Abstract
Background
Many circulating biomarkers have been reported for the diagnosis of breast cancer, but few, if any, have undergone rigorous credentialing using prospective cohorts and blinded evaluation.
Methods
The NCI Early Detection Network (EDRN) has created a prospective, multicenter collection of plasma and serum samples from 832 subjects designed to evaluate circulating biomarkers for the detection and diagnosis of breast cancer. These samples are available to investigators who wish to evaluate their biomarkers using a set of blinded samples. The breast cancer reference set is comprised of blood samples collected using a standard operating procedure at four U.S. medical centers from 2008–2010 from women undergoing either tissue diagnosis for breast cancer or routine screening mammography. The reference set contains samples from women with incident invasive cancer (n=190), carcinoma in situ (n=55), benign pathology with atypia (n=63), benign disease with no atypia (n=231), and women with no evidence of breast disease by screening mammography (BI-RADS 1 or 2, n=276). Using a subset of plasma samples (n=505) from the reference set, we analyzed 90 proteins by multiplexed immunoassays for their potential utility as diagnostic markers.
Results
We found that none of these markers is useful for distinguishing cancer from benign controls. However, elevated CA-125 does appear to be a candidate marker for ER negative cancers.
Conclusions
Markers that can distinguish benign breast conditions from invasive cancer have not yet been found.
Impact
Availability of prospectively collected samples should improve future validation efforts.
Introduction
In order to assess circulating biomarkers for the detection of cancer, high quality case and control blood specimens must be available. There have been several papers dealing with the pipeline for biomarker discovery and validation with the ultimate benchmark being a prospective trial to determine if use of the biomarker reduces disease-specific mortality (or morbidity), i.e. clinical utility (1, 2). Typically, markers are first discovered and tested on small convenience sets of cases and controls. Without validation on more carefully controlled sets, highly misleading results that demonstrate significant differences between cases and controls are not uncommon. The most rigorous test sets (prior to the final determination of clinical benefit) adhere to PRoBE design (prospective collection with retrospective blinded evaluation) wherein samples are collected from cohorts that match the intended use of the biomarkers (3).
To address the need for publically available resources for biomarker discovery and validation, we previously reported on the creation and deposit of pooled sets of serum samples designed to test markers for breast and gynecologic malignancies (4). These sets are deposited at the NCI-Frederick facility for distribution, but the use of pooled cases has inherent limitations for markers that are altered in subsets of subjects and markers that exhibit dramatic outliers in cases or controls. To address these shortcomings and to create a fully PRoBE-compliant resource, the NCI Early Detection Research Network (EDRN) created a prospectively collected standard reference set of blood samples from women with breast cancer and matching controls, termed the Breast Cancer Reference Set (BCRS). The BCRS can be used for late stage discovery and early stage validation of biomarkers of breast cancer (5) and is now available for distribution from the biorepository at NCI-Frederick to qualifying investigators upon approval by a review committee.
The prospectively collected samples were donated by women being examined at two distinct clinical venues related to breast cancer diagnosis: 1) Screening mammography and 2) Diagnostic radiology where tissue sampling occurs to determine the type of breast abnormality found by imaging or clinical exam. Women were recruited from both settings and blood was collected prior to diagnosis. Since the samples were collected at different stages of breast cancer diagnosis, we consider these to be two different PRoBE compliant sets (screening versus diagnostic). Demographic and clinical data were also collected using common questionnaires and data abstraction approaches.
In this communication, we describe the parameters and criteria that were used in assembling these reference sets and the detailed composition of the sets. Further, we present an analysis of 90 protein biomarkers in plasma from the Diagnostic Set (n=505) of cases and controls that highlights the potential for serious confounders when PRoBE design is not rigorously adhered to.
Materials and Methods
Subjects, Enrollment, and Accrual
We restricted the composition to women based on the fact that if included, men would constitute a rare subset of the cases (and controls). Overall criteria for inclusion were as follows: 1) female, 2) over 18 years of age, 3) not pregnant (self-reported) or breast feeding at the time of participation, 4) no prior history of invasive cancer except basal or squamous cell carcinoma of the skin, and 5) undergoing screening or diagnosis for breast cancer, 6) diagnosis occurring within 30 days after the blood draw for incident benign or cancer cases. All four participating sites obtained local IRB approval for the study with specific indication that a portion of the blood sample would be provided to the NCI for storage and distribution.
At the time of enrollment, a questionnaire was administered to each subject (supplemental materials, Participant Form) and after final pathologic diagnosis, information regarding the cancer or benign condition was abstracted from medical records (supplemental materials, Clinical Form). Questionnaire and medical data were entered into an online system administered by the EDRN Data Management and Coordinating Center (DMCC at the Fred Hutchinson Cancer Research Center (FHCRC)). FHCRC also obtained and maintains an IRB protocol that covers handling of data associated with the reference set. Based on these data, final eligibility was determined All eligible cases and a subset of controls matched to cases on age, race, and date of blood draw were selected for inclusion from each site. Supplemental Table 1 shows subjects included in the reference set by site and disease category.
