Abstract
Parasites, defined as eukaryotic microbes and parasitic worms that cause global diseases of human and veterinary importance, span many lineages in the eukaryotic Tree of Life. Historically challenging to study due to their complicated life-cycles and association with impoverished settings, their inherent complexities are now being elucidated by genome sequencing. Over the course of the last decade, projects in large sequencing centers, and increasingly frequently in individual research labs, have sequenced dozens of parasite reference genomes and field isolates from patient populations. This “tsunami” of genomic data is answering questions about parasite genetic diversity, signatures of evolution orchestrated through anti-parasitic drug and host immune pressure, and the characteristics of populations. This brief review focuses on the state of the art of parasitic protist genomics, how the peculiar genomes of parasites are driving creative methods for their sequencing, and the impact that next-generation sequencing is having on our understanding of parasite population genomics and control of the diseases they cause.
The State of Parasite Whole Genome Sequencing
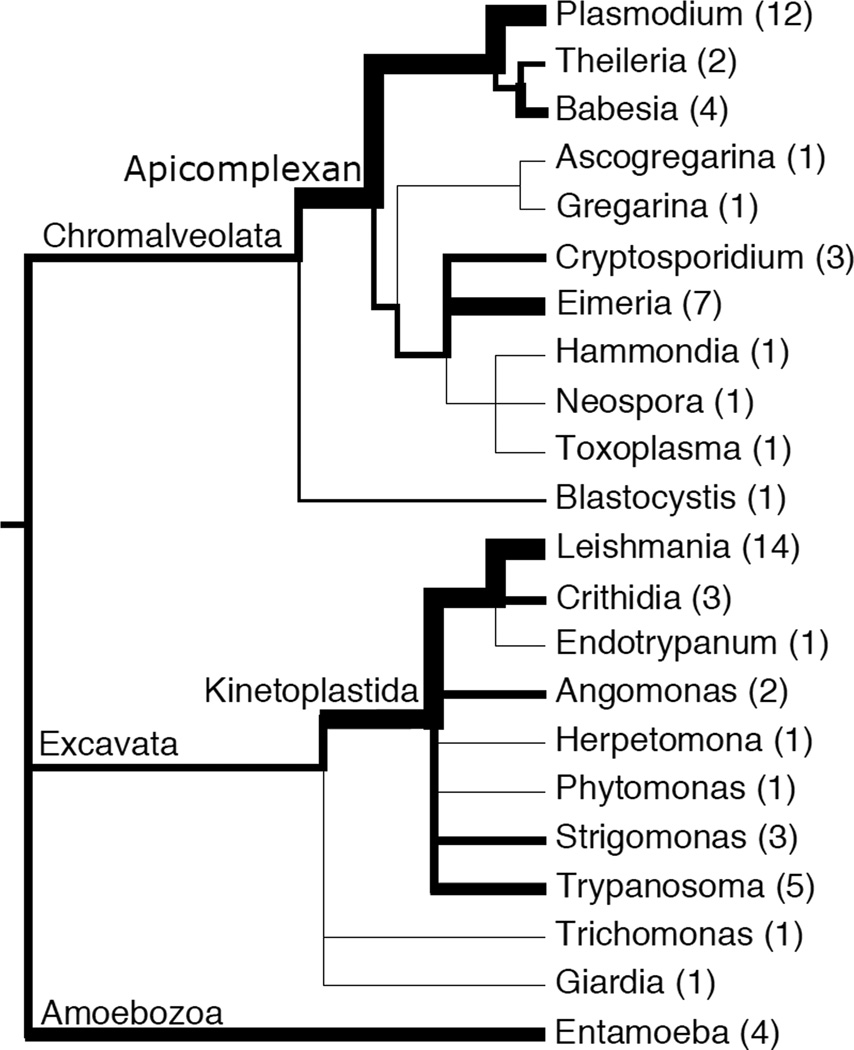
As of August 2014, sixty-five reference genomes of parasitic protists and their close relatives have been deposited in GenBank (Figure 1). The majority of these genomes fall within two phyla: (1) the Kinetoplastidae, which contains parasites responsible for diseases ranging from African sleeping sickness (Trypanosoma brucei) and Chagas disease (Trypanosoma cruzi) to visceral leishmaniasis (Leishmania spp.); and (2) the Apicomplexa, including the genus Plasmodium, whose transmission by the Anopheles mosquito causes malaria in more than 100 countries. Accordingly, research utilizing “comparative genomics” of parasite genomes has been focused within the Plasmodium [1], Leishmania [2] and Trypanosoma [3] clades. The majority of these genomes were sequenced using first-generation Sanger technology, and some have taken many years to complete assembly, gene finding and annotation [4]. More recently, the advent of cheaper, faster and more accurate next-generation sequencing platforms such as those provided by Illumina (e.g., HiSeq series), Roche 454 (e.g., GS Junior) and Life Technologies (e.g., Ion Torrent Personal Genome Machine), has enabled whole genome sequencing of multiple field isolates from patients. These unassembled genomes are deposited in GenBank's Sequence Read Archive or the European Bioinformatics Institute European Nucleotide Archive (http://www.ncbi.nlm.nih.gov/sra and http://www.ebi.ac.uk/ena, respectively) as well as in organism-specific databases such as those hosted by EuPathDB [5]. This new wave of parasite genome sequence data is revolutionizing the study of population genetics of parasites in two major ways: (1) by generating preliminary descriptions of the population genetics of commonly used lab strains, and (2) by enabling “real-time” population genetics of patient field isolates. Examples of these are given below.
Figure 1.
A cartoon phylogenetic tree of parasite genera (and some closely-related free-living relatives) with reference genomes, with branch widths weighted according to the number of genomes (indicated in parentheses) in GenBank as of August 2014. Monophyletic supergroup and phylum name are labeled above their branches.
The Challenges to Genomics Posed by Parasite Biology
