Abstract
There has been increasing recognition of the importance of cellular metabolism and metabolic substrates for the function and differentiation of immune cells. Here, for the first time, we investigate the metabolic requirements for production of IFN-γ by freshly isolated NK cells. Primary murine NK cells mainly utilize mitochondrial oxidative phosphorylation at rest and with short-term activation. Remarkably, we discovered significant differences in the metabolic requirements of murine NK cell IFN-γ production depending upon the activation signal. Stimulation of NK cell IFN-γ production was independent of glycolysis or mitochondrial oxidative phosphorylation when cells were activated with IL-12+IL-18. By contrast, stimulation via activating NK receptors required glucose-driven oxidative phosphorylation. Prolonged treatment with high-dose, but not low dose, IL-15 eliminated the metabolic requirement for receptor stimulation. In summary, this study demonstrates that metabolism provides an essential second signal for induction of IFN-γ production by activating NK cell receptors that can be reversed with prolonged high-dose IL-15 treatment.
INTRODUCTION
NK cells are innate immune lymphocytes that provide a first-line of defense against infection, particularly viruses, and can recognize and kill tumor cells that have down-regulated self-MHC or express activating ligands (1–3). NK cell effector functions can be triggered by inflammatory cytokines, such as IL-12, IL-15, and IL-18; or by engaging germline-encoded activating NK receptors whose ligands are displayed by infected and/or tumor cells (3–5). In response, NK cells produce inflammatory cytokines, principally IFN-γ, and kill target cells. While many triggers of NK cell activation and subsequent NK cell effector responses have been well-characterized, the metabolic fuels required to drive NK cell functional responses are largely unknown.
Metabolism is the biochemical process used by cells to breakdown fuels for energy production (i.e., ATP) or to generate critical biomolecules. There are two primary and overlapping metabolic pathways for generating ATP from metabolic fuels, anaerobic glycolysis and mitochondrial oxidative phosphorylation (OXPHOS) (6). Glucose fuels both pathways, and is first metabolized in the cytoplasm via glycolysis to produce two molecules of pyruvate, for a net yield of 2 ATP molecules. Pyruvate can then be converted to lactate (anaerobic metabolism), or, in the presence of oxygen, transported into the mitochondria to fuel OXPHOS for a net yield of 30+ ATP molecules. Other metabolic substrates that can fuel mitochondrial OXPHOS include fatty acids (fatty acid oxidation) and glutamine (glutaminolysis). Metabolic pathways, including glycolysis, are attractive pharmacologic targets for disease therapy, particularly cancers (7,8). Furthermore, metabolic fuels are globally altered in many disease states such as metabolic syndrome and sepsis, and probably locally altered in certain microenvironments including sites of infection and tumors (9–11). Thus, it is important to consider the effect of changes in metabolic fuels on the immune system.
Metabolic pathways in immune cells, including dendritic cells, neutrophils, and T cells, have been shown to be critical for cellular activation and differentiation (11–13). For example, differentiation of memory T cells is critically dependent on a switch in energy metabolism from primarily OXPHOS to glycolysis, while quiescent memory cells revert back to OXPHOS (11,14). In CD4+ effector T cells, a glycolytic switch is required for synthesis of IFN-γ protein via the release of a post-transcriptional block in IFN-γ processing (15).
Here, for the first time, we investigate the basic metabolic requirements for resting NK cell IFN-γ production via two different pathways, cytokine- and receptor-mediated activation. We hypothesized that NK cells would require metabolic fuels for activation and production of IFN-γ. Our results demonstrate that, unlike T cells, NK cells do not require a glycolytic switch for efficient IFN-γ production. Rather, we observed activation-specific metabolic requirements for NK cell IFN-γ production.
MATERIALS AND METHODS
Mice and NK cell isolation
All mice were on the C57BL/6 background. Wild-type (wt) mice were purchased from the NCI and Rag-2−/−γc−/− mice were purchased from Taconic. Mice expressing the congenic CD45.1 receptor (CD45.1+) and Rag-1−/− animals were obtained from The Jackson Laboratory and bred to obtain CD45.1+Rag−/− mice. All animals were used between 6–14 weeks of age. Animals were housed in specific pathogen free conditions and studies were approved by the Washington University Animal Studies Committee. NK cells were enriched from spleens using a negative-selection magnetic bead kit (Miltenyi Biotec). NK cell purity ranged from 66–98%, with an average purity of 85.5 ± 5.9% (standard deviation) and contaminating cells were generally CD3 negative.
Extracellular flux and ATP assays
Extracellular flux assays were performed using a XF96 Analyzer (Seahorse Bioscience). 96-well flux plates were coated with 10µl of poly-L-lysine (Sigma) and NK cells (5 × 105 cells/well) were adhered to plates for 30 min prior to the assay. NK cells were assayed in triplicate in non-buffered RPMI (Sigma) supplemented with 2mM L-glutamine, 1% FBS (Sigma), glucose to a final concentration of 25mM, and 1mM sodium pyruvate. OCR and ECAR readings were taken every 2.5–3 minutes and OCR and ECAR results shown represent the average over 60 minutes or average readings at each timepoint. Intracellular ATP was measured in lysed NK cells by a luminescence assay (ATPLite, Perkin Elmer) as recommended by the manufacturer.
Culture media and metabolic inhibitors
Complete media consisted of RPMI1640 (Mediatech) supplemented with 10% FBS (Sigma), 2mM L-glutamine (Sigma; final concentration 4mM), 0.05mM 2-mercaptoethanol (BME, Sigma), and antibiotics (penicillin/streptomycin). Glutamine-free media was prepared with glutamine-free RPMI1640 (Sigma). Glucose-free media was prepared with a combination of PBS and glucose-free RPMI1640 (Gibco) (15% PBS, 85% RPMI) and supplemented with 10% dialyzed FBS containing <50ug/ml glucose by enzymatic assay [Glucose (HK) assay, Sigma], 0.05mM BME, 2mM L-glutamine (final concentration 4mM), and antibiotics. For culture of cells in glucose or glutamine free media, cells were washed in an excess of PBS and then brought up in the appropriate media prior to culture. Inhibitors were obtained from Sigma including oligomycin A, antimycin A, etomoxir, and 2DG, and added to cultures at the indicated concentrations. Galactose, galactose oxidase, and H202 were obtained from Sigma and added at the indicated concentrations.
