Abstract
The phosphate clamp is a distinct mode of ligand-DNA binding where the molecular is manifest through (“non-covalent”) hydrogen-bonding from am(m)ines of polynuclear platinum complexes to the phosphate oxygens on the oligonucleotide backbone. This third mode of DNA is unique from the “classical” DNA intercalators and minor groove binding agents and even the closely related covalently binding mononuclear and polynuclear drugs. 2D 1H NMR studies on the Dickerson Drew Dodecamer (DDD, d(CGCGAATTCGCG)2) showed significant A-T contacts mainly on nucleotides A6, T7 and T8 implying a selective bridging from C9G10 in the 3' direction to C9G10 of the opposite strand. {1H, 15N} HSQC NMR Spectroscopy using the fully 15N-labelled compound ([{trans-Pt(NH2)3(H2N(CH2)6NH3}2μ-(H2N(CH2)6NH2)2(Pt(NH3)2]8+ (TriplatinNC) showed at pH6 significant chemical shift and 1J(195Pt-15N) coupling constants from free drug and DDD-TriplatinNC at pH 7 indicative of formation of the phosphate clamp. 31P NMR results are also reported for the hexamer d(CGTACG)2 showing changes in 31P NMR chemical shifts indicative of changes around the phosphorous center. The studies confirm the DNA binding modes by substitution-inert (non-covalent) polynuclear platinum complexes and help to further establish the chemotype as a new class of potential anti-tumor agents in their own right with a distinct profile of biological activity.
Introduction
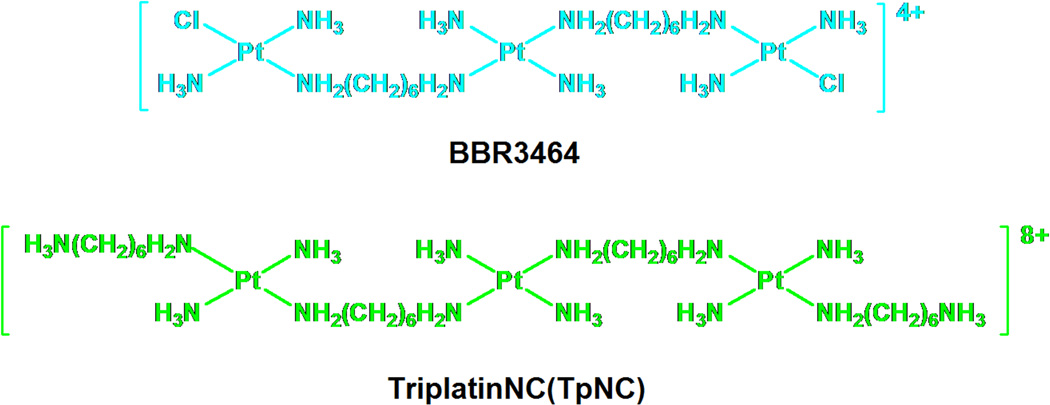
DNA binding is the mechanistic paradigm by which cytotoxic platinum complexes are believed to exert their antitumour activity. In general, the associated processes of DNA function such as transcription and repair may be affected by DNA-targeted drugs. The development of polynuclear platinum complexes (PPCs) represents an approach to systematically alter the cellular response induced by cisplatin by changing the nature and structure of the DNA lesion induced.1 The development of BBR3464 (Fig. 1), the only platinum compound not based on the mononuclear cisplatin chemotype to have entered human clinical trials, validated this approach.1,2 Unique features of the DNA adducts induced by BBR3464 and other di/trinuclear complexes include long-range {Pt,Pt} inter- and intrastrand crosslinks where the sites of platination may be 4–6 base pairs apart1 and the observation of “walking” of bifunctional adducts on DNA dictated in part by thermodynamic stability of the crosslinks.3 Directional isomers of 5'→5' and 3'→3' interstrand crosslinks isomers have also been isolated and characterized.4,5 In the latter case, the pre-association of the central Pt moiety in the minor groove may strongly influence the formation and the structure of the cross-links.6,7
Fig. 1.
Structures of BBR3464 and its substitution-inert analogue TriplatinNC (TpNC).
To study solely the electrostatic component of the DNA binding, we replaced the chloride leaving groups of BBR3464 in several polynuclear platinum compounds with substitution-inert NH3 and “dangling amine -NH2(CH2)nNH3+ carrier ligands (Fig. 1).
The thus created “non-covalently” binding platinum drugs exhibit some unique features in several biophysical studies, with high DNA binding affinity and very effective condensation of DNA.8,9 DNA binding by these compounds also protects against minor groove alkylation, in a manner similar to minor-groove binders such as Adriamycin.10 The most studied compound among this new series of potential platinum drugs, a sub-set of the large polynuclear platinum class, is TriplatinNC (TpNC) where the leaving groups are displaced by two diaminohexanes resulting in an 8+ charged complex.
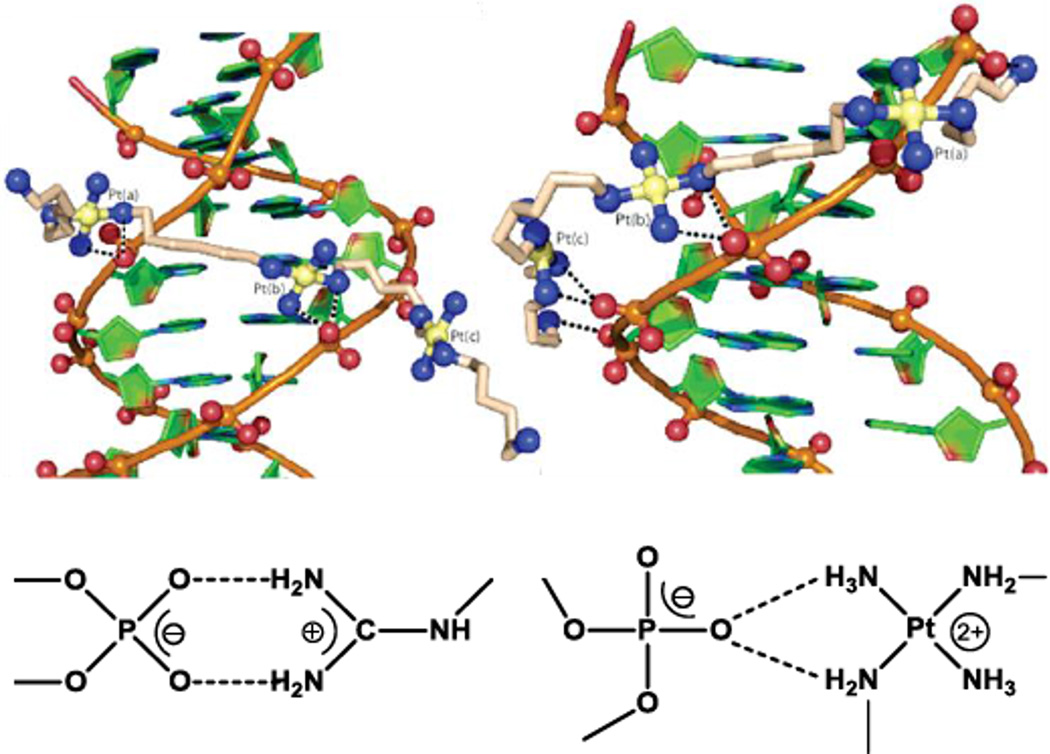
The mode of DNA binding of the substitution-inert compounds was revealed in the crystal structure of TriplatinNC with the Dickerson-Drew Dodecamer (DDD, d(CGCGAATTCGCG)2), (Fig. 2).11 The platinum compound was associated to the DNA either through backbone tracking where the complex extends along the nucleic acid backbone or minor groove spanning involving interactions with both strands of the oligonucleotide. The binding is manifest through the hydrogen bonding of the am(m)ines of all three platinum units with the phosphate oxygens in a clamp like fashion. The binding pattern resembles the interaction of arginine groups with the nucleic acid backbone in protein RNA recognition, the arginine fork, Fig. 2. The generality of the motif has been confirmed by a second crystal and molecular structure with AH44 – that is [{Pt(NH3)3}2μ-(H2N(CH2)6NH2)2(Pt(NH3)2]6+ with NH3 replacing the Cl− ligand of BBR3464.12 This third DNA binding motif, the phosphate clamp, is distinct from the classical intercalation and minor-groove binding modes. Indeed, the phosphate clamp-DNA interactions further result in displacement of intercalated ethidium bromide, and facilitate cooperative binding of Hoechst 33258 at the minor groove.8,9,13
Fig. 2.
Crystal structure of TriplatinNC associated with the Dickerson-Drew Dodecamer showing minor-groove spanning and backbone tracking (top), and representation of arginine fork (bottom, left) and phosphate clamp molecular recognition motifs (bottom, right).
