Abstract
The secondary cell wall constitutes a rigid frame of cells in plant tissues where rigidity is required. Deposition of the secondary cell wall in fiber cells contributes to the production of wood in woody plants. The secondary cell wall is assembled through co-operative activities of many enzymes, and their gene expression is precisely regulated by a pyramidal cascade of transcription factors. Deposition of a transmuted secondary cell wall in empty fiber cells by expressing selected gene(s) in this cascade has not been attempted previously. In this proof-of-concept study, we expressed chimeric activators of 24 transcription factors that are preferentially expressed in the stem, in empty fiber cells of the Arabidopsis nst1-1 nst3-1 double mutant, which lacks a secondary cell wall in fiber cells, under the control of the NST3 promoter. The chimeric activators of MYB46, SND2 and ANAC075, as well as NST3, reconstituted a secondary cell wall with different characteristics from those of the wild type in terms of its composition. The transgenic lines expressing the SND2 or ANAC075 chimeric activator showed increased glucose and xylose, and lower lignin content, whereas the transgenic line expressing the MYB46 chimeric activator showed increased mannose content. The expression profile of downstream genes in each transgenic line was also different from that of the wild type. This study proposed a new screening strategy to identify factors of secondary wall formation and also suggested the potential of the artificially reconstituted secondary cell walls as a novel raw material for production of bioethanol and other chemicals.
Keywords: Arabidopsis, Fiber cell, Secondary cell wall, Synthetic biology, Transcription factor
Introduction
Plant carbohydrates are important natural resources for industrial uses, and have recently become a major resource for bioethanol production. Current industrial uses are limited largely to glucose, sucrose and starch extracted from crop plants, such as corn, rice straw and sugar cane (Kim and Dale 2004, Hahn-Hagerdal et al. 2006). The increasing demand for edible crops has created strong competition for these plants between industrial consumption and food supply, which endangers global food security (Hahn-Hagerdal et al. 2006, Ajanovic 2011). However, lignocellulose from woody and herbaceous biomass is now about to be established as an alternative and abundant resource for industrial purposes. Lignocellulose, which is a common component of secondary cell walls in plants, has a complex structure mainly composed of cellulose, hemicellulose and lignin (Albersheim et al. 2010). Although the structural complexity contributes to normal development of plant tissues by providing structural strength and stability against environmental stresses (Mitsuda et al. 2005, Mitsuda et al. 2007, Hirano et al. 2013, Xu et al. 2014), the rigidity hampers the utilization of lignocellulose for industrial purposes. Hence, reduced deposition and modification of lignocellulose composition in plants by genetic engineering has long been attempted by many researchers.
The classical approach to overcome the recalcitrance of lignocellulose to degradation is to reduce lignin and hemicellulose content. Improved enzymatic saccharification efficiency by reduction of lignin or hemicellulose content has been reported in a number of transgenic plants (Chen and Dixon 2007, Lee et al. 2009, Fu et al. 2011, Xu et al. 2011, Cook et al. 2012, Mansfield et al. 2012, Petersen et al. 2012, Van Acker et al. 2013, Yang et al. 2013). However, the survival of such plants is usually adversely affected, as exemplified by reduced mechanical strength and/or increased susceptibility to pathogens (Lao et al. 2003, Brown et al. 2007, Pena et al. 2007, Persson et al. 2007, Quentin et al. 2009, Huang et al. 2010). Furthermore, lignin recovered using a hydrolysis process is usually used for boiler fuel to distill ethanol after yeast fermentation and therefore reduction of lignin from lignocellulose requires additional consumption of a carbon source as boiler fuel. In addition, fermentation of xylose derived from xylan, which is a major component of hemicellulosic polysaccharides, has been developed for bioethanol production (Johansson et al. 2001, Sonderegger et al. 2004). Therefore, reduction of specific cell wall components is not necessarily a desirable strategy for overcoming the recalcitrance of lignocellulose in plants. More recently, monolignol engineering, which modifies the side chain structure of monolignol, has been used successfully to enhance cell wall degradability by cellulase without reduction of total lignin amount and abnormal plant growth (Eudes et al. 2012, Zhang et al. 2012). Although modification of the molecular structure of specific components in lignocellulose may be a superior alternative strategy, all of these transgenic strategies are based on a wild type and are intended to change one component of lignocellulose by manipulating up to a few genes.
To change lignocellulose more dynamically, zero-based reconstruction of lignocellulose by combining building blocks is considered a possible strategy. Given that master transcriptional regulators of lignocellulose biosynthesis in Arabidopsis thaliana have been identified (Kubo et al. 2005, Mitsuda et al. 2005, Zhong et al. 2006, Mitsuda et al. 2007), lignocellulose can be synthesized in ectopic tissues, such as the leaf epidermis, by overexpression of these master regulators. For example, overexpression of NAC SECONDARY WALL THICKENING PROMOTING FACTOR1 (NST1), NST2 and NST3/SECONDARY CELL WALL ASSOCIATED NAC DOMAIN PROTEIN1 (SND1), or VASCULAR-RELATED NAC-DOMAIN PROTEIN6 (VND6) and VND7, which belong to the NAC transcription factor family, induces ectopic formation of secondary cell walls in a variety of cell types (Kubo et al. 2005, Mitsuda et al. 2005, Zhong et al. 2006, Mitsuda et al. 2007). Double knockout of NST1 and NST3 showed complete loss of secondary cell wall deposition in fiber cells of the inflorescence stem, and plants expressing the dominant-negative form of VND6 or VND7 showed seriously defective vessel formation in A. thaliana; therefore, these proteins are considered to be top-level master regulators in the pyramidal transcriptional network in fiber cells and vascular vessels. In addition, MYB46 and MYB83 are also key regulators of secondary cell wall formation at a level just below NSTs and VNDs (Zhong et al. 2007, McCarthy et al. 2009). Although overexpression of these MYB proteins also induces ectopic secondary cell wall formation, the downstream genes preferentially activated differ from those activated by NSTs (Zhong et al. 2006, Ko et al. 2009, McCarthy et al. 2009) and therefore characteristics of the resultant secondary cell wall should differ from those in fiber cells and those induced by overexpression of NSTs. However, it is difficult to determine the characteristics of ectopically produced secondary cell wall because the formation of secondary cell walls severely affects plant growth (Mitsuda et al. 2005, Zhong et al. 2006, Mitsuda et al. 2007). Furthermore, the epidermal cell environment differs from that of fiber cells, and the ectopically induced secondary cell wall in epidermal cells is deposited similarly to vessel elements, showing a striated pattern, even in the case of NST transcription factors, which are master regulators in fiber cells (Mitsuda et al. 2005, Mitsuda et al. 2007). This suggests that the ectopic secondary cell wall formation in epidermal cells may not reflect the original secondary cell wall induced by such transcription factors. However, a vessel-like cell wall was formed in fiber cells of the nst1-1 nst3-1 double mutant by the expression of VND7 under the control of the NST3 promoter, suggesting that fiber cells have an environment that allows gene products related to secondary cell wall formation to work properly (Yamaguchi et al. 2011). Therefore, the zero-based reconstruction of lignocellulose in fiber cells is an ideal system to identify and characterize transcription factors involved in secondary cell wall formation. In this proof-of-concept study, we expressed 24 transcription factors fused with the VP16 transcriptional activation domain in the A. thaliana nst1-1 nst3-1 double-knockout mutant, in which fiber cells lack a secondary cell wall, to isolate transcription factors that reconstitute the secondary cell wall in fiber cells by partially activating the regulatory network under NST master regulators. As a result, some of the transcription factors partially restored the pendent phenotype of the nst1-1 nst3-1 double-knockout mutant. Detailed analysis of the cell wall components of these plants revealed that the secondary cell walls reconstituted by these transcription factors differ from the secondary cell wall in the wild type. Our findings indicated that this approach is a powerful tool to identify novel transcription factors that potentially regulate the gene set for secondary cell wall formation and develop an innovative technology for ‘made to order’ wood production.
Results
Chimeric activators of some NAC and MYB transcription factors restored the pendent phenotype of the nst1-1 nst3-1 double-knockout mutant.