Antibodies and NK cell stimulation assays
Fluorochrome-conjugated antibodies recognizing NK1.1 (PK136, BD Biosciences), NKp46 (29A1.4, BD Biosicences), CD3 (145-2C11, Biolegend), and IFN-γ (XMG1.2, Biolegend) were used to identify NK cell production of IFN-γ. Viability was assessed by staining cells with Zombie Yellow Stain (Biolegend) or Fixable Yellow Dead Stain (Life Technologies). NK cells were stimulated with 10ng/ml murine IL-12 (Peprotech), 10 or 100ng/ml murine IL-15 (Peprotech), 50ng/ml murine IL-18 (MBL), 10ng/ml PMA (Sigma), and/or 500ng/ml calcimycin (Sigma). Where indicated, NK cells were cultured in low-dose IL-12 (1ng/ml) and IL-18 (1ng/ml). Antibody stimulation was performed by culturing enriched NK cells in plates coated with 20µg/ml of purified anti-NK1.1 (PK136, BioXcell), anti-Ly49D (4E4, courtesy of W.M. Yokoyama and prepared by the Rheumatic Diseases Core Center), or IgG control (BioXcell). Intracellular IFN-γ production was assayed using BD Cytofix/Cytoperm™ (BD Biosciences) as recommended, with brefeldin A added to cells after 1hr of culture to inhibit secretion of IFN-γ. For in vivo priming of NK cells, wt mice were injected intra-peritoneally with 300µg of polyinosine-polycytidylic acid [poly(I:C) HMW; InvivoGen] or control PBS and splenocytes harvested 14–16h later.
Quantitative RT-PCR
NK cells (2–3×105) were activated with 1ng/ml IL-12 and IL-18 or plate-bound anti-NK1.1 in the presence or absence of oligomycin (100nM). Cells were harvested between 0.5 and 6h. Control cells were cultured in media or with plate-bound isotype control IgG. RNA was isolated from NK cells (TRIzol®, Life Technologies or RNeasy Mini, Qiagen) and cDNA generated with random hexamers (Promega). Ifng primers/probe (16) were from IDT, and β-actin primer/probe set was from Applied Biosystems. Copy numbers of transcript were quantitated by generation of plasmid clones of Ifng and β-actin amplicons for use as standards and quantitated by real-time qPCR (TaqMan®, 7500 Fast Real-time PCR instrument, Life Technologies).
NK cell proliferation assays
NK cells were labeled with 1µM CFSE or VioletTrace (Invitrogen) and cultured in 96-well plates with the indicated concentrations of muIL-15. For in vivo proliferation, splenocytes from CD45.1+ Rag-1−/− mice (2–5×105/mouse) were labeled with CFSE and adoptively transferred by tail vein injection into congenic CD45.2+ Rag-2−/−γc−/− hosts and assayed 3 days later.
Flow cytometric analysis and statistics
Flow cytometric analysis was performed on a Cytek-modified (Cytek Development Inc.) 8-color BD FACScan or BD FACSAria Fusion (BD Biosciences). Analysis was performed using FlowJo software (Tree Star Inc.). Statistical analysis was performed with GraphPad Prism 6 (GraphPad Software, Inc.). Student’s paired t test was used to compare 2 matched groups or ANOVA analysis was performed for more than two groups with a p value <0.05 considered significant.
RESULTS
NK cells primarily use glucose-fueled OXPHOS at rest and with activation
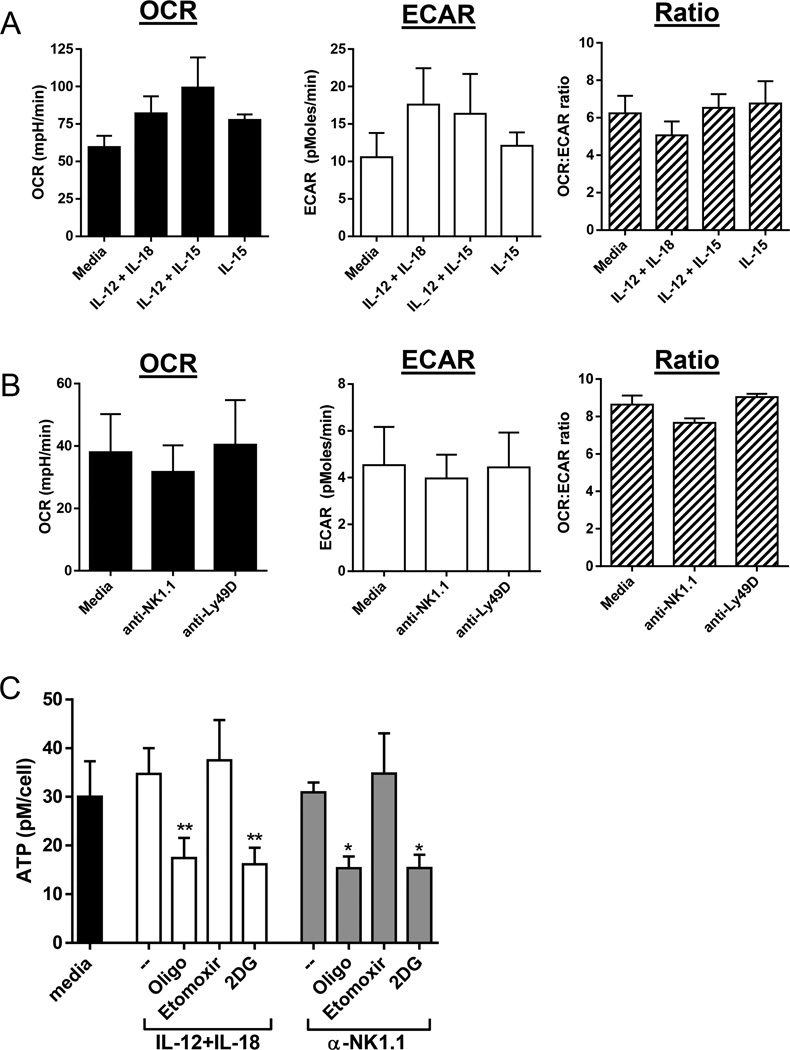
To determine the basic metabolic profile of NK cells we used an extracellular flux assay to measure oxygen consumption rate (OCR, a measure of OXPHOS) and extracellular acidification rate (ECAR, a measure of lactate and anaerobic glycolysis) of freshly isolated murine splenic NK cells (Fig. 1A&B). Baseline metabolic activity of resting splenic NK cells was relatively low, consistent with another recent report (17). At rest, NK cells preferentially utilize OXPHOS as shown by the OCR:ECAR ratio. Short-term activation (4–6hr) with cytokines or antibodies recognizing the activating receptors NK1.1 or Ly49D did not induce substantial changes in energy pathway usage. NK cell intracellular ATP was also stable following activation with IL-12+IL-18 or anti-NK1.1 (Fig. 1C), suggesting that these activation signals do not significantly increase or deplete ATP. Inhibition of OXPHOS with the ATP synthase inhibitor oligomycin, or inhibition of glucose metabolism by 2-deoxy-glucose (2DG), a competitive inhibitor of glycolysis, significantly reduced ATP in activated NK cells (Fig. 1C). These results suggest that glucose is the primary OXPHOS fuel used during NK cell activation, since blockade of glucose metabolism reduced intracellular ATP to the same degree as global inhibition of OXPHOS. Consistent with this hypothesis, inhibition of fatty acids with etomoxir, a fatty acid oxidation inhibitor that blocks carnitine palmitoyltransferase-1 (CPT1), had no effect on NK cell ATP (Fig. 1C).
Figure 1. Metabolism of resting and activated NK cells.
