Abstract
Rapid and robust induction of type I interferon (IFN-I) is a critical event in host antiviral innate immune response. It has been well demonstrated that cyclic GMP-AMP (cGAMP) synthase (cGAS) plays an important role in sensing cytosolic DNA and triggering stimulator of interferon genes (STING)-dependent signaling to induce IFN-I. However, it is largely unknown how cGAS itself is regulated during pathogen infection and IFN-I production. Here, we show that pattern-recognition receptor (PRR) ligands including lipidA, LPS, polyI:C, polydA:dT, and cGAMP induce cGAS expression in a IFN-I-dependent manner in both mouse and human macrophages. Further experiments indicate that cGAS is an IFN-stimulated gene (ISG), and two adjacent IFN-sensitive response elements (ISREs) in the promoter region of cGAS mediate the induction of cGAS by IFN-I. In addition, we show that optimal production of IFNβ triggered by polydA:dT or HSV-1 requires IFNAR signaling. Knockdown of the constitutively expressed DNA sensor DDX41 attenuates polydA:dT-triggered IFNβ production and cGAS induction. By analyzing the dynamic expression of polydA:dT-induced IFNβ and cGAS transcripts, we have found that induction of IFNβ is earlier than cGAS. Furthermore, we have provided evidence that induction of cGAS by IFN-I meditates the subsequent positive feedback regulation of DNA-triggered IFN-I production. Thus, our study not only provides a novel mechanism of modulating cGAS expression, but also adds another layer of regulation in DNA-triggered IFN-I production by induction of cGAS.
Keywords: PRR, cGAS, cGAMP, STING, DDX41, Type I IFN, ISG, DNA sensor
Introduction
Type I interferon (IFN-I) has been recognized as the first line of defense against viral infection. Rapid and robust induction of IFN-I is a key event during host antiviral responses (1, 2). The binding of IFN-I to its receptor IFNAR initiates a signaling cascade, which leads to the induction of more than 300 IFN-stimulated genes (ISGs) (3-5). Some ISGs encode proteins with potential for direct antiviral activity, including ISG15 (IFN-stimulated protein of 15 kDa), the GTPase Mx1 (myxovirus resistance 1), ribonuclease L (RNaseL), and protein kinase R (PKR; also known as EIF2αK2). However, more ISGs encode pattern-recognition receptors (PRRs) that detect viral molecules, modulate signaling pathways, and form an amplification loop resulting in increased IFN production (1, 2). For example, the central members of the mammalian RIG-I-like receptors (RLRs) RIG-I (retinoic acid-inducible gene I) and MDA5 (melanoma differentiation factor 5) are found in the cytosol of the most cell types and strongly induced by IFNs in a positive feedback loop of RNA virus detection (2, 6). AIM2-like receptors (ALRs) including IFI16 are another family of cytosolic DNA sensors that are induced by IFN and positively regulate IFN-I production by sensing more viral DNA (7-9). Numerous DNA sensors have been identified in the past several years. These sensors recognize intracellular or pathogenic DNA and trigger IFN-I production signaling (10, 11). However, it is unclear whether these DNA sensors are also regulated by IFN signaling and play roles in the positive regulation of IFN-I production.
Cyclic GMP-AMP (cGAMP) synthase (cGAS) is a newly identified DNA sensor that triggers IFN-I production (12-15). cGAS binds to cytosolic pathogenic DNA as well as self DNA in a sequence-independent manner, which may allow this DNA sensor to detect any DNA that invades the cytoplasm. Upon DNA binding, cGAS is activated to catalyze the synthesis of a unique isomer of cGAMP that contains G (2’ 5’)pA and A (3’ 5’)pG phosphodiester linkages from ATP and GTP (16-19). This cGAMP, termed 2’3’-cGAMP, functions as a second messenger that binds to the endoplasmic reticulum membrane protein STING (also known as TMEM173, MITA, MPYS, or ERIS) (15, 19, 20). cGAMP binding induces a conformational change of STING, which then recruits the kinases TBK1 to activate IRF3 and trigger IFN-I production (21, 22). Recent genetic studies have validated the essential role of cGAS in sensing cytosolic DNA in multiple cell types and in immune defense against DNA viruses including HSV-1 and MHV68 in vivo (14, 23). In addition, cGAS has been shown to be an innate immune sensor of RNA viruses, including HIV and WNV (13, 23). cGAS is also essential for induction of IFN-I during Chlamydia trachomatis and Listeria monocytogenes infections (24, 25). Although the functions of cGAS and cGAS-mediated innate immune responses have been extensively studied, the regulation of cGAS itself during pathogen infection is largely unknown. In addition, the crosstalk between cGAS and other DNA sensors is also still unclear.
Here, we provide data to show that cGAS is specifically induced by IFN-I through two adjacent IFN-sensitive response elements (ISREs) in the cGAS promoter. Positive feedback regulation loop is required for optimal production of DNA-triggered IFN-I production. Knockdown of the constitutively expressed DNA sensor DDX41 attenuates both polydA:dT-triggered IFNβ production and cGAS induction. We further show that induction of cGAS by the first wave of IFN-I plays a role in the subsequent positive feedback regulation of DNA-triggered IFN-I production. Our study not only demonstrates that cGAS is positively regulated by IFN-I, but also indicates that the induction of cGAS plays a role in IFN-I positive feedback loop.
Material and Methods
Mice and Reagents
Wild type C57BL/6 (6~8 weeks of age) and age-matched Ifnar1−/−, Stat1−/−, Myd88−/−, Trif−/−, Cardif−/−, Sting-gt/gt, and Irf3−/− male mice were either bred at the UCLA animal facility or purchased from the Jackson Laboratory. All mice experiments were performed in accordance with guidelines from the University of California, Los Angeles Institutional Animal Care and Use Committee. cGAMP, polyI:C, and polydA:dT were purchased from InvivoGen (San Diego, CA). LipidA was from Enzo life sciences (Farmingdale, NY). LPS (Escherichia coli 0111:B4), anti-α-tubulin antibody, human cGAS antibody (anti-C6ORF150), and anti-p204 antibody were from Sigma-Aldrich (St. Louis, MO). Anti-Ddx41 (H00051428) antibody was from Novus Biologicals (Littleton, CO). Anti-GAPDH (GT239) was from GeneTex (Irvine, CA). Recombinant human and mouse IFNα was from PBL interferon source (Piscataway, NJ) and recombinant mouse IFNγ was from R&D systems (Minneapolis, MN).
Cell culture and activation
HEK293T, RAW264.7, and THP-1 cell lines were obtained from American Type Culture Collection (Manassas, VA). HEK293T and RAW264.7 cells were maintained in DMEM containing 10% FBS and 1% penicillin/streptomycin. THP-1 cells were cultured in RPMI1640 supplemented with 5% FBS and 1% penicillin/streptomycin. For bone marrow-derived macrophage (BMM) differentiation, bone marrow cells were harvested from wild type or indicated gene-deficient C57B/L6 mice and differentiated in DMEM+10%FBS for 7 days with 10 ng/ml of M-CSF. The cell culture medium was replaced on day 3 and day 6, and on day 7 the cells were used for experiments as BMMs. For J2 virus-immortalized macrophages (J2-BMMs), a cell line (called GG2EE) transformed by retrovirus expressing v-raf and c-myc, was established and grown in RPMI1640 (10mM HEPES PH7.8, 10%FBS, 1% penicillin/streptomycin). Supernatant containing J2 viruses was harvested and filtered through 0.22 μM filter. Bone marrow cells were infected with the J2 virus and immortalized as described previously (26, 27). Femur and tibia from Irf7−/− mice (8 weeks, male, C57BL/6 background) were overnight shipped from Michael S. Diamond's lab (Washington University). Irf7−/− bone marrow cells were differentiated into BMMs and immortalized as Irf7−/− J2-BMMs. To activate BMMs or J2-BMMs, 100 ng/ml LPS was added into culture medium, or indicated amount of cGAMP, polyI:C, or polydA:dT was transfected into cells by Lipofectamine 2000 (Life Technologies). The ratio of transfection reagent to ligands was 2.5 (μl/μg). Detail Lipofectamine 2000 transfection protocol was followed as described in previous study and manufacturer's instructions (28). Prior to being activated by stimulation with IFNα or transfection with polyI:C or polydA:dT, THP-1 cells were differentiated into macrophages by incubating with 50 nM phorbol 12-myristate 13-acetate (PMA; Sigma) for 16 h and further cultured for an additional 48 h without PMA.