Parasite genomes have highly diverse architectures. They vary in properties such as nucleotide bias, for example the extremely AT-rich Plasmodium falciparum genome [6], or the “isochore” structure of Plasmodium vivax chromosomes that have GC-rich cores but AT-rich subtelomeric regions [7]. Many genomes are highly repetitive or replete with transposable elements, for example the amoebic dysentery-causing parasite Entamoeba histolytica [8]. Genome sizes of parasites also vary widely. The first eukaryotic parasite genome to be published, from the microsporidian Encephalitozoon cuniculi, was found to be 2.3 Mb [9], whereas the sexually transmitted parasite Trichomonas vaginalis has a ~160 Mb genome that has undergone recent genome expansion [10].
Such diversity poses unique challenges to whole genome sequencing, including attaining adequate genome coverage, identifying polymorphisms, and obtaining reliable estimates of population genetic parameters. These challenges have fostered new sequencing strategies for sampling patient isolates, such as the “reduced representation” methods [11] that are being used to develop genetic markers for population genomic surveys. One such method, “restriction-site associated DNA sequencing” uses either one (RAD) or a pair (ddRAD) of restriction enzymes in combination with partial sequencing [12,13], and has been employed to resequence ~180 T. vaginalis genomes ([14]; M. Bradic, New York University, unpublished). A second new technology adopted by the parasite genomics community is “hybrid selection”, which uses biotinylated RNA baits designed from a parasite reference genome sequence to capture parasite DNA from a host-parasite DNA mixture [15]. Starting from exceedingly small quantities of patient material, Plasmodium DNA has been purified and enriched up to 40-fold [16,17] -- – a key achievement in our ability to undertake population genetic surveys of parasites that cannot be grown in culture or are grossly contaminated with host genetic material.
Using Genome Sequence Data to Investigate Parasite Population Genetics
Prior to the era of fast and cheap next-generation sequencing (NGS), population studies of parasites were limited to a few genetic loci because of the lack of parasite genome sequence data. These initial studies using small numbers of microsatellite (MS) markers across chromosomes and single nucleotide polymorphisms (SNPs) in single-copy genes provided a preliminary glimpse of the genetic diversity, local and global population structure, and gene flow within and between populations of several different parasite species (see for example [18–20]), and identified loci suitable for classifying patient isolates [21,22]. Such genotyping has also been invaluable in epidemiology studies and disease classification [23,24]. Single-locus studies have also been used to identify mutations associated with parasite phenotypes such as virulence and drug resistance (reviewed for species of the malaria parasite in [25]). More recently, whole genome sequence data have enabled a genome-wide approach to population studies of commonly used parasite lab-adapted strains, and also of natural isolates taken directly from patients. In many instances, this has improved initial estimates of important population genetic parameters (see Glossary). We concentrate here on those parasites for which NGS data are now available that illustrate some of the impact that NGS data are having on population genetic studies of these parasites.