The overall final composition of the reference set includes incident invasive cancer (n=190, carcinoma in situ (n=55), benign pathology with atypia (n=63), benign disease with no atypia (n=231), and women with no evidence of breast disease by screening mammography (BI-RADS 1 or 2, n=276). In the screening set, 17 women were consented who later developed cancer of varying types.
Sample, Data Handling, and Application Procedure
Blood was collected into EDTA and serum collection tubes before cytoreductive surgery and in the absence of systemic anesthesia. For the Diagnostic Set, most of the blood was drawn immediately after the diagnostic biopsy was performed except at UCSF where biopsies were performed an average of 2.8 days after blood collection. For the Screening Set and BI-RADS 1 and 2 samples from the Diagnostic Set, blood was drawn immediately after the screening mammogram. Blood was processed within 5 hours of collection. Blood was centrifuged at 3000 × g for 10 minutes and the serum or plasma removed by pipette. Serum and plasma were dispensed into 1 ml aliquots and stored at −80°C at each institution. White blood cells were also banked at each institution but do not constitute part of the central reference set. Time to processing was recorded in each case.
For each selected sample, 4 × 1 ml of serum and 4 × 1 ml of plasma were shipped on dry ice to NCI-Frederick. At NCI-Frederick, each subject’s sera/plasma aliquots were thawed, pooled, centrifuged, and distributed into 200 µl final aliquots. Each individual 200 µl aliquot was barcoded and the link to the sample identity retained only by the DMCC. Therefore, only the individual study sites know the identity of the subjects and only the DMCC knows the link between the deposited aliquots and de-identified subject information including case-control status. Biomarker data generated on the reference set is linked by the barcoded identifier and thus can only be analyzed with respect to subject information by the DMCC. The protocol is currently designed so that case-control status is never unblinded any of the biomarker scientists who are the end users of this resource.
An application form outlining the required information for obtaining the reference set samples is available at http://edrn.nci.nih.gov/resources/sample-reference-sets. Depending on the level of preliminary evidence for a given biomarker and its potential clinical application (screening or diagnosis), small preliminary validation cohorts or full sets can be requested. EDRN Investigators and NCI program staff work with individual applicants to determine the most efficient approach. Applications are reviewed by EDRN scientific and statistical investigators.
Biomarker Analysis
Two vials of each sample (400 µl total volume) were provided to Meso Scale Diagnostics, LLC. (MSD®) for testing using a selection of MSD’s multiplexed assay panels. The samples were thawed and further aliquoted to strip-tubes before freezing on dry ice and storage at −80°C. This approach minimized the number of freeze-thaw cycles to no more than two for each sample.
The assays used in this study are shown in Supplemental Table 2 (www.mesoscale.com). A number of assays contained in panels 4 and 5 were developed in work supported in part by the National Cancer Institute (NCI) through SBIR Phase I and II contracts (Topic 238), HHSN261200700032C and HHSN261200900042C, using antibodies and proteins developed through the Clinical Proteomic Technologies for Cancer (CPTC) initiative at the National Institutes of Health. Assays were performed using electrochemiluminescence (ECL) detection in an array-based multiplexed format (6). The samples and calibrator dilutions were assayed in duplicate. The plates additionally contained replicates of an internal quality control (QC) plasma pool for evaluation of plate-to-plate assay reproducibility.
For each of the 90 assays, calibration curves were established from the serial dilutions of calibrators (8-point calibration curves), and the data were fitted with a weighted 4-parameter logistic curve fit. The assay detection limits (analytical sensitivities) were determined based on the calibration curves and standard deviations of background measurements. The calibration curves were also used to estimate the upper end of the linear range of each assay. Concentrations of biomarkers in each sample were calculated from the calibrator curves taking into account sample dilutions. The mean of two measurements was derived for each analyte in each sample. Calculated concentrations were reported to the DMCC for analysis.
Statistical Analysis
Data were split into a training data set and a test data set. The training data set was comprised of half the invasive cancers, half the benign without atypia controls and half the normal screening controls. The test data set was comprised of the remaining data from these categories. Since the goal of this study was to determine if there was any association between marker and case-control status (as opposed to verifying if the marker had utility for a specific clinical application) we used AUC as a general measure of discrimination and corresponding p-value derived from the Wilcoxon ranksum test. ROC curves and AUCs were estimated non-parametrically. Markers were ranked in the training set according to their p-value. For markers that had p-values <0.06 we developed linear 2 and 3 marker combinations using logistic regression analysis and evaluated empirical estimates of the corresponding AUCs. Those markers with statistically significant p-values alone or in combination were examined in the independent test data set.
