Abstract
The crosstalk between tumor cells and cells of the tumor stroma dictate malignant progression and represent an intriguing and viable anticancer therapeutic target. The successful development of therapeutics targeting tumor-stroma interactions is tied to the insight provided by basic research on such crosstalk. Tumor-stroma interactions can be transient and dynamic, and they occur within defined spatiotemporal contexts among genetically and compositionally heterogeneous populations of cells, yet methods currently applied to study the said crosstalk do not sufficiently address these features. Emerging imaging and genetic methods, however, can overcome limitations of traditional approaches and provide unprecedented insight into tumor-stroma crosstalk with unparalleled accuracy. The comprehensive data obtained by applying emerging methods will require processing and analysis by multidisciplinary teams, but the efforts will ultimately rejuvenate hope in developing novel therapies against pro-tumorigenic tumor-stroma crosstalk.
Keywords: tumor microenvironment, tumor stroma, tumor heterogeneity, RNAseq, omic analysis, intravital imaging, deep-tissue imaging, tissue clearing, personalized therapy
Introduction
Tumors are multicellular microcosms that develop and evolve through constant interactions between cancer cells and the tumor stroma (or tumor microenvironment/TME)1. It is increasingly appreciated that cellular members of the tumor stroma - immune cells2, vascular cells3, and fibroblasts4 - are not innocent bystanders to a neighboring outlaw, but are more often co-conspirators in malignant progression or law enforcing residents that promote tissue normalcy5.
The study of the tumor-stroma interactions that influence tumor development is met with significant challenges, including spatiotemporal tissue heterogeneity and the dynamic nature of cellular crosstalk. The greater biological scientific community has encountered similar difficulties in studying multicellular biological systems. Innovation has spurred the development of multiple novel approaches that overcome the aforementioned challenges, yet they have not been extensively applied to cancer research. Herein, we will first revisit prevailing methods used to study tumor-stromal cell interactions. More importantly, we will detail the challenges tumor-stroma researchers face and describe a myriad of emerging methods that should be applied to study tumor-stroma interaction studies to obtain exceptionally accurate, in-depth insights of tumor-stroma crosstalk.
Exploring the Role of the Tumor Microenvironment: The Past & Present
Although researchers and clinicians now view the TME to play a pivotal role in tumorigenesis, only four decades ago a tumor-centric view guided how cancer was studied and treated, leaving non-malignant cells of the tumor stroma neglected in cancer research6. Studies conducted between 1975 and 1985 by Mintz et al.7 and Dolberg et al.8,9 provided strong evidence that the fate of a cancer cell was not cancer cell autonomous, but could be decided by its environment6. Although now considered landmark studies that offer proof of concept of the importance of the TME in tumorigenesis, these research efforts did not overturn the tumor-centric view at the time. Rather, the cause of the paradigm shift toward the influential role of the TME in tumor progression can be traced to observations made on increased stromal cell infiltration in patient tumor samples10 and research on the necessity of angiogenesis11 for tumor outgrowth from mid-1980s through the late 1990s. Today, it is fully recognized that multiple microenvironmental factors influence seven of Hanahan and Weinberg’s eight “Hallmarks of Cancer”12,13. Thus, logically, the TME and it’s interactions with the tumor are viewed as therapeutically targetable prospects1.
The majority of early evidence for the role of tumor-stroma interactions in cancer progression came from clinical observations rather than the wet lab bench. Between 1985 and 1995, several pathologists observed a seemingly increased infiltration of immune cells in routine immunostained patient tumor biopsy sections. Subsequent quantification of immune cell infiltration suggested a positive correlation between the extent of immune cell infiltration and the degree of malignancy14. Today, immunostaining remains one of the fundamental methods to analyze tumor-stroma interactions and continues to reveal correlative roles for different types of stromal cells in either promoting or reverting tumorigenesis15,16.
With the advent of high-throughput genetic profiling methods such as Serial Analysis of Gene Expression (SAGE) and cDNA microarrays, unbiased genetic and transcriptome profiling of tumors followed by correlative analyses have been increasingly performed. To separate tumor cells from their associated stroma and profile genetic changes in the TME, mechanical cell separation techniques have been utilized prior to profiling, as traditionally accomplished by manual dissection, laser capture microdissection (LCM), or flow cytometry17–25. In an early, foundational profiling study using SAGE, a sequencing-based profiling method, Allinen et al. found that most genetic changes occurred within cancer cells, not stromal cells, whereas both the tumor and stromal compartments underwent transcriptome changes during malignant progression18. This study additionally noted that certain genes up-regulated in the stroma were secreted factors that could promote cancer cell proliferation18. Following Allinen and colleagues’ study, other groups identified intriguing transcriptome changes within both cells of the tumor and the associated stroma occurring during tumorigenesis using cDNA microarrays25,24,22,21,4. Retrospectively correlating clinical prognosis with collective genetic and transcriptome signatures of patient tumors has allowed clinicians to stratify patients and tailor personalized therapy. At the same time, the few microarray studies on the TME have also implied a profound role of the TME in tumor progression.
While highly informative, correlative analyses have limited power to definitively resolve the functional consequences of observed TME phenotypes and profiling signatures. Therefore, soon after strong correlations between TME alterations and malignancy status were established in the clinic, researchers embarked to characterize the functional role of stromal cells during malignant progression. Scientists have used loss-of-function systems, including conditional knockouts4 and immune cell-specific antibody blockade26, to explore the functional impact of tumor-stroma interactions on tumor initiation, progression, and metastasis. These functional studies resulted in the identification of numerous cellular and molecular participants in tumor-stroma crosstalk26,4. In addition to gene targeting methods, less specific methods that deactivate or deplete certain TME cells, such as clodronate liposome treatment to deactivate immune cells27, have also been used to demonstrate the critical role of stromal cells during tumorigenesis. Regardless of how histology, imaging, and sequencing techniques evolve, cellular functionality studies will remain the gold standard in studying the role of tumor-stroma crosstalk in tumor development.
