Abstract
The origin, structure, and function of the claustrum, as well as its role in neural computation, have remained a mystery since its discovery in the 17th century. Assessing the in vivo connectivity of the claustrum may bring forth useful insights with relevance to model the overall functionality of the claustrum itself. Using structural and diffusion tensor neuroimaging in N = 100 healthy subjects, we found that the claustrum has the highest connectivity in the brain by regional volume. Network theoretical analyses revealed that (a) the claustrum is a primary contributor to global brain network architecture, and that (b) significant connectivity dependencies exist between the claustrum, frontal lobe, and cingulate regions. These results illustrate that the claustrum is ideally located within the human central nervous system (CNS) connectome to serve as the putative “gate keeper” of neural information for consciousness awareness. Our findings support and underscore prior theoretical contributions about the involvement of the claustrum in higher cognitive function and its relevance in devastating neurological disease. Hum Brain Mapp 36:827–838, 2015. © 2014 Wiley Periodicals, Inc.
Keywords: claustrum, connectivity, cortex, graph theory, magnetic resonance imaging, diffusion tensor imaging, brain networks, consciousness, cognition, neurology
INTRODUCTION
Considerable mystery has surrounded the origin, structure, and function of the claustrum—a diminutive bilateral anatomical structure of the brain whose very name means “hidden away” [Crick and Koch, 2005]. The structure was identified in humans as early as 1672 as shown by the drawings of Thomas Willis [Bayer and Altman, 1991], though was first described (under the name “vormauer”) by Karl Friedrich Burdach in his seminal work, Von Baue und Leben des Gehirns in the early 19th century. Burdach himself, however, credited the discovery to the 1,786 drawings by Félix Vicq‐d'Azyr . Despite this long history, knowledge of the claustrum, its organization, afferents, and efferents has been studied intermittently and relatively poorly explored until recently [Edelstein and Denaro, 2004; Smythies et al., 2014].
In animals, extensive neuronal projections have been noted between the claustrum and numerous cortical regions [Buchanan and Johnson, 2011; Crick and Koch, 2005; Minciacchi et al., 1995; Tanne‐Gariepy et al., 2002; Wilhite et al., 1986] with white matter connections to multiple cortical layers [Carey et al., 1980] (although see Crick and Koch [2005]). Likewise, the claustrum exhibits many connections to subcortical structures [Arikuni and Kubota, 1985; Berke, 1960; Buchanan and Johnson, 2011; Crick and Koch, 2005; Salerno et al., 1984; Tanne‐Gariepy et al., 2002]. The structure is present in all mammals with species such as tree shrews and cats having relatively large claustra by volume in contrast to monotremes, who apparently lack claustra altogether [Butler et al., 2002].
In electrophysiological studies, the claustrum has been shown to send signals to four thalamic nuclei: the medial geniculate nucleus, the lateralis posterior, centrum medianum, and ventralis lateralis [Chachich and Powell, 2004; Cortimiglia et al., 1987, 1991; Crescimanno et al., 1989; Spector et al., 1970; see also Sherk 2013, Chapter 5]. Excitotoxic lesioning in the rat has also shown evidence of frontostriatal connectivity as well [Grasby and Talk 2013]. Both the medial ectosylvian gyrus (A2) and the anterior ectosylvian gyrus (S2) also have been reported as transmitting information to the claustrum [Edelstein and Denaro, 2004; Hassmannova, 1977]. Moreover, the claustrum is topologically organized with frontal cortex being linked to the claustrum's anterior portion, the parietal cortex with its central and posterior parts, and the occipital and temporal cortices are linked to its posterior and inferior margins [Druga, 2014; Markowitsch et al., 1984]. Nontopographic projections to other parts of the same cortical area exist, but there is complete segregation between distinct cortical areas [Minciacchi et al., 1995]. Fiber bundles such as the corona radiata, uncinate fasciculi, and inferior occipitofrontal fasciculi project to the claustrum from the superior frontal, precentral, postcentral, superior parietal, and parietooccipital regions [Fernandez‐Miranda et al., 2008a, b]. Only a relatively small number of claustral cells appear to project to the contralateral hemisphere [Markowitsch et al., 1984]. Opinions have differed, however, as to whether the claustrum receive inputs via brain stem and spinal afferents [Arikuni and Kubota, 1985; Edelstein and Denaro, 2004].
In humans, the volume of the claustrum is one quarter of 1% of the volume of the cerebral cortex [Crick and Koch, 2005; Edelstein and Denaro, 2004]. As illustrated in the Talairach and Tournoux [1988] human brain atlas, the claustrum extends 22 mm inferior‐to‐superior and 38 mm anterior‐to‐posterior. The right claustrum has an average volume of 828.83 mm3, while the left has a volume of 705.82 mm3 and lies approximately 1 mm from the insular cortex and about 1 mm from the putamen [Kapakin, 2011]. Bilaterally, it displays asymmetry in shape and anisotropy between the hemispheres [Cao et al., 2003] (as shown schematically in Fig. 1A). It projects downward and crosses the rhinalis fissure—though it may be interrupted by fibers from the uncinate fasciculus in some places—before extending to the lateral rhinencephalon. A thorough review of claustral gross anatomy can be found in Druga [2014]. Historically, the claustrum was often considered to be the innermost layer of the insula [Landau, 1919], though has also been associated with the basal ganglia as a potential pallial derivative [Puelles, 2014]. The claustrum contains both fusiform cells and pyramidal somata, which are indicative of cortical areas, but also contains subcortical cell types. Due to the variation in cell morphometry, the generally nonlayered structure of the claustrum cannot formally be pronounced as strictly cortical or subcortical [Mathur et al., 2009]. This might explain its only modest description in early as well as modern neuroscience texts and resources (e.g., http://neurolex.org/wiki/Category:Claustrum).
Figure 1.
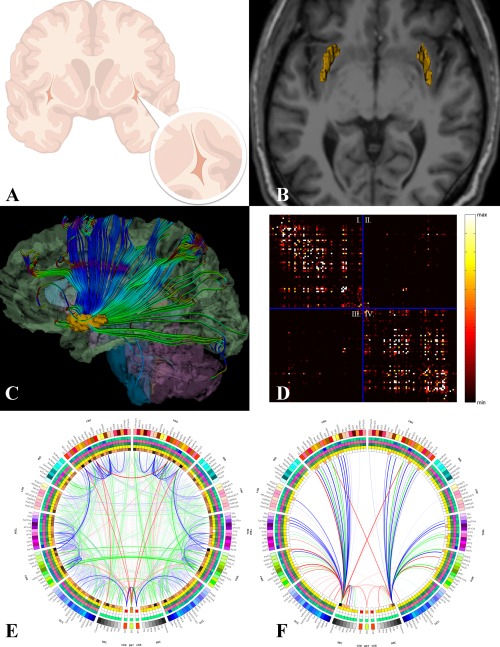
(A) In the human brain, the claustrum is a thin band of cells located medial to the insular cortex and lateral to the putamen. (B) Three‐dimensional models of the bilateral claustra were obtained from T1‐weighted MPRAGE structural MRI image volumes. (C) White matter fiber tractography was performed and inter‐regional connectivity was computed by determining the relative proportion of extracted fibers initiated or terminated within the boundaries of each anatomical parcel. Here, fibers linking the claustrum to other brain regions are illustrated in an example subject. (D) These measures are aggregated into an M × M connectivity matrix, shown here as an image in which black refers to no or relatively low inter‐regional connectivity between region i and region j, varying to white reflecting a greater extent of region. (E) Regional morphometrics for the entire brain are represented as a circular “connectogram.” The measurements for each region are shown as “heat rings” including, from the outside inward, cortical thickness, regional volume, surface area, and connectivity density. The information contained in the computed connectivity matrix is used here to illustrate the pattern and strength of connections between brain regions. Line opacity is proportional to connection density, whereas color represents the average FA integrated along all pathways comprising that connection. Red is high average FA, green in medium average FA, and blue if low mean FA, according to the upper, middle, and lowest thirds of the FA distribution. Further information on connectogram construction and interpretation can be found in Irimia, Chambers et al. [2012b]. (F) The connectogram showing only those fiber pathways which eminate from or are connected to the left and/or right claustra. These fibers were subject to the graph theoretical analysis described in the body of the text.