We examined associations between biomarker values and a variety of demographic/clinical factors including age, race, body mass index, and use of hormone replacement therapy. We also examined marker levels with respect to length of sample storage. A linear regression model for the biomarker that simultaneously included case-control status and these demographic factors was fit to the training data. A likelihood ratio test p-value was calculated for each factor. For those factors that were statistically significant in the training data we used the same strategy to obtain a p-value in the test data set.
We applied unsupervised clustering to summarize the correlation structure amongst the analytes in the combined sample set (n=505) using an absolute correlation distance metric to identify groups of mutually correlated analytes and depicted the hierarchical structure evident in the data in a dendrogram.
Results
Construction and Composition of the Breast Cancer Reference Set
Four member institutions of the EDRN (Dana Farber Cancer Institute (DFCI), Duke University Medical Center (DUMC), Fox Chase Cancer Center (FCCC), and University of California, San Francisco Medical Center (UCSF)) enrolled subjects for the purpose of evaluating blood-based biomarkers for breast cancer detection. Two collection strategies were employed based upon the clinical venue in which subjects were consented and enrolled (Supplemental Figure 1). Women undergoing screening mammography constitute the “Screening Set” and women undergoing tissue sampling followed by pathology review constitute the “Diagnostic Set”. A small supplementary set of normal controls were enrolled in the mammography clinics at the same institutions where diagnostic samples were collected. Consent, enrollment, and blood draw occurred prior to the subject being informed of their imaging findings or tissue diagnosis. Subjects were accrued over a 3 year time frame (2008–2010). In 2011, the EDRN Data Management and Coordinating Center (DMCC) selected a series of cases and controls from each site to comprise the two reference sets that are now available blinded in 200 µl aliquots from the biorepository at NCI-Frederick.
The specific breakdown of cases and controls by collection strategy is shown in Table 1. Samples from FCCC were collected at the time of screening mammography and they included a large number of women who participated in a longitudinal screening mammography (pre-clinical samples) study. Tissue diagnoses (benign, DCIS, invasive cancer) for the screening set occurred within 30 days of the blood draw. Some of these individuals (n=17) were later diagnosed with various types of cancer (9 breast cancers including DCIS) and were no longer included as controls. The other three sites (Duke, UCSF, and DFCI) enrolled their subjects in a diagnostic radiology clinic at the time of tissue sampling (core biopsy or needle aspirate). These three sites also enrolled a limited number of women at screening mammography and a subset of those with BI-RADS score of 1 or 2 (normal, no elevated risk (7)) were contributed to the final reference set. These normal controls are not considered to be part of the PRoBE designed “Diagnostic Set” as they were collected from a population of subjects from a different clinical venue. However, given the prevalent use of such controls in biomarker studies, these samples were considered to be a relevant aspect of the reference set and represent the type of controls used in many studies.
Table 1.
Composition of the Breast Cancer Reference Set (n=831)
| Sample Type | Screening (PRoBE 1) | Diagnostic (PRoBE 2) |
|---|---|---|
| Normal | 176 | 1001 |
| Benign w/out atypia | 72 | 159 |
| Benign w/atypia | 11 | 52 |
| Carcinoma in situ | 15 | 40 |
| Invasive Carcinoma | 35 | 1543 |
| Cancer (pre-clinical2) | 17 | 0 |
Women with normal mammograms not referred for biopsy collected at the same institutions as the diagnostic samples
Women with normal screening mammograms later diagnosed with cancer including DCIS, invasive breast cancer, and cancers of other organs.
These numbers are for plasma. There is one more invasive cancer (n=155) that has serum deposited.
Subject characteristics for the reference set are shown in Supplemental Tables 3A and B (Screen and Diagnostic, respectively) separated into the major breast diagnostic categories. In the diagnostic set, subjects with invasive cancer were similar to their relevant controls, namely women with benign conditions. In the screening set subjects with invasive cancer were also similar to their relevant controls who in this case were normal. This was achieved in part by selecting controls that were matched to cases on demographic factors.
Characteristics of the invasive cancers are shown in Table 2 categorized by inclusion in the Screening versus Diagnostic sets. Hormone receptor status and disease stage were comparable in the two groups. Screening cancers from FCCC had a slightly higher rate of HER2 positivity (p=0.06) than the Diagnostic Set but otherwise, the groups have similar distributions of clinical parameters.
Table 2.