New Routes to Study Tumor-Stromal Cell Interactions: The Future
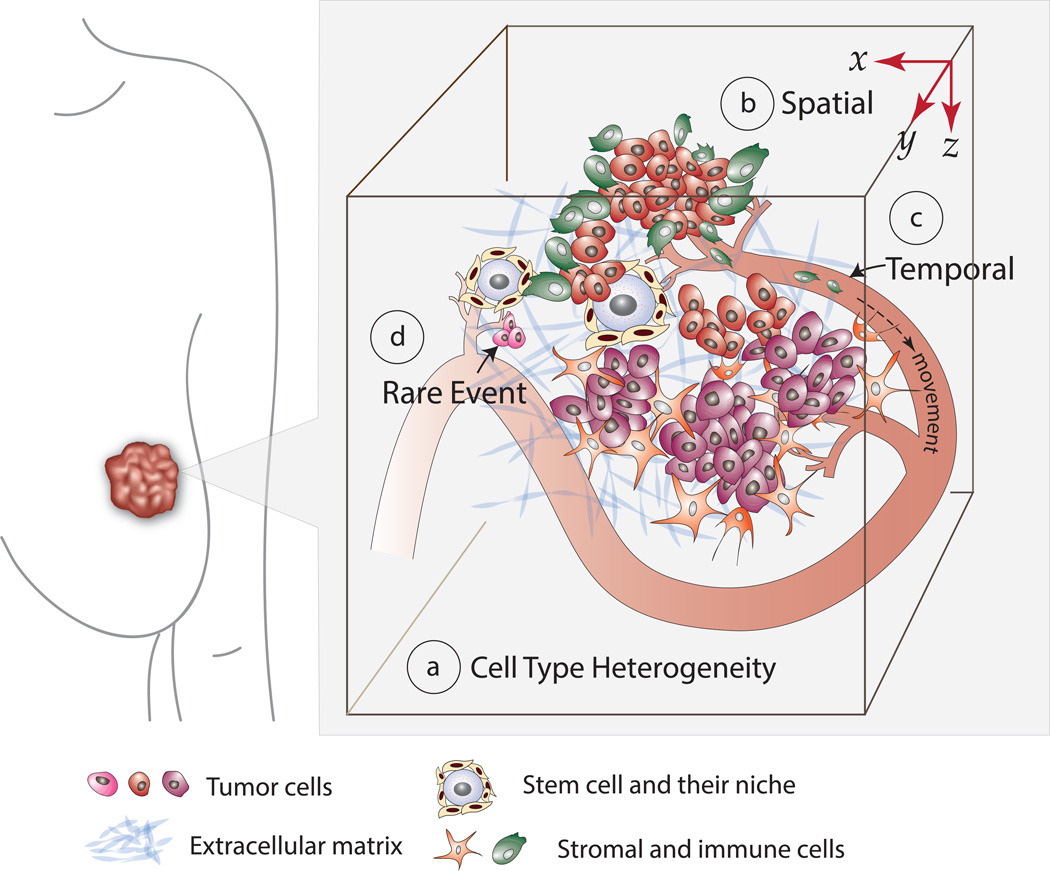
Current knowledge of tumor-stromal cell interactions is largely based on studies using the traditional approaches described above. However, traditional methods lack the capability to (1) provide a spatial perspective of the tumor and TME, (2) capture the temporal dynamics within the same tissue, (3) visualize rare/transient cellular and molecular events, and (4) dissect specific cell type population dynamics and single cell behavior (Figure 1). Emerging innovative approaches, as we will focus on below, hold great potential to overcome these multifaceted technical challenges, thereby allowing us to fully delineate the role of tumor-stroma crosstalk in tumor progression.
Figure 1. The Complex Features of the Tumor and TME Under-addressed by Traditional Approaches.
Tumor-stroma crosstalk occurs among a (a) compositionally and genetically heterogeneous population of tumor and stromal cells. The tumor and TME have (b) unique spatial features that influence crosstalk and tumor progression. The crosstalk has a (c) temporal aspect that ranges from transient to extensive, and sometimes consists of (d) rare events, such as metastasis seeding.
Imaging Approaches
Seeing is believing. For this reason, imaging is practically an indispensible method in demonstrating the role of certain cells or proteins in biological events. With technological advances in imaging modalities, it has become possible to visualize dynamic biological events in real-time with deep tissue penetration.
Gaining a Spatial Perspective & Capturing Rare Events by Deep Tissue Imaging
Traditional thin tissue section-based immunostaining provides extremely limited spatial insight. Spatial perspective is fundamentally important to study certain cell types and accurately measure distances. For example, it is impossible to obtain a faithful morphological perspective of vasculature or microenvironmental cells with long protrusions, such as microglia or astrocytes, which cannot be completely captured in a two-dimensional plane. Furthermore, many biological phenomena occur within spatially defined niches28, sometimes spanning long distances29 that cannot be fully captured in a single thin tissue slice. It has been shown, for instance, that brain neurogenesis occurs within spatially defined niches with extensive boundaries that exceed ten microns29. Similarly, with the proposal of a cancer niche30, it is likely that tissue beyond that which is represented in a thin tissue section of a tumor plays a role in determining tumor outgrowth. Finally, as recently reported, there is spatial heterogeneity within individual tumors, and the spatial arrangement of different tumor cells is an important determinant of tumor outgrowth31. This spatial heterogeneity of tumors underpins the importance of utilizing methods that enable a comprehensive three-dimensional view of the tumor and its associated stroma.