Recombination is an important population genetic parameter to consider in parasites, since the ability of a species to undergo sexual recombination directly impacts the spread of important genes through populations, such as those involved in virulence or drug resistance. Population genetic studies of several parasite species have indicated that genetic exchange is likely to occur or has taken place evolutionarily recently in the species (see for example in T. vaginalis [18], Giardia [26] and other parasite species reviewed in [27]). In the enteric pathogen E. histolytica, analysis of the first reference genome revealed a complement of genes necessary for meiosis, pointing to the possibility of sex in natural populations [28]. However, more substantial evidence was not available until the generation of a large NGS dataset of ten lab-cultured lines from Mexico, Bangladesh, Italy, United Kingdom, Korea and Venezuela [29]. By random sampling of pairs of polymorphic sites on the same reference genome scaffold, the authors found strong evidence that sexual reassortment of chromosomes and meiotic recombination had occurred, suggesting that E. histolytica may reproduce sexually and that a reevaluation of the life-cycle of this species may be warranted. These findings were extended by Gilchrist et al. [30] who used 16 marker loci identified from next generation sequencing datasets of 12 E. histolytica strains to genotype 84 samples. The study noted the extreme diversity present in the species indicative of regular and recent recombination, and was able to link specific loci to clinical phenotypes, supporting the existence of a relationship between the genotype of an E. histolytica strain and its virulence.
In two back-to-back papers, sequencing of four P. vivax isolates adapted to growth in monkeys [31], and three strains of the closely related monkey malaria Plasmodium cynomolgi isolated from macaques in Cambodia and Malaysia [32], showed how NGS data is impacting population genetic studies of species of the monkey malaria clade. P. cynomolgi shares many of the biological and phenotypic traits of its sister taxon P. vivax and has been assumed to be a model system for the study of P. vivax, which cannot be cultured in vitro. Tachibana et al. [32] for the first time identified ~60 genes with dN (the number of nonsynonymous changes per nonsynonymous site) greater than dS (the number of synonymous changes per synonymous site), and ~3,200 genes with dS > dN, over ~4,000 pairs of orthologs within two P. cynomolgi strains, providing clues as to the types of genes subject to different selective pressures in the species. Similarly, whether P. cynomolgi is a good model system for P. vivax was investigated by exploring the degree to which evolution of orthologs between the two species had been constrained over evolutionary time. Of ~4,600 pairs of orthologs analyzed between the two species, less than 2% were found to be under positive selection and at least 81% to be under strong selective constraint, indicating that the genome of P. cynomolgi is highly conserved in single-locus genes compared to P. vivax and emphasizing the value of the monkey malaria species as a biomedical and evolutionary model for studying P. vivax.
Next generation sequencing has improved the ability to detect loci undergoing lineage-specific changes that previously would have been overlooked. In the trypanosomatid Leishmania for example, the generation and refinement of four reference genomes for species within that genus has helped identify chromosomal and gene copy number variation [33]. Large-scale variation in chromosome copy number between species found up to nine supernumerary chromosomes (small accessory chromosomes with high heterochromatic content) within individuals. In contrast, comparisons between species found little difference at the gene level, with only a limited number (2–67) lineage-specific genes identified. At a population level, a recent study compared the genome sequences of 16 clinical isolates of Leishmania donovani that display a gradient in drug susceptibility [34]. The increased resolution of next generation sequencing highlighted nine loci that differ in copy number between drug resistant and susceptible populations of L. donovani. More recent genomic studies in Kinetoplastidiae have explored recombination within the Trypanosoma genus by sequencing subspecies of Trypanosoma brucei [35]. Two isolates, sourced from different geographic regions of Africa, were whole-genome sequenced, revealing a heterozygous ~2.5 Mb region that may underlie the differences in virulence observed between the two subspecies. These explorations have shown that structural changes, such as altering copy number, are an important mechanism for modifying virulence in parasite populations.
