Abstract
Consumer use of herbal and dietary supplements has recently grown in the United States and, with increased use, reports of rare adverse reactions have emerged. One such supplement is green tea extract, containing the polyphenol epigallocatechin gallate (EGCG), which has been shown to be hepatotoxic at high doses in animal models. The Drug-Induced Liver Injury Network has identified multiple patients who have experienced liver injury ascribed to green tea extract consumption and the relationship to dose has not been straightforward, indicating that differences in sensitivity may contribute to the adverse response in susceptible people. The Diversity Outbred (DO), a genetically heterogeneous mouse population, provides a potential platform for study of interindividual toxicity responses to green tea extract. Within the DO population, an equal exposure to EGCG (50 mg/kg; daily for three days) was found to be tolerated in the majority of mice; however, a small fraction of the animals (16%; 43/272) exhibited severe hepatotoxicity (10–86.8% liver necrosis) that is analogous to the clinical cases. The data indicate that the DO mice may provide a platform for informing risk of rare, adverse reactions that may occur in consumer populations upon ingestion of concentrated herbal products.
Keywords: Green tea, Epigallocatechin gallate, Hepatotoxicity, Population variability, Herbal, Diversity outbred
1. Introduction
In the United States, there is growing consumer interest in the use of herbal and dietary supplements (HDS), with the American Botanical Council estimating over $5 billion in total sales in 2009 (Cavaliere et al., 2010). A recent U.S. Food and Drug Administration survey found in 2002 that 72% of the respondents used supplements, of which half were herbals (Navarro and Seeff, 2013; Timbo et al., 2006). In part, the rise in HDS sales is due to a common public perception that these products are safe and effective. The current regulatory framework for natural products differs from that of conventional pharmaceuticals in that the Food and Drug Administration (FDA) lacks the authority to approve herbal products prior to marketing. Thus, nonclinical and clinical assessments of safety and efficacy are not required to bring herbal products into the marketplace and safety concerns are not known until post-marketing surveillance (Ehrenpreis et al., 2013; Navarro and Seeff, 2013).
The Drug-Induced Liver Injury Network (DILIN) involves eight academic centers in the U.S. that identify cases of liver injury induced by conventional pharmaceutical drugs or HDS products. Causality assessment for the implication of HDS products in DILI is often complicated by co-exposure with other herbals or pharmaceutical products, determination of latency time, and exclusion of other common causes of hepatic injury (e.g. viral hepatitis). Despite these limitations, the DILIN Study Group has established a scoring system to assess causality in each case (Navarro et al., 2013). Between 2003 and 2011, 679 patients with liver injury were enrolled in the DILIN study, of which 109 cases (16%) were attributed to HDS products (Navarro and Seeff, 2013).
Within the DILIN and elsewhere, multiple cases of herbal-induced liver injury have been specifically attributed to green tea extract (GTE) (Lambert et al., 2010; Mazzanti et al., 2009; Navarro et al., 2013; Teschke et al., 2012). Concentrated GTE or its constituent components are commonly included in products marketed for weight reduction and body building (Panza et al., 2008). Consumer interest in GTE has increased in recent years, owing to several epidemiologic and basic research studies that have demonstrated the benefits of green tea on cardiovascular disease, neurodegenerative disorders and in lowering cancer incidence.
Notable examples of previous formulations containing GTE that lead to cases of liver injury are Hydroxycut™ (Dara et al., 2008) and X-elles™ (Teschke et al., 2012). Patients with hepatotoxicity attributed to GTE present with characteristic hepatocellular injury and elevated aminotransferase levels (Bjornsson and Olsson, 2007; Bonkovsky, 2006; Jimenez-Saenz and Martinez-Sanchez Mdel, 2006; Shim and Saab, 2009). The assumption that these cases of hepatotoxicity were precipitated by a GTE supplement was supported by the fact that liver injury ceased when product use was discontinued (Bonkovsky, 2006; Jimenez-Saenz and Martinez-Sanchez Mdel, 2006; Mazzanti et al., 2009). In at least one instance, causality was established when hepatotoxicity reemerged during resumed supplement utilization by the patient (Mazzanti et al., 2009). In one DILIN-sponsored study, 40% HDS products associated with cases of herbal-induced injury were found to contain components of GTE when chemically analyzed, even when not indicated among the ingredients on the product label (Navarro et al., 2013). While a finding of unreported inclusion of GTE in the product does not establish causality, the finding suggests that cases of GTE-induced liver injury may be underreported when a supplement not known to contain GTE was implicated as the causal factor.
The hepatotoxicity potential of GTE has been attributed to epigallocatechin gallate (EGCG), the most abundant polyphenol in green tea (Lambert et al., 2010). While EGCG has proven to be a dose-dependent hepatotoxin in mice, the relationship between catechin dosage and clinically important liver injury may be less clear (Lambert et al., 2010). In the DILIN-sponsored study investigating GTE-induced liver injury in 16 patients, there was no correlation found between daily or total catechin dose and causality score, peak liver enzyme values, or disease severity (the highest dose taken was 40 mg/kg) (Navarro et al., 2013). Because of the rarity of cases, the hepatotoxic threshold dose for consumer use of GTE or its catechin components has not been clearly established. A 2006 review of reported cases found that liver injury could result from as little as 5.9 g over 5 days to as much as 240 g over 120 days (Bonkovsky, 2006). The lack of an observed dose–response relationship may be mediated by several factors; these include co-ingested herbal or conventional pharmaceutical products, co-morbidities, lifestyle factors, or inherent factors such as genetic variation. Current data do not support a gender bias in the frequency of DILI due to green tea extract (Navarro et al., 2013).
Occurrences of GTE-induced hepatotoxicity are observed in only rare, susceptible patients suggesting that genetic variation may contribute to inter-individual sensitivity to green tea extract. While a variety of extrinsic and intrinsic factors may influence individual susceptibility to an adverse reaction, recent evidence suggests that genetic host factors may play a role in the injury outcome and relative risk of toxicity for several pharmaceutical drugs (Hughes et al., 2008; Kaniwa and Saito, 2013; Monshi et al., 2013). In the present study, we hypothesized that utilizing a mouse population-based approach to model the hepatotoxicity of EGCG may clarify whether genetic variation affects the liver injury outcome. Previous in vitro data suggest that mice may be a better comparator species to humans in terms of the biotransformation of EGCG than the rat. Methylation, sulfation, and glucuronidation of EGCG in liver cytosol were more similar between human and mouse than to rat (Lambert et al., 2003). In vivo studies indicate that humans and mice have a more similar high EGCG bioavailability than rats (Lambert et al., 2003).
