Abstract
Cell dedifferentiation is an integral component of post-traumatic regeneration in echinoderms. Since dedifferentiated cells become multipotent, we asked if this spontaneous broadening of developmental potential is associated with the action of the same pluripotency factors (known as Yamanaka factors) that were used to induce pluripotency in specialized mammalian cells. In this study, we investigate expression of orthologs of the four Yamanaka factors in regeneration of two different organs, the radial nerve cord and the digestive tube, in the sea cucumber Holothuria glaberrima. All four pluripotency factors are expressed in uninjured animals, although their expression domains do not always overlap. In regeneration, the expression levels of the four genes were not regulated in a coordinated way, but, instead, showed different dynamics for individual genes and also were different between the radial nerve and the gut. SoxB1, the ortholog of the mammalian Sox2, was drastically downregulated in the regenerating intestine, suggesting that this factor is not required for dedifferentiation/regeneration in this organ. On the other hand, during the early post-injury stage, Myc, the sea cucumber ortholog of c-Myc, was significantly upregulated in both the intestine and the radial nerve cord, and is, therefore, hypothesized to play a central role in dedifferentiation/regeneration of various tissue types.
Keywords: Regeneration, Dedifferentiation, Pluripotency factors, Gene Expression, Echinodermata
Introduction
Echinoderms have emerged as important models in which to study cellular and molecular mechanisms of post-traumatic tissue regeneration. These animals are capable of regenerating different parts of their body after various types of injuries. An important phylogenetic position, as a sister group to chordates within the monophyletic group Deuterostomia, makes studies on echinoderms particularly relevant for understanding what we can learn from spontaneously regenerating animals to develop better treatment options for human patients.
Dedifferentiation of specialized cells is a crucial step in echinoderm regeneration. Upon injury, specialized cells of mature tissues are able to simplify their phenotype, initiate active mitotic divisions and give rise to new specialized cell. There is evidence suggesting that dedifferntiated cells become multipotent, as they not only acquire the ability of giving rise to the cells of their original cell type, but can also generate other cell types. For example, radial glial cells of the injured sea cucumber central nervous system (CNS) give rise to both new glial cell and neurons (Mashanov et al, 2013). Mesothelial cells of the regenerating digestive tube show even deeper levels of dedifferentiation and can transdifferentiate across germ layer boundaries to produce cells of the luminal digestive epithelium (Mashanov et al, 2005; Mosher, 1956). Even though the ability of differentiated cells in echinoderm tissues to acquire a broadened developmental potential has been known for at least a decade, the molecular mechanisms underlying this phenomenon have remained completely unexplored. This knowledge would not only be significant from the point of view of fundamental biology, but also could have an impact on the development of new therapeutic approaches.
In mammals, including humans, the limited capacity of specialized cells to undergo in vivo dedifferentition correlates with generally poor spontaneous regeneration (Mashanov et al, 2014a). However, as has been famously shown, forced expression of a combination of just four transcription factors (now known as the Yamanaka factors), including Sox2, Klf4, c-Myc, and Oct4 can induce transformation of differentiated mammalian flbroblasts into induced pluripotent stem (iPS) cells in vitro (Takahashi and Yamanaka, 2006). In spite of the great biomedical value of these data, it has remained unknown if these four transcription factors constitute a universal recipe for reprogramming of all differentiated cell types. In particular, how close are the mechanisms of experimental in vitro reprogramming to spontaneous in vivo cell dedifferentiation in regeneration-competent animals? Forced expression of Yamanaka factors was achieved under controlled conditions in cell culture and with very low efficiency, whereas large numbers of cells are known to undergo dedifferentiation in vivo in injured tissues, and these cells remain within the tissue, being affected by the whole gamut of signals from other cells and the extracellular matrix. Further comparative studies of the two phenomena would eventually contribute to deeper understanding of the mechanisms underlying the naturally occurring in vivo dedifferentation and promote development of more efficient approaches of iPS cell generation.
Expression of Yamanaka factors in post-traumatic regeneration has been previously described in various organs of regeneration-competent vertebrates, such fish and amphibians (Christen et al, 2010; Maki et al, 2009; Perry et al, 2013), showing variation of expression patterns in different experimental settings. For example, Oct4 was found to be required for fin regeneration in zebrafish (Christen et al, 2010), but inhibited lens regeneration in newts (Bhavsar and Tsonis, 2014). This variation can be explained in part by the fact that regeneration-competent vertebrates, in particular amphibians, might have evolved a number of taxon-specific regeneration mechanisms, which are absent in other animals (Brockes and Gates, 2014; Garza-Garcia et al, 2010). Therefore, in order to build a complete picture of the the role that pluripotency factors play in spontaneous in vivo dedifferentiation and regeneration, the studies should be extended to other model organisms beyond vertebrates.
Here, we describe the temporal and spacial expression patterns of homologs of the four Yamanaka factors in the sea cucumber Holothuria glaberrima. In order to determine whether dedifferentiation/reprograming processes in the same species involve the same molecular mechanisms in different contexts, expression of those genes was investigated in two regenerating organs: the digestive tube and the radial nerve cord. All four genes are transcriptionally active in the adult uninjured tissues of both organ systems, although their expression domains in the digestive tube do not always overlap. In regenerating animals, individual genes showed different expression dynamics, which was also dependent on the tissue type. Our study suggests that SoxB1, the sea cucumber homolog of the mammalian Sox2, is not required for cell dedifferentiation and early regeneration in the digestive tube. On the other hand, stereotypical overexpression of Myc in regeneration of both the gut and the radial nerve indicates that it might play an important role in post-traumatic response in various echinoderm tissues.
Materials and methods
Animal collection, maintenance, and sampling procedure
Adult individuals of the sea cucumber Holothuria glaberrima Selenka, 1867 were collected from the intertidal zone of Puerto Rico. The animals were kept in well aerated seawater in indoor aquaria at room temperature. To study visceral regeneration, autotomy of internal organs (evisceration) was induced by intracoelomic injection of 2–4 ml of 0.35M KCl. The injury paradigm in the central nervous system involved surgical transection of the midventral radial nerve cord at about the mid-body level, as described previously (Mashanov et al, 2012b, 2013, 2014b). At different time points after evisceration or nerve cord transcection, the animals were anesthetized by immersion into seawater containing 0.2% solution of chlorobutanol (Sigma) for 10–30 min. Samples of normal and regenerating tissues were excised and used for quantitative PCR (qPCR) or in situ hybridization as described below.
Sequence retrieval and analysis
The sequences of the H. glaberrima orthologs of Yamanaka factors were retrieved from the radial nerve reference transcriptome database (http://dx.doi.org/10.6070/H4PN93J1) (Mashanov et al, 2014b) by local TBLASTN search run on a Bio-Linux (Field et al, 2006) (http://environmentalomics.org/bio-linux/) workstation. Protein domains were identified by Pfam (http://pfam.xfam.org) and Interpro (https://www.ebi.ac.uk/interpro/) database search.
Phylogenetic analysis
Phylogenetic trees were constructed to determine the phylogenetic relationships between the sea cucumber genes and their homologs from other animals. The reference homologous sequences (listed in Electronic Supplementary Material, Table S1) were retrieved from the Uniprot and NCBI’s nr databases by BLAST search with H. glaberrima translated ORF sequences. Multiple sequence alignments were performed with ClustalW. These alignments then served as input to construct phylogenetic trees with the MEGA software (version 5 or 6) (Tamura et al, 2013) using the neighbor-joining method and 2,000 bootstrap replicates.
