Abstract
Neutrophil serine proteases (NSPs) are critical for the effective functioning of neutrophils and greatly contribute to immune protection against bacterial infections. Thanks to their broad substrate specificity, these chymotrypsin-like proteases trigger multiple reactions that are detrimental to bacterial survival such as direct bacterial killing, generation of antimicrobial peptides, inactivation of bacterial virulence factors and formation of neutrophil extracellular traps. Recently, the importance of NSPs in antibacterial defenses has been further underscored by discoveries of unique bacterial evasion strategies to combat these proteases. Bacteria can indirectly disarm NSPs by protecting bacterial substrates against NSP cleavage, but also produce inhibitory molecules that potently block NSPs. Here we review recent insights in the functional contribution of NSPs in host protection against bacterial infections and the elegant strategies that bacteria use to counteract these responses.
Introduction
Neutrophils are the most abundant circulating leukocytes [1] and are the first cells of the innate immune system to migrate to an infection site [1]. Neutrophils can rapidly kill bacteria using three mechanisms that all depend on their antimicrobial granular components (Fig. 1) [2]. First, neutrophils can engulf bacteria (phagocytosis) and subsequently kill them inside the phagocytic vacuole after fusion with granules. Second, they can release their granular content into the extracellular milieu via exocytosis (degranulation) [1]. Third, they can release neutrophil extracellular traps (NETosis), which contain the antimicrobial granule proteins, to entrap and kill bacteria [3]. It is now evident that neutrophil serine proteases (NSPs) play key roles in each of these antibacterial responses.
Figure 1. Locations where bacteria encounter NSPs.
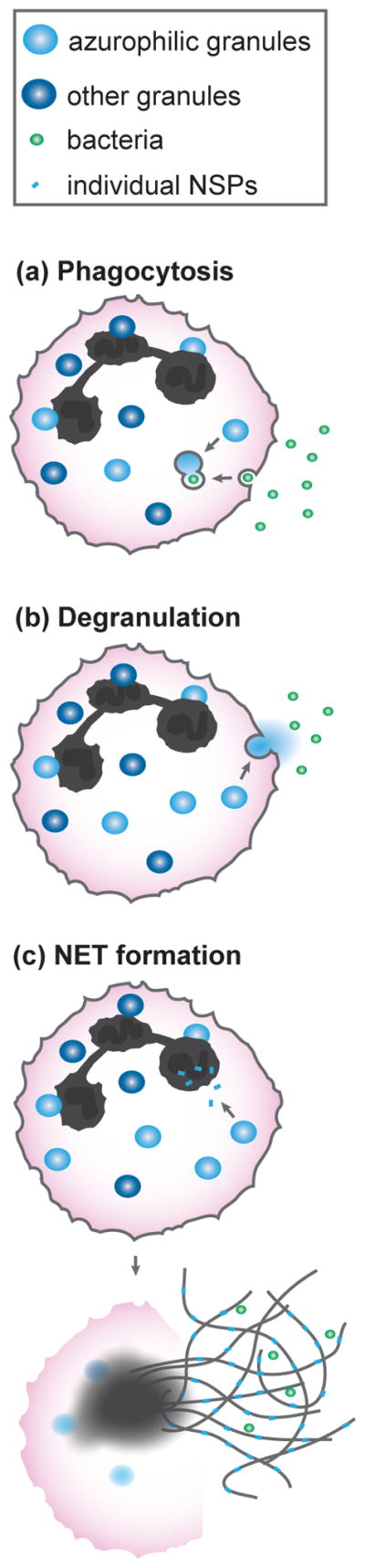
(a) After neutrophils ingest opsonized bacteria, the granules fuse with the phagocytic vacuole to release NSPs and antimicrobial components. (b) During degranulation, neutrophils exocytose their granule contents into the extracellular space. (c) NSPs can translocate to the nucleus and induce NET formation. NSPs are embedded within NETs that capture bacteria.
This protease family consists of neutrophil elastase (NE), proteinase 3 (PR3), cathepsin G (CG) and the recently discovered neutrophil serine protease-4 (NSP4) [4]. NSPs are stored within the acidic granules tightly bound to proteoglycans that inactivate them [5]. They only become active after their release into the phagocytic vacuole [2,6] where their concentrations are believed to reach as high as 50 mg/ml (based on calculations for MPO [5,7,8]). In addition to their intracellular role, NSPs are also important components of neutrophil degranulation fluid and NETs [9]. NSPs belong to the chymotrypsin family of serine proteases, in which a charge-relay system of His-Asp-Ser forms the catalytic site (for excellent reviews on NSP biochemistry please read [10] and [11]). Despite their similar sequences (35–56 % identical) and tertiary structures, however, they display different substrate specificities. Together they have the ability to cleave a wide variety of substrates. This broad substrate specificity, and the fact that they act at multiple locations (intracellular and extracellular), often complicates detailed understanding of NSP contributions to anti-bacterial host defense. Here we discuss recent insights into how NSPs contribute to the defense against bacteria and illustrate how bacteria can effectively antagonize NSP activity.