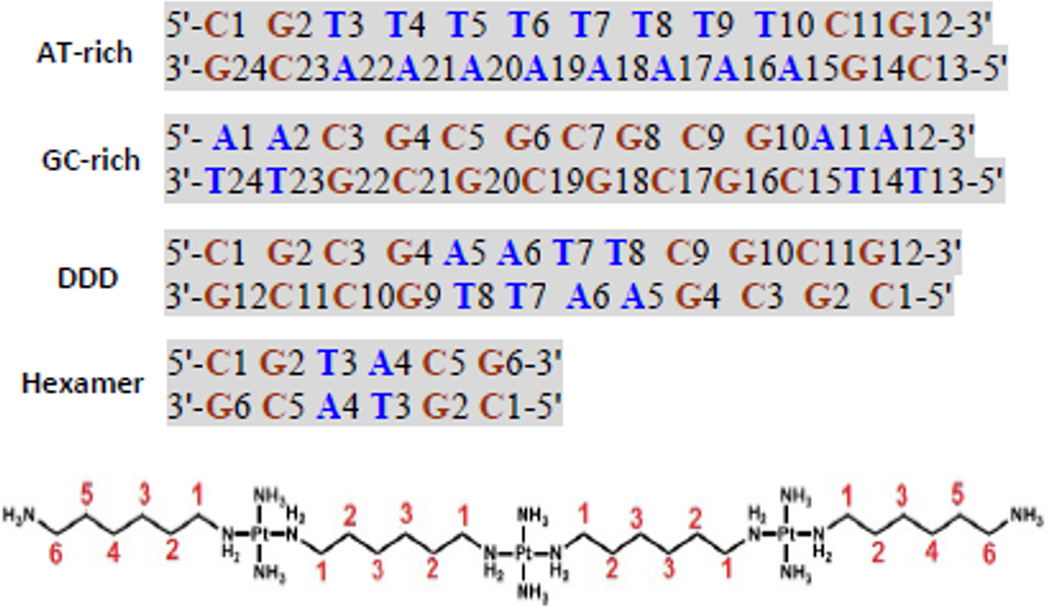
A general question is how the solution binding properties reflect the crystallographically determined modes of groove spanning and backbone tracking. NMR is the best analytical method to achieve structural characterization in solution at atomic resolution. In this contribution we examine by NMR spectroscopic techniques the interactions of TriplatinNC with three sequence-defined oligomers, an AT-rich and a GC-rich dodecamer as well as the Dickerson-Drew dodecamer itself, Fig. 3. The use of the DDD served two purposes. Firstly it represents an oligomer with an intermediate extent of AT nucleobases and secondly allows for a direct comparison with the crystal structure of the DDD-TpNC adduct. Further, 15N labeling of the TriplatinNC allows use of {1H, 15N} HSQC NMR Spectroscopy to further examine details of the DNA interactions. The use of {1H, 15N} HSQC NMR Spectroscopy has been especially useful in delineating the mechanism of DNA binding of a wide range of mononuclear and polynuclear Pt complexes.14 The combined evidence is consistent with maintenance of the crystallographically determined binding modes in solution and helps explain the consequences of suchbinding.
Fig. 3.
Naming and sequences of the oligomer duplexes studied, and numbering for the protons of the TriplatinNC (TpNC) for chemical shift and NOE assignments. The linker protons are referred to as LH1 through LH6. The very similar chemical shifts of LH2 and LH5 as well as LH3 and LH4 are assigned together as TpNC-LH2/LH5 and TpNC-LH3/LH4.
Results and discussion
The assignments of the resonances for the free oligomers and in the presence of TriplatinNC were conducted by 2D {1H, 1H} COSY, TOCSY and NOESY NMR in the standard manner as described in previous reports.15,16,17 In this paper we emphasize the changes on the DDD – studies on the AT-rich and GC-rich oligonucleotides have been summarized.9 The DDD assignments were in agreement with published results.18,19 The {1H, 15N} HSQC NMR assignments were consistent with those previously published for BBR3464 and congeners.6,7
Dickerson-Drew Dodecamer
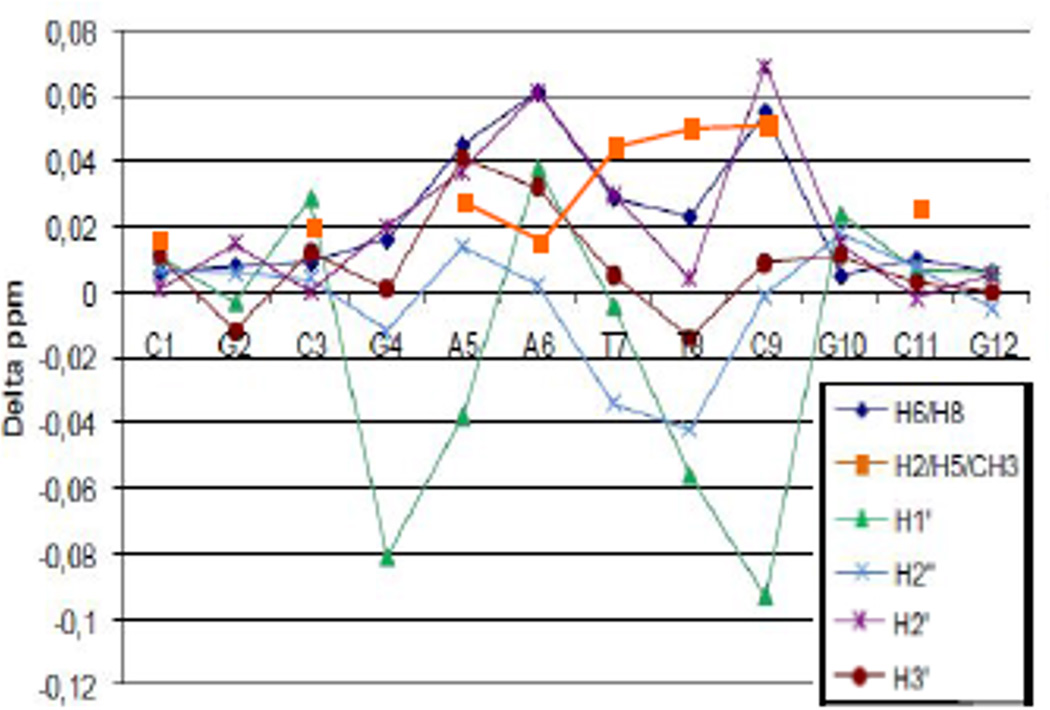
The changes in the DDD chemical shifts caused by the association with TriplatinNC are most prominent in the central AT-rich part of the oligomer between G4 and C9 (Fig. 4).
Fig. 4.
Changes in chemical shifts for non-exchangeable protons of Dickerson Drew Dodecamer (DDD), δ(DDD+TpNC) – δ(DDD).