Quantitative real-time PCR (qPCR)
Quantitative real-time PCR was used to determine relative expression levels of each of the four transcription factors in normal and regenerating sea cucumbers. In order to prevent possible RNA degradation, all tissue sampling manipulations were perfomed as quickly as possible. From non-eviscerated animals, small pieces of about the same size and wet weight were taken from each of the five regions (i.e., esophagus, three regions of the intestine proper, and the cloaca) of the digestive tube and then combined together prior to RNA extraction to represent the normal gut. From regenerating animals, the entire gut rudiment was used as the source of total RNA. Both normal and regenerating digestive tubes were sampled together with the supporting mesentery. To extract RNA from the radial nerve cord, we followed the same sampling protocol as in our previous studies (Mashanov et al, 2012b, 2014b). Briefly, the tissue samples consisted of the region of the injury gap measuring 3–4 mm across plus ~3 mm of the flanking stump regions on either side of the wound. Pieces of the similar size were also excised from uninjured animals to represent the normal radial nerve. During dissection, every effort was made to surgically separate the radial nerve cord from the surrounding tissues. After excision, the tissue samples were immediately homogenized in Trizol reagent (Sigma). Total RNA was extracted following the manufacturer’s instructions and then treated with DNAse I (Qiagen). The RNA samples isolated from the normal and regenerating digestive tube were directly used in the first strand cDNA synthesis reaction. In the case of the RNA samples derived from the radial nerve cord, we found out that we had to perform an extra step of poly(A) RNA purification using the Poly(A)Purist kit (Ambion), since the samples contained some inhibitors of downstream reactions, which could not be removed even after repeated rounds of phenol-chloroform extraction and ethanol precipitation. Template cDNA was synthesized from 1 µg of total RNA (digestive tube samples) or from 250 ng of poly(A) RNA (radial nerve cord samples) with random hexamer primers and SuperScript II reverse transcriptase (Invitrogen).
PCR primers were designed using Primer Premier 5.0 software (PREMIER Biosoft International), and their sequences are shown in Electronic Supplementary Material, Table S2. qPCR reactions were performed in 25 µl volumes using Brilliant II SYBR Green QPCR Master Mix (Agilent) and were run on a Mx3005P qPCR System (Stratagene). Each cDNA sample was assayed twice (technical replicates), and each of the conditions was represented by three independent RNA samples (biological replicates). Threshold Ct value calling, melting curve analysis, and calculations of PCR efficiency were performed using MxPro QPCR software (Stratagene) supplied with the qPCR instrument. The raw data produced by qPCR assays were considered acceptable for further analysis if the difference between technical replicates was less than 0.5×Ct and the slope values corresponding to PCR efficiencies was between −3.2 and −3.5 with the R2 value above 0.98 (Nolan et al, 2006).
Statistical analysis of qPCR data was performed using the MCMC.qpcr R package (Matz et al, 2013; R Core Team, 2014) in the ”classic” mode, which uses a normalization procedure relative to ”control” genes. The following control genes were used to normalize the qPCR expression values in the radial nerve cord dataset: elongation factor 2 (EF2), ribosomal protein rpL18a, Mn-superoxide dismutase (Sod), and V-type proton AT-Pase 16 kDa proteolipid subunit (ATP6L). These genes have been previously shown as stably expressed in both normal and regenerating radial organs in a high-throughput RNAseq assay (Mashanov et al, 2012b, 2014b). To normalize the expression values obtained for the normal and regenerating digestive tube, we used NADH dehydrogenase subunit 5 as a reference gene (Mashanov et al, 2010). P-values were adjusted for multiple comparisons using the Benjamini & Hochberg methods implemented in the p.adjust function in R.
In situ hybridization
Antisense digoxigenin-labeled riboprobes for in situ hybridization were transcribed from PCR-generated DNA templates using Roche DIG-labeling mix. The sequences of the primers used to generate the templates are listed in Electronic Supplementary Material, Table S2. Identity of all probes was confirmed by Sanger sequencing of the templates. Tissue samples were immersion fixed overnight in 4% paraformaldehyde in 0.01 PBS (pH 7.4, 1030 mOsm), cryoprotected in graded sucrose solutions and frozen in the OCT medium (Takara). In situ hybridization was performed on 10 µm-thick cryosections as described previously (Mashanov et al, 2010, 2012a,c).
Results
Identification and characterization of the H. glaberrima homologs of Yamanaka factors
The sequences coding for sea cucumber homologs of the vertebrate Yamanaka factors were retrieved from the H. glaberrima reference transcriptome database (Mashanov et al, 2014b) by tblastn search. Since this database consists of contigs that were derived from an automatic assembly of next-generation sequencing reads, the sequences were validated by re-sequencing using Sanger technology. The verified sequences were deposited in the Gen-Bank under accession numbers KM281936 – KM281939.
Oct1/2/11
Oct proteins are a group of developmentally important POU domain-containing transcription factors that recognize a specific 8 nt DNA sequence (octamer) (Tantin, 2013). The vertebrate Oct3/4 is a core transcription factor that activates protein-coding genes and non-coding RNAs required for pluripotency (Shi and Jin, 2010). In mammalian stem cells, Oct3/4 is co-expressed together with Oct1, a related transcription factor that is also known to be involved in maintenance of the stem cell phenotype (Maddox et al, 2012; Tantin, 2013). For example, Oct1 is required for radial glia formation in the CNS and is also strongly expressed in Lgr5-positive intestinal stem cells (Kiyota et al, 2008; Maddox et al, 2012).
Relatively little is known about the roles of Oct proteins in non-mammalian animal models. Unlike in mammals, the sea urchin genome, the most thoroughly characterized echinoderm genome so far, contains a single Oct gene, which has been shown to be most closely related to the vertebrate Oct1 and Oct2 genes. In early sea urchin embryogenesis, this gene is involved in dorsoventral axis specification (Range and Lepage, 2011). The role of Oct in echinoderm regeneration has not been investigated so far.
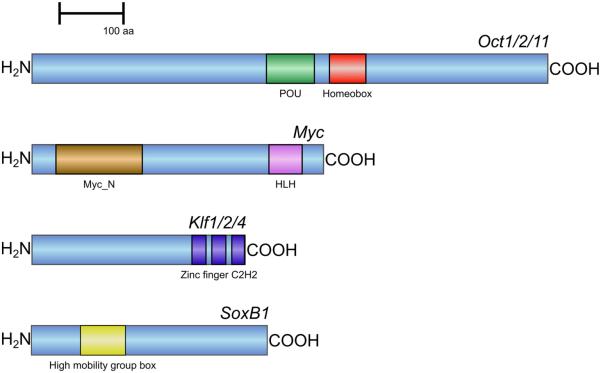
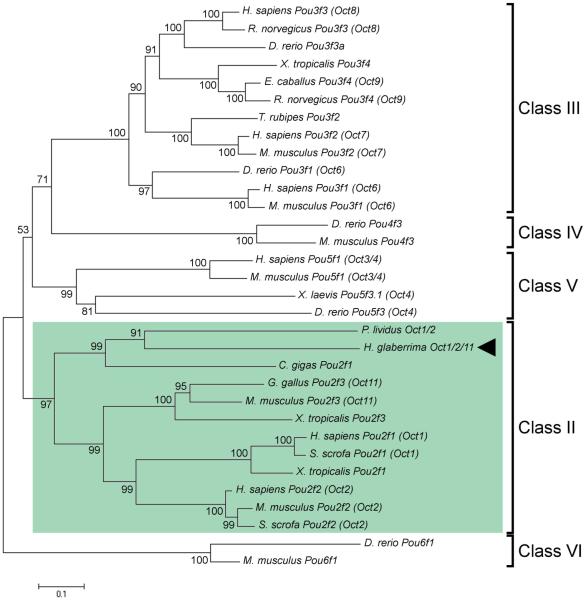
The H. glaberrima ortholog of the vertebrate Oct3/4 has an open reading frame coding for a 779 amino acid-long protein, which contains both motifs characteristic of Pou transcripton factor proteins, a Pou domain and a homeobox domain, separated by a 21-amino acid linker region (Fig. 1). Phylogenetic analysis (Fig. 2) shows that H. glaberrima Oct protein belongs to class II POU proteins and clusters together with Oct1/2 of the sea urchin P. lividus and Pou2f1 of the bivalve C. gigas to form an outgroup to vertebrate Oct1, 2, and 11. Therefore, the H. glaberrima protein was designated as Oct1/2/11.