To depict the spatially defined and dictated tumor-stroma interactions, there is an imperative need for whole tissue imaging by deep tissue optical sectioning. Successful whole tissue optical sectioning is often impeded by two practical challenges: the natural density of a tissue32 and the light scattering properties of the tissue33. While the density of the tissue limits the permeability of macromolecules, such as antibodies32, that permit visualization of cells and proteins, the different light scattering properties of the tissue (refractory index mismatch) prevent high resolution imaging deep into the tissue sample even with cutting edge imaging technology33. Recent advances in fluorescence-compatible optical tissue clearing techniques overcome the above tissue-imposed obstacles associated with deep tissue immunofluorescence imaging. Generally, there are two broad means of achieving tissue optical transparency - physical clearing and chemical clearing - both of which are typically used in tandem to render a tissue more permeable to macromolecules and optically transparent to reduce light scattering34. In the last three years, four efficient, user friendly, and generally fluorescent-labeling-compatible optical tissue clearing methods have been developed: Scale35 and SeeDB36, chemical clearing agents, and CLARITY37, CUBIC38, and PACT/RIMS39, consisting of both physical clearing approaches and chemical clearing agents. CLARITY has additional utility in that it allows for multiple rounds of antibody staining, destaining, and restaining without tissue destruction due to its infusion and subsequent polymerization of tissues with a hydrogel compound37,40–42. While the clearing, staining, and imaging of whole, intact organs have been the emphasis of the aforementioned methods, staining and imaging whole organs is intrinsically time-consuming. Depending on an individual researcher’s research question, practicality can be increased by sectioning the whole organ into tissue blocks and following with passive clearing39,42,43. As demonstrated by Kim et al.44, the imaging of 200 micron cortical brain slices was sufficient to determine that a lack of Glycogen Synthase Kinase-3α activity resulted in reduced axon growth. This study is proof that imaging whole organs may not be necessary to answer the biological question posed, and thus it is at the discretion of the researcher to choose the sample thickness that is optimal to test a hypothesis. Optical tissue clearing of whole or thick tissue samples, coupled with confocal, multiphoton, or lightsheet microscopy and three-dimensional reconstruction, have demonstrated an unprecedented power to resolve extensive neural networks37 and transient biological events45. Studying the structurally complex and spatially vast TME and tumor-stroma interactions are natural extensions of these optical tissue clearing methods. Deep tissue imaging and three-dimensional reconstruction will depict a global structural and spatial perspective of tumor-stroma interactions. In addition, sequential, multiplexed immunostaining permitted by the infusion of tissues with hydrogel, as in the CLARITY37 and CUBIC38 approaches, will allow for the analysis of a virtually unlimited set of markers within a single tissue sample. Such multiplexed staining has been performed on thin tissue sections and has revealed signaling pathway heterogeneity that would have been impossible to uncover by staining for a limited set of markers46. Building upon classic thin tissue immunostaining studies, these cutting-edge tissue clearing and imaging approaches open a new avenue to explore spatially defined biological events with extensive detail.
Obtaining a Temporal Perspective & Observing Dynamic Events with Intravital Microscopy (IVM)
IVM, the imaging of tissues within live animals, provides an unmatched capability to observe dynamic biological events at a cellular resolution in the same animal with an extensive temporal perspective47. IVM has already been utilized to image tumor-stroma interactions in real-time, including cancer cell blood vessel cooption48 and ligand-induced natural killer cell-mediated tumor regression49, revealing cellular behaviors that would, at best, be postulated by those observing immunostained sections. Some groups have taken more elaborate approaches combining IVM with functional studies to undisputedly show the roles of certain molecules in cancer cell motility and intravasation50–52. As an example, one research group used shRNA to silence N-WASP, a protein involved in invadopodia formation, in breast cancer cells, and subsequently used IVM to demonstrate that the functionality of this protein is important for cancer cells to migrate or invade surrounding tissue51.
Although fluorescence-based IVM has been applied to tumor-stroma studies for a number of years, we include this technique as an emerging method because it has not been applied to its full potential. One roadblock to maximally implementing IVM is the fluorescent tagging of cells. To date, the number of cell types or proteins tagged in single tumor-stroma IVM experiments is typically limited to two cell types/proteins53,48. There is a dire need for multiplexed cell/protein tagging methods in IVM that is more time and cost effective than the creation of transgenic mouse models or cell lines with endogenous fluorescent proteins. A recent study demonstrated that such multiplexed tagging in IVM can be accomplished by injecting antibodies conjugated to fluorophores into the living tissue under observation54. Label-free imaging approaches, such as Coherent anti-Stokes Raman Scattering (CARS), offers another solution to the IVM labeling problem. CARS relies on the unique vibrational properties of proteins for an imaging readout, thereby, in principle, enabling highly multiplexed protein visualization at a single cell resolution without fluorescent taggging55,56. Although still in its infancy in IVM applications, CARS has already demonstrated a capability to monitor subcellular events and cellular interactions with a temporal perspective57,58.
Traditionally, IVM has been used to monitor cellular level behaviors, such as immune responses. However, simple whole cell labeling of cell-specific fluorescent protein tags provides little information on cell signaling kinetics or spatial insight into specific molecular interactions guiding cellular communications and interactions, representing a second significant challenge to IVM studies. Applying fluorescence-based imaging methods such as fluorescence resonance energy transfer (FRET), fluorescence lifetime imaging microscopy (FLIM), and bimolecular fluorescence complementation (BiFC) to IVM would enable specific protein-protein interactions to be better visualized and quantified59. The general principal behind these three fluorescence-based imaging methods is similar: when fluorescently tagged proteins come in near proximity to each other, a fluorescence shift occurs. In the case of BiFC, the fluorescent shift is due to the physical interaction of two tagged proteins that anneals two halves of a fluorophore whereas in FRET and FLIM, the fluorescence shift occurs as the photon emission from the fluorophore of one protein is absorbed by that of another nearby tagged protein59. FLIM, FRET, and BiFC fluorescence biosensors are gradually being incorporated into IVM studies, and have already led to the finding of events, such as the spatial regulation of RhoA activity during cancer cell invasion, that would not have been revealed in vitro or without the use of such imaging modalities60.
Lastly, functional assays combined with IVM have been rare and have not addressed interactions between tumor cells and stromal cells, signifying a key feature missing in tumor-stroma IVM studies. A real-time view of altered communications due to functional manipulations would be highly informative and likely become the ultimate means of analyzing tumor-stroma relations. With IVM finding a commonplace in tumor-stroma interaction studies, it is likely that in the near future researchers will address the current weaknesses of IVM studies, incorporating functional assays and integrating novel fluorescence-based and label-free imaging strategies.
Genetic Approaches
High-throughput, unbiased profiling of genetic and transcriptomic changes of tumor samples is the fundamental approach used in The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC) project. Next generation sequencing has replaced cDNA microarrays as the primary high-throughput genetic profiling approach, which have indeed led to the identification of genetic alterations during the tumor progression. With accumulating evidence of intratumoral heterogeneity and the spatiotemporal nature of tumor-stroma crosstalk, standard sequencing procedures based on bulk tumor tissue are insufficient. In this section, we will focus on novel genetic approaches that will enable in situ genetic dissection of dynamic genetic changes within defined cell types.