The profile is similar to that of the AT-rich oligomer where NOE cross peaks are also observed with all A-H2 protons suggesting that TriplatinNC binds in the minor groove.9 The changes in chemical shift are also similar to those observed between AH44 and the self-complementary “high-affinity” sequence for minor groove binders, d(GGTAATTACC)2.10 In contrast to the AT-duplex the changes in the GC-rich dodecamer upon binding of TriplatinNC are considerably smaller, with changes mainly occurring for the H1' chemical shifts.9 The fact that no connections with G-NH2 or C-NH2 are detected implies that binding via major or minor groove spanning can be excluded suggesting that TriplatinNC binds predominantly in a backbone tracking manner towards GC-rich DNA.9
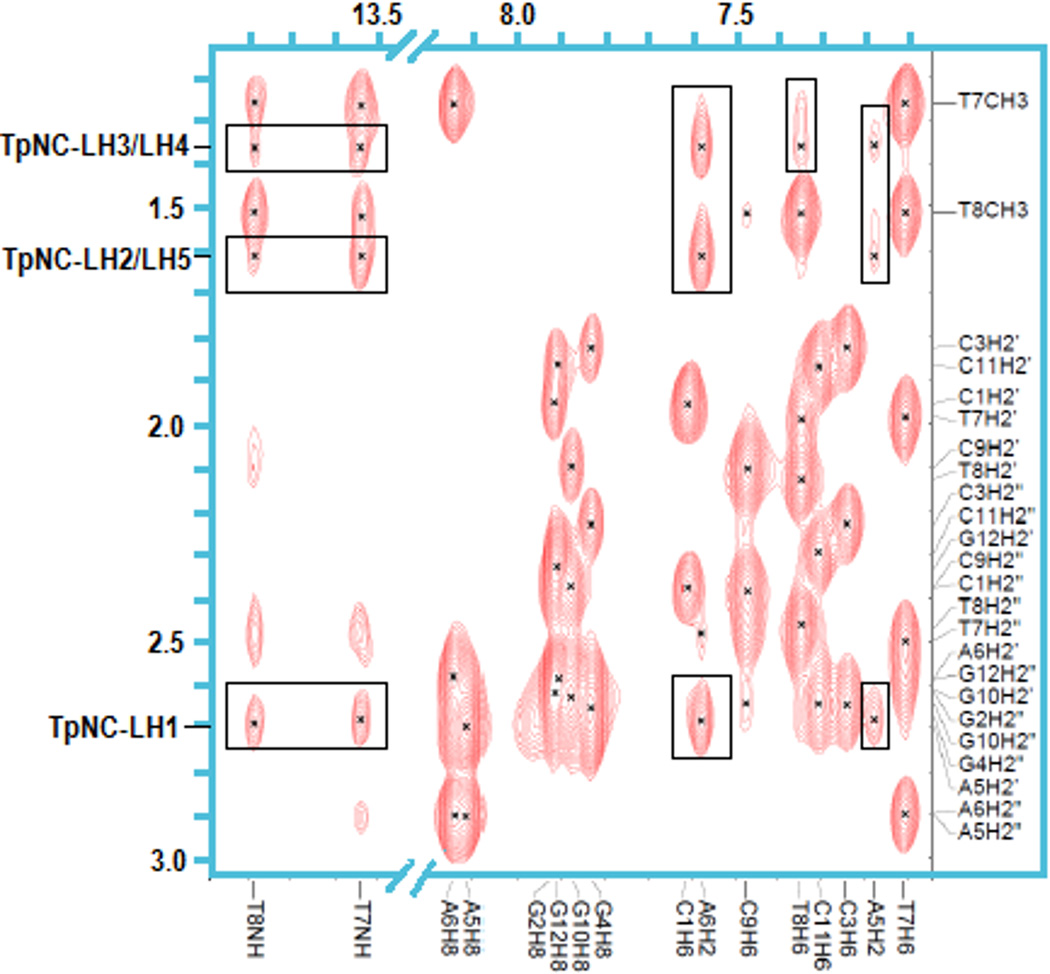
2D NOESY experiments allow determination of interactions between nuclei that are separated by multiple bonds but still in close proximity through space.15 The principal detected NOE contacts are in agreement with the observed chemical shift changes. Strong NOE cross-peaks are observed between TpNC-LH1, TpNC-LH2/LH5 and TpNC-LH3/LH4 and A6-H2 (Fig. 5). In contrast, the connectivities with the adjacent A5-H2 are observed but weaker. At the same time the interstrand connectivities between A6-H2 and the CH3’s of T8 and T9 as well as the H2'/2" of these bases are noticeably weakened compared to the free dodecamer.
Fig. 5.
NOE cross-peaks between TriplatinNC(TpNC)-LH1 through LH5 with T7 and T8 imino protons and H2 of A5 and A6 of DDD. Note that the intensity level for the imino proton region is increased compared to the H6/H8 area.
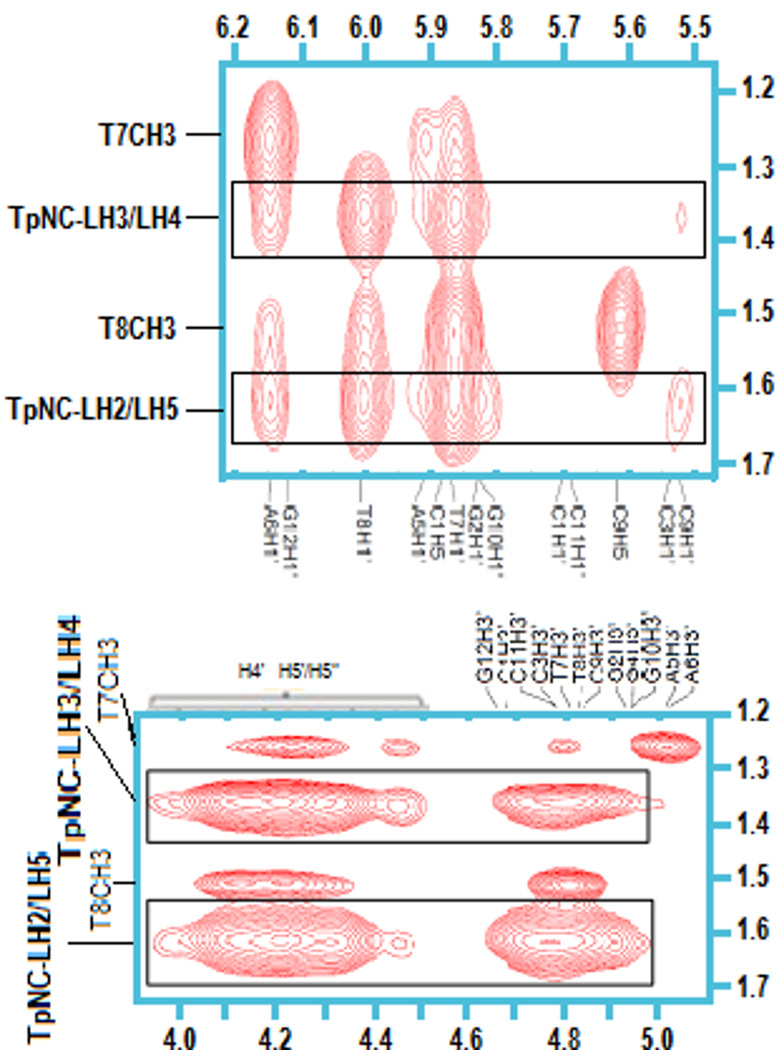
Strong NOE connections are also seen between the TriplatinNC linker protons and H1' of T7, T8 and A6, whereas no connectivity to H1' of A5 is detected (Fig. 6, top). The strong resonance shifts observed for these particular protons are likely due to changes in the secondary structure of the dodecamer. Weak contacts with C9-H1' are registered but no interactions with G4-H1' are observed. Further strong NOE signals of TpNC-LH2/LH5 and TpNC-LH3/LH4 are detected in the H3'/H4'/H5'/H5" region (Fig. 6, bottom). Due to the overlap of the sugar protons the contacts are not distinguishable. It was obvious however, that no cross-peaks with A5-H3' nor A6-H3' existed. Cross signals for TpNC-LH1 could not be confirmed unambiguously in this area. Connectivities between the protons of TpNC-LH6 and the dodecamer are not observed.
Fig. 6.
NOE cross-peaks between TpNC-LH2/LH5 and –LH3/LH4 with H1' of DDD (top) and H3'/H4'/H5'/5" (bottom).
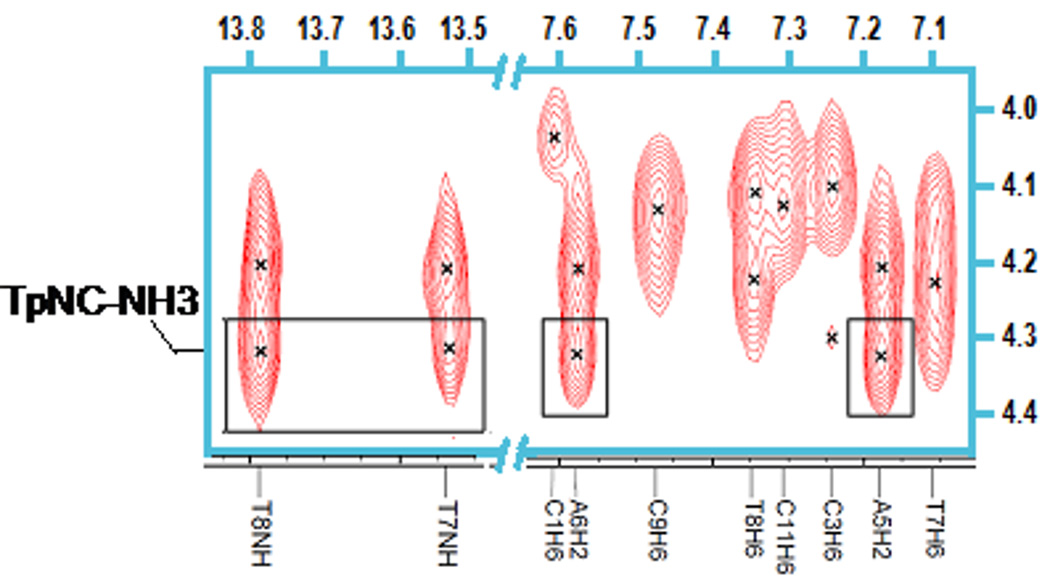
The NOESY spectrum recorded in 8% D2O/92% H2O further reveals strong connectivities of the TpNC-NH3 groups with A5-H2 andA6-H2, as well as weak cross-peaks with T8-NH and T7-NH (Fig. 7). Cross peaks with TpNC-NH2 are not observed, possibly because the signals are suppressed by water presaturation. No further connectivities are detected for TpNC-NH3. The study of the exchangeable protons additionally shows weaker NOE contacts of the LH1 through LH5 with T7-NH and T8-NH (Fig. 7). Similar connectivities are not registered in the spectra of the AT-duplex.9
Fig. 7.