We have previously demonstrated that a genetically diverse mouse population offers advantages to modeling clinical variability to liver or kidney toxicities induced by acetaminophen and the drug candidate DB289 (Harrill et al., 2009, 2012). In the current study, we utilized Diversity Outbred (DO) mice, a newly derived heterogeneous stock of mice developed to maximize genetic variation within the population. The DO was initially seeded via 144 recombinant inbred lines generated through the Collaborative Cross (CC) (Churchill et al., 2004; Threadgill et al., 2002). A rationally designed random breeding scheme is utilized to maintain an active breeding colony of 175 pairs (Churchill et al., 2012). Both the CC inbred lines and the DO stock contain approximately 45 million segregating single nucleotide polymorphisms (SNPs) and structural genetic variations (Roberts et al., 2007). Importantly, due to the randomization of alleles in the offspring, the frequency of minor alleles averages 12.5%, allowing for detection of “rare” susceptible individuals with smaller sample sizes than would be needed in a comparable human population (Svenson et al., 2012). Following the identification of a genomic interval of interest in the DO population, CC strains, or the F1 progeny of two CC strains, can be utilized to reproduce loci of interest for downstream mechanistic analyses.
2. Materials and methods
2.1. Animals
Male DO mice (approximately 8 weeks in age, generations 7 and 8 for a dose-ranging study and expanded pharmacogenomics study respectively) were purchased from The Jackson Laboratory (Bar Harbor, ME) and housed in polycarbonate cages on a 12-hour (h) light–dark cycle at North Carolina State University. Mice were provided Purina LabDiet 5058 rodent chow ad libitum, with the exception of an approximately 18 h fast immediately prior to euthanasia. Reverse osmosis water was available ad libitum. All animal use was approved by the North Carolina State University Institutional Animal Care and Use Committee.
2.2. EGCG treatment and tissue collection
Mice were acclimated for at least one week prior to experimentation. One day prior to the start of dosing, tail biopsies were performed and blood was collected from the tail vein for measurement of alanine aminotransferase (ALT). Tissue and serum were stored for genotyping and baseline serum ALT measurements, respectively.
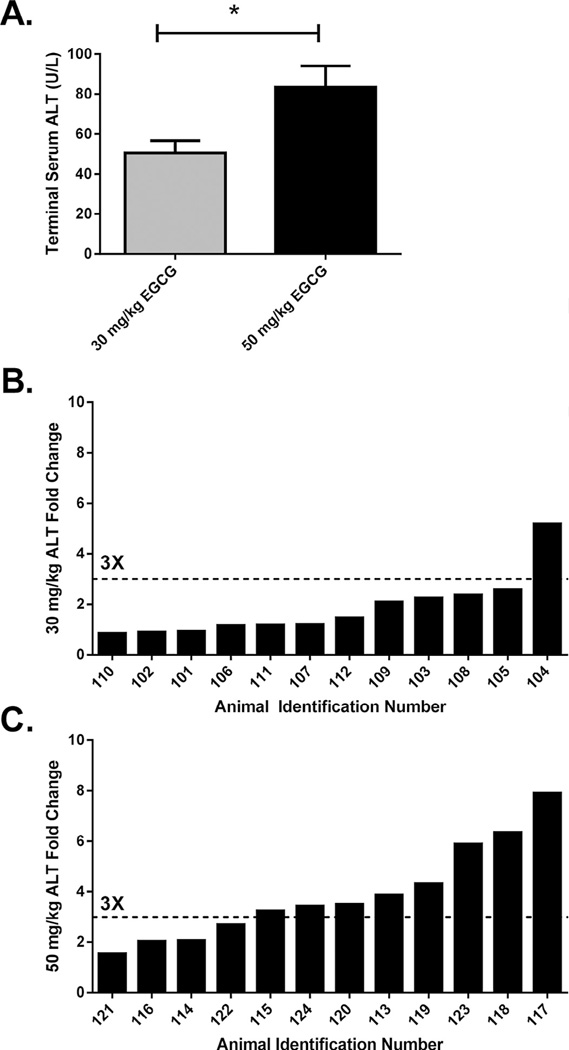
A small subset of animals was utilized for an initial dose response analysis to determine the optimal dose and duration of treatment with EGCG. Beginning on the day after the blood draw, mice received daily i.p. administrations of either 30 mg/kg/day (N = 12/dosing regimen) or 50 mg/kg/day (N = 12/dosing regimen) EGCG (Cayman Chemical, Ann Arbor, MI) dissolved in sterile 0.9% saline at a volume of 8 mL/kg for either three or seven consecutive days. Time of dosing was standardized to 0900 ±1 h. From this pilot study, it was determined that a range of hepatic injury (based on elevations in serum ALT between pre-dose and terminal measurements) was observable following three daily administrations of EGCG (Fig. 3).
Fig. 3.
(A) Terminal serum ALT concentrations are shown for individual DO mice treated with 30 mg/kg/day or 50 mg/kg/day EGCG for three consecutive days. Error bars represent the SEM. *P < 0.05. ALT fold change (terminal to baseline) in DO animals treated with (B) 30 mg/kg/day EGCG or (C) or 50 mg/kg/day EGCG is shown with the dotted line indicating a three-fold change.
Animals in the expanded genomics study were administered 50 mg/kg/day EGCG for three consecutive days only. Due to the large size of the cohort, animals were administered compound in groups (25 animals at a time; n = 2 Vehicle- and n = 23 EGCG-treated) and ranged in age from 10 to 26 weeks at the initiation of dosing. Beginning the following day, mice were dosed at 0900 ± 1 h, once daily (i.p.) with 50 mg/kg EGCG dissolved in sterile 0.9% saline (8 mL/kg; n = 272) or saline alone (n = 25). Animals were euthanized 24 h after the final dose following an 18 h fast via CO2 inhalation and exsanguination via cardiac puncture into serum separation tubes (Fisher Scientific, Pittsburgh, PA). Serum was isolated from pre-dose and terminal blood via centrifugation at room temperature for 20 minutes at maximum speed (13.2 × 1000 rpm). Sections were taken from the left liver lobes for histological analysis.
2.3. Clinical chemistry
Isolated baseline and terminal serum was utilized for analysis of ALT. Concentrations were quantified using the Infinity ALT Liquid Stable Reagent (Thermo Scientific, Hudson, NH) on a SpectraMax microtiter plate reader (Molecular Devices, Sunnyvale, CA) following the manufacturer’s protocol.