Invasive Cancer Characteristics
| Diagnostic (PRoBE 2) |
Screening (PRoBE 1) |
|
|---|---|---|
| ER Positive | 114 (76%) | 29 (83%) |
| yes (% of known) | 37 | 6 |
| no | 3 | 0 |
| Unknown | ||
| PR Positive | 95 (64%) | 23 (66%) |
| yes (% of known) | 51 | 12 |
| no | 8 | 0 |
| Unknown | ||
| HER2 Positive | 19 (13%) | 8 (25%) |
| yes (% of known) | 120 | 20 |
| no | 10 | 4 |
| equivocal | 5 | 3 |
| Unknown | ||
| Stage | 48 (51%) | 15 (45%) |
| I | 36 (38%) | 9 (27%) |
| IIA | 11 (12%) | 6 (18%) |
| IIB | 0 | 1 |
| IIIA | 0 | 2 |
| IIIC | 59 | 2 |
| Unstaged | ||
Biomarker Analysis of the Diagnostic Set
In order to establish baseline procedures for the use of the reference set and explore relationships between markers and disease state, we applied for use of the Diagnostic Set to quantitate levels of 90 markers by commercial multiplexed ELISA assays. The intent was to follow-up this discovery phase with validation on the Screening Set if useful results were found. Our written application was formally reviewed by the EDRN Breast Cancer subcommittee. After responding to this review, our application was approved and we received 2 × 200 µl aliquots of plasma from 405 subjects in the Diagnostic Set plus 100 screening controls collected at the three institutions that contributed to the Diagnostic Set (n=505, all combined subjects from Duke, UCSF, DFCI in Table 1). The identity of the biomarkers is shown in Supplemental Table 2 with their inclusion in specific multiplexed panels as indicated. Each assay was performed in duplicate and the raw data were returned to the EDRN Data Management Center for analysis.
Analysis was performed in a two-step process whereby data from half of the subjects (the training set) were analyzed for significant associations with disease state (benign, invasive cancer, normal). The training phase excluded subjects with DCIS and atypical hyperplasia in order to enhance our ability to find markers that discriminate invasive cancer from benign conditions. In this phase, we tested each individual marker and linear combinations of the best markers (pairs, trios, quartets) for their ability to discriminate case from control. In the training phase we explored whether there were consistent differences between cases and both types of controls (benign and mammographically normal) and also whether there were significant differences between the two control groups. During the training phase, we also analyzed association of biomarker levels with subsets of cancers defined by hormone receptor and HER2 status and the impact of co-variates including age, race, menopausal status, use of hormone replacement therapy, body mass index, and sample storage time.
A number of markers demonstrated statistically significant (p<0.05) discrimination in the training phase comparing benign disease (without atypia) to invasive cancer without correction for multiple testing (Table 3, Training). These included the known cancer marker CEA and a series of circulating markers that have not been associated with presence of disease: PPP2R4, RAC1, Sclerostin, IL-12, and IL-2. It should be noted that for PPP2R4, RAC1, and Sclerostin levels were higher in the controls compared to cases. None of these reached significance after correction for multiple comparisons with or without adjustment for co-variates. Two and three way combinations of the top performing markers did result in additional discrimination in the training phase between cases and controls with the pairs of PP2R4+Sclerostin, PPP2R4+IL-2, and PPP2R4+CEA reaching AUC values close to 0.7 (data not shown). Using the other half of the invasive cancers and benign controls (without atypia) in a validation phase, we found that none of these individual markers or marker combinations were significant with our without correction for multiple testing (Table 3, Validation).
Table 3.
Invasive Cancer versus Benign w/out Atypia
| Training | Validation | ||||
|---|---|---|---|---|---|
| Marker | AUC | p value1 | Invert2 | AUC | p value |
| CEA | 0.615 | 0.01292 | 0.51 | 0.82743 | |
| CA 125 | 0.586 | 0.06225 | 0.567 | 0.14861 | |
| PPP2R4 | 0.633 | 0.00375 | Yes | 0.508 | 0.86407 |
| RAC1 | 0.61 | 0.01718 | Yes | 0.53 | 0.51718 |
| Sclerostin | 0.607 | 0.02106 | Yes | 0.567 | 0.1496 |
| IL−12 p70 | 0.604 | 0.02416 | 0.54 | 0.38493 | |
| IL−2 | 0.604 | 0.02385 | 0.583 | 0.07337 |
All markers with unadjusted p values less than 0.05 in the training phase are shown.
If “yes”, then the values were higher in the controls compared to cases.