Fig. 1.
Diagram showing domain organization for the predicted Oct1/2/11, Myc, Klf1/2/4, and SoxB1 proteins of the sea cucumber Holothuria glaberrima
Fig. 2.
Evolutionary relationships between Oct1/2/11 of H. glaberrima (arrowhead ) and homologous POU proteins in other animals. Green rectangle indicates class II of POU transcription factors. The numbers at branch nodes indicate the percentage of recovery of the branch in replicate trees in the bootstrap test. The tree is drawn to scale. The evolutionary distances were calculated using maximum composite likelihood method and are represented as number of base substitutions per site. The database accession numbers of the sequences used in the phylogenetic analysis are listed in Electronic Supplementary Material, Table S1
Myc
Another of the four Yamanaka factors, mammalian c-Myc, is known to directly control transcription of as many as ~15% of all protein-coding genes and, therefore, mediates a wide range of biological processes. In addition to c-Myc, mammals have other functionally redundant Myc genes, including N-Myc, L-Myc, and S-Myc (Mahani et al, 2013). Unlike mammals, many invertebrates, such as tunicates, amphioxus, sea urchins, and Drosophila, have a single Myc gene in their genome (Gallant, 2006; Morita et al, 2009). Among the many biological functions of Myc proteins, the control of growth and cell proliferation have been conserved between invertebrates and vertebrates (Gallant, 2006).
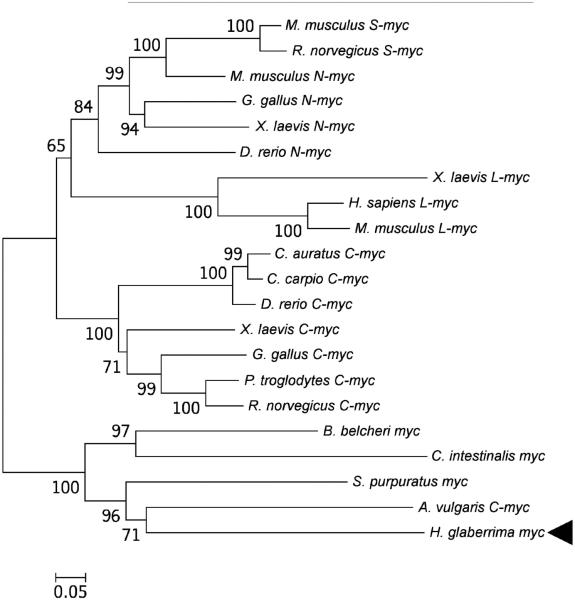
The sea cucumber homolog of Myc contains a predicted 441 amino acid-long open reading frame, which includes two conserved motifs that define Myc proteins, the N-terminal Myc domain and the C-terminal basic helix-loop-helix domain (Fig. 1). Our phylogenetic analysis (Fig. 3) showed that the H. glaberrima Myc did not belong to any of the vertebrate myc subfamilies, but instead clustered together with Myc proteins from urochordates, cephalochordates, echinoids, and asteroids. This cluster received strong bootstrap support and formed an outgroup to Myc proteins of vertebrates
Fig. 3.
Evolutionary relationships between Myc of H. glaberrima (arrowhead ) and homologous proteins in other Deuterostomia
Klf1/2/4
Klf proteins are zinc-finger containing transcription factors. Klf2 and Klf4 are known to inhibit growth, DNA synthesis and cell cycle progression and, therefore, are regarded as tumor suppressor genes. Klf4 functions in reprogramming of iPS cells by promoting their survival through inhibition of c-Myc-induced programmed cell death. Simultaneously, c-Myc suppresses the anti-proliferation function of Klf4. (McConnell and Yang, 2010; Takahashi and Yamanaka, 2006).
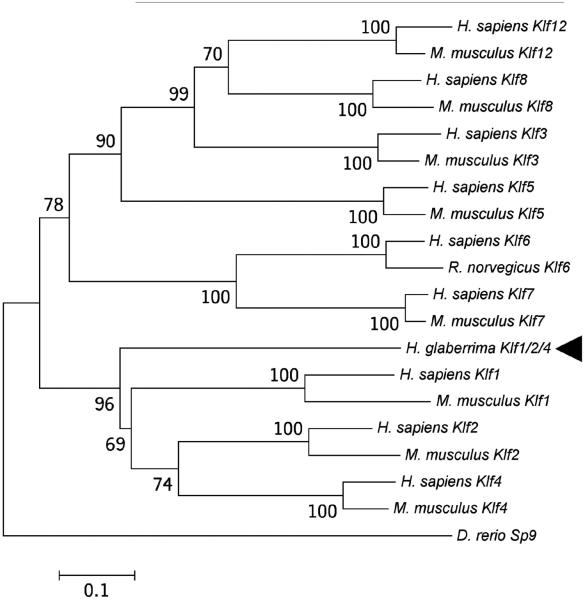
Studies of the sea urchin genome have identified several Klf genes (Materna et al, 2006), whose functions in ehinoderms remain largely unstudied. The sea cucumber homolog of Klf4 codes for a 323 animo-acid long polypeptide with three zinc finger C2H2 motifs at the carboxy-terminal end of the protein (Fig. 1). Such organization is typical for all members of the Klf family (McConnell and Yang, 2010). Phylogenetic analysis (Fig. 4) clusters the sea cucumber protein as an outgroup to the clade containing mammalian Klf1, Klf2, and Klf4 with a strong bootstrap support. Therefore, this sequence was designated as Klf1/2/4.
Fig. 4.
Evolutionary relationships between Klf1/2/4 of H. glaberrima (arrowhead ) and homologous proteins in other Deuterostomia. The tree was rooted using the Sp9 transcription factor of Danio rerio
SoxB1
Sox genes code for a diverse family of transcription factors (26 members and 11 members in the human and sea urchin genome, respectively) (Howard-Ashby et al, 2006) that contain the high-mobility group (HMG) domain and are known to be involved in various aspects of development. Mammalian Sox2 protein has been shown to play a key role in acquisition and maintenance of pluripotency (Lefebvre et al, 2007; Pevny and Placzek, 2005; Takahashi and Yamanaka, 2006). Likewise, a related sea urchin gene, SoxB1, is thought to be involved in maintenance of the enteric neuronal precursors (Wei et al, 2011).
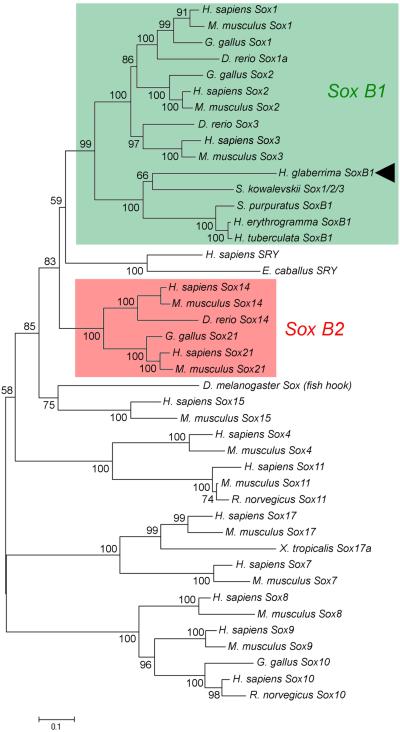
Our phylogenetic analysis (Fig. 5) shows that the sea cucumber homolog of mammalian Sox2 clusters together with the sea urchin SoxB1 proteins, as well as with the hemichordate protein designated as Sox1/2/3. Together, these proteins from invertebrates deuterostomes are most closely related to mammalian Sox1, Sox2, and Sox3, which constitute the so-called SoxB1 subgroup. Therefore, the H. glaberrima homolog of Sox2 was designated as SoxB1. This transcript codes for a 357 amino acid-long peptide containing the signature HMG domain (Fig. 1).