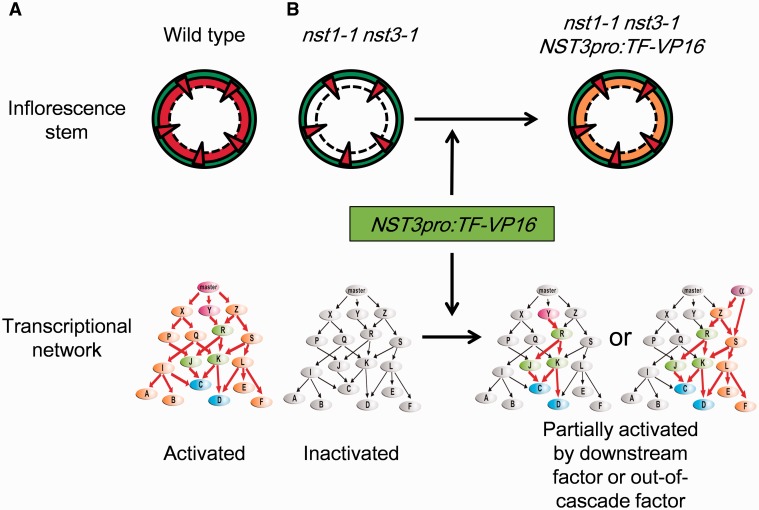
To isolate transcription factors which can promote secondary cell wall formation in the nst1-1 nst3-1 double-knockout mutant, in which fiber cells lack a secondary cell wall, we focused on transcription factors in this study because transcription factors regulate expression of many genes and therefore introduction of one gene could reconstitute the secondary cell wall efficiently by activating part of the regulatory network under NST master regulators (Fig. 1). From a microarray data analysis, we selected 23 genes that were preferentially expressed by at least 2-fold more in the inflorescence stem than the average expression level of all tissues examined (Schmid et al. 2005), and were induced by at least 1.5-fold in the leaves of NST3 overexpressors and/or repressed by at least 0.5-fold in the stem of the nst1-1 nst3-1 double mutant as candidate transcription factors (Supplementary Table S1), in addition to NST3 as a positive control. To examine their ability to stimulate secondary cell wall formation, we expressed these 24 genes fused with the sequence encoding the VP16 transcriptional activation domain (hereafter referred to as the ‘chimeric activator’) in interfascicular fiber cells of the nst1-1 nst3-1 double mutant under control of the NST3 promoter, which induces gene expression specifically in interfascicular fiber cells in A. thaliana (Mitsuda et al. 2007).
Fig. 1.
Schematic illustration of the experimental strategy. (A) Normal secondary cell wall is deposited in fiber cells of the inflorescence stem and the whole regulatory network is activated in the wild-type plant. (B) The nst1-1 nst3-1 double mutant lacks a secondary cell wall in fiber cells and the entire regulatory network is inactivated. Introduction of chimeric activator functioning downstream of NSTs or out of the cascade driven by the NST3 promoter into the nst1-1 nst3-1 double mutant could reconstitute a transmuted secondary cell wall by partially activating the regulatory network.
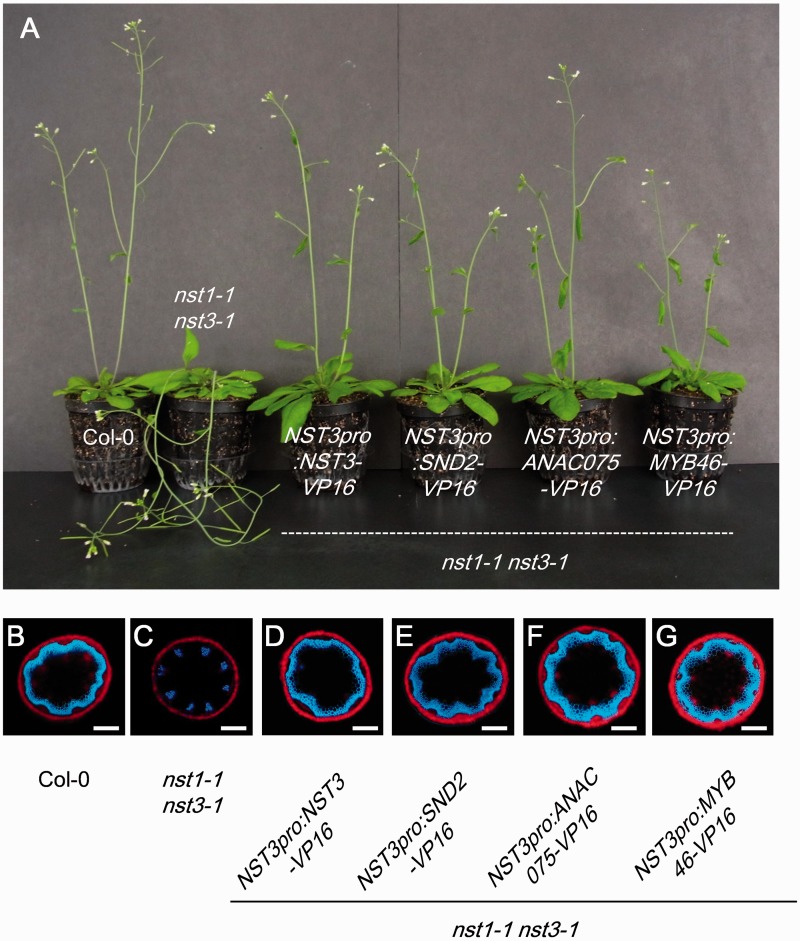
We observed the extent of restoration of the pendent stem phenotype of 17 T1 plants and also examined the distribution of autofluorescence emitted from lignified secondary cell walls under UV illumination in stem cross-sections in at least three T1 plants which showed partial or full restoration of the pendent stem phenotype. From this screening, we identified four chimeric activators, namely NST3, SND2, ANAC075 and MYB46, which restored the pendent stem phenotype and loss of lignin in stem fiber cells of the nst1-1 nst3-1 double mutant (Table 1; Fig. 2A–G). SND2 and ANAC075 share high sequence homology and belong to the NAC transcription factor family, which also includes NST transcription factors. However, the sequence homology between SND2 (and ANAC075) and NSTs was considerably lower than the homology between SND2 and ANAC075 and therefore their target downstream genes were presumed to differ (Supplementary Fig. S1). We also examined their ability to restore the phenotypes of the nst1-1 nst3-1 mutant without fusion to the VP16 transcriptional activation domain. NST3, ANAC075 and MYB46, but not SND2, restored the pendent phenotype of the nst1-1 nst3-1 mutant in a similar manner to the chimeric activators (Supplementary Fig. S2).
Table 1.
Phenotypic restoration of the pendent stem phenotype of the nst1-1 nst3-1 double mutant
| Locus | Alias | Phenotype |
|---|---|---|
| At1g32770 | NST3 | Restored |
| At4g28500 | SND2 | Restored |
| At4g29230 | ANAC075 | Restored |
| At2g40470 | ASL11 | Not restored |
| At4g00220 | ASL19 | Dwarf, petiole-less |
| At1g66230 | ATMYB20 | Not restored |
| At5g12870 | MYB46 | Restored |
| At1g17950 | MYB52 | Not restored |
| At1g73410 | MYB54 | Not restored |
| At1g16490 | ATMYB58 | Not restored |
| At1g79180 | ATMYB63 | Not restored |
| At4g33450 | ATMYB69 | Not restored |
| At4g22680 | ATMYB85 | Not restored |
| At1g63910 | AtMYB103 | Not restored |
| At3g51080 | GATA6 | Not restored |
| At5g25830 | GATA12 | Not restored |
| At4g08150 | KNAT1 | Not restored |
| At1g62990 | KNAT7, IXR11 | Not restored |
| At1g66810 | AtC3H14 | Not restored |
| At2g44745 | WRKY12 | Not restored |
| At4g34610 | BLH6 | Not restored |
| At1g74660 | MIF1 | Not restored |
| At1g62360 | WAM1 | Not restored |
| At1g80730 | ZFP1 | Not restored |
Fig. 2.
Phenotypic restoration of the nst1-1 nst3-1 double mutant by chimeric activators. (A) The pendent phenotype of the nst1-1 nst3-1 double mutant was restored by chimeric activators. (B–G) Autofluorescence of lignin in interfascicular fiber cells in the wild type (B) was not observed in the nst1-1 nst3-1 double mutant (C). Autofluorescence was restored in nst1-1 nst3-1 NST3pro:NST3-VP16 (D), nst1-1 nst3-1 NST3pro:SND2-VP16 (E), nst1-1 nst3-1 NST3pro:ANAC075-VP16 (F) and nst1-1 nst3-1 NST3pro:MYB46-VP (G) plants. Scale bars in (B–G) = 200 µm.
We then tested the transcriptional activation ability of SND2 by fusing the gene to the DNA-binding domain of a yeast GAL4 transcription factor and co-bombarding it into Arabidopsis rosette leaves with the reporter construct driven by a promoter carrying repeated GAL4-binding sites. As shown in Supplementary Fig. S3A, SND2 lacked sufficient transcriptional activation ability, whereas ANAC075 possessed sufficient ability. Thus, the strategy to fuse the transcriptional activation domain to the candidate transcription factor was advantageous to identify potential secondary cell wall regulators lacking sufficient transcriptional activation ability. We also overexpressed NST3, SND2, ANAC075, MYB46 and SND2–VP16 under the control of the Cauliflower mosaic virus (CaMV) 35S promoter in Arabidopsis. NST3, ANAC075, MYB46 and SND2–VP16 induced stunted growth because of ectopic secondary cell wall formation in rosette leaves (Supplementary Fig. S3B), while SND2 alone did not (data not shown). Of the 24 candidate transcription factors examined in this study, ASYMMETRIC LEAVES2-LIKE19 (ASL19)/LATERAL ORGAN BOUNDARY DOMAIN30 (LBD30) have been shown previously to induce ectopic secondary cell wall formation when overexpressed (Soyano et al. 2008). However, we observed that all 17 T1 plants of the nst1-1 nst3-1 NST3pro:ASL19-VP16 mutant showed a dwarf phenotype and petiole-less rosette leaves without any phenotypic restoration of the nst1-1 nst3-1 double mutant in the late-bolting inflorescence stem (Supplementary Fig. S4). The observed phenotypes were similar to those observed in ASL19-overexpressing plants (Soyano et al. 2008), therefore the NST3 promoter used in the present study may show activity in the petiole and/or other unexpected tissues.