2.4. Histopathology and percent necrosis measurement
Liver tissue was paraffin-embedded, cut into 5 µm sections, and stained with hematoxylin and eosin (H&E). Stained liver sections were used for quantification of liver necrosis percentages via quantitative stereology. A centralized picture of the left liver lobe was taken at 200× magnification and an unbiased point counting technique was utilized to evaluate percent liver necrosis as previously described (Harrill et al., 2009; Mouton, 2002). Briefly, images were overlaid with a grid containing equally spaced points. At each point that fell within the tissue (excluding points that fell within blood vessels) it was visually determined whether the cell was necrotic vs. normal based on histological appearance. The percentage of liver necrosis was calculated as the percentage of “necrotic” points among the total number of points included in the count.
2.5. TUNEL assay and image analysis
The extent of cell death in the liver sections of control and EGCG-treated mice was evaluated by terminal deoxynucleotidyl transferase (TdT) mediated dUTP nick end labeling (TUNEL) of DNA fragments (ApopTag Peroxidase In Situ Apoptosis Detection Kit, Millipore Corporation, Billerica, MA). TUNEL-stained liver sections were scanned and digital images were obtained with Aperio Scanscope System (Aperio Technologies, Inc., Vista, CA). Due to the presence of necrotic areas in the livers of some treated mice it was impossible to count individual apoptotic bodies in these liver sections. Therefore, the proportion of TUNEL-stained area (=apoptosis + necrosis) was evaluated for each animal using a Positive Pixel Count Algorithm (Aperio). The percent of TUNEL-positive area was calculated in four randomly chosen regions (4 × 1 mm2) of two liver sections present on each slide. Animals #319, 320, 321, 323, 324, 325, 326, 328, and 329 were excluded from the image analysis due to nonspecific staining of stromal cells.
2.6. DNA isolation and SNP genotyping in mice
DNA was extracted from biopsied tail samples using the Qiagen DNeasy Blood and Tissue Kit following the manufacturer’s instructions (Qiagen, Valencia, CA). SNP genotyping was performed utilizing the Mouse Universal Genotyping Array (MUGA; GeneSeek, Lincoln, NE) an Illumina Infinium Platform array with 7851 evenly spaced SNP markers.
2.7. Genome reconstruction and QTL mapping in mice
DO genome reconstruction and QTL mapping were carried out as previously described (Svenson et al., 2012). Briefly, DO genomes were reconstructed using a hidden Markov model in which the MUGA genome intensity values and marker spacing served as input and the output consisted of unphased genotype probabilities. A programming algorithm was then used to calculate the maximum-likelihood founder assignment for each chromosome. Following reconstruction of haplotypes, QTL mapping for association with ALT fold change was conducted in EGCG-treated animals with date of necropsy as a covariate to control for batch effects. Phenotype data were rankZ transformed and an additive model with kinship correction was fit at each locus. The model produces estimates of the eight founder allele contributions at each marker. The mapping statistic was the log of the odds ratio (LOD) and the most significant QTL was utilized for further analysis.
2.8. Mouse phenotyping data analysis
Unless stated otherwise, all statistical analyses were performed using GraphPad Prism statistical software (GraphPad Software Inc., La Jolla, CA) and significance was determined using a Student’s t test and P < 0.05. A paired t test was used when comparing pre-dose and post-dose ALT levels within the same cohort of mice. Correlation analysis was performed using the Spearman test because the analyzed data were not normally distributed, as assessed by the D’Agostino and Pearson omnibus normality test (P > 0.05; GraphPad Prism).
2.9. Clinical study participants
Cases (n = 26) were recruited from the Drug-Induced Liver Injury Network (DILIN) between August 2004 and November 2011 from eight DILIN clinical sites (Fontana et al., 2009). Of these, we utilized data from 15 cases that were of European ancestry; of these, 9 were male and 6 were female. All participants provided written informed consent, and each study was approved by the appropriate institutional review boards. Details of recruitment and the inclusion and exclusion criteria for these networks have been published previously (Lucena et al., 2011). Genotyped control individuals available at the Duke Center for Human Genome Variation (n = 4432) were used for comparison.
2.10. Clinical cohort genotyping
DNA was prepared as described previously (Lucena et al., 2011). Genome-wide exome-centric genotyping of EGCG-induced hepatic injury cases and controls was carried out by the Center for Human Genome Variation, Duke University. All patients were genotyped using the Illumina Human Exome BeadChip v1.1 (Illumina Inc., San Diego, CA, USA), which contains 245,907 markers, 428 of which were located in human orthologs of genes implicated by the mouse QTL studies, and passed variant-level quality filters. The 15 EGCG-cases and 4432 controls analyzed passed quality control carried out as described previously (Ge et al., 2009).
2.11. Clinical SNP variant analysis
A logistic regression model was fitted to the data in order to assess association between each of the 428 SNPs across the 46 genes with QTL evidence and the outcome. To control for gender effect and population structure, gender and the top four principal components were included as covariates in addition to SNPs. SNPs were determined to have a trend association where P < 0.05 without multiple test correction.