While Plasmodium and Leishmania are clear focal points for next generation sequencing, another parasitic protist, Toxoplasmsa gondii, has also undergone a recent wave of NGS data generation. The population structure of T. gondii is unique, with the majority of strains isolated in Europe and North America belong to three distinct clonal haplotypes (types I, II, and III) with very little genetic variation between (1–3%) and within (<0.01%) them, and a fourth clonal lineage prevalent in North American wild animals. Sequencing of the first single isolate quickly revealed ~1,250 novel SNPs divergent between the reference genome and an isolate from Uganda [36]. These SNPs had the potential to impact coding sequences, highlighting the need for population level NGS efforts of the species. Towards this goal, sequencing ten T. gondii strains from Europe and the Americas generated data that was used to improve the inferred ancestry of the species by creating a haplotype map for the genome [37], thereby further defining the origin and spread of the species. Most recently, whole-genome sequencing of isolates from the Type I clonal lineage of T. gondii identified a cohort of ~1,400 SNPs that differ between Type I strains, and which may explain observed phenotypic differences between isolates [38]. Thus these last three studies exemplify how a stepwise increase in resolution of population level genomic information through the use of NGS data has resolved not only the T. gondii demographic history but also identified disease-relevant loci in the parasite.
By far the most extensively sequenced parasite species is the malaria pathogen Plasmodium falciparum. Whole genome Sanger sequencing of several lab clones [39,40] and the first patient isolate [41] provided initial estimates for a range of population genetic values, including nucleotide diversity (π), a key parameter that describes recent mutations within a genome. Understanding the scope of genetic diversity within parasite genomes provides insight into both natural and anthropogenic influences on parasite populations. The discrepancy in the average π estimated by these three papers most likely reflects the different types of data collected and the differences in SNP calling [42], illustrating the importance of generating high-quality, standardized data sets. More recently, NGS has enabled population genomic analyses of deeply sampled local populations of P. falciparum field isolates. For example, analysis of NGS data from 25 culture-adapted P. falciparum isolates from three sites in Senegal showed little population structure among the sites (located less than 250 miles from each other), and estimated a 60-fold expansion of this population ~20,000–40,000 years ago [43]. Linkage disequilibrium (LD) was found to decay rapidly to a baseline of ~ 1kb, indicative of high levels of recombination in the Senagalese parasite population, and using Tajima’s D, 29 genes were identified with skewed neutrality scores including several not identified in previous studies. A subsequent study by the same group analyzing 45 Senagalese genomes used a test for natural selection that identifies areas in the genome where resistant parasites show much longer haplotypes than sensitive parasites (indicative of recent positive selection on the resistant population) to identify loci associated with drug resistance [44]. This approach was possible because drug treatment provides a strong selective pressure on the P. falciparum genome. Loci identified included several genes not previously implicated in drug resistance, including those in the ubiquitination pathway. On a more global scale, NGS data of 227 patient isolates of P. falciparum from Africa, Asia and Oceania have provided genome-wide estimates of allele frequency distribution, population structure and LD [45]. For example, FST values in this study confirmed that P. falciparum shows a clear division by continent, and that the parasite’s population structure is increased in regions of low transmission.
However, the most noteworthy recent impact that genomics has had on population genetic studies has been to reveal a locus involved in resistance to artemisinin. P. falciparum has developed resistance to every antimalarial drug manufactured, and the recent findings of clinical resistance to artemisinin and its derivatives in patients in Cambodia, Vietnam and Thailand has sent shockwaves through the malaria community. Initial genotyping studies of 91 clones from Laos, Cambodia and Thailand by Cheeseman et al., showed significant population differentiation and a region on chromosome 13 under strong selection [46]. Using additional genetic markers and screening 715 isolates, a 35 kb region within a selective sweep was identified as containing multiple candidate resistance genes. Concurrently, Takala-Harison et al. [47] also used population genetic analysis of SNP array data from ~330 patient isolates from clinical trials of artesunate monotherapy in Southeast Asia to identify several genomic regions containing SNPs associated with artemisinin resistance phenotypes, among them a region of chromosome 13. Subsequently, Miotto et al. [48] used a population genomics approach through analysis of 414 West African and 411 Southeast Asian P. falciparum genomes to provide a “population-level genetic framework” that could assist in investigating the biological origins of the resistance and to define molecular markers. They found a high level of genetic differentiation of Cambodian parasites in their whole genome data, with multiple distinct but sympatric parasite subpopulations identified, indicating founder effects and recent population expansion. No correlation between admixture proportions and parasite artemisinin resistance assays was found, suggesting that the acquisition of artemisinin resistance depends upon inheriting a small number of genetic loci from resistant ancestors. Ultimately, a strong candidate artemisinin resistance marker was identified through NGS sequencing of P. falciparum lab clones made resistant to artemisinin in vitro and analysis of sequence polymorphisms in Cambodian isolates [49]. Together, these papers exemplify how population genetic studies of NGS data is being used by the malaria community to study new life-threatening parasite traits such as drug resistance.
