Abstract
All aerobic cells and organisms must synthesize heme from the amino acid glycine and the tricarboxylic acid (TCA) cycle intermediate succinyl Coenzyme A for incorporation into hemoproteins such as the cytochromes needed for oxidative phosphorylation. Most studies on heme regulation have been done in erythroid cells or hepatocytes; however much less is known about heme metabolism in other cell types. The feline leukemia virus subgroup C receptor (FLVCR) is a 12 transmembrane domain surface protein that exports heme from cells and was previously shown to be required for erythroid development. Here we show that deletion of Flvcr in murine hematopoietic precursors caused a complete block in αβ T cell development at the CD4+CD8+ double-positive stage, though other lymphoid lineages were unaffected. Moreover, FLVCR was required for the proliferation and survival of peripheral CD4+ and CD8+ T cells. These studies identify a novel and unexpected role for FLVCR, a major facilitator superfamily (MFS) metabolite transporter, in T cell development and suggest that heme metabolism is particularly important in the T lineage.
INTRODUCTION
The role of heme as a prosthetic group in proteins involved in oxygen transport, electron transfer, and catalysis has been long-appreciated. Heme is critical for mitochondrial oxidative phosphorylation, and heme deficiency impairs assembly of the electron chain subunits (1). Heme also has important regulatory functions. Heme regulates erythroid lineage differentiation by binding transcriptional (2) and translational regulators of globin synthesis (3). On the organismal level, heme synthesis and the circadian clock are reciprocally regulated (4) and heme plays a role in integrating the internal circadian clock with metabolic states such as the fasting and fed states (5, 6).
While the enzymatic steps of heme synthesis and degradation have been well-characterized (Supplemental Fig. 1), less is known about intra- and intercellular heme trafficking (7). Free heme causes lipid peroxidation and oxidative damage and must be carefully regulated (8). The feline leukemia virus subgroup C receptor (FLVCR), a 12 transmembrane domain protein in the major facilitator superfamily (MFS) (9), was previously shown by our group to export heme (10). The gene encoding FLVCR is referred to as FLVCR1 in humans and Mfsd7b in mouse; in order to avoid confusion and maintain consistency with the existing literature, we refer to the gene and protein as Flvcr and FLVCR here. Conditional deletion of Flvcr in neonatal or adult mice caused severe anemia (11), similar to the erythroid failure seen in cats viremic with feline leukemia virus subgroup C (FeLV-C) in which cell surface expression of FLVCR is inhibited by viral interference (12, 13).
Older studies had noted that cats viremic with FeLV-C had thymic aplasia and lymphopenia (14), though it was not known whether lymphopenia was due to cell-intrinsic loss of FLVCR or secondary to chronic viremia and/or anemia. To answer this question, we developed and studied several models in which FLVCR function could be knocked out in lymphoid cells or more specifically in T cells during development.
MATERIALS AND METHODS
Mice
Flvcrflox/flox;Mx-cre mice and controls were previously described (11) and were backcrossed to C57BL/6 mice (The Jackson Laboratory) for 10 generations. C57BL/6, CD45.1 (Pep3b) and Rag1−/− mice (15) were obtained from The Jackson Laboratory. Homozygous Lck-cre and CD4-cre mice (16) were obtained from Taconic and crossed to Flvcrflox/flox mice to generate Flvcrflox/flox;Lck-cre and Flvcrflox/flox;CD4-cre mice. OT-I (17) and OT-II (18) mice were crossed to Flvcrflox/flox;Mx-cre mice. OT-I; Flvcrflox/flox mice were bred to Lck-cre mice to obtain OT-I; Flvcrflox/flox;Lck-cre. All mice were bred and maintained in a specific pathogen-free barrier facility at the University of Washington. Animal studies were performed according to protocols approved by the IACUC of the University of Washington.
Non-competitive and competitive transplants
Flvcr+/+, Flvcr+/−, and Flvcr−/− mice were generated by treating 6–12 week-old Flvcr+/+;Mx-cre, Flvcr+/flox;Mx-cre, or Flvcrflox/flox;Mx-cre mice with i.p. injection of 0.15 mg polyinosinic:polycytidylic acid (polyI:C) (Amersham) x 3 doses every other day. 8–9 days later, bone marrow mononuclear cells (BM) from the femurs and/or tibias of polyI:C-treated mice was harvested and 2.5×106 BM were injected i.v. into sublethally irradiated (6.5 Gy) Rag1−/− mice. For competitive transplants, Flvcr+/+;Mx-cre, Flvcr+/flox;Mx-cre, or Flvcrflox/flox;Mx-cre and CD45.1 mice were treated with i.p. injection of 0.15 mg polyI:C x 3 doses every other day. 8–9 days later, BM from the femurs and/or tibias of polyI:C-treated mice was harvested and 5×106 BM from Flvcr+/+;Mx-cre or control was mixed with 5×106 BM CD45.1 BM and injected i.v. into lethally irradiated (11 Gy) Rag1−/− mice or C57BL/6 x Pep3b F1 (CD45.1/CD45.2) mice.
Blood cell analysis
Mice were bled retroorbitally into EDTA anti-coagulated microtainer tubes (Becton Dickenson). Complete blood counts were performed on a Hemavet HV950FS analyzer (Drew Scientific) programmed for mouse blood.
Flow cytometric analysis and sorting
Flow cytometric analysis was performed using FACSCantoII and LSRII (BD Biosciences); cells were sorted using BD FACS Aria (BD Biosciences) at the Cell Analysis Facility, Department of Immunology, University of Washington. Flow data were analyzed with FlowJo7.6 (Tree Star Inc.).
Antibodies and reagents
Fluorochrome-conjugated antibodies were purchased from BD Biosciences and eBioscience.
Tissue staining
Whole thymus was fixed in 10% buffered formalin. Fixed thymus was paraffin-embedded, sectioned, and stained with hematoxylin and eosin staining by the Experimental Histopathology Facility at the FHCRC.
Quantitative RT-PCR (qRT-PCR) analysis
RNA was isolated from sorted cells using RNeasy Plus Mini Kit (Qiagen) and reverse-transcribed using iScript reverse transcription (Biorad) as per the manufacturers’ instructions. qRT-PCR was performed on cDNA or genomic DNA using either Taqman assay or SybrGreen assay with gene-specific primers obtained from Integrated DNA Technology (IDT). Primer sequences: Flvcr (F 5′-ATCTGGAACCTGTGCAGAAACA-3′, R 5′-ATTGAATAAAATGCTCCAGTCATGAT-3′, Probe 5′-CCCCTTTGTTCTCCTGCTGGTCAGTTATG-3′); Hmox1 (F 5′-CTGCTAGCCTGG TGCAAGATACT-3′, R 5′-GTCTGGGATGAGCTAGTGCTGAT-3′, Probe 5′-AGACACCCCGAGGGAAACCCCA-3′); Alas1 (F 5′-TGGTCGGTTTAGCGTCCTC-3′, R 5′-GGGATAAGAATGGGCATCGG-3′, Probe 5′-CGAGTGCCTACCGCCGCTTC-3′); Cox6a2 (F 5′-CCAGAGTTCATCCCGTATCAC-3′, R 5′-CAGACATCAAGGGTGCTCATAAC-3′); β-actin (F 5′-ACCTTCTACAATGAGCTGCG-3′, R 5′-CTGGATGGCTACGTACATGG-3′, Probe 5′ TCTGGGTCATCTTTTCACGGTTGGC-3′) Gene expression was quantified as fold-change expression using the Pffafl method (19) using β-actin as the reference gene and either C57BL/6 splenic CD8+ T cells or whole thymus as the reference sample. Genomic Flvcr deletion was quantified from genomic DNA using cloned and purified wild-type and FlvcrΔexon 3 DNA to generate standard curves. Primer sequences: genomic wild-type Flvcr (F 5′-ATCTGGAACCTGTGCAGAAACA-3′, R 5′-GAGTCTAGTGCTCAAAGGTCACCAA-3′, Probe 5′-CCCCTTTGTTCTCCTGCTGGTCAGTTATG-3′); genomic deleted Flvcr (F 5′-CCCACCCAAGTAGGATACAGAGA-3′, R 5′-GATCATATTCAATAACCCTTAATATAACTTCG-3′, Probe 5′-ACTTGCAGGACAGACACAAGGCCT-3′).