Fig. 5.
Evolutionary relationships between SoxB1 of H. glaberrima (arrowhead) and Sox proteins of other animals. Green and red rectangles indicate the SoxB1 and SoxB2 subgroups of Sox transcription factors, respectively
Temporal expression of Oct1/2/11, Myc, Klf1/2/4, and SoxB1 in visceral and neural regeneration
To determine if and how the sea cucumber orthologs of the mammalian pluripotency factors change their expression in the regenerating digestive and nervous systems, we ran quantitative real-time RT-PCR assays on RNA samples isolated from uninjured animals and from regenerating tissues at different time points after the injury (Fig. 6).
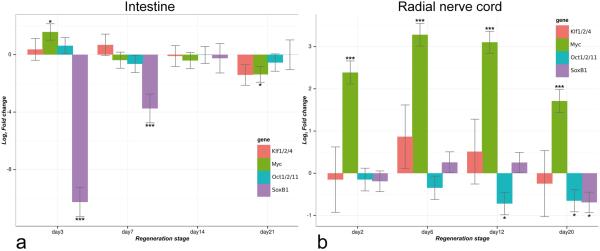
Fig. 6.
Temporal expression pattern of Klf1/2/4, Myc, Oct1/2/11, and SoxB1 in the regenerating digestive tube (A) and radial nerve cord (B) as determined by quantitative real time RT-PCR. Three animals were used per time point. Expression values are plotted as fold change relative to the uninjured tissues in log2 scale. Error bars show standard deviation. ∗P < 0.05,∗∗∗ P < 0.001. The full output of the MCMC.qpcr R package (Matz et al, 2013) containing relative expression values and corresponding p-values can be found in Electronic Supplementary Material, Text S3, and Text S4
In the regenerating digestive tube, SoxB1 showed a surprisingly prominent down-regulation at the early post-evisceration stages (over 1,000-fold and ~ 13 fold on day 3 and 7, respectively). Myc was moderately (~3-fold) upregulated on day 3 post-evisceration, but then returned to its basal expression level, before being slightly (~1.5-fold) downregulated on day 21 post-evisceration. The two other genes, Klf1/2/4 and Oct 1/2/11, were transcriptionally active in both non-autotomized and regenerating digestive tube, but did not show any significant differences in expression level across the conditions (Fig. 6A, Electronic Supplementary Material Text S3).
The expression profile of the pluripotency factors in the radial nerve cord was markedly different from that in the intestine (Compare Fig. 6A and B, Electronic Supplementary Material Text S4). First, Myc was up-regulated significantly (3–10-fold) and continuously throughout the duration of the experiment. Second, unlike in the digestive tube, SoxB1 expression remains at the normal levels in the regenerating radial nerve cord, before becoming slightly downregulated during the late regeneration phase (~1.6 fold, day 20 post-injury). Moreover, Oct1/2/11, after remaining at the normal levels during the early stages of neural regeneration (days 3–6), also becomes slightly downregulated during late stages of regeneration (~1.6-fold, days 12–21). Like in the regenerating intestine, Klf1/2/4 maintains the same level of expression both under normal conditions and during neural regeneration.
Expression of the pluripotency factors at the tissue level
Non-eviscerated digestive tube
Anatomically, the digestive tube of sea cucumbers consists of several regions including the eosphagus, three subdivisions of the intestine proper (first descending intestine, ascending intestine, second descending intestine), and the cloaca (Feral and Massin, 1982). Along its entire length, the digestive tube has the same basic architecture and is composed of three tissue layers: the luminal epithelium, the connective tissue partition, and the mesothelium. However, in each of those regions, the tissue components show different organization (Fig. 7a, f, k, p, u). Likewise, all four pluripotency factors show differential expression along the length of the digestive tube.
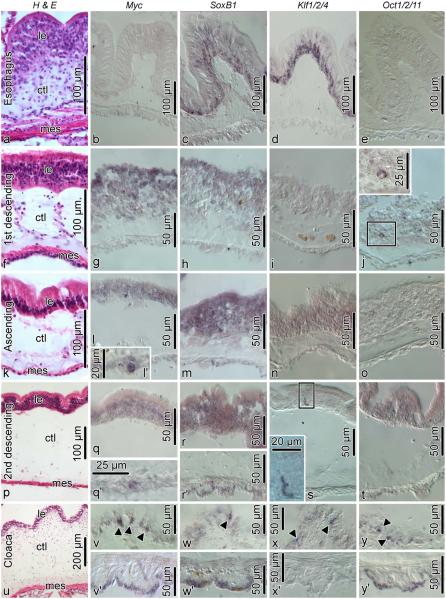
Fig. 7.
Spatial expression of Myc, SoxB1, Klf1/2/4, and Oct1/2/11 in the non-eviscerated digestive tube. Each horizontal row of images corresponds to a different region of the digestive tube: a–e, esophagus; f–j, first descending intestine; k–o, ascending intestine; p–t, second descending intestine; u–y’, cloaca. The leftmost column of images (a, f, k, p, u) show reference paraffin sections (hematoxylin and eosin staining) for each of the regions of the digestive tube (a, esophagus; f, 1st descending intestine; k, ascending intesine; p, 2nd descending intestine; u, cloaca). The second thru fifth columns show in situ hybridization signal with riboprobes for each of the transcripts. All micrographs show transverse sections with the luminal epithelium to the top and the mesothelium to the bottom. In cases when two micrographs are shown for a particular gut region (l and l’; q and q’; r and r’, etc.), the upper micrograph (l, q, r, etc) shows the luminal epithelium and the bottom micrograph (l’, q’, r’, etc) shows the mesothelium. Insets in j and s show higher magnification views of the boxed areas. arrowheads indicate positively stained cells in the luminal epithelium of the cloaca. ctl, connective tissue layer; le, luminal epithelium; mes, mesothelium.
Myc is silent in the esophagus (Fig. 7b), but extensively expressed in the luminal epithelium of the intestine proper (Fig. 7g, l, q), as well as in scattered individual cells of the mesothelium (Fig. 7l’, q’).
SoxB1, like Myc, is broadly expressed in the luminal epithelium of the intestine (Fig. 7h, m, r), but also in the luminal epithelium of the esophagus (Fig. 7c).
Klf1/2/4 is also strongly and broadly expressed in the luminal epithelium of the esophagus (Fig. 7d), but shows no transcriptional activity in the first descending intestine (Fig. 7i) and is very weakly expressed in the luminal epithelium of the ascending intestine (Fig. 7n). In the second descending intestine, positive in situ hybridization signal is seen in extremely rare, but strongly labeled cells of the luminal epithelium (Fig. 7s).
Oct1/2/11 is not transcriptionally active in the esophagus and ascending intestine (Fig. 7e, o), but is strongly expressed in scattered cells of the luminal epithelium in the first descending intestine (Fig. 7j) and shows very weak, but broad expression in the luminal epithelium of the second descending intestine (Fig. 7t).
The cloaca is the only organ of the digestive system, where all four transcription factors show similar expression patterns in scattered enterocytes of the luminal epithelium (Fig. 7v, w, x, y). Three of the four pluripotency genes, Myc, SoxB1, Oct1/2/11, are also broadly expressed in the cell bodies of peritoneal cells in the apical region of the cloacal mesothelium (Fig. 7v’, w’, y’).