Chimeric activators induce substantial accumulation of cell wall components in the plant without secondary cell wall
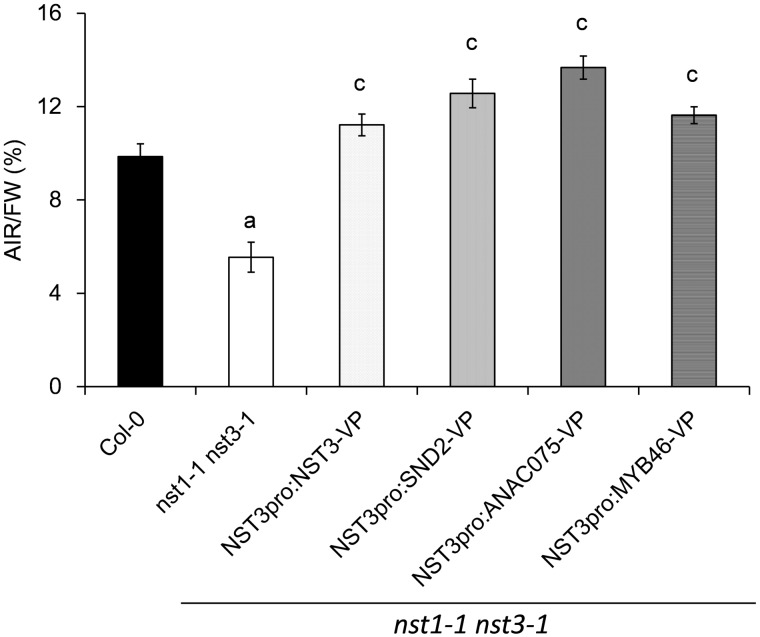
To evaluate restoration of the secondary cell wall in the nst1-1 nst3-1 double mutant quantitatively, the percentage of alcohol-insoluble residues (AIRs) of the total fresh weight (AIRs/FW; w/w) in the inflorescence stem of each plant line was measured. The amount of AIRs of the double mutant (5.6 ± 0.2%) was almost half that of the wild type (9.9 ± 0.5%) (Fig. 3). On the other hand, that of plants restored by the chimeric activator of NST3 (11.2 ± 0.6%), SND2 (12.5 ± 0.7%), ANAC075 (13.7 ± 0.5%) and MYB46 (11.6 ± 0.5%) was much higher than that of the double mutant and even slightly higher than that of the wild type. In particular, the conspicuous increase in AIRs in nst1-1 nst3-1 NST3pro:SND2-VP16 and nst1-1 nst3-1 NST3pro:ANAC075-VP16 was noted. Therefore, we measured the area of secondary cell walls by measuring an area of UV autofluorescence in stem cross-sections in restored lines and found that the area of the secondary cell walls was slightly larger in the restored lines than in the wild type, especially for the nst1-1 nst3-1 NST3pro:SND2-VP16 and the nst1-1 nst3-1 NST3pro:ANAC075-VP16 lines (Supplementary Fig. S5). This result is consistent with the increased AIRs of these lines, indicating that cell wall components were substantially accumulated in the inflorescence stem of the lines restored by the chimeric activators. The overaccumulation observed in the nst1-1 nst3-1 NST3pro:SND2-VP16 and the nst1-1 nst3-1 NST3pro:ANAC075-VP16 lines could be caused by the fusion of a strong transcriptional activation domain.
Fig. 3.
Cell wall residues of plants phenotypically restored by chimeric activators. Alcohol-insoluble residues (AIRs) were purified from the lowermost portion (10 cm) of the inflorescence stem and expressed as a percentage of fresh weight (FW). Error bars indicate the SD [n = 4, except for NST3pro:SND2-VP16 (n = 3)]. The letter above each bar indicates a statistically significant difference (Dunnett’s test, P < 0.05) from the wild type (a) and from both the wild type and the nst1-1 nst3-1 double mutant (c).
The amount of lignin is not necessarily fully recovered by chimeric activators
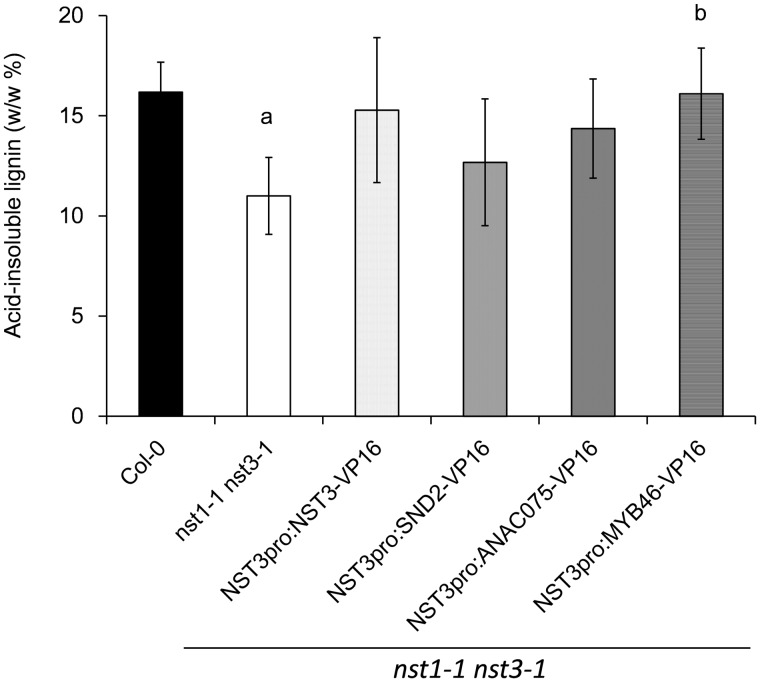
To examine lignin accumulation in the restored lines, the level of acid-insoluble lignin in the inflorescence stem was determined. The level of acid-insoluble lignin in the nst1-1 nst3-1 double mutant (11.0 ± 1.9%) was significantly (32%) lower than that in the wild type (16.2 ± 1.5%) (Fig. 4), which was consistent with the results of a previous study (Iwase et al. 2009). In general, the level of acid-insoluble lignin in the restored lines was 15–30% higher than that of the double mutant; the difference was non-significant for nst1-1 nst3-1 NST3pro:NST3-VP16 (15.3 ± 3.6%, P = 0.07), nst1-1 nst3-1 NST3pro:SND2-VP16 (12.7 ± 3.2%, P = 0.46) and nst1-1 nst3-1 NST3pro:ANAC075-VP16 (14.4 ± 2.5%, P = 0.14), but was significant for nst1-1 nst3-1 NST3pro:MYB46-VP16 (16.1 ± 2.3%, P = 0.04). It should be noted that restoration of lignin accumulation in nst1-1 nst3-1 NST3pro:SND2-VP16 and nst1-1 nst3-1 NST3pro:ANAC075-VP16 showed a weaker tendency than that of the other restored lines. These data suggest that the amount and/or characteristics of lignin in nst1-1 nst3-1 NST3pro:SND2-VP16 and nst1-1 nst3-1 NST3pro:ANAC075-VP16 might be different from those of the original in the wild type.
Fig. 4.
Acid-insoluble lignin of plants phenotypically restored by chimeric activators. The inflorescence stem was hydrolyzed with sulfuric acid and the resultant residues were weighed as acid-insoluble lignin. The data are presented as acid-insoluble lignin per dry weight of destarched residue (w/w %). Error bars indicate the SD [n = 4, except for NST3pro:SND2-VP16 (n = 3)]. The letter above each bar indicates a statistically significant difference (Dunnett’s test, P < 0.05) from the wild type (a) and from the nst1-1 nst3-1 double mutant (b).