RNA isolation and quantitative PCR (qPCR)
Total RNA was extracted with TRIzol reagent (Life Technologies) according to manufacturer's instructions. 1 μg RNA from each sample was reverse transcribed by using iScript One-Step RT-PCR Kit with SYBR Green Dye (Bio-Rad). Real-time quantitative RT-PCR analysis was performed by using SensiFAST SYBR & Fluorescein Kit (Bioline) and CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Relative mRNA expression level of genes was normalized to the internal control ribosomal protein gene Rpl32 by using 2−ΔΔCt cycle threshold method (29). Primer sequences for qPCR were obtained from primer bank and are available upon request (30).
Microarray and RNA sequencing (RNA-Seq)
Microarrays were perform on Affymetrix Mouse Genome 430.2 array at UCLA Genotyping and Sequencing Core as described in our previous study (5). The data were deposited in GEO (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE35825 accession no. GSE35825). Briefly, wild type BMMs were stimulated with 62.5 U/ml IFNα or 1 U/ml IFNγ for 2.5 h. Total RNA was extracted for microarray experiment. In this study, we further analyzed our published microarray data and focused on the regulation of cGAS by IFN. For RNA sequencing experiment, day 7 BMMs differentiated from wild type or Ifnar1−/− mice were stimulated with 100 ng/ml lipidA for 4 h or 12 h. Total RNA was extracted, cDNA libraries were constructed by using TruSeq SBS Kit v3 (FC-401-3001) according to manufacturer's guidelines (Illumina, San Diego, CA). Next-generation sequencing was performed by using Illumina HiSeq2000 with 100 bp single end reads at High Throughput Sequencing Core of the UCLA Broad Stem Cell Research Center. Details of RNA-Seq data analysis was described as previous study (27).
ELISA and Immunoblot
IFNα and IFNβ in culture supernatant was quantified with VeriKine Mouse Interferon Alpha and Beta ELISA Kit (PBL interferon source), respectively, according to manufacturer's instructions. For immunoblot analysis, cells were collected in Triton lysis buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 and 5% glycerol) containing complete protease inhibitors (Roche). Protein concentrations of the extracts were measured with BCA assay (Thermo Scientific) and equalized with the lysis buffer. Equal amounts of the extracts were loaded and subjected to SDS-PAGE, transferred onto PVDF membrane (Millipore), and then blotted with enhanced chemiluminescence (Pierce) or Odyssey Imaging Systems (LI-COR Biosciences).
cGAS promoter reporter and dual-luciferase reporter assay
The potential transcription (TF) binding sites in the mouse cGas gene promoter region were predicted by MatInspector (Genomatix, Ann Arbor, MI) (31). Conservation analysis of the TF binding sites among the mammalian species was analyzed and viewed by UCSC genome browser (http://genome.ucsc.edu/). Different length of cGAS promoters were amplified from C57BL/6 genome DNA and subcloned into the pGL4.20 [luc2/Puro] vector (Promega, Madison, WI) to generate WT-luc and Δ#3-luc reporter constructs. The IRES#2, IRES#1, and Stat1 binding site of Δ#3-luc reporter were mutated to generate Δ#3-luc-mut#2-luc, Δ#3-luc-mut#1-luc, and Δ#3-luc-mutStat1-luc reporter constructs via QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA), respectively. The indicated cGas promoter reporter construct was co-transfected with Renilla-luciferase reporter into RAW264.7 cells by Amaxa Cell Line Nucleofector Kit V (Lonza). At 12 h post transfection, the cell culture medium was replaced and stimulated with 100ng/ml LPS for another 12 h. The cells was lysed by passive lysis buffer, the firefly luciferase activity of the cGAS reporters was measured and normalized by Renilla luciferase activity according to manufacturer's instructions of the Dual-Luciferase Reporter Assay System (Promega, Madison, WI). The transfection of cGAS reporters constructs in HEK293T cells was according to manufacturer's instructions of Jet-PEI (Polyplus-transfection). At 24 h post transfection, the cells were lysed and the relative luciferase activity was measured as in RAW264.7 cells.
Chromatin Immunoprecipitation Sequencing (ChIP-Seq) data analysis
Stat1 ChIP-Seq raw data from BMMs were downloaded from the Gene Expression Omnibus (accession no. GSE33913). BMMs cells differentiation and activation were described previously (32). Briefly, BMMs were differentiated with M-CSF, and treated with IFNβ or IFNγ for 6 h before cross-linking for chromatin isolation. ChIP reactions were performed with anti-STAT1α antibody from Santa Cruz, and libraries were generated by standard Illumina protocols. Sequenced reads were aligned to mouse genome (mm9) allowing up to two mismatches using Bowtie (33). The data were processed as previously described (34). For peak calling, mouse genome was divided into 100 bp windows. A P value for Poisson distribution of enriched ChIPed DNA over input DNA for each window was calculated. Significant peaks were defined as the windows with significant P value less than 10−3 and with two neighboring windows at the same significance.
Ddx41 and p204 siRNA knockdown
2×105 BMMs were differentiated for 7 days in a 12-well plate. On day 7, cell culture medium was replaced and the cells were transfected with 20 nM non-targeting control, Ddx41-spcific siRNA (Dharmacon RNAi & gene expression, SMARTpool: siGENOME Ddx41 siRNA), or p204-spcific siRNA (Dharmacon RNAi & gene expression, SMARTpool: siGENOME p204 siRNA) by using INTERFERin transfection reagent (Polyplus-transfection) according to manufacturer's instructions. At 36 h post transfection, the knockdown efficiency was measured by western blot.
Lentivirus packaging and lentiviral transduction
Full length of mouse cGAS were cloned into the lentiviral vector pCDF1-CMV-MCS2-EF1-copGFP (CD111B-1; System Bioscience) to make the expression constructs LV-cGAS. LV-Ctrl or LV-cGAS vector was cotransfected into HEK293T cells with the pPACKF1 Packaging plasmids mix (LV100A-1, System Bioscience). Control or cGAS-overexpressing lenti-viruses were produced, and WT or Ifnar1−/− J2-BMM cells were transduced by these lenti-viruses according to user's manual (System Bioscience) and previous study (35).
Software and Graphing
Microarray analysis was performed by using Bioconductor Affy package (http://www.bioconductor.org/). RNA-Seq data was analyzed on UCLA galaxy server (http://galaxy.hoffman2.idre.ucla.edu/root). All graphs were generated with GraphPad Prism and Photoshop.
