Abstract
The incidence of diabetes and its associated micro- and macrovascular complications is greatly increasing worldwide. The most prevalent vascular complications of both type 1 and type 2 diabetes include nephropathy, retinopathy, neuropathy and cardiovascular diseases. Evidence suggests that both genetic and environmental factors are involved in these pathologies. Clinical trials have underscored the beneficial effects of intensive glycaemic control for preventing the progression of complications. Accumulating evidence suggests a key role for epigenetic mechanisms such as DNA methylation, histone post-translational modifications in chromatin, and non-coding RNAs in the complex interplay between genes and the environment. Factors associated with the pathology of diabetic complications, including hyperglycaemia, growth factors, oxidant stress and inflammatory factors can lead to dysregulation of these epigenetic mechanisms to alter the expression of pathological genes in target cells such as endothelial, vascular smooth muscle, retinal and cardiac cells, without changes in the underlying DNA sequence. Furthermore, long-term persistence of these alterations to the epigenome may be a key mechanism underlying the phenomenon of ‘metabolic memory’ and sustained vascular dysfunction despite attainment of glycaemic control. Current therapies for most diabetic complications have not been fully efficacious, and hence a study of epigenetic mechanisms that may be involved is clearly warranted as they can not only shed novel new insights into the pathology of diabetic complications, but also lead to the identification of much needed new drug targets. In this review, we highlight the emerging role of epigenetics and epigenomics in the vascular complications of diabetes and metabolic memory.
Keywords: Diabetic complications, Epigenetics, Epigenomics, Metabolic memory, DNA methylation, Histone modifications, Non-coding RNA, Review
Introduction
The increased incidence of diabetes worldwide has greatly augmented the risk for numerous associated complications that lead to reduced quality of life and increased mortality. A high percentage of patients with type 1 and type 2 diabetes develop microvascular complications such as diabetic retinopathy, nephropathy and neuropathy, as well as life-threatening macrovascular diseases such as atherosclerosis, stroke and hypertension [1-6]. Several common biochemical mechanisms and genetic factors have been implicated in the pathology of diabetes and its complications. They have a profound impact on cellular transcription programs in target organs, leading to aberrant expression of growth promoting, proinflammatory, pro-apoptotic, and pro-fibrotic genes [6-9]. However, it is increasingly evident that, because of the influences of gene–environment interactions, epigenetic mechanisms may also play a key role. Epigenetics refers to heritable changes in gene expression and ensuing phenotypes that occur without changes in the DNA sequence. Epigenetic changes include DNA cytosine methylation, histone post-translational modifications (PTMs) in chromatin, and non-coding RNAs (ncRNAs), all of which can affect gene expression individually or co-operatively and modulate disease states [8]. Current therapies for most diabetic complications have not been fully efficacious, and hence a study of epigenetic mechanisms is clearly warranted as they can offer novel and valuable insights into the pathophysiology of diabetic complications, and can also identify much needed new drug targets. Recent advances in human genome sequencing have yielded unprecedented information about chromatin states and the epigenome under normal and disease states [10, 11] which can be exploited to enhance our understanding of the molecular mechanisms involved in diabetic complications.
In this review, we highlight the role of epigenetic mechanisms in vascular complications of diabetes and metabolic memory, their potential use for the development of new biomarkers and therapeutic targets, and recent technological advances that have helped accelerate the field.
Biochemical mechanisms and factors involved in diabetic vascular complications
Diabetes and diabetogenic agents such as high glucose, AGEs, angiotensin II (Ang II), TGF-β and oxidised lipids have adverse effects in major target cells involved in vascular dysfunction, including endothelial cells, vascular smooth muscle cells (VSMCs), monocytes and retinal, neural and renal cells [1-8, 12]. Multiple signalling pathways and kinases activated by these agents lead to the activation of key transcription factors, such as nuclear factor-κB (NF-κB) and Smads, leading to increased expression of growth factors, inflammatory chemokines and cytokines involved in endothelial dysfunction, vascular cell growth, macrophage infiltration, fibrosis, inflammation and organ dysfunction [3-9, 13-17].
Whereas many of these mechanisms are common to most vascular complications of diabetes, the specific genes regulated, phenotype and pathology can vary depending on the target cell and organ. For example, levels of pro-fibrotic TGF-β are increased in most renal cells in diabetes, which mediates glomerular hypertrophy, accumulation of extracellular matrix (ECM) proteins and podocyte apoptosis, leading to diabetic nephropathy [2, 8, 12, 17]. Endothelial dysfunction and inflammation caused by macrophage infiltration is seen in most diabetic complications [6]. In diabetic retinopathy, there is retinal cell apoptosis and angiogenesis, while diabetic cardiac disease is manifested as myopathy and hypertrophy [6].
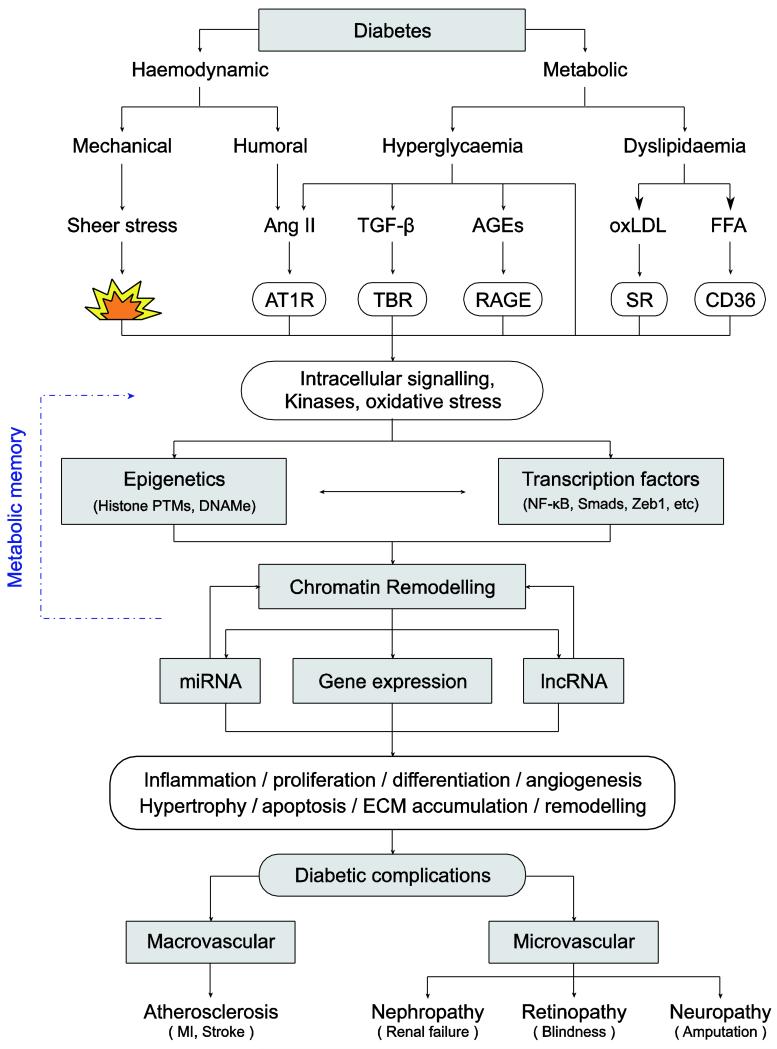
Crosstalk between the actions of various diabetogenic factors can amplify and perpetuate the expression of pathological genes associated with the progression of complications (Fig. 1). Recently, microRNA (miRNA)- and long ncRNA (lncRNA)-mediated mechanisms have also been implicated [8] (Fig. 1). Further understanding of these novel epigenetic mechanisms could help improve the efficacy of currently available therapies for overt and irreversible complications, and could also be used in the clinical management of metabolic memory implicated in the continued development of the diabetic complications despite glycaemic control.
Fig. 1.