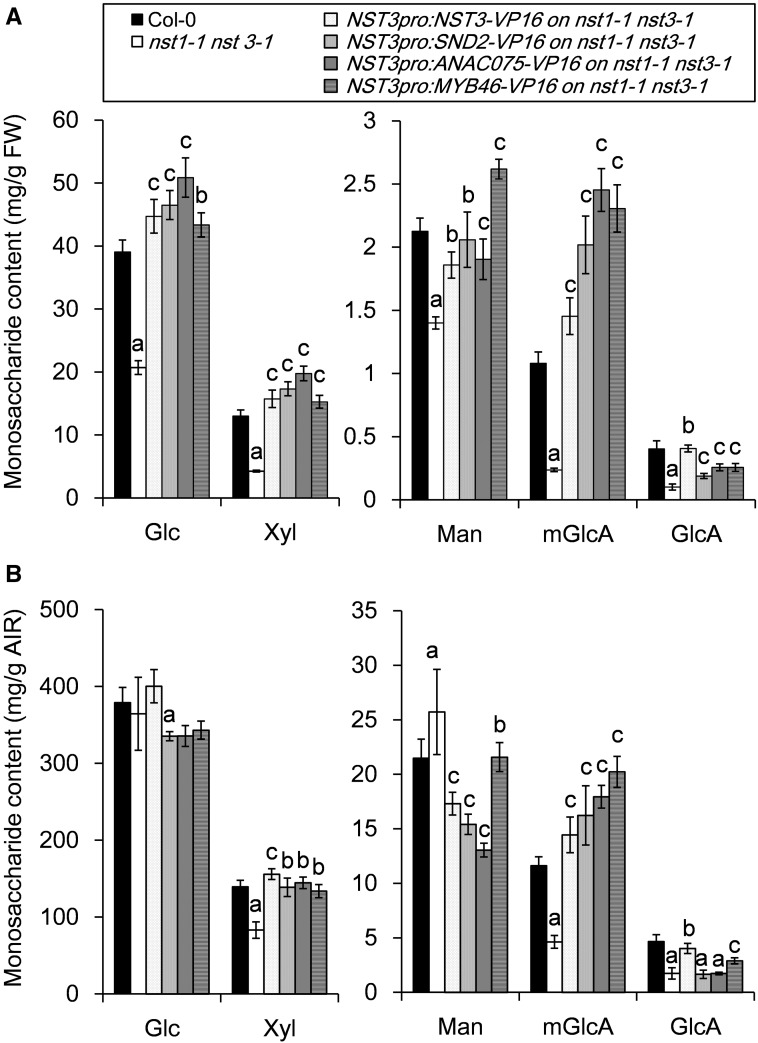
Monosaccharide composition in cell walls reconstituted by the chimeric activators is different from that of the wild type
To investigate further differences in cell wall composition, the monosaccharide composition in the cell wall restored by the chimeric activators was determined (Supplementary Tables S2, S3). Even though any notable difference of plant growth was not observed among the wild type, restored lines and the nst1-1 nst3-1 mutant, AIRs/FW of the nst1-1 nst3-1 double mutant is much reduced due to the lack of secondary cell walls (Fig. 3). The contents of five monosaccharides associated with the secondary cell wall, namely glucose (Glc), xylose (Xyl), mannose (Man), 4-O-methyl glucuronic acid (mGlcA) and glucuronic acid (GlcA), on a fresh weight basis, was decreased in cell walls of the inflorescence stem of the nst1-1 nst3-1 double mutant as expected (Fig. 5A). In contrast, the contents of other monosaccharides, such as arabinose (Ara), galacturonic acid (GalA), fucose (Fuc), galactose (Gal) and rhamnose (Rha), listed in Supplementary Table S2 and S3, which are mostly incorporated in primary cell walls, were relatively increased in the stem of the double mutant owing to the absence of secondary cell walls even though we described the amount as mg g FW–1. As expected, the monosaccharide composition (both per fresh weight and per AIRs) of the cell walls restored by the NST3 chimeric activator was largely similar to that of the wild type (Fig. 5A, B; Supplementary Tables S2, S3). Because SND2 and ANAC075 share high sequence homology, cell walls of nst1-1 nst3-1 NST3pro:SND2-VP16 and nst1-1 nst3-1 NST3pro:ANAC075-VP16 plants showed similar monosaccharide compositions (both per fresh weight and per AIRs) (Fig. 5A, B; Supplementary Tables S2, S3). More specifically, contents of Glc, Xyl and mGlcA per fresh weight, which are major monosaccharide constituents of secondary cell walls, were higher in nst1-1 nst3-1 NST3pro:SND2-VP16 and nst1-1 nst3-1 NST3pro:ANAC075-VP16 plants than in the other restored lines and the wild type, reflecting more substantial accumulation of secondary cell walls in these lines (Fig. 5A; Supplementary Table S3). However, among them, only mGlcA was conspicuously increased in these lines compared with the wild type when we described monosaccharide contents per AIRs, and, instead, the Man content decreased in these lines (Fig. 5B; Supplementary Table S2). In contrast, in nst1-1 nst3-1 NST3pro:MYB46-VP16 plants, the increase of Glc and Xyl contents (per fresh weight) was less pronounced, whereas a distinct increase in Man content per fresh weight was observed, compared with those of the other restored lines and the wild type (Fig. 5A). However, per AIRs, the Man content was similar to the wild type, but the mGlcA content was almost twice that of the wild type (Supplementary Table S2). All of these results suggested that the composition of the cell walls reconstituted by the chimeric activators differed from that of the wild type.
Fig. 5.
Monosaccharide composition of plants phenotypically restored by chimeric activators. The data are presented as monosaccharide content per fresh weight (mg g FW–1, A) and per alcohol-insoluble residue (mg g AIR–1, B). Error bars indicate the SD [n = 4, except for NST3pro:SND2-VP16 (n = 3)]. The letter above each bar indicates a statistically significant difference (Dunnett’s test, P < 0.05) from the wild type (a), from the nst1-1 nst3-1 double mutant (b) and from both the wild type and nst1-1 nst3-1 (c).
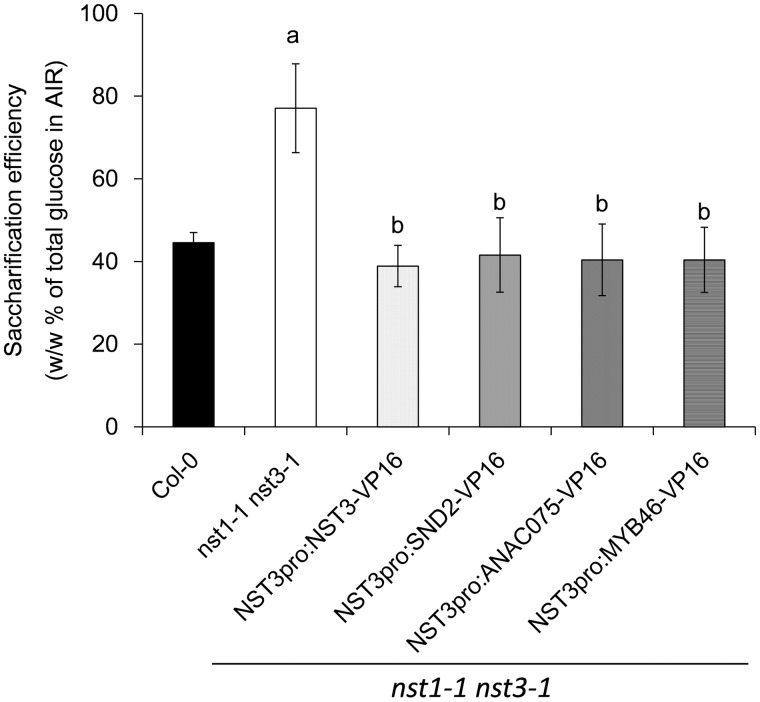
Enzymatic saccharification efficiency of the reconstituted secondary cell wall does not change significantly compared with the wild type
Monosaccharide composition and lignin analysis demonstrated that the reconstituted secondary cell wall was different from that of the wild-type secondary cell wall. We hypothesized that this might influence the saccharification efficiency of the cell wall. The saccharification efficiency was calculated as the percentage Glc released by cellulase hydrolysis from the AIRs of the total Glc in the AIRs (w/w). The saccharification efficiency of the nst1-1 nst3-1 double mutant (77.1 ± 10.7%) was significantly higher than that of the wild type (44.5 ± 2.5%) (Fig. 6), which is consistent with the results of a previous report (Iwase et al. 2009). The restored lines, namely nst1-1 nst3-1 NST3pro:NST3-VP16 (38.9 ± 5.0%), nst1-1 nst3-1 NST3pro:SND2-VP16 (41.6 ± 9.0%), nst1-1 nst3-1 NST3pro:ANAC075-VP16 (40.4 ± 8.7%) and nst1-1 nst3-1 NST3pro:MYB46-VP16 (40.4 ± 7.9%), showed similar levels of saccharification efficiency to the wild type. These data suggested that the changes induced by the chimeric activators did not affect saccharification efficiency, at least under our sampling strategy and assay conditions.
Fig. 6.
