Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that regulate various biological pathways. As their role in phagocytosis remains poorly understood, we investigated their impact on phagocytosis in myeloid inflammatory cells. Seven miRNAs (miR-24, -30b, -101, 142-3p, -652-3p, -652-5p, and -1275) that were differentially expressed during monocyte to macrophage (Mφ) and monocyte to dendritic cells (DC) differentiation were screened for their potential role in phagocytosis. Among these, overexpression of miR-24, miR-30b and miR-142-3p in human monocyte-derived Mφ, DC, monocytes and peripheral blood mononuclear cells (PBMCs) significantly attenuate phagocytosis of E. coli and S. aureus, as well as the secretion of inflammatory mediators including TNF-α, IL-6, and IL-12p40. MiRNA-mediated changes in cytokine profiles were observed at transcriptional and/or post-transcriptional levels and importantly exhibit miRNA specific impact. To examine the underlying mechanism, we monitored the expression of phagocytosis pathway associated genes and identified several genes that were altered in Mφ and DC transfected with miR-24, miR-30b and miR-142-3p mimics. Some of these genes with altered expression also harbor putative miRNA binding sites. We show that miR-142-3p directly regulates protein kinase C alpha (PKCα), a key gene involved in phagocytosis. Interestingly, miR-142-3p and PKCα exhibit antagonistic expression during Mφ and DC differentiation. SiRNA-mediated knockdown of PKCα in Mφ leads to reduced bacterial uptake further highlighting the role of the gene in phagocytosis. Overall, these results demonstrate that miR-24, miR-30b and miR-142-3p regulate phagocytosis and associated cytokine production in myeloid inflammatory cells through modulation of various genes involved in the pathway.
Keywords: microRNA, myeloid cell, phagocytosis, cell differentiation, immunity
Introduction
Monocytes, Mφ, and DC are myeloid inflammatory cells capable of recognizing a plethora of microorganisms via conserved pathogen-recognition receptors (PRRs). Ligation of these PRRs results in a pattern of altered gene expression that enhances pathogen recognition, uptake, and removal (1, 2). This response is fine-tuned by various factors, including the class of pathogen present, the local cytokine milieu, concomitant signaling via damage-associated molecular patterns (DAMPs) (3), as well as site-specific mechanisms of immune regulation.
Critical for innate immunity and host survival is the ability of myeloid inflammatory cells to engulf microorganisms, activate anti-microbial functions, and to degrade, process and present foreign antigen to the adaptive immune system. Each type of myeloid inflammatory cell has a role for which it is best evolved. For example, while Mφ are the most potent at killing engulfed microrganisms, DC are most effective at presenting antigen to lymphocytes (2, 4). In monocytes, phagocytosis induces autocrine and paracrine signaling that influences local monocyte differentiation, monocyte production in the bone marrow, and extravasation into tissues (5).
MiRNAs represent a class of regulatory, non-protein coding RNAs that bind to mRNA transcripts bearing complementary sequences leading to target degradation or translation suppression (6-8), and have recently emerged as potent regulators of diverse biological pathways including cell differentiation, apoptosis and immunity (9, 10). For instance, miR-146a and miR-155 regulate the function of various immune cells, including Mφ and DC, by targeting genes that comprise key components of immune function (11-13). Various miRNAs including cluster miR-17 ~ 92, miR-223 and miR-142-3p have been associated with the differentiation of myeloid and granulocytic cells suggesting lineage specific functions (14, 15).
Recent reports demonstrate that pathogens modulate host miRNA profiles to escape host responses. Mycobacterial infection of Mφ and DC affect phagocytosis, production of cytokines, and results in differential expression of miRNAs (16, 17). Singh et al. (17) showed induced expression of miR-99b in both Mφ and DC infected with M. tuberculosis strain H37Rv which contributed towards impaired cytokine responses. However, miRNA involvement in phagocytosis is poorly studied. In this study, we identify miR-24, miR-30b and miR-142-3p as regulators of phagocytosis in Mφ, DC and monocytes. Overexpression of these miRNAs modulates secretion of TNF-α, IL-6 and IL-12p40 and expression of various genes involved in pathogen recognition and downstream signaling. We further show that miR-142-3p directly regulates PKCα in Mφ and DC and depletion of PKCα had adverse impact on bacterial uptake. Taken together, our data demonstrate that miR-24, miR-30b and miR-142-3p regulate phagocytosis and associated innate responses by targeting genes involved in the pathway.
Materials and Methods
Primary human monocyte isolation and differentiation
Freshly prepared buffy coats were collected from healthy donors (n ≥ 3, Sylvan N. Goldman Oklahoma Blood Institute, Oklahoma City, OK, USA) by density gradient centrifugation as described earlier (18). Briefly, PMBCs were purified using Ficoll Paque™ (GE Healthcare, Piscataway, NJ, USA) based density centrifugation. PBMCs were incubated with magnetic labeled CD14 beads (Miltenyi Biotech, Cologne, Germany) according to manufacturer's instructions. The purity of CD14+ cells was >95% as determined by flow cytometry. For generation of M1 and M2 Mφ, monocytes were plated at 2×106/ml in DMEM supplemented with penicillin (100U/ml) and streptomycin (100μg/ml). After 2 hours, media was removed and replaced with media containing 10% FBS (Life Technologies, Grand Island, NY, USA), and either 1000U/ml rhGM-CSF or 50 ng/ml rhM-CSF (both from Peprotech, Rocky Hill, NJ, USA) for generation of M1 and M2 Mφ, respectively. At day 7, cells were harvested and surface expression of CD14, CD163 and HLA-DR was examined by flow cytometric analysis. For DC, monocytes were cultured in RPMI-1640 supplemented with 10% FBS and rhGM-CSF (1000U/ml) and rhIL-4 (500U/ml) (both from Peprotech). Media was replaced every 72 h.
Transient miRNA transfections
MiScript miRNA mimics (miR-24, -30b, -101, 142-3p, -652-3p, -652-5p, and -1275) and inhibitors were purchased from Qiagen (Germantown, MD, USA). For control, all stars negative mimics (Qiagen) were used. For PKCα knockdown, gene specific and control siRNA were purchased from Sigma (St. Louis, MO, USA). Transient transfections were performed using Lipofectamine 2000 (Life Technologies) according to manufacturer's instructions. Mφ were transfected with mimics or inhibitors at a final concentration of 50 nM while DC, monocytes and PBMCs were transfected at a final concentration of 100 nM. Red siGLO oligos (ThermoScientific, Waltham, MA, USA) were used to determine transfection efficiency.
Flow cytometry
Cells were harvested after treatments and washed in ice-cold phosphate-buffered saline supplemented with 1% (v/v) FBS and 0.08% sodium azide. Cellular debris and detritus was excluded based on size (forward scatter; FSC) and granularity (side scatter; SSC). The FSC/SSC gate for Mφ, DC and monocytes comprised ~60%, ~80-90%, and ~90% of total events, respectively. Couplets were excluded based on SSC vs FSC and SSC vs Pulse width measurements. Fluorescence minus-one samples constituted controls for cells treated with bioparticles. Samples were analyzed using a FACScan or BD Cyan flow cytometer using CellQuest software (BD Biosciences, San Jose, CA, USA). Further analysis was performed using FlowJo software (Tree Star Inc., Ashland, OR, USA).
MTS assay
Cell viability was determined using the CellTiter 96 AQueous Cell Proliferation Assay Kit (Promega, Madison, WI, USA). Briefly, 4×105 cells (Mφ, DC and monocytes) grown in 96-well plates were transfected with miRNA mimics or inhibitors at final concentration mentioned above and assays were performed after 24 h according to manufacturer's instructions.