Results
cGAS is IFN-I-inducible while DDX41 is constitutively expressed in BMMs
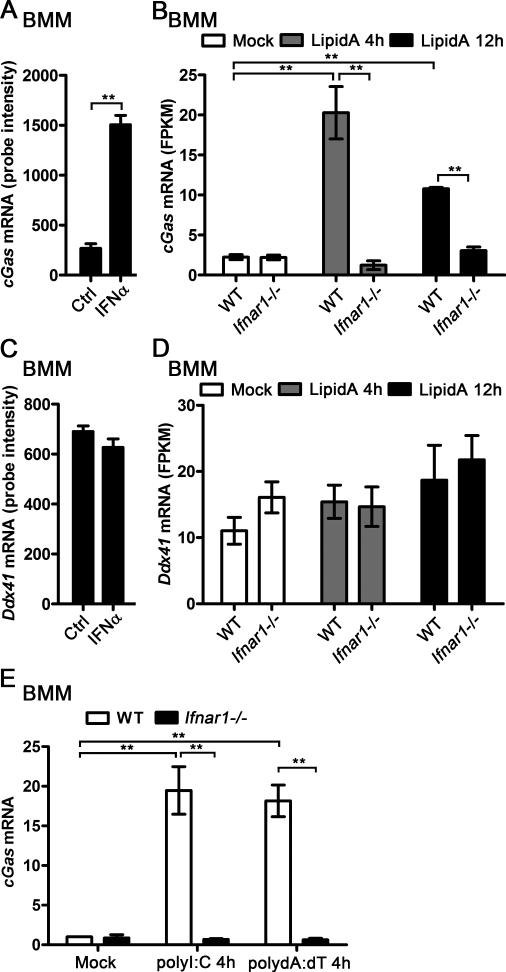
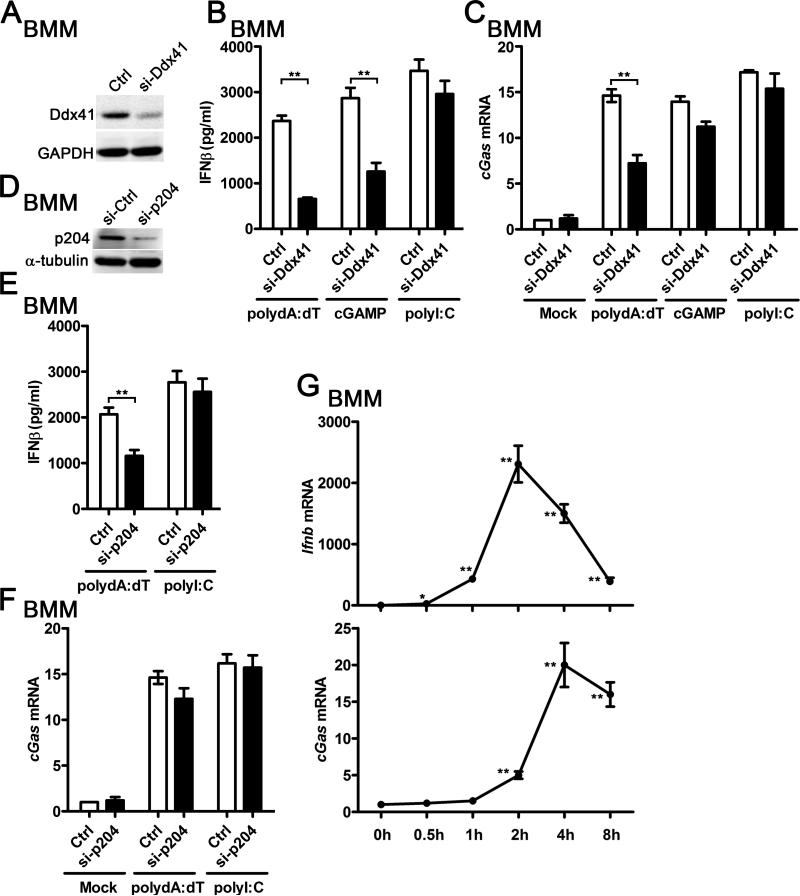
By analyzing the gene expression profile of the IFN-I- and IFN-II-stimulated bone marrow-derived macrophages (BMMs) (5), we found that cGAS mRNA expression was significantly upregulated in IFNα-treated BMMs (Fig. 1A). We compared the cGAS mRNA level in wild type (WT) and Ifnar1−/− BMMs activated by TLR4 ligand lipidA, our RNA-Seq data indicated that cGAS was significantly induced by lipidA in WT BMMs but not in Ifnar1−/− BMMs (Fig. 1B). Higher cGAS mRNA level was detected in WT BMMs than in Ifnar1−/− BMMs when the cells were activated by lipidA (Fig. 1B). However, the expression of another DNA sensor, DDX41, was not affected by IFNα treatment in BMMs (Fig. 1C). No significant difference of DDX41 mRNA level was detected between WT and Ifnar1−/− BMMs, neither in resting condition nor in lipidA-activated condition (Fig. 1D). Furthermore, both polyI:C and polydA:dT significantly induced cGAS expression in WT BMMs, however, the induction of cGAS was completely abolished in Ifnar1−/− BMMs (Fig. 1E). These data suggest that cGAS is an ISG and DDX41 is constitutively expressed in BMMs.
Figure 1.
cGAS and DDX41 expression in BMMs during response to IFNα and lipidA. A, BMMs were treated with 62.5 U/ml IFNα for 2.5 h, RNA was extracted and gene expression profile was detected by Affimetrix430.2 Chip, cGas mRNA level was shown as probe intensity from microarray. B, WT or Ifnar1−/− BMMs were stimulated with 100 ng/ml lipidA for indicated time points, RNA was extracted and gene expression profile was detected by RNA sequencing (RNA-seq). cGas mRNA level was shown as FPKM (fragments per kilobase of transcript per million fragments mapped). C, Ddx41 mRNA level was shown as probe intensity from the microarray data as described in (A). D, Ddx41 mRNA level was shown as FPKM from the RNA-seq data as described in (B). E, WT and Ifnar1−/− BMMs were transfected with 1 μg/ml polyI:C or polydA:dT for 4 h, cGas mRNA level in these cells was detected by qPCR and normalized to Rpl32. **p<0.01 (Student's t-test). Data are from three independent experiments (mean ± s.e.m).
cGAS is specifically induced by IFN-I
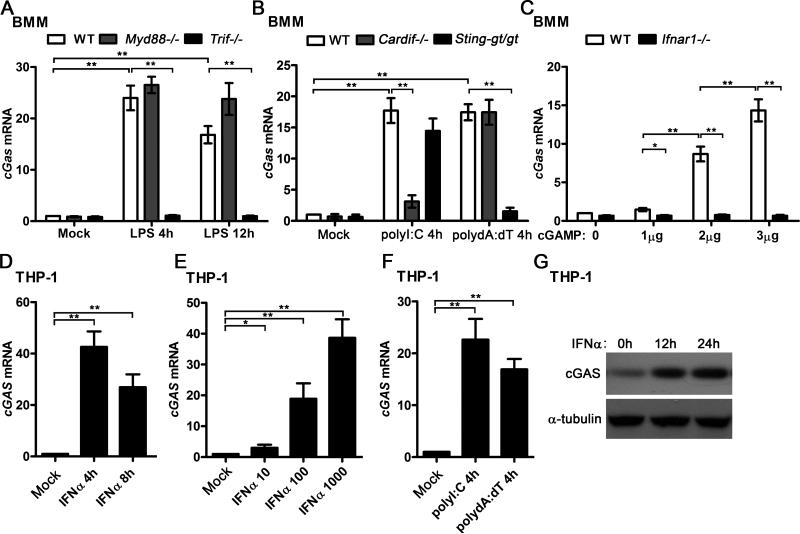
To determine the specificity of cGAS induction by IFN-I, we treated the WT and several gene-deficient BMMs with more different PRR ligands. It is well known that LPS activates NF-κB and MAPK signaling through MyD88-depedent pathway, and triggers IFN-I production through TRIF-dependent pathway (6). We found that LPS stimulation significantly induced cGAS expression in WT and Myd88−/− BMMs, but not in Trif−/− BMMs, which indicated that LPS could induce cGAS expression through the TRIF-dependent pathway (Fig. 2A). Transfection of polyI:C triggers IFN-I production mainly through RIG-I-CARDIF-dependent pathway while transfection of polydA:dT triggers IFN-I production through a STING-dependent pathway(2, 6, 20). We found that both polyI:C and polydA:dT induced cGAS expression in WT BMMs. However, the induction of cGAS was significantly impaired in Cardif−/− BMMs activated by polyI:C transfection but not in Cardif−/− BMMs activated by polydA:dT transfection. Attenuated polydA:dT-triggered IFN-I production was observed in Sting-gt/gt macrophages which fail to produce detectable STING protein (36, 37). We found that the induction of cGAS was significantly impaired in Sting-gt/gt BMMs activated by transfection of polydA:dT but not in Sting-gt/gt BMMs activated by transfection of polyI:C (Fig. 2B). It has been shown that cGAS coverts DNA to cGAMP to trigger the STING-dependent IFN-I production (12). Interestingly, our results indicated that cGAMP, in turn, could induce cGAS mRNA in a dose-dependent manner in WT BMMs but not in Ifnar1−/− BMMs (Fig. 2C). Taken together, these data suggest that multiple PRR ligands could induce cGAS expression by triggering IFN-I production and activating the IFNAR signaling, while activating other signaling such as the MyD88-dependent pathways does not seem to affect cGAS expression. To test whether cGAS is also induced by IFN-I in human cells, we treated THP-1 cells with IFNα and different PRR ligands. As shown in Fig. 2D, cGAS was significantly induced by IFNα in THP1 cells at the both time points we examined. IFNα triggered cGAS expression in a dose-dependent manner (Fig. 2E). Similar as the data from mouse BMMs, both polyI:C and polydA:dT induced cGAS in THP-1 cells (Fig. 2F). In addition, by using a commercial antibody specifically against human cGAS, we found IFNα significantly induced cGAS protein expression in THP-1 cells (Fig. 2G). Therefore, our data indicate that cGAS could be induced by IFN-I specifically in both mouse and human macrophages.