Extracellular flux assays were used to measure resting and activated NK cell oxygen consumption rate (OCR), a measure of mitochondrial OXPHOS, and extracellular acidification rate (ECAR), a measure of glycolysis. (A) Cytokine activation (4h) or (B) receptor stimulation (6h) did not significantly change OCR, ECAR or the OCR:ECAR ratio. Results represent the mean +/−SEM of triplicate wells from 3 independent experiments. (C) Intracellular ATP (pM/cell) after 6hr culture of NK cells with cytokines or plate-bound anti-NK1.1 in complete media (--) or with the metabolic inhibitors oligomycin (oligo, 1uM), etomoxir (300µM), and 2DG (50mM). Statistics represent the comparison between stimulation alone versus stimulation with the indicated inhibitor for IL-12+IL-18 or anti-NK1.1 activated NK cells (one-way paired ANOVA). Results represent the mean +/−SEM of triplicate wells from 4 independent experiments. ns, not significant; *p≤0.05; **p≤0.01.
OXPHOS is required for receptor-stimulated NK cell IFN-γ
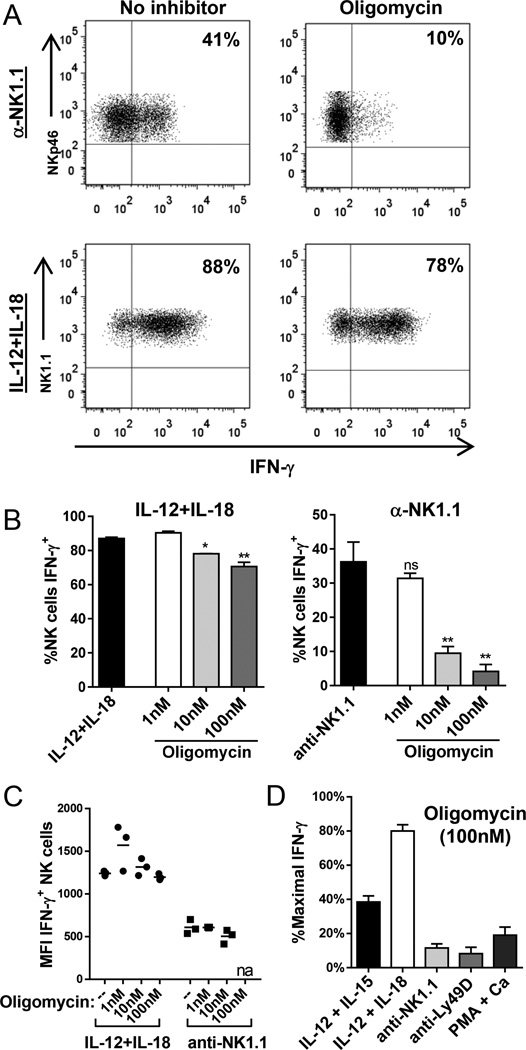
Since NK cells primarily use OXPHOS, we hypothesized that inhibition of this pathway might alter NK cell function. We tested the ability of fresh splenic NK cells to produce IFN-γ when stimulated with cytokines or activating receptors in the presence of increasing concentrations of oligomycin. NK cell survival was intact during all assays, but was significantly reduced if NK cells were cultured in the presence of oligomycin for greater than 12hr (data not shown). Remarkably, treatment with 10nM oligomycin almost completely abolished receptor-stimulated NK cell IFN-γ production, but had minimal effect on IL12+IL-18-stimulated IFN-γ production (Fig. 2A). Inhibition of receptor-stimulated IFN-γ by oligomycin was dose dependent (Fig. 2B), while there was only minimal decrease in IFN-γ production by IL-12+IL-18-stimulated NK cells when cultured with up to 1µM of oligomycin (Fig. 2B and data not shown). The mean fluorescence intensity of IFN-γ in cytokine-producing NK cells was similar between untreated and treated cells, suggesting that metabolic inhibition did not alter strength of activation with IL-12+IL-18 (Fig. 2C). In addition, there was no decrease in NK1.1 expression with oligomycin that might account for the inability to stimulate NK cells via this receptor (data not shown).
Figure 2. Inhibition of mitochondrial OXPHOS significantly impairs receptor-stimulated NK cell IFN-γ production.
(A–C) NK cells were stimulated for 6h with anti-NK1.1 or IL-12+IL-18 with or without the OXPHOS inhibitor oligomycin. (A) Representative flow plot of IFN-γ production by NK cells in response to anti-NK1.1 (top) or IL-12+IL-18 (bottom) stimulated without (left) or with (right) 10nM oligomycin. (B) Percentage of NK cells producing IFN-γ measured by intracellular flow cytometry in response to IL-12+IL-18 (left) or anti-NK1.1 (right) in the presence of increasing doses of oligomycin. (C) Geometric mean fluorescence intensity (MFI) of IFN-γ measured in IFN-γ+ NK cells in the presence of oligomycin compared to no inhibitor (--). (D) Percentage of maximal IFN-γ positive NK cells following 4–6h culture with 100nM oligomycin normalized to percentage of IFN-γ+ NK cells without inhibitor for each stimulus [cytokines (IL-12+IL-15 or IL-12+IL-18), receptors (anti-NK1.1 or anti-Ly49D), or PMA+calcimycin]. Results represent the mean +/−SEM or individual data points in (C), from 3–4 independent experiments. na, not available (too few events); ns, not significant; *p≤0.05, **p≤0.01.
Preservation of IFN-γ response with OXPHOS inhibition was unique to IL-12+IL-18 stimulation, since activation with another cytokine combination (IL-12+IL-15) or with PMA and calcimycin, a stimulus that results in a percentage and strength of IFN-γ response similar to IL-12+IL-18, was impaired. However, the most profound defects in NK cell activation were in response to activating receptors, NK1.1 and Ly49D (Fig. 2D, normalized to maximum signal for each stimulus). Similar results were obtained with antimycin, an inhibitor of the electron transport chain and OXPHOS (data not shown), confirming that OXPHOS is critical for receptor-mediated activation of NK cells.
OXPHOS results in the production of reactive oxygen species (ROS), which have been shown to be signaling molecules, including for antigen-specific expansion of T cells and IL-2 production (18). While oligomycin inhibits OXPHOS, it has also been shown to increase ROS (19). We therefore tested whether addition of ROS affected receptor-mediated NK cell activation. There was no change in the percentage of IFN-γ positive NK cells after NK1.1 stimulation when the culture media was supplemented with ROS (Supplemental Fig. 1) with or without oligomycin. These data suggest that ROS do not inhibit or enhance receptor-mediated NK cell activation.