Examining Tumor Heterogeneity by In Situ Isolation of Genetic Material From Specific Cell Types
The sample processing preceding transcriptome profiling, inclusive to the isolation of specific cell types and subsequent RNA isolation from these cells, is a major challenge that has a significant influence on the accuracy of profiling results. Separation of specific cell types from tissues has been traditionally accomplished by flow cytometry, manual dissection, and LCM61. However, for solid tumors, mechanical or enzymatic separation before flow cytometry cell sorting sacrifices tissue integrity and significantly alters the transcriptome profile prior to sequencing62,61. It is also impractical to use LCM to isolate particular cell types from a highly invasive tumor in which cancer cells and stromal cell are intermixed. Ideally, the transcripts from specific cells of interest would be isolated in situ, thereby largely maintaining tissue integrity. One means of accomplishing this is by use of species-specific microarrays. Applying the bulk RNA pool derived from a human-in-mouse xenograft tumor, which contains tumor cells of human origin and murine TME cells, to a human or mouse-specific microarray negates the need for mechanical cell separation22. While a clever approach, such methodology cannot be applied to immune-competent animal models and it only profiles the microenvironment in bulk rather than specific cell populations22.
Recently, the development of innovative transcriptome tagging methods has enabled in situ transcriptome profiling in heterogeneous tissue. One of the first RNA tagging-isolation methods developed was Thiouracil (TU) Tagging63,64. In TU Tagging, cell lines or mice are genetically modified to express uracil phosphoribosyltransferase (UPRT), an enzyme that adds 4-thiouracil (4-TU) to nascent RNA strands, thereby “TU tagging” the transcript63. Spatiotemporal control over TU tagging is exerted by placing the transgene under inducible cell type-specific promoters so that following induction of UPRT expression, newly transcribed RNA in the certain cell types is pulse labeled63. Upon total RNA extraction from a whole tissue sample of mixed cell types, TU tagged RNA is biotintylated and affinity purified from the mixed pool of RNA63. TU tagging has been demonstrated to be very sensitive, capable of identifying transcripts in a specific cell type that makes up only five percent of the total tissue63. Furthermore, RNA extracted by TU tagging appears to be extremely specific without contamination from other cell types63. Another cell type-specific RNA isolation technique, Translating Ribosome Affinity Purification (TRAP), is conceptually similar to TU tagging. TRAP methodology places EGFP-tagged large ribosomal subunit protein L10 under cell-specific promoters. EGFP-tagged ribosomes can be affinity purified, thereby enabling isolation of mRNA undergoing translation from a specific cell type among a mixture of cell types65. While the capture of RNA under the process of translation by TRAP may be more desirable than the pull down of freshly transcribed mRNA as achieved by TU-Tagging, TRAP is susceptible to greater contamination61. Finally, Transcriptome In Vivo Analysis (TIVA) is the most recently developed RNA isolation method for analysis of heterogeneous cell populations66. By TIVA, cells within intact tissue are transfected with a photocleavable, biotin-conjugated tag. Using a confocal microscope, researchers photoactivate the photocleavable tag in a single cell of interest, allowing the tag to anneal to mRNA within the target cell. Subsequently, the mRNA can be affinity purified66. With the above transcriptome tagging tools, sophisticated cell type-specific RNA analysis can be achieved. For example, TIVA is well suited to be combined with TU tagging or TRAP in which the RNA of a single cancer cell could be isolated for analysis by TIVA while the RNA of a population of specific environmental cells could be isolated by TU tagging or TRAP. There is little doubt that the application of new RNA isolation methods as described above will provide new perspectives in the study of heterogeneous and dynamic tumor-stroma crosstalk. Cell type specific in situ genetic profiling will allow for more reliable transcriptome profiling of the tumor and its microenvironment with precise cell lineage, spatial, and temporal control at a single cell resolution.
Studying Single Cell Transcriptomics with RNAseq
With the recent appreciation of the heterogeneous composition of tumors and divergent cellular responses to microenvironmental cues and therapeutic treatments, it is essential to investigate the transcriptome dynamics at a single cell level in this post-microarray era. Fortunately, the three primary shortcomings of microarrays – the requirement of high RNA input, high background noise, and an ability to only profile genes on the array – can be largely overcome by RNAseq technology67,68. RNAseq reduces the RNA input requirement from micrograms, as required by microarrays, to picograms, thereby making single cell sequencing possible69,70. Recent applications of single cell RNAseq dramatically change the dogmatic view of cell signaling. Single cell RNAseq has revealed that a seemingly homogeneous population of cells is very heterogeneous at the transcriptional level, as in the case of glioblastoma cells which display a diversity of gene expression patterns71. While our current knowledge of tumor-stroma transcriptomes is largely based on the cell population level studies22,18, single cell sequencing could provide unprecedented insight to reveal a heterogeneity in the responses of individual tumor cells to microenvironmental stimuli as well as very rare cancer cells, such as those that initially seed tissue during metastasis.
The Next Impasse: The Data Processing and Analysis Challenges
The data obtained from deep tissue imaging, IVM, and RNAseq will be more comprehensive than that provided by traditional imaging and transcriptome profiling methods. The extraction of maximal information from studies applying the techniques described herein will further rely on multidisciplinary data processing and analysis approaches. In the case of imaging studies, although optical tissue clearing methods and multiphoton microscopy will combine to provide outstanding three-dimensional datasets, there will still be issues to overcome in terms of background noise and data registration and segmentation that commercial imaging data processing packages cannot fully address or resolve with total accuracy72,73. Such issues related to imaging data sets are unlikely to be resolved by the biologists capturing the imaging data, necessitating collaborations with researchers in fields including, but not limited to, bioinformatics, computer sciences, and mathematics. Indeed, needs such as cell segmentation and image correction74,75 and deconvolution76 have been satisfied by groups of computer scientists to produce images that resolve single cells and maintain spatial resolution which, more importantly, permit accurate biological quantifications. Likewise, transcriptome profiling data processing and analysis has demanded collaborations between biologists and mathematicians to overcome noise and biological variability issues, to correctly map transcripts of different species to the correct genome, and to identify causal relationships between transcriptome changes in the tumor and the microenvironment77,78.