Summary and conclusions
Understanding the genetic structure of parasite populations and the process of genome evolution and adaptation within parasites is essential for crafting effective control strategies against the diseases that they cause. Population genomic data have revealed the patterns of evolution within human pathogens, most noticeably in the Plasmodium genus, and form a solid framework that other neglected and understudied parasites can aspire to in the coming years. As sequencing costs continue to fall and bioinformatic tools improve, addressing population genetic questions of parasitic diseases will become accessible to laboratories large and small.
Highlights.
We summarize the influence of genomics on parasite population genetics
The explosion of genomic data has enabled new types of investigations
Population genomic data has refined estimates of LD and heterozygosity in parasites
Plasmodium population genomics is a foundation for studies of neglected parasites
Acknowledgments
We thank Steven Sullivan for his excellent manuscript editing. This work was supported by National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH) grant U19AI089676 to JMC, and DNH was supported under Bioinformatics Administrative Supplement 5U19AI089676-04 REVISED and MB under R01AI097080 to P. Kissinger. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Glossary
- Admixture
interbreeding of individuals issued from two or more distinct populations or species
- Coalescent theory
a theory describing the genealogy of chromosomes or genes. The genealogy is constructed backwards-in-time, starting with the present-day sample. Lineages coalesce until the most recent common ancestor (MRCA) of the sample is reached.
- FST
the mean fixation index is a measure of population differentiation due to genetic structure
- Linkage disequilibrium
when a genotype present at one locus is dependent on the genotype at a second locus. LD decays each generation at a rate determined by the degree of recombination
- Population genetics
the study of the interrelated patterns of phenotypic, genotypic and allelic frequencies within populations, and how these frequencies change due to influences like selection and chance
- Selective sweep
the fixation of an advantageous mutation that reduces levels of linked silent polymorphism. The size of the chromosomal region impacted by a selective sweep is determined by the level of local recombination
- Tajima’s D
a test that distinguishes between an allele evolving neutrally i.e randomly, and one evolving under a non-random process such as positive selection, balancing selection, or selective sweep
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Daniel N. Hupalo, Email: dh123@nyu.edu.
Martina Bradic, Email: mb3188@nyu.edu.
Jane M. Carlton, Email: jane.carlton@nyu.edu.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Carlton JM, Angiuoli SV, Suh BB, Kooij TW, Pertea M, Silva JC, Ermolaeva MD, Allen JE, Selengut JD, Koo HL, et al. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature. 2002;419:512–519. doi: 10.1038/nature01099. [DOI] [PubMed] [Google Scholar]
- 2.Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, Quail MA, Peters N, Adlem E, Tivey A, Aslett M, et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nature Genetics. 2007;39:839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Sayed NM, Myler PJ, Blandin G, Berriman M, Crabtree J, Aggarwal G, Caler E, Renauld H, Worthey EA, Hertz-Fowler C, et al. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309:404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- 4.Forrester SJ, Hall N. The revolution of whole genome sequencing to study parasites. Mol Biochem Parasitol. 2014 doi: 10.1016/j.molbiopara.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Aurrecoechea C, Barreto A, Brestelli J, Brunk BP, Cade S, Doherty R, Fischer S, Gajria B, Gao X, Gingle A, et al. EuPathDB: the eukaryotic pathogen database. Nucleic Acids Res. 2013;41:D684–D691. doi: 10.1093/nar/gks1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, Amedeo P, Roncaglia P, Berriman M, Hirt RP, Mann BJ, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 9.Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 10.Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, Wortman JR, Bidwell SL, Alsmark UC, Besteiro S, et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luca F, Hudson RR, Witonsky DB, Di Rienzo A. A reduced representation approach to population genetic analyses and applications to human evolution. Genome Res. 2011;21:1087–1098. doi: 10.1101/gr.119792.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One. 2008;3:e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One. 2012;7:e37135. doi: 10.1371/journal.pone.0037135. *This article presents a reduced-representation protocol that can be used for for sampling diversity from large, complex, parasite genomes using the ddRAD approach.