Phagocytosis assay and imaging
For Mφ (M1 and M2) and DC, cells at a density of 400,000/well (96-well plate) were transfected on day 7 with miScript miRNA mimics, inhibitors or control miRNA mimics (Qiagen). Monocytes and PBMCs were transfected immediately after isolation. Transfection was performed as described above. After 24 h, phagocytosis assay was performed with pHrodo™ Red E. coli BioParticles® conjugate (Invitrogen, Carlsbad, CA, USA) according to manufacturer's instructions. Briefly, the labeled bioparticles were resuspended in Live Imaging Buffer (Life Technologies) and homogenized by sonication for 2 minutes. Culture media was replaced with resuspended labeled E. coli and incubated for 1 h. As a negative control, cells were treated with 5 μM cytochalasin D (Sigma) prior to adding bioparticles. The cells were washed three times with PBS, fixed with 4% PFA and analyzed by flow cytometry. Images were captured using on a Zeiss LSM 710 confocal microscope with 40x/1.2 Water DIC C-Apochromat objective and 2X zoom or EVOS® florescent microscope (Life Technologies) at 20X magnification. Confocal images were processed on ZEN lite software. Experiments were conducted on three independent, randomly selected fields for each donor.
Cytokine analysis
Supernatants were collected after a 4 h and 18 h challenge and analyzed for levels of IL-6, IL-10, IL-12p40 and TNF-α by ELISA (Cytoset, Invitrogen). Cellular levels of cytokines were measured in E. coli challenged Mφ (M1 and M2), DC and monocytes after 4 or 18 h. Supernatants and cell lysates were diluted (1:10) before use. Absorbance was measured at 450nm on a SpectraMax® M2 (Molecular Devices, Sunnyvale, CA, USA) plate reader.
Immunofluorescence and imaging
Intracellular levels of TNF-α were monitored in miRNA, inhibitor or control transfected Mφ challenged with E. coli for 4h. Cells were fixed in 4% paraformaldehyde for 10 minutes and incubated in 10% sheep serum in 0.1% PBS-Tween for 2h. Staining was performed with TNF-α antibody (1:100 dilution; ab1793, Abcam, Cambridge, England) for overnight at 4°C in PBS containing 1% BSA and 0.1% Tween. Cells were washed five times before incubation with DyLight-488 goat anti-mouse antibody (Biolegend, San Diego, CA, USA) at 1:250 dilution for 1 h. After washing five times, cells were counterstained with Hoescht nuclear staining dye. Images were captured on the EVOS® microscope at 20X magnification and processed on Adobe CS5 Photoshop (Adobe Systems Incorporated, San Jose, CA, USA).
Total RNA isolation, cDNA synthesis and PCR array
Mφ and DC transfected with miRNA mimic or inhibitor were harvested after 36h. Total RNA was isolated using the miRNeasy micro kit (Qiagen) according to manufacturer's instructions. First strand cDNA was synthesized from 500 ng total RNA using the RT kit (Qiagen). A custom PCR array plate (96-well) containing 88 different genes involved in phagocytosis was used to assess gene expression (Qiagen). One microgram of cDNA was aliquoted onto each well and real-time PCR performed using a StepOne 7500 thermocycler (Applied Biosystems, Carlsbad, CA, USA). Expression levels were normalized with respect to beta-2 microglobulin as it demonstrated the most consistent levels across all transfections. Next, the fold change was calculated with respect to the negative miRNA control. Finally, the ratio of miRNA inhibitor and miRNA mimic was calculated. Real-time PCR for three randomly selected genes were also carried out in two independent donors.
Total RNA was isolated from E. coli challenged Mφ and DC and the expression of proinflammatory cytokine mRNA analyzed by RT-PCR. To monitor PKCα mRNA levels, total RNA was also isolated from monocytes, differentiating Mφ and DC at various time points. The data was presented as normalized fold change with standard deviation.
Reporter constructs and dual luciferase assays
The cloning of PKCα 3′UTR was performed as described previously (19). Briefly, PKCα 3′UTR was amplified (Forward Primer- GCACTCGAGGAAAGGCCGGGATAAACCTA; Reverse primer- ATGCGGCCGCACACACAGCAAGAGGGGAG) using genomic DNA isolated from freshly prepared PBMCs. The amplicons were digested with XhoI and NotI and cloned into the psiCHECK™-2 vector (Promega). The 3′UTR of PKCα was PCR amplified using Phusion Taq polymerase (New England Biolabs, Ipswich, MA, USA). Dual-luciferase assays were carried out in a 96-well format. In brief, HEK293 cells were seeded at the density of 2×104 in DMEM supplemented with 10% fetal bovine serum. All the transfections were performed in quadruplicate using 0.3 μL Lipofectamine 2000 (Life Technologies), 60 ng dual luciferase reporter plasmids, and a final concentration of 2 pmol or 5 pmol synthetic miRNA mimics or scramble (Qiagen). After 36h post-transfection, cells were lysed in passive lysis buffer (Promega) and dual luciferase assays (Promega) were performed using the Lumat plate reader (Turner BioSystems, Sunnyvale, CA, USA). For each reporter 3′UTR construct, the Rluc/Fluc value obtained was normalized to the value obtained for psiCHECK™-2 no-insert control (EV) cotransfected with the same miRNA mimic. The values obtained were plotted as histograms, where EV is set at 1.
Western blotting
Mφ or DC transfected with miRNA mimics, inhibitors or control mimics were harvested after 36h and lysed in cell lysis buffer (Cell Signaling Technology, Davers, MA, USA) supplemented with protease inhibitors (Roche, Basel, Switzerland). Lysates were incubated on ice for 30 min and were clarified using centrifugation at 14,000 rpm for 15 min at 4° C, and protein content was estimated using the Bradford assay (BioRad Laboratories, Hercules, CA, USA). Equal amounts of protein were resolved in 10% Mini-PROTEAN® TGX™ (BioRad) gels and electrotransferred to nitrocellulose membranes (GE Healthcare). Membranes were blocked with 5% skimmed milk for 2h and incubated with primary antibodies against PKCα or GAPDH (1:1000; Cell Signaling) overnight at 4° C. The blots were washed with 0.05% TBS/Tween20 for five times before incubating with secondary antibody (1:10,000). Blots were washed five times with 0.05% TBS/Tween20 and protein detection was performed using enhanced chemiluminescence (GE Healthcare). ImageJ software (http://rsbweb.nih.gov/ij/) was used to quantify the results. The values for each lane were normalized with respect to endogenous control GAPDH and compared with the negative control mimic transfection.
Statistical Analysis
Data were analyzed on GraphPad Prism (GraphPad Software, Inc., La Jolla, CA, USA). The results were presented as standard deviation or ±SEM from three independent replicates and experiments were conducted at least three times. P-values were calculated using Student's t-test. P<0.05 were considered significant.
Results
Overexpression of miR-24, miR-30b and miR-142-3p attenuate phagocytosis by primary human Mφ
Previously, our miRNA profiling of monocytes during differentiation to Mφ and DC identified differential expression of numerous miRNAs including miR-24, -30b, -101, -142-3p, -652-3p, -652-5p, and -1275 (GEO reference number GSE60839). To examine their function, we first screened these miRNAs in monocyte-derived Mφ for their potential role in phagocytosis. Using labeled oligonucleotides we achieved ~90% transfection efficiency in Mφ, as assessed by flow cytometry and fluorescent microscopy (Supplemental Fig. 1A, 1B). Primary monocyte-derived Mφ were transiently transfected with miRNA mimics, corresponding inhibitors, or control mimic. Rhodamine-labeled E. coli bioparticles were added after 24 h and their uptake was observed by fluorescent microscopy and quantified by flow cytometry. Overexpression of miR-24, -30b and 142-3p mimics but not others (miR-101, -652-3p, -652-5p, and -1275) significantly decreased the uptake of bacteria, compared to either miRNA inhibitors or control mimic (Fig. 1A, Supplemental Fig. 2A) indicating an inhibitory role of these miRNAs on phagocytosis. Cells treated with cytochalasin D, an inhibitor of actin polymerization, blocked the uptake of labeled bacteria and served as negative controls (Supplemental Fig. 2A). Importantly, this miRNA-mediated effect on phagocytosis was not specific to E. coli (Gram negative) as similar results were observed with S. aureus (Gram positive) bioparticles (Supplemental Fig. 2B) suggesting that these miRNAs regulate phagocytosis independent of the pathogen. We further examined possible cooperative effects of miRNA mimics on phagocytosis by co-transfecting Mφ with the three miRNA mimics. Compared to the individual miRNA mimics, no further reduction in phagocytosis was observed (Supplemental Fig. 2C). Therefore, subsequent experiments were focused on individual miRNA transfections.