Saccharification efficiency of plants phenotypically restored by chimeric activators. Saccharification efficiency was measured as the percentage glucose content released by cellulase hydrolysis of the total glucose content in the alcohol-insoluble residues determined with the UPLC-ABEE system (w/w %). Error bars indicate the SD [n = 4, except for NST3pro:SND2-VP16 (n = 3)]. The letter above each bar indicates a statistically significant difference (Dunnett’s test, P < 0.05) from the wild type (a) and from the nst1-1 nst3-1 double mutant (b).
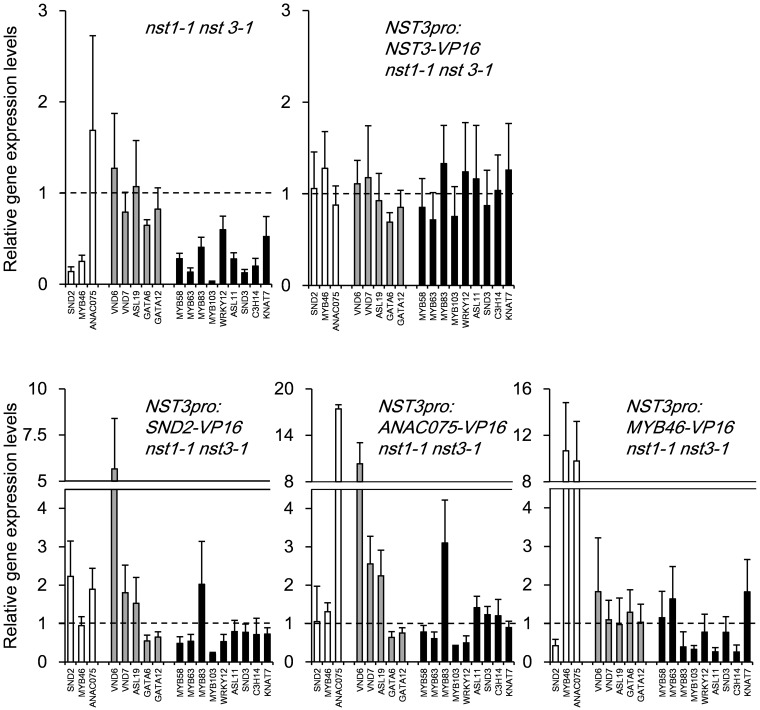
The expression of secondary cell wall-associated transcription factor genes in each restored line shows a distinct pattern
To gain more insight into how the above-described changes in cell walls are established, we analyzed the expression of secondary cell wall-associated transcription factor genes in the restored lines using quantitative real-time PCR (RT-PCR) (Fig. 7). As expected, most down-regulated genes in the nst1-1 nst3-1 mutant were restored to wild-type levels by the introduction of NST3–VP16. The induction patterns of most of the tested genes in nst1-1 nst3-1 NST3pro:SND2-VP16 and nst1-1 nst3-1 NST3pro:ANAC075-VP16 were similar, but were induced more in nst1-1 nst3-1 NST3pro:ANAC075-VP16. It should be noted that the expression of VND6, VND7 and ASL19 was induced in these lines. These data suggested that, because VNDs and ASL19 induce ectopic differentiation of vascular vessels (Kubo et al. 2005, Soyano et al. 2008), restoration of the pendent phenotype was achieved through vascular vessel formation in fiber cells in these lines. The data also suggested that at least ANAC075 could be an upstream regulator of VNDs and/or ASL19, which is mostly consistent with the conclusion of a recent study in which ANAC075 was suggested to function as an upstream regulator of VNDs (Endo et al. 2015). However in the nst1-1 nst3-1 NST3pro:MYB46-VP16 line, even though the expression of ANAC075 was highly induced, the expression of VNDs and ASL19 was not induced, which is consistent with the RT-PCR data shown in the previous study (Endo et al. 2015). These data suggest that VP16 fusion to ANAC075 is required to activate VNDs and ASL19. On the other hand, the expression of SND2, MYB83, MYB103, ASL11 and C3H14 was not fully restored in the nst1-1 nst3-1 NST3pro:MYB46-VP16 line, suggesting that MYB46–VP16 partially or differently activates a downstream network of transcription factors under the regulation of NSTs.
Fig. 7.
Gene expression pattern of transcription factors associated with secondary cell wall formation in plants that were phenotypically restored by chimeric activators. Quantitative RT-PCR was carried out to analyze the expression of the gene of interest in the 10 cm inflorescence stem (1–2 weeks after bolting) of the wild type, the nst1-1 nst3-1 double mutant and plants phenotypically restored by each chimeric activator. The expression level of the gene of interest was normalized with that of UBQ1, and the normalized expression level in the wild type was set to 1. The examined genes are classified into introduced genes except for NST3 (white), VND-related transcription factor genes (gray) and others (black). The error bar indicates the SD (n = 5).
Discussion
The secondary cell wall of fiber cells constitutes a large portion of plant dry weight, but is not necessarily essential for plant survival; therefore, it offers great potential for exploitation with appropriate genetic manipulations. In this study, we demonstrated that new artificial xylem can be produced by expression of certain transcription factors in a mutant that lacks secondary cell walls in fiber cells. The transcription factors might function downstream of the master regulators or in different pathways; therefore, it can be hypothesized that they activate the secondary wall formation pathway differently from the master regulator, NSTs, and, as a result, secondary cell walls that show different characteristics were synthesized.
Interestingly, the chimeric activator of SND2 restored the pendent phenotype of the nst1-1 nst3-1 double mutant, whereas SND2 alone did not. On the other hand, ANAC075 restored the pendent phenotype of the nst1-1 nst3-1 double mutant without fusion to the VP16 domain. Their chimeric activators clearly activated the expression of VND6 and VND7. These results indicated that SND2 and ANAC075 might regulate the same target genes, acting as a negative and positive regulator, respectively. On the other hand, expression of SND2 was reduced in the nst1-1 nst3-1 double mutant, but was recovered by complementation with the chimeric activator of NST3 or MYB46, while the expression of ANAC075 was not reduced in the nst1-1 nst3-1 double mutant and was up-regulated in nst1-1 nst3-1 NST3pro:MYB46-VP16. These data suggested that SND2 and ANAC075 function at different levels of the regulatory cascade of secondary cell wall formation and/or vascular vessel formation even though they share high sequence homology and the same target genes. From the point of view of cell wall characteristics, even though the AIRs/FW showed a higher increase in the restored lines by the chimeric activators of SND2 and ANAC075 than that induced by the other chimeric activators and exceeded that of the wild type, lignin accumulation was not restored to the wild-type level. Furthermore, the Glc content was higher than those of the other restored lines and the wild type. These data suggested that secondary cell wall induced by the chimeric activators of SND2 and ANAC075 through activation of VNDs could have different characteristics from those of the original in fiber cells, even though we could not exclude the possibility that the chimeric activators of SND2 and ANAC075 partly contributed to reconstituted secondary cell wall through a pathway independent from VNDs. It would be intriguing to examine cell wall characteristics of NST3pro:VND7 nst1-1 nst3-1, in which vessel-like secondary cell wall was deposited in fiber cells (Yamaguchi et al. 2011), to have clearer knowledge of how the secondary cell wall in vascular vessels is different from that in fiber cells.
The cell wall characteristics and expression pattern in the plants restored by the chimeric MYB46 activator were also different from those of other restored lines. In particular, the expression of SND2, MYB83, MYB103, ASL11 and C3H14, which are known downstream transcription factors of NSTs, was not restored by the introduction of a chimeric activator of MYB46, suggesting that these transcription factors are not located in the regulatory network governed by MYB46, but within the regulatory network under the control of NSTs. However, in the case of C3H14, the expression in mesophyll protoplasts was induced by the transient overexpression of MYB46 (Ko et al. 2009). This discrepancy may reflect the different cell types used, suggesting again that the regulatory system of secondary wall formation in fiber cells is different from ectopic secondary wall formation in mesophyll cells. In terms of cell wall characteristics, the Man content was increased in the restored plants by the MYB46 chimeric activator, which was in contrast to the other restored lines and the wild type. Recent studies revealed that MYB46 directly regulates the expression of the cellulose synthase-like A 9 (CSLA9) gene, which encodes mannan synthase (Ko et al. 2009, Kim et al. 2014a, Kim et al. 2014b). This finding is consistent with our observation of the increase in Man content in the nst1-1 nst3-1 NST3pro:MYB46-VP16 plants.
In the cases of both SND2 (and ANAC075) and MYB46, mGlcA content in the reconstituted secondary cell wall was notably higher than that of the wild type and nst1-1 nst3-1 NST3pro:NST3-VP16 plants. Glucuronoxylan methyltransferase (GXM) catalyzes the methylation of GlcA attached to xylan, and the expression of GXMs is under the regulation of NST transcription factors (Lee et al. 2012, Urbanowicz et al. 2012). However, this regulation is not proven to be direct, and therefore might be through the function of SND2, ANAC075 and MYB46. Further study is needed to establish the regulation of hemicellulose-related gene expression.