3. Results
3.1. EGCG-induced liver injury varies across a genetically diverse population
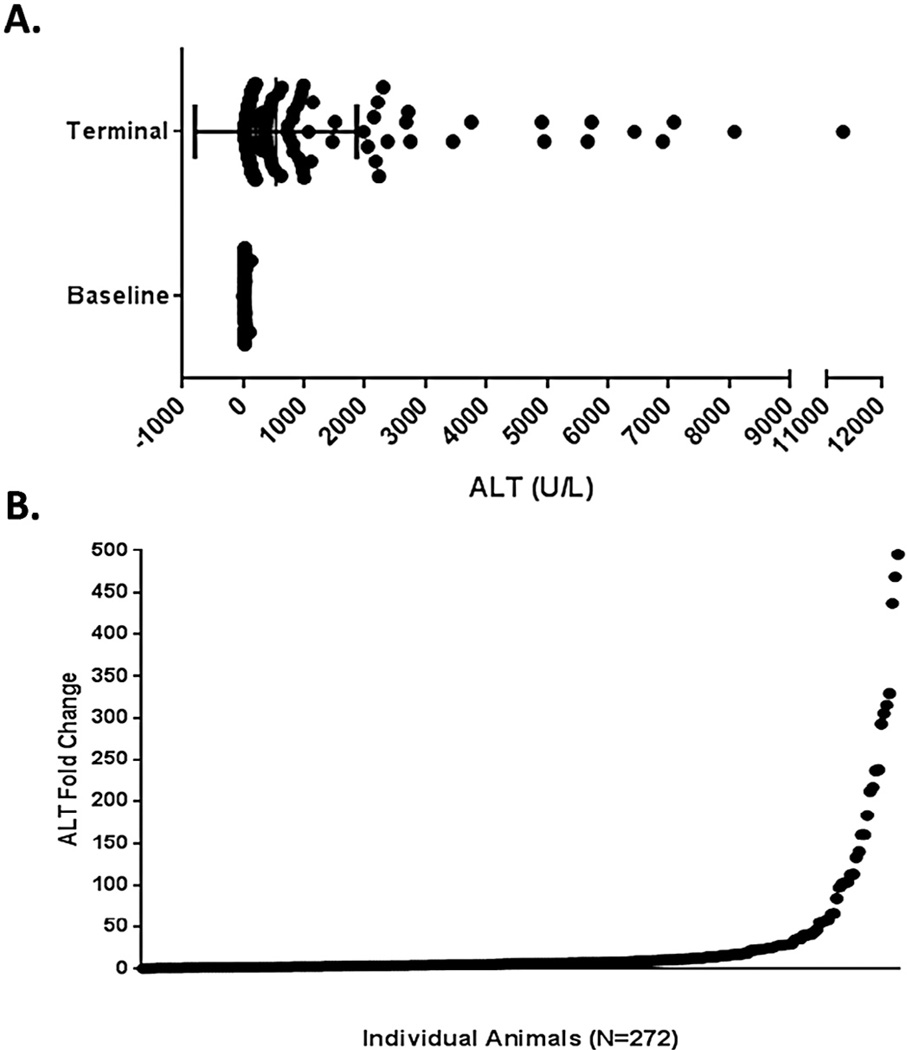
To determine whether genetic background affects sensitivity to EGCG-induced liver injury, a large population of DO mice was exposed to 50 mg/kg/day for three days. The 50 mg/kg dose was chosen because it had been reported in the literature to cause hepatotoxicity in mice (Goodin et al., 2006). ALT was measured in serum for each individual at baseline and at necropsy. Baseline ALT for the cohort was measured as 24.36 ± 0.78, while terminal ALT was significantly elevated as a group at 537.8 ± 80.84 (mean ± SEM for both measurements; P < 0.05; Fig. 1A). There was a high degree of variability between individual ALT values post dosing – the coefficient of variation for the terminal ALT values was 248.01%. ALT fold change measurements were calculated for each animal as the terminal value divided by the baseline value derived from blood sampled prior to the start of dosing (Fig. 1B). The data demonstrated significant phenotypic variation in response to EGCG administration. ALT elevations from baseline ranged from 0.15 to 495.5 fold (27.15 ± 4.20 fold, mean ± SEM).
Fig. 1.
Baseline and terminal ALT concentrations (A) and resulting fold change (B, terminal concentration to pre-dose concentration) were quantified in serum samples obtained from DO mice treated with 50 mg/kg/day EGCG for three consecutive days. Each black dot represents an individual EGCG-treated animal. Bars shown in (A) represent the mean ± SEM ALT concentration and significance between groups was P < 0.05.
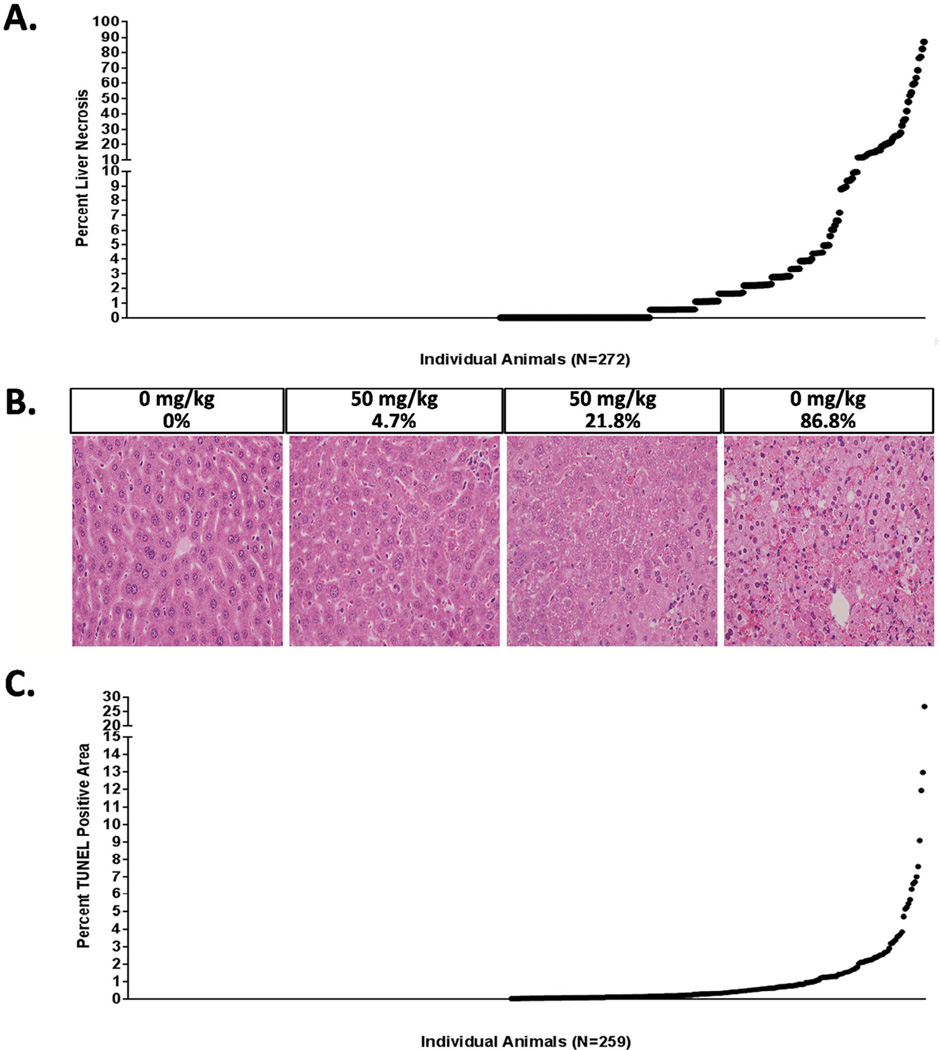
Histopathological findings corroborated the ALT data and were typified by hepatocellular necrosis. Liver necrosis varied widely between DO animals treated with EGCG (0% to 86.8%; 6.47 ± 0.82%, mean ± SEM) (Fig. 2A). No observed liver necrosis occurred in 35% (100/272) of the mice tested. By comparison, 49% of the mice exhibited a mild degree of injury, ranging from 0.55 to 9.94% necrosis (133/272). A small percentage of the animals tested (16%; 43/272) exhibited severe liver injury as defined by greater than 10% liver necrosis. Examination of H&E stained liver sections showed variable degrees of hepatic necrosis, inflammation, hypertrophy, and hemorrhage (Fig. 2B). ALT fold change correlated with liver necrosis in the DO mice exposed to EGCG (ρ = 0.70, P < 0.05).