Signalling and epigenetic networks mediating the pathogenesis of diabetic complications and metabolic memory. Diabetes and its consequent metabolic disorders can upregulate several growth factors and lipids that can act via their receptors and other mechanisms to trigger multiple signalling pathways, transcription factors (TFs) and crosstalk with epigenetic networks. These events can lead to chromatin remodelling and changes in the transcriptional regulation of key pathological genes in cells from target tissues relevant to various complications of diabetes. Persistence of such epigenetic aberrations (including histone PTMs, DNAme and ncRNAs) may lead to metabolic memory, which is implicated in an increased risk for developing diabetic complications even after normalisation of hyperglycaemia. AT1R, Ang II type 1 receptor; MI, myocardial infarction; oxLDL, Oxidized-LDL; RAGE, Receptor for AGEs; SR, scavenger receptors; TBR, TGF-β receptor
Metabolic memory: a unique phenomenon
Hyperglycaemia is a major pathological factor involved in diabetic complications. Although it can be controlled through medication, dietary modifications and exercise, many patients continue to experience numerous life-threatening complications. This could be due to a memory of prior exposure of target cells to high glucose, leading to persistence of its harmful effects long after attainment of glycaemic control [15, 18] (Fig. 1). This memory phenomenon has been observed in experimental models as well as in clinical trials such as the Diabetes Control and Complications Trial (DCCT) and the follow-up observational Epidemiology of Diabetes Interventions and Complications (EDIC) study. The results of the DCCT indicated that patients with type 1 diabetes placed on intensive glycaemic control had a much lower incidence or severity of various complications, including nephropathy and neuropathy, relative to those on standard/conventional therapy [19]. After the DCCT, both groups were placed on intensive therapy and were followed over the long term in the EDIC study phase. Despite attainment of similar levels of HbA1c in both groups during the EDIC study, patients in the original DCCT intensive treatment group had significantly lower risks of developing microvascular and macrovascular complications relative to the original DCCT conventional treatment group [20, 21], a phenomenon termed ‘metabolic memory’. Other trials with type 2 diabetic patients have also found that the benefits of intensive glycaemic control lasted long after cessation of intervention, suggesting a ‘legacy effect’ [22].
Metabolic memory has been demonstrated in several experimental models. Cell culture studies have implicated metabolic memory in the persistently aberrant expression of fibrotic, anti-oxidant and inflammatory genes in VSMCs [18, 23-25], endothelial cells [26, 27] and retinal endothelial cells (RECs) [28-31], despite glucose normalisation. Furthermore, metabolic memory has also been studied in animal models of complications such as atherosclerosis [27], diabetic nephropathy and diabetic retinopathy [28-33]. Thus, both in vivo and in vitro studies demonstrate the beneficial effects of good glycaemic control and, conversely, that the deleterious effects of prior hyperglycaemic exposure, on target organs last even after subsequent glycaemic control. Metabolic memory remains a major impediment to the effective management of diabetic complications, and, in recent years, epigenetic mechanisms have been implicated.
Epigenetics and the epigenome: rationale for study in diabetic complications
Epigenetic control of gene regulation plays an important role in development, cell identity, stable inheritance of gene expression patterns in differentiated cells, genomic imprinting, X chromosome inactivation, stem cell plasticity, differential disease susceptibility between monozygotic twins, and cellular responses to environmental signals [34, 35].
In mammalian cells, chromosomal DNA is tightly packaged into chromatin, which is made up of arrays of subunits called nucleosomes. Each nucleosome consists of an octamer protein complex containing two copies each of core histone proteins H2A, H2B, H3 and H4, with 147 bp of chromosomal DNA wrapped around it. PTMs of nucleosomal histones and DNA methylation (DNAme) represent epigenetic modifications (Fig. 2) [34, 35]. These modifications, along with ncRNAs, including short non-coding miRNAs and lncRNAs, regulate chromatin structure/function and cell-type-specific gene expression patterns and collectively constitute the epigenome. Recent advances in high-throughput genome-wide profiling and sequencing have significantly enhanced our knowledge of various aspects of the epigenome and its correlations to phenotype [10, 36]. Alterations in epigenome states have a profound effect on gene regulation and biological outcomes and are thus associated with the pathogenesis of various disorders, including cancer [37, 38]. In addition, long-term persistence of epigenetic modifications long after the removal of the original stimuli might contribute to chronic diseases such as diabetic complications and resistance to conventional therapies. Over- or under-nutrition, physical activity and environmental factors can influence epigenetic mechanisms in adults and offspring, leading to aberrant expression of genes involved in metabolic and cardiovascular disorders [39, 40]. The heritable nature of epigenetic marks could also predispose future generations to metabolic abnormalities [40], and are likely to make them prone to diabetic complications later in life.
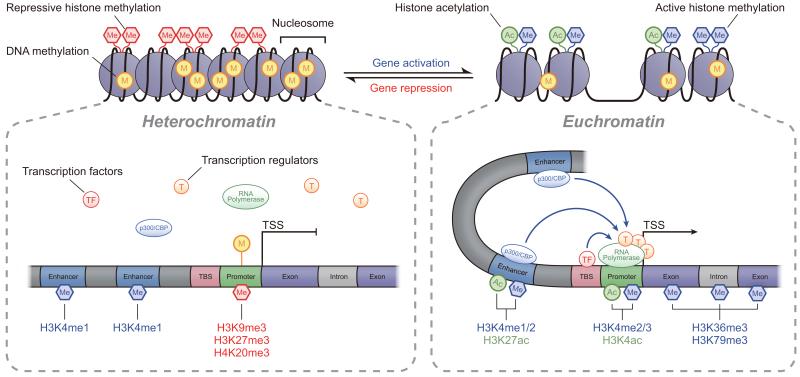
Fig. 2.
Chromatin structure and transcription regulation. Chromatin is made up of nucleosome subunits, each of which consists of an octamer of histones wrapped by chromosomal DNA. Lysine residues in the amino-terminal tails of nucleosomal histones are susceptible to several PTMs, including KAc and Kme, which regulate chromatin accessibility to transcription factors (TFs) and RNA polymerase II (labelled in the figure as RNA polymerase). Heterochromatin is marked by repressive histone PTMs, and increased DNAme at promoter CpG islands, leading to chromatin condensation and transcription repression. In contrast, euchromatin is enriched with permissive histone PTMs (H3/H4Kac and H3K4me1-3), and reduced promoter DNAme, leading to open chromatin and increased transcription. In addition, enhancers, which regulate promoters up to several thousand base pairs away, are marked by H3K4me1. Active enhancers are enriched with H3K27ac and the co-activator histone acetyl transferase p300. Actively transcribed gene bodies are enriched with H3K36me3/H3K79me3, whereas H3K27me3 is associated with reduced transcription. In addition, lncRNAs and miRNAs can also regulate gene expression by various post-transcriptional and epigenetic mechanisms (not shown here). TBS, TF binding site; TSS, transcription start site
Several complications of diabetes appear to have a genetic pre-disposition and there have been intense efforts to identify causal genes or related single-nucleotide polymorphisms (SNPs) [41-43]. Since these efforts and genome-wide association studies (GWAS) have uncovered only limited candidate loci, the evaluation of epigenotypes by epigenome-wide association studies (EWAS) can provide critical new information about the pathogenesis of diabetic complications and metabolic memory, which in turn could identify newer therapeutic modalities and diagnostic biomarkers for early intervention. Furthermore, most disease-associated SNPs are present in non-coding or other regulatory regions of the genome such as enhancers [44], which can affect gene expression by altering transcription factor binding.
DNAme and its role in diabetic complications
DNAme, the most well established epigenetic mark, occurs at 5′ cytosines of CpG dinucleotides, although non-CpG methylation can also occur. Both DNA methyl transferase (DNMT)3A and DNMT3B mediate de novo DNAme, whereas DNMT1 acts as a maintenance methyltransferase [35, 45]. Emerging evidence also shows the occurrence of cytosine 5-hydroxymethylation enzymatically mediated by oxidases (the Tet proteins) and active DNA demethylation, suggesting that DNAme is quite dynamic [35]. In general, DNAme at promoter regions leads to gene repression, whereas at gene bodies it might regulate transcription elongation and alternative splicing. DNAme is recognised by methyl binding proteins that can recruit transcriptional co-repressors and other proteins via protein–protein interactions to alter gene expression [35].
The role of DNAme has been studied in the transgenerational inheritance of metabolic diseases, suggesting that environment and diet may influence epigenetic modifications that predispose individuals to diabetes [46]. Aberrant DNAme has also been reported in the reduced expression of genes involved in diabetes and metabolism, and DNAme variations have also been noted near diabetes susceptibility genes and enhancers [15, 47].