Regenerating digestive tube
During evisceration (visceral autotomy) in H. glaberrima, the entire intestine detaches from the supporting mesentery and from the esophagus and cloaca at the anterior and posterior ends, respectively. The rejected organ is then expelled from the body cavity through the rupture in the cloacal wall and then through the anus (García-Arrarás et al, 1998). Subsequent regeneration involves a number of events including (a) healing of the ruptured ends of the esophagus and cloaca, (b) formation of a solid connective tissue thickening at the free margin of the supporting mesentery, and (c) ingression of the digestive epithelia of the cloaca and esophagus into this solid rudiment to restore the gut lumen. In H. glaberrima, the majority of the luminal epithelium of the newly regenerated intestine is derived from the cloaca, whereas the contribution of the esophagus to regeneration is limited.
The details of the anatomical organization and histological architecture of gut rudiments at different stages of visceral regeneration in H. glaberrima are described elsewhere (García-Arrarás et al, 1998). Here, we provide only a brief description of the anatomical structures and cellular events that are pertinent to our discussion of gene expression patterns.
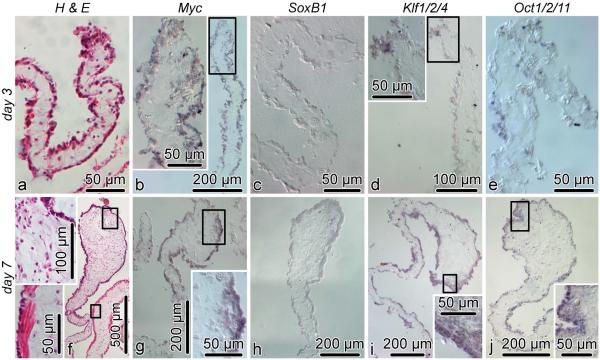
On day 3 post-evisceration, dedifferentiation is initated in the the mesothelium of the free distal margin of the mesentery. At the cellular level, dedifferentiation involves remodeling of the cytoskeleton (Candelaria et al, 2006; Mashanov et al, 2005) and simplification of the epithelial architecture, but no changes are yet visible at the gross level (Fig. 8a). In line with the qRT-PCR data, there is no in situ hybridization signal for SoxB1 on day 3 post-evisceration (Fig. 8c). The other three pluripotency factors are transcriptonally active in the mesothelium of the gut rudiment, with Myc being extensively expressed in most of the mesothelial cells (Fig. 8b), whereas Klf1/2/4 and Oct1/2/11 were detected only in some scattered cells (Fig. 8d, e).
Fig. 8.
Spatial expression of Myc, SoxB1, Klf1/2/4, and Oct1/2/11 during early stages of visceral regeneration. The two horizontal rows of images (a thru e, f thru j) show cross sections of the regenerating intestine on day 3 and 7 post-evisceration, respectively. The first column (a and f ) shows hematoxylin and eosin stained reference sections. The second thru fifth columns (b–e, g–j) show in situ hybridization labeling with respective riboprobes. Insets show high magnification view of the boxed areas in the respective main micrographs. ctl, connective tissue layer; mes, mesothelium.
By day 7 post-injury, the mesothelium of the free distal margin of the mesentery is completely dedifferentiated. It has the organization of a simple epithelium with no muscular component, in contrast to more proximal regions of the mesentery (Fig. 8f). This dedifferentiated mesothelium surrounds a notable swelling in the distal mesentery, which constitutes the early rudiment of the new intestine. At this stage, very weak in situ hybridization signal for SoxB1 is detected in the mesothelium of the regenerating gut (Fig. 8h), whereas the other three transcription factors are extensively expressed in the mesothelial epithelial cells and also in few mesenchymal cells directly beneath the mesothelium at the anti-mesenterial side of the developing gut rudiment (Fig. 8g, i, j).
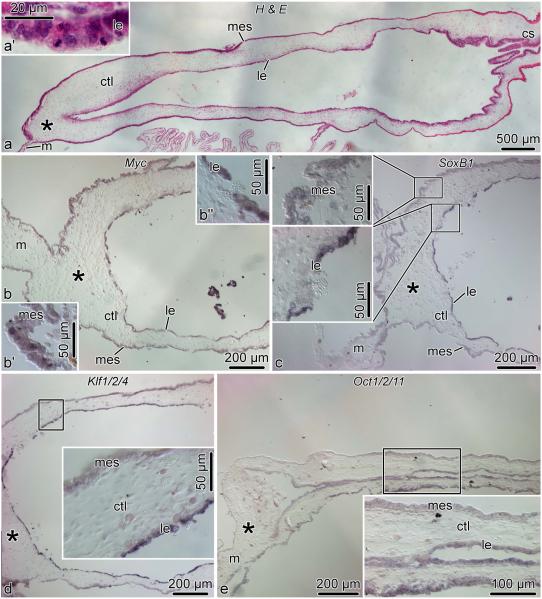
On day 14 post-evisceration, the luminal epithelium of the cloaca starts invading the connective tissue of the solid gut rudiment (Fig. 9a). This epithelial invasion involves extensive mitotic division in the regenerating tissues (Fig. 9a’) (García-Arrarás et al, 1998). All four sea cucumber orthologs of the pluripotency factors are expressed at moderate to high levels in both the luminal epithelium and the mesothelium of the regenerating intestine. In situ hybridization signal in the luminal epithelium is often stronger than in the mesothelium. (Fig. 9).
Fig. 9.
Spatial expression of Myc, SoxB1, Klf1/2/4, and Oct1/2/11 in the regenerating intestine on day 14 post-evisceration. All micrographs show longitudinal sections of the regenerating intestine. The growing anterior tip of the gut rudiment (asterisk ) is on the left, whereas the cloacal stump (cs) is on the right. a, reference section stained with hematoxylin and eosin. a’, high-magnification view of the luminal epithelium of the growing intestinal rudiment showing abundant mitotic figures. b, Myc expression in the intestinal rudiment. b’ and b” show high-magnification views of the mesothelium and luminal epithelium, respectively. c, d, and e show expression of SoxB1, Klf1/2/4, and Oct1/2/11, respectively. Insets in c–e are high magnification views of the boxed areas in the respective main micrographs. cs, cloacal stump; ctl, connective tissue layer; le, luminal epithelium; m, mesentery; mes, mesothelium.
By day 21 post-evisceration, the regenerating digestive tube of H. glaberrima resumes its normal anatomical and histological organization (Fig. 10a), but the cells in the newly regenerated intestine are still mitotically active (Fig. 10a’), as the organ is still growing to restore its normal size and looping. All four genes are extensively expressed in most of the cells of the luminal epithelium. On the other hand, the mesothelium of the new gut shows very weak or no in situ hybridization signal (Fig. 10b–e).
Fig. 10.
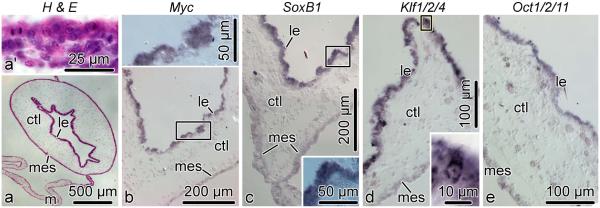
Expression of Myc, SoxB1, Klf1/2/4, and Oct1/2/11 in the tissues of the newly regenerated intestine on day 21 post-evisceration. All micrographs are cross sections. a, Low-magnification view of the newly regenerated intestine; reference paraffin section stained with hematoxylin and eosin. a’, high-magnification view of the luminal epithelium showing mitotic figures. b–e, In situ hybridization with riboprobes for Myc, SoxB1, Klf1/2/4, and Oct1/2/11, respectively. Insets show high magnification views of the boxed areas in the respective main micrographs. ctl, connective tissue layer; le, luminal epithelium; m, mesentery; mes, mesothelium
None of the four transcription factors were expressed in the cells of the connective tissue partition of the gut wall.
Radial nerve cord
Five radial nerve cords (RNCs) constitute the major parts of the echinoderm central nervous system (Mashanov et al, 2006). Unlike in mammals, these regions of the echinoderm nervous system readily regenerate after injury. The key cellular event underlying this post-traumatic neural regeneration is activation of radial glial cells in the vicinity of the wound. These cells undergo dedifferentiation, form an early glial scaffold that constitutes the rudiment of the regenerating segment of the radial nerve cord, and extensively proliferate to give rise to new glial cells and, most importantly, new neurons (Mashanov et al, 2008, 2013).