In the present study, four chimeric activators, including NST3 as a positive control, restored the pendent phenotype of the nst1-1 nst3-1 double mutant. In addition, all of the chimeric activators induced ectopic secondary cell wall formation in rosette leaves when they were overexpressed under the control of the CaMV 35S promoter. Conversely, ASL19, which is known to induce ectopic secondary cell wall, did not restore the pendent phenotype of the nst1-1 nst3-1 double mutant. Overexpression of MYB58 and MYB63 is known to induce ectopic lignification without deposition of cellulose in wild-type Arabidopsis (Zhou et al. 2009). No such lignin deposition was observed in the fiber cells of nst1-1 nst3-1 NST3pro:MYB58 and nst1-1 nst3-1 NST3pro:MYB63 plants. In addition, OsMYB103, OsMYB55/61, OsMYB42/85 and OsBLH6 induce ectopic lignification in wild-type rice (Hirano et al. 2013), but chimeric activators of their orthologs in Arabidopsis did not restore the phenotype of the nst1-1 nst3-1 double mutant (Table 1). These data suggested that these transcription factors might require a particular molecule(s) and/or environment that are not present in empty fiber cells in order to induce proper deposition of the reconstituted secondary cell wall. Otherwise, some transcription factors that did not restore any secondary wall formation in this study may be transcriptional repressors. In this case, fusion to the VP16 domain impedes their activity (Fujiwara et al. 2014) and therefore chimeric repressors should be expressed to induce the gain-of-function phenotype, namely secondary cell wall formation (Mitsuda et al. 2009). Employment of a chimeric repressor is also appropriate if the original transcription factor is a transcriptional activator but functions as a negative regulator that suppresses the deposition of secondary cell walls by activating target genes. In this case, the chimeric repressor induces the loss-of-function phenotype, namely secondary cell wall formation.
In summary, we have succeeded in reconstituting transmuted secondary cell walls in NST master-regulator mutants, which lack a secondary cell wall in fiber cells, by expressing chimeric activators of certain transcription factors. Because the target gene differed among the chimeric activators, secondary cell walls with different characteristics were synthesized. Although improvement of saccharification efficiency was not observed in Arabidopsis under our sampling strategy and assay condition, we confirmed the potential to produce cell wall with transmuted characteristics in the mutant lacking a secondary cell wall. Further screening of chimeric activators and repressors of additional transcription factors, which are not necessarily expressed in the stem, is also of interest to identify other new candidate transcription factors that may be involved in secondary cell wall formation and might induce formation of secondary cell walls with more valuable characteristics. We believe our strategy opens up a new avenue for synthesizing artificial secondary cell walls for human benefit.
Materials and Methods
Chemicals
The standard monosaccharides Gal, Man, Ara, Xyl, Fuc and Rha were purchased from Kanto Chemical Inc., GlcA, GalA and dGlc were purchased from Sigma-Aldrich Inc., and Glc was purchased from Wako Inc. p-Aminobenzoic ethyl ester (ABEE) was purchased from Ark Pharm Inc. and sodium cyanoborohydride was obtained from MP Biomedicals Inc. mGlcA purified from sap of the lac tree (Nakamura et al. 1984, Kuroyama et al. 2001) was kindly gifted by Professor Y. Tsumuraya (Saitama University, Saitama, Japan).
Plant materials
The nst1-1 nst3-1 double mutant (Mitsuda et al. 2007) of A. thaliana ecotype Columbia-0 was grown in soil at 22°C under a 16 h/8 h (light/dark) photoperiod (illumination 60–70 µmol m−2 s−1). Two-month-old double-mutant plants were transformed using the floral dip method (Clough and Bent 1998). Transgenic T1 seeds were sown on 1/2 Murashige and Skoog agar supplemented with hygromycin and grown for 3 weeks. Hygromycin-resistant plants were transferred to soil. After 4–5 weeks, all inflorescence stems of the transgenic plants were harvested. The lowermost 10 cm portion of the stem was used for the following analyses. To ensure a sufficient quantity of tissue for the various analyses, the lower stem portions harvested from 4–5 plants were combined as one sample.
Plasmid construction
To generate transgenic plants expressing the chimeric activators, the coding sequence of each transcription factor was transferred from the Gateway Entry clone (Mitsuda et al. 2010) into the pDEST_NST3p_VP16_HSP_GWB5 vector using the Gateway LR reaction (Life Technologies Inc.). The resultant plasmid was introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. The pDEST_NST3p_VP16_HSP_GWB5 vector was constructed by inserting the approximately 3 kb NST3 upstream region, which was amplified with the primer pair NST3pF_Kpn (5′-AAATTTGGTACCACTATATATAATTTACCCAACAAACAC-3′) and NST3pR_Xho (5′-AAATTTCTCGAGTAACGAAGATAGCAATATATTTTTGG-3′) and inserted into the pDEST_VP16_HSP_GWB5 vector. The pDEST_VP16_HSP_GWB5 vector was constructed by inserting the Gateway cassette followed by the VP16 activation domain and partial HSP terminator, which was amplified from the pDEST_35S_VP16_HSP_GWB5 vector (Fujiwara et al. 2014), into the pMCS_delHSP_GWB5 vector. The pMCS_delHSP_GWB5 vector was constructed by inserting annealed oligonucleotides (5′-AGCTGGGTTAATTAACCGGTACCTTACTAGTTTCTCGAGAAGATATCAATCTAGAGGA-3′ and 5′-AGCTTCCTCTAGATTGATATCTTCTCGAGAAACTAGTAAGGTACCGGTTAATTAACCC-3′) into the HindIII-digested pDEST_35S_VP16_HSP_GWB5 vector (Fujiwara et al. 2014) from which the 35S promoter followed by the VP16 domain and partial HSP terminator were removed. To generate transgenic plants expressing the specific transcription factor driven by the 35S or NST3 promoter, the coding sequence of each transcription factor was amplified and inserted into the SmaI site of the p35SG (Mitsuda et al., 2005) or pNST3_EntG vector (Mitsuda et al. 2007) and then transferred to the binary vector pBCKH (Mitsuda et al. 2006). For transient effector–reporter analysis, the coding sequence of SND2 and ANAC075 was ligated into the p430T1.2 vector, which possesses the GAL4 DNA-binding domain (Ohta et al. 2000), and used as the effector plasmid.
Transient effector–reporter analysis
The effector plasmid (0.8 µg) was bombarded into Arabidopsis leaves together with 0.6 µg of a reporter plasmid, p35S-GAL4-TATA-LUC (Hiratsu et al. 2002). As an internal control, 0.1 µg of pRL, which included the Renilla luciferase gene (Promega Inc.) under the control of the 35S promoter, was co-bombarded with both reporter and effector plasmids. After bombardment, samples were maintained for 12 h in darkness, after which luciferase activity was measured.
Microscopic observation of stem cross-sections
The series of T1 transgenic plants were grown for 5–6 weeks. The lowermost 3 cm of the inflorescence stem was harvested and embedded in 5% agar. Micro-slice sections (50 µm) were prepared with a vibrating microtome (HM-650V; Microm Inc.). For observation of lignin autofluorescence, we used a filter set with the following specifications: excitation filter, 365 nm short pass; dichroic mirror, 395 nm; emission filter, 400 nm long pass. All light and fluorescence microscopic observations were made with an Axioscop2 Plus system (Zeiss Inc.).
Preparation of cell wall residues
Cell wall residues were prepared using a modified procedure described in previous reports (Sakurai et al. 1979, York et al. 1986, Iwase et al. 2009). The inflorescence stem from 4–5 individuals of 8-week-old A. thaliana plants were cut into 1 cm long segments, fixed in methanol, and stored at room temperature until use. The fixed stems were treated twice with methanol at 80°C for 10 min, twice with acetone at 70°C for 5 min, twice with methanol/chloroform (1 : 1, v/v) at 70°C for 5 min, and then dried at 65°C for 18 h. The dried stem segments were transferred to 2 ml tubes with a screw cap and ground into powder using a stainless steel bead (6 mm, Biomedical Science Inc.) with three zirconia beads (3 mm, Nikkato Inc.) using a Shake Master NEO (Biomedical Science Inc.). The resultant powder was gelatinized in 0.1 M sodium malate buffer (pH 6.0) at 65°C for 10 min and then starch was digested with α-amylase solution, containing 500 U ml−1 α-amylase from porcine pancreas (Megazyme Inc.) and 0.33 U ml−1 amylo-glucosidase from Aspergillus niger (Megazyme) in 0.1 M sodium malate buffer (pH 6.0) at 37°C for 18 h with shaking at 150 r.p.m. The destarched residue was washed three times each with ultrapure water and 100% ethanol, and then dried at 65°C.