The present push for the use of big data demands the eventual cross reference of data from “old school” studies and “new age” studies as well as the integration of any two studies applying seemingly disparate methods. As a result, it will be imperative to further develop data integration methods. Two studies in particular by Bindea et al.79 and Han et al.80 serve as exemplars to the data integration that must be accomplished to extract the most information from dissimilar data sets. Bindea et al.79 used an integrative approach to combine tissue microarray data with standard immunostaining to illustrate the immune landscape of colorectal tumors with a spatiotemporal perspective. In their integrative approach, Han et al.80 correlated morphological information of cells within 3D culture obtained by phase contrast microscopy with that of gene expression information to establish a morphological predictor cancer cell subtype, cancer progression, and therapeutic response. With the realization of the need for integrative analysis tools, those in fields such as bioinformatics are developing models, such as the Bayesian Multiple Dataset Integration model81, to address a broad variety of data integration needs. New studies will continuously reveal new problems to be solved in creative ways. The meaning and reliability of the data of future studies will only be as good as the successful multidisciplinary collaborations that collect and analyze the data.
Computer-driven data analysis and integration may just be the beginning of an even greater reliance on computers in biological studies. It is conceivable that experiments will move from bench top to in silico in the form of computational modeling. Computer modeling-based tumor-stroma studies have already demonstrated the significant impact of the stroma on tumor growth and have been used to predict the prognosis associated with various microenvironmental cellular compositions and conditions82. One computer modeling-based study demonstrated the ability of a low level immune response to have a slight cytotoxic effect on a tumor, unexpectedly resulting in increased room for the expansion of cancer stem cells and the formation of a freeway by which they can metastasize83. Given the complexity of manipulating stromal cells in vivo, computer modeling can serve as a starting point to validate eventual animal experiments or even replace animal models under some circumstances.
Concluding Remarks
As the world begs for new anti-cancer therapies, researchers are increasingly looking toward tumor-stroma interactions to find the next targets of novel treatment methods. We are in the midst of a period of methodological advancement, and the emerging techniques described in this review hold promise for giving a more comprehensive, accurate understanding of tumor-stroma interactions (Figure 2). New paths are being laid in science, and now it is time to venture to unchartered territory, and in the act take the first steps toward developing future break-through treatments for cancer patients.
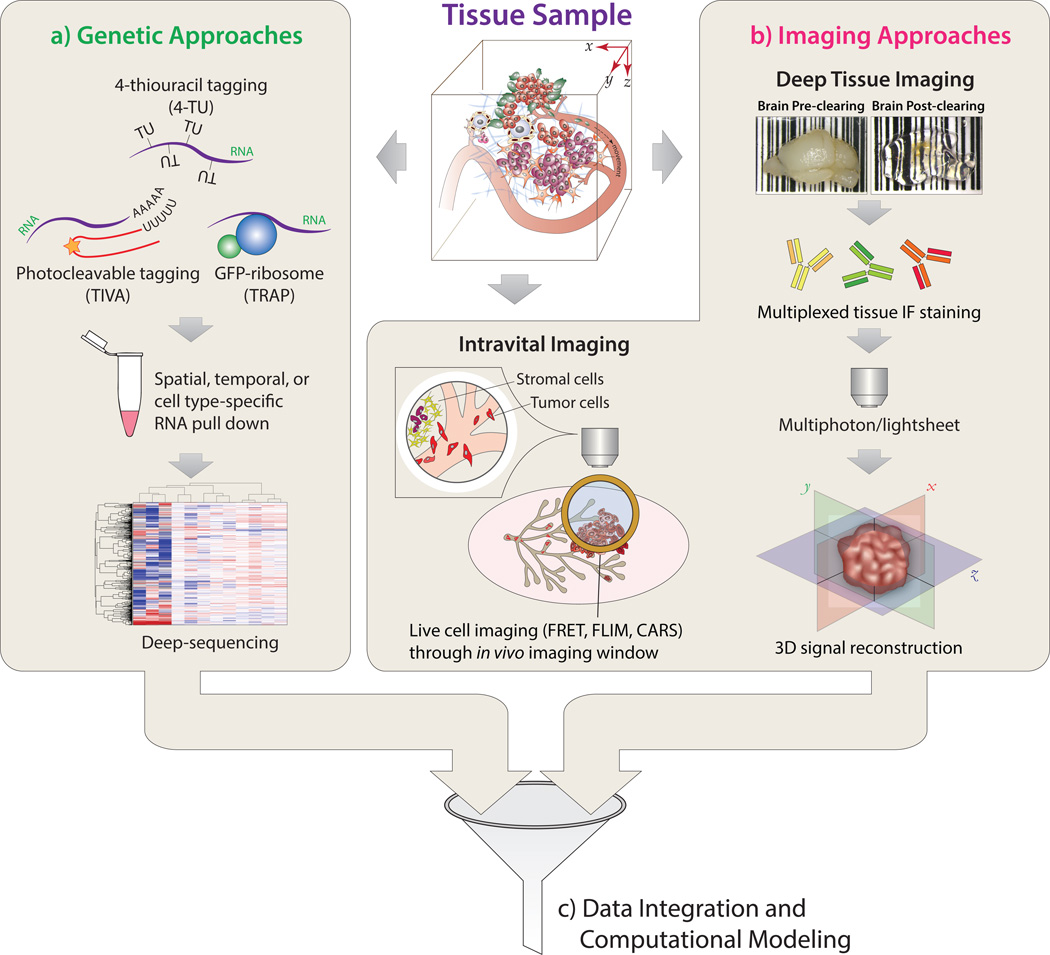
Figure 2. New Technical Frontiers in Studying Tumor-Stromal Cell Interactions.
The compositional and genetic complexity of tumor-stromal cell interactions can be studied via (a) Genetic Approaches: gene expression revealed by transcriptome sequencing after spatial, temporal, or cell type-specific RNA pull down. (b) Imaging Approaches: structural features of tissues and behavioral dynamics of cellular constituents are revealed by intravital imaging and tissue clearing-based deep tissue imaging. (c) Integrative Analysis: integration of multimodal datasets through statistical data analysis and computational modeling.