Rescue transplant experiments
PCR cloning was used to create a DNA fragment containing the human FLVCR coding region without the stop codon fused in-frame with the enhanced green fluorescent protein (EGFP) coding sequence (Clontech). The FLVCR-EGFP DNA was ligated into the pMSCVneo retroviral vector (Clontech) to generate pMSCV FLVCR-EGFP. This vector and control pMXIG (20) containing EGFP only were pseudotyped with ecotropic murine retrovirus (Plat-Eco packaging line (21) (Cell Biolabs, Inc.)). BM was harvested from 6–12 week-old, Flvcrflox/flox;Lck-cre mice and lineage-depleted using biotinylated-lineage-specific antibodies and magnetic separation (Dynal). Lineage-depleted BM cells were pre-stimulated with 20 ng/ml murine IL-3, 100 ng/ml human IL-6, 50 ng/ml murine stem cell factor (all cytokines from Peprotech) in IMDM with 10% FBS for two days, then cultured with viral supernatant plus cytokines and 4 μg/ml of protamine sulfate on viral-preloaded RetroNectin-coated dishes (r-fibronectin CH296, Takara Bio Inc.) for 8 hours. Viral supernatants were replaced by fresh media and the cells were allowed to recover overnight, and the treatment was repeated once. Transduced cells were then harvested and 2×106 transduced cells were transplanted into sublethally irradiated Rag1−/− mice (6.5 Gy).
Adoptive cell transfer
Thymocytes from Flvcrflox/flox;CD4-cre or Flvcr+/+;CD4-cre were mixed 2:1 with CD45.1 thymocytes and incubated with 10 μM carboxyfluorescein succinimidyl ester (CFSE; Invitrogen) in serum-free HBSS for 15 min at 37°C. The reaction was quenched with pure FCS and the cells were washed twice with serum-free medium and then 1.5×107 cells of the CFSE-labeled mixed thymocytes were injected i.v. into sublethally irradiated Rag1−/− mice (6.5 Gy). A sample of the CFSE-labeled mix was also analyzed by flow for CD4 and CD8 staining so the starting CD45.2:CD45.1 ratio of the CD4SP and CD8SP could be determined. Recipient mice were sacrificed at the specified time points and splenocytes counted and analyzed by flow cytometry.
Microarray sample preparation
3 separate competitive transplant cohorts were generated as described above using lethally irradiated Rag1−/− mice (11 Gy) transplanted with polyI:C-treated Flvcr+/+;Mx-cre, or Flvcrflox/flox;Mx-cre BM mixed with polyI:C-treated CD45.1 BM. 12 weeks post-transplant, thymocytes were harvested from 3 recipients per group, pooled, and CD45.2+CD4+CD8+ DP cells sorted. This was repeated for the 3 cohorts resulting in 3 Flvcr+/+ and 3 Flvcr−/− replicates. After sorting, cells were washed twice with cold PBS and cell pellets frozen and stored at −80°C until all samples were collected. RNA was isolated using Qiagen RNeasy Plus Mini Kit per the manufacturer’s instructions and the yield was determined on a NanoDrop ND-1000 spectrophotometer (Thermo Scientific). All samples had an appropriate yield (>100ng total RNA) and were analyzed for RNA integrity using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.). RNA determined to be of high quality was converted to cDNA and biotin-labeled for microarray analysis using Ambion’s Illumina TotalPrep RNA Amplification kit (Life Technologies). Labeled cDNA was analyzed using the MouseWG-8v2 Expression BeadChip Kit (Illumina, Inc.). Sample labeling, array hybridization, and array scanning were performed by the Genomics Shared Resource at the Fred Hutchinson Cancer Research Center.
Microarray data analysis
The dataset consisting of all arrays was processed using the Bioconductor package lumi by employing quantile normalization (22). The data were initially filtered by flagging probes that were below a defined signal “noise floor,” which was calculated as the 75th percentile of the negative control probe signals within each array. A probe was retained if at least 2 of 3 samples in at least one condition were not flagged by the intensity filter. We further filtered the data through the application of a variance filter, using the “shorth” function of the Bioconductor package genefilter. Differential gene expression was determined using the Bioconductor package limma (23), and a false discovery rate (FDR) method was used to correct for multiple testing (24). Significant differential gene expression was defined as |log2 (ratio)| ≥ 0.585 (± 1.5-fold) and FDR = 5%.
Statistical analyses
Statistical analyses were performed with Prism version 5.0 (GraphPad Software), using unpaired two-tailed Student’s t tests. A p-value of <0.05 was considered statistically significant.
Accession data
The microarray data have been submitted to the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/gds) under accession number GSE50202.
RESULTS
FLVCR is required for T cell development beyond the DP stage
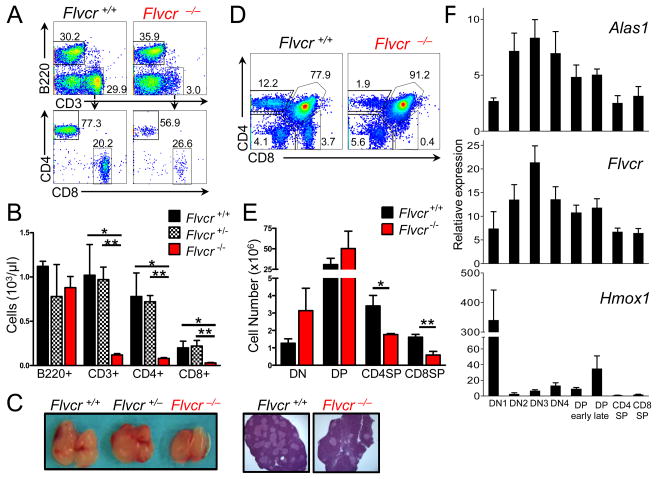
To determine whether FLVCR plays a role in lymphocyte development, we transplanted bone marrow mononuclear cells (BM) from polyinosinic:polycytidylic acid (polyI:C)-treated Flvcrflox/flox;Mx-cre, Flvcr+/flox;Mx-cre mice, or Flvcr+/+;Mx-cre mice into sublethally irradiated Rag1−/− mice, referred to henceforth as Flvcr−/−, Flvcr+/− or Flvcr+/+ mice. Cells with the Mx-cre transgene express cre-recombinase after exposure to IFNα or β, and the double-stranded synthetic RNA polyI:C induces IFN production and cre-mediated gene excision in Mx-cre BM (25). The use of sublethal irradiation permitted reconstitution of endogenous Rag1−/− hematopoiesis; therefore, the mice that received Flvcr−/− BM were not anemic (data not shown), and any T or B cells present derived from the transplanted marrow. Strikingly, we found very few T cells in the blood (Fig. 1, A and B), spleen, and lymph nodes (Supplemental Fig. 2, A and B) of Flvcr−/− mice as compared to controls. Both CD4+ and CD8+ T cells were similarly affected. B cells were present in normal numbers (Fig. 1B), suggesting that FLVCR loss selectively blocked development downstream of the common lymphoid progenitor stage.