FIGURE 1.
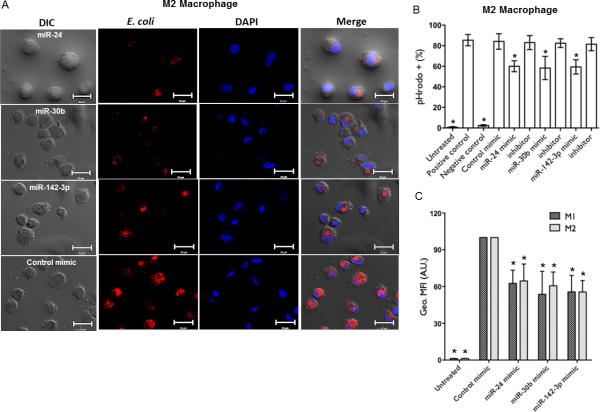
Overexpression of miRNA mimics attenuate phagocytosis in human Mφ. (A) Day 6 primary monocyte-derived Mφ were transfected with 50 nM miR-24, miR-30b and miR-142-3p mimics or control mimic and phagocytosis assays performed by replacing media with pHrodo™ E. coli bioparticles, as described in Materials and Methods. After 1h incubation, images from three different fields were captured on a confocal microscope and are representative of assays on 4 independent donors. Scale bar, 20 μm. Mφ were harvested after phagocytosis assays and analyzed by FACS. (B) Histograms showing percent E. coli positive Mφ. As a negative control, Mφ were treated with cytochalasin D (5μM) for 30 min prior to conducting phagocytosis assays. Mφ treated with E. coli were used as a positive control. (C) M1 and M2 Mφ were transfected with miRNA mimics or control and challenged with E. coli. Comparison of geometric mean florescence intensity (Geo. MFI) as assessed by FACS. Geometric mean florescence intensity (Geo. MFI) values were obtained and normalized to control mimic transfected cells. Data are presented as ±SEM from five donors. *p<0.01 is considered significant.
Flow cytometric analysis of Mφ showed ~20-30% reduction in the number of cells positive for bioparticle uptake in the presence of miR-24, miR-30b and miR-142-3p mimics, but not with inhibitors, nor control mimic (p<0.01; Fig. 1B). Further, comparing the geometric mean fluorescent intensity (geo. MFI) of the cells, being semi-quantitative for the number of bioparticles taken up on a per cell basis, indicated a larger degree of inhibition (~40-60%) than the percentage uptake alone, and similar to that observed by microscopy (Fig. 1C). Bacterial challenge to miRNA mimic transfected cells had no effect on viability as assessed by MTS assay (Supplemental Fig. 1C).
Depending on the activation status, Mφ phenotype can be broadly classified into two categories viz., classically (M1) or alternatively (M2) activated. The M1 Mφ are primarily involved in Th1 responses, lymphokine production and degradation of intracellular pathogens while, M2 Mφ trigger Th2 responses, immunotolerance and tissue remodeling. Incubation of Mφ with different cytokines can alter their activation status (20). In the presence of M-CSF and GMCSF monocytes differentiate into Mφ with M2 and M1 phenotype, respectively. In our hands, M1 Mφ were HLA-DR high, CD163 low and CD14 low compared to donor matched M2 Mφ and correlates with reported phenotyping (Supplemental Fig. 3A). To investigate whether miR-24, miR-30b and miR-142-3p impact bacterial uptake by M1 Mφ, cells were transfected with miRNA mimics and assessed for phagocytic potential. Overexpression of mRNA mimics inhibited uptake of bacteria with respect to control mimics (Fig. 1C, Supplemental Fig. 3B). We also noticed that M1 Mφ were less efficient in phagocytosis compared to M2 Mφ, as expected with the former phenotype (Supplemental Fig. 3B). Nonetheless, flow cytometry analysis show that percent MFI values were similarly reduced by miRNA mimics in M1 and M2 Mφ (~40- 60%; Fig. 1C). Together, these results demonstrate that overexpression of miR-24, miR-30b and miR-142-3p attenuate phagocytosis by primary M1 or M2 Mφ.
miR-24, miR-30b and miR-142-3p inhibit phagocytosis by DC and monocytes
Having demonstrated a role of miR-24, miR-30b or miR-142-3p in regulating phagocytosis in Mφ, we investigated whether these miRNAs regulate this process in other myeloid inflammatory phagocytes viz., DC and monocytes. Cells were transiently transfected with labelled oligonucleotides and transfection efficiency assessed by microscopy and flow cytometry. We were able to achieve ~85% and ~60% transfection efficiency in DC and monocytes, respectively (Supplemental Fig. 1A, 1B).
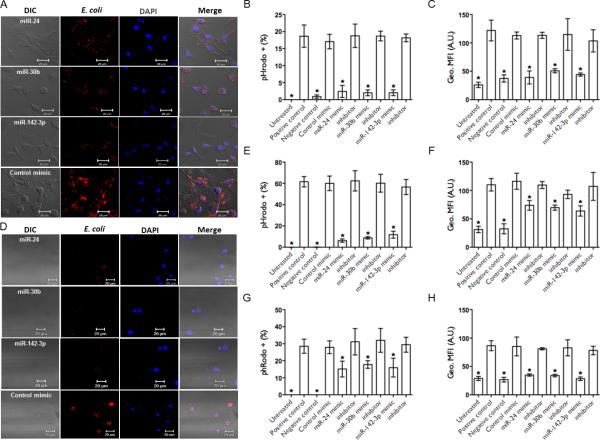
DC transfected with mimics, but not with inhibitors or control mimic showed reduced E. coli uptake as observed under microscopy (Fig. 2A, Supplemental Fig. 3D). Quantitative analysis on flow cytometry showed ~4-6 folds decrease in bacterial uptake by mimic transfected DC (Fig. 2B). The geo. MFI data further confirmed reduced uptake (Fig. 2C). In monocytes, overexpression of miRNA mimics significantly reduced phagocytosis of bioparticles (Fig. 2D) while miRNA inhibitors or control mimic had no adverse effect (data not shown). Flow cytometric analysis showed 5-8 fold less bacterial uptake in miRNA mimic transfected monocytes (Fig. 2E). However, compared with Mφ and DC, we noticed fewer uptake of bioparticles during the assay incubation (1 h) which increased over time (data not shown). Nonetheless, miRNA mimic mediated affects were still evident as determined by the geo. MFI data that reflect reduced E. coli uptake (Fig. 2F).
FIGURE 2.