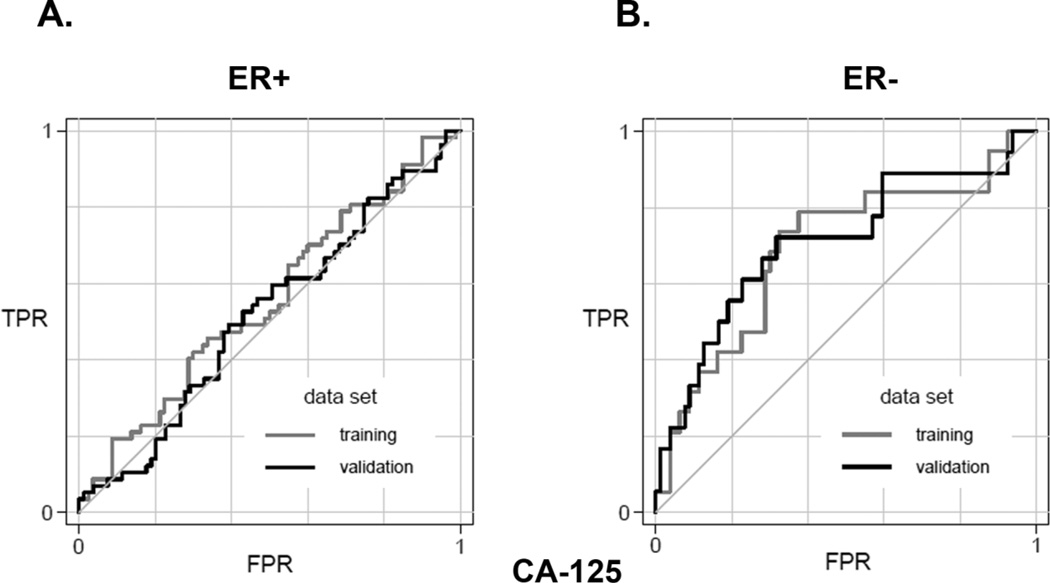
Examining cancer subsets defined by receptor status, we found that CEA, PPP2R4, and Sclerostin levels were associated with ER+ cancers and RAC1 was associated with HER2+ cancers in the training set. None of these associations survived the validation phase (not shown). For ER- cancers, CA-125 (MUC16) was the most discriminating marker in the training set followed by IL-2 and IL-12 (Table 4). CA-125 retained its relatively strong significance in the validation set with an AUC of ~0.7. ROC curves demonstrating this in the subset of ER+ (Figure 1A) and ER- cancers (Figure 1B) are shown.
Table 4.
ER-Cancers versus Benign w/out Atypia
| Training | Validation | ||||
|---|---|---|---|---|---|
| Marker | AUC | p value1 | Invert2 | AUC | p value |
| CA 125 | 0.692 | 0.0095 | 0.707 | 0.00619* | |
| IL−2 | 0.679 | 0.0152 | Yes | 0.574 | 0.32858 |
| IL−12 p70 | 0.663 | 0.0279 | Yes | 0.586 | 0.2574 |
| IL−10 | 0.662 | 0.0288 | 0.507 | 0.92234 | |
| CHGA | 0.652 | 0.0401 | 0.637 | 0.07037 |
Markers that retained significance in validation phase.
All markers with unadjusted p values less than 0.05 in the training phase are shown.
If “yes”, then the values were higher in the controls compared to cases.
Figure 1.
Performance of CA-125 (MUC16) in discriminating invasive cancers from benign controls in the breast cancer reference set. A) ROC curves showing the training and validation data for estrogen receptor positive cancers based on CA-125 levels. The comparison is invasive cancers versus benign breast conditions without atypia. B) ROC curves showing the training and validation data for estrogen receptor negative cancers based on CA-125 levels. The comparison is invasive cancers versus benign breast conditions without atypia.
We also tested whether there were significant differences between invasive cancers and subjects with BI-RADS 1 or 2 mammograms. In the training phase, we found a number of markers that demonstrated very significant differences even after correction for multiple testing between these two groups (Table 5 shows all markers that had p<0.05 in the training phase). The top 5 markers (bFGF, NME2, GLO1, hS100A6, and hS100A4) had AUC values >0.7 but all were higher in controls compared to invasive cancers. In the validation phase, all of these top markers remained significant and continued to demonstrate higher levels in control subjects compared to women with invasive breast cancer. Comparing the two control populations, we found that many of these same markers are significantly different between benign and normal (Supplemental Table 4) indicative of systematic differences between the two control groups.
Table 5.