Glucose-dependent NK cell activation
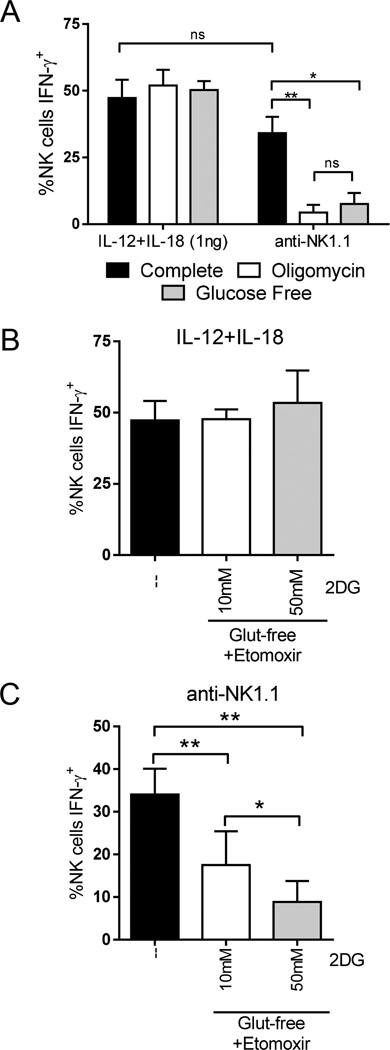
Our findings suggest that metabolism functions as a second signal for receptor activation. We considered the possibility that the differences observed in IL-12+IL-18 versus receptor stimulation with metabolic inhibitors were due to the high number of IFN-γ positive cells with this cytokine combination (>90% IFN-γ+) as compared to receptor stimulation (~30–40% IFN-γ+). NK cells were therefore cultured in low concentrations (1ng/ml) of IL-12 and IL-18. This resulted in a similar percentage of IFN-γ producing NK cells as anti-NK1.1 stimulation, but similar to data with higher dose cytokines, there were no defects in IFN-γ production with oligomycin (Fig. 3A).
Figure 3. Receptor stimulation is also dependent on glycolysis, while IL-12+IL-18 activation is metabolism independent.
(A) NK cells were stimulated for 6h with low dose IL-12+IL-18 (1ng/ml each) or anti-NK1.1 in complete media (black), in the presence of 100nM oligomycin (white), or in glucose free media (grey). (B&C) NK cells were stimulated for 6h with (B) low-dose IL-12+IL-18 (1ng/mL each) or (C) anti-NK1.1 in complete media (--) or in glutamine-free (Glut-free) media with 10 or 50mM 2DG plus an inhibitor of fatty acid oxidation (etomoxir). Results represent the mean +/−SEM of 4 independent experiments. *p≤0.05, **p≤0.01, and ns, not significant.
Since we observed minimal effect of OXPHOS inhibition, even with low dose IL-12+IL-18-activation, we hypothesized that NK cells might require aerobic glycolysis to respond to this stimulus. However, culture of NK cells in glucose-free (GF) media resulted in minimal decreases in IFN-γ producing NK cells with low-dose IL-12+IL-18, but significant defects with anti-NK1.1 (Fig. 3A) or anti-Ly49D (Supplemental Fig. 2). In contrast to OXPHOS inhibition, glycolytic inhibition more selectively inhibited receptor-stimulated IFN-γ since there were only modest decreases in PMA/calcimycin or IL-12+IL-15 activation (Supplemental Fig. 2). We tested the ability of NK cells to utilize other OXPHOS fuels by extracellular flux assay. Indeed, inhibition of glucose metabolism with 2DG resulted in decreased glycolysis (measured by ECAR) with compensatory increased non-glucose fueled OXPHOS (measured by OCR) (Supplemental Fig. 2). However, inhibition of alternative OXPHOS fuels, fatty acids and glutamine, had minimal effect on receptor activation (Supplemental Fig. 3). These findings suggest that glucose is the primary OXPHOS fuel required for receptor activation.
IL-12+IL-18 stimulation is metabolism-independent
Having confirmed that IL-12+IL-18 stimulation is independent of glucose metabolism or OXPHOS at low or high dose stimulation, we next evaluated whether cytokines, but not receptors, induce metabolic plasticity and allow NK cells to efficiently utilize either glycolysis or mitochondrial OXPHOS. To test this, NK cells were cultured with inhibitors of both glycolysis and OXPHOS. Since ATP is required for cell survival, NK cells did not live when cultured in the presence of both oligomycin and 2DG (data not shown). Therefore, NK cells were activated with low concentrations of IL-12+IL-18 in the presence of two different doses of the glycolytic inhibitor 2DG (10 or 50mM) plus etomoxir in glutamine-free media to simultaneously block glycolysis and the other two major mitochondrial OXPHOS fuels (Fig. 3B). Consistent with the hypothesis that NK cells can produce IFN-γ relatively independent of metabolism, NK cell IFN-γ production with IL-12+IL-18 was completely preserved when both glycolysis and each of the other major OXPHOS fuels, fatty acids or glutamine, were blocked (Fig. 3B). By contrast, inhibition of glycolysis and the other major fuels of OXPHOS led to a significant impairment in anti-NK1.1-mediated NK cell activation (Fig. 3C), which was not significantly different than oligomycin or glucose-free media alone (Fig. 3A and C).
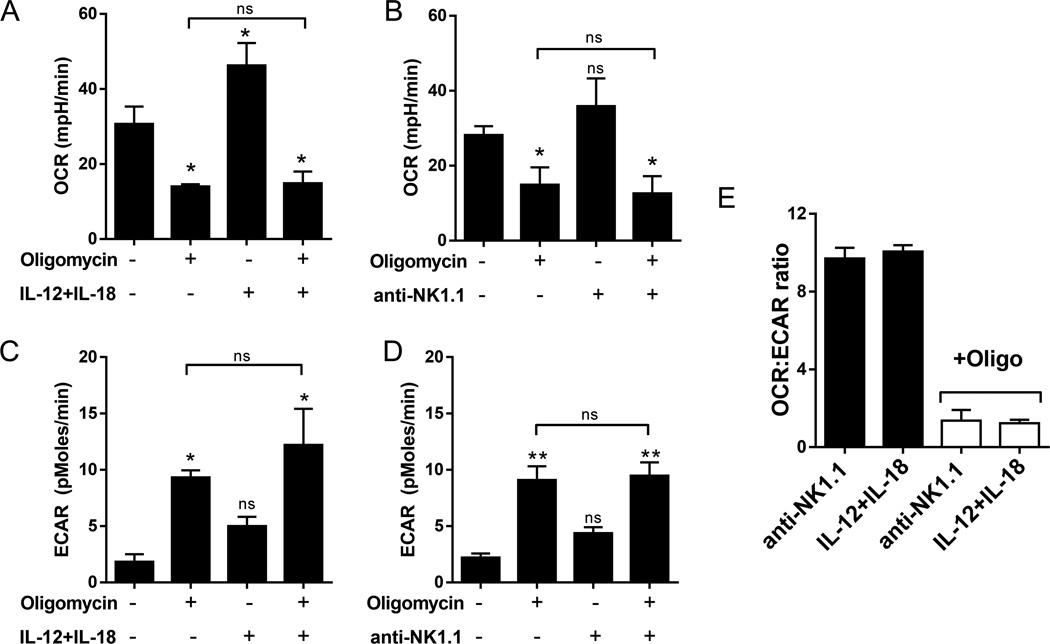
We next tested whether IL-12+IL-18 induced a glycolytic switch upon treatment with oligomycin that was absent in receptor-stimulated NK cells. Oxygen consumption (OCR) decreased and glycolysis (ECAR) increased with oligomycin treatment as expected (Fig. 4A–D). However, there was no difference in OCR or ECAR readings from oligomycin treated NK cells with or without stimulation (IL-12+IL-18 or NK1.1). Similarly, the OCR:ECAR ratios of oligomycin treated cells stimulated with IL-12+IL-18 or anti-NK1.1 was equivalent (Fig. 4E). Together, these data suggest that IL-12+IL-18 does not induce significant metabolic plasticity that would account for the observed metabolism-independent activation.
