innate triggers. Here we report the expansion of cells of the NK cell lineage with atypical characteristics in the context of mouse models of lupus. We show that either repeated TLR activation, or expression of multiple copies of the Tlr7 gene, induce the expansion of this atypical NK cell population. However these cells also appear in substantial numbers in FcγRIIB-KO, a lupus-prone mouse strain bearing WT levels of TLR7.
Given their multiple functional abilities, ascribing mouse NK1.1+CD11c+ cells to a developmental lineage has been controversial. Our full characterization of these atypical cells, defined as NK1.1+CD11c+CD122+, suggests that they arise from immature NK cells because they require IL15 and express E4BP4 (NFIL3), essential for generation of the NK cell lineage (45-47). Additionally, we demonstrate that these atypical cells are distinct from the DC lineage because they lack expression of the DC-restricted transcription factors Zbtb46 (BTBD4) and Tcf4 (E2-2). Functionally these cells are both cytotoxic and efficient APCs in a way that they resemble the previous described IKDCs (15-21). However the cells we characterize in our lupus models seems to differ slightly in surface expression of some markers (MHC, B220) and importantly, our transfer experiments show that they survive for months and in large numbers upon adoptive transfer and don’t become mature immune cells as they never acquire CD49b or DX5 expression. This difference might be due to specific activation of lupus-associated NK cells, either because of the inflammatory environment or through TLR7-dependent activation. This type of chronic activation might lead to an oncogenic-like transformation of these cells, which would explain the highly proliferative expression profile that we uncovered in our full genome and inflammatory-associated RNA analysis.
Expansion of NK1.1+CD11c+ cells correlates with the inflammatory pathology and inversely correlates with the number of mature NK cells (NKp46+DX5+) in TLR7tg mice (data not shown). This is consistent with the view that activation of immature NK cells through TLR7, combined with an inflammatory environment, thwarts differentiation of these cells into mature NK cells and leads to the acquisition of markers nonspecific for NK cell lineages like MHC class II, and the integrin CD11c. The combination of unusual surface-expressed markers and novel capabilities relative to mature NK cells, combined with their block in maturation, makes this expanded population atypical, perhaps primarily linked to chronic inflammatory conditions. NFκB activation in immature NK cells, either by TLR engagement or through an inflammatory environment, can lead to extended survival and proliferation. Our gene expression analyses showed the expected pattern of NF-kB activation and interferon signature gene expression in NK1.1+CD11c+ cells (Figure 6). These expression analyses also showed pronounced changes in genes related to cell cycle regulation, in a pattern that recalls that of oncogenic transformation and would explain the high survival rate upon adoptive transfer of these cells.
NK cells have been suggested to be both regulatory and inducers of autoimmune disease (48). Several studies showed lower percentages and absolute numbers of NK cells in SLE patients peripheral blood compared with normal individuals (49, 50), and this correlated with their impaired activity and killing capacity (51), a process known as NK-cell degeneration. Even with a decreasing number of NK cells in SLE patients, Schepis et al. reported that the CD56bright subpopulation was increased (52). Interestingly, this population has high potential to produce cytokines, and is located mainly in secondary lymphoid organs and inflamed sites and thus could represent an expanded intermediate as we observe in mice.
Our data shows that chronic TLR7 activation induces the survival and proliferation of atypical NKs in detriment of mature immune cells. This finding highlights the need to reevaluate immune cell lineages in chronic inflammatory conditions, as early precursor cells might proliferate and acquire new functionalities that make them pathogenic and should be taken into consideration in the search for therapeutic targets.