Invasive Cancer versus Screen Normals
| Training | Validation | ||||
|---|---|---|---|---|---|
| Marker | AUC | p value1 | Invert2 | AUC | p value |
| bFGF | 0.769 | 3.1e−07 | Yes | 0.724 | 2.1e−05* |
| NME2 | 0.759 | 8.2e−07 | Yes | 0.772 | 2.4e−07* |
| GLO1 | 0.746 | 3.0e−06 | Yes | 0.784 | 6.9e−08* |
| hS100A6 | 0.733 | 9.4e−06 | Yes | 0.803 | 9.0e−09* |
| S100A4 | 0.709 | 7.0e−05 | Yes | 0.737 | 6.9e−06* |
| CEA | 0.697 | 0.0002 | 0.583 | 0.117 | |
| ErbB2 | 0.678 | 0.001 | 0.551 | 0.336 | |
| MBD1 | 0.67 | 0.001 | Yes | 0.556 | 0.279 |
| E−cadherin | 0.669 | 0.001 | 0.625 | 0.011* | |
| AKR1B1 | 0.668 | 0.001 | Yes | 0.651 | 0.004* |
| Eotaxin | 0.666 | 0.002 | Yes | 0.626 | 0.016* |
| MCP−4 | 0.666 | 0.002 | Yes | 0.55 | 0.343 |
| ICAM | 0.654 | 0.003 | 0.643 | 0.007* | |
| CA125 | 0.651 | 0.004 | 0.589 | 0.091 | |
| IL−1β | 0.643 | 0.006 | Yes | 0.583 | 0.109 |
| GSTM1 | 0.64 | 0.008 | Yes | 0.619 | 0.023 |
| MCP−1 | 0.639 | 0.008 | Yes | 0.621 | 0.022* |
| TARC | 0.637 | 0.009 | Yes | 0.658 | 0.003* |
| GPI | 0.637 | 0.009 | Yes | 0.645 | 0.006* |
| ODC1 | 0.633 | 0.011 | Yes | 0.6 | 0.057 |
| RAC1 | 0.631 | 0.012 | Yes | 0.665 | 0.002 |
| IL−10 | 0.63 | 0.013 | 0.504 | 0.933 | |
| SERPINB3 | 0.624 | 0.018 | Yes | 0.521 | 0.693 |
| Osteoprotegerin | 0.622 | 0.02 | 0.537 | 0.483 | |
| GSTM2 | 0.622 | 0.02 | Yes | 0.588 | 0.091 |
| TNFRI | 0.619 | 0.024 | 0.583 | 0.114 | |
| TNFRII | 0.616 | 0.027 | 0.554 | 0.309 | |
| VEGF−C | 0.613 | 0.031 | Yes | 0.651 | 0.004* |
| LBP | 0.612 | 0.033 | 0.697 | 0.0002 | |
| SAT | 0.612 | 0.034 | Yes | 0.606 | 0.043 |
| IL−12p70 | 0.611 | 0.035 | 0.575 | 0.152 | |
| VEGF | 0.608 | 0.041 | Yes | 0.585 | 0.108 |
| SFN | 0.607 | 0.042 | Yes | 0.521 | 0.697 |
| Eotaxin−3 | 0.604 | 0.049 | Yes | 0.626 | 0.016 |
Also significant for benign vs. normal
All markers with unadjusted p values less than 0.05 in the training phase are shown.
If “yes”, then the values were higher in the controls compared to cases.
Biomarker Correlation Structure
Given the large data set measured for 90 protein biomarkers, we also examined how the levels of these markers were correlated with each other and with major population variables. The absolute linkage clustering of the data (Supplemental Figure 2) shows the most highly correlated markers branching closest to 0 (one minus the absolute value of the correlation coefficient, ‘1-|rho|’) at the bottom of the figure. The most significant correlations were observed within groups of cytokines suggesting inflammatory or immune related processes. Analytes that are highly correlated with one another are likely to show evidence of associations with the same phenotypes, reflecting a common underlying mechanistic signal. For example, CRP and SAA (rho = 0.835) are both associated with BMI (Supplemental Table 5).
We also examined whether biomarker levels were significantly associated with common demographic variables including age, race, BMI, and HRT use and whether length of sample storage time affected specific analytes. This analysis was performed using the same two-step training and validation approach that we employed above. Associations that were significant in both training and validation groups are shown in Supplemental Table 5. We found a number of markers that were significantly associated with these variables, only some of which have been previously described. Age and BMI were strong factors with 22 and 16 markers showing significant correlations, respectively. Among the stronger associations with age were Osteoprotegerin, MCP1, and Eotaxin. BMI was most strongly associated with CRP, SAA, Adiponectin, and HGF. Race (white versus non-white) was strongly associated with VCAM-1 and P-Cadherin levels whereas HRT use showed relatively weak associations with only two markers. While some of the markers also show up in the list of markers that discriminate BI-RADS 1–2 from cancer (and benign), there is little overlap indicating that these demographic variables do not account for the differences observed. Finally, longer storage time was associated with lower levels of two markers (GLO1 and S100A6) and higher levels of two markers (E-Cadherin and IL-8) indicating that most of the biomarkers were not affected by length of time at −80°C.
Discussion
Testing or validating the performance of promising cancer related biomarkers is an uneven enterprise at best. The NCI Early Detection Research Network has made a concerted effort to provide useful resources collected in a rigorous manner to support clinical research. To this end, a series of standard reference specimen sets related to the detection of different solid malignancies have been developed and are available to researchers following submission of an application that is assessed by a formal review process (5). In this communication, we describe the creation and use of the breast cancer reference set for late stage discovery and validation of blood based biomarkers.
Developing a blood test for the detection of breast cancer remains a potentially important but unfulfilled goal. There are intrinsic hurdles for bringing such a marker to the clinic including the widespread implementation of a screening test that provides a physical location for suspected malignancy (mammography), other non-invasive modalities that can refine or provide additional information to the screening test (ultrasound and MRI), and the relatively low threshold for performing tissue sampling procedures. These common clinical approaches may reduce breast cancer specific mortality (8, 9) but there is likely room for improvement and blood-based biomarkers could further reduce the disease burden if they performed adequately. Another major hurdle is that discovery and early testing of biomarkers are commonly conducted using convenience samples that do not mirror the intended use of the marker. We believe that this is one of the primary reasons that most biomarkers which show initial promise fail to progress towards clinical application.