NSP functions in antibacterial defense
Although NSPs can also indirectly modulate the immune response, for instance by functioning as chemoattractants or cleaving chemokines (see [12], [13] and [14] for recent reviews), we will here focus on the more direct interactions of NSPs with bacteria (Fig. 2).
Figure 2. Antimicrobial functions of NSPs.
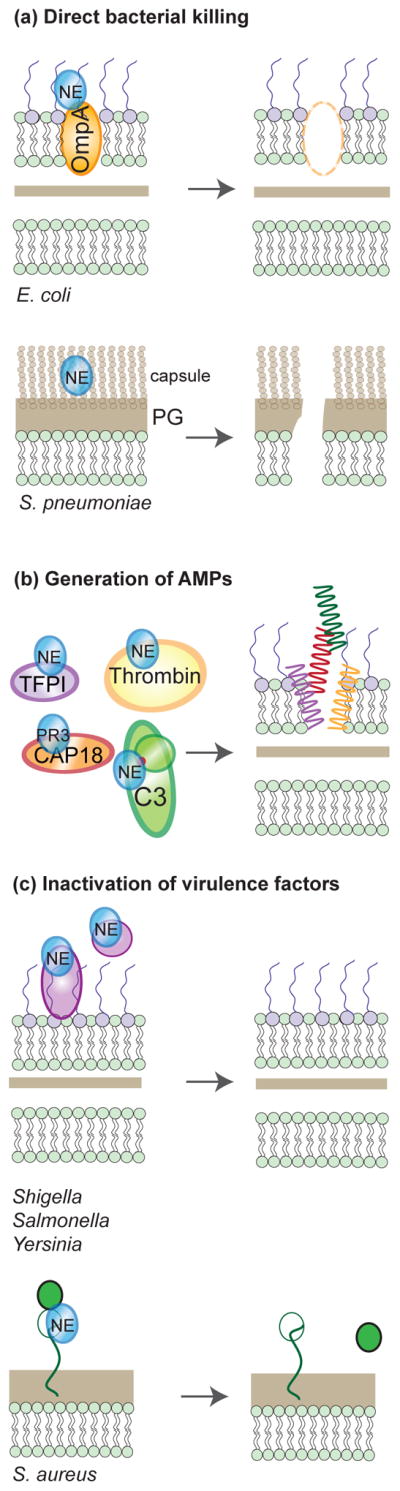
(a) NSPs can directly kill bacteria by attacking membrane associated (E. coli) or capsule proteins (S. pneumonia), which leads to loss of membrane integrity. (b) NSPs can cleave host immune proteins to generate antimicrobial peptides that can directly kill bacteria. (c) NSPs can target and inactivate bacterial virulence factors, resulting in attenuated bacteria.
Direct killing
The best-known antibacterial function of NSPs is direct killing of bacterial cells. While NE has been shown to directly kill the Gram-negative bacteria Klebsiella pneumoniae and Escherichia coli, only for the latter it has been shown to depend on cleavage of its outer membrane protein A (OmpA), resulting in loss of membrane integrity and cell death [15,16] (Fig. 2a). Separately, the Gram-positive Streptococcus pneumoniae is known to be killed by the concerted action of NE, CG, and PR3 within the phagocytic vacuole, which was also demonstrated in vivo [17,18]. This process requires the presence of pneumococcal capsule, although the mechanism is yet unknown [19] (Fig. 2a). Surprisingly, NE seems trivial for killing of the closely related organism, Staphylococcus aureus, nor has the role of CG been well established [5,16,20]. In addition to the afore mentioned examples, NSPs might also be bactericidal independent from their proteolytic activity, but the exact mechanism of action is unknown [21]. Taken together, direct anti-microbial activity of NSPs is only demonstrated for a very limited amount of bacterial species.