Acknowledgements
The authors thank Bethany Scott for technical assistance with animal care and adoptive transfer experiment; Dr. Elizabeth R. Fischer (NIAID Microscopy Facility) for help with electron microscopy, Timothy Myers (NIAID Microarray Facility) for help with microarray experiment and analysis; Austin Athman (NIAID, Visual & Medical Arts) for help preparing Figure 7; Steven Crampton and Hemanta Kole (LIG/NIAID) for critical review of the manuscript. This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
References
- 1.Galli SJ, Borregaard N, Wynn TA. Phenotypic and functional plasticity of cells of innate immunity: macrophages, mast cells and neutrophils. Nature immunology. 2011;12:1035–1044. doi: 10.1038/ni.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagai Y, Garrett KP, Ohta S, Bahrun U, Kouro T, Akira S, Takatsu K, Kincade PW. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 2006;24:801–812. doi: 10.1016/j.immuni.2006.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green NM, Marshak-Rothstein A. Toll-like receptor driven B cell activation in the induction of systemic autoimmunity. Seminars in immunology. 2011;23:106–112. doi: 10.1016/j.smim.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak-Rothstein A. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 5.Lau CM, Broughton C, Tabor AS, Akira S, Flavell RA, Mamula MJ, Christensen SR, Shlomchik MJ, Viglianti GA, Rifkin IR, Marshak-Rothstein A. RNA-associated autoantigens activate B cells by combined B cell antigen receptor/Toll-like receptor 7 engagement. The Journal of experimental medicine. 2005;202:1171–1177. doi: 10.1084/jem.20050630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pisitkun P, Deane JA, Difilippantonio MJ, Tarasenko T, Satterthwaite AB, Bolland S. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312:1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- 7.Fossati L, Sobel ES, Iwamoto M, Cohen PL, Eisenberg RA, Izui S. The Yaa gene-mediated acceleration of murine lupus: Yaa- T cells from non-autoimmune mice collaborate with Yaa+ B cells to produce lupus autoantibodies in vivo. European journal of immunology. 1995;25:3412–3417. doi: 10.1002/eji.1830251231. [DOI] [PubMed] [Google Scholar]
- 8.Bolland S, Yim YS, Tus K, Wakeland EK, Ravetch JV. Genetic modifiers of systemic lupus erythematosus in FcgammaRIIB(−/−) mice. The Journal of experimental medicine. 2002;195:1167–1174. doi: 10.1084/jem.20020165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Izui S, Higaki M, Morrow D, Merino R. The Y chromosome from autoimmune BXSB/MpJ mice induces a lupus-like syndrome in (NZW × C57BL/6)F1 male mice, but not in C57BL/6 male mice. European journal of immunology. 1988;18:911–915. doi: 10.1002/eji.1830180612. [DOI] [PubMed] [Google Scholar]
- 10.Deane JA, Pisitkun P, Barrett RS, Feigenbaum L, Town T, Ward JM, Flavell RA, Bolland S. Control of toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity. 2007;27:801–810. doi: 10.1016/j.immuni.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh ER, Pisitkun P, Voynova E, Deane JA, Scott BL, Caspi RR, Bolland S. Dual signaling by innate and adaptive immune receptors is required for TLR7-induced B-cell-mediated autoimmunity. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:16276–16281. doi: 10.1073/pnas.1209372109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rowland SL, Riggs JM, Gilfillan S, Bugatti M, Vermi W, Kolbeck R, Unanue ER, Sanjuan MA, Colonna M. Early, transient depletion of plasmacytoid dendritic cells ameliorates autoimmunity in a lupus model. The Journal of experimental medicine. 2014;211:1977–1991. doi: 10.1084/jem.20132620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stary G, Bangert C, Tauber M, Strohal R, Kopp T, Stingl G. Tumoricidal activity of TLR7/8-activated inflammatory dendritic cells. The Journal of experimental medicine. 2007;204:1441–1451. doi: 10.1084/jem.20070021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drobits B, Holcmann M, Amberg N, Swiecki M, Grundtner R, Hammer M, Colonna M, Sibilia M. Imiquimod clears tumors in mice independent of adaptive immunity by converting pDCs into tumor-killing effector cells. The Journal of clinical investigation. 2012;122:575–585. doi: 10.1172/JCI61034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan CW, Crafton E, Fan HN, Flook J, Yoshimura K, Skarica M, Brockstedt D, Dubensky TW, Stins MF, Lanier LL, Pardoll DM, Housseau F. Interferon-producing killer dendritic cells provide a link between innate and adaptive immunity. Nature Medicine. 2006;12:207–213. doi: 10.1038/nm1352. [DOI] [PubMed] [Google Scholar]