Fig. 2.
(A) Percent liver necrosis was scored for each animal using an unbiased point scoring method. (B) Representative photomicrographs are shown for the left liver lobe stained with H&E sections at 200× magnification. The dose administered and the percent liver necrosis score are indicated above each image. (C) Percent TUNEL positive area is shown for each animal from the left liver lobe. In panels (A) and (C), each dot represents the endpoint value for an individual DO mouse treated with 50 mg/kg EGCG.
3.2. TUNEL positive staining correlated with liver necrosis
To investigate the extent to which DNA strand breaks contribute to downstream EGCG-induced liver necrosis in genetically sensitive DO mice, TUNEL staining was performed. Although this technique typically detects the presence of apoptosis, DNA strand breaks and positive TUNEL staining is also observed following necrosis (Grasl-Kraupp et al., 1995). Because immunohistochemical staining failed on a small subset of the slides, the analysis was based on 259 DO mice. Percent positive area ranged from 0.01% to 26.7% (1.11% ± 0.15%, mean ± SEM; Fig. 2C). TUNEL staining was observed in less than 0.5% of the stained area in over half of the DO mice tested (58% of the population; 149/259) and in less than 1% of the area in the majority of DO mice (73% of mice; 189/259). Consistent with the ALT and necrosis data, TUNEL staining indicated a genetically sensitive subset of mice with a 1.01–26.7% TUNEL positive area (27% of DO mice, 70/259). Percent TUNEL staining correlated with both percent liver necrosis and ALT fold change in the DO mice exposed to EGCG (ρ = 0.75, P < 0.05 and ρ = 0.70, P < 0.05) respectively.
3.3. Effect of EGCG dose within diversity outbred mice
Owing to the inherent genetic variation of the Diversity Outbred and the potential for inclusion of uniquely sensitive individuals, a dose response experiment was conducted to determine whether a lower dose of EGCG could elicit a measurable hepatotoxic effect. Two dose levels were tested: 30 mg/kg/day (N = 12) and 50 mg/kg/day (N = 12). As expected, DO mice treated with 50 mg/kg EGCG exhibited an ALT elevation that, as a group, was greater than that of the lower dosed animals (30 mg/kg EGCG; P < 0.05; Fig 3A). Of the animals tested in this phase, 66.7% (8/12) exhibited an ALT elevation that exceeded three-fold higher than the respective pre-dose values (Fig. 3B).
While as a group, the data were not statistically significant, one animal dosed with 30 mg/kg/day (#104) exhibited an ALT elevation greater than three-fold baseline (fold change of 5.2; Fig. 3C). As a group, the absolute ALT values following treatment ranged from 28 U/L to 86 U/L (50 ± 6 U/L, mean ± SEM).
3.4. Identification of gene regions associated with EGCG-dependent ALT elevations
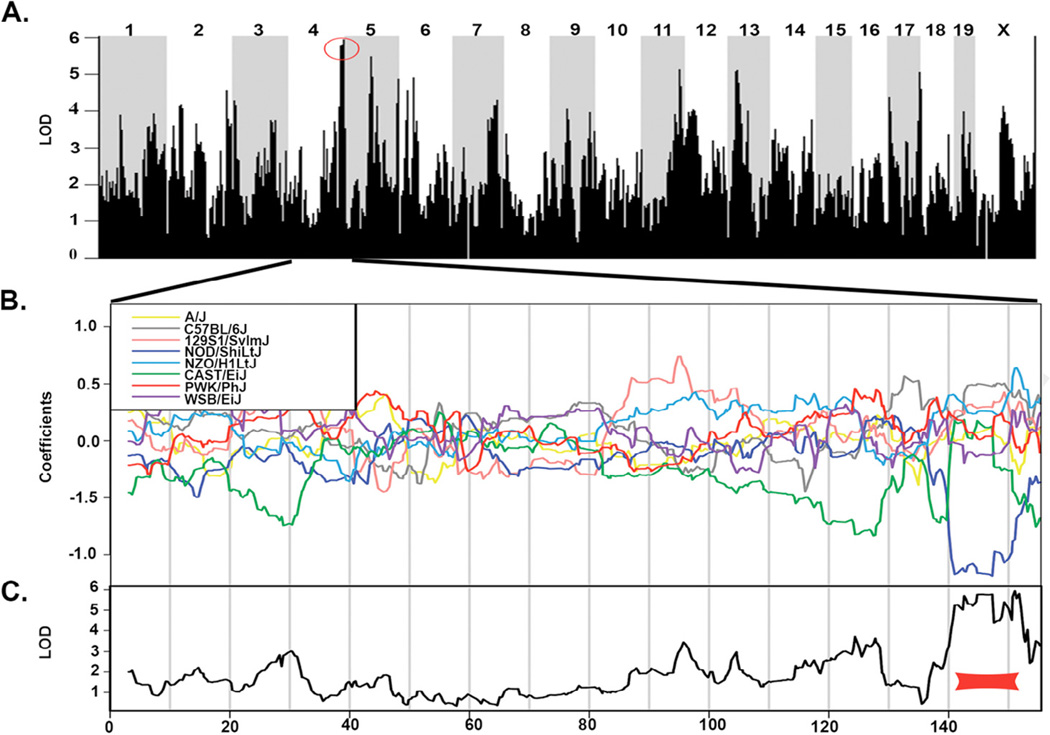
The significant toxicity variation observed in the DO population supported the hypothesis that genetic variability may potentiate the adverse liver response to EGCG. QTL mapping was conducted to identify genomic loci that contributed to the hepatotoxic outcome. Because ALT measurements are used clinically to confirm liver injury and would provide a translational marker, ALT fold changes in EGCG-treated mice were mapped to the genome. QTL analysis identified a peak on chromosome (Chr) 4 as having the highest association score with EGCG-induced ALT fold changes with a LOD score of 5.94 (Fig. 4A). This QTL was the sole peak that surpassed the LOD significance threshold of 5.85. Among the SNPs with the highest LOD scores, 22 of the top 25 (88%) fell within the QTL identified on Chr 4. A coefficient effect plot was constructed to determine the relative effect of genetic founder alleles at the locus of interest on Chr 4. The coefficient effect plot for the gene region on Chr 4 indicated that allelic variation inherited from seven of the eight founder strains conferred a higher degree of injury, while alleles inherited from the NOD/ShiLtJ founder strain conferred resistance to the injury (Fig. 4B). The most significant LOD scores for the genomic association mapping fell within an approximately 9 Mb region, spanning 142.591722– 151.777278 Mb on Chr 4 (Fig. 4C). Within this region, 49 genes were determined to harbor sequence variants (SNPs, insertions, or deletions) that are exclusive to the NOD/ShiLtJ haplotype (as identified by the Sanger Mouse Genomes Project, mouse assembly NCBI m37; Supplementary Table S1). Of these, 46 genes had human orthologs that could be examined for variation that might underlie the idiosyncratic effect of EGCG observed in the clinic.