Supplementary Material
Acknowledgements
This works is partially supported by NIH Pathway to Independence Award CA158066-04 (SZ). S.Z. is a Nancy Dee Assistant Professor in Cancer Research at the University of Notre Dame and Harper Cancer Research Institute.
List of abbreviations
- TME
Tumor Microenvironment
- SAGE
Serial Analysis of Gene Expression
- LCM
Laser Capture Microdissection
- IVM
Intravital Microscopy
- CARS
Coherent anti-Stokes Raman Scattering
- FRET
Fluorescence Resonance Energy Transfer
- FLIM
Fluorescence Lifetime Imaging Microscopy
- BiFC
Bimolecular Fluorescence Complementation
- TCGA
The Cancer Genome Atlas
- ICGC
International Cancer Genome Consortium
- TU
Thiouracil
- UPRT
Uracil Phosphoribosyltransferase
- 4-TU
4-Thiouracil
- TRAP
Translating Ribosome Affinity Purification
Footnotes
Competing interests
No potential competition of interest is disclosed.
Authors' contributions
IG and SZ developed the original concept and wrote the manuscript.
References
- 1.Quail DF, Joyce JA. Nat. Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Visser KE, Eichten A, Coussens LM. Nat. Rev. Cancer. 2006;6:24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 3.Carmeliet P, Jain RK. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 4.Bhowmick NA, Neilson EG, Moses HL. Nature. 2004;432:332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller MM, Fusenig NE. Nat. Rev. Cancer. 2004;4:839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 6.Schuldt A. Nat. Rev. Cancer. 2006;6:S15. doi: 10.1038/nrc3011. [DOI] [PubMed] [Google Scholar]
- 7.Mintz B, Illmensee K. Proc. Natl. Acad. Sci. U. S. A. 1975;72:3585–3589. doi: 10.1073/pnas.72.9.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolberg DS, Bissell MJ. Nature. 1984;309:552–556. doi: 10.1038/309552a0. [DOI] [PubMed] [Google Scholar]
- 9.Dolberg DS, Hollingsworth R, Hertle M, Bissell MJ. Science. 1985;230:676–678. doi: 10.1126/science.2996144. [DOI] [PubMed] [Google Scholar]
- 10.Wolf GT, Hudson JL, Peterson KA, Miller HL, McClatchey KD. Otolaryngol.--Head Neck Surg. Off. J. Am. Acad. Otolaryngol.-Head Neck Surg. 1986;95:142–152. doi: 10.1177/019459988609500203. [DOI] [PubMed] [Google Scholar]
- 11.Kim KJ, Li B, Winer J, Armanini M, Gillett N, Phillips HS, Ferrara N. Nature. 1993;362:841–844. doi: 10.1038/362841a0. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan D, Coussens LM. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan D, Weinberg RA. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 14.Svennevig JL, Lunde OC, Holter J, Bjørgsvik D. Br. J. Cancer. 1984;49:375–377. doi: 10.1038/bjc.1984.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maturu P, Overwijk WW, Hicks J, Ekmekcioglu S, Grimm EA, Huff V. Transl. Oncol. 2014 doi: 10.1016/j.tranon.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tjin EPM, Krebbers G, Meijlink KJ, van de Kasteele W, Rosenberg EH, Sanders J, Nederlof PM, van de Wiel BA, Haanen JBAG, Melief CJM, Vyth-Dreese FA, Luiten RM. Cancer Immunol. Res. 2014;2:538–546. doi: 10.1158/2326-6066.CIR-13-0097. [DOI] [PubMed] [Google Scholar]
- 17.Moinfar F, Man YG, Arnould L, Bratthauer GL, Ratschek M, Tavassoli FA. Cancer Res. 2000;60:2562–2566. [PubMed] [Google Scholar]
- 18.Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, Porter D, Hu M, Chin L, Richardson A, Schnitt S, Sellers WR, Polyak K. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 19.Creighton CJ, Bromberg-White JL, Misek DE, Monsma DJ, Brichory F, Kuick R, Giordano TJ, Gao W, Omenn GS, Webb CP, Hanash SM. Mol. Cancer Res. 2005;3:119–129. doi: 10.1158/1541-7786.MCR-04-0189. [DOI] [PubMed] [Google Scholar]
- 20.Finak G, Sadekova S, Pepin F, Hallett M, Meterissian S, Halwani F, Khetani K, Souleimanova M, Zabolotny B, Omeroglu A, Park M. Breast Cancer Res. 2006;8:R58. doi: 10.1186/bcr1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma X-J, Dahiya S, Richardson E, Erlander M, Sgroi DC. Breast Cancer Res. BCR. 2009;11:R7. doi: 10.1186/bcr2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park ES, Kim SJ, Kim SW, Yoon S-L, Leem S-H, Kim S-B, Kim SM, Park Y-Y, Cheong J-H, Woo HG, Mills GB, Fidler IJ, Lee J-S. Proc. Natl. Acad. Sci. U. S. A. 2011;108:17456–17461. doi: 10.1073/pnas.1114210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knudsen ES, Ertel A, Davicioni E, Kline J, Schwartz GF, Witkiewicz AK. Breast Cancer Res. Treat. 2012;133:1009–1024. doi: 10.1007/s10549-011-1894-3. [DOI] [PubMed] [Google Scholar]
- 24.Vargas AC, McCart Reed AE, Waddell N, Lane A, Reid LE, Smart CE, Cocciardi S, da Silva L, Song S, Chenevix-Trench G, Simpson PT, Lakhani SR. Breast Cancer Res. Treat. 2012;135:153–165. doi: 10.1007/s10549-012-2123-4. [DOI] [PubMed] [Google Scholar]