NOE cross-peaks between TpNC-NH3 and T7 and T8 imino protons and H2 of A5 and A6 of DDD. Note that the intensity level for the imino proton region is increased compared to the H6/H8 area.
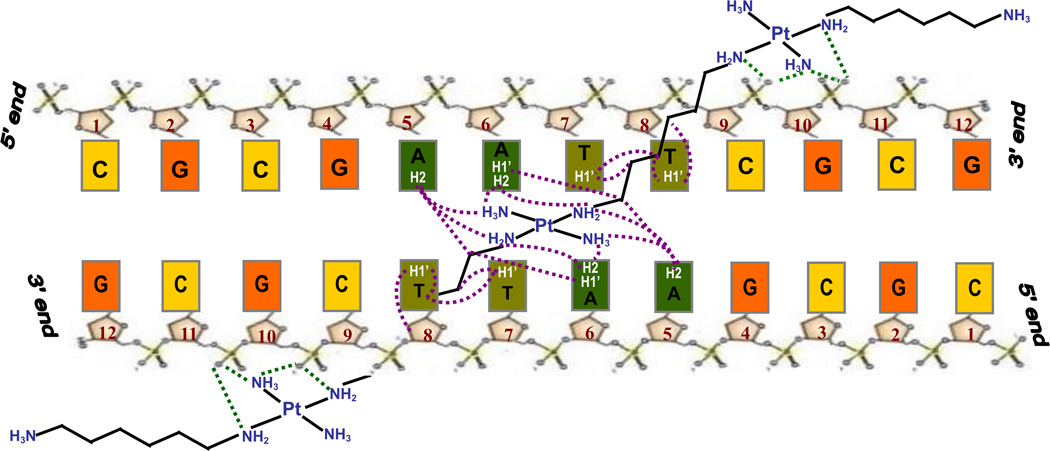
In summary, the results strongly suggest a groove spanning binding mode across the AATT center of the Dickerson-Drew Dodecamer, Scheme 1. Moreover, since the contacts in the minor groove concentrate mainly on nucleotides A6, T7 and T8 the findings imply a selective bridging from C9G10 in the 3' direction to C9G10 of the opposite strand. It is possible that interactions between the central PtN4 unit and the oxygen of thymine additionally aid the selective binding. The specificity of the drug association is surprising considering that TriplatinNC exhibits a relatively simple structure and was not explicitly designed to fit a certain DNA sequence. The AT-rich preference is consistent with that reported for the AT-rich oligomer whereas, in contrast, backbone tracking is suggested for the GC-rich oligomer.9
Scheme 1.
Proposed scheme for the Phosphate Clamp, groove spanning and minor groove binding in Solution by 2D {1H, 15N} HSQC experiments.
The results to date support the concept that the substitution-inert trinuclear platinum drugs bind preferably towards the DNA minor groove and are able to differentiate between AT and GC-rich DNA. To gain further insights into the selectivity as well as the binding geometry of TriplatinNC on a molecular level the interactions with different oligomers were studied using {1H,15N} HSQC NMR Spectroscopy. With the association of the drug taking place via the am(m)ine ligands of the platinum compounds this technique could be especially useful in evaluating binding preferences and motifs.14 For this purpose the fully 15N-labeled TriplatinNC was synthesized.
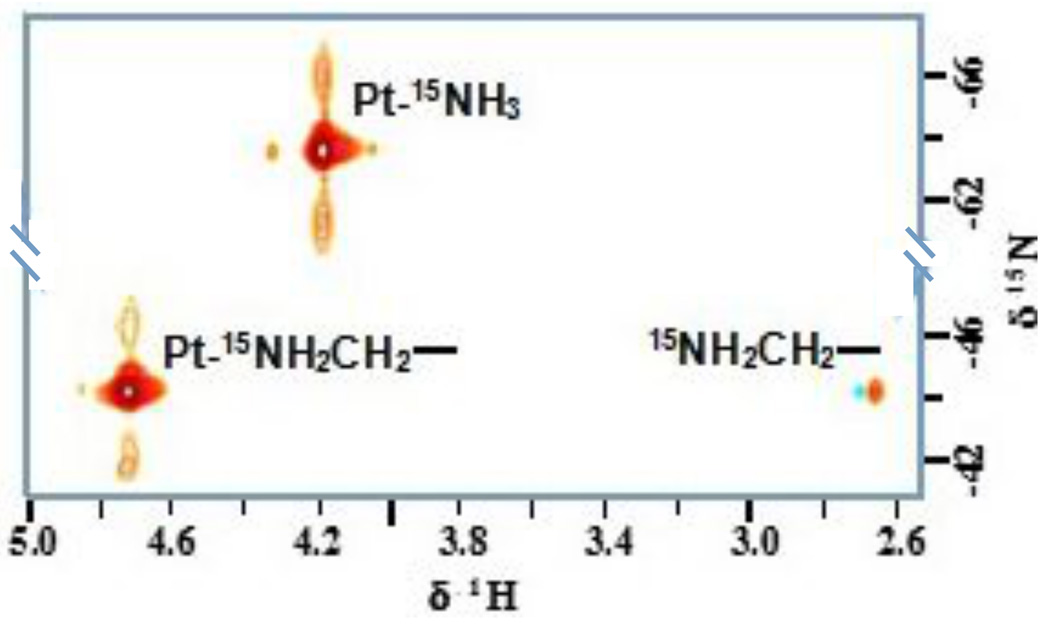
The 2D {1H,15N} HSQC spectrum of 15N-labeled TpNC shows only two cross-peaks (Fig. 8). Due to the symmetry of the molecule the three PtN4 units are chemically equivalent and hence are not distinguishable by NMR. The peaks with 1H/15N chemical shifts of 4.18/−63.8 ppm were assigned to the 15NH3 groups and accordingly the signals at 4.72/−44.7 ppm and 4.72/−44.4 ppm were assigned to the 15NH2 units.6 The downfield shift of the latter in both dimensions is in agreement with the deshielding effect of the hydrocarbon chain of the linker. Further verification for the assignment is the occurrence of a cross-peak between the NH2 and its neighboring CH2-group at 2.67 ppm. The absence of a signal for the terminal protonated amine can be ascribed to the fast proton exchange at pH 7. The results were consistent with those found for 15N-BBR3464.6,2
Fig. 8.
2D {1H,15N} HSQC NMR spectrum of fully 15N-labeled TriplatinNC.
Upon addition of the dodecamers a downfield shift between 0.07–0.13 ppm for both peaks is observed in the proton dimension at pH7, Table 1. The fact that the protons of the Pt-15NH3 as well as the Pt-15NH2CH2- units are affected to the same extent indicates that both groups are equally involved in the binding process. The shifts are, however, rather small and it is possible that the high salt concentration leads to an augmented competition between the sodium ions and the drug. The chemical shifts in the 15N dimension are likewise not significantly influenced by the presence of the DNA which also contradicts a close contact to the phosphate oxygens. 1J(195Pt-15N) coupling constants for the Pt-NH2CH2- system were not available because of proximity with the H2O peak.
Table 1.
Chemical shifts 1H/15N of TriplatinNC (TpNC) and with oligomers at pH7.
| Pt-NH3 | Pt-NH2CH2- | |||
|---|---|---|---|---|
| δ(1H/15N) | 1J(195Pt-15N) | δ(1H/15N) | δ(1H/15N) | |
| TpNC | 4.18/−63.8 | 291 | 4.72/−44.7 | 4.72/−44.4 |
| TpNC-DDD | 4.31/−63.6 | 294 | 4.79/−44.3 | 4.79/−44.3 |
| TpNC-AT | 4.30/−63.5 | 290 | 4.84/−45.0 | 4.84/−44.4 |
| TpNC-GC | 4.28/−63.6 | 297 | 4.82/−44.7 | 4.82/−44.4 |
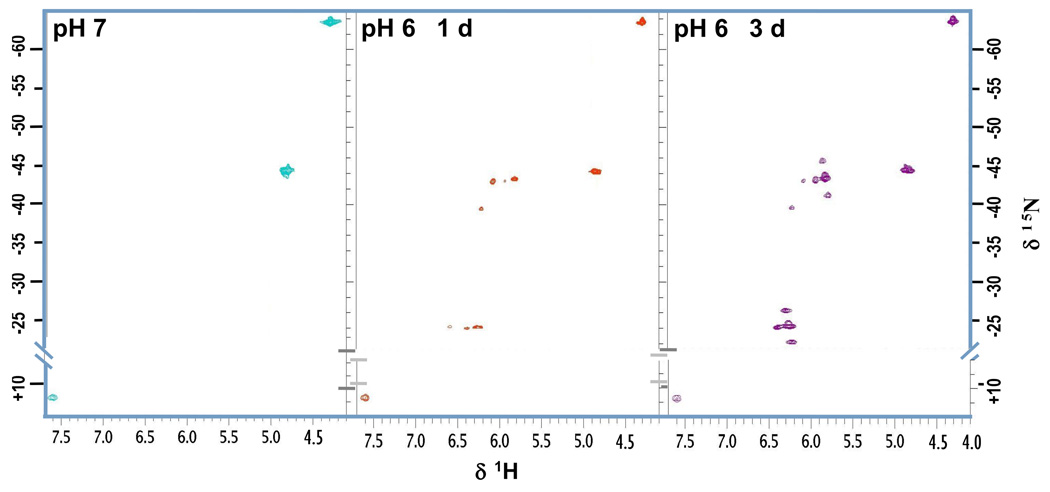
We next repeated the experiments with the oligomers at pH 6 because the lower pH helps to shift the NH2/NH3+ equilibrium towards the site of the protonated amine. Consequently, the corresponding signal which appears at 7.58 ppm/11.6 ppm in the HSQC spectra can be assigned to the dangling amine (Fig. 9). Initially, comparison with the free platinum drug at pH 6 reveals small downfield shifts between 0.10 ppm and 0.13 ppm in the proton dimension (Table 2). The 15NH3 and 15NH2shifts also undergo small changes (up to 0.8 ppm in the case of Pt-NH2CH2- in the GC duplex). Additional signals appear in the spectra within hours of changing the pH of the drug-treated DNA but not in the spectrum of the free platinum compound. Two major sets of peaks are observed, each with the same distribution pattern in the spectral region, centered round 5.8 ppm/−43 ppm and 6.3 ppm/−24 ppm, respectively (Fig. 9).