FIGURE 1.
FLVCR is required for T cell development beyond the DP stage. (A) Flow cytometric analysis of peripheral blood leukocytes by B220 and CD3 staining (upper) and CD4 and CD8 staining (lower) of cells within the CD3+ gate. Numbers indicate the percentage of cells within the gate. (B) Absolute numbers of B220+, CD3+, CD3+CD4+, and CD3+CD8+ peripheral blood lymphocytes. Bars show the mean and s.e.m. of 3–4 mice per group. *p<0.03 **p<0.009. (C) Representative gross thymuses from Flvcr+/+, Flvcr+/−, and Flvcr−/− mice (left) with representative thymus sections from Flvcr+/+ and Flvcr−/− mice stained with hematoxylin and eosin (10x magnification) (right). (D) CD4 and CD8 flow staining of Flvcr+/+ and Flvcr−/− thymocytes. (E) Enumeration of thymocyte subsets. *p=0.05 **p=0.02. (F) Heme regulatory gene expression quantified by qRT-PCR in thymocyte subsets relative to peripheral splenic CD8+ T cells. Mean and s.e.m. from 3 mice are shown. Data are representative of 2 (C, F) or 3 (A, B, D, E) separate experiments with 3–4 mice per group.
We next examined the thymuses in Flvcr−/− mice and found that they appeared grossly normal (Fig. 1C, left). Early thymic progenitors derived from BM hematopoietic stem cells enter the thymus as CD4-CD8- double-negative (DN) cells that subsequently progress through 4 sub-stages, DN1-DN4 (26). Mice had similar total numbers of DN (Fig. 1, D and E). Because Rag1−/− mice have DN thymocytes, we examined Flvcr−/− DN thymocytes obtained from mice that were lethally-irradiated and transplanted with a mixture of congenically-marked control or Flvcr-deleted BM and wild-type BM (described in greater detail in the next section) to analyze the DN1-DN4 subsets. Though there was a deficit of Flvcr−/− DN1, similar numbers of Flvcr-deleted and control cells were present at each of the subsequent 3 stages (Supplemental Fig. 2, C and D). During DN3, thymocytes that have successfully re-arranged the TCRβ chain (β-selection checkpoint) undergo a proliferative burst before becoming CD4+CD8+ double-positive (DP) cells (26). The number of CD4+CD8+ double-positive (DP) thymocytes was unaffected by FLVCR loss (Fig. 1, D and E); thus, FLVCR is not required for β-selection or subsequent late DN proliferation.
DP thymocytes re-arrange their TCRα chain and undergo positive and negative selection to identify thymocytes capable of recognizing peptide-MHC complexes on APC without excessive auto-reactivity before finally differentiating into CD4+ or CD8+ single-positive (SP) cells (27). CD4+ and CD8+ single-positive (SP) thymocytes were severely decreased in Flvcr−/− mice (Fig. 1, D and E). On thymic histology there were fewer medullary regions in Flvcr−/− mice (lighter regions in Fig. 1C, right), correlating with the decreased number of SP, typically found in the thymic medulla.
We examined Flvcr gene expression in sorted normal thymic subsets and found that Flvcr gene expression peaked during the DN3 stage and then declined (Fig. 1F, middle). The expression of Flvcr during thymic development mirrored that of the aminolevulinic acid synthase1 gene (Alas1) (Fig. 1F, upper), encoding the rate-limiting enzyme in heme synthesis, consistent with FLVCR’s role in maintaining intracellular heme homeostasis. Heme oxygenase1 (Hmox1), which encodes an enzyme that breaks down heme, was present at high levels early during thymic development but down-regulated after the DN stage (Fig. 1F, lower); thus later-stage thymocytes may rely more on FLVCR to regulate intracellular free heme and prevent toxicity.
Requirement for FLVCR is T cell-intrinsic
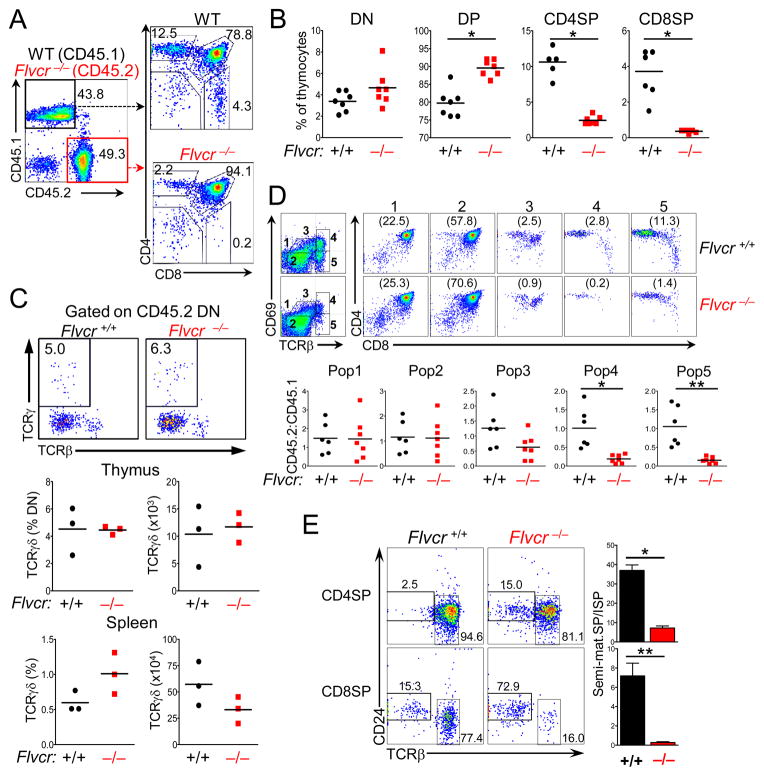
To determine whether the block was due to FLVCR loss in antigen presenting cells (BM and non-BM-derived) or stromal cells in the thymus rather than to T cell-intrinsic FLVCR loss, we performed competitive repopulation studies, transplanting poly I:C-treated Flvcrflox/flox;Mx-cre or Flvcr+/flox;Mx-cre (CD45.2) BM mixed 1:1 with congenically-marked (CD45.1) wild-type (WT) BM into lethally irradiated Rag1−/− mice. Few CD45.2+ Flvcr−/− CD4SP or CD8SP were found in recipients, though CD45.1+ WT CD4SP and CD8SP were found in expected proportions (Fig. 2A). Therefore, the loss of SP T cells was not due to thymic stromal cell FLVCR deficiency. The proportion of Flvcr−/− DP thymocytes was increased while CD4SP and CD8SP thymocytes were markedly decreased (Fig. 2B). We sorted CD45.2+ (Flvcrflox/flox;Mx-cre-derived) B220+, Gr-1+, CD4+, and CD8+ peripheral blood cells and quantified Flvcr deletion through quantitative RT-PCR (qRT-PCR). While donor BM and donor-derived peripheral B220+ and Gr-1+ blood cells were nearly completely deleted (BM mean 97.5%, S.D. 2.1, n=3; B220+ mean 99.3, S.D.=0.9, n=5; Gr-1+ mean =98.1%, S.D.=1.6, n=5), donor-derived peripheral CD4+ and CD8+ cells were only partially deleted (CD4+ mean 68.9%, S.D.=19.2 n=5; CD8+ mean =58.7% S.D.=12.6, n=5). Thus the few peripheral T cells present from poly I:C-treated donor Flvcrflox/flox;Mx-cre BM likely expanded from a few precursors in which only one Flvcr allele was deleted (resulting in 50% deletion), reinforcing the conclusion that FLVCR is absolutely required for CD4+ and CD8+ T cell development.