Genomic DNA from diabetic patients with nephropathy relative to those without displayed differential methylation at several genes, including UNC13B, which had previously been linked to diabetic nephropathy [48]. In another study, a number of regions with differential DNAme were identified in saliva samples of patients with end-stage renal disease relative to chronic kidney disease (CKD) alone [49]. Interestingly, DNAme was found to play a role in a model of renal fibrosis and TGF-β actions [50]. A recent study implicated DNAme in crosstalk between podocytes and proximal tubules in diabetic nephropathy that was associated with increased expression of claudin-1 in podocytes and reduced expression of sirtuin 1 in tubules [51]. Of note, an examination of DNAme profiles in microdissected tubuli obtained from patients with CKD and diabetic nephropathy vs controls revealed significant differences in their genome-wide DNAme (methylomes), with key differentially methylated genes being related to fibrosis [52], indicating direct connections between epigenetics and human diabetic nephropathy. High glucose treatment induced alterations in DNAme at key genes involved in dysfunction of endothelial cells and neuronal cells [53, 54]. Additional studies in cell culture and in relevant tissues of animal models and humans at various stages of disease are needed to specifically link abnormalities in DNAme patterns with diabetic complications.
Histone PTMs and their role in diabetic complications and inflammation
Covalent PTMs of nucleosomal histone proteins in chromatin also regulate gene expression via epigenetic mechanisms [34, 36]. Several histone PTMs (mostly at exposed amino-terminal tails) have been identified, including lysine acetylation (Kac) and lysine methylation (Kme). In general, histone Kac (such as H3K9ac, H3K14ac, H4K5ac) at gene promoters correlates with transcriptional activation, whereas its removal is associated with gene repression. Histone Kme can be associated with either gene activation or repression depending on the amino acid residue modified and the extent of methylation, i.e. mono (Kme1), di (Kme2) or tri (Kme3) methylation. H3K4me1/2/3 and H3K36me2/3 are generally associated with transcriptionally active genome regions, whereas H3K9me3, H3K27me3 and H4K20me3 are associated with repressed domains [34]. Genome-wide profiling of histone PTMs has been instrumental in demonstrating that distinct patterns of specific histone modifications can distinguish key regulatory regions, including promoters, enhancers, gene bodies and repetitive elements [10, 36] (Fig. 2). Transcriptionally active gene promoters are enriched with H3K9ac, H3K4me2 and H3K4me3, while gene bodies and transcribed regions are enriched with H3K36me3 and H3K79me3. On the other hand, inactive or silent gene promoters are enriched with repressive marks H3K9me3 and H3K27me3 [10, 36]. Enhancers are typically enriched with H3K4me1 (poised) and H3K27ac (active) [55] (Fig. 2). Histone Kac is enzymatically mediated by histone acetyltransferases (HAT) such as p300, CREB-binding protein (CBP) and Tat-interactive protein 60 kDa (Tip60), which also act as transcription co-activators. Conversely, histone deacetylases (HDAC), including HDAC1–11 and sirtuins, remove acetylation marks and in general act as co-repressors with some exceptions [34]. Kme is mediated by histone lysine methyltransferases (HMTs) and removed by lysine demethylases (KDMs) [36, 56]. Histone modifying enzymes (HMEs) can also modify lysine residues on non-histone proteins, including transcription factors [57]. Therefore, newer nomenclature has been proposed for HMEs based on their enzymatic activity and the order of discovery [57].
Differential regulation of HME activity, recruitment and expression under various conditions determines the epigenome landscape. They can be recruited to promoters or enhancers by binding to specific DNA sequences, or by binding to pre-existing modifications or via interaction with RNA polymerase II and transcription factors [58] or via lncRNAs [59]. Because HMEs use metabolites such as acetyl-CoA (HATs), S-(5′-adenosyl)-l-methionine (HMTs) and α-ketoglutarate (KDMs) as cofactors, they might act as metabolic sensors. Therefore, mis-regulation of their functions can lead to metabolic abnormalities [60]. The role of DNAme in epigenetic transmission is generally more widely studied than that of histone PTMs, although histone modifiers like Polycomb repressor complexes and histone recycling proteins have been implicated [61]. Overall, the crosstalk and interplay between histone PTMs, DNAme and ncRNAs provides another layer of epigenetic regulation to impact gene expression [62, 63], which, if dysregulated, can result in diabetes and associated complications.
Changes in histone PTMs in target organs such as pancreas, liver and adipose tissue can affect the expression of numerous genes associated with obesity and diabetes [15]. Such epigenetic changes may also directly or indirectly influence key genes in target organs affected by vascular complications. Cell culture and animal models have demonstrated the involvement of histone PTMs in the expression of genes associated with the pathogenesis of diabetic nephropathy. TGF-β signalling plays an important role in the expression of key fibrotic and ECM genes and cell cycle inhibitor genes in renal cells, which contribute to diabetic nephropathy [2, 6, 8, 12]. TGF-β regulates gene expression mostly through activation of the transcription factors Smads2/3/4, which can collaborate with HATs and chromatin remodelling factors [12, 17, 64]. Recent studies have examined these mechanisms as well as histone Kac and Kme in rat mesangial cells treated with TGF-β and high glucose. TGF-β increased H3K9/14ac near Smad and SP1 binding sites by recruiting the HATs p300 and CBP to the promoters of the genes encoding plasminogen activator inhibitor type 1 (PAI-1) and p21 [65]. Induction of expression of fibrotic genes in rat mesangial cells by TGF-β was also associated with enrichment of active Kme marks (H3K4me1/2/3) and reduced levels of repressive marks (H3K9me2/me3) at their promoters [66]. Furthermore, SET domain-containing (lysine methyltransferase) 7 (SET7), a H3K4 methyltransferase, was found to play a key role in fibrotic gene expression. High glucose treatment of RMC also led to similar changes in histone PTMs (H3Kac and H3K4me) and increased SET7 recruitment at fibrotic and cell cycle gene promoters [65, 66]. Interestingly, these effects of high glucose were blocked by a TGF-β antibody, demonstrating that TGF-β acts as a mediator in high glucose-induced epigenetic effects [65, 66]. Together, these studies support a critical role for epigenetic mechanisms in TGF-β- and high glucose-induced pathological gene expression in mesangial cells which are relevant to diabetic nephropathy (Fig. 3).
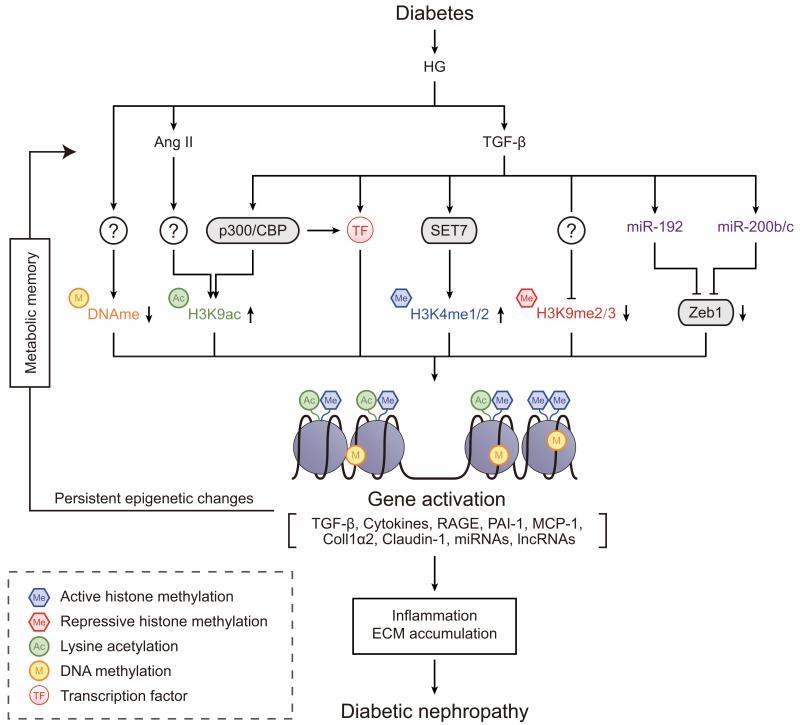
Fig. 3.