Cell wall hydrolysis with sulfuric acid
Cell wall hydrolysis was performed by the two-step sulfuric hydrolysis method based on the NREL protocol (Sluiter et al. 2008). Briefly, approximately 2–3 mg of destarched cell wall residue was depolymerized with 50 µl of 72% sulfuric acid (w/v) for 1 h with gentle shaking at room temperature and then diluted to 4% sulfuric acid (w/v) with 1.4 ml of ultrapure water. The resultant suspension was hydrolyzed at 121°C for 1 h and then cooled at room temperature. After addition of 10 µl of 2-d-Glc solution (1 mg ml−1) as an internal standard, the supernatant was neutralized with calcium carbonate powder and adjusted to around pH 5 using pH paper.
Monosaccharide quantification
Chromatographic separation and detection of monosaccharides labeled with ABEE was performed with a modified method described by Kumagai et al. (2012). The ABEE-labeled monosaccharides were subjected to liquid chromatography (ACQUITY UPLC H-Class system, Waters Inc.) equipped with an ACQUITY UPLC BEH C18 column (Waters) and fluorescence detector (ACQUITY UPLC FLR Detector, Waters). The mixture of 200 mM potassium borate buffer (pH 8.9) and pure acetonitrile was used as the mobile phase. The ABEE-labeled monosaccharides were detected by a fluorescence detector using emission and excitation wavelengths of 305 and 360 nm, respectively. The peak assignment and quantification were made with a mixture of 10 monosaccharides as an external standard. The detailed procedure will be published elsewhere.
Quantification of acid-insoluble lignin
Insoluble residues after hydrolysis of destarched cell wall residues with sulfuric acid were washed with ultrapure water, 50% ethanol and 100% ethanol. After drying for 18 h at 65°C, the remaining residues in 2 ml microtubes were weighed. The level of acid-insoluble lignin was calculated on the basis of dry weight of destarched residue and expressed as percentage weight (w/w %).
Saccharification efficiency analysis
The saccharification efficiency of the amylase-treated sample was measured by enzymatic reaction with Celluclast 1.5L and Novozyme 188 (Novozymes Inc.) as described in Iwase et al. (2009). Approximately 2–3 mg of destarched cell wall residue was weighed in a 2 ml microtube and treated with 1 ml of cellulose mixture containing a 0.8 filter paper unit of Celluclast 1.5L and a 1.6-cellobiase unit of Novozyme 188 and then incubated at 50°C for 24 h with shaking at 200 r.p.m. After heating to inactivate cellulase for 10 min at 95°C, the reaction mixture was centrifuged at 20,000 × g for 5 min and the glucose content in the supernatant was determined with the Glucose test kit (Wako Inc.).
RNA extraction and quantitative RT-PCR analysis
Total RNA was extracted from 10 cm inflorescence stems (1–2 weeks after bolting) of at least five independent T2 transgenic lines, using an RNeasy Plant Mini Kit (Qiagen Inc.). Genomic DNA contamination was removed using DNase I, following the manufacturer’s instructions. First-strand cDNA was synthesized using a PrimeScript RT reagent Kit (TAKARABIO INC.). Quantitative RT-PCR analysis was performed by the SYBR green method, using an ABI7300 real-time PCR system (Life Technologies). The relative expression level of each gene was calculated by the absolute quantification method and expressed as the value divided by the expression level of the UBQ1 gene. Primer sets for quantitative RT-PCR are listed in Supplementary Table S4.
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the Japan Science and Technology Agency (JST) [Advanced Low Carbon Technology Research and Development Program (ALCA) (to N.M.)].
Supplementary Material
Acknowledgments
The authors are grateful to Professor Yoichi Tsumuraya (Saitama University) for kindly gifting mGlcA, Dr. Koki Yoshida (Taisei Inc.) for technical advice and fruitful discussions, and laboratory members for discussion and encouragement. We also thank Ms. Aeni Hosaka (AIST), Ms. Akiko Kuwazawa (AIST), Ms. Fumie Tobe (AIST), Ms. Miyoko Yamada (AIST), Ms. Yoshimi Sugimoto (AIST) and Ms. Yukie Kimura (AIST) for their skillful technical assistance.
Glossary
Abbreviations
- ABEE
p-aminobenzoic ethyl ester
- AIR
alcohol-insoluble residue
- Ara
arabinose
- CaMV
Cauliflower mosaic virus
- CSLA
cellulose synthase-like A
- Fuc
fucose
- Gal
galactose
- GalA
galacturonic acid
- Glc
glucose
- GlcA
glucuronic acid
- GXM
glucuronoxylan methyltransferase
- Man
mannose
- mGlcA
4-O-methyl glucuronic acid
- NST
NAC secondary wall-thickening promoting factor
- Rha
rhamnose
- RT-PCR
real-time PCR
- SND
secondary cell wall-associated NAC domain protein
- UBQ
ubiquitin
- VND
vascular-related NAC-domain protein
- Xyl
xylose
Disclosures
The authors have no conflicts of interest to declare.
References
- Ajanovic A. Biofuels versus food production: does biofuels production increase food prices? Energy. 2011;36:2070–2076. [Google Scholar]
- Albersheim P, Darvill A, Roberts K, Sederoff R, Staehelin A. New York: Garland Science; 2010. Plant Cell Walls. From Chemistry to Biology; pp. 67–118. [Google Scholar]
- Brown DM, Goubet F, Wong VW, Goodacre R, Stephens E, Dupree P, et al. Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant J. 2007;52:1154–1168. doi: 10.1111/j.1365-313X.2007.03307.x. [DOI] [PubMed] [Google Scholar]
- Chen F, Dixon RA. Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 2007;25:759–761. doi: 10.1038/nbt1316. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cook CM, Daudi A, Millar DJ, Bindschedler LV, Khan S, Bolwell GP, et al. Transcriptional changes related to secondary wall formation in xylem of transgenic lines of tobacco altered for lignin or xylan content which show improved saccharification. Phytochemistry. 2012;74:79–89. doi: 10.1016/j.phytochem.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo H, Yamaguchi M, Tamura T, Nakano Y, Nishikubo N, Yoneda A, et al. Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 2015;56:242–254. doi: 10.1093/pcp/pcu134. [DOI] [PubMed] [Google Scholar]
- Eudes A, George A, Mukerjee P, Kim JS, Pollet B, Benke PI, et al. Biosynthesis and incorporation of side-chain-truncated lignin monomers to reduce lignin polymerization and enhance saccharification. Plant Biotechnol. J. 2012;10:609–620. doi: 10.1111/j.1467-7652.2012.00692.x. [DOI] [PubMed] [Google Scholar]
- Fu C, Mielenz JR, Xiao X, Ge Y, Hamilton CY, Rodriguez M, et al. Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc. Natl Acad. Sci. USA. 2011;108:3803–3808. doi: 10.1073/pnas.1100310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Sakamoto S, Kigoshi K, Suzuki K, Ohme-Takagi M. VP16 fusion induces the multiple-knockout phenotype of redundant transcriptional repressors partly by med25-independent mechanisms in Arabidopsis. FEBS Lett. 2014;588:3665–3672. doi: 10.1016/j.febslet.2014.08.010. [DOI] [PubMed] [Google Scholar]
- Hahn-Hagerdal B, Galbe M, Gorwa-Grauslund MF, Liden G, Zacchi G. Bio-ethanol–the fuel of tomorrow from the residues of today. Trends Biotechnol. 2006;24:549–556. doi: 10.1016/j.tibtech.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Hirano K, Kondo M, Aya K, Miyao A, Sato Y, Antonio BA, et al. Identification of transcription factors involved in rice secondary cell wall formation. Plant Cell Physiol. 2013;54:1791–1802. doi: 10.1093/pcp/pct122. [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Ohta M, Matsui K, Ohme-Takagi M. The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett. 2002;514:351–254. doi: 10.1016/s0014-5793(02)02435-3. [DOI] [PubMed] [Google Scholar]
- Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou YH, et al. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010;153:1526–1538. doi: 10.1104/pp.110.157370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase A, Hideno A, Watanabe K, Mitsuda N, Ohme-Takagi M. A chimeric NST repressor has the potential to improve glucose productivity from plant cell walls. J. Biotechnol. 2009;142:279–84. doi: 10.1016/j.jbiotec.2009.05.011. [DOI] [PubMed] [Google Scholar]
- Johansson B, Christensson C, Hobley T, Hahn-Hagerdal B. Xylulokinase overexpression in two strains of Saccharomyces cerevisiae also expressing xylose reductase and xylitol dehydrogenase and its effect on fermentation of xylose and lignocellulosic hydrolysate. Appl. Environ. Microbiol. 2001;67:4249–4255. doi: 10.1128/AEM.67.9.4249-4255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Dale BE. Global potential bioethanol production from wasted crops. Biomass Bioenergy. 2004;26:361–375. [Google Scholar]