Recent in vivo examinations in humans using diffusion tensor imaging (DTI) have shown the claustrum to possess long‐range projections to many prominent Brodmann's areas (BA) [Milardi et al., 2013], including connections to visual cortex (BA 17, 18, 19, and 39), frontal areas (BA 8, 9, 10, 11, 12, and 34), as well as superior regions (BA 7, 5, 1/2/3, 4, 6, and 8) and language areas (e.g., BA 44, 45, 31, etc). Fernandez‐Miranda et al. [2008a, b, 2012; Smythies et al., 2014] have traced the extensive, intricate claustrocortical system using both microsurgery in concert with DTI mapping. For their investigation, they examine these connections in subdivisions of the claustrum. They demonstrated that the claustrocortical fibers connect the dorsal claustrum with the superior frontal, precentral, postcentral, and posterior parietal cortices with a regular topographic organization. Additionally, they reiterate that the ventral portion of the claustrum is connected with the orbitofrontal, prefrontal, and temporal cortex and with the amygdala. These results confirm the patterns of connectivity previously observed in nonhuman species (see Supporting Information, Table 1).
Because of its cellular composition and wide‐ranging connectivity, the claustrum has been proposed as the cornerstone of sensory integration—putatively receiving, assimilating, integrating, and channeling information throughout the brain from each sensory cortex [Edelstein and Denaro, 2004; Remedios et al., 2014]. Moreover, it has been posited that the claustrum assesses the congruence of information while binding multiple facets of perception into a whole, such as assigning motion, direction, and sound to a singular visual object [Naghavi et al., 2007]. Indeed, in a thorough series of reviews, Crick and Koch [2003, 2005] and Smythies et al. [2012a, b] have argued that the claustrum might form the foundation for the neural locus of consciousness. With little doubt, these important papers are principally responsible for the recent resurgence of interest in the claustrum, its structure, connectivity, and its function [Mathur, 2014]. The mechanisms of its role in brain networks, however, is not well understood especially in light of ongoing debates concerning claustral neurons as multisensory processors [Braak and Braak, 1982; Edelstein and Denaro, 2004; Remedios et al., 2010; Smith and Alloway, 2010].
Despite its consideration in a modest number of largely descriptive neuroimaging studies, however, the relative importance or strengths of the diverse and wide‐ranging pattern of claustral connectivity have not, to date, been thoroughly explored using large human samples. Quantitative methods for the assessment of white matter networks offer an opportunity to explore the connectivity of the claustrum in vivo and to measure its relative influence on brain subnetworks. Indeed, graph theoretical analytic techniques have been widely applied to connectomics data from human neuroimaging studies to characterize the novel properties of structural and functional brain networks [Bassett and Bullmore, 2009; Bullmore and Sporns, 2009]. Graph‐wide as well as nodal connectivity characteristics provide insights into the integrity and efficiency of information flow through a network and can shed light on the effects of injury or disease. They can also highlight the macrolevel importance of certain regions on the overall structure of a network and how those influence the connectivity of brain subnetworks.
With this in mind, we sought to quantitatively examine the relative importance of putative subnetworks, which involve the claustrum—assessed using DTI, structural neuroimaging, and graph theoretical analysis—in a cohort large enough to establish a reliable and comprehensive model of the full macroscale connectivity of this so often over‐looked brain structure.
METHODS
Subjects and Data Acquisition
The study cohort included N = 100 healthy adult subjects (47 males and 53 females) with ages between 18.6 and 61.1 (mean and SD: 32.71 ± 11.6 years). Subjects provided informed written consent as required by the Declaration of Helsinki, U.S. 45 CFR 46, and neuroimage volume acquisition was conducted with the approval of the local ethics committees at the respective research institutions where data were acquired. Participants were recruited by advertisements in local newspapers and campus flyers. Subjects were all healthy and had no history of neurological or psychiatric illnesses. No participant had a current or past psychiatric diagnosis (including substance abuse) or was taking medications for any medical reasons. Additional exclusion criteria for all participants included left‐handedness, hypertension, neurological illness, metal implants, and a history of head trauma with loss of consciousness for more than 5 min. Neuroimaging datasets were fully anonymized, and no linked coding or keys to subject identity were maintained. For these reasons, in compliance with the U.S. Health Insurance Portability and Accountability Act (HIPAA; http://www.hhs.gov/ocr/privacy), this study does not qualify as involving human subjects' materials.
Structural T1‐weighted magnetic resonance imaging (MRI) and DTI volumes were acquired from each patient using a Siemens Trio Tim 3.0 Tesla whole body scanning system with a 12‐channel head coil. For MRI, a Turbo MP‐RAGE sequence (repetition time [TR] = 20 ms, echo time [TE] = 3 ms, flip angle = 25°, slice thickness = 1 mm, acquisition matrix = 256 × 256 × 256) was obtained. A DTI protocol having the following parameters was obtained: TR = 9.4 s, TE = 88 ms, flip angle = 90°, slice thickness = 2 mm, number of gradient directions = 68, acquisition matrix = 128 × 128 × 128. Two nondiffusion‐weighted volumes were acquired for each subject (b‐values: 0 and 1,000).
Neuroimage Data Processing
Data processing workflows were created using the Laboratory of Neuro Imaging (LONI) Pipeline Workflow Environment (http://pipeline.loni.ucla.edu; version 5.9.1). The LONI Pipeline is a graphical environment for construction, validation and execution of advanced neuroimaging data analysis protocols [Dinov et al., 2010]. It enables the creation of heterogeneous tool interactivity, automated data format conversion, allows grid utilization, facilitates data provenance, and provides a significant library of computational tools. It is built as a distributed grid computing environment and permits efficient tool integration, protocol validation, and broad resource distribution. Further details are available via the LONI Pipeline website.
A set of customized LONI Pipeline workflow was created to perform basic preprocessing of all subject data as a prelude to the application of regionally focused processing, atlas creation, and connectomics estimation. This initial workflow performed FreeSurfer 5.3 reconstruction [Dale et al., 1999; Fischl et al., 1999], stripped the skulls and other nonbrain tissues out of the image volumes, bias‐corrected the T1‐weighted structural images, performed eddy current correction on the DTI, and then reconstructed the diffusion image into 3D tracts in both .vtk and .trk file formats so that the white matter can be visually examined using commonly available tractography software. Specifically, DTI fractional anisotropy (FA) computation and fiber reconstruction was performed using the TrackVis Diffusion Toolkit (http://trackvis.org) via streamline tractography using a subject‐specific B0 white‐matter mask and a turning angle threshold of 35°. The workflow also carried out the spatial registration of the T1 and DTI images, and likewise applied the transformation to the cortical parcels of the FreeSurfer labelmap. Finally, connectivity matrices were computed to quantify the relative number of extracted DTI fibers and average FA values over them which connected each pair of cortical regions. These matrices are then combined with the cortical region morphometric analyses to generate whole‐brain connectograms (see below). The LONI Pipeline workflows described herein, along with additional supporting workflows, are available as Supporting Information.
Identification and Segmentation of the Claustrum
While the claustrum has been assigned labelmap values in the context of the FreeSurfer implementation of the Destrieux et al. [2010] brain atlas, the version of FreeSurfer we used (version 5.3) does not, in fact, assign voxels to this structure. So, following the application of the initial image processing workflow, 3D Slicer (http://slicer.org) was used to manually delineate left and right claustrum masks in the sample of healthy subjects. For each subject, an axially resampled T1 anatomical was examined in a superior‐to‐inferior manner to identify the body of the claustrum. As described above and illustrated in Figure 1A, the claustrum is located between the external and internal capsules, medially from the insular cortex and laterally from the body of the putamen. Separate left and right binary masks of this region were manually drawn (by Carinna M. Torgerson (CMT)) in the axial plane and then edited in the sagittal plane for accuracy, before being visually reviewed by trained raters (by Andrei Irimia (AI) and John D. Van Horn (JDVH)), and the regional delineation refined as necessary.
Pooling and Applying Claustral Masks
Each subject's pair of claustral masks were then used as input into an “Atlas Creation” LONI Pipeline workflow, which spatially warped the claustrum label masks into Montreal Neurological Institute (MNI) Atlas space and, using FSL modules, spatially averaged them, before returning the average back into the native space of each individual subject via inverting the spatial warping transform. This averaging was performed to make claustral delineation consistent with the FreeSurfer mapping technique, which uses averaged data to generate accurate labelmaps. These newly applied masks of the claustrum (now in native space for all 100 subjects), the “Adding Labelmaps” workflow was used to spatially merge the left and right claustrum labels into the FreeSurfer parcellation and segmentation labelmap. At this stage, new connectivity matrices were generated for each subject based upon whole‐brain labelmaps, which now included the spatially averaged and unwarped claustra, using the “Connectivity Calculation Workflow.”