Fig. 9.
Monitoring the formation of the phosphate clamp by HSQC for the reaction of TriplatinNC and DDD: pH 7 (left), 1d (mid) and 3d (right) after adjusting the pH to 6.
Table 2.
Chemical shifts (1H / 15N in ppm) and 1J(195Pt-15N) coupling constants of TriplatinNC (TpNC) and TpNC-oligomer adducts at pH 6. PC indicates shifts arising from the formation of the phosphate clamp.
| Pt-NH3 | Pt-NH2CH2- | Pt-NH3(PC) | Pt-NH2CH2-(PC) | −CH2NH3 | |||||
|---|---|---|---|---|---|---|---|---|---|
| δ(1H/15N) | 1J(195Pt-15N) | δ(1H/15N) | δ(1H/15N) | δ(1H/15N) | 1J(195Pt-15N) | δ(1H/15N) | 1J(195Pt-15N) | δ(1H/15N) | |
| TpNC | 4.19/−63.5 | 293 | 4.72/−44.2 | 4.72/−44.6 | 7.58/11.6 | ||||
| TpNC-DDD | 4.30/−63.9 | 303 | 4.84/−44.3 | 4.84/−44.3 4.85/−44.6 |
5.82/−43.3 5.94/−43.0 |
270 259 |
6.25/−24.2 6.39/−24.0 |
249 241 |
7.61/11.4 |
| TpNC-AT | 4.31/−64.2 | 290 | 4.85/−44.9 | 4.84/−44.4 | 5.86/−44.0 5.99/−43.9 |
269 256 |
6.30/−24.9 6.45/−24.6 |
256 257 |
7.66/10.8 |
| TpNC-GC | 4.29/−64.3 | 296 | 4.83/−45.0 | 4.82/−44.4 | 5.85/−44.1 5.98/−43.7 |
264 | 6.28/−24.8 6.43/−24.7 |
242 260 |
7.64/10.7 |
These new peaks are significantly shifted in both the 1H and 15N dimension and grow and persist over time while the initial peaks for NH3 and NH2CH2- become smaller but remain observable. Upon increasing the temperature from 15 to 26 °C the new cross-peaks also remain unchanged while the Pt-H2N(CH2)6NH3+ signal for the dangling amine disappears due to the faster proton exchange. Only one set of peaks is unambiguously assigned. The appearance of the new cross-peaks due to the formation of platinum aqua species or covalent binding to the nucleobases as a result of ligand (diamine) displacement on the platinum center can be excluded, as the chemical shifts do not fall in the usually observed ranges for either Pt-Gua(N7) or Pt-H2O species14,21, and loss of diamine would result in loss of signal anyway. 1H-NMR control experiments showed that TriplatinNC is stable even at pH 2.5 and does not show any signs of disintegration. Thus, the downfield shift of approximately 1.5 ppm in the proton dimension and 20 ppm in the nitrogen dimension for both cross-peaks indicates a dramatic change in the chemical environment of the amino/ammine hydrogens rather than the platinum. We therefore conclude that the signals are caused by NHn-groups bound to platinum and not by protonated amines. The new signals centered at 6.3 ppm/−24 ppm also show coupling to the protons of the linker CH2-group at 2.77 ppm and are therefore assigned as Pt-15NH2-(CH)6. Accordingly, the peaks centered at 5.8 ppm/−43 ppm are attributed to Pt-15NH3.
Typical coupling constants are ~300 Hz for 195Pt-N4 complexes or for Pt-15NH3 trans to another NH3 (Z), for example [Pt(15NH3)2(Z2)n+ complexes.14,22,23 The new signals exhibit satellite peaks with a lower coupling frequency of 1J(195Pt-15N) 240–270 Hz for both 15NH3 and 15NH2 ligands. Interestingly, these new peaks correlate with the significantly lesser shielding in the 15N dimension (Tables 1 & 2). This trend has been previously observed within a series of platinum amine complexes. The major contribution to the one-bond coupling constant 1J(195Pt-15N) is usually explained in terms of the Fermi contact interaction involving Pt 6s and N 2s orbitals – thus, classically, the coupling constants are approximately 1.2 – 1.5 times smaller for Pt(IV) than Pt(II) based on d2+sp3 and dsp2 hybridization respectively.14,22,24
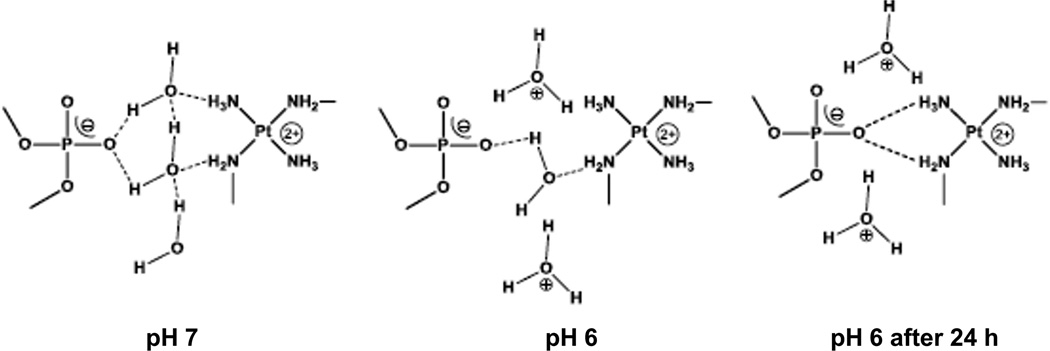
The observed changes in chemical shift and coupling constants are consistent with formation of the phosphate clamp binding motif. Hydrogen-bonding to highly electronegative phosphate oxygen will reduce electron density on the nitrogen am(m)ine. The “delocalization” of the NH bond may be considered deshielding for both 1H and 15N. At neutral pH the association of TriplatinNC to the DNA backbone is most probably water mediated. Minor-groove binding in the DDD mediated by water has been studied extensively25,26, and our results are consistent with the fact that only a small shift is observed upon binding to the oligomer at pH7 in the present case. We suggest that when lowering the pH to 6 the hydrogen-bond network and with it the interface between the backbone and the drug is partly disrupted. Hence, the association of TriplatinNC is no longer mediated by water but can interact directly with the negatively charged oxygen (Scheme 2). The coexistence of the initial and the new set of cross-peaks in the HSQC spectra is in agreement with the phosphate clamp motif where only one pair of NH3/NH2CH2- per PtN4 unit is involved in the binding process (Scheme 2).
Scheme 2.
Proposed scheme for the pH dependence of phosphate clamp formation.