Overexpression of miRNA mimics attenuate phagocytosis in human DC, monocytes and PBMCs. Monocyte-derived DC and monocytes were transfected with miR-24, miR-30b and miR-142-3p miRNA mimic, inhibitors or control mimics for 24 h. Phagocytosis assays were performed with pHrodo™ E. coli bioparticles. Representative confocal images of corresponding (A) DC and (D) monocytes with internalized E. coli are shown. Images were captured using settings mentioned above. Scale bar, 20 μm. Cells were harvested and analyzed by flow cytometry. Histograms show percent E. coli positive (B) DC, (E) monocytes and (G) PBMCs. Geometric mean florescence intensity (geo. MFI) assessed by FACS in (C) DC, (F) monocytes and (H) PBMCs. Data are presented as ± SEM from four donors. *p<0.01 is considered significant.
Finally, we examined the regulatory role of these miRNAs in PBMCs. The composition of PBMCs isolated from buffy coats is ~70-90% lymphocytes, 10-30% monocytes and 1-2% DC (personal observation). We show that PBMCs transfected with miRNA mimics also exhibit significant decrease in phagocytosis (Fig. 2G and 2H). Overall, these results unequivocally show that miR-24, miR-30b and miR-142-3p regulate phagocytosis in myeloid cells.
MiRNA mimics decrease TNF-α production
Secretion of TNF-α is associated with active phagocytosis (21). Mφ, DC, and monocytes were transiently transfected with miRNA mimics, inhibitors, or control mimic and incubated with E. coli. Supernatant levels of TNF-α were significantly lower in cells transfected with miR-24, miR-30b and miR-142-3p mimics compared to inhibitors or control mimics (Fig. 3). In M2 Mφ, miRNA mimic-mediated reduction of TNF-α secretion was more pronounced at 18 h while, in DC and monocytes the secretion of TNF-α was affected more significantly at 4 h (Fig. 3A, 3C and 3D). Moreover, time-dependent increase in TNF-α secretion was observed for M2 Mφ and monocytes but not DC suggesting cell specific innate responses. In addition, we compared the impact of miRNA mimics on the release of TNF-α in M1 and M2 Mφ. We noted that transfection of M1 Mφ with miR-30b and miR-142-3p resulted in a greater inhibition of secreted TNF-α levels compared to M2 Mφ at 4 h. Conversely, M2 Mφ transfected with miR-24 mimic had a greater impact on M2 Mφ indicating subtle differences likely associated with the respective phenotype (Fig. 3B). Taken together, TNF-α secretion profiles correlated with the uptake of bacteria indicating that miRNA mimics also affect phagocytosis-associated innate immune responses.
FIGURE 3.
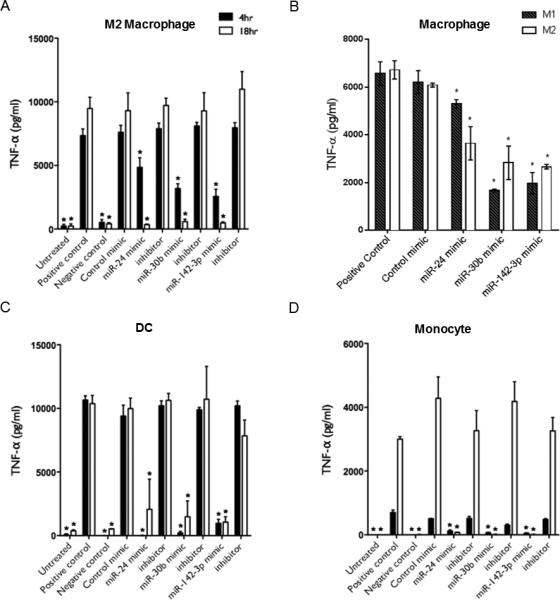
MiRNA mimics modulate secretion of proinflammatory cytokine TNF-α. Production of TNF-α was assessed in myeloid inflammatory cells stimulated by E. coli subsequent to transfection with miR-24, miR-30b and miR-142-3p mimics, inhibitors, or control mimic. Mφ and DC were transfected at day 7 of culture; monocytes at day 0, and pHrodo™ E. coli bioparticles were added 24 h later. Production of TNF-α detected in (A) M2 Mφ, (C) DC, and (D) monocytes was assayed by ELISA after 4 h and 18 h of stimulation. (B) Secreted TNF-α level in M1 and M2 Mφ challenged with E. coli for 4 h. N=3-6 biological replicates. Mean and SD are shown. *p<0.01 is considered significant.
MiRNA mimics modulate IL-6 and IL-12p40 production
During phagocytosis, antigen is processed for presentation to lymphocytes and the cytokines produced influence the type of adaptive response that is mounted, for example bacterial responses include cytokines that promote Th1 and Th17 differentiation. We assayed the levels of the Th1/Th17-promoting cytokines IL-6 and IL-12p40, being a shared component of (Th1-promoting) IL-12 and (Th17-promoting) IL-23. Transfected Mφ and DC were incubated with E. coli and supernatant levels of IL-6 and IL-12p40 assayed by ELISA. After 4 and 18 h, IL-6 and IL-12p40 were reduced in the presence of miRNA mimics, but not inhibitors or control mimic (Fig. 4A and 4B). We also compared the levels of IL-6 in M1 and M2 Mφ after 4 h time point. The miRNA mimics downregulated the IL-6 secretion in supernatants in both M1 and M2 Mφ (Fig. 4C). However, we noticed relatively less impact of miR-24 on IL-6 secretion by M1 as observed with TNF-α earlier. Compared to DC, secretion of IL-12p40 in M2 Mφ showed marked reduction at both time-points further highlighting the differences in miRNA mediated effects on cell function. Concurrently, secretion of the anti-inflammatory/immunosuppressive cytokine IL-10 by both M2 Mφ and DC was detectable at 18 h but not 4 h and was comparable in miRNA mimics, inhibitors and control transfected cells (Fig. 4D, Mφ data shown). These results show that the effects of miRNA mimics are not only confined to the local and immediate innate response, but also extend to the release of factors that orchestrate adaptive immunity.
FIGURE 4.
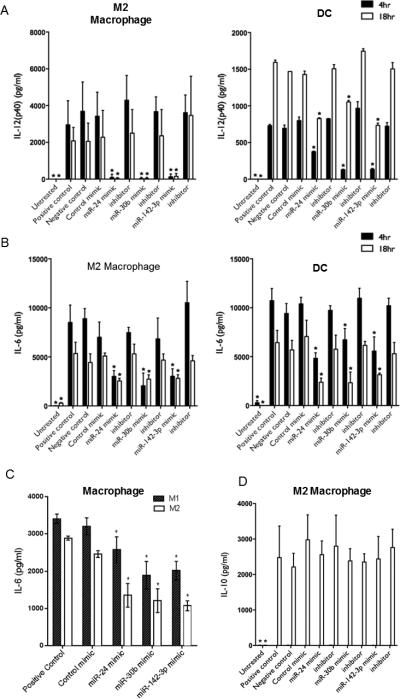
MiRNA mimics modulate secretion of IL-6 and IL-12p40 but not IL-10. Mφ and DC were transfected at day 7 of culture with miR-24, miR-30b and miR-142-3p mimics, inhibitors, or control mimic and stimulated by E. coli pHrodo™ bioparticles 24 h later. Supernatant levels of (A) IL-12p40, (B) IL-6 and (D) IL-10 were assayed after 4 h and 18 h in supernatants of Mφ and DC by ELISA. (C) Comparison of supernatants IL-6 levels in M1 and M2 Mφ challenged with E. coli for 4 h. N=3-6 biological replicates. Mean and SD are shown. *p<0.01 is considered significant.