Subsequently, the fully integrated labelmaps, which include the left and right claustra, were used as input into three different workflows. First, the “Whole Brain Plus Claustrum” workflow took the full labelmaps and generated connectograms depicting the whole, macroscale pattern of DTI connectivity across the complete collection of segmented brain regions. Finally, the “Only Claustrum” workflow used the connectivity matrices from the whole‐brain plus claustrum analysis resulting in new files created for each subject depicting the specific connectivity patterns of the left and right claustrum in each subject. Values of all connections originating or terminating in the insular cortex an putamen were set to zeroes to avoid misattribution due to proximity, as well as double‐counting errors.
Connectogram Generation
Cortical parcellations were represented as a circular arrangement of 165 positioned elements representing the connectivity matrix of the left (quadrant IV of Fig. 1D) and right (quadrants I of Fig. 1D) cerebral hemispheres, and their jointly connecting fiber pathways (quadrants II and III), each positioned symmetrically with respect to the vertical axis. This graphical representation—termed a “connectogram”—has been described extensively elsewhere [Irimia et al., 2012a, b; Van Horn et al., 2012]. The connectogram is becoming widely used as a method for showing patterns of connectivity in a variety of research domains, as is demonstrated by the variety of topics mentioned on its Wikipedia page (http://en.wikipedia.org/wiki/Connectogram). Briefly, the connectograms represent cortical region labeling and morphometric measurements as color‐coded rings and illustrate inter‐regional connectivity as lines between lobar and regional segments [Irimia et al., 2012a, b]. Line opacity is proportional to connection density, while in these figures, specifically, red indicates high average FA (aggregated over fibers connecting those regions), green represents moderate FA, and blue represents low FA. FA measures the degree of preferred water diffusivity, ranging between zero (no preferred diffusion) and unity (strong directional diffusion) [Basser et al., 2000]. A complete list of parcellations, their abbreviations and associated Red‐Green‐Blue (RGB) codes are provided in the supplemental materials of Van Horn et al. [2012]. Parcellations were arranged within each lobe in the order of their location along the anteroposterior axis of the cortical surface associated with the published FreeSurfer normal population atlas [Destrieux et al., 2010].
Network Theoretical Analyses
To examine the network contributions of the claustrum, we used graph theoretical methods, which have been demonstrated to provide useful constructs in characterizing neuroimaging‐derived brain networks [Sporns, 2012]. Graph theoretical metrics are useful for describing the properties of network architecture and have been successfully applied to patterns of inter‐regional brain connectivity. Additionally, using a method for systematic removal of white matter connections in conjunction with graph theoretical analysis [Irimia and Van Horn, 2014], the left and right claustra were analyzed for their role in major brain networks (Fig. 1D). Such methods have provided a convenient approach for the computational modeling of the effects of brain injury [Van Horn et al., 2012] and for the assessment of nodal clustering and assortativity which can illustrate the presence of various brain sub‐networks [van den Heuvel et al., 2008].
All connectivity metrics were determined using the Brain Connectivity Toolbox (https://sites.google.com/site/bctnet/). Specifically, the graph‐wide metrics used here included network density, diameter, and assortativity, as well as the nodal metric betweeness centrality. To simplify interpretation, particularly short (<5 mm) connections were not included and, notably, connections to the insula and putamen were disregarded as their close proximity to the claustra would increase the likelihood of Type I errors in our analyses. A statistical MANOVA analysis of the claustrum's “topological neighborhood” subnetwork, with the claustrum excision being considered as a treatment, was then performed. This analysis was accompanied by a leave‐one‐out modeling strategy to determine which graph‐wide metrics were most statistically influential.
RESULTS
In our large sample of N = 100 healthy subjects, the claustrum was found to possess the highest density of fiber connections per unit volume out of all examined brain regions (Table 1; listed by name according to the FreeSurfer aparc.a2009 naming scheme, which can be found at https://surfer.nmr.mgh.harvard.edu/fswiki/FsTutorial/AnatomicalROI/FreeSurferColorLUT following the list of aparc.a2005 labels). The normalization by regional volume is commonly recommended practiced when comparing fiber tract densities [Bassett et al., 2011; Cahalane et al., 2012; Varkuti et al., 2011]. The complete pattern of inter‐regional connectivity between all regions is illustrated as a connectogram in Figure 1E, while the specific pattern of claustral connectivity is depicted in Figure 1F. The claustra had their densest DTI fiber tract connections to frontal cortices, with more modest degrees of connectivity to parietal, temporal, and occipital lobes, respectively. Connections appeared the fewest involving limbic structures in agreement with the findings of Markowitsch et al. [1984]. Evidence was also found for fiber tracts between the claustra and the brain stem, in contrast to prior reports which hypothesized that the claustrum communicates with the brain stem and autonomic spinal neurons via intermediaries, such as the medial prefrontal cortex [Edelstein and Denaro, 2004; Hatam et al., 2013].
Table 1.
Volumes (mm3) and Connectivity of Extracted Brain Structures Sorted by Connection Density per cm3
| Both hemispheres | Right hemisphere | Left hemisphere | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Volume [mm3] | # of connections per cm3 | Volume [mm3] | # of connections per cm3 | Volume [mm3] | # of connections per cm3 | |||||||
| Structure | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD |
| Region | ||||||||||||
| Clau | 654.67 | 78.88 | 34.202 | 10.259 | 694.94 | 68.86 | 30.723 | 8.136 | 614.41 | 67.01 | 37.681 | 10.996 |
| G_cingul‐Post‐ventral | 620.06 | 169.08 | 26.493 | 12.445 | 667.08 | 177.61 | 23.576 | 9.853 | 573.04 | 146.47 | 29.409 | 14.038 |
| G_subcallosal | 780.43 | 317.13 | 19.553 | 9.747 | 801.32 | 253.14 | 19.837 | 8.508 | 759.55 | 370.41 | 19.269 | 10.882 |
| S_suborbital | 830.12 | 352.35 | 17.902 | 8.274 | 542.69 | 191.35 | 21.879 | 9.256 | 1117.54 | 214.57 | 13.925 | 4.469 |
| G_front_inf‐Orbital | 848.24 | 223.04 | 16.808 | 10.023 | 913.35 | 237.38 | 16.681 | 10.177 | 783.13 | 187.29 | 16.934 | 9.916 |