Epigenetic mechanisms in Diabetic nephropathy Signal transduction events downstream of high glucose (HG), AGEs and growth factors (TGF-β and Ang II) alter the expression/recruitment of epigenetic factors that mediate or remove histone PTMs and DNA methylation leading to chromatin remodelling. This alters promoter/enhancer access to transcription factors (TFs) such as Smads, SP1 and NF-κB, which are involved in the expression of genes mediating the pathogenesis of diabetic nephropathy. Recent studies also reveal regulatory roles for several miRNAs in diabetic nephropathy, including those that promote fibrotic gene expression in renal cells by targeting transcription repressors (Zeb1/2). Certain lncRNAs have also been shown to modulate fibrotic genes. Inhibitors of TGF-β, Ang II type 1 receptor signalling or miRNAs can block some but not all the events involved in the pathogenesis of diabetic nephropathy, suggesting the need for novel combination therapeutic approaches. Coll1α2, collagen, type I, α2; RAGE, receptor for AGEs
In an in vivo study [67], increases in RNA polymerase II recruitment and H3K4me2, but decreases in H3K27me3 levels, were associated with the expression of diabetic nephropathy-related genes in mouse and rat models of diabetic nephropathy, with some differences between the two species. In other studies, high glucose increased the expression of the redox-regulating protein p66Shc by inhibiting promoter DNAme and increasing H3K9ac in podocytes in vitro and in experimental diabetic nephropathy [68]. Another report implicated endoplasmic reticulum stress upstream of SET7-mediated regulation of monocyte-chemoattractant protein-1 (MCP-1) expression in kidneys from db/db mice [69].
Since chromatin status around expressed genes is likely to be affected by a code of multiple histone PTMs, another recent study used Matrix chromatin immunoprecipitation (ChIP) assays to profile several histone PTMs in vivo in mice glomeruli [70]. Relative to db/+ mice, glomeruli from diabetic db/db mice exhibited increased RNA polymerase II recruitment, enhanced levels of key activation marks and decreased levels of key repressive marks at the promoters of the genes encoding PAI-1 and receptor for AGEs (RAGE). These results suggest that epigenetic histone PTMs regulated by diabetes in vivo can co-operate to promote permissive chromatin states around these and other promoters, enhanced access to transcription machinery and gene expression. Interestingly, treatment of db/db mice with losartan, an Ang II type 1 receptor blocker (ARB), ameliorated key indices of diabetic nephropathy, and reversed key changes in epigenetic enzymes and H3K9ac enrichment at promoters of genes encoding PAI-1 and RAGE, but did not reverse all the diabetes-induced epigenetic changes [70]. Thus, the relative inefficiency of drugs commonly used for diabetic nephropathy, such as ARBs to prevent progression to renal failure, in many patients could be due to the incomplete reversal of diabetic nephropathy-associated epigenetic changes [71].
Changes in histone PTMs at key retinal genes have also been demonstrated in RECs treated with high glucose and tissues from animal models of diabetic retinopathy. Inhibition of superoxide dismutase (SOD2), and concomitant increase in oxidant stress in RECs are key events in diabetic retinopathy. High glucose-induced downregulation of Sod2 mRNA was accompanied by enrichment of the repressive histone mark H4K20me3 and corresponding HMT SUV420H2 at the Sod2 promoter [31], as well as reduced levels of activation marks H3K4me1/2, and increased occupancy of lysine-specific demethylase 1, which erases H3K4me1/2 [29]. Upregulation of matrix metalloproteinase gene Mmp9, which encodes another enzyme implicated in diabetic retinopathy, was associated with reduced promoter H3K9me2 and increased H3K9ac levels, along with increased recruitment of NF-κB in RECs from diabetic rats [30]. Furthermore, thioredoxin-interacting protein, a pro-oxidant factor induced by high glucose and RAGE ligands, upregulates inflammatory genes in RECs by inhibiting repressive H3K9me3 and increasing active H3K9ac at their promoters [72]. Mass spectrometry studies demonstrated that hyperglycaemia causes acetylation of retinal histones, which was associated with increases in proinflammatory proteins [73]. The HAT p300 was implicated in endothelial fibronectin expression related to diabetic retinopathy [74], and in gene expression relevant to diabetic cardiac hypertrophy [75]. With respect to diabetic neuropathy, several biological mechanisms have been studied, although the role of epigenetics has only recently been suggested [5, 6]. Epigenetic mechanisms have also been evaluated in the impaired wound healing associated with diabetes [76].
Chronic inflammation is a hallmark of the majority of the vascular complications of diabetes, with increased macrophage infiltration and inflammatory gene expression being observed in the kidney, blood vessels, eyes and other target organs. High glucose-mediated activation of NF-κB is a major mechanism of inflammatory gene expression in vascular cells and monocytes, and several studies have demonstrated the involvement of epigenetic modifications and histone PTMs in these events [15]. In monocytes, high glucose treatment increased the recruitment of co-activator HATs CBP and p300, and augmented the levels of active marks H3Kac and H4Kac at inflammatory gene promoters to enhance chromatin relaxation and gene expression [77]. Notably, profiling approaches with ChIP linked to microarrays (ChIP-on-chip) revealed differential enrichment of H3K4me2 (active) and H3K9me2 (repressive) marks at gene bodies of several genes in high glucose-treated THP-1 monocytes [78]. Similar changes in H3K9/14ac, H3K4me2 and H3K9me2 at key genes were also observed in blood monocytes obtained from diabetic patients, demonstrating direct relevance to diabetes [77, 78]. Furthermore, ChIP-on-chip epigenome profiling of blood lymphocytes from type 1 diabetic patients vs healthy controls demonstrated significant variations in the repressive H3K9me2 mark at a subset of genes associated with type 1 diabetes, inflammation and autoimmunity [79]. Key variations in monocyte H3K9ac were also observed at two HLA genes with SNPs that are closely linked to type 1 diabetes [80], suggesting a crosstalk between epigenetic and genetic variations—a concept being increasingly investigated. Other studies noted that SET7 was required for the maximal activation of a subset of NF-κB-inducible inflammatory genes in monocytes [81], besides its role in regulating fibrotic genes in the kidney.
In vascular endothelial cells, high glucose treatment increased the expression of the NF-κB active subunit (p65) and inflammatory genes by promoting SET7 recruitment and H3K4me1 enrichment at their promoters [26], while also directly promoting nuclear localisation and activity of SET7 [82]. In studies with VSMCs, high glucose was found to upregulate various inflammatory genes via decreases in the repressive mark H3K9me3 and corresponding HMT SUV39H1 at their promoters [24]. Together, these studies demonstrate a direct role of high glucose in the regulation of histone PTMs, and SET7 has emerged as one of the key epigenetic regulators of gene expression in many cell types relevant to diabetic complications, including mesangial cells, monocytes/macrophages and endothelial cells [26, 66, 81, 82] (Figs 3, 4). Thus, SET7 inhibitors could be evaluated for the treatment of diabetic complications. A recent report observed the downregulation of gene expression related to IL-6 and activator protein-1 (AP-1) signalling pathways in endothelial cells treated with two arginine methyltransferase inhibitors, one of which also had putative inhibitory effects on SET7 [83].
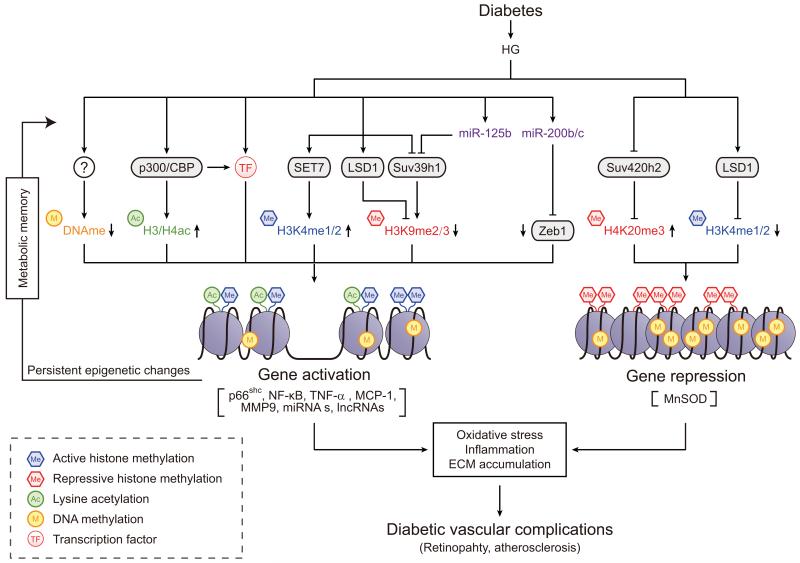
Fig. 4.