Generation of antimicrobial peptides
More indirectly, NSPs can cleave host proteins to generate antimicrobial peptides. The best-known example is the extracellular cleavage of hCAP-18 by PR3 to generate the antimicrobial peptide LL-37 [22]. Separately from this, extracellular NSPs can also cleave serum proteins of the complement and coagulation systems to generate distinct antimicrobial peptides. For example, NE cleaves the central complement protein C3 to generate a peptide that mimics the natural C3a anaphylatoxin. As with C3a, this NE-derived peptide of C3 shows antimicrobial activity against Enterococcus faecalis and Pseudomonas aeruginosa [23] (Fig. 2b). NE and CG can also cleave thrombin and release peptides that are antimicrobial to E. coli [24]. Lastly, NE cleaves the tissue-factor pathway inhibitors (TFPI-1 and TFPI-2) into peptides that kill a wide range of bacteria, or bind E. coli, respectively [25,26].
Virulence attenuation
Furthermore, NSPs may attenuate bacterial virulence by inactivating factors required for pathogenesis. For example, NE, but not CG, cleaves the invasins IpaA–C and the mobility protein IcsA of Shigella flexneri to prevent bacterial dissemination into the cytoplasm of neutrophils [27] (Fig. 2c). Virulence factors of the related enterobacteria Salmonella typhimurium and Yersinia enterocolitica were also cleaved [27]. Such effects are not limited to Gram-negatives, however, as CG cleaves the S. aureus adhesin clumping factor A (ClfA) and removes its active domain (Fig. 2c) [28]. Judging from the broad substrate specificity of NSPs and the relatively low concentrations needed to target virulence factors [27], it seems likely that many more bacteria are attenuated in this way.
NET formation
The role of NSPs during NET formation is perhaps best illustrated by the absolute requirement of active NE to form NETs. Upon NET induction, NE translocates to the nucleus and cleaves histones to facilitate the DNA decondensation central to this process [29,30] (Fig. 1c). In addition, all NSPs are found within the NETs [9]. NETs are currently believed to have three functions. First, they catch the extracellular NSPs, and other antimicrobial agents released from neutrophils, to prevent host damage at distal sites [31]. Second, they ensnare bacteria to prevent them from disseminating to other body sites [32]. Third, they can kill the captured bacteria via the antimicrobial agents found within them [33].
Besides the large debate about the direct bacterial killing by NETs in general [34], it is unlikely that the NSPs contribute to killing within NETs since even S. pneumonia survived entrapment [32], despite its sensitivity to NSPs within the phagocytic vacuole. Moreover, activities of NSPs within NETs are decreased, which for NE and PR3 can be restored upon DNAse treatment of NETs [35]. In addition, NET-bound NSPs may also be inactivated by high concentration of NSP inhibitors in serum [36]. Altogether, the main role for NSPs probably lies in the induction of NETs, so that they can confine the bacterial infection.
This bacterial entrapment might be an interplay with the coagulation system. The coagulation inhibitor TFPI-1 is bound by NET DNA and inactivated by NET-associated NE, which promotes thrombus formation. During systemic E. coli infection, this thrombus prevented bacterial dissemination into other tissues [37].
Lessons from patients and in vivo studies
The fact that neutrophils are crucial in human antimicrobial defense is evident from the numerous recurrent infections in patients with neutrophil deficiencies [38,39]. For long it was believed that reactive oxygen species (ROS), generated in the phagocytic vacuole by NADPH oxidase and myeloperoxidase, are the major bactericidal molecules responsible for neutrophil killing of bacteria. However, the direct antibacterial effects of ROS seem to be overstated and it is currently believed that a concerted action of NADPH oxidase and NSPs is necessary to effectively eradicate bacteria [6]. This is demonstrated by studies with NE−/− and CG−/− mice that showed that NSPs are crucial for clearance of E. coli, K. pneumoniae and S. pneumoniae [15,16,18].
Bacterial mechanisms to block NSPs
Increasing evidence now shows that bacterial pathogens have evolved strategies to counteract human NSPs. The mechanisms identified thus far range from protecting bacterial substrates against proteolytic cleavage to the production of protease inhibitors that directly block NSPs (Fig. 3).