Differential regulation of proinflammatory cytokine expression by miRNA mimics
Since we performed miRNA profiling in M2 Mφ and that overexpression of miR-24, miR-30 and miR-142-3p show similar impact on the phagocytosis and associated innate responses by both M1 and M2 Mφ, subsequent experiments were conducted on M2 Mφ alone. Unless otherwise mentioned, the Mφ in context will be M2 phenotype. To determine if the reduced secretion of proinflammatory cytokines is a consequence of transcriptional regulation, we monitored mRNA levels of IL-6, IL-12p40 and TNF-α in Mφ and DC transfected with miRNA mimics and inhibitors. We observed that IL-12p40 mRNA levels were remarkably reduced (2-8 fold) in the presence of miRNA mimics and correlated with the ELISA data. Mφ transfected with miRNA inhibitors exhibited relatively decreased expression of TNF-α compared to miRNA or control mimics (Fig. 5A). Similarly, expression of IL-6 was significantly downregulated in Mφ transfected with miR-24 and miR-30b inhibitors but not miR-142-3p. However, comparable expression of TNF-α and IL-6 transcript was observed in mimic and inhibitor transfected DC (Fig. 5A). Of note, comparing the expression profiles with the unchallenged controls clearly show that enhanced expression of miRNA mimics does not compromise the signaling leading to induction of cytokines (data not shown). These observations suggest that enhanced expression of miRNA mimics attenuate innate responses but does not compromise the signaling leading to induction of cytokines examined.
FIGURE 5.
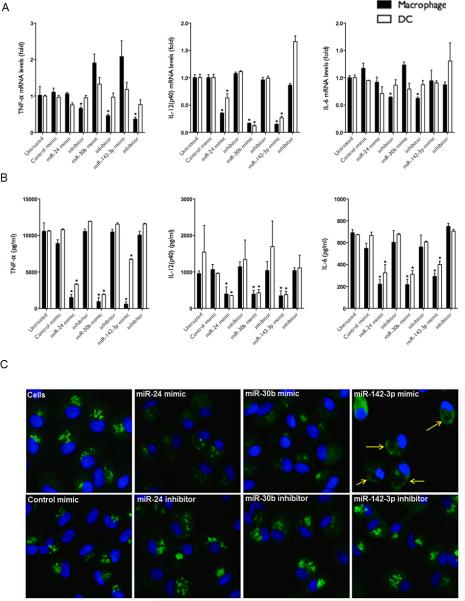
MiRNA mimics differentially modulate transcription and intracellular accumulation of cytokines. MiRNA mimics, inhibitors or control transfected cell were challenged with E. coli and cytokine mRNA levels were examined by qRT-PCR after 4 h. (A) Expression levels of TNF-α, IL-6 and IL-12p40 are shown for Mφ and DC. (B) ELISA showing intracellular cytokine expression in Mφ and DC. (C) Immunostaining shows decreased intracellular TNF-α in miRNA mimic overexpressing Mφ. Images were captured using a fluorescent microscope and a 20x objective lens. Green signals indicate intracellular TNF-α. Cells were counterstained with Hoechst nuclear dye. Scale bar, 200μm. The yellow arrows point to accumulation of TNF-α localized to the cell periphery. The data represents three different fields captured from each well from at least three independent experiments. Student's t-test was conducted to calculate p values. *p< 0.01 was considered as significant.
These observations tempted us to investigate whether miRNA mimics interfere with the production of proinflammatory cytokines. Intracellular levels of TNF-α, IL-6 and IL-12p40 were examined in cell lysates prepared from transfected Mφ and DC challenged with E. coli for 4h. We observed a significant decrease in cellular TNF-α, IL-6 and IL-12p40 in Mφ and DC transfected with miRNA mimics compared to controls which corroborates with the secretory profiles (Fig. 5B). These results show that while transcript levels of TNF-α and IL-6 are increased to a degree, in the presence of mimics, the corresponding proteins are downregulated indicating possible post-transcriptional interference by these miRNAs.
To further confirm these findings, we examined the cellular levels of TNF-α by immunostaining. E. coli challenged Mφ transfected with miRNA inhibitors or control mimics exhibit sharp punctate structures, however, diffuse and relatively reduced staining was observed with corresponding miRNA mimics (Fig. 5C). This corroborates with the intracellular cytokine profiles (Fig. 5B). Further, we observed marked differences in the staining pattern of TNF-α in the presence of mimics. Unlike inhibitor or control transfected cells, miRNA mimics do not show a consistent tapering pattern as expected for secreted proteins; from the nucleus towards the cell periphery. Interestingly, we also noticed accumulation of TNF-α localized to the cell periphery in miR-142-3p mimic transfected cells (Fig. 5C, yellow arrows). These results suggest that protein translation and/or secretion pathways are rendered less functional by these miRNAs.
Genes involved in phagocytosis are downregulated by miRNA mimics
To examine the underlying mechanism of miRNA-mediated impact on phagocytosis, we next focused on identifying genes altered by overexpression of miRNA mimics. To this end, we selected 88 genes associated with phagocytosis and analyzed their levels by quantitative RTPCR. Mφ and DC were transfected with miRNA mimics, inhibitors or control mimic and gene expression levels monitored after 36 h. PCR array profiling showed that mRNA levels were modulated in the presence of miRNA mimics compared with control mimic. Dramatic modulation of gene expression was observed in DC but only a few genes were affected in Mφ. In DC, a total of 29, 25 and 40 genes were altered in presence of miR-24, miR-30b and miR-142-3p, respectively (Supplemental Table I). On the other hand, altered expression of 1, 6 and 4 genes was noted in Mφ transfected with miR-24, miR-30b and miR-142-3p, respectively (Supplemental Table I). Venn diagram shows that most of the altered genes were common to these miRNAs while, only few genes were specific to each miRNA (Fig. 6A). Intriguingly, the majority of the deregulated genes that were common targets of the miRNAs are receptors for pathogen recognition, however, miR-142-3p uniquely impacted signaling associated genes in DC. The PCR array results were confirmed by an independent RT-PCR analysis of three genes altered by miRNA mimics on two additional donors (data not shown).
FIGURE 6.
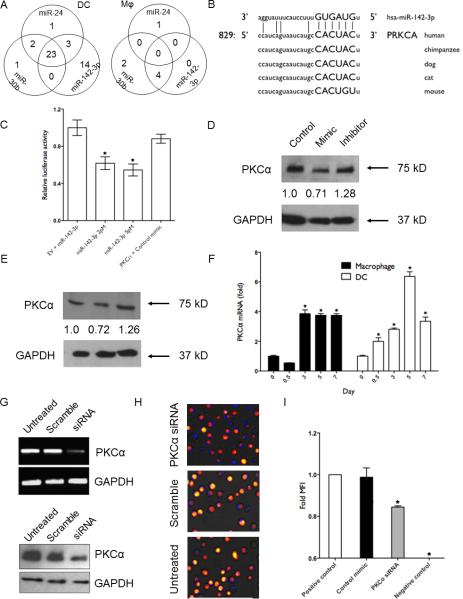
PKCα is a novel target of miR-142-3p. (A) Venn diagram showing the distribution of unique and overlapping genes downregulated by miR-24, miR-30b and miR-142-3p mimics in DC and Mφ. (B) Sequence alignment of predicted miR-142-3p binding site in the 3′UTR of PKCα of various mammals. (C) HEK293 cells were cotransfected with PKCα 3′UTR construct and with either miR-142-3p or control mimic. Renilla activity was normalized to firefly activity and the ratios subsequently normalized to empty vector transfected with miR-142-3p mimic set as 1. Data are expressed as ± SEM of four independent transfections. Western blot analysis of PKCα levels in miR-142-3p overexpressing (D) Mφ and (E) DC. Day 7 differentiated cells were transfected with miR-142-3p mimic, inhibitor or control mimics. Cell lysates were prepared after 36 h and PKCα levels were detected by western blotting. GAPDH was used as internal control. Densitometric analysis of bands was performed with ImageJ. The normalized PKCα in control-transfected cells was set as 1. (F) Expression kinetics of PKCα in Mφ and DC. Expression levels of PKCα transcript were monitored at indicated time points during the monocyte to Mφ and monocyte to DC differentiation. (G) siRNA mediated knockdown of PKCα as observed by RT-PCR and immunoblotting in Mφ. GAPDH served as endogenous control. Phagocytosis assays were performed with labelled E. coli in Mφ transfected with PKCα or scramble siRNA and untreated cells. (H) Representative images were captured using a florescence microscope and (I) Geo. MFI values obtained from flow cytometry values. Scale bar, 200 μm. The data presented are representative of three independent experiments in each cell type. Student's t-test was conducted to calculate p values. *p<0.01 was considered significant.