FIGURE 2.
The requirement for FLVCR is T cell-intrinsic. (A) Representative flow analysis of thymocytes from mice transplanted with a 1:1 mix of Flvcr−/−:CD45.1 BM with CD4 and CD8 expression shown on gated CD45.1 or CD45.2 thymocytes. (B) Quantification of CD45.2 thymocyte subsets from competitively-repopulated (CR) mice that received Flvcr+/+:CD45.1 (black circles) or Flvcr−/−:CD45.1 (red squares). Each symbol represents an individual mouse. *p<0.0001. (C) Representative flow analysis with TCRγδ and TCRβ expression shown on gated CD45.2 Flvcr+/+ (left) and Flvcr−/− (right) DN (upper) from CR mice. Numbers indicate the percentage of TCRγδ + cells within the gate. Enumeration of the proportion and absolute number of Flvcr+/+ and Flvcr−/− TCRγδ-expressing cells in the thymus (middle) and spleen (lower) are shown. (D) Representation flow analysis with CD69 and TCRβ expression on Flvcr+/+ and Flvcr−/− thymocytes (gated on CD45.2) from CR mice with gates indicating successive stages of development (upper left). CD4 and CD8 expression on gated populations with the percentage of cells in the population shown in parentheses (upper right). CD45.2:CD45.1 ratios for the gated populations are shown below for individual mice (lower). *p=0.02 **p=0.001. (E) CD24 and TCRβ staining on gated CD4SP and CD8SP thymocytes from representative mice (left). Ratio of mature (CD24loTCRβhi) SP to immature SP (ISP, CD24hiTCRβlo), which are precursors to the DP stage, is shown in the bar graphs. *p<0.0001 **p=0.0006. Data are representative of 4 separate experiments (A, B, and D) with 4–5 mice per group and 2 separate experiments (C) with 3 mice per group.
FLVCR is not required for post-natal γδ T cell development
Given that FLVCR loss impacted T cell development specifically, we also looked at the numbers and early thymic development of γδ T cells. TCRβ, γ, and δ gene re-arrangements occur during the DN stage, and thymocytes that express a functional TCRγδ receptor on the surface diverge from αβ T cells before CD4 and CD8 up-regulation and the DP stage (28). We examined the proportion of γδ T cells in CD45.2 DN thymocytes from lethally-irradiated mice transplanted with poly I:C-treated Flvcrflox/flox;Mx-cre or Flvcr+/flox;Mx-cre (CD45.2) BM mixed 1:1 with wild-type (WT) (CD45.1) BM. The proportion and numbers of γδ TCR-expressing DN thymocytes were similar between Flvcr−/− and Flvcr−/− DN (Fig. 2C, upper and middle). Moreover, the proportion and numbers of γδ T cells in the spleen were also similar (Fig. 2C, lower). Thus FLVCR is required specifically for αβ and not γδ T cell development.
Flvcr-deleted thymocytes fail selection
We next analyzed the expression of CD69 and TCRβ on Flvcr−/− or Flvcr+/+ thymocytes from competitively-transplanted mice to more precisely identify the point at which Flvcr−/− thymocytes failed. While the proportion of cells in immature TCRloCD69lo (Pop1, DN and early DP) and pre-selection TCRintCD69lo (Pop2) populations was similar between Flvcr−/− and control, the proportion of Flvcr−/− thymocytes in the TCRβintCD69hi (Pop3) stage and beyond was greatly diminished (Fig. 2D). Up-regulation of CD69 signifies TCR signaling and positive selection; thus, the decrease in CD69+ cells from the DP stage suggested that Flvcr-deleted thymocytes were failing selection. In addition, Flvcr−/− CD4SP and CD8SP failed to undergo maturation as indicated by down-regulation of CD24. The majority of Flvcr−/− CD8 SP as well as a greater proportion of CD4SP as compared to control SP were in fact immature SP (ISP, CD24hiTCRβlo), the precursor stage to the DP stage, (Fig. 2E).
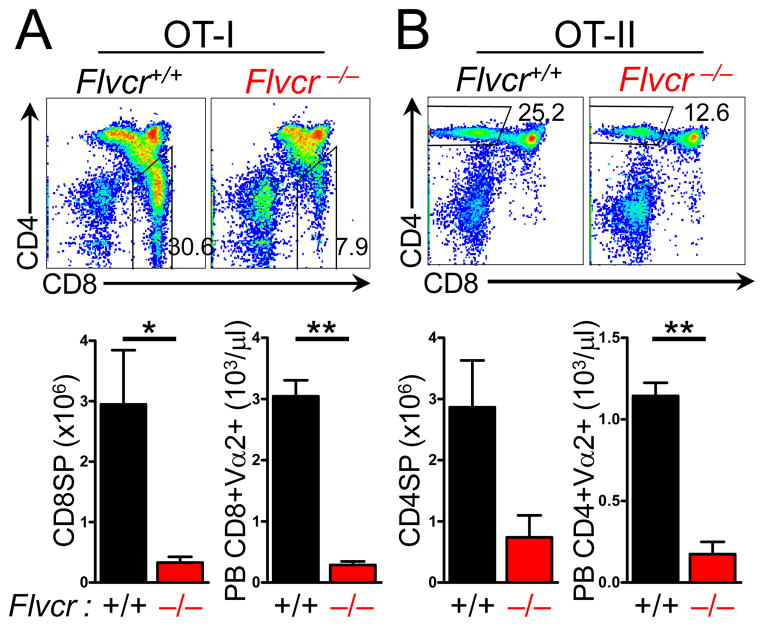
To test the hypothesis that the failure of Flvcr-deleted thymocytes to undergo positive selection was due to their inability to re-arrange and express a functional TCRα chain, we crossed Flvcrflox/flox;Mx-cre, Flvcr+/flox;Mx-cre mice, and Flvcr+/+;Mx-cre mice to mice expressing transgenes encoding an MHC class I-restricted (OT-I) (17) or MHC class II-restricted (OT-II) (18) αβTCR. We then transplanted BM from polyI:C-treated OT-I or OT-II;Flvcrflox/flox;Mx-cre; mice (referred to as OT-I and OT-II Flvcr−/−) and controls into sublethally irradiated Rag1−/− mice. In spite of the fact that both TCR are capable of providing a positive selection signal in C57BL/6 mice, both OT-I and OT-II Flvcr−/− mice had a marked reduction in the number of CD8SP or CD4SP in the thymus and the periphery (Fig. 3). The failure of transgenic TCR expression to rescue positive selection in Flvcr-deleted thymocytes demonstrates that the effect of FLVCR loss is downstream of TCRα re-arrangement.
FIGURE 3.
Positive selection is impaired in Flvcr−/− thymocytes expressing transgenic TCR. CD4 and CD8 expression on thymocytes from Rag1−/− mice transplanted with Flvcr+/+ and Flvcr−/− BM expressing transgenes encoding the MHC class I-restricted OT-I TCR (A), or the MHC class II-restricted OT-II TCR (B). Enumeration of CD8SP and CD4SP thymocytes (lower left in A and B) and of CD8+ Vα2+ peripheral blood (PB) lymphocytes in OT-I mice (A, lower right) and CD4+ Vα2+ PBL in OT-II mice (B, left lower). *p=0.02 **p<.0001. Data are representative of 2 separate experiments with 3–4 mice per group.