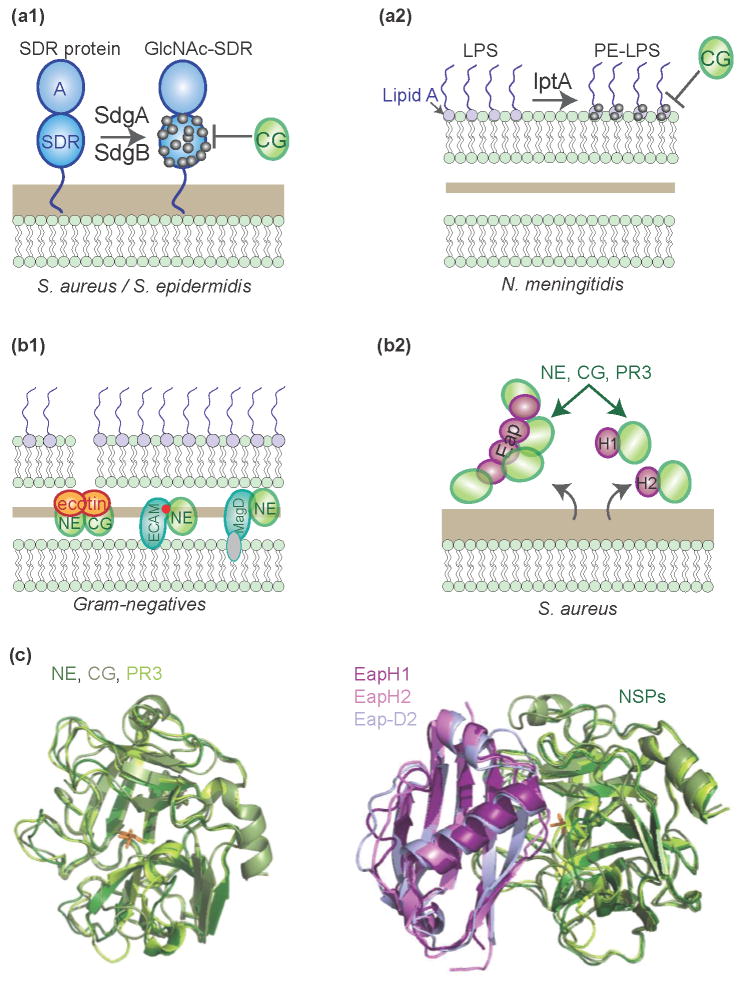
Figure 3. Bacterial mechanisms to block NSPs.
(a) Modification of bacterial substrates. (1) Glycosyltransferases SdgA and SdgB modify staphylococcal virulence factors with N-acetylglucosamine (GlcNAc) to prevent them being cleaved by CG. (2) LptA modifies Neisserial lipid A to prevent killing by CG. (b) Bacterial inhibitors of chymotrypsin-like serine proteases. (1) Gram-negative bacteria express ecotin, or the bMG proteins ECAM (E. coli) or MagD (P. aeruginosa) in their periplasmic space to prevent activity of a broad range of serine proteases. (2) S. aureus secretes the Eap family of proteins, consisting of Eap, EapH1, and EapH2, that specifically inhibits NSPs. Full-length Eap consists of multiple domains (resembling EapH1 and EapH2) that each bind one NSP molecule. (c) Model of NE, CG, and PR3 binding to different Eap proteins (EapH1, EapH2, and the second domain of Eap (Eap-D2)), inferred from the co-crystal structure of NE with EapH1 (PDB code 4NZL).
Modifications of bacterial NSP substrates
The opportunistic pathogens S. epidermidis and S. aureus express glycosyltransferases (SdgA and SdgB) that decorate cell surface-bound proteins with N-acetylglucosamine (GlcNAc) moieties on their serine-aspartate dipeptide (SDR) repeats [28]. These SDR repeats are, among others, found in the virulence factors ClfA and ClfB, SdrC, SdrD, SdrE (S. aureus) and SrdF, SdrG and SdrH (S. epidermidis). The GlcNAc modification protects them from proteolytic degradation by CG (Fig. 3a).
Another mechanism is used by the Gram-negative human pathogen Neisseria meningitidis. Like all Gram-negatives, the outer membrane of N. meningitidis contains LPS molecules that are anchored to the membrane via lipid A. This can be modified by Neisserial phosphoethanolamine transferase (lptA) with phosphoethanolamine to prevent proteolysis-independent killing by CG [40] (Fig. 3a).
Bacterial protease inhibitors
Bacteria can also directly counteract NSPs by producing high-affinity inhibitors. Many Gram-negative bacteria express the dimeric, periplasmic protein ecotin (16 kDa) that inhibits NSPs by forming heterotetrameric complexes [41]. The ecotin orthologues of pathogenic E. coli, Yersinia pestis and P. aeruginosa all potently block NE and CG (Fig. 3b). Ecotin is not a specific NSP inhibitor, however, since it also potently inhibits a wide range of other chymotrypsin-like proteases, like trypsin and chymotrypsin [41].