We next focused on identifying genes with altered expression that are direct targets of miRNAs. For this, 3′ untranslated regions (UTRs) were scanned for potential miRNA binding sites using miRWalk (22). We noted that among the genes exhibiting altered expression, only a few possess corresponding miRNA binding sites (data not shown). Further, a small fraction of predicted targets exhibited altered expression indicating that these genes may be subject to post-transcriptional regulation by miRNA. Taken together, these results show that miR-24, miR-30b and miR-142-3p modulate phagocytosis by directly and indirectly targeting key genes linked to phagocytosis.
MiR-142-3p targets protein kinase C, alpha (PKCα)
PKCα was among the list of altered genes with a potential miR-142-3p binding site. The 3′UTR region comprising 829-851 nts harbor miR-142-3p interacting sequences which are highly conserved in mammals (Fig. 6B). To validate the predicted miRNA-target interaction, we performed dual luciferase assays. HEK293 cells transfected with miR-142-3p (2 pmol) showed reduced (~50%) luciferase activity when cotransfected with plasmids containing 3′UTR of PKCα, but not with control mimic (p≤0.001; Fig. 6C). Increasing the concentration of miRNA mimic to 5 pmol, however, had no further significant effect on luciferase activity. This suggests that miR-142-3p can bind target sites with high affinity leading to efficient silencing of the reporter gene. We then investigated the functional consequence of this miRNA-target interaction in primary Mφ and DC. Overexpression of miR-142-3p decreased PKCα levels by ~30% compared to control mimic in both the primary cells (Fig. 6D, 6E). However, more pronounced differences (~42% decrease) in the levels of PKCα was observed comparing miRNA mimic and corresponding inhibitor as the later blocks the miRNA function leading to increased target gene translation. Overall, these results validate that PKCα is a bona-fide target of miR-142-3p.
Our previous miRNA profiling data of differentiating Mφ and DC showed that the expression of miR-142-3p is maximal at day 3 and decreased thereafter (Gene Expression Omnibus submission GSE60839). We monitored the expression of its novel target PKCα during the course of differentiation. Indeed, in both Mφ and DC, a significant increase (~3 fold) in the expression levels of PKCα was noted after day 1 which peaked around day 3 in Mφ and day 5 in DC (Fig. 6F). These results establish the antagonistic expression of miR-142-3p and its novel target, PKCα. We further examined the role of PKCα in bacterial phagocytosis. Mφ were transiently transfected with PKCα-targeting siRNA. Fig. 6F shows significant knockdown of PKCα at the mRNA and protein level. Moreover, cells transfected with PKCα siRNA exhibit reduced phagocytosis of labelled E. coli as observed by florescent microscopy (Fig. 6H). Flow cytometry analysis shows approximately 20% reduction in MFI in PKCα transfected cells compared to control siRNA (p<0.05; Fig. 6I). These results further support direct involvement of PKCα in bacterial phagocytosis.
Discussion
The importance of miRNA for the function of myeloid inflammatory cells has been revealed in recent years, particularly in the area of pathogen recognition and innate signaling; however, very little has been reported linking miRNA expression with phagocytosis. In this study we show that overexpression of miR-24, miR-30b and miR-142-3p, which are downregulated during Mφ and DC differentiation, attenuate phagocytosis by myeloid inflammatory cells. Moreover, both M1 and M2 Mφ show similar defects in bacterial uptake upon forced expression of the tested miRs. These results demonstrate a conserved role of miRNA in regulating the key myeloid inflammatory cell function that bridges innate and adaptive arms of immunity.
Recent studies highlight the role of miRNA in phagocytosis. For instance, mice Mφ J774.A1 challenged with non-virulent strain, Mycobacterium smegmatis show induced expression of miR-142-3p (23), lending support to our data that overexpression of miR-142-3p inhibits uptake of bacteria in myeloid cells. miR-142-3p regulates expression of actin-binding protein N-wasp, thereby disturbing actin dynamics which is required for efficient phagocytosis (23). Our PCR array results also show reduced N-wasp levels in miR-142-3p overexpressing DC (Supplemental Table I). In another report, induced expression of miR-615-3p in hypersplenic Mφ was shown to promote phagocytosis by targeting ligand-dependent nuclear receptor corepressor (LCoR) expression, a negative regulator of PPARγ (24). Taken together, these findings indicate that dysregulation of miRNAs may render cells susceptible to pathogen invasion and disease.
All three miRNAs (miR-24, miR-30b and miR-142-3p) identified in this study as regulator of phagocytosis were down-regulated during differentiation. A growing body of evidence indicates that expression patterns of miRNAs permits cells to enter a new functional state and it is possible that dysregulation at any point can result in aberrant phagocytic function. For instance, Ma et al. (25) demonstrated that downregulation of miR-29 in T-cells allows for enhanced expression of IFN-γ, which promotes classical Mφ activation and Th1-dominant adaptive responses appropriate to bacterial infection. Similarly, we have previously reported that LPS-mediated reduction of miR-29b regulates expression of its target, IL-6 receptor (IL-6R), a key component of innate immune responses (19). These findings signify a role of miRNA in shaping immune responses. Indeed, during phagocytosis it was observed that cells overexpressing miRNA mimics secrete significantly less IL-6, IL-12p40 and TNF-α compared to inhibitors or control, while the anti-inflammatory cytokine IL-10 was unaffected. Interestingly, miRNA mimic transfected cells show modulation of cytokines with the decrease in IL-12p40 and increase, albeit less, in IL-6 and TNF-α mRNA levels.
Transcription of TNF-α and IL-6 expression is predominantly regulated by NF-κB and MAPK (in particular p38) in LPS treated Mφ and other cells (26-28). Our PCR array data did not show significant differences in the expression of NF-κB and p38 transcripts. IL-12p40 expression, besides regulated by NFκB and Jun, is also regulated by transcription factor nuclear factor of activated T cells (NFAT), which in turn is activated by CLEC7A/Dectin-1 (29). MiR-24, miR-30b and miR-142-3p mimics significantly decreased (>15 fold) the levels of CLEC7A and likely block IL-12p40 transcription. However, other factors and post-transcriptional mechanisms including mRNA synthesis, transcript stability, translation and protein secretion could also control production of these cytokines. This is supported by the observation that despite the differences in mRNA profiles, cellular and secreted levels of both TNF-α and IL-6 was downregulated. TNF-α imunostaining in E. coli challenged Mφ also show reduced cellular staining in the presence of miRNA mimic further validating the cellular cytokine profiles. Interestingly, remarkable differences in the TNF-α staining pattern were observed among the miRNA mimics with miR-142-3p transfected Mφ exhibiting accumulation of TNF-α localized to the cell periphery. These findings suggest that besides modulating shared pathways, each miRNA mimic also modulate unique pathways that direct immune responses.