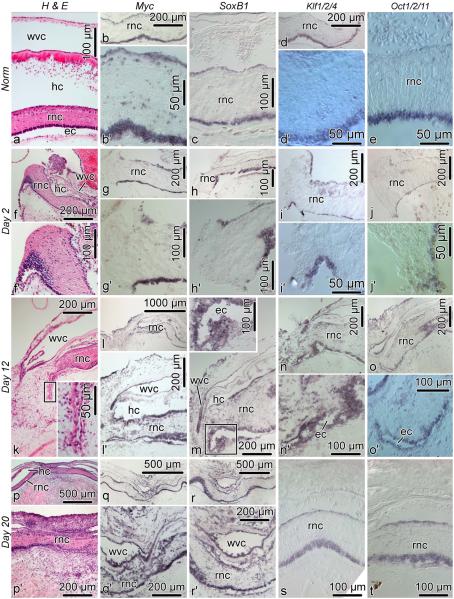
Unlike in the digestive tube, the spatial distribution of the four pluripotency factors was found to be similar in both the uninjured and regenerating radial nerve cord (Fig. 11). In the normal RNC, the genes are expressed in the apical region of both the ectoneural and hyponeural neuroepithelia (Fig. 11b–e). Myc is also expressed in scattered cell bodies throughout the neural parenchyma (Fig. 11b’). During regeneration, the four genes continue to be transcriptionally active in the apical regions of the neuroepithelia of the stumps (Fig. 11g–o), but are also expressed in glial tubes, which constitute the early rudiments growing across the wound gap (Fig. 11k–o’). After the wound gap has been bridged, all four genes continue to be expressed in the newly created region of the RNC (Fig. 11q– t).
Fig. 11.
Expression of Myc, SoxB1, Klf1/2/4, and Oct1/2/11 in the uninjured and regenerating radial nerve cord. All sections are longitudinal. Horizontal rows of images correspond to the normal animals and three different stages of regeneration. The first vertical column shows reference paraffin sections stained with hematoxylin and eosin. The second thru fifth columns show in situ hybridization labeling with respective riboprobes. Insets show high magnification view of the boxed areas in the respective main micrographs. When two micrographs are shown for a particular condition (e.g., b and b’, the upper (e.g., b) and lower (e.g., b’) images show an overview and high-magnification view, respectively). ec, epineural canal; en, ectoneural neuroepithelium; hc, hyponeural canal; hn, hyponeural neuroepithelium; wvc, water-vacular canal; rnc, radial nerve cord.
Discussion
In this study, we investigated expression of homologs of the four Yamanaka factors in spontaneous post-traumatic regeneration of two different body parts, the digestive tube and the central nervous system, in the sea cucumber H. glaberrima. Previous morphological studies (Candelaria et al, 2006; Mashanov et al, 2005, 2008, 2013) indicated that regeneration in both of these organs is preceded by extensive dedifferentiation of specialized cells. Dedifferentiating cells undergo activation of nuclear heterochromatin, cytoskeleton reorganization, and complete or partial loss of their specialized cytoplasmic features. Once dedifferentiation is completed, the cells contribute to regeneration through extensive cell division. Their progeny undergo cell migration and rearrange themselves to reconstruct the lost body part. The available data indicate that the dedifferentiated cells may become at least transiently multipotent, as they are able to give rise not only to new cells of the same cell type, but also to other cell types. For example, dedifferentiated mesothelial cells can transdifferentiate into enterocytes of the gut luminal epithelium (Mashanov et al, 2005), whereas radial glial cells of the regenerating central nervous system generates neurons (Mashanov et al, 2013). Therefore, we wanted to see if the four pluripotency Yamanaka factors were expressed in spontaneously regenerating tissues, whose specialized cells undergo dedifferentiation to broaden their developmental potential.
Our results show that dedifferentiation of specialized tissues and subsequent regeneration in H. glaberrima do not involve coordinated upregulation of all four pluripotency factors. For example, Klf1/2/4 and Oct1/2/11 remained expressed at about the same level as in the normal tissues. Moreover, those genes that change their expression do so in different ways depending on the tissue type, suggesting existence of multiple gene regulatory scenarios operating in different context in the same species. The most notable difference between the gut and the RNC is the absence of SoxB1 expression during early stages of intestinal regeneration, whereas it remains expressed at the normal level in the injured radial nerve. This observation suggests that cell dedifferentiation in the gut mesothelium does not require expression of SoxB1, which is puzzling, as Sox2, the homolog of H. glaberrima SoxB1, is not merely expressed in various vertebrate regeneration models (Christen et al, 2010; Luz-Madrigal et al, 2014; Maki et al, 2009; Perry et al, 2013), but is absolutely required for regeneration (Christen et al, 2010). The functional significance of SoxB1 down-regulation in the regenerating sea cucumber intestine is unknown and can only be determined in future experiments involving forced ectopic over-expression.
One of the common features between the regenerating gut and CNS in H. glaberrima is significant upregulation of Myc during early post-injury stages, when both organs undergo extensive cell dedifferentiation (Candelaria et al, 2006; Mashanov et al, 2013). Myc proteins play a central role in dedifferentiation and regeneration even in poorly regenerating mammals. For example, it has been shown that expression of c-Myc was required for regeneration of the intestinal epithelium (Ashton et al, 2010). In the mammalian CNS, experimental forced expression of v-Myc, a viral homolog of c-Myc, stabilized undifferentiated state of embryonic radial glia. When injected into the injured adult spinal cord, these cells contributed to increased neuroprotection and axonal regrowth (Hasegawa et al, 2005). Therefore, our observations combined with known roles of Myc proteins in regeneration and control of cell differentiation in various organisms (Mahani et al, 2013) make the sea cucumber Myc gene a promising candidate for further studies. Our ongoing efforts are aimed at determining the functional role of Myc over-expression in dedifferentiating tissues of H. glaberrima.
The fact that some pluripotency factors are present in the regenerating sea cucumber tissues and may even be significantly up-regulated in response to injury (e.g., Myc), whereas other factors (such as SoxB1 ) are absent and obviously not involved in regeneration, suggests that even though there might be some parallels between in vivo dedifferentiation and in vitro iPS cell generation, the mechanisms are not exactly the same. One possible explanation why these two processes can be different is that the cells that dedifferentiate in vivo, unlike iPS cells, may not be pluripotent, but at best only multipotent (Christen et al, 2010; Maki et al, 2009). Alternatively, we cannot rule out that some other, yet unknown, factors can contribute to in vivo dedifferentiation in regeneration-competent animals.
Conclusions
In the sea cucumber H. glaberrima, the orthologs of mammalian Yamanaka pluripotency factors, Myc, SoxB1, Oct1/2/11, and Klf1/2/4, are expressed in the adult uninjured central nervous system and in the digestive tube, although in the latter, their expression domains do not overlap (except in the cloaca).
Expression of individual pluripotency factors is not coordinated in injured/regenerating tissues. Some of these genes (Oct1/2/11 and Klf1/2/4 ) keep being expressed at about the same levels as in uninjured animals, while others (SoxB1 and Myc) undergo considerable changes in expression.
There are differences in pluripotency factor expression dynamics between the regenerating radial nerve cord and the regenerating intestine, suggesting that cell dedifferentiation in different regenerating organs may involve organ- and/or tissue-specific mechanisms.
Drastic down-regulation of SoxB1 in the regenerating intestine during the early post-evisceration phase suggests that expression of this gene is not required for cell dedifferentiation in this organ.
Consistent up-regulation of Myc in both intestinal and neural regeneration in H. glaberrima suggests that this gene may play an important role in dedifferentiation/regeneration in various tissues and organs.