Another interesting group of protease inhibitors are the Gram-negative homologues of the mammalian alpha-macroglobulin (MG) family. MGs are large (>170 kDa) glycoproteins that inactivate a wide variety of proteases [42,43] (Fig. 2c). These bacterial MG-like proteins (bMGs), supposedly acquired via horizontal gene transfer from their metazoan hosts [44], display remarkable structural and functional homology to human α2-MGs. Like αMG, proteolytic cleavage of a bait region in bMGs results in a conformational change that allows the inhibitor to covalently capture the protease. The E. coli protein ECAM has a similar secondary structure to human plasma α2-MG and likewise contains an internal thioester to form covalent complexes with NE, trypsin and chymotrypsin [45].
Intriguingly, both ecotin and bMGs are expressed in the periplasmic space of Gram-negative bacteria. Thus, they can only function against proteases that can breach the bacterial outer cell membrane. Since ecotin was found to protect E. coli from killing by purified NE [41], this suggests that NE gains access to the periplasm following degradation of OmpA and disrupts the membrane integrity. Still, the question remains whether these broad-range protease inhibitors evolved specifically to protect bacteria from NE, or whether they mainly serve to block pancreatic digestive proteases in the mammalian gut or bacterial aggressors that inject proteases in the periplasm [45].
These broad-range protease inhibitors aside, a very recent study indicates that S. aureus has evolved a family of highly specific NSP inhibitors. Our group identified three related proteins that potently inhibit NE, CG and PR3 but not the closely related thrombin, plasmin and kallikrein [46]. This family includes the extracellular adherence protein (Eap, 53 kD) and its smaller homologues EapH1 (12 kD) and EapH2 (13 kD) (Fig. 3b). EapH1 forms a non-covalent, 1:1 complex with NE, occluding the NE active site and preventing substrate cleavage. Since NE, CG and PR3 show high structural resemblance [47], structural interpretations of the NE/EapH1 complex are directly relevant to CG and PR3 complexes as well (Fig. 3c). Although expression of these three staphylococcal proteins is upregulated in the presence of neutrophil granules [48], it remains unknown whether they are expressed inside phagocytic vacuoles or rather serve to block extracellular NSPs of degranulated neutrophils or within NETs. Eap proteins were shown to also affect outcome of infection in vivo. The fact that S. aureus evolved three very potent and specific inhibitors of NSPs implies a crucial role for NSPs in the defense against S. aureus.
Conclusions
Recent advances in understanding the molecular interplay between NSPs and bacteria now indicate that the role of NSPs in antibacterial host defense is much more diverse than simply directly killing of bacteria. Considering the few species it this has been proven for, a directly bactericidal activity of NSPs seems very much overstated. On the contrary, a large body of evidence shows that these proteases can diminish bacterial virulence in many ways and their specific activities may even differ depending on the exact location where they encounter bacteria (within the phagocytic vacuole or in the extracellular space). In our opinion, future work should also address whether NSPs collaborate with other antibacterial host defense components such as membrane-perturbing granular components. Still, the fact that bacteria evolved inhibitors of NSPs, which are highly specific in case of S. aureus, indicates that their role in anti-bacterial defense is indispensable. The endogenous production of these inhibitors has probably complicated previous studies analyzing the role of NSPs in defense in vivo and we feel that future work on NSP host defense functions should take these evasion strategies into account. Overall, these insights will contribute to a better understanding of the roles of NSPs in host defense against bacteria.
Highlights.
Neutrophil serine proteases (NSPs) support bacterial clearance by neutrophils.
NSPs can kill bacteria, generate antimicrobial peptides or induce NET formation.
The anti-bacterial activities of NSPs are underlined by the discovery of bacterial inhibitors.
Gram-negative bacteria block chymotrypsin-like proteases like NSPs within the periplasm.
Staphylococcus aureus secretes a family of three highly specific NSP inhibitors.