Figure 4. IL-12+IL-18 stimulation does not induce a significant metabolic switch in NK cells.
Extracellular flux analysis was used to measure oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) of NK cells following 6h culture with (A&C) IL-12+IL-18 or (B&D) anti-NK1.1, with or without oligomycin (100nM). There was no significant difference in OCR or ECAR of oligomycin treated NK cells in the presence or absence of cytokine or receptor stimulation. Statistics over error bars represent comparison to untreated cells (first bar), statistics above the line represents comparison of oligomycin treatment to oligomycin plus NK cell activation (one-way ANOVA). *p≤0.05, **p≤0.01, and ns, not significant. (E) OCR:ECAR ratio of receptor or cytokine-stimulated NK cells. Results represent the mean +/−SEM of 3 independent experiments.
IL-12+IL-18 activation of NK cell IFN-γ production is transcriptionally regulated
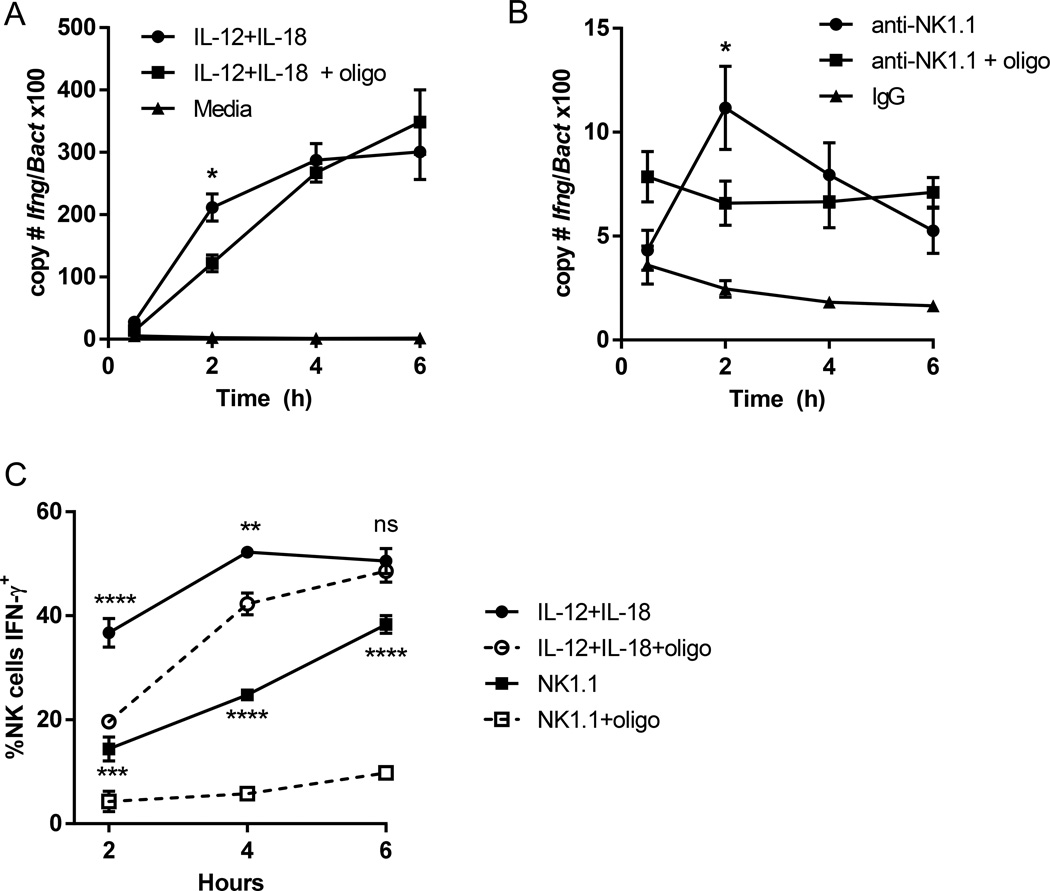
To determine whether OXPHOS inhibition affected receptor-mediated transcription of Ifng, we measured levels of transcript in IL-12+IL-18 or anti-NK1.1 stimulated NK cells with or without oligomycin at different timepoints by quantitative RT-PCR (Fig. 5A & B). There was delayed transcription of Ifng in low-dose IL-12+IL-18 stimulated NK cells with oligomycin, which normalized by 4h (Fig 5A). However, while the percentage and MFI of IFN-γ producing NK cells was similar between low-dose IL-12+IL-18 and anti-NK1.1 activated NK cells at 6h (Fig. 5C and data not shown), receptor stimulation resulted in very little upregulation of Ifng transcript (Fig. 5B). The low levels of Ifng transcription were delayed with oligomycin in NK1.1-activated NK cells, and, similar to cytokine stimulation, normalized by 4h. These data suggest that production of IFN-γ protein is transcriptionally regulated with cytokine activation, but post-transcriptionally regulated with receptor stimulation. Consistent with this hypothesis, IL-12+IL-18 stimulated NK cells had delayed production of IFN-γ protein with oligomycin, that normalized by 6h. By contrast, there was very little production of IFN-γ protein with oligomycin treatment of anti-NK1.1-stimulated NK cells, even at 6h when transcript levels of Ifng were the same. Thus, while oligomycin had a similar effect on Ifng transcription for both stimuli (i.e., delayed upregulation), there are significant differences in the transcriptional and post-transcriptional control of IFN-γ protein production between the two stimuli.
Figure 5. Differences in Ifng transcription with oligomycin treatment.
NK cells were stimulated with low-dose IL-12+IL-18 (1ng/ml each) or anti-NK1.1 in the presence or absence of oligomycin (oligo). (A&B) Absolute copy number of Ifng and β-actin (Bact) were quantitated by real-time PCR between 0.5–6h. Results represent the ratio of Ifng to Bact × 100 and the mean +/−SEM of duplicate wells from 3–4 independent experiments. Statistics represent differences between activated NK cells without or with oligomycin. (C) Intracellular IFN-γ protein in the presence of brefeldin A was measured by flow cytometry at the same timepoints. Results represent the mean +/−SEM of 5 independent experiments. Statistics represent differences between activated NK cells without or with oligomycin for each stimulus (IL-12+IL18 or anti-NK1.1). *p≤0.05; **p≤0.01, ***p≤0.001, ****p≤0.0001 (two-way ANOVA analysis).