- 16.Taieb J, Chaput N, Menard C, Apetoh L, Ullrich E, Bonmort M, Pequignot M, Casares N, Terme M, Flament C, Opolon P, Lecluse Y, Metivier D, Tomasello E, Vivier E, Ghiringhelli F, Martin F, Klatzmann D, Poynard T, Tursz T, Raposo G, Yagita H, Ryffel B, Kroemer G, Zitvogel L. A novel dendritic cell subset involved in tumor immunosurveillance. Nat Med. 2006;12:214–219. doi: 10.1038/nm1356. [DOI] [PubMed] [Google Scholar]
- 17.Blasius AL, Barchet W, Cella M, Colonna M. Development and function of murine B220+CD11c+NK1.1+ cells identify them as a subset of NK cells. The Journal of experimental medicine. 2007;204:2561–2568. doi: 10.1084/jem.20070991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vosshenrich CA, Lesjean-Pottier S, Hasan M, Richard-Le Goff O, Corcuff E, Mandelboim O, Di Santo JP. CD11cloB220+ interferon-producing killer dendritic cells are activated natural killer cells. The Journal of experimental medicine. 2007;204:2569–2578. doi: 10.1084/jem.20071451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caminschi I, Ahmet F, Heger K, Brady J, Nutt SL, Vremec D, Pietersz S, Lahoud MH, Schofield L, Hansen DS, O'Keeffe M, Smyth MJ, Bedoui S, Davey GM, Villadangos JA, Heath WR, Shortman K. Putative IKDCs are functionally and developmentally similar to natural killer cells, but not to dendritic cells. The Journal of experimental medicine. 2007;204:2579–2590. doi: 10.1084/jem.20071351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guimont-Desrochers F, Boucher G, Dong Z, Dupuis M, Veillette A, Lesage S. Redefining interferon-producing killer dendritic cells as a novel intermediate in NK-cell differentiation. Blood. 2012;119:4349–4357. doi: 10.1182/blood-2011-11-395954. [DOI] [PubMed] [Google Scholar]
- 21.Guimont-Desrochers F, Lesage S. Revisiting the Prominent Anti-Tumoral Potential of Pre-mNK Cells. Front Immunol. 2013;4:446. doi: 10.3389/fimmu.2013.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christensen SR, Shupe J, Nickerson K, Kashgarian M, Flavell RA, Shlomchik MJ. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25:417–428. doi: 10.1016/j.immuni.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 23.Berland R, Fernandez L, Kari E, Han JH, Lomakin I, Akira S, Wortis HH, Kearney JF, Ucci AA, Imanishi-Kari T. Toll-like receptor 7-dependent loss of B cell tolerance in pathogenic autoantibody knockin mice. Immunity. 2006;25:429–440. doi: 10.1016/j.immuni.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 24.Lee PY, Kumagai Y, Li Y, Takeuchi O, Yoshida H, Weinstein J, Kellner ES, Nacionales D, Barker T, Kelly-Scumpia K, van Rooijen N, Kumar H, Kawai T, Satoh M, Akira S, Reeves WH. TLR7-dependent and FcgammaR-independent production of type I interferon in experimental mouse lupus. The Journal of experimental medicine. 2008;205:2995–3006. doi: 10.1084/jem.20080462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savarese E, Steinberg C, Pawar RD, Reindl W, Akira S, Anders HJ, Krug A. Requirement of Toll-like receptor 7 for pristane-induced production of autoantibodies and development of murine lupus nephritis. Arthritis and rheumatism. 2008;58:1107–1115. doi: 10.1002/art.23407. [DOI] [PubMed] [Google Scholar]
- 26.Schnorrer P, Behrens GM, Wilson NS, Pooley JL, Smith CM, El-Sukkari D, Davey G, Kupresanin F, Li M, Maraskovsky E, Belz GT, Carbone FR, Shortman K, Heath WR, Villadangos JA. The dominant role of CD8+ dendritic cells in cross-presentation is not dictated by antigen capture. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10729–10734. doi: 10.1073/pnas.0601956103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Team RC. R: A language and environment for statistical computing. 2014.