- Kim WC, Kim JY, Ko JH, Kang H, Han KH. Identification of direct targets of transcription factor MYB46 provides insights into the transcriptional regulation of secondary wall biosynthesis. Plant Mol. Biol. 2014a;85:589–599. doi: 10.1007/s11103-014-0205-x. [DOI] [PubMed] [Google Scholar]
- Kim WC, Reca IB, Kim Y, Park S, Thomashow MF, Keegstra K, et al. Transcription factors that directly regulate the expression of CSLA9 encoding mannan synthase in Arabidopsis thaliana. Plant Mol. Biol. 2014b;84:577–587. doi: 10.1007/s11103-013-0154-9. [DOI] [PubMed] [Google Scholar]
- Ko JH, Kim WC, Han KH. Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant J. 2009;60:649–665. doi: 10.1111/j.1365-313X.2009.03989.x. [DOI] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, et al. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005;19:1855–1860. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai Y, Usuki H, Yamamoto Y, Yamasato A, Mukaihara T, Hatanaka T. Preparation of hemicellulolic oligosaccharides from chamaecyparis obtuse (hinoki) slurry using commercial enzymes. Front. Chem. Sci. Eng. 2012;6:224–231. [Google Scholar]
- Kuroyama H, Tsutsui N, Hashimoto Y, Tsumuraya Y. Purification and characterization of a beta-glucuronidase from Aspergillus niger. Carbohydr. Res. 2001;333:27–39. doi: 10.1016/s0008-6215(01)00114-8. [DOI] [PubMed] [Google Scholar]
- Lao NT, Long D, Kiang S, Coupland G, Shoue DA, Carpita NC, et al. Mutation of a family 8 glycosyltransferase gene alters cell wall carbohydrate composition and causes a humidity-sensitive semi-sterile dwarf phenotype in Arabidopsis. Plant Mol. Biol. 2003;53:647–661. doi: 10.1023/B:PLAN.0000019074.60542.6c. [DOI] [PubMed] [Google Scholar]
- Lee C, Teng Q, Huang W, Zhong R, Ye ZH. Down-regulation of poGT47C expression in poplar results in a reduced glucuronoxylan content and an increased wood digestibility by cellulase. Plant Cell Physiol. 2009;50:1075–1089. doi: 10.1093/pcp/pcp060. [DOI] [PubMed] [Google Scholar]
- Lee C, Teng Q, Zhong R, Yuan Y, Haghighat M, Ye ZH. Three Arabidopsis DUF579 domain-containing GXM proteins are methyltransferases catalyzing 4-O-methylation of glucuronic acid on xylan. Plant Cell Physiol. 2012;53:1934–1949. doi: 10.1093/pcp/pcs138. [DOI] [PubMed] [Google Scholar]
- Mansfield SD, Kang KY, Chapple C. Designed for deconstruction—poplar trees altered in cell wall lignification improve the efficacy of bioethanol production. New Phytol. 2012;194:91–101. doi: 10.1111/j.1469-8137.2011.04031.x. [DOI] [PubMed] [Google Scholar]
- McCarthy RL, Zhong R, Ye ZH. MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 2009;50:1950–1964. doi: 10.1093/pcp/pcp139. [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Hiratsu K, Todaka D, Nakashima K, Yamaguchi-Shinozaki K, Ohme-Takagi M. Efficient production of male and female sterile plants by expression of a chimeric repressor in Arabidopsis and rice. Plant Biotechnol. J. 2006;4:325–332. doi: 10.1111/j.1467-7652.2006.00184.x. [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Ikeda M, Takada S, Takiguchi Y, Kondou Y, Yoshizumi T, et al. Efficient yeast one-/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana. Plant Cell Physiol. 2010;51:2145–2151. doi: 10.1093/pcp/pcq161. [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, et al. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell. 2007;19:270–280. doi: 10.1105/tpc.106.047043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Ohme-Takagi M. Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol. 2009;50:1232–1248. doi: 10.1093/pcp/pcp075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell. 2005;17:2993–3006. doi: 10.1105/tpc.105.036004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Tsumuraya Y, Hashimoto Y, Yamamoto S. Arabinogalactan-proteins reacting with eel anti-H agglutinin from leaves of cruciferous plants. Agric. Biol. Chem. 1984;48:753–760. [Google Scholar]
- Ohta M, Ohme-Takagi M, Shinshi H. Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. Plant J. 2000;22:29–38. doi: 10.1046/j.1365-313x.2000.00709.x. [DOI] [PubMed] [Google Scholar]
- Pena MJ, Zhong R, Zhou GK, Richardson EA, O’Neill MA, Darvill AG, et al. Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell. 2007;19:549–563. doi: 10.1105/tpc.106.049320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Caffall KH, Freshour G, Hilley MT, Bauer S, Poindexter P, et al. The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell. 2007;19:237–255. doi: 10.1105/tpc.106.047720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen PD, Lau J, Ebert B, Yang F, Verhertbruggen Y, Kim JS, et al. Engineering of plants with improved properties as biofuels feedstocks by vessel-specific complementation of xylan biosynthesis mutants. Biotechnol. Biofuels. 2012;5:84. doi: 10.1186/1754-6834-5-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin M, Allasia V, Pegard A, Allais F, Ducrot PH, Favery B, et al. Imbalanced lignin biosynthesis promotes the sexual reproduction of homothallic oomycete pathogens. PLoS Pathog. 2009;5:e1000264. doi: 10.1371/journal.ppat.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai N, Nishitani K, Masuda Y. Auxin-induced changes in the molecular weight of hemicellulosic polysaccharides of the avena coleoptile cell wall. Plant Cell Physiol. 1979;20:1349–1357. [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, et al. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Sluiter A, Hames B, Ruiz R, Scarlata C, Sluiter J, Templeton D. Golden, CO: National Renewable Energy Laboratory; 2008. Determination of Sugars, Byproducts, and Degradation Products in Liquid Fraction Process Samples. [Google Scholar]
- Sonderegger M, Jeppsson M, Larsson C, Gorwa-Grauslund MF, Boles E, Olsson L, et al. Fermentation performance of engineered and evolved xylose-fermenting Saccharomyces cerevisiae strains. Biotechnol. Bioeng. 2004;87:90–98. doi: 10.1002/bit.20094. [DOI] [PubMed] [Google Scholar]
- Soyano T, Thitamadee S, Machida Y, Chua NH. ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell. 2008;20:3359–3373. doi: 10.1105/tpc.108.061796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowicz BR, Pena MJ, Ratnaparkhe S, Avci U, Backe J, Steet HF, et al. 4-O-methylation of glucuronic acid in Arabidopsis glucuronoxylan is catalyzed by a domain of unknown function family 579 protein. Proc. Natl Acad. Sci. USA. 2012;109:14253–14258. doi: 10.1073/pnas.1208097109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Acker R, Vanholme R, Storme V, Mortimer JC, Dupree P, Boerjan W. Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol. Biofuels. 2013;6:46. doi: 10.1186/1754-6834-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Escamilla-Trevino LL, Sathitsuksanoh N, Shen Z, Shen H, Zhang YH, et al. Silencing of 4-coumarate:coenzyme A ligase in switchgrass leads to reduced lignin content and improved fermentable sugar yields for biofuel production. New Phytol. 2011;192:611–625. doi: 10.1111/j.1469-8137.2011.03830.x. [DOI] [PubMed] [Google Scholar]
- Xu B, Ohtani M, Yamaguchi M, Toyooka K, Wakazaki M, Sato M, et al. Contribution of NAC transcription factors to plant adaptation to land. Science. 2014;343:1505–1508. doi: 10.1126/science.1248417. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Mitsuda N, Ohtani M, Ohme-Takagi M, Kato K, Demura T. VASCULAR-RELATED NAC-DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J. 2011;66:579–590. doi: 10.1111/j.1365-313X.2011.04514.x. [DOI] [PubMed] [Google Scholar]
- Yang F, Mitra P, Zhang L, Prak L, Verhertbruggen Y, Kim JS, et al. Engineering secondary cell wall deposition in plants. Plant Biotechnol. J. 2013;11:325–335. doi: 10.1111/pbi.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York WS, Darvill AG, McNeil M, Stevenson TT, Albersheim P. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 1986;118:3–40. [Google Scholar]
- Zhang K, Bhuiya MW, Pazo JR, Miao Y, Kim H, Ralph J, et al. An engineered monolignol 4-O-methyltransferase depresses lignin biosynthesis and confers novel metabolic capability in Arabidopsis. Plant Cell. 2012;24:3135–3152. doi: 10.1105/tpc.112.101287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Demura T, Ye ZH. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell. 2006;18:3158–3170. doi: 10.1105/tpc.106.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell. 2008;20:2763–2782. doi: 10.1105/tpc.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Ye ZH. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell. 2007;19:2776–2792. doi: 10.1105/tpc.107.053678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lee C, Zhong R, Ye ZH. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell. 2009;21:248–266. doi: 10.1105/tpc.108.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.