Short-term cytokine priming cannot compensate for the absence of an OXPHOS signal for receptor activation
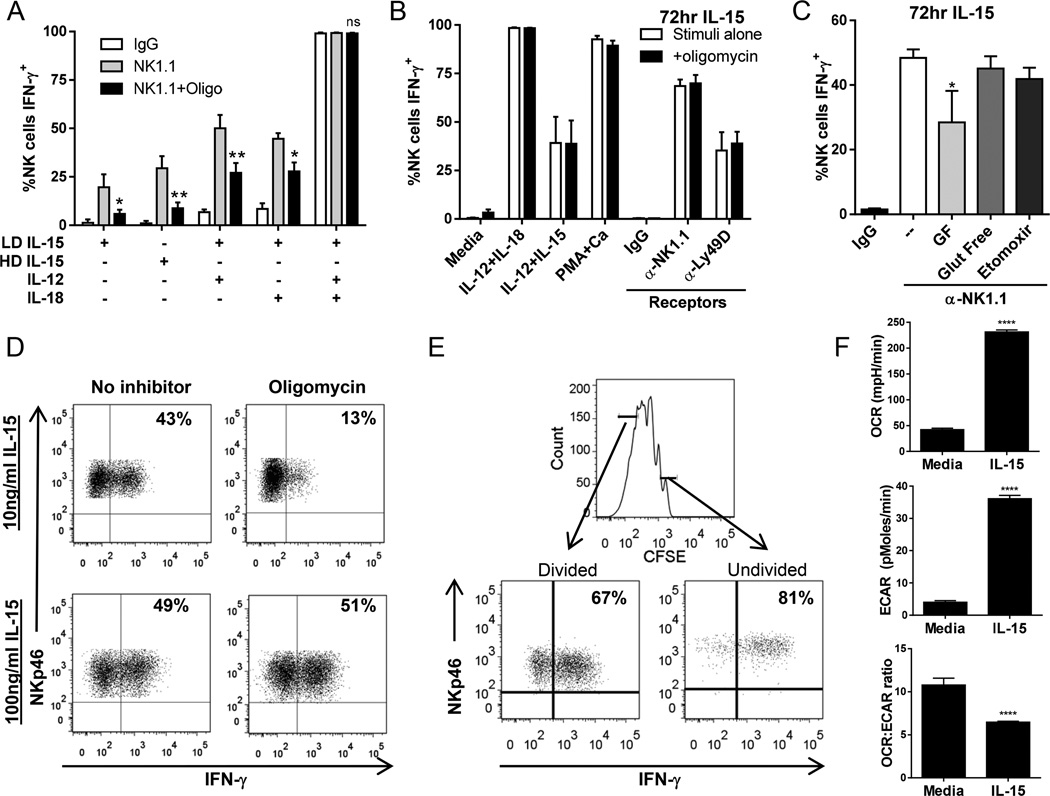
To determine whether IL-12 or IL-18 could prime cells to respond to receptors independent of metabolism, fresh NK cells were pre-treated with different combinations of cytokines, followed by activation with anti-NK1.1 with or without oligomycin (Fig. 6A). All cells were cultured with IL-15 (10ng/ml low-dose or 100ng/ml high-dose) to maintain survival. While the baseline IFN-γ production with NK1.1 stimulation was higher with cytokine priming, there was still a defect in activation in the presence of oligomycin (Fig. 6A). Similar results were obtained when cytokines were added at the start of receptor stimulation (data not shown). Thus, these results suggest that receptor-mediated activation of NK cells requires a metabolically-derived second signal that is not required for, or stimulated by, IL-12 or IL-18 activation.
Figure 6. Exposure to high-dose IL-15, but not proliferation, alters the metabolic requirements for receptor-stimulated IFN-γ production.
(A) NK cells were incubated for 14–16hr with IL-15 (10 or 100ng/ml), IL-12, and/or IL-18, washed, and then stimulated for 6h with anti-NK1.1 or control IgG in the presence or absence of oligomycin (Oligo). Results represent the mean +/−SEM from 4 independent experiments; statistics represent comparison of anti-NK1.1 versus anti-NK1.1 plus oligomycin. (B&C) NK cells were cultured for 72hr with 100ng/ml IL-15, washed, and activated (6hr) with cytokines, PMA + calcimycin (PMA+CA), or receptors (anti-NK1.1, anti-Ly49D or control IgG) in the presence of (B) oligomycin (100nM) or (C) glucose-free (GF) media, glutamine-free media, etomoxir, or glutamine-free media. Results represent the mean +/−SEM from 3 independent experiments. (D) NK cells were cultured for 72h with 10ng/ml or 100ng/ml IL-15 followed by anti-NK1.1 stimulation in the presence or absence of oligomycin. (E) Production of IFN-γ by highly divided and undivided NK cells, as measured by CFSE dilution following IL-15 activation (100ng/ml). Results for D&E are representative of 3 independent experiments. (F) Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) of NK cells treated with IL-15 (100ng/ml) for 72h compared to freshly isolated NK cells rested in media for 4hr. Results represent the mean +/−SEM of 3–6 replicate wells from 2 independent experiments. *p≤0.05; **p≤0.01, ***p≤0.001.
NK cell metabolic requirements for receptor stimulation are reversed with prolonged IL-15 priming
IL-15 is critical for NK cell differentiation and survival and also primes NK cells for cytotoxicity and in vivo effector functions (4,20–22). Long-term treatment with high-dose IL-15 or IL-2 differentiates lymphokine-activated killer (LAK) cells that have enhanced anti-tumor responses (23). We hypothesized that one mechanism by which IL-15 might enhance and prime NK cell function is by altering the metabolic requirements for NK cell activation. Indeed, following 72hr culture with 100ng/ml IL-15, NK cells had no defect in IFN-γ production with OXPHOS inhibition in response to cytokine, receptor, or PMA and calcimycin activation (Fig. 6B). A small difference in receptor stimulation without glucose persisted, and activation continued to be independent of fatty acid oxidation or glutamine (Fig. 6C). High dose, 100ng/ml, IL-15 was required to induce metabolism-independent activation as lower dose (10ng/ml) IL-15 had no effect on the metabolic requirement for receptor activation (Fig. 6D). Short-term stimulation was not sufficient to induce this effect, since there was no change in the metabolic requirements after overnight treatment with 100ng/ml IL-15 (Fig. 6A), and at least 48hr of stimulation was required (data not shown).
One major difference between stimulation with 10 versus 100ng/ml IL-15 is that cells proliferate only with the higher dose (data not shown). Thus, it is possible that the changes in metabolic requirements were due to proliferation and not IL-15 treatment. However, when comparing IFN-γ production by the least divided versus most-divided NK cells, as marked by CFSE dilution, both populations produced normal amounts of IFN-γ with oligomycin, and in fact, we consistently observed higher IFN-γ production in undivided cells (Fig. 6E). Furthermore, adoptive transfer of CFSE-labeled splenocytes into alymphoid Rag2−/−γc−/− hosts and induction of homeostatic proliferation had no effect on the metabolic requirement for IFN-γ production (Supplemental Fig. 4). We also tested whether the TLR3 analog poly(I:C), which upregulates DC IL-15/IL-15rα (4), could prime NK cell metabolic independence. However, defects persisted in receptor stimulation of metabolically-inhibited NK cells after in vivo activation overnight with poly(I:C) (Supplemental Fig. 4D), although to a lesser degree than naïve NK cells.