2D {1H, 31P} HMQC experiments
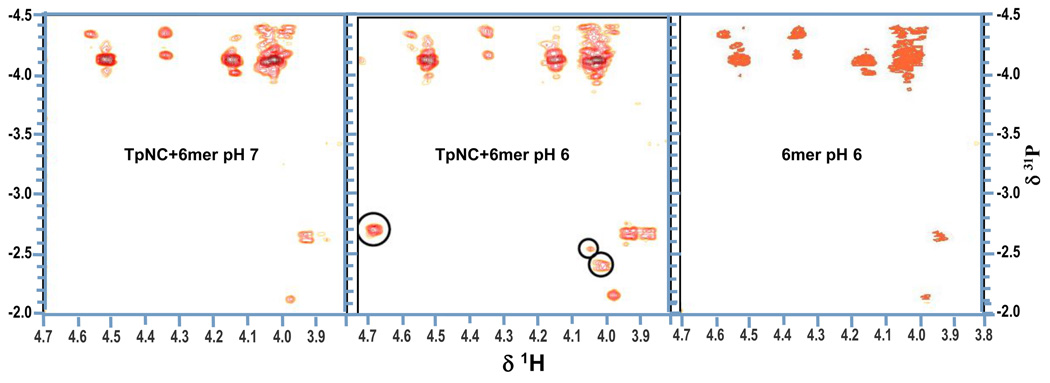
We attempted to find complementary evidence of the phosphate clamp by recording 2D {1H,31P} HMQC spectra of the Pt-DNA samples to examine changes in the 31P NMR of the phosphate backbone. The DDD samples were not concentrated enough to yield good results and overlapping signals complicated the spectra. Higher concentrations could not be applied because of an increased precipitation of the DNA-drug adduct. We therefore employed the hexamer duplex d(CGATCG)2 (Fig. 3) instead which gave a clearer spectrum. The duplex:drug ratio was increased to 3:1 in order to reduce precipitation. 1H chemical shift assignments are given in Supplemental Information, Table S1. The {1H,15N} HSQC NMR spectral behavior of the hexamer was identical to that of the DDD and further confirmed the formation of the phosphate clamp (Fig. S1). Upon lowering pH to 6 (right), the spectrum showed additional peaks at δ1H (5.82) /15N (−43.0) with 1J(195Pt-15N) = 268 Hz and δ1H (6.28) / 15N (−24.0) with 1J(195Pt-15N) = 247 Hz.
For the 2D {1H,31P} HMQC spectra, a sequential assignment was not possible for either the free or the TriplatinNC-treated hexamer. The H5'/H5" signals are overlapping and cannot be distinguished and several of the H3' as well as the H4' peaks are missing. This might be the result of a too low DNA concentration or an inadequate NMR parameter setup. The amount of oligomer duplex could not be increased further, again because of enhanced precipitation in the presence of the platinum drug. Furthermore, the variation of the relevant experimental HMQC parameters did not yield any improvement. There are no significant differences between the spectra of the free and the modified oligomer at pH 7. The spectrum of free hexamer at pH6 is also unchanged from that at the higher pH (Fig. 10). In contrast, at pH 6 a considerable downfield shift in the 31P dimension is observed for three new peaks in the spectrum of the TriplatinNC-hexamer adduct (Fig. 10 mid). The 31P shifts for all three signals are different indicating that three different phosphate groups are involved. The strong cross-peak at δ1H (4.67)/31P(−2.71) can be assigned to the H3' of C5 due to its proton shift. The two other signals at δ1H (4.04)/31P(−2.55) and d1H(4.00)/31P(−2.41) cannot be attributed unequivocally since several proton resonances in the crowded H4’ and H5'/5" region come into consideration. In principle, since the DNA hexamer is not phosphorylated at the 3' or 5' ends, each phosphate group, situated between two sugar rings, should show at least two correlations with the H3' of its 3' neighbor and H5'/H5" in the 5' direction. As mentioned before the lack of the additional cross-peaks might be due to low concentration or suboptimal parameter settings and therefore further investigations are needed. Nonetheless, the fact that downfield shifts for several 31P resonance signals are observed only at the lower pH agrees with the findings from the 2D {1H,15N} HSQC NMR experiments even though an unambiguous identification of the phosphates involved is not possible at this point.
Fig. 10.
Consequences of the formation of the phosphate clamp on the chemical shifts in the 2D {1H,31P} HMQC spectra of d(CGTACG)2 modified with TpNC: spectra at pH7 (left) and after adjusting the pH to 6 (mid), and free d(CGTACG)2 at pH 6 (right).
Experimental
Synthesis of 15N-labeled TriplatinNC
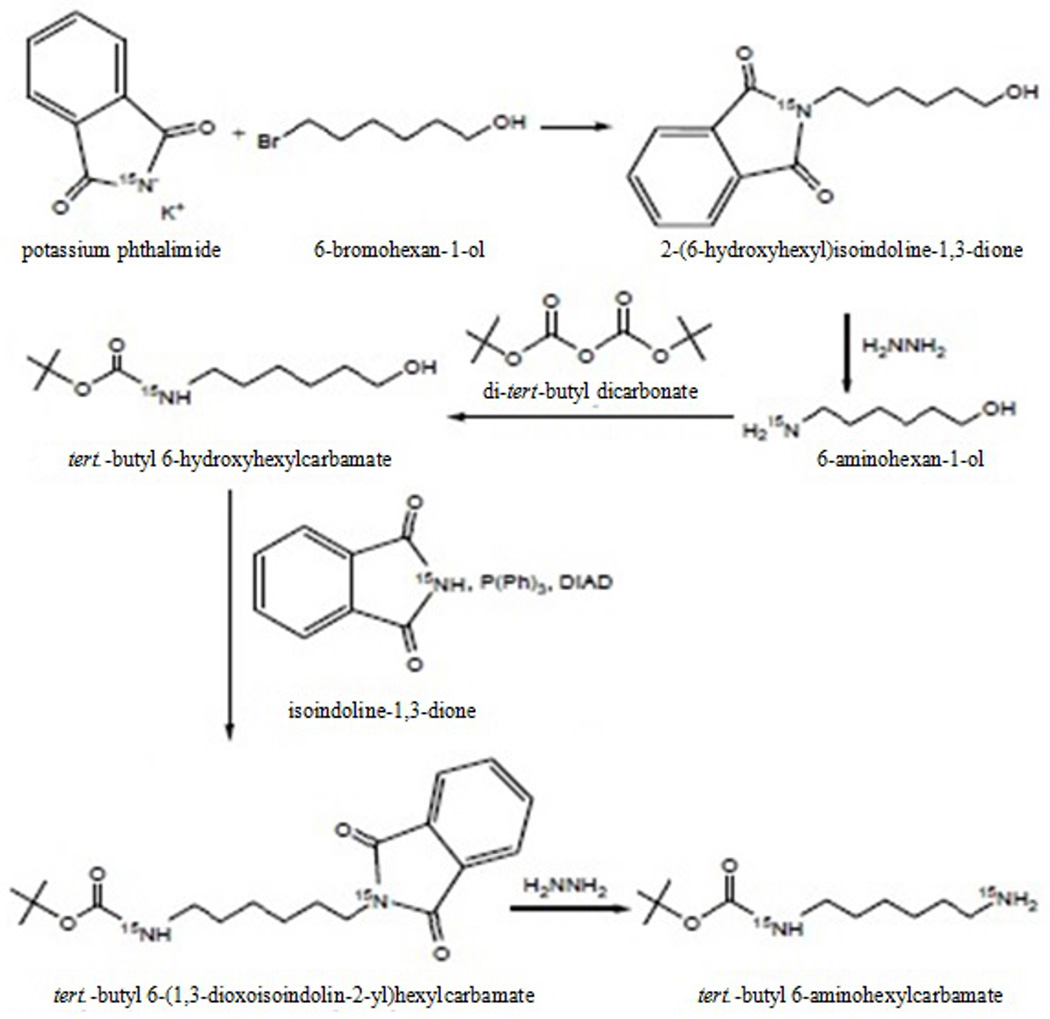
We developed a more efficient and cost effective alternative synthetic pathway to the critical mono-Boc-substituted linker. The general idea was to introduce the two amino groups at different stages of the synthesis, Scheme 3. By this the formation of a free diaminohexane would be avoided and a selective protection and deprotection would be enabled.The alcohol was chosen because it could be easily converted into an amine by a Mitsunobu reaction. After protection of the aminoalcohol with BOC the hydroxo group was converted to the free amine by a Mitsunobu reaction followed by a hydrazinolysis. The product and the intermediates were characterized by 1H, 13C and 15N NMR Spectroscopy. The overall yield was 54.3%
Scheme 3.
Procedure for the preparation of 15N labeled tert-butyl 6-aminohexyl-carbamate.
Step 1: Phthalimide reaction
2.50 g (13.50 mmol) of 15N-labeled potassium phthalimide and 2.21 ml (16.87 mmol) 1-bromohexan-6-ol were stirred under reflux in 150 ml dry acetonitrile for 24 h. After cooling to room temperature the potassium bromide precipitate was filtered off and the filtrate was evaporated under reduced pressure to give a slightly yellow colored oil. The crude product was purified by flash chromatography (SiO2, CHCl3/EtOAc 95/5 to CHCl3/EtOAc 50/50), yield 3.11 g (12.56 mmol), 93% .1H-NMR (CDCl3) [ppm]: 7.77 (d, 4H), 3.64 (m, 4H), 1.51 (m, 6H); 13C-NMR (CDCl3) [ppm]: 168.7, 134.1, 132.2, 62.7, 38.0, 32.7, 28.7, 26.7, 25.4; 15N-NMR (CDCl3) [ppm]: 140.0.