| S_temporal_transverse | 504.74 | 126.55 | 16.103 | 8.148 | 474.36 | 125.12 | 17.092 | 8.224 | 535.12 | 121.14 | 15.115 | 7.991 |
| S_pericallosal | 1575.07 | 414.08 | 15.47 | 4.643 | 1788.36 | 398.21 | 14.254 | 3.92 | 1361.77 | 306.87 | 16.686 | 4.998 |
| S_collat_transv_post | 621.63 | 209.86 | 13.794 | 6.271 | 727.31 | 224.52 | 13.172 | 5.638 | 515.94 | 124.72 | 14.416 | 6.818 |
| G_cingul‐Post‐dorsal | 1408.08 | 299.4 | 13.392 | 3.236 | 1389.35 | 300.92 | 13.568 | 2.991 | 1426.81 | 298.19 | 13.215 | 3.471 |
| S_orbital_lateral | 609.08 | 201.94 | 13.275 | 7.014 | 642.71 | 234.49 | 12.738 | 7.418 | 575.45 | 157.16 | 13.812 | 6.579 |
| G_temp_sup SUP‐G_T_TRANSV | 967.53 | 256.05 | 12.03 | 5.43 | 868.14 | 219.44 | 12.965 | 5.724 | 1066.93 | 252.34 | 11.095 | 4.974 |
| S_interm_prim_Jensen | 637.81 | 318.43 | 11.721 | 6.453 | 727.84 | 333.35 | 11.095 | 4.204 | 547.78 | 276.25 | 12.348 | 8.077 |
| G_temp_sup‐Plan_polar | 1684.57 | 416.15 | 11.114 | 4.946 | 1798.09 | 426.7 | 10.198 | 4.187 | 1571.05 | 374.17 | 12.031 | 5.472 |
| G_occipital_sup | 2998.26 | 640.62 | 10.973 | 4.79 | 3262.08 | 694.47 | 10.63 | 4.996 | 2734.44 | 449.49 | 11.316 | 4.575 |
| S_orbital_med‐olfact | 1256.38 | 232.41 | 10.238 | 4.128 | 1238.48 | 186.38 | 10.952 | 4.547 | 1274.28 | 270.53 | 9.524 | 3.542 |
| G_cuneus | 2867.94 | 492.31 | 9.498 | 3.264 | 2970.91 | 505.31 | 9.469 | 3.182 | 2764.97 | 458.73 | 9.527 | 3.359 |
| G_and_S_transv_frontopol | 2064.46 | 577.44 | 8.917 | 4.246 | 2436.06 | 518.8 | 7.998 | 3.283 | 1692.86 | 349.46 | 9.836 | 4.873 |
| G_temp_sup‐Plan_tempo | 1646.52 | 446.23 | 8.685 | 3.874 | 1504.76 | 389.7 | 9.78 | 4.073 | 1788.27 | 455.84 | 7.589 | 3.34 |
| S_occipital_ant | 1230.93 | 386.32 | 8.561 | 3.009 | 1275.57 | 407.45 | 8.009 | 2.498 | 1186.3 | 360.49 | 9.113 | 3.367 |
| S_cingul‐Marginalis | 1745.48 | 336.55 | 8.425 | 2.905 | 1900.88 | 338.84 | 8.029 | 2.729 | 1590.09 | 253.14 | 8.82 | 3.034 |
| G_front_inf‐Triangul | 2637.3 | 573.8 | 8.371 | 3.859 | 2602.16 | 597.02 | 8.354 | 4.201 | 2672.45 | 550.35 | 8.388 | 3.505 |
| G_and_S_paracentral | 2407.92 | 420.44 | 8.366 | 2.271 | 2254.69 | 375.69 | 8.406 | 2.192 | 2561.15 | 408.36 | 8.327 | 2.357 |
| S_subparietal | 1816.81 | 445.35 | 8.269 | 3.094 | 1917.39 | 479.9 | 8.341 | 3.085 | 1716.23 | 384.62 | 8.197 | 3.117 |
| G_rectus | 2110.24 | 496.87 | 8.19 | 3.343 | 1757.5 | 298.17 | 8.764 | 3.46 | 2462.98 | 394.95 | 7.617 | 3.135 |
| G_and_S_frontomargin | 1945.5 | 402.95 | 7.786 | 4.024 | 1818.95 | 386.96 | 8.434 | 4.07 | 2072.05 | 379.84 | 7.139 | 3.889 |
| G_and_S_cingul‐Mid‐Post | 2647.99 | 445.02 | 7.31 | 1.594 | 2788.45 | 472.08 | 7.039 | 1.392 | 2507.54 | 367.94 | 7.581 | 1.738 |
| G_precuneus | 5798.45 | 971.41 | 7.234 | 2.604 | 5798.64 | 967.05 | 7.298 | 2.625 | 5798.27 | 980.63 | 7.169 | 2.595 |
| G_front_inf‐Opercular | 3186.14 | 585.49 | 7.135 | 2.405 | 3060.85 | 548.06 | 7.302 | 2.304 | 3311.42 | 597.48 | 6.968 | 2.502 |
| S_oc_sup_and_transversal | 1991.96 | 489.98 | 6.868 | 2.942 | 2162.72 | 488.62 | 6.548 | 2.859 | 1821.21 | 430.03 | 7.188 | 3.003 |
| S_oc‐temp_lat | 1539.9 | 394.26 | 6.799 | 2.905 | 1589.85 | 358.64 | 6.504 | 2.707 | 1489.95 | 422.84 | 7.093 | 3.076 |
| G_and_S_occipital_inf | 2666.47 | 591.09 | 6.761 | 2.7 | 2533.03 | 552.68 | 6.28 | 2.231 | 2799.91 | 600.73 | 7.242 | 3.035 |
| S_collat_transv_ant | 1762.77 | 451.41 | 6.728 | 3.636 | 1763.61 | 419.99 | 6.797 | 3.824 | 1761.93 | 482.92 | 6.659 | 3.457 |
| S_parieto_occipital | 2846.92 | 581.03 | 6.636 | 2.44 | 2918.64 | 615.71 | 6.574 | 2.641 | 2775.19 | 537.7 | 6.698 | 2.231 |
| Pole_occipital | 3699.11 | 972.86 | 6.627 | 2.877 | 4466.21 | 697.5 | 5.725 | 2.15 | 2932.01 | 476.66 | 7.529 | 3.221 |
| G_oc‐temp_med‐Lingual | 4787.69 | 758.04 | 6.382 | 1.998 | 4787.5 | 711.83 | 6.174 | 1.802 | 4787.88 | 805.2 | 6.591 | 2.166 |
| G_and_S_cingul‐Mid‐Ant | 2878.24 | 506.94 | 6.209 | 1.812 | 3032.81 | 481.31 | 5.967 | 1.649 | 2723.67 | 486.47 | 6.451 | 1.939 |
| G_parietal_sup | 5605.55 | 1153.54 | 6.175 | 2.134 | 5014.18 | 877.72 | 6.557 | 2.277 | 6196.91 | 1094.46 | 5.793 | 1.917 |
| S_calcarine | 2948.55 | 554.52 | 6.116 | 1.656 | 2897.59 | 530.37 | 5.936 | 1.461 | 2999.5 | 575.82 | 6.296 | 1.819 |
| G_postcentral | 3766.18 | 719.69 | 6.114 | 1.9 | 3595.87 | 687.68 | 6.329 | 1.923 | 3936.5 | 713.89 | 5.899 | 1.862 |
| S_temporal_inf | 2084.3 | 547.48 | 5.97 | 2.597 | 2055.57 | 470.68 | 5.623 | 2.125 | 2113.03 | 615.87 | 6.317 | 2.966 |
| G_temp_sup‐Lateral | 5216.72 | 991.97 | 5.574 | 1.965 | 4972.02 | 905.54 | 5.619 | 2.067 | 5461.42 | 1018.33 | 5.53 | 1.866 |
| S_oc_middle_and_Lunatus | 1444.58 | 425.37 | 5.561 | 2.598 | 1461.36 | 468.49 | 5.52 | 2.417 | 1427.79 | 379.01 | 5.603 | 2.78 |
| G_occipital_middle | 4776.68 | 919.82 | 5.537 | 2.043 | 5102.78 | 953.46 | 4.953 | 1.869 | 4450.58 | 759.46 | 6.121 | 2.052 |
| G_oc‐temp_med‐Parahip | 3684.89 | 719.73 | 5.505 | 1.957 | 3882.85 | 747.5 | 5.481 | 1.895 | 3486.92 | 635.08 | 5.53 | 2.027 |
| G_and_S_subcentral | 2879.21 | 603.96 | 5.492 | 1.594 | 2735.02 | 548.09 | 5.795 | 1.713 | 3023.4 | 625.16 | 5.19 | 1.411 |
| Pole_temporal | 5539.3 | 870.77 | 5.253 | 2.451 | 5493.25 | 829.15 | 5.476 | 2.551 | 5585.35 | 912.34 | 5.03 | 2.338 |
| S_oc‐temp_med_and_Lingual | 3151.12 | 568.64 | 5.007 | 1.676 | 2919.38 | 488.4 | 5.1 | 1.566 | 3382.86 | 550.4 | 4.914 | 1.782 |
| G_and_S_cingul‐Ant | 4810.7 | 788.3 | 4.703 | 1.149 | 5177.32 | 754.95 | 4.63 | 1.093 | 4444.09 | 638.47 | 4.776 | 1.204 |
| G_precentral | 5998.47 | 866.49 | 4.534 | 1.115 | 6019.14 | 881.32 | 4.676 | 1.241 | 5977.79 | 855.34 | 4.392 | 0.959 |
| G_orbital | 6264.03 | 884.88 | 4.422 | 1.577 | 6514.48 | 873.41 | 4.473 | 1.664 | 6013.58 | 827.27 | 4.37 | 1.493 |
| G_oc‐temp_lat‐fusifor | 4553.95 | 791.07 | 4.35 | 1.525 | 4476.53 | 783.59 | 4.29 | 1.437 | 4631.38 | 794.85 | 4.41 | 1.613 |
| S_precentral‐sup‐part | 2055.31 | 544.84 | 4.086 | 2.073 | 2103.49 | 626.22 | 4.236 | 2.007 | 2007.13 | 447.07 | 3.936 | 2.136 |
| S_postcentral | 3541.86 | 828.88 | 3.945 | 1.561 | 3247.51 | 766.41 | 4.265 | 1.476 | 3836.21 | 786.51 | 3.625 | 1.586 |
| S_central | 3438.17 | 584.79 | 3.729 | 1.849 | 3417.99 | 571.37 | 3.922 | 2.004 | 3458.35 | 600.1 | 3.535 | 1.667 |
| S_orbital | 2294.95 | 411.28 | 3.692 | 2.116 | 2354.25 | 430.9 | 3.623 | 2.311 | 2235.64 | 383.69 | 3.761 | 1.911 |
| S_precentral‐inf‐part | 2440.52 | 579.99 | 3.689 | 1.365 | 2523.19 | 605.96 | 3.501 | 1.27 | 2357.85 | 543.31 | 3.876 | 1.435 |
| S_intrapariet_and_P_trans | 4418.54 | 718.41 | 3.666 | 1.436 | 4435.42 | 686.32 | 4.049 | 1.382 | 4401.66 | 752.22 | 3.283 | 1.392 |
| S_front_middle | 2797.12 | 771.43 | 3.519 | 1.856 | 3235.95 | 700.26 | 3.085 | 1.592 | 2358.29 | 562.88 | 3.953 | 2.003 |