FLVCR is required for peripheral T cell survival
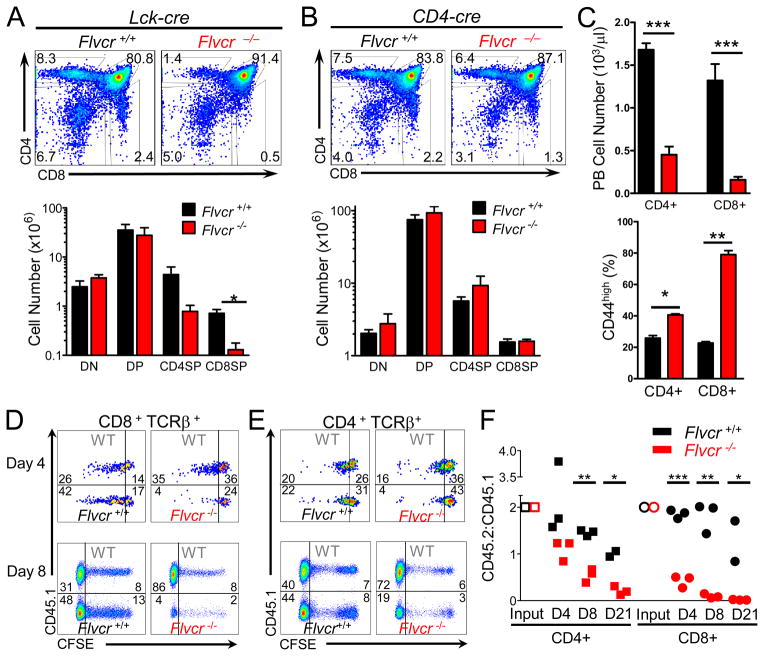
To characterize the temporal requirement for FLVCR during T cell development, Flvcrflox/flox mice were crossed to Lck-cre or CD4-cre transgenic mice (16). Lck-cre mice express cre in the T cell lineage during the early DN stage, leading to early DN Flvcr deletion. The FLVCR protein has a long half-life (29), but as there is a strong proliferative burst at the end of the DN stage, we predicted that in Flvcrflox/flox;Lck-cre mice, FLVCR protein would be largely diluted by multiple cell divisions, resulting in little FLVCR at the DP stage and consequently a block in T cell development. In CD4-cre mice, cre is expressed in the late DN/early DP stage, and there is little subsequent proliferation (30). Therefore we predicted that thymic development in Flvcrflox/flox;CD4-cre mice would proceed normally through to the SP stage.
Flow analysis of Flvcrflox/flox;Lck-cre and control thymocytes showed that early Flvcr deletion blocked development beyond the DP stage, and Flvcrflox/flox;Lck-cre mice had decreased numbers of CD4SP and CD8SP cells (Fig. 4A). Few peripheral CD4+ or CD8+ T cells were seen in Flvcrflox/flox;Lck-cre mice (data not shown). Thus FLVCR was required for primary T cell development and not only for T cell development after bone marrow transplantation.
FIGURE 4.
FLVCR is required for peripheral T cell survival. (A) CD4 and CD8 staining of thymocytes from Flvcrflox/flox;Lck-cre and control mice (upper) with absolute cell numbers of thymus subsets shown in lower panel. *p=0.007. (B) CD4 and CD8 staining of thymocytes of Flvcrflox/flox;CD4-cre and control mice (upper) with absolute cell numbers of thymus subsets shown below. (C) Absolute numbers of CD4+ and CD8+ peripheral blood T lymphocytes in Flvcrflox/flox;CD4-cre and control mice (upper). Percentages of CD4+ and CD8+ peripheral blood T lymphocytes from Flvcrflox/flox;CD4-cre and control mice with high CD44 surface expression (lower). *p=0.003 **p=0.0005 ***p≤0.0001. (D–E) CFSE-labeled Flvcrflox/flox;CD4-cre or Flvcr+/+;CD4-cre thymocytes were mixed at a ratio of 2:1 with CD45.1 (WT) control thymocytes and adoptively transferred into sublethally irradiated Rag1−/− mice. 4 and 8 days later, spleens were harvested and CFSE dilution of transferred thymocytes was analyzed by flow cytometry. Dot plots are gated on CD8 + TCRβ + (D) or CD4 + TCRβ+ cells (E). Inset numbers show frequency of cells in each gate. (F) CD45.2:CD45.1 ratio of adoptively-transferred SP thymocytes and splenic lymphocytes at days 4 and 8 post-transfer. CD45.2:CD45.1 ratios were normalized to the CD45.2:CD45.1 ratio of the input CD4SP or CD8SP. *p<0.03 **p<0.0008 ***p<0.0001. Data are representative of 4 separate experiments with 4–6 mice per group (A–C). Data are representative of 3 separate experiments with 10–12 mice per group total (D–F).
In contrast to Flvcrflox/flox;Lck-cre mice, Flvcrflox/flox;CD4-cre mice had relatively normal thymic T cell development (Fig. 4B). In spite of normal thymic T cell development, Flvcrflox/flox;CD4-cre mice had few peripheral CD4+ or CD8+ T cells (Fig. 4C upper). The few CD8+ T cells present were nearly all CD44-high (Fig. 4C lower), and when Flvcrflox/flox;CD4-cre peripheral T cells were sorted and genomic Flvcr deletion assessed, CD8+ T cells showed 49.3% deletion (S.D.=0.03, n=3). Thus, the few CD44highCD8+ T cells present in Flvcrflox/flox;CD4-cre mice were likely due to homeostatic proliferation of a small number of incompletely-deleted CD8+ T cells.
To determine whether Flvcrflox/flox;CD4-cre SP were maturing normally, we examined surface expression of CD24 and CD69 on Flvcrflox/flox;CD4-cre and control SP. Few CD24-high cells (ISP, DP precursors) were found among either Flvcrflox/flox;CD4-cre or control CD4 or CD8SP (Fig. S2E). CD69 down-regulation is required for Sphingosine-1-phosphate (S1P) receptor 1 (S1PR1) expression and function (31). The S1P1/S1PR1 interaction mediates SP thymic emigration in response to S1P1 gradients from peripheral blood (31). Flvcrflox/flox;CD4-cre CD4 and CD8SP evinced similar down-regulation of CD69 as compared to controls (Fig. S2E). Thus deletion of Flvcr late during the DP stage allowed for the production of normal numbers of phenotypically normal SP.
To determine the fate of Flvcrflox/flox;CD4-cre SP thymocytes in the periphery, we adoptively transferred CFSE-labeled CD45.2 Flvcrflox/flox;CD4-cre or Flvcr+/+;CD4-cre thymocytes mixed with WT CD45.1 thymocytes into sublethally irradiated Rag1−/− mice. CFSE analysis of recipient splenocytes is shown (Fig. 4, D and E) with each plot showing CFSE dilution by CD45.1 WT competitors above CD45.1-negative Flvcr+/+ (left plots) or Flvcr−/− thymocytes (right plots). At day 4 and day 8 post-transfer, Flvcr−/− CD8+ T cells showed little CFSE dilution as compared to co-transferred CD45.1 WT CD8+ T cells or WT littermate Flvcr+/+ CD8+ T cells transferred into another host (Fig. 4, D and F). Similarly, only WT CD4+ T cells had proliferated by day 8 (Fig. 4, E and F). These results suggest that FLVCR is not only required for the DP to SP transition during T cell development, but also for the survival and/or proliferation of T cells in the periphery under lymphopenic conditions.