Acknowledgments
The authors were supported by grants of the Netherlands Organization for Scientific Research (NWO-Vidi #91711379 to S.H.M.R.) and the U.S. National Institutes of Health (NIH#AI071028, to B.V.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
**of outstanding interest
* of special interest
- 1.Korkmaz B, Horwitz MS, Jenne DE, Gauthier F. Neutrophil Elastase, Proteinase 3, and Cathepsin G as Therapeutic Targets in Human Diseases. Pharmacol Rev. 2010;62:726–759. doi: 10.1124/pr.110.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borregaard N, Cowland JB. Granules of the human neutrophilic polymorphonuclear leukocyte. Blood. 1997;89:3503–3521. [PubMed] [Google Scholar]
- 3.Fuchs TA, Abed U, Goosmann C, Hurwitz R, Schulze I, Wahn V, Weinrauch Y, Brinkmann V, Zychlinsky A. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. 2007;176:231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perera NC, Schilling O, Kittel H, Back W, Kremmer E, Jenne DE. NSP4, an elastase-related protease in human neutrophils with arginine specificity. Proc Natl Acad Sci U S A. 2012;109:6229–6234. doi: 10.1073/pnas.1200470109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reeves EP, Lu H, Jacobs HL, Messina CGM, Bolsover S, Gabella G, Potma EO, Warley A, Roes J, Segal AW. Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature. 2002;416:291–297. doi: 10.1038/416291a. [DOI] [PubMed] [Google Scholar]
- 6.Segal AW. How neutrophils kill microbes. Annu Rev Immunol. 2005;23:197–223. doi: 10.1146/annurev.immunol.23.021704.115653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell EJ, Campbell MA, Owen CA. Bioactive Proteinase 3 on the Cell Surface of Human Neutrophils: Quantification, Catalytic Activity, and Susceptibility to Inhibition. J Immunol. 2000;165:3366–3374. doi: 10.4049/jimmunol.165.6.3366. [DOI] [PubMed] [Google Scholar]
- 8.Campbell EJ, Silverman EK, Campbell MA. Elastase and cathepsin G of human monocytes. Quantification of Cellular Content, Release in Response to Stimuli, and Heterogeneity in Elastase-Mediated Proteolytic Activity. J Immunol. 1989;143:2961–2968. [PubMed] [Google Scholar]
- 9.Urban CF, Ermert D, Schmid M, Abu-Abed U, Goosmann C, Nacken W, Brinkmann V, Jungblut PR, Zychlinsky A. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 2009;5:e1000639. doi: 10.1371/journal.ppat.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellman L, Thorpe M. Granule proteases of hematopoietic cells, a family of versatile inflammatory mediators - an update on their cleavage specificity, in vivo substrates, and evolution. Biol Chem. 2014;395:15–49. doi: 10.1515/hsz-2013-0211. [DOI] [PubMed] [Google Scholar]
- 11.Korkmaz B, Moreau T, Gauthier F. Neutrophil elastase, proteinase 3 and cathepsin G: physicochemical properties, activity and physiopathological functions. Biochimie. 2008;90:227–242. doi: 10.1016/j.biochi.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 12.Meyer-Hoffert U, Wiedow O. Neutrophil serine proteases: mediators of innate immune responses. Curr Opin Hematol. 2011;18:19–24. doi: 10.1097/MOH.0b013e32834115d1. [DOI] [PubMed] [Google Scholar]
- 13.Kessenbrock K, Dau T, Jenne DE. Tailor-made inflammation: how neutrophil serine proteases modulate the inflammatory response. J Mol Med. 2011;89:23–28. doi: 10.1007/s00109-010-0677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nauseef WM, Borregaard N. Neutrophils at work. Nat Immunol. 2014;15:602–611. doi: 10.1038/ni.2921. [DOI] [PubMed] [Google Scholar]
- 15.Belaaouaj AA, McCarthy R, Baumann M, Gao Z, Ley TJ, Abraham SN, Shapiro SD. Mice lacking neutrophil elastase reveal impaired host defense against gram negative bacterial sepsis. Nature. 1998;4:615–618. doi: 10.1038/nm0598-615. [DOI] [PubMed] [Google Scholar]
- 16.Belaaouaj AA, Kim KS, Shapiro SD. Degradation of Outer Membrane Protein A in Escherichia coli Killing by Neutrophil Elastase. Science. 2000;289:1185–1187. doi: 10.1126/science.289.5482.1185. [DOI] [PubMed] [Google Scholar]
- 17.Standish AJ, Weiser JN. Human neutrophils kill Streptococcus pneumoniae via serine proteases. J Immunol. 2009;183:2602–2609. doi: 10.4049/jimmunol.0900688. [DOI] [PubMed] [Google Scholar]