Step 2: 1st Hydrazinolysis
3.0 g (12.13 mmol) of the 2-(6-hydroxyhexyl)isoindoline-1,3-dione was dissolved in 225 ml absolute ethanol and 2 ml (63.72 mmol) anhydrous hydrazine were added. The reaction mixture was stirred under reflux for 16 h. The white precipitate formed was filtered off and washed with ethanol. The filtrates were combined and after removing the solvent and the excess of hydrazine under reduced pressure the product was obtained as a yellowish oil. The product was used in the next synthesis step without further purification, yield, 1.41 g (12.01 mmol), 99%. 1H-NMR (CDCl3) [ppm]: 3.62 (t, 2H), 2.69 (t, 2H), 1.76 (s, NH2), 1.57 (m, 2H), 1.46 (m, 2H), 1.37 (m, 4H) ; 13C-NMR (CDCl3) [ppm]: 62.5, 41.9, 33.5, 32.7, 26.6, 25.6; 15N-NMR (CDCl3) [ppm]: 24.2.
Step 3: BOC protection
1.40 g (11.95 mmol) 6-aminohexan-1-ol and 1.84 ml (13.19 mmol) triethylenamine were dissolved in 50 ml methanol. A solution of 3.14 g (14.41 mmol) di-tert-butyl dicarbonate in 25 ml methanol was added drop wise within 30 min. The mixture was stirred for 16 h at room temperature. The solvent was evaporated and the resulting yellow oil was purified by flash chromatography (SiO2, hexane/EtOAc 95:5 à 30:70), yield, 2.47 g (11.35 mmol), 95%. 1H-NMR (CDCl3) [ppm]: 3.64 (2H), 3.11(2H), 1.44 (m, 17 H); 13C-NMR (CDCl3) [ppm]: 156.2, 79.1, 62.5, 40.4, 32.6, 30.0, 28.4, 26.4, 25.3; 15N-NMR (CDCl3) [ppm]: 61.3.
Step 4: Mitsunobu reaction
1.71 g (11.64 mmol) 15N-labeled phthalimide, 3.05 g (11.64 mmol) triphenylphosphine and 2.30 g 10.58 mmol) tert.-butyl 6-hydroxyhexylcarbamate were dissolved in dry THF under nitrogen at 0°C. A solution of 2.35 g (11.64 mmol) DIAD (diisopropyl azodicarboxylate) in dry THF was added drop wise within 30 min. The reaction mixture was stirred at 0°C for 2h, then for another 10h at room temperature. The solvent was evaporated under vacuum and the resulting yellow oil was purified by flash chromatography (chloroform/ethyl acetate), yield 3.31 g (9.55 mmol) slightly yellow colored oil, 90 %. 1H-NMR (CDCl3) [ppm]: 7.78 (d, 4H), 3.67 (t, 2H), 3.10 (t, 2H), 1.68 (m, 2H), 1.40 (m, 15H); 13C-NMR (CDCl3) [ppm]: 167.9, 155.9, 132.2, 132.0, 123.7, 79.5, 40.2, 40.0, 30.2, 28.4, 26.4; 15N-NMR (CDCl3) [ppm]: 138.8, 59.4
Step 5: Hydrazinolysis
3.25 g (9.38 mmol) tert.-butyl 6-(1,3-dioxoisoindolin-2-yl)hexylcarbamate was dissolved in 250 ml ethanol. After addition of 1.46 ml (36.32 mmol) hydrazine the reaction mixture was heated under reflux for 10h. Upon cooling to room temperature a white solid precipitated which was removed by filtration. The filtrate was concentrated under reduced pressure to give a yellow oil and some white precipitate. The mixture was suspended in chloroform, filtered and the filtrate was purified by flash chromatography using a chloroform/methanol/ammonia gradient system, yield 1.39 g (6.43 mmol) yellow oil/semi-solid, 69%. 1H-NMR (CDCl3) [ppm]: 3.11 (t, 2H), 2.68 (t, 2H), 1.40 (m, 17H); 13C-NMR (CDCl3) [ppm]: 156.0, 79.0, 42.0, 40.5, 33.4, 30.0, 28.4, 26.6; 15N-NMR (CDCl3) [ppm]: 60.9, 24.0
Preparation of 15N-labeled TpNC
The preparation of 15N-labeled TpNC and the corresponding intermediates followed the standard procedures of synthesis.1,27 TriplatinNC, yield 138 mg (82.6 μmol) white powder, 65.9 %. 1H-NMR (D2O) [ppm]: 2.97 (t, 4H), 2.64 (t, 12H), 1.62 (m, 16H), 1.34 (m, 16H); 15N-NMR (D2O) [ppm]: 11.8, −44.2, −63.7.
Sample preparation
The oligonucleotides were purchased from the Midland Certified Reagent (Midland, TX, USA). Annealing of the oligonucleotides was achieved by heating the aqueous solutions to 80°C for 15 minutes, then slow cooling to 15°C over 6 hours. All samples, 1 mM DDD and 1 mM DDD with 1 mM TriplatinNC were prepared in 8% D2O / 92% H2O with 10 mM PO4, 300 mM NaCl and adjusted to pH 6 with HNO3/NaOH. Solutions were lyophilized twice from 99.999% D2O and dissolved in D2O for the acquisition of non-exchangeable protons 2D NMR.
NMR spectroscopy
The NMR spectra were recorded on a Bruker AVANCE III 600 MHz spectrometer (1H, 600.1 MHz; 15N, 60.8 MHz, 31P, 242.9 MHz) fitted with a pulsed field gradient module and 5mm inverse quadruple resonance (QXI) probe. The 1H NMR chemical shifts are internally referenced to TSP, the 15N chemical shifts externally referenced to 15NH4NO3 and 31P chemical shifts externally referenced to trimethylphosphine. The two-dimensional [1H,1H] COSY and NOESY spectra were acquired with water suppression using the excitation sculpting, TOCSY spectra were recorded with the watergate 3-9-19 pulse sequence. The two-dimensional [1H,15N] HSQC spectra were recorded using phase sensitive with gradients in back-inept pulse sequence. The two-dimensional [1H,31P] HMQC spectra were acquired using standard Bruker phase sensitive HMQC pulse sequence.
The oligonucleotides were analyzed by COSY, TOCSY, NOESY and HSQC experiments in the presence and absence of 15N labeled TriplatinNC at 15°C. HSQC and the exchangeable proton NOESY experiments were carried out in H2O:D2O ratio of 92:8. Solutions were lyophilized twice from 99.999% D2O and dissolved in D2O for the acquisition of non-exchangeable protons by NOESY and TOCSY.
Summary and Conclusions
Substitution-inert complexes containing only PtN4 coordination spheres are emerging as a distinct sub-set of the wide class of structures within the polynuclear platinum chemotype1. Within this class of cationic molecules, possible systematic changes to give overall charge diversity include dinuclear complexes such as [{Pt(NH3)3}2-μ-spermidine]5+ and [{Pt(NH3)3}2-μ-spermine]6+ connected via polyamine central linkers rather than the trinuclear {Pt(tetraamine)} unit, and the use of NH3 rather than the 'dangling' amine of TriplatinNC. The crystallographic description of the DNA binding through the phosphate clamp mediated through purely electrostatic and hydrogen-bonding interactions is a unique feature of this sub-set, a feature that has so far been exclusively a domain of organic DNA binders. This motif is distinct from intercalation and the “classic” minor groove binding and produces an overall high binding affinity to CTDNA. The strong binding has implications for inhibition of DNA-protein interactions.8,9,28
The AT contacts observed in solution by NMR spectroscopy suggest at first glance very similar binding to those elucidated for the Dickerson Drew Dodecamer with minor groove binders by NMR and crystallography.25 Thus, Berenil, Hoechst 33258, pentamidine and netropsin all show some AT selectivity and a dominant feature of their DNA recognition is through AT contacts.17,25,29,30 The distinction between these minor groove agents and the polynuclear platinum complexes such as Triplatin NC is two-fold. Firstly the molecular description of the contacts is clearly different with the minor groove binders mostly binding to the exocyclic groups of the nucleic acid bases rather than the phosphate backbone. As a consequence there is less shape-dependence for phosphate clamping than exists for minor-groove binders. Secondly, for the substitution-inert polynuclear platinum compounds, binding is accompanied by compaction of the DNA, significantly more efficient that that induced by spermine.8,9,28 The two features combined then reinforce the description of the phosphate clamp as a distinct third mode of ligand-DNA binding. The results here also indicate that TriplatinNC binds predominantly in a selective fashion to isolated AT base-pairs of the Dickerson-Drew dodecamer. Complementary studies also strongly propose a minor groove spanning binding mode of TriplatinNC for the AT-rich oligomer while, in contrast, the drugs seem to preferably stretch along the backbone in GC group without bridging the grooves.9
Evidence for the existence of the phosphate clamp in solution has been accumulate using {1H, 15N} HSQC NMR spectroscopy. At pH 6 the large downfield shifts in both dimensions for the resonances of the Pt-NH3 and Pt-NH2 are consistent with the close contact to an anionic species such as the phosphate oxygen. The reduced 1J(195Pt-15N) coupling constants and 31P chemical shifts in the TriplatinNC-bound hexamer are also consistent with this interpretation. The role of water molecules in DNA-ligand minor groove recognition has been much discussed.25,26 The pH dependency of the chemical shifts observed here may also reflect this point and water-mediated binding may explain the lack of major shifts at pH7, as outlined by the schematic. Once formed, the phosphate clamp is very stable.