Figure 2.
cGAS is specifically induced by IFN-I in mouse and human macrophages. A, WT, Myd88−/−, and Trif−/− BMMs were stimulated with 100 ng/ml LPS for indicated time points, cGas mRNA level in these cells was detected by qPCR and normalized to Rpl32. B, WT, Cardif−/−, and Sting-gt/gt BMMs were transfected with 1 μg/ml polyI:C or polydA:dT for 4 h, cGas mRNA level in these cells was detected by qPCR and normalized to Rpl32. C, WT and Ifnar1−/− BMMs were transfected with indicated amount of cGAMP for 4 h, cGas mRNA level in these cells was detected by qPCR and normalized to Rpl32. D, THP-1-differentiated macrophages were treated with 500 U/ml human IFNα for indicated time points, RNA was extracted from these cells and cGAS mRNA level was detected by qPCR and normalized to RPL32. E, THP-1 cells were treated with indicated amount of human IFNα (10 to 1000 U/ml) for 4 h, RNA was extracted from these cells and cGAS mRNA level was detected by qPCR and normalized to RPL32. F, THP-1 cells were transfected with 1 μg/ml polyI:C or polydA:dT for 4 h, RNA was extracted from these cells and cGAS mRNA level was detected by qPCR and normalized to RPL32. G, THP-1 cells were treated with 500 U/ml human IFNα for indicated time points, cGAS protein level was detected by western blot. α-tubulin was shown as a loading control, *p<0.05, **p<0.01 (Student's t-test). Data of (A-F) are from three independent experiments (mean ± s.e.m). Data of (G) is one representative of three independent experiments.
ISREs in cGAS promoter are critical for IFN-I triggered cGAS expression
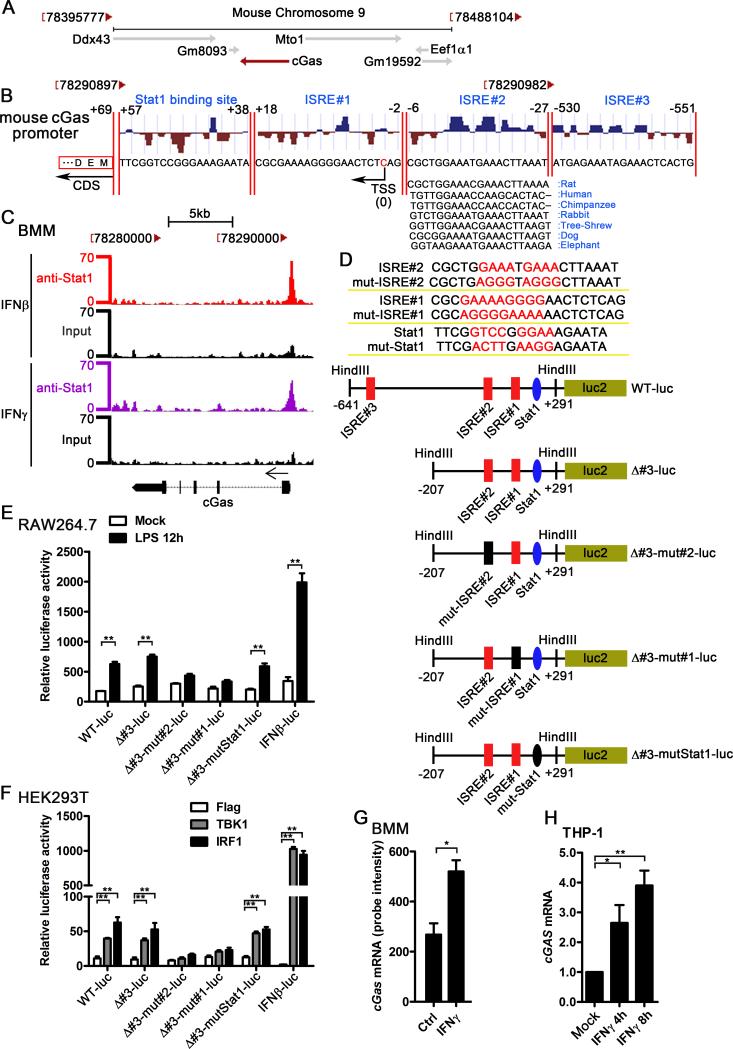
To determine how cGAS is induced by IFN-I, we analyzed the potential transcription factor (TF) binding sites in cGAS 5’UTR region. Mouse cGas locates on chromosome 9, and is encoded by the negative strand of DNA (Fig. 3A). Among all the predicted TF binding sites around the cGas transcription start site (TSS), there are three ISREs and one STAT1 binding site that are potentially responsible for the induction of cGAS by IFN-I. The sequence of ISRE#2 is very conserved in multiple mammalian cGAS 5’UTR (Fig. 3B). Given that ISRE is the motif bound by ISGF3, a tripartite complex of tyrosine-phosphorylated STAT1/STAT2 and IRF9 (38), we analyzed the STAT1 chromatin immunoprecipitation sequencing (ChIP-Seq) data from BMMs to determine if STAT1 could bind to these predicted ISREs and the Stat1 binding site. According to the STAT1 ChIP-Seq data from Tom Maniatis's group (32), we noticed a significant STAT1 binding peak in the promoter region of cGas in BMMs treated with IFNβ or IFNγ. Both ISRE#1 and ISRE#2 were in the middle region of the peak while ISRE#3 was not in the peak region. Although predicted Stat1 binding site was within the peak region, however, much less STAT1-ChIPed reads were aligned in the predicted Stat1 binding site than ISRE#1 and ISRE#2 (Fig. 3C). To verify the potential functions of these TF binding sites, several reporter constructs were made and luciferase reporter assay were performed (Fig.3D). As shown in Fig. 3E, IFNβ-luc reporter was significantly activated by LPS in RAW264.7 cells. Using the similar experimental system, we found that LPS activated WT, Δ#3, and Δ#3-mutStat1 luciferase reporters, but not the Δ#3-mut#1 and Δ#3-mut#2 reporters in RAW264.7 cells (Fig. 3E), which suggested that ISRE#1 and ISRE#2 played a major role for regulating cGAS expression by IFN-I. It has been known that TBK1 and IRF1 trigger IFN-I production in HEK293T cells (39, 40). Consistent with the results from RAW264.7 cells, both TBK1 and IRF1 activated WT, Δ#3, and Δ#3-mutStat1 luciferase reporters, but not the Δ#3-mut#1 and Δ#3-mut#2 reporters in HEK293T cells (Fig. 3F). Considering that multiple common ISGs could be induced by both IFN-I and IFN-II (5), we checked the cGAS mRNA level in IFNγ-stimulated BMMs. Comparing the induction of cGAS by IFN-I (Fig.1A and 3G-H), IFNγ stimulation only modestly upregulated cGAS mRNA in BMMs and THP1 cells (Fig. 3G-H), which was consistent with less STAT1 binding in cGAS promoter during IFNγ treatment than IFNβ treatment based on the analysis of STAT1 ChIP-Seq data (Fig. 3C). These data further suggested that induction of cGAS is mainly mediated by ISREs rather than the IFN-gamma-activated sites (GAS).