- 18.Hahn I, Klaus A, Janze A-K, Steinwede K, Ding N, Bohling J, Brumshagen C, Serrano H, Gauthier F, Paton JC, et al. Cathepsin G and Neutrophil Elastase play critical and nonredundant roles in lung-protective immunity against Streptococcus pneumoniae in mice. Infect Immun. 2011;79:4893–4901. doi: 10.1128/IAI.05593-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van der Windt D, Bootsma HJ, Burghout P, van der Gaast-de Jongh CE, Hermans PWM, van der Flier M. Nonencapsulated Streptococcus pneumoniae resists extracellular human neutrophil elastase- and cathepsin G-mediated killing. FEMS Immunol Med Microbiol. 2012;66:445–448. doi: 10.1111/j.1574-695X.2012.01028.x. [DOI] [PubMed] [Google Scholar]
- 20.MacIvor DM, Shapiro SD, Pham CT, Belaaouaj A, Abraham SN, Ley TJ. Normal neutrophil function in cathepsin G-deficient mice. Blood. 1999;94:4282–4293. [PubMed] [Google Scholar]
- 21.Bangalore N, Travis J, Onunka VC, Pohl J, Shafer WM. Identification of the primary antimicrobial domains in human neutrophil Cathepsin G. J Biol Chem. 1990;265:13584–13588. [PubMed] [Google Scholar]
- 22.Sørensen OE, Follin P, Johnson AH, Calafat J, Tjabringa GS, Hiemstra PS, Borregaard N. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood. 2001;97:3951–3959. doi: 10.1182/blood.v97.12.3951. [DOI] [PubMed] [Google Scholar]
- 23.Nordahl EA, Rydengård V, Nyberg P, Nitsche DP, Mörgelin M, Malmsten M, Björck L, Schmidtchen A. Activation of the complement system generates antibacterial peptides. Proc Natl Acad Sci U S A. 2004;101:16879–16884. doi: 10.1073/pnas.0406678101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papareddy P, Rydengård V, Pasupuleti M, Walse B, Mörgelin M, Chalupka A, Malmsten M, Schmidtchen A. Proteolysis of human thrombin generates novel host defense peptides. PLoS Pathog. 2010;6:e1000857. doi: 10.1371/journal.ppat.1000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papareddy P, Kalle M, Sørensen OE, Lundqvist K, Mörgelin M, Malmsten M, Schmidtchen A. Tissue factor pathway inhibitor 2 is found in skin and its C-terminal region encodes for antibacterial activity. PLoS One. 2012;7:e52772. doi: 10.1371/journal.pone.0052772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papareddy P, Kalle M, Kasetty G, Mörgelin M, Rydengård V, Albiger B, Lundqvist K, Malmsten M, Schmidtchen A. C-terminal peptides of tissue factor pathway inhibitor are novel host defense molecules. J Biol Chem. 2010;285:28387–28398. doi: 10.1074/jbc.M110.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinrauch Y, Drujan D, Shapiro SD, Weiss J, Zychlinsky A. Neutrophil elastase targets virulence factors of enterobacteria. Nature. 2002;417:91–94. doi: 10.1038/417091a. [DOI] [PubMed] [Google Scholar]
- 28.*Hazenbos WLW, Kajihara KK, Vandlen R, Morisaki JH, Lehar SM, Kwakkenbos MJ, Beaumont T, Bakker AQ, Phung Q, Swem LR, et al. Novel Staphylococcal Glycosyltransferases SdgA and SdgB Mediate Immunogenicity and Protection of Virulence-Associated Cell Wall Proteins. PLoS Pathog. 2013;9:e1003653. doi: 10.1371/journal.ppat.1003653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papayannopoulos V, Metzler KD, Hakkim A, Zychlinsky A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol. 2010;191:677–691. doi: 10.1083/jcb.201006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30**.Metzler KD, Goosmann C, Lubojemska A, Zychlinsky A, Papayannopoulos V. A Myeloperoxidase-Containing Complex Regulates Neutrophil Elastase Release and Actin Dynamics during NETosis. Cell Rep. 2014 doi: 10.1016/j.celrep.2014.06.044. This study provides the first insights into the mechanism of NE translocation to the nucleus that is required for NET formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papayannopoulos V, Zychlinsky A. NETs: a new strategy for using old weapons. Trends Immunol. 2009;30:513–521. doi: 10.1016/j.it.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 32.Beiter K, Wartha F, Albiger B, Normark S, Zychlinsky A, Henriques-Normark B. An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr Biol. 2006;16:401–407. doi: 10.1016/j.cub.2006.01.056. [DOI] [PubMed] [Google Scholar]
- 33.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 34.Menegazzi R, Decleva E, Dri P. Killing by neutrophil extracellular traps: fact or folklore? Blood. 2012;119:1214–1216. doi: 10.1182/blood-2011-07-364604. [DOI] [PubMed] [Google Scholar]