- 14.Conrad MD, Bradic M, Warring SD, Gorman AW, Carlton JM. Getting trichy: tools and approaches to interrogating Trichomonas vaginalis in a post-genome world. Trends Parasitol. 2013;29:17–25. doi: 10.1016/j.pt.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bright AT, Tewhey R, Abeles S, Chuquiyauri R, Llanos-Cuentas A, Ferreira MU, Schork NJ, Vinetz JM, Winzeler EA. Whole genome sequencing analysis of Plasmodium vivax using whole genome capture. BMC Genomics. 2012;13:262. doi: 10.1186/1471-2164-13-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Melnikov A, Galinsky K, Rogov P, Fennell T, Van Tyne D, Russ C, Daniels R, Barnes KG, Bochicchio J, Ndiaye D, et al. Hybrid selection for sequencing pathogen genomes from clinical samples. Genome Biol. 2011;12:R73. doi: 10.1186/gb-2011-12-8-r73. *This study adapted the hybrid selection methodology developed for human exome sequencing to parasite genomes, enabling genomic sequencng directly from clinical isolates and the sequencing of endosymbiotic parasites.
- 18.Conrad MD, Gorman AW, Schillinger JA, Fiori PL, Arroyo R, Malla N, Dubey ML, Gonzalez J, Blank S, Secor WE, et al. Extensive genetic diversity, unique population structure and evidence of genetic exchange in the sexually transmitted parasite Trichomonas vaginalis. PLoS Negl Trop Dis. 2012;6:e1573. doi: 10.1371/journal.pntd.0001573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gelanew T, Kuhls K, Hurissa Z, Weldegebreal T, Hailu W, Kassahun A, Abebe T, Hailu A, Schonian G. Inference of population structure of Leishmania donovani strains isolated from different Ethiopian visceral leishmaniasis endemic areas. PLoS Negl Trop Dis. 2010;4:e889. doi: 10.1371/journal.pntd.0000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunawardena S, Karunaweera ND, Ferreira MU, Phone-Kyaw M, Pollack RJ, Alifrangis M, Rajakaruna RS, Konradsen F, Amerasinghe PH, Schousboe ML, et al. Geographic structure of Plasmodium vivax: microsatellite analysis of parasite populations from Sri Lanka, Myanmar, and Ethiopia. Am J Trop Med Hyg. 2010;82:235–242. doi: 10.4269/ajtmh.2010.09-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howe DK, Honore S, Derouin F, Sibley LD. Determination of genotypes of Toxoplasma gondii strains isolated from patients with toxoplasmosis. J Clin Microbiol. 1997;35:1411–1414. doi: 10.1128/jcm.35.6.1411-1414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viriyakosol S, Siripoon N, Petcharapirat C, Petcharapirat P, Jarra W, Thaithong S, Brown KN, Snounou G. Genotyping of Plasmodium falciparum isolates by the polymerase chain reaction and potential uses in epidemiological studies. Bull World Health Organ. 1995;73:85–95. [PMC free article] [PubMed] [Google Scholar]
- 23.Caccio SM, Thompson RC, McLauchlin J, Smith HV. Unravelling Cryptosporidium and Giardia epidemiology. Trends Parasitol. 2005;21:430–437. doi: 10.1016/j.pt.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 24.Archie EA, Luikart G, Ezenwa VO. Infecting epidemiology with genetics: a new frontier in disease ecology. Trends Ecol Evol. 2009;24:21–30. doi: 10.1016/j.tree.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 25.Conway DJ. Molecular epidemiology of malaria. Clin Microbiol Rev. 2007;20:188–204. doi: 10.1128/CMR.00021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper MA, Adam RD, Worobey M, Sterling CR. Population genetics provides evidence for recombination in Giardia. Curr Biol. 2007;17:1984–1988. doi: 10.1016/j.cub.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 27.Schurko AM, Neiman M, Logsdon JM., Jr Signs of sex: what we know and how we know it. Trends Ecol Evol. 2009;24:208–217. doi: 10.1016/j.tree.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 28.Stanley SL., Jr The Entamoeba histolytica genome: something old, something new, something borrowed and sex too? Trends Parasitol. 2005;21:451–453. doi: 10.1016/j.pt.2005.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weedall GD, Clark CG, Koldkjaer P, Kay S, Bruchhaus I, Tannich E, Paterson S, Hall N. Genomic diversity of the human intestinal parasite Entamoeba histolytica. Genome Biol. 2012;13:R38. doi: 10.1186/gb-2012-13-5-r38. **The first population genomic study of a small set of global E. histolytica isolates, providing strong evidence for recombination sometime in the recent evolutionary history of the parasite.