Fig. 4.
(A) A Manhattan plot is shown for significance scores resulting from QTL mapping of ALT fold change in DO mice dosed with EGCG. The red circle indicates a QTL identified on Chr 4. (B) The coefficient effect plot indicates the contribution of each cofounder haplotype to the QTL support interval. (C) Expanded Manhattan plot is shown for Chr 4. The red bar shows the QTL falling between 142.591722 and 151.777278 Mb. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5. Mouse genes associated with EGCG-induced liver injury have suggestive associations in human EGCG-induced DILI cases
To evaluate the human relevance of candidate susceptibility genes identified using the DO, polymorphisms in the 46 orthologous genes identified on mouse Chr 4 were examined. The data were derived from exome-centric SNP genotyping analyses performed for 15 patients enrolled by the DILIN who were judged to have experienced GTE-induced hepatotoxicity. SNP variants for the candidate genes (428 total SNPs) within the QTL support interval identified in the DO were analyzed against gene frequencies in 4432 control individuals (Supplementary Table S2).
Within the DILIN patients, suggestive associations (P < 0.05 before multiple test correction) were observed between genotype and EGCG hepatotoxicity within three SNPs, representing three genes: mitofusin 2 (MFN2, P = 0.0067), period circadian clock 3 (PER3, p = 0.004937), and vacuolar protein sorting 13 d (VPS13D, P = 0.043) (Table 1).
Table 1.
EGCG hepatotoxicity risk alleles in human clinical cohorts.
| Gene symbol | SNP (Array) | Gene name | Chr | Position (bp) | P value | Risk/Protective allele | Effect |
|---|---|---|---|---|---|---|---|
| PER3 | exm10762 | Period circadian clock 3 | 1 | 7 887 234 | 0.004937 | T/C | Missense (R/W) |
| MFN2 | exm15928 | Mitofusin 2 | 1 | 12 069 692 | 0.0067 | A/G | Missense (I/V) |
| VPS13D | exm16480 | Vacuolar protein sorting 13 homolog D (S. cerevisiae) |
1 | 12 343 493 | 0.043064 | A/T | Missense (R/S) |
4. Discussion
The individual mice within the DO population displayed highly variable hepatotoxicity following EGCG administration. Surprisingly, while 65% of the DO mice developed some degree of hepatic injury, the majority of these animals experienced only mild liver damage, comprising less than 10% necrosis. The implication of this finding is that a potentially larger proportion of consumers taking GTE-containing supplements may develop mild hepatotoxicity that goes undiagnosed due to a lack of severity which would necessitate clinical recognition. It should be noted, however, that the EGCG dose utilized in this study is approximately four-to-five times greater than the maximum recommended human daily dose (i.e. 750– 856.8 mg/day or up to 12.2 mg/kg for a 70 kg person (Hsu et al., 2011)). While phase II biotransformation enzyme activity has been reported to be similar between mice and humans, plasma EGCG is more abundant in the conjugated form in mice whereas plasma EGCG in humans is largely in the free form (Lambert et al., 2003). In addition, while one animal administered 30 mg/kg EGCG in this study exhibited an ALT elevation greater than three times its baseline value, the potential hepatotoxic effects across a larger population of DO mice at lower doses were not investigated.
The diverse genetic backgrounds of the DO allowed for the identification of a small subset of animals that were highly sensitive to EGCG-induced hepatotoxicity, exhibiting greater than 10% liver necrosis. The livers of sensitive animals exhibited inflammatory infiltrates, consistent with clinical findings observed when liver biopsies were conducted (Bjornsson and Olsson, 2007; Bonkovsky, 2006; Mazzanti et al., 2009). The finding that 16% of the DO mice within the population developed severe liver toxicity despite an equivalent EGCG dose may imply that the DO has the potential to be a useful tool for population-based safety assessment of xenobiotic agents in general. Specifically, these data suggest that the DO may be an effective animal model for the study of rare toxicity resulting from EGCG-containing supplements.
Surprisingly, the significance score of the QTL support interval for ALT elevations in the mice was relatively low (peak LOD score 5.94; P ≤ 0.63) as compared to other traits that have been mapped using the DO population, including behavioral traits and plasma cholesterol levels (Churchill et al., 2012; Logan et al., 2013). The reasons for this finding are unclear, but there are several possible contributing factors, including: (1) use of ALT as a mapping phenotype for which levels may be affected by inter-individual differences in hepatic expression or clearance half-life and (2) the complexity of the hepatic response to EGCG, which may involve multiple interacting genes of small effect. Interestingly, the founder haplotype of the largest effect in the QTL interval indicated that seven of the founder strains contributed allelic variation that was associated with ALT elevations, while the NOD/ShiLtJ founder alleles contributed to resistance. Further experimental work is needed to elucidate the mechanisms by which variants in this region are protective.
The genomic support interval contained 46 genes that contained both genomic variation that is specific to the NOD/ShiLtJ background and a human ortholog, making these genes particularly interesting for identifying toxic pathways translatable to humans. Of these, three genes contained suggestive variants in clinical cases of EGCG-induced hepatotoxicity, although these associations failed to pass multiple test correction. The weak associations observed in this population likely reflect the small patient cohort available (n = 15 cases) for which there was no sufficient statistical power to detect a robust association. Another key consideration for the interpretation of the data is that it is unknown whether the control subjects in the human genetic study had been exposed to green tea containing supplements. An important future direction would be to examine EGCG-induced liver injury in both cases and in controls that had ingested similar amounts of green tea supplement, allowing for investigation of genetic factors that modulate the toxic response. In addition, repeating these studies in DO mice using the oral ingestion route would ensure a greater degree of translation for the dose responsive injury data to human populations.