The current breast cancer reference set contains samples for two distinct applications as they were collected from women having different types of clinical evaluation: 1) a screening set from women undergoing routine mammography and 2) a diagnostic set from women referred for biopsy. From a practical standpoint, accrual of incident cancers was much higher when enrolling women undergoing tissue diagnosis which led to most of the cancers residing in the diagnostic set. The subjects with cancer that were enrolled in these two settings had similar clinical and demographic characteristics.
The current biomarker study was designed primarily to test the reference set utilization protocol and provide a survey of the levels of plasma protein biomarkers to assist in future analyses of the set. These plasma “demographics” are now permanently associated with the Diagnostic Set of samples and any future biomarker measurements can be informed by these data. A number of established cancer biomarkers were included in the survey but none that had been shown to have high sensitivity or specificity for breast cancer. Ninety biomarkers were measured using a series of multiplexed immunoassays and results analyzed by the EDRN data management center splitting the cases and controls into training and validation sets, excluding subjects with DCIS or atypical hyperplasia from the training phase. The most promising results from the training phase were tested in the validation phase and the results mirror our previous similar but smaller study conducted on a different set of subjects (10). Specifically, we found little evidence that any of these markers can discriminate women with invasive cancer from those with benign breast conditions. However, a number of markers demonstrated significant differences (that remained significant in validation) between women with no evidence of breast abnormality by screening mammography (BI-RADS 1 or 2) and those with either benign or malignant conditions of the breast. Based on these results and those from our prior study, we conclude that there are systematic differences in circulating biomarker levels between women undergoing screening mammography and those undergoing a diagnostic biopsy highlighting the critical importance of using controls derived from the same clinical or population setting as cases, a key condition of PRoBE design (3). We consider that a possible source of these systematic differences may be related to the level of stress in individuals undergoing a diagnostic biopsy compared to a screening mammogram. Another possibility may be related to lifestyle or diet changes prompted by an impending diagnostic biopsy for breast cancer. Finally, since we obtained blood immediately after mammography, there is the possibility that breast compression could induce an acute inflammatory reaction in some women leading to increased cytokine levels.
Regarding disease specific marker associations, while no markers were useful in discriminating breast cancer from benign disease, CA-125 is elevated in a subset of estrogen receptor negative cancers specifically. This is consistent with the shared biology of triple negative breast cancers and serous ovarian cancers, commonly connected by their occurrence in BRCA1 mutation carriers (11). Given that CA-125 is elevated in many other conditions, it could only be useful in conjunction with other markers of triple negative disease.
Having detailed demographic information related to the subjects also allowed us to explore other types of associations. In particular, we examined a series of common parameters that could influence biomarker levels including age, race, body mass index (BMI), and use of hormone replacement therapy (HRT). Significant associations were observed, many having been reported previously in other settings including age related levels of osteoprotogerin, MCP-1, eotaxin and CEA (12–15) race related levels of VCAM-1 (16), and BMI related levels of inflammatory cytokines and growth factors (17–20). These confirmed associations support the quality of the assays and the absence of significant population biases in the reference set subjects.
The EDRN breast cancer reference set of plasma and serum annotated with demographic, clinical, and common protein biomarker levels should allow for the rapid testing and validation of candidate blood-based markers for the detection of disease. This valuable resource is available to any investigator with potentially useful markers provided that they are willing to comply with the standard procedures developed along with the reference set.
Supplementary Material
Acknowledgements
We thank the commitment and dedication of the women who voluntarily participated in this study. We would like to acknowledge the research and clinical coordinators at the participating sites including Elizabeth Wildermann, Nicole Ryabin, JoEllen Weaver, Erin Bowlby, Pamela Tsing, Amada Romani, and Stig Kreps.
Funding Sources: This work was supported by the NCI Early Detection Research Network (U01 CA117374 to K.Anderson, U01 CA113916 to P.Engstrom and A. Godwin, UO1 CA084955 to J.Marks, UO1 CA111234 to L.Esserman and U24 CA086368 to M.Pepe).
Footnotes
Conflict of Interest Statement: Anu Mathew is an employee of Meso Scale Diagnostics, LLC. (MSD). MSD manufactures and commercializes the assays and equipment for the measurement of biomarker levels that are described herein. All other authors declare that they have no conflicts based on the content of this manuscript.