Re-expression of FLVCR in deleted BM rescues T cell development
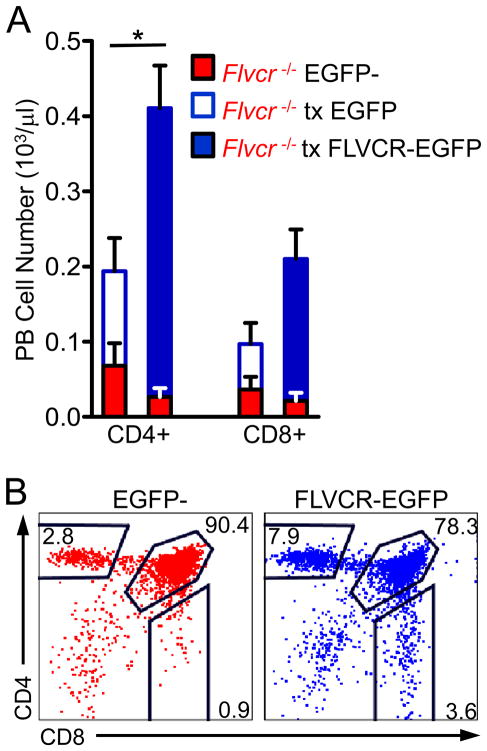
To confirm that the failure of Flvcr-deleted hematopoietic cells to generate T cells was due to loss of FLVCR expression and not to an off-target effect, we performed rescue transplants with Flvcrflox/flox;Lck-cre BM transduced with vectors encoding full-length FLVCR-EGFP fusion protein (FLVCR-EGFP) or EGFP alone. Re-expression of FLVCR led to increased numbers of peripheral CD4+ and CD8+ T cells as compared to EGFP-transduced controls (Fig. 5A), and thymic analysis showed that while there were few CD4SP or CD8SP among the EGFP-negative Flvcr-deleted thymocytes, there were normal numbers of CD4SP and CD8SP in the EGFP+ thymocytes in the same mouse (Fig. 5B). A recent report has suggested that an alternate splice form of Flvcr leads to mitochondrial targeting of a partial FLVCR (FLVCR1b), and that FLVCR1b exports heme from the mitochondria (32). The rescue construct we used encodes full-length FLVCR without intronic sequences and is unlikely to be spliced; these data suggest that full-length FLVCR is necessary and sufficient for T cell development.
FIGURE 5.
FLVCR re-expression in Flvcrflox/flox;Lck-cre BM rescues T cell development. (A) Absolute numbers of CD4+ and CD8+ peripheral blood lymphocytes. Red bars show numbers of EGFP-negative (non-transduced) Flvcrflox/flox;Lck-cre lymphocytes while blue bars show numbers of EGFP-positive lymphocytes transduced (tx) either with EGFP only (open) or FLVCR-EGFP (filled). The bars show the mean and s.e.m. of 3–4 mice per group. *p=0.02 (comparison between EGFP-transduced and FLVCR-EGFP-transduced). (B) Representative flow plots of thymocytes from a recipient of Flvcrflox/flox;Lck-cre BM transduced with FLVCR-EGFP. The left plot shows CD4 versus CD8 staining of the EGFP-negative (non-transduced) thymocytes while the right plot shows EGFP-positive (FLVCR-EGFP-transduced) thymocytes from the same mouse. The data are representative of 2 experiments with 3–5 mice per group.
Flvcr loss does not cause global transcriptional changes in DP thymocytes
The finding that a heme export protein is required for T cell development is surprising because in contrast to developing erythroid cells, whose main function is to synthesize heme and hemoglobin, there is no known specific role for heme in T cell development or function. Heme binds to and inhibits the transcription factor BACH2, a negative regulator of BLIMP1, the master regulator of plasma cell differentiation, thus affecting humoral immunity (33). A recent study showed loss of regulatory T cells in Bach2−/− mice (34) though other CD4+ and CD8+ T cell subsets were present in normal numbers. Thus FLVCR function during T cell development is unlikely to be mediated by BACH2. However, this raised the question whether FLVCR loss/heme dysregulation alters transcription in DP thymocytes. Therefore, we carried out transcriptional profiling on sorted Flvcr−/− and Flvcr+/+ DP thymocytes, but surprisingly, there were very few transcriptional differences (Supplemental Fig. 3A). The increased expression of Cox6a2 seen by profiling (Supplemental Table 1 and Supplemental Fig. 3B) was validated in independent cohorts by qRT-PCR (Supplemental Fig. 3C). COX6A2 is a subunit of respiratory cytochrome c oxidase (complex IV) expressed in striated muscle and shown to play a role in respiratory uncoupling in muscle (35). Studies in yeast demonstrated that COX6 expression is up-regulated by heme (36), thus the increase in Cox6a2 expression in Flvcr-deleted thymocytes may result from increased heme levels. The lack of transcriptional alterations in Flvcr-deleted DP subsets implies that FLVCR loss abruptly disrupts development in the late DP stage, rather than the block occurring at this stage due to cumulative deleterious effects during earlier stages.
DISCUSSION
In this paper, we demonstrate that FLVCR, an MFS protein previously shown to transport heme and to be required for erythroid development, is absolutely required for αβ T cell development beyond the DP stage. Through competitive transplantation, we showed that FLVCR’s role in developing thymocytes is T cell-intrinsic. Flvcr-deleted thymocytes failed selection, as shown by the failure of FLVCR-deficient thymocytes to up-regulate CD69, and by the failure of TCR-transgene expression to rescue development of Flvcr-deleted thymocytes. Through the use of mouse strains in which FLVCR could be deleted early or late during thymic T cell development, we showed that early deletion of Flvcr during the DN stage led to a block in T cell development, while late deletion of Flvcr during the early DP stage allowed for the formation of CD4 and CD8SP. However, these SP were unable to proliferate and/or survive in the periphery.
All aerobic cells require heme for oxidative phosphorylation and other key functions; however excess intracellular free heme is toxic and must be carefully controlled. We analyzed expression of the genes encoding key heme regulators during T cell development and found that expression of Flvcr parallels that of Alas1, supporting the notion that FLVCR may act as a safety valve to export excess heme. Moreover, HMOX1, which breaks down free heme, is down-regulated after the DN stage; therefore FLVCR may be particularly critical for heme regulation after the DN stage in thymocytes.
The finding that FLVCR is required for T cell development beyond the DP stage and peripheral T cell survival is surprising because it supports a novel and specific role for heme metabolism in T cell development. Because heme can regulate gene expression by binding directly to transcription factors, we carried out transcriptional profiling. One explanation for the paucity of transcriptional alterations in Flvcr-deleted DP cells as compared to controls is that FLVCR loss blocks T cell development by non-transcriptional means. Metabolite profiling and heme measurement on whole thymus from deleted or control mice revealed no significant differences (data not shown), and metabolite and heme levels from sorted Flvcr-deleted DP and control thymus were below the limit of detection, likely because the DP subset is metabolically and transcriptionally quiescent due to c-myc down-regulation (37). We are working to develop more sensitive reporter assays to measure heme in vivo and in ex vivo cells in order to determine heme levels during T cell development. Nonetheless, the increased expression of Cox6a2 seen in Flvcr-deleted DP suggests that heme levels are increased in Flvcr-deleted thymocytes. This raises the interesting question of why careful heme regulation might be required in T cells but not other lymphoid lineages. One avenue for future studies is to leverage the differential requirement for FLVCR in αβ but not γδ T cells to identify specific functions or characteristics of αβ thymocytes/T cells that require more stringent heme metabolism.