Phagocytosis can be broadly categorized into three steps: ligand binding, signal transduction, and internalization. Dysregulation at any point can result in aberrant phagocytic function. TLRs are the largest family involved in pathogen recognition. Our PCR array data showed miRNA mimic mediated downregulation of various TLRs including TLR1, 2, 3, 4, 5, 6 and 9. While TLR1, 2, 4, 5 and 6 are present on the cell surface TLR3 and 9 are located on endosomal membranes (30). It is thus likely that these miRNAs can regulate recognition and removal of both extracellular and intracellular pathogens. In addition to recognition, these TLRs are intimately involved in phagocytosis, the production of inflammatory mediators, activation of the adaptive immune system, and enhancement of microbicidal mechanisms (31, 32). Besides TLRs, opsonic receptors, which include receptors for immunoglobulins (Ig) and complement components, also participate in pathogen recognition. We noticed modulation of Fc receptor (FcR) genes specifically, FcRα, FcεRI, FcεRII, FcεRIγ and complement component 3. Opsonins circulate in the bloodstream detecting and binding to foreign antigens; increasing phagocytic uptake via Fc receptors that include FcγRI (CD64), FcγRII (CD32) and FcγRIII (CD16) (33, 34). Interestingly, we also found that similar to E. coli and S. aureus uptake, overexpression of miR-24, miR-30b and miR-142-3p mimics have profound effect on opsonin-mediated phagocytosis (Naqvi et al., manuscript in preparation). The miRNA-mediated impact on phagocytosis appears independent of the receptors involved and signifies their key role in the process.
PKCα belongs to family of serine/threonine kinases that regulate various cellular functions including cell differentiation, apoptosis, and LPS signaling (35, 36). Employing dual-luciferase assays we show PKCα is a novel target of miR-142-3p. Enforced expression of miR-142-3p down-regulated PKCα expression in Mφ and DC. The siRNA-mediated knockdown of PKCα led to reduced uptake of bacteria supporting its key role in the process. This observation is in agreement with previous studies demonstrating a requirement of PKCα during phagocytosis (37-39). However, we noted that siRNA knockdown of PKCα alone had relatively less impact (~20%) on phagocytosis compared to miRNA overexpression (~70%). This is most likely attributed to the capability of miRNAs to simultaneously regulate a wide range of targets and/or functional redundancy among PKC isoforms.
Initial reports on PKCα showed that inhibiting its activity blocked secretion of various proinflammatory cytokines in LPS challenged Mφ and was associated with defects in phagocytosis (40). Consistent with these findings, we observed significant reduction in IL-6, IL-12p40 and TNF-α release in miR-142-3p transfected cells. Interestingly, PKCα is also reported to regulate TLR signaling by interacting with TLRs. Unlike other conventional PKCs, however, PKCα can trigger both MyD-dependent (TLR2) and –independent (TLR3) signaling (41, 42). TLR2 signaling terminates with the activation of NFκB and MAPK while TLR3 activation leads to IRF3-mediated IFNβ expression, thus, PKCα can affect both early and late innate responses. Our studies demonstrate that miR-142-3p expression is downregulated during myeloid cell differentiation while its novel target PKCα level increases. The role of PKC isoforms in differentiation of monocyte-derived Mφ is well established. In line with earlier reports, we support the significance of PKCα in both Mφ and DC differentiation. Importantly, this functional miRNA-target interaction highlights the regulatory potential of miRNA in both differentiation and function of immune cells.
In conclusion, we present miRNAs that significantly inhibit phagocytosis and the production of inflammatory mediators in myeloid inflammatory cells including monocytes, M1/M2 Mφ and DC. These miRNAs target pathways associated with and phagocytosis, cytokine production and inflammation, and the productive pairing of the innate to the adaptive immune system. This study opens new avenues to explore novel roles of miRNAs identified here in diseases with impaired phagocytosis and related inflammatory disorders.
Supplementary Material
Acknowledgements
Portions of this work were carried out in the Confocal Microscopy Facility via the Research Resources Center at the University of Illinois at Chicago.
This study was supported by the NIH/NIDCR [DE021052].
Abbreviations
- miRNA
micro-RNA
- siRNA
short-interfering RNA
- TNF-α
tumor necrosis factor-α
- IL-6
interleukin-6
- IL-12p40
interleukin -12 subunit p40
- IL-10
interleukin- 10
- Mφ
macrophages
- DC
dendritic cells
- ELISA
enzyme-linked immunosorbent assay
- PCR
polymerase chain reaction
- RT-PCR
Reverse transcription-PCR
- TLR
toll-like receptor
- FcR
fragment c receptor
- PKCα
protein kinase C alpha
- NF-kB
nuclear factor kappa-light-chain-enhancer of activated B cells
- LPS
lipopolysaccharide
References
- 1.Soehnlein O, Lindbom L. Phagocyte partnership during the onset and resolution of inflammation. Nat. Rev. Immunol. 2010;10:427–439. doi: 10.1038/nri2779. [DOI] [PubMed] [Google Scholar]
- 2.Kubach J, Becker C, Schmitt E, Steinbrink K, Huter E, Tuettenberg A, Jonuleit H. Dendritic Cells: Sentinels of immunity and tolerance. Int. J. Hemat. 2005;81:197–203. doi: 10.1532/IJH97.04165. [DOI] [PubMed] [Google Scholar]
- 3.Matt U, Sharif O, Martins R, Furtner T, Langeberg L, Gawish R, Elbau I, Zivkovic A, Lakovits K, Oskolkova O, Doninger B, Vychytil A, Perkmann T, Schabbauer G, Binder CJ, Bochkov VN, Scott JD, Knapp S. WAVE1 mediates suppression of phagocytosis by phospholipid-derived DAMPs. J. Cli. Invest. 2013;123:3014–3024. doi: 10.1172/JCI60681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auffray C, Sieweke MH, Geissmann F. Blood Monocytes: Development, heterogeneity, and relationship with dendritic cells. Ann. Rev. Immunol. 2009;27:669–692. doi: 10.1146/annurev.immunol.021908.132557. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Didiano D, Hobert O. Molecular architecture of a miRNA-regulated 3′ UTR. RNA. 2008;14:1297–1317. doi: 10.1261/rna.1082708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makeyev EV, Maniatis T. Multilevel Regulation of Gene Expression by MicroRNAs. Science. 2008;319:1789–1790. doi: 10.1126/science.1152326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 10.Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat. Rev. Immunol. 2008;8:120–130. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- 11.O'Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Ann. Rev. Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 12.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jurkin J, Schichl YM, Koeffel R, Bauer T, Richter S, Konradi S, Gesslbauer B, Strobl H. miR-146a is differentially expressed by myeloid dendritic cell subsets and desensitizes cells to TLR2-dependent activation. J. Immunol. 2010;184:4955–4965. doi: 10.4049/jimmunol.0903021. [DOI] [PubMed] [Google Scholar]
- 14.Croce CM. MicroRNA dysregulation in acute myeloid leukemia. J. Clin. Onco. 2013;31:2065–2066. doi: 10.1200/JCO.2013.48.8239. [DOI] [PubMed] [Google Scholar]
- 15.Wang XS, Gong JN, Yu J, Wang F, Zhang XH, Yin XL, Tan ZQ, Luo ZM, Yang GH, Shen C, Zhang JW. MicroRNA-29a and microRNA-142-3p are regulators of myeloid differentiation and acute myeloid leukemia. Blood. 2012;119:4992–5004. doi: 10.1182/blood-2011-10-385716. [DOI] [PubMed] [Google Scholar]