Epigenetic mechanisms of gene regulation in vascular inflammatory complications, atherosclerosis, retinopathy and metabolic memory. Schematic diagram showing the role of multiple epigenetic mechanisms, including those mediated by miRNAs and lncRNAs, associated with the regulation of genes involved in vascular inflammatory complications. These epigenetic mechanisms increase the expression of NF-κB-regulated inflammatory genes and inhibit the expression of protective anti-oxidant genes, leading to the development of vascular complications such as atherosclerosis and diabetic retinopathy. Studies on in vitro and in vivo models of diabetic complications suggest that persistence of variations in epigenetic histone PTMs and chromatin states at key pathological genes, as well as ncRNA expression, long after the removal from diabetic stimuli play key roles in metabolic memory implicated in an increased risk for diabetic complications. LSD1, lysine-specific demethylase 1; MCP-1, monocyte chemoattractant protein-1; MMP9, matrix metalloproteinase 9
Overall, these emerging studies demonstrate the involvement of histone PTMs and corresponding enzymes in gene regulation in target cells related to diabetic complications. They also highlight the complexity of profiling tissues from animal models of chronic progressive complications like diabetic nephropathy or retinopathy, and the need to study several stages of disease progression.
Epigenetic mechanisms and metabolic memory
There is intense interest in determining the molecular mechanisms underlying metabolic memory, especially those related to epigenetics. In recent years, the potential persistence of histone Kme variations at key genes related to complications has been evaluated as a mechanism for their chronic mis-regulation and metabolic memory. In one study, enhanced inflammatory gene expression and migration in VSMCs obtained from db/db mice relative to control db/+ mice despite being cultured in vitro for several passages was associated with decreased enrichment of the repressive mark H3K9me3 and the corresponding H3K9me3 methyltransferase SUV39H1 at inflammatory gene promoters. The expression levels of SUV39H1 were also down-regulated in parallel, due in part to the upregulation of miR-125b (which targets SUV39H1) in db/db mice, indicating crosstalk between these epigenetic layers (ncRNAs and chromatin) in diabetes [24, 84]. Similarly, sustained changes in H3K4me1 and SET7 were implicated in the prolonged upregulation of p65 in endothelial cells previously cultured in high glucose for short time periods [26]. In a rat model of diabetic retinopathy and metabolic memory, sustained downregulation of Sod2 was attributed to persistent promoter enrichments of the repressive mark H4K20me3 and decreases in the activation mark H3K4me2 [29, 31]. Together, these reports strongly suggest that alterations in epigenetic histone PTMs might establish a metabolic memory of diabetic complications (Figs 1-4). Further studies are needed to understand how high glucose and diabetes provoke these epigenetic events, and how they can be reversed to prevent the progression of complications despite glycaemic control.
Importantly, investigations with appropriate human diabetic individuals can help extrapolate these observations to clinical metabolic memory and glycaemic variations. In the first report of epigenome profiling of patients with type 1 diabetes experiencing metabolic memory [85], several histone PTMs were compared in white blood cells collected from a case group of EDIC study participants selected from the former DCCT conventional treatment group experiencing progression of nephropathy and retinopathy vs a control group from the former DCCT intensive treatment group not showing progression. Results showed significant enrichment in H3K9ac at promoters of key inflammatory genes and genes related to diabetic complications in monocytes from cases compared with controls. Monocyte H3K9ac was significantly associated with the mean HbA1c during long-term periods of DCCT and EDIC.
These novel findings suggest a potential epigenetic explanation for metabolic memory in humans [85]. Further studies with bigger cohorts of patients will help provide additional support for the role of various epigenetic marks in the metabolic memory phenomenon.
ncRNAs: miRNAs and lncRNAs in diabetic complications
Emerging data from whole transcriptome sequencing (RNA-seq) have revealed that the majority of the genome is transcribed into RNA, much of which is non-coding (i.e. unlike mRNA which codes for protein) [86]. The role of ncRNAs in diabetic complications has elicited great interest because they are also epigenetic regulators that can modulate the expression of genes via various transcriptional and post-transcriptional mechanisms and thus fine-tune the actions of diabetogenic stimuli. ncRNAs include not only small ncRNAs such as miRNAs (about 22 nucleotides long), but also lncRNAs, which are >200 nucleotides, up to 100 kb, in length. Recent evidence demonstrates that miRNAs and lncRNAs can also regulate the expression of genes and modulate the actions of growth factors and inflammatory factors related to diabetic complications [8]. These reports have been described in several reviews [8, 87-91] and are only briefly discussed here.
Numerous recent reports have demonstrated abnormal expression of various miRNAs in renal, vascular and retinal cells under diabetic conditions, and in vivo models of related diabetic complications [8, 87-91]. Notably, the functional relevance of these miRNAs has been highlighted by the fact they target key genes associated with the progression of, or protection against, these complications. In particular, the role of miRNAs in diabetic nephropathy has been extensively studied, including in the actions of TGF-β related to fibrosis and other key renal outcomes in vitro and in vivo [8, 87-90]. In diabetic retinopathy, several miRNAs have been reported to modulate the disease by targeting factors associated with angiogenesis, inflammation, and oxidant stress in RECs and in diabetic retinas [88, 89]. Reports have also implicated various miRNAs in the aberrant expression of genes associated with diabetic cardiomyopathy [88, 91]. In addition, effective in vivo targeting of miRNAs has now been demonstrated thanks to advances in nucleotide chemistry and the design of nuclease-resistant anti-miRNAs, which suggest future translational potential of miRNA-based therapies for human diabetic complications [8]. Importantly, since miRNAs are stable in biological fluids such as urine and serum [8], they are being assessed in samples from various clinical cohorts as valuable biomarkers for the early detection of diabetic complications, for which there is a major unmet clinical need. It is clear that research in the field of miRNAs and diabetic complications will continue at a rapid pace.
LncRNAs are long transcripts similar to mRNAs, but lack protein-coding (translation) potential [63, 92]. Most lncRNAs are expressed at much lower levels than protein-coding genes and exhibit tissue-specific expression. They can affect gene expression by various epigenetic mechanisms, including acting as scaffolds to bring protein complexes together, recruiting chromatin-modifying complexes to key gene loci, acting as sponges of miRNAs and as host genes for miRNA [59, 63]. A few reports have now demonstrated roles for lncRNAs in diabetic complications, especially diabetic nephropathy [8]. Key miRNAs were induced by TGF-β, together with their host gene, RP23, an ncRNA, in mesangial cells [93]. One lncRNA, plasmacytoma variant translocation 1, was identified as potential locus for end-stage renal disease, and implicated in the pathogenesis of diabetic nephropathy [94, 95]. In VSMCs, Ang II induced several lncRNAs, of which one novel lncRNA was a host gene of miR-221/222, and could regulate VSMC proliferation [96]. Furthermore, lncRNA E330013P06 (also known as MIR143HG) was recently reported to be induced in macrophages under diabetic conditions in mice and human cells [97]. Interestingly, this lncRNA could modulate the expression of macrophage inflammatory genes and foam cell formation [97], thus implicating lncRNAs in vascular complications such as atherosclerosis. Research into the role of lncRNAs in human disease and approaches to harness their therapeutic potential are still in their infancy. Clearly, this emerging field is expected to show rapid growth in the upcoming years, aided by technological advances, including the CRISPR-Cas9 system for genome editing [98]
Epigenomic approaches: applications in diabetic complications research
Epigenetic studies in human disease have been greatly accelerated as a result of advances in whole-genome and epigenome profiling technologies as well as bioinformatics and genomic data analysis platforms [99, 100]. DNAme is analysed using bisulfite conversion of genomic DNA, immunoprecipitation of methylated DNA, followed by hybridisation to arrays or next-generation sequencing to obtain genome-wide distribution of DNAme [100]. Histone PTMs are analysed using ChIP assays in which DNA in cross-linked chromatin is immunoprecipitated with antibodies against specific histone modifications, transcription factors or other chromatin factors. ChIP-enriched DNA is analysed by PCR to identify candidate enriched genome locations or by hybridisation to microarrays (ChIP-on-chip) or next-generation sequencing (ChIP-Seq) for genome-wide localisation analyses [99]. Formaldehyde Assisted Isolation of Regulatory Elements (FAIRE)-Seq detects accessible chromatin and regulatory elements based on differential cross-linking efficiency of nucleosome enriched and depleted regions [101]. Recently, in order to determine whether obesity alters chromatin accessibility, FAIRE-seq was used for the first time to examine chromatin variations in mouse livers induced by a high-fat diet [102]. Such genomic approaches can be used to evaluate variations in chromatin accessibility associated with diabetic complications.