| S_front_inf | 3342.05 | 733.61 | 3.507 | 1.43 | 3138.64 | 725.79 | 3.63 | 1.418 | 3545.47 | 686.62 | 3.385 | 1.439 |
| G_pariet_inf‐Angular | 6348.84 | 1177.87 | 3.489 | 1.335 | 6949 | 1050.44 | 3.231 | 1.193 | 5748.68 | 978.62 | 3.747 | 1.422 |
| G_pariet_inf‐Supramar | 6123.03 | 1251.57 | 3.294 | 0.861 | 6010.84 | 1225.92 | 3.43 | 0.876 | 6235.23 | 1272.94 | 3.158 | 0.829 |
| G_temporal_middle | 7423.52 | 1479.97 | 3.216 | 0.918 | 7667.88 | 1516.67 | 3.102 | 0.902 | 7179.17 | 1407.78 | 3.33 | 0.925 |
| G_front_sup | 16504.56 | 2260.64 | 3.038 | 0.885 | 15933.01 | 2206.4 | 3.137 | 0.897 | 17076.11 | 2178.18 | 2.94 | 0.867 |
| G_temporal_inf | 6709.9 | 1263.14 | 3.001 | 1.195 | 6570.02 | 1279.14 | 2.963 | 1.211 | 6849.77 | 1237.52 | 3.039 | 1.185 |
| S_front_sup | 4343.69 | 871.01 | 2.764 | 1.195 | 4119.55 | 769.28 | 2.831 | 1.27 | 4567.82 | 911.98 | 2.697 | 1.118 |
| G_front_middle | 9808.58 | 1799.06 | 2.48 | 0.872 | 9376.61 | 1737.47 | 2.452 | 0.875 | 10240.56 | 1763.55 | 2.509 | 0.872 |
| S_temporal_sup | 9567.53 | 1470.56 | 2.158 | 0.568 | 10107.04 | 1449.78 | 1.996 | 0.475 | 9028.02 | 1287.25 | 2.32 | 0.609 |
Our statistical analyses of the claustrum's topological neighborhood subnetwork, as measured using graph theoretical metrics with claustrum excision being considered as a level of treatment, were as follows.
Analysis of Graph‐Level Metrics
The three specific network feature variables included in a MANOVA statistical model were assortativity, density, and graph diameter. We note that of the variety of available graph‐based metrics, these were found to be only ones which were negligibly intercorrelated. But, importantly, they are particularly useful for assessing overall network structure and sensitivity to network alteration [Sporns, 2011]. These are defined as follows: (a) density is the ratio of present connections to all possible connections; (b) assortativity measures the correlation coefficient between the degrees of all nodes on two opposite ends of a link. A positive assortativity coefficient indicates that nodes tend to link to other nodes with the same or similar degree; while (c) graph diameter refers to the maximum network eccentricity, where eccentricity is the maximal shortest path length between a node and any other node. Under the MANOVA, it was found that the removal of the claustrum led to a statistically significant change in the network measure feature vector at the omnibus level, F(3,196) = 3.49, P ≤ 0.017 (FDR corrected). Systematic removal of individual network measures in the MANOVA was performed in order to identify their relative contribution to the overall model. The results, indicated by which variable had been removed, are as follows: assortativity—F(2,197) = 2.62; P ≤ 0.07; diameter—F(2,197) = 4.73, P ≤ 0.001; and density—F(2,197) = 1.87, P ≤ 0.16.
Analysis of Node‐Level Metrics:
Betweenness centrality assesses the fraction of all shortest paths in the network which contain a given node. Of particular interest at the nodal level is the notion of the betweenness centrality of brain regions connecting to the claustrum and how betweenness centrality of nodes is altered when the connectivity to the claustrum is removed. Nodes with high values of betweenness centrality participate in a large number of shortest paths. Assessing this measure of a node's relative “position” in a network is often a more informative measure than measuring connectivity density alone [Sporns, 2011]. Measurement of betweenness centrality has become a useful strategy for understanding the elements of complex networks, notably in the brain [Mirzasoleiman and Jalili, 2011]. It was found that the systematic removal of claustral connections led to significant changes (P ≤ 0.05, FDR corrected) in the ANOVA main‐effects models of nodal betweenness centrality involving the structures listed in Table 2.
Table 2.
Influence of the Claustrum Removal on Nodal Betweenness Centrality (P < 0.05, FDR)
| With claustrum | Claustrum removed | |||||||
|---|---|---|---|---|---|---|---|---|
| Structure | Hemisphere | Mean | SD | Mean | SD | F | Student's t‐test | P |
| G_front_inf‐Opercular | Right | 59.98 | 17.42 | 64.89 | 15.57 | 4.4168 | 2.1016 | 0.0369 |
| G_precentral | Right | 37.95 | 12.94 | 44.17 | 9.38 | 3.8984 | 1.9744 | 0.0497 |
| G_postcentral | Right | 42.98 | 15.82 | 47.90 | 15.23 | 5.0053 | 2.2373 | 0.0264 |
| G_front_middle | Right | 47.47 | 11.91 | 51.49 | 14.95 | 4.4243 | 2.1034 | 0.0367 |
| G_front_middle | Left | 45.92 | 10.89 | 49.41 | 11.78 | 4.7387 | 2.1769 | 0.0312 |
| G_postcentral | Left | 40.52 | 14.21 | 44.57 | 13.34 | 4.2994 | 2.0735 | 0.0394 |
| G_precentral | Left | 39.92 | 11.27 | 42.79 | 8.74 | 4.0488 | 2.0122 | 0.0456 |
| G_front_inf‐Opercular | Left | 55.62 | 14.76 | 65.25 | 17.65 | 4.1853 | 2.0458 | 0.0421 |
DISCUSSION
As examined using tract‐tracing and other methods [Berke, 1960; Markowitsch et al., 1984], claustral neuroanatomy is noteworthy across multiple species for receiving input from almost all regions of cortex and then directly projecting back to them [Buchanan and Johnson, 2011; Edelstein and Denaro, 2004; Kowianski et al., 1999; Smythies et al., 2012a, b]. Yet, while prior descriptive or small‐sample human neuroimaging examinations of the claustrum's white matter architecture have been performed, to our knowledge, ours is the first population‐level study to quantitatively examine the structural connectivity of the claustrum using in vivo brain mapping and graph analytic methods.
Here, using DTI tractography and graph theoretical analytics, we computationally verify in a large human sample (N = 100) that the human claustrum is widely connected as assessed by its contribution to graph‐wide diameter. Dense connectivity exists to most cortical areas although the structure is only modestly connected to limbic and occipital regions. While many contemporary examinations of the claustrum subdivide the claustrum [Fernandez‐Miranda et al., 2008a, b, 2012; Gattass et al., 2014; Smith and Alloway, 2014; Smith et al., 2012], we did not feel that any such divisions would be necessary for our study, as boundaries are generally drawn according to perceived function or previously reported cortical region connectivity. In contrast, this study sought to solidify understanding of the entire connectome of the claustrum and attempt to better comprehend how this connectivity shapes networks; theoretically, one could use our network analysis in the future to subdivide the region according to network participation. Careful and accurate delineation of the claustrum itself, however, is already difficult at standard imaging resolution, and therefore, research that seeks to understand single networks that the claustrum participates in ought to be carefully designed to achieve fine resolution.