Figure 3.
The ISREs in the cGAS promoter mediate the induction of cGAS by IFN-I. A, the chromosome location of mouse cGas and its nearby genes, the diagram was modified from NCBI gene (Gene ID: 214763). B, the potential ISREs and STAT1 binding site in the promoter of cGas, transcription factor binding site prediction was performed by MatInspector. The location of the ISREs and STAT1 binding site and the conservation score were shown. The conservation comparison of ISRE#2 between human, rat, chimpanzee, rabbit, tree-shrew, dog, and elephant was according to the sequence from UCSC Genome Browser. TSS: transcription start site; CDS: coding DNA sequence. C, BMMs were treated with 100 U/ml IFNβ and IFNγ for 6 h, STAT1 ChIP-Seq data were analyzed and the Stat1 binding region in mouse cGas promoter was shown. The Stat1 ChIP-Seq raw data were downloaded from GEO (accession no. GSE33913). D, the sequence of the mutated ISRE#2, mutated ISRE#1, mutated Stat1, and the schematic diagram of the cGAS promoter reporter plasmids. E, indicated cGAS promoter reporter constructs or IFNβ luciferase reporter (IFNβ-luc) which expressing firefly luciferase was transfected into RAW264.7 cells by nucleofection system. pRL-TK-luc vector expressing Renilla luciferase was cotransfected as a control for transfection efficiency. Data were shown as the relative luciferase activity. F, Flag, TBK1, or IRF1 was cotransfected with indicated promoter reporter constructs and pRL-TK-luc vector. Data were shown as the relative luciferase activity. G, BMMs were treated with 1 U/ml IFNγ for 2.5 h, RNA was extracted and gene expression profile was detected by Affimetrix430.2 Chip, cGas mRNA level was shown as probe intensity from microarray. H, THP-1 cells were treated with human recombinant IFNγ (100 U/ml) for indicated time points. cGAS mRNA level in these cells was measured by qPCR and normalized to RPL32. Data of (E) and (F) are from one representative experiment (mean ± s.d., n=6), **p<0.01 (Student's t-test). Similar results were obtained in three independent experiments. Data of (G) and (H) are from three independent experiments (mean ± s.e.m), *p<0.05, **p<0.01 (Student's t-test).
Optimal production of viral DNA-triggered IFN-I requires IFNAR signaling
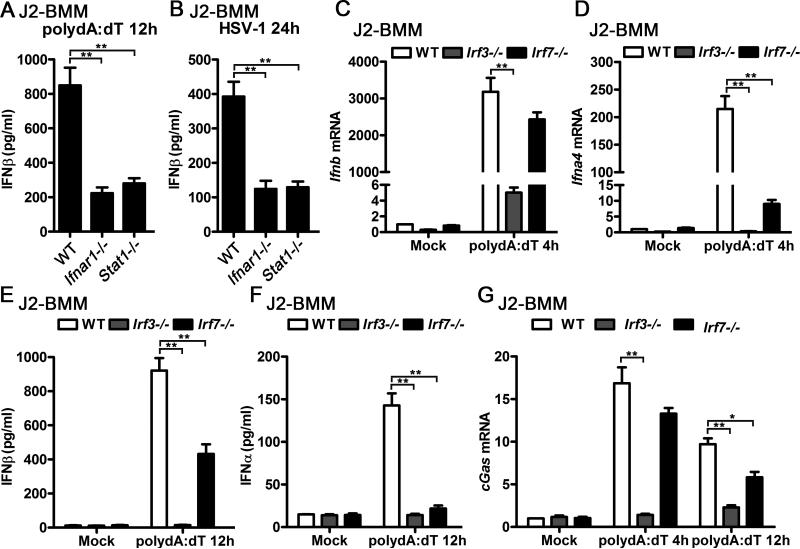
In response to viral DNA, DDX41 and cGAS recognize viral DNAs and activate STING-TBK1-IRF3 signaling axis by directly binding to STING or producing the endogenous CDN, cGAMP (12, 41). Host cells can produce large amount of IFN-I to defense the DNA viral infections upon the activation of STING-TBK1-IRF3 dependent pathway. Interestingly, less production of IFNβ transcript and protein could be detected in polydA:dT-transfected or HSV-1-infected WT J2-BMMs than Ifnar1−/− and Stat1−/− J2-BMMs (Fig. 4A-B), which suggested that optimal production of viral DNA-triggered IFN-I requires IFNAR signaling. IFN-I-inducible transcription factor IRF7 is a well-known ISG that mediates IFN-I positive feedback loop through the IRF3-IFNβ-IRF7-IFNα/β axis during viral infection (42-44). To test if IRF7 is also required for viral DNA-triggered IFN-I induction in macrophages, we compared the IFN-I transcripts and supernatant IFN-I protein from polydA:dT-triggered WT, Irf3−/− and Irf7−/− J2-BMMs. Significantly attenuated induction of IFNβ mRNA was observed in Irf3−/− but not in Irf7−/− J2-BMMs activated by transfection of polydA:dT for 4 hours, while IFNα4 mRNA induction was impaired in both Irf3−/− and Irf7−/− J2-BMMs at this time point (Fig. 4C-D). Significant less supernatant IFNβ and IFNα were detected in Irf3−/− and Irf7−/− J2-BMMs activated by transfection of polydA:dT for 12 hours, although the downregulation of IFNβ was not as dramatic as IFNα in Irf7−/− J2-BMMs (Fig. 4E-F). Consistent with the IFN-I production results, induction of cGAS was impaired in Irf3−/− J2-BMMs activated by transfection of polydA:dT for 4 hours and 12 hours, while modest but significant downregulation of cGAS induction in Irf7−/− J2-BMMs activated by transfection of polydA:dT for 12 hours but not 4 hours (Fig. 4G). These data indicate that IRF7 is critical for viral DNA-triggered IFNα production and modestly regulates IFNβ production at the later stage. However, IRF7 is dispensable for the early stage of IFNβ induction during viral DNA activation. Comparing to WT cells, Irf7−/− J2-BMMs could produce as much as 50% IFNβ while Ifnar1−/− and Stat1−/− J2-BMMs only produced 25% IFNβ (Fig. 4A and 4E), which suggested that defect of IRF7 induction in Ifnar1−/− and Stat1−/− J2-BMMs could not account for less viral DNA-triggered IFN-I in these cells. Given that cGAS induction is IFNAR-dependent but IRF7-independent at the early stage of viral DNA activation, no induction of DNA sensors such as cGAS in Ifnar1−/− and Stat1−/− cells is an alternative explanation of attenuated viral DNA-triggered IFN-I in these cells.