- 25.Neman J, Termini J, Wilczynski S, Vaidehi N, Choy C, Kowolik CM, Li H, Hambrecht AC, Roberts E, Jandial R. Proc. Natl. Acad. Sci. U. S. A. 2014;111:984–989. doi: 10.1073/pnas.1322098111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pyonteck SM, Akkari L, Schuhmacher AJ, Bowman RL, Sevenich L, Quail DF, Olson OC, Quick ML, Huse JT, Teijeiro V, Setty M, Leslie CS, Oei Y, Pedraza A, Zhang J, Brennan CW, Sutton JC, Holland EC, Daniel D, Joyce JA. Nat. Med. 2013;19:1264–1272. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halin S, Rudolfsson SH, Van Rooijen N, Bergh A. Neoplasia N. Y. N. 2009;11:177–186. doi: 10.1593/neo.81338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 29.Riquelme PA, Drapeau E, Doetsch F. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2008;363:123–137. doi: 10.1098/rstb.2006.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barcellos-Hoff MH, Lyden D, Wang TC. Nat. Rev. Cancer. 2013;13:511–518. doi: 10.1038/nrc3536. [DOI] [PubMed] [Google Scholar]
- 31.Almendro V, Cheng Y-K, Randles A, Itzkovitz S, Marusyk A, Ametller E, Gonzalez-Farre X, Muñoz M, Russnes HG, Helland A, Rye IH, Borresen-Dale A-L, Maruyama R, van Oudenaarden A, Dowsett M, Jones RL, Reis-Filho J, Gascon P, Gönen M, Michor F, Polyak K. Cell Rep. 2014;6:514–527. doi: 10.1016/j.celrep.2013.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gleave JA, Lerch JP, Henkelman RM, Nieman BJ. PloS One. 2013;8:e72039. doi: 10.1371/journal.pone.0072039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ntziachristos V. Nat. Methods. 2010;7:603–614. doi: 10.1038/nmeth.1483. [DOI] [PubMed] [Google Scholar]
- 34.Zhu D, Larin KV, Luo Q, Tuchin VV. Laser Photonics Rev. 2013;7:732–757. doi: 10.1002/lpor.201200056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, Miyawaki A. Nat. Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- 36.Ke M-T, Fujimoto S, Imai T. Nat. Neurosci. 2013;16:1154–1161. doi: 10.1038/nn.3447. [DOI] [PubMed] [Google Scholar]
- 37.Chung K, Wallace J, Kim S-Y, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, Pak S, Bernstein H, Ramakrishnan C, Grosenick L, Gradinaru V, Deisseroth K. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, Yokoyama C, Onoe H, Eguchi M, Yamaguchi S, Abe T, Kiyonari H, Shimizu Y, Miyawaki A, Yokota H, Ueda HR. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 39.Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen C-K, Lubeck E, Shah S, Cai L, Gradinaru V. Cell. 2014;158:945–958. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung K, Deisseroth K. Nat. Methods. 2013;10:508–513. doi: 10.1038/nmeth.2481. [DOI] [PubMed] [Google Scholar]
- 41.Kim S-Y, Chung K, Deisseroth K. Trends Cogn. Sci. 2013;17:596–599. doi: 10.1016/j.tics.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 42.Tomer R, Ye L, Hsueh B, Deisseroth K. Nat. Protoc. 2014;9:1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poguzhelskaya E, Artamonov D, Bolshakova A, Vlasova O, Bezprozvanny I. Mol. Neurodegener. 2014;9:19. doi: 10.1186/1750-1326-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim W-Y, Zhou F-Q, Zhou J, Yokota Y, Wang Y-M, Yoshimura T, Kaibuchi K, Woodgett JR, Anton ES, Snider WD. Neuron. 2006;52:981–996. doi: 10.1016/j.neuron.2006.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, Ragozzino D, Gross CT. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- 46.Gerdes MJ, Sevinsky CJ, Sood A, Adak S, Bello MO, Bordwell A, Can A, Corwin A, Dinn S, Filkins RJ, Hollman D, Kamath V, Kaanumalle S, Kenny K, Larsen M, Lazare M, Li Q, Lowes C, McCulloch CC, McDonough E, Montalto MC, Pang Z, Rittscher J, Santamaria-Pang A, Sarachan BD, Seel ML, Seppo A, Shaikh K, Sui Y, Zhang J, Ginty F. Proc. Natl. Acad. Sci. U. S. A. 2013;110:11982–11987. doi: 10.1073/pnas.1300136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gligorijevic B, Condeelis J. Cell Adhes. Migr. 2009;3:313–315. doi: 10.4161/cam.3.4.9581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kienast Y, von Baumgarten L, Fuhrmann M, Klinkert WEF, Goldbrunner R, Herms J, Winkler F. Nat. Med. 2010;16:116–122. doi: 10.1038/nm.2072. [DOI] [PubMed] [Google Scholar]
- 49.Deguine J, Breart B, Lemaître F, Di Santo JP, Bousso P. Immunity. 2010;33:632–644. doi: 10.1016/j.immuni.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 50.Engelhardt JJ, Boldajipour B, Beemiller P, Pandurangi P, Sorensen C, Werb Z, Egeblad M, Krummel MF. Cancer Cell. 2012;21:402–417. doi: 10.1016/j.ccr.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gligorijevic B, Wyckoff J, Yamaguchi H, Wang Y, Roussos ET, Condeelis J. J. Cell Sci. 2012;125:724–734. doi: 10.1242/jcs.092726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giampieri S, Manning C, Hooper S, Jones L, Hill CS, Sahai E. Nat. Cell Biol. 2009;11:1287–1296. doi: 10.1038/ncb1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boissonnas A, Licata F, Poupel L, Jacquelin S, Fetler L, Krumeich S, Théry C, Amigorena S, Combadière C. Neoplasia N. Y. N. 2013;15:85–94. doi: 10.1593/neo.121572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abdulreda MH, Faleo G, Molano RD, Lopez-Cabezas M, Molina J, Tan Y, Echeverria OAR, Zahr-Akrawi E, Rodriguez-Diaz R, Edlund PK, Leibiger I, Bayer AL, Perez V, Ricordi C, Caicedo A, Pileggi A, Berggren P-O. Proc. Natl. Acad. Sci. 2011;108:12863–12868. doi: 10.1073/pnas.1105002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saar BG, Freudiger CW, Xu X, Huttner A, Kesari S, Young G, Xie XS. Cold Spring Harb. Protoc. 2014;2014 doi: 10.1101/pdb.top081695. [DOI] [PubMed] [Google Scholar]