Minor groove binders and DNA-targeted ligands have provided a rich source of potential anti-cancer drugs.31,32 The nature of the PPC chemotype means it is a discrete and modular DNA binding device with high potential as a drug-design scaffold. Specifically, TriplatinNC has demonstrated interesting biological activity in its own right. The complex is cytotoxic at micromolar concentrations, similar to cisplatin, in a range of human tumor cell lines sensitive and resistant to cisplatin but is unaffected by serum degradation, unlike cisplatin or BBR3464.33,34 Other notable features of the biological activity are the rapid nucleolar localization and nucleic acid condensation observed in cells.34,35 The induction of apoptosis in tumor cells suggests that covalent Pt-DNA bond formation is not a prerequisite for antitumor activity for compounds with high DNA affinity, a further significant shift in the structure-activity paradigm of platinum antitumor agents. In conclusion our investigations emphasize the distinct modes of DNA binding by non-covalent polynuclear platinum complexes and the discrete nature of the phosphate clamp motif. It is axiomatic that this binding mode – combined with high cellular accumulation - should lead to a distinct profile of biological activity.
Supplementary Material
Acknowledegments
This work was supported by NIH RO1CA78754.
Footnotes
Electronic Supplementary Information (ESI) available: [Table S1 1H chemical shift assignments for of6mer in the absence and presence of AH78 at pH7 and pH6; Fig. S12D {1H,15N} HSQC spectrum of 15N-labeled TpNC (left) and with duplex hexamer at pH 7 (mid). And 24 hours after changing the pH to 6 (right), the HSQC showed the additional peaks at δ1H (5.82) / 15N (−43.0) and δ1H (6.28) / 15N (−24.0).]. See DOI: 10.1039/b000000x/
Notes and References
- 1.Mangrum JB, Farrell NP. J. Chem. Soc. Chem. Comm. 2010;46:6640–6650. doi: 10.1039/c0cc01254h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farrell NP. Drugs of the Future. 2012;37:795–806. [Google Scholar]
- 3.Malina J, Kasparkova J, Farrell JNP, Brabec V. Nuc. Acids Res. 2011;39:720–728. doi: 10.1093/nar/gkq803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kasparkova J, Zehnulova J, Farrell N, Brabec V. J. Biol. Chem. 2002;277:48076–48086. doi: 10.1074/jbc.M208016200. [DOI] [PubMed] [Google Scholar]
- 5.Malina J, Farrell NP, Brabec V. Chem. Asian. J. 2011;6:1566–1574. doi: 10.1002/asia.201000935. [DOI] [PubMed] [Google Scholar]
- 6.Hegmans A, Berners-Price SJ, Davies MS, Thomas DS, Humphreys AS, Farrell N. J. Am. Chem. Soc. 2004;126:2166–2180. doi: 10.1021/ja036105u. [DOI] [PubMed] [Google Scholar]
- 7.Ruhayel RA, Moniodis JJ, Yang X, Kasparkova J, Brabec V, Berners-Price SJ, Farrell NP. Chem.−Eur. J. 2009;15:9365–9374. doi: 10.1002/chem.200900958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malina, J J, Farrell NP, Brabec V. Angew. Chem. 2014 doi: 10.1002/anie.201408012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prisecaru A, Molphy Z, Kipping RG, Peterson EJ, Kellett A, Farrell NP. Nuc. Acids Res. doi: 10.1093/nar/gku1157. NAR-02733-F-2014. Accepted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qu Y, Moniodis JJ, Harris AL, Yang X, Hegmans A, Povirk LF, Berners-Price SJ, Farrell NP. In: Polyamine Drug Discovery. Woster PM, editor. Cambridge: Royal Society of Chemistry; 2012. pp. 191–204. [Google Scholar]
- 11.Komeda S, Moulaei T, Chikuma M, Odani A, Kipping R, Farrell NP, Williams LD. Nucleic Acids Res. 2011;39:325–336. doi: 10.1093/nar/gkq723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komeda S, Moulaei T, Woods KK, Chikuma M, Farrell NP, Williams LD. J. Am. Chem. Soc. 2006;128:16092–16103. doi: 10.1021/ja062851y. [DOI] [PubMed] [Google Scholar]
- 13.Harris AL, Qu Y, Farrell NP. Inorg. Chem. 2005;44:1196–1198. doi: 10.1021/ic048356p. [DOI] [PubMed] [Google Scholar]
- 14.Berners-Price SJ, Ronconi L, Sadler PJ. Prog. Nucl. Magn. Reson. Spectrosc. 2006;49:65–98. [Google Scholar]
- 15.Wüthrich K. NMR of Proteins and Nucleic Acids. New York, NY: Wiley; 1986. [Google Scholar]
- 16.Qu Y, Scarsdale NJ, Tran M-C, Farrell NP. J. Biol. Inorg. Chem. 2003;8:19–28. doi: 10.1007/s00775-002-0383-x. [DOI] [PubMed] [Google Scholar]
- 17.Qu Y, Scarsdale NJ, Tran M-C, Farrell NP. J. Inorg. Biochem. 2004;98:1585–1590. doi: 10.1016/j.jinorgbio.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 18.Lane AN, Jenkins TC, Brown T, Neidle S. Biochemistry. 1991;30:1372–1385. doi: 10.1021/bi00219a030. [DOI] [PubMed] [Google Scholar]
- 19.Jenkins TC, Lane AN. Biochim. Biophys. Acta. 1997;1350:189–204. doi: 10.1016/s0167-4781(96)00160-1. [DOI] [PubMed] [Google Scholar]
- 20.Moniodis JJ, Thomas DS, Davies MS, Berners-Price SJ, Farrell NP. J. Chem. Soc. Dalton Trans. doi: 10.1039/c4dt02942a. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies MS, Thomas DS, Hegmans A, Berners-Price SJ, Farrell N. Inorg. Chem. 2002;41:1101–1109. doi: 10.1021/ic010851n. [DOI] [PubMed] [Google Scholar]
- 22.Appleton TG, Hall JR, Ralph SF. Inorg. Chem. 1985;24:4685–4693. [Google Scholar]
- 23.Appleton TG, Connor JW, Hall JR, Prenzler PD. Inorg. Chem. 1989;28:2030–2037. [Google Scholar]
- 24.Pregosin PS, Omura H, Venanzi LM. J. Am. Chem. Soc. 1973;75:2047–2048. [Google Scholar]
- 25.Geierstanger BH, Wemmer DE. Ann. Rev. Biophys. Biomol. Struct. 1995;24:463–493. doi: 10.1146/annurev.bb.24.060195.002335. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen B, Neidle S, Wilson WD. Acc. Chem. Res. 2009;42:11–21. doi: 10.1021/ar800016q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris AL, Yang X, Hegmans A, Povirk LF, Ryan JJ, Kelland LR, Farrell NP. Inorg. Chem. 2005;44:9598–9600. doi: 10.1021/ic051390z. [DOI] [PubMed] [Google Scholar]
- 28.Malina, J J, Farrell NP, Brabec V. Inorg. Chem. 2014;53:1662–1671. doi: 10.1021/ic402796k. [DOI] [PubMed] [Google Scholar]
- 29.Teng MK, Usman N, Frederick CA, Wang AH. Nuc. Acids Res. 1988;16:2671–2690. doi: 10.1093/nar/16.6.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coll M, Aymami J, van der Marel GA, van Boom JH, Rich A, Wang AH-J. Biochemistry. 1989;28:310–320. doi: 10.1021/bi00427a042. [DOI] [PubMed] [Google Scholar]
- 31.Sheng J, Gan J, Huang Z. Med. Res. Rev. 2013;33:1119–1173. doi: 10.1002/med.21278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai X, Gray PJ, Jr, Von Hoff DD. Cancer Treat. Rev. 2009;35:437–450. doi: 10.1016/j.ctrv.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 33.Benedetti, B.T BT, Peterson EJ, Kabolizadeh P, Martínez A, Kipping RG, Farrell NP. Mol. Pharmaceutics. 2011;8:940–948. doi: 10.1021/mp2000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterson EJ, Menon VR, Gatti L, Kipping RG, Perego P, Povirk LF, Farrell NP. Mol. Pharmaceutics. doi: 10.1021/mp5006867. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wedlock LE, Kilburn MR, Liu R, Shaw JA, Berners-Price SJ, Farrell NP. J. Chem. Soc. Chem. Comm. 2013;49:6944–6946. doi: 10.1039/c3cc42098a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.