RNA-Seq has revolutionised transcriptome analysis in diverse cell types and disease conditions and led to the genome-wide detection of known and novel transcripts, including lncRNAs [92, 103]. Integration of transcriptome (RNA-seq), DNA-methylome and ChIP-seq data can yield comprehensive information about the epigenome state and its outcomes on gene expression under pathophysiological conditions [99]. The Encyclopedia of DNA Elements (ENCODE) project completed high-quality whole-genome functional annotations of the human and mouse genomes [10, 104]. These publicly available data, including from NIH Roadmap Epigenomics consortium (www.roadmapepigenomics.org), are valuable reference tools to accelerate and catalyse new research into the epigenomics of human disease.
As discussed earlier, these high-throughput approaches are already being implemented in diabetic complications research. They have been complemented with systems biology and systems genetics efforts to effectively identify new players in and drug targets for diabetic complications [105]. There are also ongoing efforts to systematically profile epigenetic marks in tissues, cells and archived genomic DNA from various clinical trials. The major challenge, however, is expected to be in the analysis of the ensuing large datasets, the complexity of bioinformatics/biostatistics and in silico modelling. If these hurdles can be overcome, these efforts are likely to yield novel insights into epigenome variations linked with diabetic complications.
Summary
Increasing evidence shows that, besides the well-described biochemical mechanisms, epigenetic mechanisms might also participate by fine-tuning gene expression to modulate the aetiology of diabetic complications. Persistence of epigenetic modifications triggered by diabetic stimuli could be one of the key mechanisms underlying metabolic memory. However, the involvement of many epigenetic factors and mechanisms involved in the regulation of the modifications by upstream signal transduction pathways remains unknown. However, this is a rapidly expanding and dynamic field and it is likely that other epigenetic factors related to diabetic complications will soon be uncovered. Epigenomics may also aid in determining the functional roles of complications-associated genetic variants. It would be worthwhile to assess whether lifestyle modifications such as exercise and healthy diets can reduce diabetic complications by altering epigenetic marks. A recent study showed the beneficial effects of exercise on epigenetic marks related to diabetes [106]. Because epigenetic changes are potentially reversible in nature, combination therapies with epigenetic drugs (epidrugs) [38] and antagomirs (miRNA inhibitors) [8] could be considered to complement the current treatments for complications. However, there are also key challenges. Since epigenetic patterns are cell specific, data from heterogeneous tissue samples and biopsies could be difficult to interpret. Furthermore, apart from hyperglycaemia, other factors associated with diabetes, including insulin resistance, obesity, dyslipidaemia, environment, lifestyles and genetics, can work independently or co-operatively to also promote epigenetic changes in various affected target tissues. Because inflammation is closely associated with most diabetic complications, epigenetic variations could be examined non-invasively in inflammatory cells like blood monocytes and lymphocytes. Overall, it is anticipated that further research in the field of epigenetics could lead to the identification of much needed new biomarkers and drug targets for the early detection and treatment of the debilitating vascular complications of diabetes.
Acknowledgments
Funding
The authors are supported by funding from National Institutes of Health (R01 DK081705, R01 DK058191, R01 DK065073, R01 HL106089) (to RN) the Juvenile Diabetes Research Foundation (to RN) and China Scholarship Council (to EZ).
Abbreviations
- Ang II
Angiotensin II
- ARB
Angiotensin II type1 receptor blocker
- CBP
CREB-binding protein
- ChIP
Chromatin immunoprecipitation
- CKD
Chronic kidney disease
- DNAme
DNA methylation
- ECM
Extracellular matrix
- EDIC
Epidemiology of Diabetes Interventions and Complications
- HAT
Histone acetyltransferase
- HME
Histone-modifying enzymes
- HMT
Histone methyltransferase
- Kac
Lysine acetylation
- KDM
Lysine demethylase
- Kme
Lysine methylation
- lncRNA
long non-coding RNA
- miRNA
microRNA
- ncRNA
Non-coding RNA
- NF-κB
Nuclear factor-κB
- PTM
Post-translational modifications
- REC
Retinal endothelial cell
- SET7
SET domain-containing (lysine methyltransferase) 7
- SNP
Single-nucleotide polymorphism
- VSMC
Vascular smooth muscle cell
Footnotes
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
References
- [1].Beckman JA, Creager MA, Libby P. Diabetes and atherosclerosis: epidemiology, pathophysiology, and management. JAMA. 2002;287:2570–2581. doi: 10.1001/jama.287.19.2570. [DOI] [PubMed] [Google Scholar]
- [2].Ziyadeh FN, Sharma K. Overview: combating diabetic nephropathy. Journal of the American Society of Nephrology : JASN. 2003;14:1355–1357. doi: 10.1097/01.asn.0000065608.37756.58. [DOI] [PubMed] [Google Scholar]
- [3].Fong DS, Aiello L, Gardner TW, et al. Diabetic retinopathy. Diabetes care. 2003;26:226–229. doi: 10.2337/diacare.26.1.226. [DOI] [PubMed] [Google Scholar]
- [4].Natarajan R, Nadler JL. Lipid inflammatory mediators in diabetic vascular disease. Arterioscler Thromb Vasc Biol. 2004;24:1542–1548. doi: 10.1161/01.ATV.0000133606.69732.4c. [DOI] [PubMed] [Google Scholar]
- [5].Vincent AM, Calabek B, Roberts L, Feldman EL. Biology of diabetic neuropathy. Handbook of clinical neurology. 2013;115:591–606. doi: 10.1016/B978-0-444-52902-2.00034-5. [DOI] [PubMed] [Google Scholar]
- [6].Forbes JM, Cooper ME. Mechanisms of diabetic complications. Physiological reviews. 2013;93:137–188. doi: 10.1152/physrev.00045.2011. [DOI] [PubMed] [Google Scholar]
- [7].Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414:813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- [8].Kato M, Natarajan R. Diabetic nephropathy—emerging epigenetic mechanisms. Nature reviews Nephrology. 2014;10:517–530. doi: 10.1038/nrneph.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Woroniecka KI, Park AS, Mohtat D, Thomas DB, Pullman JM, Susztak K. Transcriptome analysis of human diabetic kidney disease. Diabetes. 2011;60:2354–2369. doi: 10.2337/db10-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dunham I, Kundaje A, Aldred SF, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Human Epigenome Task Force Moving AHEAD with an international human epigenome project. Nature. 2008;454:711–715. doi: 10.1038/454711a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kanwar YS, Sun L, Xie P, Liu FY, Chen S. A glimpse of various pathogenetic mechanisms of diabetic nephropathy. Annual review of pathology. 2011;6:395–423. doi: 10.1146/annurev.pathol.4.110807.092150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Geraldes P, King GL. Activation of protein kinase C isoforms and its impact on diabetic complications. Circulation research. 2010;106:1319–1331. doi: 10.1161/CIRCRESAHA.110.217117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ramasamy R, Yan SF, Schmidt AM. Receptor for AGE (RAGE): signaling mechanisms in the pathogenesis of diabetes and its complications. Annals of the New York Academy of Sciences. 2011;1243:88–102. doi: 10.1111/j.1749-6632.2011.06320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Reddy MA, Natarajan R. Epigenetic mechanisms in diabetic vascular complications. Cardiovascular research. 2011;90:421–429. doi: 10.1093/cvr/cvr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Averill MM, Bornfeldt KE. Lipids versus glucose in inflammation and the pathogenesis of macrovascular disease in diabetes. Curr Diab Rep. 2009;9:18–25. doi: 10.1007/s11892-009-0005-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sanchez AP, Sharma K. Transcription factors in the pathogenesis of diabetic nephropathy. Expert reviews in molecular medicine. 2009;11:e13. doi: 10.1017/S1462399409001057. [DOI] [PubMed] [Google Scholar]
- [18].Villeneuve LM, Natarajan R. The role of epigenetics in the pathology of diabetic complications. American journal of physiology Renal physiology. 2010;299:F14–F25. doi: 10.1152/ajprenal.00200.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Writing Team for the DCCT/EDIC Research Group Effect of intensive therapy on the microvascular complications of type 1 diabetes mellitus. JAMA. 2002;287:2563–2569. doi: 10.1001/jama.287.19.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].EDIC study Sustained effect of intensive treatment of type 1 diabetes mellitus on development and progression of diabetic nephropathy: the Epidemiology of Diabetes Interventions and Complications (EDIC) study. JAMA. 2003;290:2159–2167. doi: 10.1001/jama.290.16.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nathan DM, Cleary PA, Backlund JY, et al. Intensive diabetes treatment and cardiovascular disease in patients with type 1 diabetes. N Engl J Med. 2005;353:2643–2653. doi: 10.1056/NEJMoa052187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chalmers J, Cooper ME. UKPDS and the legacy effect. N Engl J Med. 2008;359:1618–1620. doi: 10.1056/NEJMe0807625. [DOI] [PubMed] [Google Scholar]