Based on what is known in the literature about the function of the candidate genes, one of the three suggestive variants identified, located in the MFN2 (Mitofusin 2) gene, represents an interesting candidate gene for follow up study. MFN2 is a mitochondrial membrane protein that participates in regulation and maintenance of the mitochondrial network. While the association of EGCG-induced hepatic injury with a variation in MFN2 failed to pass multiple test correction, suggesting only a modest contribution of this protein to the phenotype, it can be speculated how variation in this gene may be advantageous. Loss of Mfn2 in mouse embryonic fibroblasts results in reduction of mitochondrial fusion (Chen et al., 2003) and reduction of mitophagy (Chen and Dorn, 2013); both are processes that prevent widespread mitochondrial damage and cellular death (Han et al., 2013). Some of the variation in toxicity observed in human populations may be explained by a synergistically toxic effect of mitochondrial stress and EGCG ingestion. One example of such an interaction is recent work demonstrating that green tea extract can potentiate acetaminophen-induced liver injury in mice and that this may result from effects on mitochondrial function (Lu et al., 2013; Salminen et al., 2012). Furthermore, EGCG has been shown to inhibit mitochondrial respiratory complexes in swelling, but not in healthy mitochondria, suggesting that environmental stressors may potentiate EGCG-induced hepatocellular injury (Weng et al., 2013). Importantly, Mfn2 has been shown to promote apoptosis by activating the intrinsic mitochondrial pathway (Papanicolaou et al., 2011) and liver necrosis observed in the present study was significantly correlated with TUNEL staining (a marker of DNA strand breaks which can accompany both apoptosis and necrosis). Interestingly, genomic mapping attempted using the TUNEL data did not yield any significant QTL. Further experimentation is necessary to clarify the contribution of the mitochondria to EGCG-induced hepatotoxicity.
In summary, we demonstrated that the DO mouse resource was successful in modeling the interindividual hepatic variability that occurs in the human population following exposure to EGCG-containing herbal supplements. The data suggest that the DO may have utility to identify subpopulations that are resistant or sensitive to the toxic effects of drugs or chemicals, thereby enabling mechanistic study. While additional examples are needed, the findings suggest that the DO population may provide an alternative model for human variability in toxicities.
Supplementary Material
Acknowledgements
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) at the National Institute of Health [1RC1DK087510-01 to P.B.W. and D.W.T] and the Drug Induced Liver Injury Network [DKUO1 065201-10 to P.B.W.]. The information in these materials is not a formal dissemination of information by FDA and does not represent agency position or policy.
Appendix: Supplementary material
Supplementary data to this article can be found online at doi:10.1016/j.fct.2014.11.008.
Footnotes
Conflict of interest
The authors declare that there are no conflicts of interest.
Transparency document
The Transparency document associated with this article can be found in the online version.
References
- Bjornsson E, Olsson R. Serious adverse liver reactions associated with herbal weight-loss supplements. J. Hepatol. 2007;47(2):295–297. doi: 10.1016/j.jhep.2007.05.010. author reply 7–8. [DOI] [PubMed] [Google Scholar]
- Bonkovsky HL. Hepatotoxicity associated with supplements containing Chinese green tea (Camellia sinensis) Ann. Intern. Med. 2006;144(1):68–71. doi: 10.7326/0003-4819-144-1-200601030-00020. [DOI] [PubMed] [Google Scholar]
- Cavaliere C, Rea P, Lynch ME, Blumenthal M. Herbal supplement sales rise in all channels in 2009. HerbalGram. 2010;86:62–65. [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003;160(2):189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340(6131):471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 2004;36(11):1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Gatti DM, Munger SC, Svenson KL. The Diversity Outbred mouse population. Mamm. Genome. 2012;23(9–10):713–718. doi: 10.1007/s00335-012-9414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dara L, Hewett J, Lim JK. Hydroxycut hepatotoxicity: a case series and review of liver toxicity from herbal weight loss supplements. World. J. Gastroenterol. 2008;14(45):6999–7004. doi: 10.3748/wjg.14.6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenpreis ED, Kulkarni P, Burke C Gastroenterology FD-RMCotACo. What gastroenterologists should know about the gray market, herbal remedies, and compounded pharmaceuticals and their regulation by the Food and Drug Administration. Am. J. Gastroenterol. 2013;108(5):642–646. doi: 10.1038/ajg.2012.348. [DOI] [PubMed] [Google Scholar]
- Fontana RJ, Watkins PB, Bonkovsky HL, Chalasani N, Davern T, Serrano J, et al. Drug-Induced Liver Injury Network (DILIN) prospective study: rationale, design and conduct. Drug Saf. 2009;32(1):55–68. doi: 10.2165/00002018-200932010-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- Goodin MG, Bray BJ, Rosengren RJ. Sex- and strain-dependent effects of epigallocatechin gallate (EGCG) and epicatechin gallate (ECG) in the mouse. Food Chem. Toxicol. 2006;44(9):1496–1504. doi: 10.1016/j.fct.2006.04.012. [DOI] [PubMed] [Google Scholar]
- Grasl-Kraupp B, Ruttkay-Nedecky B, Koudelka H, Bukowska K, Bursch W, Schulte-Hermann R. In situ detection of fragmented DNA (TUNEL assay) fails to discriminate among apoptosis, necrosis, and autolytic cell death: a cautionary note. Hepatology. 1995;21(5):1465–1468. doi: 10.1002/hep.1840210534. [DOI] [PubMed] [Google Scholar]