In our results, the claustrum appears to play a central role in linking multiple disparate structural brain networks. Conversely, the nodal betweenness centrality of predominantly frontal regions connected to the claustrum change significantly more if their connections to the claustrum are systematically removed than any other region. Traditionally, betweenness centrality increases as the relative contributions of shorter pathways come to dominate a network. Increases in betweenness centrality following removal of claustrum connectivity indicate that the white matter networks associated with them are, more heavily influenced by shorter—presumably local—pathways. This illustrates that the claustrum occupies a unique, and presumably critical, “location” in the overall architecture of network connectivity in the brain. Given the traffic converging on this hub, damage to the claustrum may be difficult to compensate for, since efficiency would be so greatly compromised by distributing such a large number of connections over a larger area.
Indeed, disruption to corticoclaustral connectivity—forcing a greater dependence among local regional connections—appears to result in a variety of neurological symptoms (Supporting Information, Table 3). Studies from a variety of human neurological and psychiatric syndromes [Smythies et al., 2014] have identified altered claustral morphometry or have found disrupted patterns of white matter connectivity. Disorders such as Wilson's Disease, for example, are noteworthy for their specificity of claustral involvement. Wilson's Disease is an autosomal recessive genetic disorder in which copper accumulates in brain tissues [Lorincz, 2010]. Copper deposition in the claustrum is notable using T1‐weighted neuroimaging and is often considered a neurological hallmark of the disease, and can lead to general executive control difficulty, as well as various symptoms of memory dysfunction [Sener, 1993]. Other neurological conditions have been examined in the claustrum as well. Negative correlations between anhedonia and metabolism in the claustrum have been shown in both patients with unipolar depression and bipolar disorder [Chen et al., 2011]. Claustral amyloid plaques accumulation has been implicated in the outcomes of Alzheimer's disease and aging [Fernandez‐Miranda et al., 2008a, b; Morys et al., 1994]. Severity of delusions in schizophrenia is correlated with the reduction in left claustral volume [Cascella et al., 2011] and schizophrenia patients with hallucinations show signs of white matter excesses [Shapleske et al., 2002]. See Supporting Information for further summary of clinical syndromes associated with claustrum dysfunction.
Our identification of the strongest network dependencies existing between the claustrum, frontal, and cingulate cortices is a particularly noteworthy result. The simulated excision of the claustrum's representation in the overall network forces these regions to effectively bias their connectivity via more spatially local pathways. Such changes do not appear in other brain areas. This finding supports previous findings of frontal lobe connectivity across many methodologies, including electrophysiology [Berke, 1960; Chachich and Powell, 2004; Cortimiglia et al., 1991; Grasby and Talk, 2013], anterograde, and retrograde tracers [Arikuni and Kubota, 1985; Bayer and Altman, 1991; Pearson et al., 1982; Sadowski et al., 1997], and neuroimaging [Fauvel et al., 2014; Milardi et al., 2013; Park et al., 2012] and highlights the importance of widening the scope of functional observations of the claustrum beyond the primary sensory and motor regions. Some authors have suggested that the claustrum may act to synchronize cortical subnetworks, which are responsible for a variety of coordinated behaviors [Smith and Alloway, 2010] possibly serving to counterbalance spatially over‐represented cortical regions [LeVay and Sherk, 1981; Minciacchi et al., 1995]. The claustrum may also be particularly useful in sensory cross‐modal matching [Arnow et al., 2002] and possibly modulate the cortical neuron receptive field properties [Shima et al., 1996]. Such putative roles emphasize the importance of inhibitory claustral connectivity in the regulation of conscious behaviors. Furthermore, it lends credence to the suggestions of Smythies et al. [2012a, b] that, unlike the thalamus, the claustrum may not be a strict multisensory processor, per se, but may be indispensable in organizing the information used by multisensory processors as relevant to the brain's executive functions. In this way, the claustrum may well serve to filter signals about the relationships of all the thousands of sensorimotor inputs from the outside world to and from frontal and cingulate processing subnetworks.
Our study is, however, not without several limitations. Due to the paucity of large sample, in vivo human neuroimaging studies of the claustrum, we chose to focus more broadly on the macroscale structural connectomics properties of the structure, rather than examine factors which influence between‐group differences. For instance, we pooled male and female subjects together to get a picture of the connectivity of the average claustrum. We did not consider handedness or age‐related effects. Handedness, gender, age, ethnicity, and a number of other phenotypic as well as genetic factors, will be very important for understanding the role of the claustrum in developmental, mental health, and age‐related disorders.
Additionally, we made no attempt to identify subdivisions of the claustrum. Already exploring one of the smaller structures of the brain, it is arguable whether conventional imaging sequences provide sufficient spatial resolution or image contrast to permit the identification of tissue subclasses or reliable spatial landmarks thereof. While earlier studies have suggested a compelling approach to parcellating claustral subdivisions using Magnetic Resonance (MR) [Pathak and Fernandez‐Miranda, 2014], further validation and testing of such approaches are likely required before they can be of practical utility. Thus, here, we chose to focus on the claustrum as a single entity, which likely limits our ability to assign claustro‐cortico projections with high specificity. Advances in ultrahigh field, high‐resolution MRI will make such studies possible.
All in all, the high‐resolution imaging of the claustrum and the exploration of phenomic variables is encouraged in future studies to determine how the structure and connectivity of the claustrum varies between genders, how it develops, and is altered due to aging and age‐related disease.
While we have discussed how our results may support some of the functional findings of previous investigations, further research using functional imaging—such as fMRI—would be necessary to confirm that these functions are in fact correlated with these putative connections. Moreover, traditional functional imaging methods may not provide sufficient spatial resolution to accurately assess such a small structure; technological advancements may be necessary to obtain an accurate picture of the behavior of the claustrum. Hagmann et al. [2008] applied community detection or modularity analysis to demonstrate a close relationship between structural connections and functional connections. Therefore, an analysis of the structural connections of the claustrum may help generate hypotheses regarding its functional role in the networks to which it contributes. Despite these caveats, our results are in line with previous neuroimaging and non‐neuroimaging findings.
Given the results of our large‐scale analysis—and in concurrence with previous insightful research [Crick and Koch, 2005]—we concur with the hypothesis that the claustrum is likely more than a simple neural relay station in the overall context of inter‐regional white matter networks. Its differential influence on brain region subnetworks typically associate with high cognitive activity, attention, and action suggest that the claustrum plays a central role in linking multiple sensory networks with those regions, which can interpret and take action on such information. In support of previous hypotheses on the role of consciousness [Smythies et al., 2012a, b], our results show that the claustrum's position in the macroscale network would be structurally capable of allowing salient sensory information to enter brain regions associated with conscious thought while keeping superfluous input out of awareness.
In conclusion, the claustrum embodies a highly connected yet still curious brain structure whose in vivo examination in humans using neuroimaging has remained challenging due to its small, thin, irregular shape, and location. Determining the functions of the claustrum in humans has formed the basis of recent empirical and theoretical exploration [Smythies et al., 2014]. Our study highlights the claustrum as a critical and “ideally located” component in brain connectomic architecture upon which frontal and cingulate regions appear particularly dependent. These results have particular relevance for neurological disorders and diseases that affect the structure and connectivity of the claustrum and provide further support for the claustrum as a putative neural filter for conscious thought.
CONFLICT OF INTEREST STATEMENT
This research was conducted in absence of any commercial or financial relationships which could be construed as a potential conflict of interest.
CONTRIBUTIONS
CMT pre‐processed all neuroimaging data, helped to prepare the manuscript and supplemental materials. AI conducted all graph theoretical connectivity analyses. SYMG helped to prepare figures and review the manuscript text for technical correctness. JDVH conceived of the examination, guided the necessary data processing and analyses, and helped to author the manuscript and supplemental materials. All authors had the opportunity to discuss the results, participated in writing/editing, and made comments on earlier versions of the manuscript.
Supporting information
Supplementary Information
ACKNOWLEDGMENTS
The authors wish to acknowledge the artistic talents of the late Mr. Carlos Mena, his creativity, friendship, and, for this article, his contribution to the graphical illustration of claustral anatomy. Finally, the authors thank the dedicated staff of the Institute for Neuroimaging and Informatics based at the University of Southern California, Los Angeles, CA.
Footnotes
Note: In our examination, due to issues of image resolution and the uncertainty in the literature over the precise intraregional boundaries for so doing, we purposefully did not attempt to segment the claustrum into putative substructures. However, see the chapter by Pathak and Fernandez‐Miranda (2014), for a potential approach for subdividing the claustrum based on MRI data.