- [23].Li SL, Reddy MA, Cai Q, et al. Enhanced proatherogenic responses in macrophages and vascular smooth muscle cells derived from diabetic db/db mice. Diabetes. 2006;55:2611–2619. doi: 10.2337/db06-0164. [DOI] [PubMed] [Google Scholar]
- [24].Villeneuve LM, Reddy MA, Lanting LL, Wang M, Meng L, Natarajan R. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9047–9052. doi: 10.1073/pnas.0803623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Reddy MA, Jin W, Villeneuve L, et al. Pro-inflammatory role of microRNA-200 in vascular smooth muscle cells from diabetic mice. Arterioscler Thromb Vasc Biol. 2012;32:721–729. doi: 10.1161/ATVBAHA.111.241109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].El-Osta A, Brasacchio D, Yao D, et al. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. The Journal of experimental medicine. 2008;205:2409–2417. doi: 10.1084/jem.20081188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Brasacchio D, Okabe J, Tikellis C, et al. Hyperglycaemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes. 2009;58:1229–1236. doi: 10.2337/db08-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kowluru RA. Effect of reinstitution of good glycaemic control on retinal oxidative stress and nitrative stress in diabetic rats. Diabetes. 2003;52:818–823. doi: 10.2337/diabetes.52.3.818. [DOI] [PubMed] [Google Scholar]
- [29].Zhong Q, Kowluru RA. Epigenetic modification of Sod2 in the development of diabetic retinopathy and in the metabolic memory: role of histone methylation. Investigative ophthalmology & visual science. 2013;54:244–250. doi: 10.1167/iovs.12-10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhong Q, Kowluru RA. Regulation of matrix metalloproteinase-9 by epigenetic modifications and the development of diabetic retinopathy. Diabetes. 2013;62:2559–2568. doi: 10.2337/db12-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhong Q, Kowluru RA. Epigenetic changes in mitochondrial superoxide dismutase in the retina and the development of diabetic retinopathy. Diabetes. 2011;60:1304–1313. doi: 10.2337/db10-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Engerman RL, Kern TS. Progression of incipient diabetic retinopathy during good glycaemic control. Diabetes. 1987;36:808–812. doi: 10.2337/diab.36.7.808. [DOI] [PubMed] [Google Scholar]
- [33].Kowluru RA, Abbas SN, Odenbach S. Reversal of hyperglycaemia and diabetic nephropathy: effect of reinstitution of good metabolic control on oxidative stress in the kidney of diabetic rats. Journal of diabetes and its complications. 2004;18:282–288. doi: 10.1016/j.jdiacomp.2004.03.002. [DOI] [PubMed] [Google Scholar]
- [34].Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- [35].Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature reviews Genetics. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- [36].Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nature reviews Genetics. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- [37].Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- [38].Baylin SB, Jones PA. A decade of exploring the cancer epigenome—biological and translational implications. Nature reviews Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ling C, Groop L. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes. 2009;58:2718–2725. doi: 10.2337/db09-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang J, Wu Z, Li D, et al. Nutrition, epigenetics, and metabolic syndrome. Antioxidants & redox signaling. 2012;17:282–301. doi: 10.1089/ars.2011.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sandholm N, Salem RM, McKnight AJ, et al. New susceptibility loci associated with kidney disease in type 1 diabetes. PLoS genetics. 2012;8:e1002921. doi: 10.1371/journal.pgen.1002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Luo J, Zhao L, Chen AY, et al. TCF7L2 variation and proliferative diabetic retinopathy. Diabetes. 2013;62:2613–2617. doi: 10.2337/db12-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].McKnight AJ, McKay GJ, Maxwell AP. Genetic and epigenetic risk factors for diabetic kidney disease. Advances in chronic kidney disease. 2014;21:287–296. doi: 10.1053/j.ackd.2014.03.010. [DOI] [PubMed] [Google Scholar]
- [44].Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chen ZX, Riggs AD. DNA methylation and demethylation in mammals. The Journal of biological chemistry. 2011;286:18347–18353. doi: 10.1074/jbc.R110.205286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nature reviews Genetics. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Simmons R. Epigenetics and maternal nutrition: nature v. nurture. The Proceedings of the Nutrition Society. 2011;70:73–81. doi: 10.1017/S0029665110003988. [DOI] [PubMed] [Google Scholar]
- [48].Bell CG, Teschendorff AE, Rakyan VK, Maxwell AP, Beck S, Savage DA. Genome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus. BMC Med Genomics. 2010;3:33–42. doi: 10.1186/1755-8794-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sapienza C, Lee J, Powell J, et al. DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics : official journal of the DNA Methylation Society. 2011;6:20–28. doi: 10.4161/epi.6.1.13362. [DOI] [PubMed] [Google Scholar]
- [50].Bechtel W, McGoohan S, Zeisberg EM, et al. Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat Med. 2010;16:544–550. doi: 10.1038/nm.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hasegawa K, Wakino S, Simic P, et al. Renal tubular Sirt1 attenuates diabetic albuminuria by epigenetically suppressing Claudin-1 overexpression in podocytes. Nat Med. 2013;19:1496–1504. doi: 10.1038/nm.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ko YA, Mohtat D, Suzuki M, et al. Cytosine methylation changes in enhancer regions of core pro-fibrotic genes characterize kidney fibrosis development. Genome biology. 2013;14:R108. doi: 10.1186/gb-2013-14-10-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Pirola L, Balcerczyk A, Tothill RW, et al. Genome-wide analysis distinguishes hyperglycaemia regulated epigenetic signatures of primary vascular cells. Genome research. 2011;21:1601–1615. doi: 10.1101/gr.116095.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kim ES, Isoda F, Kurland I, Mobbs CV. Glucose-induced metabolic memory in Schwann cells: prevention by PPAR agonists. Endocrinology. 2013;154:3054–3066. doi: 10.1210/en.2013-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Jin F, Li Y, Ren B, Natarajan R. Enhancers: multi-dimensional signal integrators. Transcription. 2011;2:226–230. doi: 10.4161/trns.2.5.17712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nature reviews Molecular cell biology. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- [57].Allis CD, Berger SL, Cote J, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- [58].Smith E, Shilatifard A. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Molecular cell. 2010;40:689–701. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Sassone-Corsi P. Physiology. When metabolism and epigenetics converge. Science. 2013;339:148–150. doi: 10.1126/science.1233423. [DOI] [PubMed] [Google Scholar]
- [61].Abmayr SM, Workman JL. Holding on through DNA replication: histone modification or modifier? Cell. 2012;150:875–877. doi: 10.1016/j.cell.2012.08.006. [DOI] [PubMed] [Google Scholar]
- [62].Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature reviews Genetics. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- [63].Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Das F, Ghosh-Choudhury N, Venkatesan B, Li X, Mahimainathan L, Choudhury GG. Akt kinase targets association of CBP with SMAD 3 to regulate TGFbeta-induced expression of plasminogen activator inhibitor-1. Journal of cellular physiology. 2008;214:513–527. doi: 10.1002/jcp.21236. [DOI] [PubMed] [Google Scholar]
- [65].Yuan H, Reddy MA, Sun G, et al. Involvement of p300/CBP and epigenetic histone acetylation in TGF-beta1-mediated gene transcription in mesangial cells. American journal of physiology Renal physiology. 2013;304:F601–613. doi: 10.1152/ajprenal.00523.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Sun G, Reddy MA, Yuan H, Lanting L, Kato M, Natarajan R. Epigenetic histone methylation modulates fibrotic gene expression. Journal of the American Society of Nephrology : JASN. 2010;21:2069–2080. doi: 10.1681/ASN.2010060633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Komers R, Mar D, Denisenko O, Xu B, Oyama TT, Bomsztyk K. Epigenetic changes in renal genes dysregulated in mouse and rat models of type 1 diabetes. Laboratory investigation; a journal of technical methods and pathology. 2013;93:543–552. doi: 10.1038/labinvest.2013.47. [DOI] [PubMed] [Google Scholar]