There is growing appreciation that metabolites modify the epigenome, thereby regulating cell development and differentiation and contributing to oncogenesis (6, 38, 39). Recently it was demonstrated that in cancers with succinate dehydrogenase mutations, the TCA intermediate and heme building block succinate accumulates and acts as an oncometabolite, causing DNA hypermethylation and transcriptional alterations (Supplemental Fig. 1) (40, 41). Epigenetic changes often precede transcriptional changes in development (42), and one possibility is that FLVCR loss and heme accumulation causes epigenetic alterations in developing thymocytes. DP thymocytes are transcriptionally and metabolically quiescent but epigenetically poised to reactivate transcription and initiate CD4+ or CD8+ T lineage differentiation once the cells pass positive and negative selection; FLVCR loss may disrupt epigenetic programming in these cells so that even DP with a TCR capable of selection cannot transition to the SP state. Even in a model in which Flvcr-deleted thymocytes could develop to the SP stage, these SP were not able to survive or proliferate in the periphery in a lymphopenic host. This finding raises the possibility that FLVCR is required for T cells to transition between environments, for example between different thymic niches or between lymphoid niches. While Flvcr-deleted SP appeared to mature normally as determined by immunophenotyping, there may be underlying gene expression changes that lead to the demise of these cells in the periphery that could be interrogated through future microarray experiments. Our current models did not allow us to conclusively determine the role of FLVCR in mature peripheral T cells, but the requirement for FLVCR at the DP-SP transition and at the SP-peripheral T cell transition raises the question of whether FLVCR is required in mature T cells, and in particular whether FLVCR is required for naïve T cell differentiation upon antigen stimulation. We are developing models in which to address these questions.
Heme synthesis has been regarded as a generic housekeeping function occurring in all cells (43); however our finding that FLVCR is specifically required for αβ T cell and erythroid development, but not that of other hematopoietic lineages, suggests that there are important lineage-specific aspects of heme metabolism. Though there is now great interest in metabolic control of cell function and differentiation, much of the focus has been on enzymatic pathways, and less is known about the intra- and intercellular transport of metabolites. Recently, it was discovered that mutations in a ubiquitously-expressed magnesium cation importer, MAGT1, caused T cell immunodeficiency and defects in T and NK cell cytotoxic function (44, 45). This revealed a more specific role for Mg2+ in T cell and NK cell signaling in addition to this cation’s universal role as a co-factor in nucleic acids and enzyme activity (46). Similarly heme may have a specific and unique function in αβ T cells in addition to its other global functions. Our finding that FLVCR specifically blocks T cell development at the DP stage, together with the temporally-regulated models of FLVCR function in T cell development we generated, will provide the tools needed to determine the unique role of FLVCR and heme in T cell development and ultimately lead to new insights into metabolic regulation of T cell development and function.
Supplementary Material
Acknowledgments
This work was supported by NIH grants K08 CA158069 (to M.P.), R01 HL031823 (to J.L.A.), R01 AI64318 (to P.J.F.), and American Society of Hematology Basic Science Fellow Award (to M.P.). M.P. was supported by T32 CA009515, J.J.D was supported by P30 CA015704-39, and J.C. was supported by P30 DK072437.
We thank R. T. Doty for technical support and helpful discussion; T. Mittelstaedt and M. Black from the UW Flow Cytometry Facility; and R. Basom and A. Dawson from the FHCRC Genomics Shared Resource.
Abbreviations used in this article
- BM
bone marrow mononuclear cells
- CR
competitive repopulation
- FeLV-C
feline leukemia virus subgroup C
- FLVCR
Feline Leukemia Virus subgroup C Receptor
- polyI
C, polyinosinic:polycytidylic acid
Footnotes
The microarray data have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50202) with accession code GSE50202.
DISCLOSURES
The authors declare no competing financial interest.
References
- 1.Kim HJ, Khalimonchuk O, Smith PM, Winge DR. Structure, function, and assembly of heme centers in mitochondrial respiratory complexes. Biochim Biophys Acta. 2012;1823:1604–1616. doi: 10.1016/j.bbamcr.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furuyama K, Kaneko K, Vargas PD. Heme as a magnificent molecule with multiple missions: heme determines its own fate and governs cellular homeostasis. Tohoku J Exp Med. 2007;213:1–16. doi: 10.1620/tjem.213.1. [DOI] [PubMed] [Google Scholar]
- 3.Chen JJ. Regulation of protein synthesis by the heme-regulated eIF2alpha kinase: relevance to anemias. Blood. 2007;109:2693–2699. doi: 10.1182/blood-2006-08-041830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaasik K, Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004;430:467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- 5.Handschin C, Lin J, Rhee J, Peyer AK, Chin S, Wu PH, Meyer UA, Spiegelman BM. Nutritional regulation of hepatic heme biosynthesis and porphyria through PGC-1alpha. Cell. 2005;122:505–515. doi: 10.1016/j.cell.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 6.Feng D, Lazar MA. Clocks, metabolism, and the epigenome. Mol Cell. 2012;47:158–167. doi: 10.1016/j.molcel.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khan AA, Quigley JG. Control of intracellular heme levels: heme transporters and heme oxygenases. Biochim Biophys Acta. 2011;1813:668–682. doi: 10.1016/j.bbamcr.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tracz MJ, Alam J, Nath KA. Physiology and pathophysiology of heme: implications for kidney disease. J Am Soc Nephrol. 2007;18:414–420. doi: 10.1681/ASN.2006080894. [DOI] [PubMed] [Google Scholar]
- 9.Reddy VS, Shlykov MA, Castillo R, Sun EI, Saier MH., Jr The major facilitator superfamily (MFS) revisited. FEBS J. 2012;279:2022–2035. doi: 10.1111/j.1742-4658.2012.08588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quigley JG, Yang Z, Worthington MT, Phillips JD, Sabo KM, Sabath DE, Berg CL, Sassa S, Wood BL, Abkowitz JL. Identification of a human heme exporter that is essential for erythropoiesis. Cell. 2004;118:757–766. doi: 10.1016/j.cell.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 11.Keel SB, Doty RT, Yang Z, Quigley JG, Chen J, Knoblaugh S, Kingsley PD, De Domenico I, Vaughn MB, Kaplan J, Palis J, Abkowitz JL. A heme export protein is required for red blood cell differentiation and iron homeostasis. Science. 2008;319:825–828. doi: 10.1126/science.1151133. [DOI] [PubMed] [Google Scholar]
- 12.Riedel N, Hoover EA, Gasper PW, Nicolson MO, Mullins JI. Molecular analysis and pathogenesis of the feline aplastic anemia retrovirus, feline leukemia virus C-Sarma. J Virol. 1986;60:242–250. doi: 10.1128/jvi.60.1.242-250.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abkowitz JL. Retrovirus-induced feline pure red blood cell aplasia: pathogenesis and response to suramin. Blood. 1991;77:1442–1451. [PubMed] [Google Scholar]