REFERENCES
- Arikuni T, Kubota K (1985): Claustral and amygdaloid afferents to the head of the caudate nucleus in macaque monkeys. Neurosci Res 2:239–254. [DOI] [PubMed] [Google Scholar]
- Arnow BA, Desmond JE, Banner LL, Glover GH, Solomon A, Lake Polan M, Lue TF, Atlas SW (2002). Brain activation and sexual arousal in healthy, heterosexual males. Brain 125(Pt 5):1014–1023. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A (2000): In vivo fiber tractography using DT‐MRI data. Magn Reson Med 44:625–632. [DOI] [PubMed] [Google Scholar]
- Bassett DS, Bullmore ET (2009): Human brain networks in health and disease. Curr Opin Neurol 22:340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Brown JA, Deshpande V, Carlson JM, Grafton ST (2011): Conserved and variable architecture of human white matter connectivity. Neuroimage 54:1262–1279. [DOI] [PubMed] [Google Scholar]
- Bayer SA, Altman J (1991): Development of the endopiriform nucleus and the claustrum in the rat brain. Neuroscience 45:391–412. [DOI] [PubMed] [Google Scholar]
- Berke JJ (1960): The claustrum, the external capsule and the extreme capsule of Macaca mulatta . J Comp Neurol 115:297–331. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E (1982): Neuronal types in the claustrum of man. Anat Embryol (Berl) 163:447–460. [DOI] [PubMed] [Google Scholar]
- Buchanan KJ, Johnson JI (2011): Diversity of spatial relationships of the claustrum and insula in branches of the mammalian radiation. Ann N Y Acad Sci 1225 Suppl 1:E30–E63. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. (2009): Complex brain networks: Graph theoretical analysis of structural and functional systems. Nat Rev Neurosci 10:186–198. [DOI] [PubMed] [Google Scholar]
- Butler AB, Molnar Z, Manger PR (2002): Apparent absence of claustrum in monotremes: Implications for forebrain evolution in amniotes. Brain Behav Evol 60:230–240. [DOI] [PubMed] [Google Scholar]
- Cahalane DJ, Charvet CJ, Finlay BL (2012): Systematic, balancing gradients in neuron density and number across the primate isocortex. Front Neuroanat 6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Whalen S, Huang J, Berger KL, DeLano MC (2003): Asymmetry of subinsular anisotropy by in vivo diffusion tensor imaging. Hum Brain Mapp 20:82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey RG, Bear MF, Diamond IT (1980): The laminar organization of the reciprocal projections between the claustrum and striate cortex in the tree shrew, Tupaia glis . Brain Res 184:193–198. [DOI] [PubMed] [Google Scholar]
- Cascella NG, Gerner GJ, Fieldstone SC, Sawa A, Schretlen DJ (2011): The insula‐claustrum region and delusions in schizophrenia. Schizophr Res 133:77–81. [DOI] [PubMed] [Google Scholar]
- Chachich ME, Powell DA (2004): The role of claustrum in Pavlovian heart rate conditioning in the rabbit (Oryctolagus cuniculus): Anatomical, electrophysiological, and lesion studies. Behav Neurosci 118:514–525. [DOI] [PubMed] [Google Scholar]
- Chen CH, Suckling J, Lennox BR, Ooi C, Bullmore ET (2011): A quantitative meta‐analysis of fMRI studies in bipolar disorder. Bipolar Disord 13:1–15. [DOI] [PubMed] [Google Scholar]
- Cortimiglia R, Crescimanno G, Salerno M, Amato G, Infantellina F (1987): Electrophysiological relationship between claustrum and contralateral area 4 and 6 pyramidal tract neurons. Neurosci Lett 74:193–198. [DOI] [PubMed] [Google Scholar]
- Cortimiglia R, Crescimanno G, Salerno M, Amato G (1991): The role of the claustrum in the bilateral control of frontal oculomotor neurons in the cat. Exp Brain Res 84:471–477. [DOI] [PubMed] [Google Scholar]
- Crescimanno G, Salerno MT, Cortimiglia R, Amato G (1989). Claustral influence on ipsi‐and contralateral motor cortical areas, in the cat. Brain Res Bull 22:839–843. [DOI] [PubMed] [Google Scholar]
- Crick F, Koch C (2003): A framework for consciousness. Nat Neurosci 6:119–126. [DOI] [PubMed] [Google Scholar]
- Crick FC, Koch C (2005): What is the function of the claustrum? Philos Trans R Soc Lond B Biol Sci 360:1271–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI (1999): Cortical surface‐based analysis. I. Segmentation and surface reconstruction. Neuroimage 9:179–194. [DOI] [PubMed] [Google Scholar]
- Destrieux C, Fischl B, Dale A, Halgren E (2010): Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. Neuroimage 53:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinov I, Lozev K, Petrosyan P, Liu Z, Eggert P, Pierce J, Zamanyan A, Chakrapani S, Van Horn J, Parker DS, Magsipoc R, Leung K, Gutman B, Woods R, Toga A (2010): Neuroimaging study designs, computational analyses and data provenance using the LONI pipeline. PLoS One 5:e13070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druga R (2014): Structure and connections of the claustrum In Smythies J, Edelstein L, Ramachandran VS, editors. The Claustrum: Structural, Functional, and Clinical Neuroscience, Vol. 1 San Diego, CA: Academic Press; pp 29–84. [Google Scholar]
- Edelstein LR and Denaro FJ (2004): The claustrum: A historical review of its anatomy, physiology, cytochemistry and functional significance. Cell Mol Biol (Noisy‐le‐grand) 50:675–702. [PubMed] [Google Scholar]
- Fauvel B, Groussard M, Chetelat G, Fouquet M1, Landeau B1, Eustache F1, Desgranges B1, Platel H2 (2014): Morphological brain plasticity induced by musical expertise is accompanied by modulation of functional connectivity at rest. Neuroimage 90:179–188. [DOI] [PubMed] [Google Scholar]
- Fernandez‐Miranda JC, Rhoton AL Jr, Alvarez‐Linera J, Kakizawa Y, Choi C, de Oliveira EP (2008a): Three‐dimensional microsurgical and tractographic anatomy of the white matter of the human brain. Neurosurgery 62(6 Suppl 3):989–1026; discussion 1026–1028. [DOI] [PubMed] [Google Scholar]
- Fernandez‐Miranda JC, Rhoton AL Jr, Kakizawa Y, Choi C, Alvarez‐Linera J (2008b): The claustrum and its projection system in the human brain: A microsurgical and tractographic anatomical study. J Neurosurg 108:764–774. [DOI] [PubMed] [Google Scholar]
- Fernandez‐Miranda JC, Pathak S, Engh J, Jarbo K, Verstynen T, Yeh FC, Wang Y, Mintz A, Boada F, Schneider W, Friedlander R (2012): High‐definition fiber tractography of the human brain: Neuroanatomical validation and neurosurgical applications. Neurosurgery 71:430–453. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM (1999): Cortical surface‐based analysis. II: Inflation, flattening, and a surface‐based coordinate system. Neuroimage 9:195–207. [DOI] [PubMed] [Google Scholar]
- Gattass R, Soares JG, Desimone R, Ungerleider LG (2014): Connectional subdivision of the claustrum: Two visuotopic subdivisions in the macaque. Front Syst Neurosci 8:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasby K, Talk A (2013): The anterior claustrum and spatial reversal learning in rats. Brain Res 1499:43–52. [DOI] [PubMed] [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O (2008): Mapping the structural core of human cerebral cortex. Plos Biol 6:1479–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassmannova J (1977): Role of the claustrum in sensory activation. Physiol Bohemoslov 26:345–352. [PubMed] [Google Scholar]
- Hatam M, Sheybanifar M, Nasimi A (2013): Cardiovascular responses of the anterior claustrum; its mechanism; contribution of medial prefrontal cortex. Auton Neurosci 179:68–74. [DOI] [PubMed] [Google Scholar]
- Irimia A, Van Horn JD (2014): Systematic network lesioning reveals the core white matter scaffold of the human brain. Front Hum Neurosci 8:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia A, Chambers MC, Torgerson CM, Filippou M, Hovda DA, Alger JR, Gerig G, Toga AW, Vespa PM, Kikinis R, Van Horn JD (2012a): Patient‐tailored connectomics visualization for the assessment of white matter atrophy in traumatic brain injury. Front Neurol 3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia A, Chambers MC, Torgerson CM, Van Horn JD (2012b): Circular representation of human cortical networks for subject and population‐level connectomic visualization. Neuroimage 60:1340–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapakin S (2011): The claustrum: Three‐dimensional reconstruction, photorealistic imaging, and stereotactic approach. Folia Morphol (Warsz) 70:228–234. [PubMed] [Google Scholar]