- Han D, Dara L, Win S, Than TA, Yuan L, Abbasi SQ, et al. Regulation of drug-induced liver injury by signal transduction pathways: critical role of mitochondria. Trends Pharmacol. Sci. 2013;34(4):243–253. doi: 10.1016/j.tips.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill AH, Ross PK, Gatti DM, Threadgill DW, Rusyn I. Population-based discovery of toxicogenomics biomarkers for hepatotoxicity using a laboratory strain diversity panel. Toxicol. Sci. 2009;110(1):235–243. doi: 10.1093/toxsci/kfp096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill AH, Watkins PB, Su S, Ross PK, Harbourt DE, Stylianou IM, et al. Mouse population-guided resequencing reveals that variants in CD44 contribute to acetaminophen-induced liver injury in humans. Genome Res. 2009;19(9):1507–1515. doi: 10.1101/gr.090241.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill AH, Desmet KD, Wolf KK, Bridges AS, Eaddy JS, Kurtz CL, et al. A mouse diversity panel approach reveals the potential for clinical kidney injury due to DB289 not predicted by classical rodent models. Toxicol. Sci. 2012;130(2):416–426. doi: 10.1093/toxsci/kfs238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CH, Liao YL, Lin SC, Tsai TH, Huang CJ, Chou P. Does supplementation with green tea extract improve insulin resistance in obese type 2 diabetics? A randomized, double-blind, and placebo-controlled clinical trial. Altern. Med. Rev. 2011;16(2):157–163. [PubMed] [Google Scholar]
- Hughes AR, Spreen WR, Mosteller M, Warren LL, Lai EH, Brothers CH, et al. Pharmacogenetics of hypersensitivity to abacavir: from PGx hypothesis to confirmation to clinical utility. Pharmacogenomics. J. 2008;8(6):365–374. doi: 10.1038/tpj.2008.3. [DOI] [PubMed] [Google Scholar]
- Jimenez-Saenz M, Martinez-Sanchez Mdel C. Acute hepatitis associated with the use of green tea infusions. J. Hepatol. 2006;44(3):616–617. doi: 10.1016/j.jhep.2005.11.041. [DOI] [PubMed] [Google Scholar]
- Kaniwa N, Saito Y. Pharmacogenomics of severe cutaneous adverse reactions and drug-induced liver injury. J. Hum. Genet. 2013;58(6):317–326. doi: 10.1038/jhg.2013.37. [DOI] [PubMed] [Google Scholar]
- Lambert JD, Lee MJ, Lu H, Meng X, Hong JJ, Seril DN, et al. Epigallocatechin-3-gallate is absorbed but extensively glucuronidated following oral administration to mice. J. Nutr. 2003;133(12):4172–4177. doi: 10.1093/jn/133.12.4172. [DOI] [PubMed] [Google Scholar]
- Lambert JD, Kennett MJ, Sang S, Reuhl KR, Ju J, Yang CS. Hepatotoxicity of high oral dose (−)-epigallocatechin-3-gallate in mice. Food Chem. Toxicol. 2010;48(1):409–416. doi: 10.1016/j.fct.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan RW, Robledo RF, Recla JM, Philip VM, Bubier JA, Jay JJ, et al. High-precision genetic mapping of behavioral traits in the diversity outbred mouse population. Genes Brain Behav. 2013;12(4):424–437. doi: 10.1111/gbb.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Sun J, Petrova K, Yang X, Greenhaw J, Salminen WF, et al. Metabolomics evaluation of the effects of green tea extract on acetaminophen-induced hepatotoxicity in mice. Food Chem. Toxicol. 2013;62:707–721. doi: 10.1016/j.fct.2013.09.025. [DOI] [PubMed] [Google Scholar]
- Lucena MI, Molokhia M, Shen Y, Urban TJ, Aithal GP, Andrade RJ, et al. Susceptibility to amoxicillin-clavulanate-induced liver injury is influenced by multiple HLA class I and II alleles. Gastroenterology. 2011;141(1):338–347. doi: 10.1053/j.gastro.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzanti G, Menniti-Ippolito F, Moro PA, Cassetti F, Raschetti R, Santuccio C, et al. Hepatotoxicity from green tea: a review of the literature and two unpublished cases. Eur. J. Clin. Pharmacol. 2009;65(4):331–341. doi: 10.1007/s00228-008-0610-7. [DOI] [PubMed] [Google Scholar]
- Monshi MM, Faulkner L, Gibson A, Jenkins RE, Farrell J, Earnshaw CJ, et al. Human leukocyte antigen (HLA)-B*57:01-restricted activation of drug-specific T cells provides the immunological basis for flucloxacillin-induced liver injury. Hepatology. 2013;57(2):727–739. doi: 10.1002/hep.26077. [DOI] [PubMed] [Google Scholar]
- Mouton PR. Principles and Practices of Unbiased Stereology: An Introduction for Bioscientists. Baltimore: Johns Hopkins University Press; 2002. p. x.p. 214. [Google Scholar]
- Navarro VJ, Seeff LB. Liver injury induced by herbal complementary and alternative medicine. Clin. Liver Dis. 2013;17(4):715–735. x. doi: 10.1016/j.cld.2013.07.006. [DOI] [PubMed] [Google Scholar]
- Navarro VJ, Bonkovsky HL, Hwang SI, Vega M, Barnhart H, Serrano J. Catechins in dietary supplements and hepatotoxicity. Dig. Dis. Sci. 2013;58(9):2682–2690. doi: 10.1007/s10620-013-2687-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panza VS, Wazlawik E, Ricardo Schutz G, Comin L, Hecht KC, da Silva EL. Consumption of green tea favorably affects oxidative stress markers in weight-trained men. Nutrition. 2008;24(5):433–442. doi: 10.1016/j.nut.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Papanicolaou KN, Khairallah RJ, Ngoh GA, Chikando A, Luptak I, O’Shea KM, et al. Mitofusin-2 maintains mitochondrial structure and contributes to stress-induced permeability transition in cardiac myocytes. Mol. Cell. Biol. 2011;31(6):1309–1328. doi: 10.1128/MCB.00911-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pardo-Manuel de Villena F, Wang W, McMillan L, Threadgill DW. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm. Genome. 2007;18(6–7):473–481. doi: 10.1007/s00335-007-9045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen WF, Yang X, Shi Q, Greenhaw J, Davis K, Ali AA. Green tea extract can potentiate acetaminophen-induced hepatotoxicity in mice. Food Chem. Toxicol. 2012;50(5):1439–1446. doi: 10.1016/j.fct.2012.01.027. [DOI] [PubMed] [Google Scholar]
- Shim M, Saab S. Severe hepatotoxicity due to Hydroxycut: a case report. Dig. Dis. Sci. 2009;54(2):406–408. doi: 10.1007/s10620-008-0353-4. [DOI] [PubMed] [Google Scholar]
- Svenson KL, Gatti DM, Valdar W, Welsh CE, Cheng R, Chesler EJ, et al. High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics. 2012;190(2):437–447. doi: 10.1534/genetics.111.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschke R, Wolff A, Frenzel C, Schulze J, Eickhoff A. Herbal hepatotoxicity: a tabular compilation of reported cases. Liver Int. 2012;32(10):1543–1556. doi: 10.1111/j.1478-3231.2012.02864.x. [DOI] [PubMed] [Google Scholar]
- Threadgill DW, Hunter KW, Williams RW. Genetic dissection of complex and quantitative traits: from fantasy to reality via a community effort. Mamm. Genome. 2002;13(4):175–178. doi: 10.1007/s00335-001-4001-Y. [DOI] [PubMed] [Google Scholar]
- Timbo BB, Ross MP, McCarthy PV, Lin CT. Dietary supplements in a national survey: prevalence of use and reports of adverse events. J. Am. Diet. Assoc. 2006;106(12):1966–1974. doi: 10.1016/j.jada.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Weng Z, Zhou P, Salminen WF, Yang X, Harrill AH, Cao Z, et al. Green tea epigallocatechin gallate binds to and inhibits respiratory complexes in swelling but not normal rat hepatic mitochondria. Biochem. Biophys. Res. Commun. 2013;443:1097–1104. doi: 10.1016/j.bbrc.2013.12.110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.