- [68].Bock F, Shahzad K, Wang H, et al. Activated protein C ameliorates diabetic nephropathy by epigenetically inhibiting the redox enzyme p66Shc. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:648–653. doi: 10.1073/pnas.1218667110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Chen J, Guo Y, Zeng W, et al. ER stress triggers MCP-1 expression through SET7/9-induced histone methylation in the kidneys of db/db mice. American journal of physiology Renal physiology. 2014;306:F916–F925. doi: 10.1152/ajprenal.00697.2012. [DOI] [PubMed] [Google Scholar]
- [70].Reddy MA, Sumanth P, Lanting L, et al. Losartan reverses permissive epigenetic changes in renal glomeruli of diabetic db/db mice. Kidney international. 2014;85:362–373. doi: 10.1038/ki.2013.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ruggenenti P, Cravedi P, Remuzzi G. The RAAS in the pathogenesis and treatment of diabetic nephropathy. Nature reviews Nephrology. 2010;6:319–330. doi: 10.1038/nrneph.2010.58. [DOI] [PubMed] [Google Scholar]
- [72].Perrone L, Devi TS, Hosoya K, Terasaki T, Singh LP. Thioredoxin interacting protein (TXNIP) induces inflammation through chromatin modification in retinal capillary endothelial cells under diabetic conditions. Journal of cellular physiology. 2009;221:262–272. doi: 10.1002/jcp.21852. [DOI] [PubMed] [Google Scholar]
- [73].Kadiyala CS, Zheng L, Du Y, et al. Acetylation of retinal histones in diabetes increases inflammatory proteins: effects of minocycline and manipulation of histone acetyltransferase (HAT) and histone deacetylase (HDAC) The Journal of biological chemistry. 2012;287:25869–25880. doi: 10.1074/jbc.M112.375204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kaur H, Chen S, Xin X, Chiu J, Khan ZA, Chakrabarti S. Diabetes-induced extracellular matrix protein expression is mediated by transcription coactivator p300. Diabetes. 2006;55:3104–3111. doi: 10.2337/db06-0519. [DOI] [PubMed] [Google Scholar]
- [75].Feng B, Chen S, Chiu J, George B, Chakrabarti S. Regulation of cardiomyocyte hypertrophy in diabetes at the transcriptional level. American journal of physiology Endocrinology and metabolism. 2008;294:E1119–E1126. doi: 10.1152/ajpendo.00029.2008. [DOI] [PubMed] [Google Scholar]
- [76].Rafehi H, El-Osta A, Karagiannis TC. Genetic and epigenetic events in diabetic wound healing. International wound journal. 2011;8:12–21. doi: 10.1111/j.1742-481X.2010.00745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Miao F, Gonzalo IG, Lanting L, Natarajan R. In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. The Journal of biological chemistry. 2004;279:18091–18097. doi: 10.1074/jbc.M311786200. [DOI] [PubMed] [Google Scholar]
- [78].Miao F, Wu X, Zhang L, Yuan YC, Riggs AD, Natarajan R. Genome-wide analysis of histone lysine methylation variations caused by diabetic conditions in human monocytes. The Journal of biological chemistry. 2007;282:13854–13863. doi: 10.1074/jbc.M609446200. [DOI] [PubMed] [Google Scholar]
- [79].Miao F, Smith DD, Zhang L, Min A, Feng W, Natarajan R. Lymphocytes from patients with type 1 diabetes display a distinct profile of chromatin histone H3 lysine 9 dimethylation: an epigenetic study in diabetes. Diabetes. 2008;57:3189–3198. doi: 10.2337/db08-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Miao F, Chen Z, Zhang L, et al. Profiles of epigenetic histone post-translational modifications at type 1 diabetes susceptible genes. The Journal of biological chemistry. 2012;287:16335–16345. doi: 10.1074/jbc.M111.330373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Li Y, Reddy MA, Miao F, et al. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-κB-dependent inflammatory genes. Relevance to diabetes and inflammation. The Journal of biological chemistry. 2008;283:26771–26781. doi: 10.1074/jbc.M802800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Okabe J, Orlowski C, Balcerczyk A, et al. Distinguishing hyperglycaemic changes by Set7 in vascular endothelial cells. Circulation research. 2012;110:1067–1076. doi: 10.1161/CIRCRESAHA.112.266171. [DOI] [PubMed] [Google Scholar]
- [83].Okabe J, Fernandez AZ, Ziemann M, Keating ST, Balcerczyk A, El-Osta A. Endothelial transcriptome in response to pharmacological methyltransferase inhibition. ChemMedChem. 2014;9:1755–1762. doi: 10.1002/cmdc.201402091. [DOI] [PubMed] [Google Scholar]
- [84].Villeneuve LM, Kato M, Reddy MA, Wang M, Lanting L, Natarajan R. Enhanced levels of microRNA-125b in vascular smooth muscle cells of diabetic db/db mice lead to increased inflammatory gene expression by targeting the histone methyltransferase Suv39h1. Diabetes. 2010;59:2904–2915. doi: 10.2337/db10-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Miao F, Chen Z, Genuth S, et al. Evaluating the role of epigenetic histone modifications in the metabolic memory of type 1 diabetes. Diabetes. 2014;63:1748–1762. doi: 10.2337/db13-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Alvarez ML, Distefano JK. The role of non-coding RNAs in diabetic nephropathy: potential applications as biomarkers for disease development and progression. Diabetes research and clinical practice. 2013;99:1–11. doi: 10.1016/j.diabres.2012.10.010. [DOI] [PubMed] [Google Scholar]
- [88].Kato M, Castro NE, Natarajan R. MicroRNAs: potential mediators and biomarkers of diabetic complications. Free radical biology & medicine. 2013;64:85–94. doi: 10.1016/j.freeradbiomed.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Kantharidis P, Wang B, Carew RM, Lan HY. Diabetes complications: the microRNA perspective. Diabetes. 2011;60:1832–1837. doi: 10.2337/db11-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Badal SS, Danesh FR. MicroRNAs and their applications in kidney diseases. Pediatric nephrology. 2014 Jun 14; doi: 10.1007/s00467-014-2867-7. doi:10.1007/s00467-014-2867-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Zhou Q, Lv D, Chen P, et al. MicroRNAs in diabetic cardiomyopathy and clinical perspectives. Frontiers in genetics. 2014;5:185. doi: 10.3389/fgene.2014.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Kato M, Putta S, Wang M, et al. TGF-β activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol. 2009;11:881–889. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Alvarez ML, DiStefano JK. Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PloS one. 2011;6:e18671. doi: 10.1371/journal.pone.0018671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Hanson RL, Craig DW, Millis MP, et al. Identification of PVT1 as a candidate gene for end-stage renal disease in type 2 diabetes using a pooling-based genome-wide single nucleotide polymorphism association study. Diabetes. 2007;56:975–983. doi: 10.2337/db06-1072. [DOI] [PubMed] [Google Scholar]
- [96].Leung A, Trac C, Jin W, et al. Novel long noncoding RNAs Are regulated by angiotensin II in vascular smooth muscle cells. Circulation research. 2013;113:266–278. doi: 10.1161/CIRCRESAHA.112.300849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Reddy MA, Chen Z, Park JT, et al. Regulation of inflammatory phenotype in macrophages by a diabetes-induced long non-coding RNA. Diabetes. 2014 doi: 10.2337/db14-0298. doi: 10.2337/db14-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Mali P, Aach J, Stranges PB, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nature reviews Genetics. 2010;11:476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nature reviews Genetics. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- [101].Simon JM, Giresi PG, Davis IJ, Lieb JD. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nature protocols. 2012;7:256–267. doi: 10.1038/nprot.2011.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Leung A, Parks BW, Du J, et al. Open chromatin profiling in mice livers reveals unique chromatin variations induced by high fat diet. The Journal of biological chemistry. 2014;289:23557–67. doi: 10.1074/jbc.M114.581439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- [104].Mouse ENCODE Consortium. Stamatoyannopoulos JA, Snyder M, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome biology. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Hodgin JB, Nair V, Zhang H, et al. Identification of cross-species shared transcriptional networks of diabetic nephropathy in human and mouse glomeruli. Diabetes. 2013;62:299–308. doi: 10.2337/db11-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Ronn T, Ling C. Effect of exercise on DNA methylation and metabolism in human adipose tissue and skeletal muscle. Epigenomics. 2013;5:603–605. doi: 10.2217/epi.13.61. [DOI] [PubMed] [Google Scholar]