- Kowianski P, Dziewiatkowski J, Kowianska J, Moryś J (1999): Comparative anatomy of the claustrum in selected species: A morphometric analysis. Brain Behav Evol 53:44–54. [DOI] [PubMed] [Google Scholar]
- Landau E (1919): The comparative anatomy of the nucleus amygdalae, the claustrum and the insular cortex. J Anat 53(Pt 4):351–360. [PMC free article] [PubMed] [Google Scholar]
- LeVay S and Sherk H. (1981): The visual claustrum of the cat. II. The visual field map. J Neurosci 1:981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz MT (2010): Neurologic Wilson's disease. Ann N Y Acad Sci 1184:173–187. [DOI] [PubMed] [Google Scholar]
- Markowitsch HJ, Irle E, Bang‐Olsen R, Flindt‐Egebak P (1984): Claustral efferents to the cat's limbic cortex studied with retrograde and anterograde tracing techniques. Neuroscience 12:409–425. [DOI] [PubMed] [Google Scholar]
- Mathur BN (2014) The claustrum in review. Front Syst Neurosci 8:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur BN, Caprioli RM, Deutch AY (2009): Proteomic analysis illuminates a novel structural definition of the claustrum and insula. Cereb Cortex 19:2372–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milardi D, Bramanti P, Milazzo C, Finocchio G, Arrigo A, Santoro G, Trimarchi F, Quartarone A, Anastasi G, Gaeta M (2013): Cortical and Subcortical Connections of the Human Claustrum Revealed In Vivo by Constrained Spherical Deconvolution Tractography. Cereb Cortex. Available at: http://cercor.oxfordjournals.org/content/early/2013/09/07/cercor.bht231.long. [DOI] [PubMed] [Google Scholar]
- Minciacchi D, Granato A, Antonini A, Tassinari G, Santarelli M, Zanolli L, Macchi G (1995): Mapping subcortical extrarelay afferents onto primary somatosensory and visual areas in cats. J Comp Neurol 362:46–70. [DOI] [PubMed] [Google Scholar]
- Mirzasoleiman B, Jalili M (2011): Failure tolerance of motif structure in biological networks. PLoS One 6:e20512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morys J, Narkiewicz O, Maciejewska B, Wegiel J, Wisniewski HM (1994): Amyloid deposits and loss of neurones in the claustrum of the aged dog. Neuroreport 5:1825–1828. [DOI] [PubMed] [Google Scholar]
- Naghavi HR, Eriksson J, Larsson A, Nyberg L (2007): The claustrum/insula region integrates conceptually related sounds and pictures. Neurosci Lett 422:77–80. [DOI] [PubMed] [Google Scholar]
- Park S, Michael Tyszka J, Allman JM (2012): The claustrum and insula in Microcebus murinus: A high resolution diffusion imaging study. Front Neuroanat 6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak S, Fernandez‐Miranda JC (2014): Structural and functional connectivity of the claustrum in the human brain In: Smythies J, Edelstein L, Ramachandra VS, editors. The Claustrum: Structural, Functional, and Clinical Neuroscience, Vol. 1 San Diego, CA: Academic Press; pp 209–224. [Google Scholar]
- Pearson RC, Brodal P, Gatter KC, Powell TP (1982): The organization of the connections between the cortex and the claustrum in the monkey. Brain Res 234:435–441. [DOI] [PubMed] [Google Scholar]
- Puelles L (2014): Development and evolution of the claustrum In: Smythies J, Edelstein L, Ramachandran VS, editors. The Claustrum: Structural, Functional, and Clinical Neuroscience, Vol. 1 San Diego, CA: Academic Press. [Google Scholar]
- Remedios R, Logothetis NK, Kayser C (2010): Unimodal responses prevail within the multisensory claustrum. J Neurosci 30:12902–12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remedios R, Logothetis NK, Kayser C (2014): A role of the claustrum in auditory scene analysis by reflecting sensory change. Front Syst Neurosci 8:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski M, Morys J, Jakubowska‐Sadowska K, Narkiewicz O (1997): Rat's claustrum shows two main cortico‐related zones. Brain Res 756:147–152. [DOI] [PubMed] [Google Scholar]
- Salerno MT, Cortimiglia R, Crescimanno G, Amato G, Infantellina F (1984): Effects of claustrum stimulation on spontaneous bioelectrical activity of motor cortex neurons in the cat. Exp Neurol 86:227–239. [DOI] [PubMed] [Google Scholar]
- Sener RN (1993): The claustrum on MRI: Normal anatomy, and the bright claustrum as a new sign in Wilson's disease. Pediatr Radiol 23:594–596. [DOI] [PubMed] [Google Scholar]
- Shapleske J, Rossell SL, Chitnis XA, Suckling J, Simmons A, Bullmore ET, Woodruff PW, David AS (2002): A computational morphometric MRI study of schizophrenia: Effects of hallucinations. Cereb Cortex 12:1331–1341. [DOI] [PubMed] [Google Scholar]
- Sherk H (2013): Physiology of the Claustrum, Chapter 5, In: Smythies J, Edelstein L, Ramachandran VS, editors. The Claustrum: Structural, Functional, and Clinical Neuroscience, Vol. 1. San Diego, CA: Academic Press. [Google Scholar]
- Shima K, Hoshi E, Tanji J (1996): Neuronal activity in the claustrum of the monkey during performance of multiple movements. J Neurophysiol 76:2115–2119. [DOI] [PubMed] [Google Scholar]
- Smith JB, Alloway KD (2010): Functional specificity of claustrum connections in the rat: Interhemispheric communication between specific parts of motor cortex. J Neurosci 30:16832–16844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JB, Alloway KD (2014): Interhemispheric claustral circuits coordinate sensory and motor cortical areas that regulate exploratory behaviors. Front Syst Neurosci 8:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JB, Radhakrishnan H, Alloway KD (2012): Rat claustrum coordinates but does not integrate somatosensory and motor cortical information. J Neurosci 32:8583–8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythies J, Edelstein L, Ramachandran, V . (2012a). The functional anatomy of the claustrum: The net that binds. WebmedCentral Neurosciences 3 http://www.webmedcentral.com/wmcpdf/Article_WMC003182.pdf. [Google Scholar]
- Smythies J, Edelstein L, Ramachandran V (2012b): Hypotheses relating to the function of the claustrum. Front Integr Neurosci 6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythies J, Edelstein LR, Ramachandran VA, editors (2014): The Claustrum. San Diego, CA: Academic Press. [Google Scholar]
- Spector I, Hassmannova Y, Albe‐Fessard D (1970): A macrophysiological study of functional organization of the claustrum. Exp Neurol 29:31–51. [DOI] [PubMed] [Google Scholar]
- Sporns O (2011): Networks of the Brain. Cambridge, MA: MIT Press. [Google Scholar]
- Sporns O (2012): From simple graphs to the connectome: Networks in neuroimaging. Neuroimage 62:881–886. [DOI] [PubMed] [Google Scholar]
- Talairach J, Tournoux P (1988): Co‐planar stereotaxic atlas of the human brain. New York: Thieme. [Google Scholar]
- Tanne‐Gariepy J, Boussaoud D, Rouiller EM (2002): Projections of the claustrum to the primary motor, premotor, and prefrontal cortices in the macaque monkey. J Comp Neurol 454:140–157. [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, Stam CJ, Boersma M, Hulshoff Pol HE (2008): Small‐world and scale‐free organization of voxel‐based resting‐state functional connectivity in the human brain. Neuroimage 43:528–539. [DOI] [PubMed] [Google Scholar]
- Van Horn JD, Irimia A, Torgerson CM, Chambers MC, Kikinis R, Toga AW (2012): Mapping connectivity damage in the case of Phineas Gage. PLoS One 7:e37454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkuti B, Cavusoglu M, Kullik A, Schiffler B, Veit R, Yilmaz Ö, Rosenstiel W, Braun C, Uludag K, Birbaumer N, Sitaram R (2011): Quantifying the link between anatomical connectivity, gray matter volume and regional cerebral blood flow: An integrative MRI study. PLoS One 6(4): e14801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhite BL, Teyler TJ, Hendricks C (1986): Functional relations of the rodent claustral‐entorhinal‐hippocampal system. Brain Res 365:54–60. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