- 16.Sharbati J, Lewin A, Kutz-Lohroff B, Kamal E, Einspanier R, Sharbati S. Integrated microRNA-mRNA-analysis of human monocyte derived macrophages upon Mycobacterium avium subsp. hominissuis infection. PLoS One. 2011;6:e20258. doi: 10.1371/journal.pone.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh Y, Kaul V, Mehra A, Chatterjee S, Tousif S, Dwivedi VP, Suar M, Van Kaer L, Bishai WR, Das G. Mycobacterium tuberculosis controls miR-99b expression in infected murine dendritic cells to modulate host immunity. J. Biol. Chem. 2012;288:5056–5061. doi: 10.1074/jbc.C112.439778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nares S, Moutsopoulos NM, Angelov N, Rangel ZG, Munson PJ, Sinha N, Wahl SM. Rapid myeloid cell transcriptional and proteomic responses to periodontopathogenic Porphyromonas gingivalis. Am. J. Path. 2009;174:1400–1414. doi: 10.2353/ajpath.2009.080677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naqvi AR, Fordham JB, Khan A, Nares S. MicroRNAs responsive to Aggregatibacter actinomycetemcomitans and Porphyromonas gingivalis LPS modulate expression of genes regulating innate immunity in human macrophages. Inn. Imm. 2013;20:540–551. doi: 10.1177/1753425913501914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murray RZ, Kay JG, Sangermani DG, Stow JL. A role for the phagosome in cytokine secretion. Science. 2005;310:1492–1495. doi: 10.1126/science.1120225. [DOI] [PubMed] [Google Scholar]
- 22.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk-database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Bettencourt P, Marion S, Pires D, Santos LF, Lastrucci C, Carmo N, Blake J, Benes V, Griffiths G, Neyrolles O, Lugo-Villarino G, Anes E. Actin-binding protein regulation by microRNAs as a novel microbial strategy to modulate phagocytosis by host cells: the case of N-Wasp and miR-142-3p. Front. Cell Infect. Microbiol. 2013;3:19. doi: 10.3389/fcimb.2013.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang A, Zhang S, Li Z, Liang R, Ren S, Li J, Pu Y, Yang J. miR-615-3p promotes the phagocytic capacity of splenic macrophages by targeting ligand-dependent nuclear receptor corepressor in cirrhosis-related portal hypertension. Exp. Biol. Med. 2012;236:672–680. doi: 10.1258/ebm.2011.010349. [DOI] [PubMed] [Google Scholar]
- 25.Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, Li N, Yao H, Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-[gamma]. Nat. Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 26.Liu H, Sidiropoulos P, Song G, Pagliari LJ, Birrer MJ, Stein B, Anrather J, Pope RM. TNF-alpha gene expression in macrophages: regulation by NF-kappa B is independent of c-Jun or C/EBP beta. J. Immunol. 2000;164:4277–4285. doi: 10.4049/jimmunol.164.8.4277. [DOI] [PubMed] [Google Scholar]
- 27.Ajizian SJ, English BK, Meals EA. Specific inhibitors of p38 and extracellular signal-regulated kinase mitogen-activated protein kinase pathways block inducible nitric oxide synthase and tumor necrosis factor accumulation in murine macrophages stimulated with lipopolysaccharide and interferon-γ. J. Infect. Dis. 1999;179:939–944. doi: 10.1086/314659. [DOI] [PubMed] [Google Scholar]
- 28.Craig R, Larkin A, Mingo AM, Thuerauf DJ, Andrews C, McDonough PM, Glembotski CC. p38 MAPK and NF-κB collaborate to induce Interleukin-6 gene expression and release: evidence for a cytoprotective autocrine signaling pathway in a cardiac myocyte model system. J. Biol. Chem. 2000;275:23814–23824. doi: 10.1074/jbc.M909695199. [DOI] [PubMed] [Google Scholar]
- 29.Goodridge HS, Simmons RM, Underhill DM. Dectin-1 stimulation by Candida albicans yeast or zymosan triggers NFAT activation in macrophages and dendritic cells. J. Immunol. 2007;178:3107–3115. doi: 10.4049/jimmunol.178.5.3107. [DOI] [PubMed] [Google Scholar]
- 30.O'Neill LA, Golenbock D, Bowie AG. The history of Toll-like receptors -redefining innate immunity. Nat. Rev. Immunol. 2013;13:453–460. doi: 10.1038/nri3446. [DOI] [PubMed] [Google Scholar]
- 31.MacLeod H, Wetzler LM. T cell activation by TLRs: a role for TLRs in the adaptive immune response. Sci STKE. 2007;402:pe48. doi: 10.1126/stke.4022007pe48. [DOI] [PubMed] [Google Scholar]
- 32.Thoma-Uszynski S, Stenger S, Takeuchi O, Ochoa MT, Engele M, Sieling PA, Barnes PF, Rollinghoff M, Bolcskei PL, Wagner M, Akira S, Norgard MV, Belisle JT, Godowski PJ, Bloom BR, Modlin RL. Induction of direct antimicrobial activity through mammalian toll-like receptors. Science. 2001;291:1544–1547. doi: 10.1126/science.291.5508.1544. [DOI] [PubMed] [Google Scholar]
- 33.Anderson CL, Guyre PM, Whitin JC, Ryan DH, Looney RJ, Fanger M. Monoclonal antibodies to Fc receptors for IgG on human mononuclear phagocytes. Antibody characterization and induction of superoxide production in a monocyte cell line. J. Biol. Chem. 1986;261:12856–12864. [PubMed] [Google Scholar]
- 34.van Spriel AB, van den Herik-Oudijk IE, van Sorge NM, Vilé HA, van Strijp JA, van de Winkel JG. Effective phagocytosis and killing of Candida albicans via targeting FcγRI (CD64) or FcαRI (CD89) on neutrophils. J. Infect. Dis. 1999;179:661–669. doi: 10.1086/314643. [DOI] [PubMed] [Google Scholar]
- 35.Dekker LV, Parker PJ. Protein kinase C – a question of specificity. Trends Biochem. Sci. 1994;19:73–77. doi: 10.1016/0968-0004(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 36.Nakashima S. Protein kinase C alpha (PKC alpha): regulation and biological function. J. Biochem. 2002;132:669–675. doi: 10.1093/oxfordjournals.jbchem.a003272. [DOI] [PubMed] [Google Scholar]
- 37.St-Denis A, Caouras V, Gervais F. Descoteaux. Role of protein kinase C-alpha in the control of infection by intracellular pathogens in macrophages. J. Immunol. 1999;163:5505–5511. [PubMed] [Google Scholar]
- 38.Ng Yan Hing JD, Desjardins M, Descoteaux A. Proteomic analysis reveals a role for protein kinase C-alpha in phagosome maturation. Biochem. Biophys. Res. Commun. 2004;319:810–816. doi: 10.1016/j.bbrc.2004.05.054. [DOI] [PubMed] [Google Scholar]
- 39.Holm A, Tejle K, Magnusson KE, Descoteaux A, Rasmusson B. Leishmania donovani lipophosphoglycan causes periphagosomal actin accumulation: correlation with impaired translocation of PKCalpha and defective phagosome maturation. Cell Microbiol. 2001;3:439–447. doi: 10.1046/j.1462-5822.2001.00127.x. [DOI] [PubMed] [Google Scholar]
- 40.St-Denis A, Chano F, Tremblay P, St-Pierre Y, Descoteaux A. Protein kinase C-alpha modulates lipopolysaccharide-induced functions in a murine macrophage cell line. J. Biol. Chem. 1998;273:32787–32792. doi: 10.1074/jbc.273.49.32787. [DOI] [PubMed] [Google Scholar]
- 41.Langlet C, Springael C, Johnson J, Thomas S, Flamand V, Leitges M, Goldman M, Aksoy E, Willems F. PKC-alpha controls MYD88-dependent TLR/IL-1R signaling and cytokine production in mouse and human dendritic cells. Eur. J. Immunol. 2010;40:505–515. doi: 10.1002/eji.200939391. [DOI] [PubMed] [Google Scholar]
- 42.Johnson J, Albarani V, Nguyen M, Goldman M, Willems F, Aksoy E. Protein kinase C alpha is involved in interferon regulatory factor 3 activation and type I interferon-beta synthesis. J. Biol. Chem. 2007;282:15022–15032. doi: 10.1074/jbc.M700421200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.