- 30.Gilchrist CA, Ali IK, Kabir M, Alam F, Scherbakova S, Ferlanti E, Weedall GD, Hall N, Haque R, Petri WA, Jr, et al. A Multilocus Sequence Typing System (MLST) reveals a high level of diversity and a genetic component to Entamoeba histolytica virulence. BMC Microbiol. 2012;12:151. doi: 10.1186/1471-2180-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Neafsey DE, Galinsky K, Jiang RH, Young L, Sykes SM, Saif S, Gujja S, Goldberg JM, Young S, Zeng Q, et al. The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum. Nat Genet. 2012;44:1046–1050. doi: 10.1038/ng.2373. **The first population genomic study of a small set of global P. vivax isolates, revealing significantly more genetic diversity in the species than P. falciparum, with implications for control and eradication of the disease.
- 32. Tachibana S, Sullivan SA, Kawai S, Nakamura S, Kim HR, Goto N, Arisue N, Palacpac NM, Honma H, Yagi M, et al. Plasmodium cynomolgi genome sequences provide insight into Plasmodium vivax and the monkey malaria clade. Nat Genet. 2012;44:1051–1055. doi: 10.1038/ng.2375. *The first comparative analysis of orthologs across two species in the monkey malaria clade, identifying a high proportion of the genes as under strong selective constraint, and confirming the status of P. cynomolgi as a good model system for the study of P. vivax.
- 33.Downing T, Imamura H, Decuypere S, Clark TG, Coombs GH, Cotton JA, Hilley JD, de Doncker S, Maes I, Mottram JC, et al. Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res. 2011;21:2143–2156. doi: 10.1101/gr.123430.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogers MB, Hilley JD, Dickens NJ, Wilkes J, Bates PA, Depledge DP, Harris D, Her Y, Herzyk P, Imamura H, et al. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res. 2011;21:2129–2142. doi: 10.1101/gr.122945.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodhead I, Capewell P, Bailey JW, Beament T, Chance M, Kay S, Forrester S, MacLeod A, Taylor M, Noyes H, et al. Whole-genome sequencing of Trypanosoma brucei reveals introgression between subspecies that is associated with virulence. MBio. 2013;4:97–137. doi: 10.1128/mBio.00197-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bontell IL, Hall N, Ashelford KE, Dubey JP, Boyle JP, Lindh J, Smith JE. Whole genome sequencing of a natural recombinant Toxoplasma gondii strain reveals chromosome sorting and local allelic variants. Genome Biol. 2009;10:R53. doi: 10.1186/gb-2009-10-5-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Minot S, Melo MB, Li F, Lu D, Niedelman W, Levine SS, Saeij JP. Admixture and recombination among Toxoplasma gondii lineages explain global genome diversity. Proc Natl Acad Sci U S A. 2012;109:13458–13463. doi: 10.1073/pnas.1117047109. **This study describes the first population genomics study of ten T. gondii strains from Europe and the Americas used to identify haplotype blocks shared between strains and construct a Toxoplasma haplotype map.