- 56.Evans CL, Potma EO, Puoris’haag M, Côté D, Lin CP, Xie XS. Proc. Natl. Acad. Sci. U. S. A. 2005;102:16807–16812. doi: 10.1073/pnas.0508282102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salameh TS, Le TT, Nichols MB, Bauer E, Cheng J, Camarillo IG. Int. J. Cancer J. Int. Cancer. 2013;132:288–296. doi: 10.1002/ijc.27672. [DOI] [PubMed] [Google Scholar]
- 58.Hellerer T, Axäng C, Brackmann C, Hillertz P, Pilon M, Enejder A. Proc. Natl. Acad. Sci. 2007;104:14658–14663. doi: 10.1073/pnas.0703594104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Conway JRW, Carragher NO, Timpson P. Nat. Rev. Cancer. 2014;14:314–328. doi: 10.1038/nrc3724. [DOI] [PubMed] [Google Scholar]
- 60.Timpson P, McGhee EJ, Morton JP, von Kriegsheim A, Schwarz JP, Karim SA, Doyle B, Quinn JA, Carragher NO, Edward M, Olson MF, Frame MC, Brunton VG, Sansom OJ, Anderson KI. Cancer Res. 2011;71:747–757. doi: 10.1158/0008-5472.CAN-10-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okaty BW, Sugino K, Nelson SB. PLoS ONE. 2011;6:e16493. doi: 10.1371/journal.pone.0016493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hines WC, Su Y, Kuhn I, Polyak K, Bissell MJ. Cell Rep. 2014;6:779–781. doi: 10.1016/j.celrep.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 63.Gay L, Miller MR, Ventura PB, Devasthali V, Vue Z, Thompson HL, Temple S, Zong H, Cleary MD, Stankunas K, Doe CQ. Genes Dev. 2013;27:98–115. doi: 10.1101/gad.205278.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cleary MD, Meiering CD, Jan E, Guymon R, Boothroyd JC. Nat. Biotechnol. 2005;23:232–237. doi: 10.1038/nbt1061. [DOI] [PubMed] [Google Scholar]
- 65.Heiman M, Kulicke R, Fenster RJ, Greengard P, Heintz N. Nat. Protoc. 2014;9:1282–1291. doi: 10.1038/nprot.2014.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lovatt D, Ruble BK, Lee J, Dueck H, Kim TK, Fisher S, Francis C, Spaethling JM, Wolf JA, Grady MS, Ulyanova AV, Yeldell SB, Griepenburg JC, Buckley PT, Kim J, Sul J-Y, Dmochowski IJ, Eberwine J. Nat. Methods. 2014;11:190–196. doi: 10.1038/nmeth.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perkins JR, Antunes-Martins A, Calvo M, Grist J, Rust W, Schmid R, Hildebrandt T, Kohl M, Orengo C, McMahon SB, Bennett DLH. Mol. Pain. 2014;10:7. doi: 10.1186/1744-8069-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Z, Gerstein M, Snyder M. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grindberg RV, Yee-Greenbaum JL, McConnell MJ, Novotny M, O’Shaughnessy AL, Lambert GM, Araúzo-Bravo MJ, Lee J, Fishman M, Robbins GE, Lin X, Venepally P, Badger JH, Galbraith DW, Gage FH, Lasken RS. Proc. Natl. Acad. Sci. U. S. A. 2013;110:19802–19807. doi: 10.1073/pnas.1319700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang F, Barbacioru C, Nordman E, Li B, Xu N, Bashkirov VI, Lao K, Surani MA. Nat. Protoc. 2010;5:516–535. doi: 10.1038/nprot.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt-Rosen O, Suvà ML, Regev A, Bernstein BE. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eliceiri KW, Berthold MR, Goldberg IG, Ibáñez L, Manjunath BS, Martone ME, Murphy RF, Peng H, Plant AL, Roysam B, Stuurman N, Stuurmann N, Swedlow JR, Tomancak P, Carpenter AE. Nat. Methods. 2012;9:697–710. doi: 10.1038/nmeth.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Walter T, Shattuck DW, Baldock R, Bastin ME, Carpenter AE, Duce S, Ellenberg J, Fraser A, Hamilton N, Pieper S, Ragan MA, Schneider JE, Tomancak P, Hériché J-K. Nat. Methods. 2010;7:S26–41. doi: 10.1038/nmeth.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Coster AD, Wichaidit C, Rajaram S, Altschuler SJ, Wu LF. Nat. Methods. 2014;11:602. doi: 10.1038/nmeth.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hovhannisyan VA, Su P-J, Chen Y-F, Dong CY. Opt. Express. 2008;16:5107–5117. doi: 10.1364/oe.16.005107. [DOI] [PubMed] [Google Scholar]
- 76.Seo J, Hwang S, Lee J-M, Park H. J. Microsc. 2014 doi: 10.1111/jmi.12141. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- 77.Kharchenko PV, Silberstein L, Scadden DT. Nat. Methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Conway T, Wazny J, Bromage A, Tymms M, Sooraj D, Williams ED, Beresford-Smith B. Bioinformatics. 2012;28:i172–i178. doi: 10.1093/bioinformatics/bts236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, Bruneval P, Fridman WH, Becker C, Pagès F, Speicher MR, Trajanoski Z, Galon J. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 80.Han J, Chang H, Giricz O, Lee GY, Baehner FL, Gray JW, Bissell MJ, Kenny PA, Parvin B. PLoS Comput Biol. 2010;6:e1000684. doi: 10.1371/journal.pcbi.1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kirk P, Griffin JE, Savage RS, Ghahramani Z, Wild DL. Bioinforma. Oxf. Engl. 2012;28:3290–3297. doi: 10.1093/bioinformatics/bts595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anderson ARA, Weaver AM, Cummings PT, Quaranta V. Cell. 2006;127:905–915. doi: 10.1016/j.cell.2006.09.042. [DOI] [PubMed] [Google Scholar]
- 83.Enderling H, Hlatky L, Hahnfeldt P. Theor. Biol. Med. Model. 2012;9:31. doi: 10.1186/1742-4682-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.