- 35.Dubois AV, Gauthier A, Bréa D, Varaigne F, Diot P, Gauthier F, Attucci uthier S. Influence of DNA on the activities and inhibition of neutrophil serine proteases in cystic fibrosis sputum. Am J Respir Cell Mol Biol. 2012;47:80–86. doi: 10.1165/rcmb.2011-0380OC. [DOI] [PubMed] [Google Scholar]
- 36.Yipp BG, Kubes P. NETosis: how vital is it? Blood. 2013;122:2784–2794. doi: 10.1182/blood-2013-04-457671. [DOI] [PubMed] [Google Scholar]
- 37*.Massberg S, Grahl L, von Bruehl M-L, Manukyan D, Pfeiler S, Goosmann C, Brinkmann V, Lorenz M, Bidzhekov K, Khandagale AB, et al. Reciprocal coupling of coagulation and innate immunity via neutrophil serine proteases. Nat Med. 2010;16:887–896. doi: 10.1038/nm.2184. This study shows a new link between NSPs, NETs and the coagulation system. Hazenbos (2013) This study shows that staphylococci actively modify their virulence factors to protect them from degradation by CG. [DOI] [PubMed] [Google Scholar]
- 38.Van de Vosse E, Verhard EM, Tool AJT, de Visser AW, Kuijpers TW, Hiemstra PS, van Dissel JT. Severe congenital neutropenia in a multigenerational family with a novel neutrophil elastase (ELANE) mutation. Ann Hematol. 2011;90:151–158. doi: 10.1007/s00277-010-1056-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bardoel BW, Kenny EF, Sollberger G, Zychlinsky A. The Balancing Act of Neutrophils. Cell Host Microbe. 2014;15:526–536. doi: 10.1016/j.chom.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 40.Lappann M, Danhof S, Guenther F, Olivares-Florez S, Mordhorst IL, Vogel U. In vitro resistance mechanisms of Neisseria meningitidis against neutrophil extracellular traps. Mol Microbiol. 2013;89:433–449. doi: 10.1111/mmi.12288. [DOI] [PubMed] [Google Scholar]
- 41.Eggers CT, Murray IA, Delmar VA, Day AG, Craik CS. The periplasmic serine protease inhibitor ecotin protects bacteria against neutrophil elastase. Biochem J. 2004;379:107–118. doi: 10.1042/BJ20031790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doan N, Gettins PGW. alpha-Macroglobulins are present in some gram-negative bacteria: characterization of the alpha2-macroglobulin from Escherichia coli. J Biol Chem. 2008;283:28747–28756. doi: 10.1074/jbc.M803127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43*.Robert-Genthon M, Casabona MG, Neves D, Couté Y, Cicéron F, Elsen S, Dessen A, Attrée I. Unique Features of a Pseudomonas aeruginosa α2-Macroglobulin Homolog. MBio. 2013;4:e00309–13. doi: 10.1128/mBio.00309-13. This study shows that Pseudomonas aeruginosa produces an inhibitor of NE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Budd A, Blandin S, Levashina EA, Gibson TJ. Bacterial alpha2-macroglobulins: colonization factors acquired by horizontal gene transfer from the metazoan genome? Genome Biol. 2004;5:R38. doi: 10.1186/gb-2004-5-6-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neves D, Estrozi LF, Job V, Gabel F, Schoehn G, Dessen A. Conformational states of a bacterial α2-macroglobulin resemble those of human complement C3. PLoS One. 2012;7:e35384. doi: 10.1371/journal.pone.0035384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46**.Stapels DAC, von Köckritz-blickwede M, Ramyar KX, Bischoff M, Milder FJ, Ruyken M, Eisenbeis J, McWhorter WJ, Herrmann M, van Kessel KPM, et al. Staphylococcus aureus secretes a unique class of neutrophil serine protease inhibitors. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1407616111. [accepted, not yet published]. This study shows that Staphylococcus aureus secretes three specific inhibitors of the NSPs, underlining the importance of NSPs in antibacterial host defenses against staphylococci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujinaga M, Chernaia MM, Halenbeck R, Koths K, James MN. The crystal structure of PR3, a neutrophil serine proteinase antigen of Wegener’s granulomatosis antibodies. J Mol Biol. 1996;261:267–278. doi: 10.1006/jmbi.1996.0458. [DOI] [PubMed] [Google Scholar]
- 48.Palazzolo-Ballance AM, Reniere ML, Braughton KR, Sturdevant DE, Otto M, Kreiswirth BN, Skaar EP, DeLeo FR. Neutrophil Microbicides Induce a Pathogen Survival Response in Community-Associated Methicillin-Resistant Staphylococcus aureus. J Immunol. 2007;180:500–509. doi: 10.4049/jimmunol.180.1.500. [DOI] [PubMed] [Google Scholar]