- 29.Smyth GK. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer; New York: 2005. Limma: linear models for microarray data; pp. 397–420. [Google Scholar]
- 30.Murphy ED, Roths JB. A Y chromosome associated factor in strain BXSB producing accelerated autoimmunity and lymphoproliferation. Arthritis and rheumatism. 1979;22:1188–1194. doi: 10.1002/art.1780221105. [DOI] [PubMed] [Google Scholar]
- 31.Kim S, Iizuka K, Kang HS, Dokun A, French AR, Greco S, Yokoyama WM. In vivo developmental stages in murine natural killer cell maturation. Nature immunology. 2002;3:523–528. doi: 10.1038/ni796. [DOI] [PubMed] [Google Scholar]
- 32.Di Santo JP. Natural killer cell developmental pathways: a question of balance. Annual review of immunology. 2006;24:257–286. doi: 10.1146/annurev.immunol.24.021605.090700. [DOI] [PubMed] [Google Scholar]
- 33.Waldmann TA, Tagaya Y. The multifaceted regulation of interleukin-15 expression and the role of this cytokine in NK cell differentiation and host response to intracellular pathogens. Annual review of immunology. 1999;17:19–49. doi: 10.1146/annurev.immunol.17.1.19. [DOI] [PubMed] [Google Scholar]
- 34.Pletneva M, Fan H, Park JJ, Radojcic V, Jie C, Yu Y, Chan C, Redwood A, Pardoll D, Housseau F. IFN-producing killer dendritic cells are antigen-presenting cells endowed with T-cell cross-priming capacity. Cancer Res. 2009;69:6607–6614. doi: 10.1158/0008-5472.CAN-09-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang KS, Ritz J, Frank DA. IL-2 induces STAT4 activation in primary NK cells and NK cell lines, but not in T cells. Journal of immunology. 1999;162:299–304. [PubMed] [Google Scholar]
- 36.Bezman NA, Chakraborty T, Bender T, Lanier LL. miR-150 regulates the development of NK and iNKT cells. The Journal of experimental medicine. 2011;208:2717–2731. doi: 10.1084/jem.20111386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cichocki F, Felices M, McCullar V, Presnell SR, Al-Attar A, Lutz CT, Miller JS. Cutting edge: microRNA-181 promotes human NK cell development by regulating Notch signaling. Journal of immunology. 2011;187:6171–6175. doi: 10.4049/jimmunol.1100835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 39.Huang V, Place RF, Portnoy V, Wang J, Qi Z, Jia Z, Yu A, Shuman M, Yu J, Li LC. Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic acids research. 2012;40:1695–1707. doi: 10.1093/nar/gkr934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin J, Jenkins RH, Bennagi R, Krupa A, Phillips AO, Bowen T, Fraser DJ. Post-transcriptional regulation of Transforming Growth Factor Beta-1 by microRNA-744. PloS one. 2011;6:e25044. doi: 10.1371/journal.pone.0025044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, Klein LO, Davis MM, Chen CZ. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 42.Bueno MJ, Gomez de Cedron M, Laresgoiti U, Fernandez-Piqueras J, Zubiaga AM, Malumbres M. Multiple E2F-induced microRNAs prevent replicative stress in response to mitogenic signaling. Molecular and cellular biology. 2010;30:2983–2995. doi: 10.1128/MCB.01372-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karin M, Lin A. NF-kappaB at the crossroads of life and death. Nature immunology. 2002;3:221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- 44.le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafre SA, Farace MG, Agami R. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. The EMBO journal. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walzer T, Vivier E. NK cell development: gas matters. Nature immunology. 2006;7:702–704. doi: 10.1038/ni0706-702. [DOI] [PubMed] [Google Scholar]
- 46.Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, Lindsten T, Reiner SL. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36:55–67. doi: 10.1016/j.immuni.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, Coles M, Kioussis D, Brady HJ. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nature immunology. 2009;10:1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 48.Tian Z, Gershwin ME, Zhang C. Regulatory NK cells in autoimmune disease. Journal of autoimmunity. 2012;39:206–215. doi: 10.1016/j.jaut.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 49.Yabuhara A, Yang FC, Nakazawa T, Iwasaki Y, Mori T, Koike K, Kawai H, Komiyama A. A killing defect of natural killer cells as an underlying immunologic abnormality in childhood systemic lupus erythematosus. The Journal of rheumatology. 1996;23:171–177. [PubMed] [Google Scholar]
- 50.Erkeller-Yuksel FM, Lydyard PM, Isenberg DA. Lack of NK cells in lupus patients with renal involvement. Lupus. 1997;6:708–712. doi: 10.1177/096120339700600905. [DOI] [PubMed] [Google Scholar]
- 51.Park YW, Kee SJ, Cho YN, Lee EH, Lee HY, Kim EM, Shin MH, Park JJ, Kim TJ, Lee SS, Yoo DH, Kang HS. Impaired differentiation and cytotoxicity of natural killer cells in systemic lupus erythematosus. Arthritis and rheumatism. 2009;60:1753–1763. doi: 10.1002/art.24556. [DOI] [PubMed] [Google Scholar]
- 52.Schepis D, Gunnarsson I, Eloranta ML, Lampa J, Jacobson SH, Karre K, Berg L. Increased proportion of CD56bright natural killer cells in active and inactive systemic lupus erythematosus. Immunology. 2009;126:140–146. doi: 10.1111/j.1365-2567.2008.02887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]