Finally, we evaluated whether treatment with high-dose IL-15 affected NK cell metabolism. Indeed, NK cells significantly upregulated OXPHOS and glycolysis as measured by OCR and ECAR (Fig. 6F) after culture in high dose IL-15, consistent with another recent study (17). While both metabolic pathways increased, there was a decreased dependence on OXPHOS in IL-15-treated NK cells as measured by a decreased ratio of OCR:ECAR.
DISCUSSION
Metabolism is critical for a wide array of cellular functions, and has been increasingly recognized to be important for immune cell function (12). This represents the first study to investigate the metabolic fuels utilized by fresh NK cells for production of IFN-γ. Remarkably, we demonstrate that a metabolism-driven second signal is required for receptor-mediated, but not IL-12+IL-18-stimulated, activation of NK cell IFN-γ.
NK cells have the ability to rapidly produce large amounts of IFN-γ protein within hours of activation. Surprisingly, NK cells were relatively metabolically inactive at baseline and had no significant increase in their OXPHOS or glycolysis after short-term activation, as measured by an extracellular flux assay. However, despite no increased metabolic activity with stimulation, inhibition of glucose/glycolysis or global OXPHOS inhibition resulted in near-complete abrogation of receptor-stimulated IFN-γ production. The receptors tested here, NK1.1 and Ly49D, both partner with ITAM-bearing adapters (24), suggesting that OXPHOS is a requisite second signal for ITAM-mediated IFN-γ production. Inhibition of other OXPHOS fuels, including glutamine or fatty acid oxidation, had no effect on receptor-stimulated IFN-γ production, and glucose appears to be the primary fuel required to drive OXPHOS.
The activation-specific metabolic requirements for NK cell IFN-γ production shown here are quite distinct from T cells, which require glycolysis, but not mitochondrial OXPHOS, for production of IFN-γ in response to receptors, PMA and ionomycin, or IL-12+IL-18 (15,25). Here, we discovered a major difference in the transcriptional upregulation of Ifng between cytokine and receptor-stimulated NK cells. It is well-described that NK cells constitutively express Ifng transcript, but not protein (26). Thus, it is likely, that one mechanism by which metabolic inhibition impairs receptor-stimulated, but not IL-12+IL-18-stimulated, IFN-γ protein production is by inhibition of translation of pre-existing transcript. In CD4 T cells, glycolysis was shown to be important for the post-transcriptional processing of IFN-γ due to consumption of the glycolytic enzyme GAPDH, which was otherwise bound to the 3’UTR of IFN-γ transcript (15). However, in contrast to our findings here, inhibition of OXPHOS with oligomycin had no effect on receptor-stimulated T cell production of IFN-γ (15). Thus, consumption of GAPDH is unlikely to explain our observed dependence on both glycolysis and OXPHOS for NK cell receptor-stimulated IFN-γ production. The results here suggest distinct cell-specific regulation of IFN-γ protein production by innate and adaptive effector cells. However, one caveat to this conclusion is that NK cells were studied directly ex vivo here, while prior work with T cells was performed after in vitro culture/activation, thus it is possible that activation alters T cell metabolic requirements for IFN-γ production.
Prolonged culture of NK cells with high-dose IL-15 led to increased NK cell metabolism and particularly glycolysis, as measured by a decrease in the OCR:ECAR ratio. Cells cultured for more than 48hr in high-dose IL-15 no longer required OXPHOS for receptor-stimulated IFN-γ production, an effect that was dose-dependent and did not require proliferation. Interestingly, IL-15 stimulated NK cells still demonstrated some dependence on glucose for IFN-γ production. Perhaps this is a reflection of their overall increased dependence on glycolysis for cellular metabolism, or, alternatively, an indication that they have switched to a regulatory mechanism more similar to T cell control of IFN-γ post-transcriptional processing. Alterations of the metabolic requirements of NK cell activation with IL-15 is clinically relevant, since there are ongoing clinical trials of IL-15 treatment of NK cells prior to adoptive immunotherapy, as well as administration of IL-15 for cancer therapies targeting NK cells (27). Our data would suggest that, in addition to priming and expansion of NK cells, IL-15 may also impart NK cells with enhanced functionality in metabolically-deprived locations such as tumor microenvironments.
IL-15 is known to prime NK cell effector functions, including cytokine production and cytotoxicity (4,20). Recently, the Walzer laboratory demonstrated that high-dose IL-15 activates mTOR which stimulates NK cell glucose uptake, proliferation, and cytolytic responses (17). Similarly, Nandagopal et al. reported that IL-15-induced mTOR is important for NK cell IFN-γ production when co-stimulated with this cytokine (28). mTOR is an important metabolic regulator of T cell function and upregulates glucose uptake and glycolysis in activated T cells (29). Another recent study also demonstrated a dependence on glycolysis, but not OXPHOS, for IL-12+IL-2 stimulated IFN-γ production by NK cells cultured in IL-15 for 7 days, and implicated IL-15-induced mTOR for glycolytic programing of NK cells (30). Thus, IL-15-induced mTOR may represent a potential mechanism whereby NK cells cultured with high-dose IL-15 upregulate glycolysis. However, it remains unclear how prolonged high-dose IL-15 signaling and increased glycolysis alters the OXPHOS requirement for receptor-mediated signaling as shown here.
In summary, these findings demonstrate that NK cell IFN-γ responses are dictated by the metabolic environment in an activation-specific manner. This suggests that NK cell function in vivo will be affected by the availability of metabolic fuels and that drugs targeting metabolism, such as glycolytic inhibitors, will impact NK cell activation. The finding that prolonged high-dose IL-15 alleviates the metabolic requirement for receptor-mediated NK cell stimulation suggests a potential mechanism to ‘reverse’ the metabolic-dependence of receptor-activation. Additional investigation into the glucose-driven OXPHOS signal that is required for receptor signaling, and how this signal interacts with known receptor signaling pathways will be important (31). Finally, it will be interesting to explore the metabolic requirements for other NK cell functions including cytotoxicity, licensing, and generation of “memory” NK cells (32–34).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank T.A. Fehniger and W.M. Yokoyama for helpful discussion and reagents.