- 38.Yang N, Farrell A, Niedelman W, Melo M, Lu D, Julien L, Marth GT, Gubbels MJ, Saeij JP. Genetic basis for phenotypic differences between different Toxoplasma gondii type I strains. BMC Genomics. 2013;14:467. doi: 10.1186/1471-2164-14-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mu J, Awadalla P, Duan J, McGee KM, Keebler J, Seydel K, McVean GA, Su XZ. Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome. Nat Genet. 2007;39:126–130. doi: 10.1038/ng1924. [DOI] [PubMed] [Google Scholar]
- 40.Volkman SK, Sabeti PC, DeCaprio D, Neafsey DE, Schaffner SF, Milner DA, Jr, Daily JP, Sarr O, Ndiaye D, Ndir O, et al. A genome-wide map of diversity in Plasmodium falciparum. Nat Genet. 2007;39:113–119. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- 41.Jeffares DC, Pain A, Berry A, Cox AV, Stalker J, Ingle CE, Thomas A, Quail MA, Siebenthall K, Uhlemann AC, et al. Genome variation and evolution of the malaria parasite Plasmodium falciparum. Nat Genet. 2007;39:120–125. doi: 10.1038/ng1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carlton JM. Toward a malaria haplotype map. Nat Genet. 2007;39:5–6. doi: 10.1038/ng0107-5. [DOI] [PubMed] [Google Scholar]
- 43. Chang HH, Park DJ, Galinsky KJ, Schaffner SF, Ndiaye D, Ndir O, Mboup S, Wiegand RC, Volkman SK, Sabeti PC, et al. Genomic sequencing of Plasmodium falciparum malaria parasites from Senegal reveals the demographic history of the population. Mol Biol Evol. 2012;29:3427–3439. doi: 10.1093/molbev/mss161. **This paper describes the first population genomics characterization of 25 P. falciparum parasites isolated from three locations within 250 miles of each other in Senegal.
- 44. Park DJ, Lukens AK, Neafsey DE, Schaffner SF, Chang HH, Valim C, Ribacke U, Van Tyne D, Galinsky K, Galligan M, et al. Sequence-based association and selection scans identify drug resistance loci in the Plasmodium falciparum malaria parasite. Proc Natl Acad Sci U S A. 2012;109:13052–13057. doi: 10.1073/pnas.1210585109. **This study utilized tests of selection across 45 P. falciparum Senegalese genomes and identified known drug resistance genes such as pfcrt dhfr, and pfmdr1 along with novel candidates involved in ubiquitination, folate and lipid metabolism.
- 45. Manske M, Miotto O, Campino S, Auburn S, Almagro-Garcia J, Maslen G, O'Brien J, Djimde A, Doumbo O, Zongo I, et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature. 2012;487:375–379. doi: 10.1038/nature11174. **This is the first global study of genetic variation in 227 P. falciparum isolates from Africa, Asia and Oceania, providing genome-wide estimates of LD, allele frequency and population structure.
- 46. Cheeseman IH, Miller BA, Nair S, Nkhoma S, Tan A, Tan JC, Al Saai S, Phyo AP, Moo CL, Lwin KM, et al. A major genome region underlying artemisinin resistance in malaria. Science. 2012;336:79–82. doi: 10.1126/science.1215966. *A key paper in the localization of a marker genes for artemisinin resistance. A selective sweep identified a region on chromosome 13.
- 47. Takala-Harrison S, Clark TG, Jacob CG, Cummings MP, Miotto O, Dondorp AM, Fukuda MM, Nosten F, Noedl H, Imwong M, et al. Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in Southeast Asia. Proc Natl Acad Sci U S A. 2013;110:240–245. doi: 10.1073/pnas.1211205110. *A population genetic analysis using SNP array data that identified several regions of the P. falciparum genome with SNPs associated with artemisin resistance.
- 48. Miotto O, Almagro-Garcia J, Manske M, Macinnis B, Campino S, Rockett KA, Amaratunga C, Lim P, Suon S, Sreng S, et al. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat Genet. 2013;45:648–655. doi: 10.1038/ng.2624. **A landmark paper describing the development of a population genetics framework to help in the identification and characterization of molecular markers of artemisinin resistance.
- 49. Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, Kim S, Duru V, Bouchier C, Ma L, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. **The identification and characterization of a molecular marker of artemisinin resistance, achieved through whole genome sequencing of a lab clone made resistant to artemisinin in vitro over five years, and polymophism analysis of the seven candidate genes in more than 100 P. falciparum isolates from Cambodia. The K13-propeller polymorphism is a useful molecular marker for large-scale surveillance efforts to contain artemisinin resistance globally.