Figure 4.
Optimal production of viral DNA-triggered IFN-I requires IFNAR signaling. A-B, WT, Ifnar1−/−, or Stat1−/− J2-BMMs were transfected with 1 μg/ml polydA:dT for 12 h (A), or infected with HSV-1 (MOI=1) for 24 h (B). Supernatant IFNβ from these cells was measured by ELISA. C-D, WT, Irf3−/−, or Irf7−/− J2-BMMs were transfected with 1 μg/ml polydA:dT for 4 h, Ifnb (C) and Ifna4 (D) mRNA level in these cells was detected by qPCR and normalized to Rpl32. E-F, WT, Irf3−/−, or Irf7−/− J2-BMMs were transfected with 1 μg/ml polydA:dT for 12 h, supernatant IFNβ (E) and IFNα (F) from these cells was measured by ELISA. G, WT, Irf3−/−, or Irf7−/− J2-BMMs were transfected with 1 μg/ml polydA:dT for indicated time points, cGas mRNA level in these cells was detected by qPCR and normalized to Rpl32. Data are from three independent experiments (mean ± s.e.m), *p<0.05, **p<0.01 (Student's t-test).
Knockdown of DDX41 attenuated polydA:dT-triggered IFN-I production and subsequent cGAS induction
Given that DDX41 is constitutively expressed and cGAS is inducible by IFN-I in BMMs, we hypothesized that the first wave of IFN-I production was triggered by DDX41-dependent signaling and induction of cGAS by IFN-I mediated the subsequent robust IFN-I production in viral DNA-activated BMMs. Consistent with our hypothesis, we found that knockdown of DDX41 significantly reduced induction of IFN-I triggered by polydA:dT and cGAMP in BMMs (Fig. 5A-B). Meanwhile, less cGAS mRNA was induced by polydA:dT but not by cGAMP in si-Ddx41-transfected BMMs (Fig. 5C). P204 is another IFN-inducible DNA sensor reported to sense polydA:dT and trigger IFN-I production (7). Knockdown of p204 significantly reduced induction of IFN-I triggered by polydA:dT but not affected cGAS induction (Fig. 5D-F). As a control, knockdown of DDX41 or p204 did not affect the induction of IFNβ and cGAS in BMMs activated by polyI:C (Fig. 5A-F). Taken together, these data suggest that DDX41 not only regulates the production of IFN-I by DNA, but also affects the induction of the IFN-inducible sensor cGAS. Furthermore, we found that the induction of IFNβ was earlier than generation of cGAS in polydA:dT-activated BMMs, which implied that the constitutively expressed DDX41 and/or basal level of cGAS mediated the first wave of IFN-I production before induction of cGAS expression (Fig. 5G).
Figure 5.
Knockdown of DDX41 attenuated polydA:dT-triggered IFN-I production and subsequent cGAS induction. A-C, BMMs were transfected with 20 nM control siRNA (si-Ctrl) or si-Ddx41 for 36h, and then then cells were activated by transfection with 1 μg/ml polydA:dT, 3 μg/ml cGAMP, or 1 μg/ml polydI:C for another 12 h. The Ddx41 protein level was measured by western blot (A), the supernatant IFNβ from these activated cells was detected by ELISA (B), the cGas mRNA in these cells was detected by qPCR and normalized to Rpl32 (C). D-F, BMMs were transfected with 20 nM si-Ctrl or si-p204 for 36h, and then then cells were activated by transfection with 1 μg/ml polydA:dT or polyI:C for another 12 h. The p204 protein level was measured by western blot (D), the supernatant IFNβ from these activated cells was detected by ELISA (E), the cGas mRNA in these cells was detected by qPCR and normalized to Rpl32 (F). G, BMMs were transfected with 1 μg/ml polydA:dT for indicated time points, Ifnb and cGas mRNA was measured by qPCR and normalized to Rpl32. Data of (A) and (D) are representative of three independent experiments. Data of (B), (C), and (E-G) are from three independent experiments (mean ± s.e.m), *p<0.05, **p<0.01 (Student's t-test).
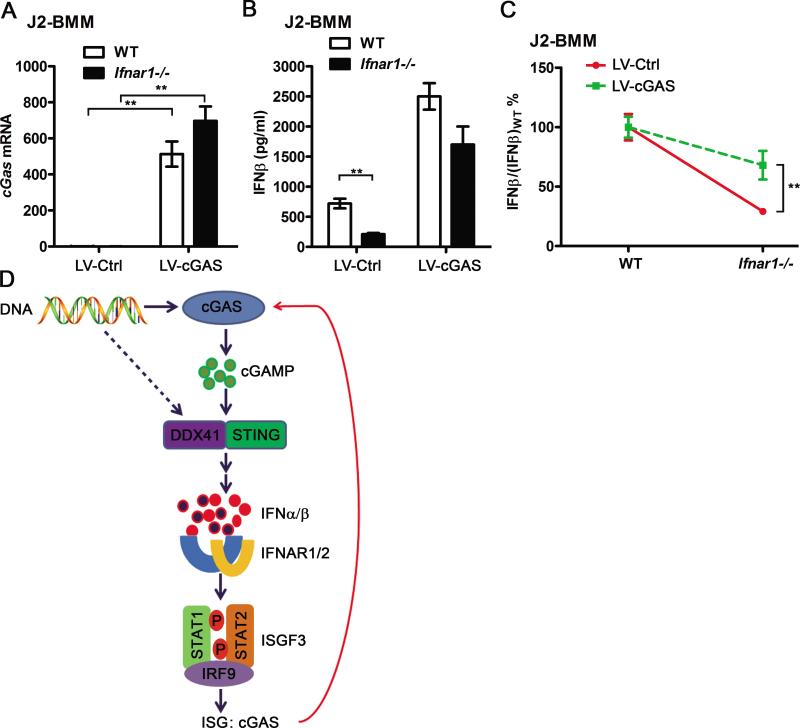
Overexpression of cGAS reduced the difference of polydA:dT-triggered IFN-I production between WT and Ifnar1−/− macrophages
To determine if the induction of cGAS by the first wave production of IFN-I plays a role in the positive feedback loop of DNA-triggered IFN-I production, we overexpressed mouse cGAS in both WT and Ifnar1−/− J2-BMMs by lentiviral gene transduction to get a similar level of cGAS expression during polydA:dT activation. cGAS mRNA was elevated dramatically after transducing with cGAS-overexpressing lentiviruses, and the mRNA expression levels of cGAS was comparable between cGAS-overexpressed WT and Ifnar1−/− J2-BMMs (Fig. 6A). Considering cGAS expression is almost saturated in cGAS-overexpressed cells, DNA-triggered IFN-I may not be able to further upregulate cGAS expression in these cells. We found polydA:dT-triggered IFNβ production has less change between cGAS-overexpressed WT and Ifnar1−/− J2-BMMs, comparing with empty lentiviral transduced WT and Ifnar1−/− J2-BMMs (Fig. 6B-C). These results indicated that overexpression of cGAS at least partially rescued the defect of IFNβ production in Ifnar1−/− macrophages. Reduced differential production of polydA:dT-triggered IFNβ between WT and Ifnar1−/− J2-BMMs suggested that induction of cGAS by IFN-I contributed to the positive feedback loop of IFN-I production (Fig. 6D). Although we cannot exclude the possibility that other ISGs may also regulate the positive feedback of polydA:dT-triggered IFN-I production, cGAS is likely to be one of the ISGs that play a role in this positive feedback loop.
Figure 6.