Supplementary Material
Database accession numbers of the sequences used to construct the phylogenetic trees
PCR primers used in the present study
Pairwise differences in expression of Oct1/2/11, Myc, Klf1/2/4, and SoxB 1 between the uneviscerated and regenerating intestine (on days 3, 7, 14, and 21 post-evisceration) calculated with the MCMC.qpcr R package (Matz et al, 2013). The upper triangle of the matrices shows log2 fold changes, whereas the lower triangle lists the corresponding p-values
Pairwise differences in expression of Oct1/2/11, Myc, Klf1/2/4, and SoxB 1 between the uninjured and regenerating radial nerve cord (on days 2, 6, 12, 20 after transection) calculated with the MCMC.qpcr R package (Matz et al, 2013). The upper triangle of the matrices shows log2 fold changes, whereas the lower triangle lists the corresponding p-values
Acknowledgements
The study was supported by grants from the NIH (1SC1GM084770-01, 1R03NS065275-01) and the NSF (IOS-0842870 , IOS-1252679), as well as by the University of Puerto Rico.
References
- Ashton GH, Morton JP, Myant K, Phesse TJ, Ridgway RA, Marsh V, Wilkins JA, Athineos D, Muncan V, Kemp R, Neufeld K, Clevers H, Brunton V, Winton DJ, Wang X, Sears RC, Clarke AR, Frame MC, Sansom OJ. Focal adhesion kinase is required for intestinal regeneration and tumorigenesis downstream of Wnt/c-Myc signaling. Dev Cell. 2010;19(2):259–269. doi: 10.1016/j.devcel.2010.07.015. DOI 10.1016/j.devcel.2010.07.015, URL http://dx.doi.org/10.1016/j.devcel.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhavsar RB, Tsonis PA. Exogenous Oct-4 inhibits lens transdifferentiation in the newt Notophthalmus viridescens. PLoS One. 2014;9(7):e102–510. doi: 10.1371/journal.pone.0102510. DOI 0, URL http://dx.doi.org/0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockes JP, Gates PB. Mechanisms underlying vertebrate limb regeneration: lessons from the salamander. Biochem Soc Trans. 2014;42(3):625–630. doi: 10.1042/BST20140002. DOI 10.1042/BST20140002, URL http://dx.doi.org/10.1042/BST20140002. [DOI] [PubMed] [Google Scholar]
- Candelaria AG, Murray G, File SK, García-Arrarás JE. Contribution of mesenterial muscle dedifferentiation to intestine regeneration in the sea cucumber Holothuria glaberrima. Cell Tissue Res. 2006;325(1):55–65. doi: 10.1007/s00441-006-0170-z. DOI 10.1007/s00441-006-0170-z, URL http://dx.doi.org/10.1007/s00441-006-0170-z. [DOI] [PubMed] [Google Scholar]
- Christen B, Robles V, Raya M, Paramonov I, Izpisuá Belmonte JC. Regeneration and reprogramming compared. BMC Biol. 2010;8:5. doi: 10.1186/1741-7007-8-5. DOI 10.1186/1741-7007-8-5, URL http://dx.doi.org/10.1186/1741-7007-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feral J, Massin C. Digestive system: Holothuroidea. In: Jangoux M, Lawrence J, editors. Echinoderm nutrition. Balkema; Rotterdam: 1982. pp. 192–212. [Google Scholar]
- Field D, Tiwari B, Booth T, Houten S, Swan D, Bertrand N, Thurston M. Open software for biologists: from famine to feast. Nat Biotechnol. 2006;24(7):801–803. doi: 10.1038/nbt0706-801. DOI 10.1038/nbt0706-801, URL http://dx.doi.org/10.1038/nbt0706-801. [DOI] [PubMed] [Google Scholar]
- Gallant P. Myc/Max/Mad in invertebrates: the evolution of the Max network. Curr Top Microbiol Immunol. 2006;302:235–253. doi: 10.1007/3-540-32952-8_9. [DOI] [PubMed] [Google Scholar]
- García-Arrarás JE, Estrada-Rodgers L, Santiago R, Torres II, Díaz-Miranda L, Torres-Avillán I. Cellular mechanisms of intestine regeneration in the sea cucumber, Holothuria glaberrima Selenka (Holothuroidea: Echinodermata) J Exp Zool. 1998;281(4):288–304. doi: 10.1002/(sici)1097-010x(19980701)281:4<288::aid-jez5>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Garza-Garcia AA, Driscoll PC, Brockes JP. Evidence for the local evolution of mechanisms underlying limb regeneration in salamanders. Integr Comp Biol. 2010;50(4):528–535. doi: 10.1093/icb/icq022. DOI 10.1093/icb/icq022, URL http://dx.doi.org/10.1093/icb/icq022. [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Chang YW, Li H, Berlin Y, Ikeda O, Kane-Goldsmith N, Grumet M. Embryonic radial glia bridge spinal cord lesions and promote functional recovery following spinal cord injury. Exp Neurol. 2005;193(2):394–410. doi: 10.1016/j.expneurol.2004.12.024. DOI 10.1016/j.expneurol.2004.12.024, URL http://dx.doi.org/10.1016/j.expneurol.2004.12.024. [DOI] [PubMed] [Google Scholar]
- Howard-Ashby M, Materna SC, Brown CT, Chen L, Cameron RA, Davidson EH. Gene families encoding transcription factors expressed in early development of Strongylocentrotus purpuratus. Dev Biol. 2006;300(1):90–107. doi: 10.1016/j.ydbio.2006.08.033. DOI 10.1016/j.ydbio.2006.08.033, URL http://dx.doi.org/10.1016/j.ydbio.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Kiyota T, Kato A, Altmann CR, Kato Y. The POU homeobox protein Oct-1 regulates radial glia formation downstream of Notch signaling. Dev Biol. 2008;315(2):579–592. doi: 10.1016/j.ydbio.2007.12.013. DOI 10.1016/j.ydbio.2007.12.013, URL http://dx.doi.org/10.1016/j.ydbio.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Dumitriu B, Penzo-Méndez A, Han Y, Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int J Biochem Cell Biol. 2007;39(12):2195–2214. doi: 10.1016/j.biocel.2007.05.019. DOI 10.1016/j.biocel.2007.05.019, URL http://dx.doi.org/10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luz-Madrigal A, Grajales-Esquivel E, McCorkle A, DiLorenzo AM, Barbosa-Sabanero K, Tsonis PA, Del Rio-Tsonis K. Reprogramming of the chick retinal pigmented epithelium after retinal injury. BMC Biol. 2014;12(1):28. doi: 10.1186/1741-7007-12-28. DOI 10.1186/1741-7007-12-28, URL http://dx.doi.org/10.1186/1741-7007-12-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox J, Shakya A, South S, Shelton D, Andersen JN, Chidester S, Kang J, Gligorich KM, Jones DA, Spangrude GJ, Welm BE, Tantin D. Transcription factor oct1 is a somatic and cancer stem cell determinant. PLoS Genet. 2012;8(11):e1003–048. doi: 10.1371/journal.pgen.1003048. DOI 10.1371/journal.pgen.1003048, URL http://dx.doi.org/10.1371/journal.pgen.1003048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahani A, Henriksson J, Wright APH. Origins of Myc proteins–using intrinsic protein disorder to trace distant relatives. PLoS One. 2013;8(9):e75–057. doi: 10.1371/journal.pone.0075057. DOI 10.1371/journal.pone.0075057, URL http://dx.doi.org/10.1371/journal.pone.0075057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki N, Suetsugu-Maki R, Tarui H, Agata K, Del Rio-Tsonis K, Tsonis PA. Expression of stem cell pluripotency factors during regeneration in newts. Dev Dyn. 2009;238(6):1613–1616. doi: 10.1002/dvdy.21959. DOI 10.1002/dvdy.21959, URL http://dx.doi.org/10.1002/dvdy.21959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashanov V, Zueva O, Heinzeller T, Dolmatov I. Ultrastructure of the circumoral nerve ring and the radial nerve cords in holothurians (Echinodermata) Zoomorphology. 2006;125(1):27–38. DOI 10.1007/s00435-005-0010-9, URL http://dx.doi.org/10.1007/s00435-005-0010-9. [Google Scholar]