References
- 1.Pepe MS, Etzioni R, Feng Z, Potter JD, Thompson ML, Thornquist M, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93:1054–1061. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
- 2.Pavlou MP, Diamandis EP, Blasutig IM. The long journey of cancer biomarkers from the bench to the clinic. Clinical chemistry. 2013;59:147–157. doi: 10.1373/clinchem.2012.184614. [DOI] [PubMed] [Google Scholar]
- 3.Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst. 2008;100:1432–1438. doi: 10.1093/jnci/djn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skates SJ, Horick NK, Moy JM, Minihan AM, Seiden MV, Marks JR, et al. Pooling of case specimens to create standard serum sets for screening cancer biomarkers. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2007;16:334–341. doi: 10.1158/1055-9965.EPI-06-0681. [DOI] [PubMed] [Google Scholar]
- 5.Feng Z, Kagan J, Pepe M, Thornquist M, Ann Rinaudo J, Dahlgren J, et al. The Early Detection Research Network's Specimen reference sets: paving the way for rapid evaluation of potential biomarkers. Clinical chemistry. 2013;59:68–74. doi: 10.1373/clinchem.2012.185140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Debad JD, Glezer EN, Wohlstadter J, Sigal GB. Clinical and Biological Applications of ECL. In: Bard AJ, Dekker M, editors. Electrogenerated Chemiluminescence. New York: 2004. pp. 359–396. [Google Scholar]
- 7.Orel SG, Kay N, Reynolds C, Sullivan DC. BI-RADS categorization as a predictor of malignancy. Radiology. 1999;211:845–850. doi: 10.1148/radiology.211.3.r99jn31845. [DOI] [PubMed] [Google Scholar]
- 8.Weedon-Fekjaer H, Romundstad PR, Vatten LJ. Modern mammography screening and breast cancer mortality: population study. Bmj. 2014;348:g3701. doi: 10.1136/bmj.g3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mandelblatt JS, Cronin KA, Berry DA, Chang Y, de Koning HJ, Lee SJ, et al. Modeling the impact of population screening on breast cancer mortality in the United States. Breast. 2011;20(Suppl 3):S75–S81. doi: 10.1016/S0960-9776(11)70299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jesneck JL, Mukherjee S, Yurkovetsky Z, Clyde M, Marks JR, Lokshin AE, et al. Do serum biomarkers really measure breast cancer? BMC Cancer. 2009;9:164. doi: 10.1186/1471-2407-9-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trofimov S, Pantsulaia I, Kobyliansky E, Livshits G. Circulating levels of receptor activator of nuclear factor-kappaB ligand/osteoprotegerin/macrophage-colony stimulating factor in a presumably healthy human population. European journal of endocrinology / European Federation of Endocrine Societies. 2004;150:305–311. doi: 10.1530/eje.0.1500305. [DOI] [PubMed] [Google Scholar]
- 13.Inadera H, Egashira K, Takemoto M, Ouchi Y, Matsushima K. Increase in circulating levels of monocyte chemoattractant protein-1 with aging. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 1999;19:1179–1182. doi: 10.1089/107999099313127. [DOI] [PubMed] [Google Scholar]
- 14.Targowski T, Jahnz-Rozyk K, Plusa T, Glodzinska-Wyszogrodzka E. Influence of age and gender on serum eotaxin concentration in healthy and allergic people. Journal of investigational allergology & clinical immunology. 2005;15:277–282. [PubMed] [Google Scholar]
- 15.Alexander JC, Silverman NA, Chretien PB. Effect of age and cigarette smoking on carcinoembryonic antigen levels. JAMA : the journal of the American Medical Association. 1976;235:1975–1979. [PubMed] [Google Scholar]
- 16.Miller MA, Sagnella GA, Kerry SM, Strazzullo P, Cook DG, Cappuccio FP. Ethnic differences in circulating soluble adhesion molecules: the Wandsworth Heart and Stroke Study. Clinical science. 2003;104:591–598. doi: 10.1042/CS20020333. [DOI] [PubMed] [Google Scholar]
- 17.Visser M, Bouter LM, McQuillan GM, Wener MH, Harris TB. Elevated C-reactive protein levels in overweight and obese adults. JAMA : the journal of the American Medical Association. 1999;282:2131–2135. doi: 10.1001/jama.282.22.2131. [DOI] [PubMed] [Google Scholar]
- 18.Hotta K, Funahashi T, Arita Y, Takahashi M, Matsuda M, Okamoto Y, et al. Plasma concentrations of a novel, adipose-specific protein, adiponectin, in type 2 diabetic patients. Arteriosclerosis, thrombosis, and vascular biology. 2000;20:1595–1599. doi: 10.1161/01.atv.20.6.1595. [DOI] [PubMed] [Google Scholar]
- 19.Rehman J, Considine RV, Bovenkerk JE, Li J, Slavens CA, Jones RM, et al. Obesity is associated with increased levels of circulating hepatocyte growth factor. Journal of the American College of Cardiology. 2003;41:1408–1413. doi: 10.1016/s0735-1097(03)00231-6. [DOI] [PubMed] [Google Scholar]
- 20.Ferri C, Desideri G, Valenti M, Bellini C, Pasin M, Santucci A, et al. Early upregulation of endothelial adhesion molecules in obese hypertensive men. Hypertension. 1999;34:568–573. doi: 10.1161/01.hyp.34.4.568. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.