Overexpression of cGAS abolishes the difference of polydA:dT-triggered IFNβ production between WT and Ifnar1−/− macrophages. A, WT and Ifnar1−/− J2-BMMs were transduced with control or cGAS-expressing lentivirus (LV-Ctrl or LV-cGAS) for 3 days. cGas mRNA was detected by qPCR and normalized to Rpl32. B, WT and Ifnar1−/− J2-BMMs transduced with LV-Ctrl or LV-cGAS for 3 days, then transfected with 3 μg/ml polydA:dT for 12 h. Supernatant IFNβ from these cells was measured by ELISA, data of (A) and (B) are from three independent experiments (mean ± s.e.m), **p<0.01 (Student's t-test). C, J2-BMMs were treated and induction of IFNβ was measured as described in (B), the IFNβ/(IFNβ)WT% calculated from LV-Ctrl- and LV-cGAS-transduced cells was compared. D, Induction of cGAS plays a role in the positive feedback loop of DNA-triggered IFN-I production. DNA is recognized by DDX41 and/or basally expressed cGAS. Both cGAMP converted from viral DNA by cGAS and DDX41 could interact with STING and trigger the STING-TBK1-IRF3 signaling axis to produce IFN-I. The first wave of IFI-I production triggered by DDX41 and/or basally expressed cGAS induces cGAS expression through the IFNAR signaling. Induction of cGAS by IFN-I contributes to the subsequent positive feedback loop of IFN-I by sensing more viral DNA and producing more cGAMP.
In summary, our study have provided a novel mechanism by which cGAS is induced by IFN-I, and suggested a role of cGAS induction in the IFN-I positive feedback regulation loop.
Discussion
cGAS (formerly C6orf150) was among the numerous ISGs according to the published microarray data sets from IFN-treated cells or tissues (45-52). Our previous IFN-treated mouse BMMs gene expression profile also showed that mouse cGAS (formerly E330016A19Rik) could be significantly induced by IFN-I and IFN-II (5). In this study, we took advantage of different PRR ligands and gene-deficient BMMs to test the signaling pathways that regulate cGAS expression. In TLR4 ligands-triggered BMMs, deficiency of Myd88 did not affect the induction of cGAS, however, deletion of Ifnar1 and Trif completely abolished the activation of cGAS expression. These data suggest that cGAS expression is not regulated by MyD88-dependent pathways. PolyI:C, polydA:dT, and cGAMP activates both IRF3 and NF-κB by recruiting the kinases TBK1 and IKK, respectively (6, 20, 21). In Ifnar1−/− BMMs, cGAS gene was not induced in response to the stimulation of polyI:C, polydA:dT, and cGAMP. Thus, our study here not only have verified that cGAS is an ISG, but also demonstrates that cGAS expression is specifically regulated by IFNAR signaling. Furthermore, we also have identified an ISRE in cGAS promoter that mediates the induction of cGAS by IFN-I. Although both IFN-I and IFN-II significantly induce cGAS expression, IFN-I induced more cGAS transcript than IFN-II in both mouse and human macrophages. It is consistent with that ISRE-dependent regulation of cGAS. Recent study shows that autophagy protein Beclin-1 suppresses cGAMP synthesis and halts IFN production by directly interacting with cGAS (53). Treatment with PMA dramatically downregulated cGAS protein level in THP-1 cells via an unknown mechanism (25). While these studies identified the potential negative regulation of cGAS protein, our study here has provided a novel mechanism by which cGAS transcription and its downstream signaling are positively regulated by IFN-I.
DDX41, IFI16, and cGAS are among the numerous described cytosolic DNA sensors in the past several years. All of them could trigger STING-dependent signaling to induce IFN-I following polydA:dT transfection or DNA virus infection (7, 12, 41). DDX41 is constitutively expressed in myeloid dendritic cells (mDCs). Knockdown of DDX41 blocked the induction of IFI16 in polydA:dT-transfected mDCs (41). Previous study has indicated that DDX41 is more important than IFI16 in the initial sensing viral DNA and triggering the early burst of the IFN-I response (41). Here, we found that DDX41 expression is not altered in both lipidA-stimulated WT and Ifnar1−/− BMMs, which indicate that DDX41 is also constitutively expressed in BMMs. Knockdown of DDX41 attenuates the induction of cGAS by polydA:dT in BMM. The results of dynamic induction of IFNβ and cGAS transcripts by polydA:dT show that induction of IFNβ is earlier than cGAS. Taken together, our data suggest that DDX41 and/or basal level of cGAS is likely to mediate the first wave of IFN-I production, and induction of cGAS by IFN-I contributes to the subsequent positive feedback loop of IFN-I. Although more evidence is required to support the model which DNA sensors may act sequentially over time, our study at least is very similar as the model has proposed for the RNA helicase DDX3, which has been suggested to act as a “sentinel sensor” for viral RNA before RIG-I (which, like IFI16 and cGAS, is also an ISG) becomes the principal RNA sensor (54).
IFNAR signaling is required for the induction of cGAS. Optimal production of IFNβ triggered by polydA:dT and HSV-1 also requires IFNAR signaling. These data further suggest that induction of cGAS by IFN-I plays a role in the IFN-I positive regulation loop. IRF3-IFNβ-IRF7-IFNα/β signaling axis is a well-established loop for IFN-I positive feedback during viral infections (42-44). In our study, we show that induction of cGAS is IRF3-dependent but IRF7-independent in polydA:dT-activated BMMs at the early stage, which suggest that cGAS-dependent IFN-I positive feedback loop is independent of the classical IRF7-depedent IFN-I positive feedback pathway, particularly at the early stage. Our current study raises a possible working model that DNA is recognized by DDX41 and/or basally expressed cGAS. Both DDX41 and cGAMP converted from DNA by cGAS could interact with STING and trigger the STING-TBK1-IRF3 signaling axis to produce IFN-I. The first wave of IFI-I production triggered by DDX41 and/or basally expressed cGAS induces cGAS expression through the IFNAR signaling. Induction of cGAS by IFN-I contributes to the subsequent positive feedback loop of IFN-I by sensing more DNA and producing more cGAMP. In addition, new synthetic IRF7 by the first wave of IFN-I activates IFNα production to initiate the classical IFN-I positive feedback loop, which may amplify the cGAS induction and the IFNα-cGAS-IFNα/β signaling at the later stage.
Highlights.
cGAS is an IFN-stimulated gene
ISREs in the cGAS promoter are critical for its induction by IFN-I
Optimal production of DNA-triggered IFN-I requires IFNAR signaling
Overexpression of cGAS abolishes the difference of DNA-triggered IFN-I production between WT and Ifnar1−/− macrophages
Acknowledgements
We thank Kislay Parvatiyar and Amir Ali Ghaffari for their helpful discussions, and thank Dr. Tadatsugu Taniguchi (University of Tykyo) and Dr. Michael S. Diamond (Washington University) for sharing Irf3−/− and Irf7−/− mice. We appreciate Neda Arora for her excellent technical support, and Jing Zhu for editing the manuscript.
This work was funded by the Tumor Immunology Training Grant (UCLA Pathology & Lab Medicine, Steven Dubinett, Director Michael Teitell, Co-Director NIH/NCI: 5T32CA009120), NIH RO1AI078389 and AI069120 grants, and the Medical Scientist Training Program.