- Mashanov VS, Dolmatov IY, Heinzeller T. Trans-differentiation in holothurian gut regeneration. Biol Bull. 2005;209(3):184–193. doi: 10.2307/3593108. [DOI] [PubMed] [Google Scholar]
- Mashanov VS, Zueva OR, Heinzeller T. Regeneration of the radial nerve cord in a holothurian: a promising new model system for studying post-traumatic recovery in the adult nervous system. Tissue Cell. 2008;40(5):351–372. doi: 10.1016/j.tice.2008.03.004. DOI 10.1016/j.tice.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Mashanov VS, Zueva OR, Rojas-Catagena C, García-Arrarás JE. Visceral regeneration in a sea cucumber involves extensive expression of survivin and mortalin homologs in the mesothelium. BMC Dev Biol. 2010;10:117. doi: 10.1186/1471-213X-10-117. URL http://dx.doi.org/10.1186/1471-213X-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashanov VS, Zueva OR, Garcia-Arraras JE. Expression of Wnt9, TCTP, and Bmp1/Tll in sea cucumber visceral regeneration. Gene Expr Patterns. 2012a;12(1-2):24–35. doi: 10.1016/j.gep.2011.10.003. DOI 10.1016/j.gep.2011.10.003, URL http://dx.doi.org/10.1016/j.gep.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashanov VS, Zueva OR, García-Arrarás JE. Posttraumatic regeneration involves differential expression of long terminal repeat (LTR) retrotransposons. Dev Dyn. 2012b;241(10):1625–1636. doi: 10.1002/dvdy.23844. [DOI] [PubMed] [Google Scholar]
- Mashanov VS, Zueva OR, García-Arrarás JE. Retrotransposons in animal regeneration: Overlooked components of the regenerative machinery? Mob Genet Elements. 2012c;2(5):244–247. doi: 10.4161/mge.22644. DOI 10.4161/mge.22644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashanov VS, Zueva OR, García-Arrarás JE. Radial glial cells play a key role in echinoderm neural regeneration. BMC Biology. 2013;11(1):49. doi: 10.1186/1741-7007-11-49. URL http://www.biomedcentral.com/1741-7007/11/49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashanov VS, Zueva O, García-Arrarás JE. Chapter Seven - Postembryonic Organogenesis of the Digestive Tube: Why Does It Occur in Worms and Sea Cucumbers but Fail in Humans? In: Galliot B, editor. Mechanisms of Regeneration, Current Topics in Developmental Biology. Vol. 108. Academic Press; 2014a. pp. 185–216. DOI 10.1016/B978-0-12-391498-9.00006-1, URL http://www.sciencedirect.com/science/article/pii/B9780123914989000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashanov VS, Zueva OR, García-Arrarás JE. Transcriptomic changes during regeneration of the central nervous system in an echinoderm. BMC Genomics. 2014b;15(1):357. doi: 10.1186/1471-2164-15-357. DOI 10.1186/1471-2164-15-357, URL http://dx.doi.org/10.1186/1471-2164-15-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna SC, Howard-Ashby M, Gray RF, Davidson EH. The C2H2 zinc finger genes of Strongy-locentrotus purpuratus and their expression in embryonic development. Dev Biol. 2006;300(1):108–120. doi: 10.1016/j.ydbio.2006.08.032. DOI 10.1016/j.ydbio.2006.08.032, URL http://dx.doi. org/10.1016/j.ydbio.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Matz MV, Wright RM, Scott JG. No control genes required: Bayesian analysis of qRT-PCR data. PLoS One. 2013;8(8):e71–448. doi: 10.1371/journal.pone.0071448. URL http://dx.doi.org/10.1371/journal.pone.0071448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell BB, Yang VW. Mammalian Krüppel-like factors in health and diseases. Physiol Rev. 2010;90(4):1337–1381. doi: 10.1152/physrev.00058.2009. DOI 10.1152/physrev.00058.2009, URL http://dx.doi.org/10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Futami K, Zhang H, Kubokawa K, Ojima Y, Okamoto N. Evolutionary analysis of amphioxus myc gene. Journal of the Tokyo University of Marine Science and Technology. 2009;5:11–16. [Google Scholar]
- Mosher C. Observation on evisceration and visceral regeneration in the sea-cucumber, Actinopyga agassizi Selenka. Zoologica (NY) 1956;41:17–26. [Google Scholar]
- Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1(3):1559–1582. doi: 10.1038/nprot.2006.236. URL http://dx.doi.org/10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- Perry KJ, Thomas AG, Henry JJ. Expression of pluripotency factors in larval epithelia of the frog Xenopus: evidence for the presence of cornea epithelial stem cells. Dev Biol. 2013;374(2):281–294. doi: 10.1016/j.ydbio.2012.12.005. DOI 10.1016/j.ydbio.2012.12.005, URL http://dx.doi.org/10.1016/j.ydbio.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny L, Placzek M. Sox genes and neural progenitor identity. Curr Opin Neurobiol. 2005;15(1):7–13. doi: 10.1016/j.conb.2005.01.016. DOI 10.1016/j.conb.2005.01.016, URL http://dx.doi.org/10.1016/j.conb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2014. URL http://www.R-project.org/ [Google Scholar]
- Range R, Lepage T. Maternal Oct1/2 is required for Nodal and Vg1/Univin expression during dorsal-ventral axis specification in the sea urchin embryo. Dev Biol. 2011;357(2):440–449. doi: 10.1016/j.ydbio.2011.07.005. DOI 10.1016/j.ydbio.2011.07.005, URL http://dx.doi.org/10.1016/j.ydbio.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Shi G, Jin Y. Role of Oct4 in maintaining and regaining stem cell pluripotency. Stem Cell Res Ther. 2010;1(5):39. doi: 10.1186/scrt39. DOI 10.1186/scrt39, URL http://dx.doi.org/10.1186/scrt39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult flbroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. DOI 10.1016/j.cell.2006.07.024, URL http://dx.doi.org/10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. Mega6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. DOI 10.1093/molbev/mst197, URL http://dx.doi.org/10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantin D. Oct transcription factors in development and stem cells: insights and mechanisms. Development. 2013;140(14):2857–2866. doi: 10.1242/dev.095927. DOI 10.1242/dev.095927, URL http://dx.doi.org/10.1242/dev.095927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Angerer RC, Angerer LM. Direct development of neurons within foregut endoderm of sea urchin embryos. Proc Natl Acad Sci U S A. 2011;108(22):9143–9147. doi: 10.1073/pnas.1018513108. DOI 10.1073/pnas.1018513108, URL http://dx.doi.org/10.1073/pnas.1018513108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Database accession numbers of the sequences used to construct the phylogenetic trees
PCR primers used in the present study
Pairwise differences in expression of Oct1/2/11, Myc, Klf1/2/4, and SoxB 1 between the uneviscerated and regenerating intestine (on days 3, 7, 14, and 21 post-evisceration) calculated with the MCMC.qpcr R package (Matz et al, 2013). The upper triangle of the matrices shows log2 fold changes, whereas the lower triangle lists the corresponding p-values
Pairwise differences in expression of Oct1/2/11, Myc, Klf1/2/4, and SoxB 1 between the uninjured and regenerating radial nerve cord (on days 2, 6, 12, 20 after transection) calculated with the MCMC.qpcr R package (Matz et al, 2013). The upper triangle of the matrices shows log2 fold changes, whereas the lower triangle lists the corresponding p-values