- 14.Anderson LJ, Jarrett WF, Jarrett O, Laird HM. Feline leukemia-virus infection of kittens: mortality associated with atrophy of the thymus and lymphoid depletion. J Natl Cancer Inst. 1971;47:807–817. [PubMed] [Google Scholar]
- 15.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 16.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 17.Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 18.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 19.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Persons DA, Allay JA, Allay ER, Ashmun RA, Orlic D, Jane SM, Cunningham JM, Nienhuis AW. Enforced expression of the GATA-2 transcription factor blocks normal hematopoiesis. Blood. 1999;93:488–499. [PubMed] [Google Scholar]
- 21.Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene therapy. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 22.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 23.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and computational biology solutions using R and Bioconductor. Springer; New York, N.Y: 2005. pp. 397–420. [Google Scholar]
- 24.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 25.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 26.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see) Nat Rev Immunol. 2014;14:377–391. doi: 10.1038/nri3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prinz I, Sansoni A, Kissenpfennig A, Ardouin L, Malissen M, Malissen B. Visualization of the earliest steps of gammadelta T cell development in the adult thymus. Nat Immunol. 2006;7:995–1003. doi: 10.1038/ni1371. [DOI] [PubMed] [Google Scholar]
- 29.Yanatori I, Yasui Y, Miura K, Kishi F. Mutations of FLVCR1 in posterior column ataxia and retinitis pigmentosa result in the loss of heme export activity. Blood Cells Mol Dis. 2012;49:60–66. doi: 10.1016/j.bcmd.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 30.Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, Marks H, Adams DJ, Schatz DG, Aragon L, Fisher AG, Krangel MS, Nasmyth K, Merkenschlager M. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476:467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cyster JG, Schwab SR. Sphingosine-1-phosphate and lymphocyte egress from lymphoid organs. Annu Rev Immunol. 2012;30:69–94. doi: 10.1146/annurev-immunol-020711-075011. [DOI] [PubMed] [Google Scholar]
- 32.Chiabrando D, Marro S, Mercurio S, Giorgi C, Petrillo S, Vinchi F, Fiorito V, Fagoonee S, Camporeale A, Turco E, Merlo GR, Silengo L, Altruda F, Pinton P, Tolosano E. The mitochondrial heme exporter FLVCR1b mediates erythroid differentiation. J Clin Invest. 2012 doi: 10.1172/JCI62422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe-Matsui M, Muto A, Matsui T, Itoh-Nakadai A, Nakajima O, Murayama K, Yamamoto M, Ikeda-Saito M, Igarashi K. Heme regulates B-cell differentiation, antibody class switch, and heme oxygenase-1 expression in B cells as a ligand of Bach2. Blood. 2011;117:5438–5448. doi: 10.1182/blood-2010-07-296483. [DOI] [PubMed] [Google Scholar]
- 34.Roychoudhuri R, Hirahara K, Mousavi K, Clever D, Klebanoff CA, Bonelli M, Sciume G, Zare H, Vahedi G, Dema B, Yu Z, Liu H, Takahashi H, Rao M, Muranski P, Crompton JG, Punkosdy G, Bedognetti D, Wang E, Hoffmann V, Rivera J, Marincola FM, Nakamura A, Sartorelli V, Kanno Y, Gattinoni L, Muto A, Igarashi K, O’Shea JJ, Restifo NP. BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis. Nature. 2013;498:506–510. doi: 10.1038/nature12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quintens R, Singh S, Lemaire K, De Bock K, Granvik M, Schraenen A, Vroegrijk IO, Costa V, Van Noten P, Lambrechts D, Lehnert S, Van Lommel L, Thorrez L, De Faudeur G, Romijn JA, Shelton JM, Scorrano L, Lijnen HR, Voshol PJ, Carmeliet P, Mammen PP, Schuit F. Mice deficient in the respiratory chain gene Cox6a2 are protected against high-fat diet-induced obesity and insulin resistance. PLoS One. 2013;8:e56719. doi: 10.1371/journal.pone.0056719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trawick JD, Wright RM, Poyton RO. Transcription of yeast COX6, the gene for cytochrome c oxidase subunit VI, is dependent on heme and on the HAP2 gene. J Biol Chem. 1989;264:7005–7008. [PubMed] [Google Scholar]
- 37.Mingueneau M, Kreslavsky T, Gray D, Heng T, Cruse R, Ericson J, Bendall S, Spitzer MH, Nolan GP, Kobayashi K, von Boehmer H, Mathis D, Benoist C, Best AJ, Knell J, Goldrath A, Jojic V, Koller D, Shay T, Regev A, Cohen N, Brennan P, Brenner M, Kim F, Rao TN, Wagers A, Heng T, Ericson J, Rothamel K, Ortiz-Lopez A, Mathis D, Benoist C, Bezman NA, Sun JC, Min-Oo G, Kim CC, Lanier LL, Miller J, Brown B, Merad M, Gautier EL, Jakubzick C, Randolph GJ, Monach P, Blair DA, Dustin ML, Shinton SA, Hardy RR, Laidlaw D, Collins J, Gazit R, Rossi DJ, Malhotra N, Sylvia K, Kang J, Kreslavsky T, Fletcher A, Elpek K, Bellemare-Pelletier A, Malhotra D, Turley S. The transcriptional landscape of alphabeta T cell differentiation. Nat Immunol. 2013;14:619–632. doi: 10.1038/ni.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katada S, Imhof A, Sassone-Corsi P. Connecting threads: epigenetics and metabolism. Cell. 2012;148:24–28. doi: 10.1016/j.cell.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Kaelin WG, Jr, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Letouze E, Martinelli C, Loriot C, Burnichon N, Abermil N, Ottolenghi C, Janin M, Menara M, Nguyen AT, Benit P, Buffet A, Marcaillou C, Bertherat J, Amar L, Rustin P, De Reynies A, Gimenez-Roqueplo AP, Favier J. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;23:739–752. doi: 10.1016/j.ccr.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 41.Killian JK, Kim SY, Miettinen M, Smith C, Merino M, Tsokos M, Quezado M, Smith WI, Jr, Jahromi MS, Xekouki P, Szarek E, Walker RL, Lasota J, Raffeld M, Klotzle B, Wang Z, Jones L, Zhu Y, Wang Y, Waterfall JJ, O’Sullivan MJ, Bibikova M, Pacak K, Stratakis C, Janeway KA, Schiffman JD, Fan JB, Helman L, Meltzer PS. Succinate dehydrogenase mutation underlies global epigenomic divergence in gastrointestinal stromal tumor. Cancer discovery. 2013;3:648–657. doi: 10.1158/2159-8290.CD-13-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 43.Bishop DF, Henderson AS, Astrin KH. Human delta-aminolevulinate synthase: assignment of the housekeeping gene to 3p21 and the erythroid-specific gene to the X chromosome. Genomics. 1990;7:207–214. doi: 10.1016/0888-7543(90)90542-3. [DOI] [PubMed] [Google Scholar]
- 44.Li FY, Chaigne-Delalande B, Kanellopoulou C, Davis JC, Matthews HF, Douek DC, Cohen JI, Uzel G, Su HC, Lenardo MJ. Second messenger role for Mg2+ revealed by human T-cell immunodeficiency. Nature. 2011;475:471–476. doi: 10.1038/nature10246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li FY, Chaigne-Delalande B, Su H, Uzel G, Matthews H, Lenardo MJ. XMEN disease: a new primary immunodeficiency affecting Mg2+ regulation of immunity against Epstein-Barr virus. Blood. 2014;123:2148–2152. doi: 10.1182/blood-2013-11-538686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaigne-Delalande B, Li FY, O’Connor GM, Lukacs MJ, Jiang P, Zheng L, Shatzer A, Biancalana M, Pittaluga S, Matthews HF, Jancel TJ, Bleesing JJ, Marsh RA, Kuijpers TW, Nichols KE, Lucas CL, Nagpal S, Mehmet H, Su HC, Cohen JI, Uzel G, Lenardo MJ C the Immunological Genome. Mg2+ regulates cytotoxic functions of NK and CD8 T cells in chronic EBV infection through NKG2D. Science. 2013;341:186–191. doi: 10.1126/science.1240094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.