Abbreviations used in this paper
- BMM
bone marrow-derived macrophage
- PRR
pattern-recognition receptor
- cGAMP
cyclic GMP-AMP
- cGAS
cGAMP synthase
- STING
stimulator of interferon genes
- ISG
interferon-stimulated gene
- ISRE
interferon-sensitive response element
- IFN-I
type I interferon
- DDX41
DEAD (Asp-Glu-Ala-Asp) box polypeptide 41
- STAT
signal transducer and activator of transcription
- HSV-1
herpes simplex virus 1
- MHV-68
murid herpesvirus 68
- HIV-1
human immunodeficiency virus 1
- WNV
West Nile virus
- TBK1
TANK-binding kinase 1
- and IRF3
interferon regulatory factor 3
- ISGF3
Interferon-stimulated gene factor 3
Reference
- 1.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nature reviews. Immunology. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu SY, Sanchez DJ, Aliyari R, Lu S, Cheng G. Systematic identification of type I and type II interferon-induced antiviral factors. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:4239–4244. doi: 10.1073/pnas.1114981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 7.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nature immunology. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunette RL, Young JM, Whitley DG, Brodsky IE, Malik HS, Stetson DB. Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. The Journal of experimental medicine. 2012;209:1969–1983. doi: 10.1084/jem.20121960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson MR, Sharma S, Atianand M, Jensen SB, Carpenter S, Knipe DM, Fitzgerald KA, Kurt-Jones EA. Interferon Gamma Inducible protein (IFI)16 transcriptionally regulates type I interferons and other interferon stimulated genes and controls the interferon response to both DNA and RNA viruses. The Journal of biological chemistry. 2014 doi: 10.1074/jbc.M114.554147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurtler C, Bowie AG. Innate immune detection of microbial nucleic acids. Trends in microbiology. 2013;21:413–420. doi: 10.1016/j.tim.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhat N, Fitzgerald KA. Recognition of cytosolic DNA by cGAS and other STING-dependent sensors. European journal of immunology. 2014;44:634–640. doi: 10.1002/eji.201344127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell reports. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, Tuschl T, Patel DJ. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Shi H, Wu J, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Molecular cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barber GN. Cytoplasmic DNA innate immune pathways. Immunological reviews. 2011;243:99–108. doi: 10.1111/j.1600-065X.2011.01051.x. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Science signaling. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, Garcia-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, Rice CM. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Yeruva L, Marinov A, Prantner D, Wyrick PB, Lupashin V, Nagarajan UM. The DNA Sensor, Cyclic GMP-AMP Synthase, Is Essential for Induction of IFN-beta during Chlamydia trachomatis Infection. J Immunol. 2014 doi: 10.4049/jimmunol.1302718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen K, Prabakaran T, Laustsen A, Jorgensen SE, Rahbaek SH, Jensen SB, Nielsen R, Leber JH, Decker T, Horan KA, Jakobsen MR, Paludan SR. Listeria monocytogenes induces IFNbeta expression through an IFI16-, cGAS- and STING-dependent pathway. The EMBO journal. 2014;33:1654–1666. doi: 10.15252/embj.201488029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palleroni AV, Varesio L, Wright RB, Brunda MJ. Tumoricidal alveolar macrophage and tumor infiltrating macrophage cell lines. International journal of cancer. Journal international du cancer. 1991;49:296–302. doi: 10.1002/ijc.2910490226. [DOI] [PubMed] [Google Scholar]
- 27.Liu SY, Aliyari R, Chikere K, Li G, Marsden MD, Smith JK, Pernet O, Guo H, Nusbaum R, Zack JA, Freiberg AN, Su L, Lee B, Cheng G. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity. 2013;38:92–105. doi: 10.1016/j.immuni.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma F, Liu SY, Razani B, Arora N, Li B, Kagechika H, Tontonoz P, Nunez V, Ricote M, Cheng G. Retinoid X receptor alpha attenuates host antiviral response by suppressing type I interferon. Nature communications. 2014;5:5494. doi: 10.1038/ncomms6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Spandidos A, Wang H, Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic acids research. 2012;40:D1144–1149. doi: 10.1093/nar/gkr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 32.Ng SL, Friedman BA, Schmid S, Gertz J, Myers RM, Tenoever BR, Maniatis T. IkappaB kinase epsilon (IKK(epsilon)) regulates the balance between type I and type II interferon responses. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:21170–21175. doi: 10.1073/pnas.1119137109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li B, Su T, Ferrari R, Li JY, Kurdistani SK. A unique epigenetic signature is associated with active DNA replication loci in human embryonic stem cells. Epigenetics : official journal of the DNA Methylation Society. 2014;9:257–267. doi: 10.4161/epi.26870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, Li N, Yao H, Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nature immunology. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 36.Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infection and immunity. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nature reviews. Immunology. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 39.Takeuchi O, Akira S. Innate immunity to virus infection. Immunological reviews. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newton K, Dixit VM. Signaling in innate immunity and inflammation. Cold Spring Harbor perspectives in biology. 2012;4 doi: 10.1101/cshperspect.a006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nature immunology. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato M, Hata N, Asagiri M, Nakaya T, Taniguchi T, Tanaka N. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS letters. 1998;441:106–110. doi: 10.1016/s0014-5793(98)01514-2. [DOI] [PubMed] [Google Scholar]
- 43.Marie I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. The EMBO journal. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 45.de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, Williams BR. Functional classification of interferon-stimulated genes identified using microarrays. Journal of leukocyte biology. 2001;69:912–920. [PubMed] [Google Scholar]
- 46.Hultcrantz M, Huhn MH, Wolf M, Olsson A, Jacobson S, Williams BR, Korsgren O, Flodstrom-Tullberg M. Interferons induce an antiviral state in human pancreatic islet cells. Virology. 2007;367:92–101. doi: 10.1016/j.virol.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 47.Leaman DW, Chawla-Sarkar M, Jacobs B, Vyas K, Sun Y, Ozdemir A, Yi T, Williams BR, Borden EC. Novel growth and death related interferon-stimulated genes (ISGs) in melanoma: greater potency of IFN-beta compared with IFN-alpha2. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2003;23:745–756. doi: 10.1089/107999003772084860. [DOI] [PubMed] [Google Scholar]
- 48.Rani MR, Shrock J, Appachi S, Rudick RA, Williams BR, Ransohoff RM. Novel interferon-beta-induced gene expression in peripheral blood cells. Journal of leukocyte biology. 2007;82:1353–1360. doi: 10.1189/jlb.0507273. [DOI] [PubMed] [Google Scholar]
- 49.Sarasin-Filipowicz M, Oakeley EJ, Duong FH, Christen V, Terracciano L, Filipowicz W, Heim MH. Interferon signaling and treatment outcome in chronic hepatitis C. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7034–7039. doi: 10.1073/pnas.0707882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He XS, Ji X, Hale MB, Cheung R, Ahmed A, Guo Y, Nolan GP, Pfeffer LM, Wright TL, Risch N, Tibshirani R, Greenberg HB. Global transcriptional response to interferon is a determinant of HCV treatment outcome and is modified by race. Hepatology. 2006;44:352–359. doi: 10.1002/hep.21267. [DOI] [PubMed] [Google Scholar]
- 51.Hilkens CM, Schlaak JF, Kerr IM. Differential responses to IFN-alpha subtypes in human T cells and dendritic cells. J Immunol. 2003;171:5255–5263. doi: 10.4049/jimmunol.171.10.5255. [DOI] [PubMed] [Google Scholar]
- 52.Indraccolo S, Pfeffer U, Minuzzo S, Esposito G, Roni V, Mandruzzato S, Ferrari N, Anfosso L, Dell'Eva R, Noonan DM, Chieco-Bianchi L, Albini A, Amadori A. Identification of genes selectively regulated by IFNs in endothelial cells. J Immunol. 2007;178:1122–1135. doi: 10.4049/jimmunol.178.2.1122. [DOI] [PubMed] [Google Scholar]
- 53.Liang Q, Seo GJ, Choi YJ, Kwak MJ, Ge J, Rodgers MA, Shi M, Leslie BJ, Hopfner KP, Ha T, Oh BH, Jung JU. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell host & microbe. 2014;15:228–238. doi: 10.1016/j.chom.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oshiumi H, Sakai K, Matsumoto M, Seya T. DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-beta-inducing potential. European journal of immunology. 2010;40:940–948. doi: 10.1002/eji.200940203. [DOI] [PubMed] [Google Scholar]