Footnotes
This research was supported by NIH/NIAID 1K08-AI085030, the Child Health Research Center at Washington University School of Medicine (K12-HD076224), and The Children’s Discovery Institute and St. Louis Children’s Hospital (to M.A.C.). A.Y.M. is supported by NIH training grant 5T32GM007200 and T.P.V. is supported by NIH training grant 2T32AR007279. Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases, part of the National Institutes of Health, under Award Number P30AR048335. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Biron CA, Brossay L. NK cells and NKT cells in innate defense against viral infections. Curr. Opin. Immunol. 2001;13:458–464. doi: 10.1016/s0952-7915(00)00241-7. [DOI] [PubMed] [Google Scholar]
- 2.French AR, Yokoyama WM. Natural killer cells and viral infections. Curr. Opin. Immunol. 2003;15:45–51. doi: 10.1016/s095279150200002x. [DOI] [PubMed] [Google Scholar]
- 3.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat. Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 4.Lucas M, Schachterle W, Oberle K, Aichele P, Diefenbach A. Dendritic cells prime natural killer cells by trans-presenting interleukin 15. Immunity. 2007;26:503–517. doi: 10.1016/j.immuni.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu. Rev. Immunol. 2013;31:227–258. doi: 10.1146/annurev-immunol-020711-075005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salway J. Metabolism at a glance. 3rd ed. Malden, MA: Blackwell Publishing; 2004. [Google Scholar]
- 7.Porporato PE, Dhup S, Dadhich RK, Copetti T, Sonveaux P. Anticancer targets in the glycolytic metabolism of tumors: a comprehensive review. Front. Pharmacol. 2011;2:49. doi: 10.3389/fphar.2011.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491:364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- 9.Bremer AA, Mietus-Snyder M, Lustig RH. Toward a unifying hypothesis of metabolic syndrome. Pediatrics. 2012;129:557–570. doi: 10.1542/peds.2011-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiolero R, Revelly JP, Tappy L. Energy metabolism in sepsis and injury. Nutrition. 1997;13:45S–51S. doi: 10.1016/s0899-9007(97)00205-0. [DOI] [PubMed] [Google Scholar]
- 11.Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013;342:1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox CJ, Hammerman PS, Thompson CB. Fuel feeds function: energy metabolism and the T-cell response. Nat. Rev. Immunol. 2005;5:844–852. doi: 10.1038/nri1710. [DOI] [PubMed] [Google Scholar]
- 15.Chang CH, Curtis JD, Maggi LB, Jr, Faubert B, Villarino AV, O'Sullivan D, Huang SC, van der Windt GJ, Blagih J, Qiu J, Weber JD, Pearce EJ, Jones RG, Pearce EL. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Overbergh L, Giulietti A, Valckx D, Decallonne R, Bouillon R, Mathieu C. The use of real-time reverse transcriptase PCR for the quantification of cytokine gene expression. J Biomol Tech. 2003;14:33–43. [PMC free article] [PubMed] [Google Scholar]
- 17.Marcais A, Cherfils-Vicini J, Viant C, Degouve S, Viel S, Fenis A, Rabilloud J, Mayol K, Tavares A, Bienvenu J, Gangloff YG, Gilson E, Vivier E, Walzer T. The metabolic checkpoint kinase mTOR is essential for IL-15 signaling during the development and activation of NK cells. Nat. Immunol. 2014;15:749–757. doi: 10.1038/ni.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sena LA, Li S, Jairaman A, Prakriya M, Ezponda T, Hildeman DA, Wang CR, Schumacker PT, Licht JD, Perlman H, Bryce PJ, Chandel NS. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He S, Kato K, Jiang J, Wahl DR, Mineishi S, Fisher EM, Murasko DM, Glick GD, Zhang Y. Characterization of the metabolic phenotype of rapamycin-treated CD8+ T cells with augmented ability to generate long-lasting memory cells. PLoS One. 2011;6:e20107. doi: 10.1371/journal.pone.0020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fehniger TA, Cai SF, Cao X, Bredemeyer AJ, Presti RM, French AR, Ley TJ. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity. 2007;26:798–811. doi: 10.1016/j.immuni.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 21.Cooper MA, Bush JE, Fehniger TA, VanDeusen JB, Waite RE, Liu Y, Aguila HL, Caligiuri MA. In vivo evidence for a dependence on interleukin 15 for survival of natural killer cells. Blood. 2002;100:3633–3638. doi: 10.1182/blood-2001-12-0293. [DOI] [PubMed] [Google Scholar]
- 22.Koka R, Burkett PR, Chien M, Chai S, Chan F, Lodolce JP, Boone DL, Ma A. Interleukin (IL)-15R[alpha]-deficient natural killer cells survive in normal but not IL-15R[alpha]-deficient mice. J. Exp. Med. 2003;197:977–984. doi: 10.1084/jem.20021836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suck G, Oei VY, Linn YC, Ho SH, Chu S, Choong A, Niam M, Koh MB. Interleukin-15 supports generation of highly potent clinical-grade natural killer cells in long-term cultures for targeting hematological malignancies. Exp. Hematol. 2011;39:904–914. doi: 10.1016/j.exphem.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 24.Yokoyama WM. Natural Killer Cells. In: Paul WE, editor. Fundamental Immunology. Philadelphia: Lippincott, Williams &Wilkins; 2012. [Google Scholar]
- 25.Cham CM, Gajewski TF. Glucose availability regulates IFN-gamma production and p70S6 kinase activation in CD8+ effector T cells. J. Immunol. 2005;174:4670–4677. doi: 10.4049/jimmunol.174.8.4670. [DOI] [PubMed] [Google Scholar]
- 26.Stetson DB, Mohrs M, Reinhardt RL, Baron JL, Wang ZE, Gapin L, Kronenberg M, Locksley RM. Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J. Exp. Med. 2003;198:1069–1076. doi: 10.1084/jem.20030630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knorr DA, Bachanova V, Verneris MR, Miller JS. Clinical utility of natural killer cells in cancer therapy and transplantation. Semin. Immunol. 2014;26:161–172. doi: 10.1016/j.smim.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nandagopal N, Ali AK, Komal AK, Lee SH. The Critical Role of IL-15-PI3K-mTOR Pathway in Natural Killer Cell Effector Functions. Front. Immunol. 2014;5:187. doi: 10.3389/fimmu.2014.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waickman AT, Powell JD. mTOR, metabolism, and the regulation of T-cell differentiation and function. Immunol. Rev. 2012;249:43–58. doi: 10.1111/j.1600-065X.2012.01152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donnelly RP, Loftus RM, Keating SE, Liou KT, Biron CA, Gardiner CM, Finlay DK. mTORC1-Dependent Metabolic Reprogramming Is a Prerequisite for NK Cell Effector Function. J. Immunol. 2014;193:4477–4484. doi: 10.4049/jimmunol.1401558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vivier E, Nunes JA, Vely F. Natural killer cell signaling pathways. Science. 2004;306:1517–1519. doi: 10.1126/science.1103478. [DOI] [PubMed] [Google Scholar]
- 32.Sun JC, Ugolini S, Vivier E. Immunological memory within the innate immune system. EMBO J. 2014 doi: 10.1002/embj.201387651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooper MA, Yokoyama WM. Memory-like responses of natural killer cells. Immunol. Rev. 2010;235:297–305. doi: 10.1111/j.0105-2896.2010.00891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, French AR, Sunwoo JB, Lemieux S, Hansen TH, Yokoyama WM. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.