Abstract
Epithelial cells line the surfaces of the body, and are on the front lines of defense against microbial infection. Like many other metazoans, the nematode C. elegans lacks known professional immune cells and relies heavily on defense mediated by epithelial cells. New results indicate that epithelial defense in C. elegans can be triggered through detection of pathogen-induced perturbation of core physiology within host cells and through autophagic defense against intracellular and extracellular pathogens. Recent studies have also illuminated a diverse array of pathogenic attack strategies used against C. elegans. These findings are providing insight into the underpinnings of host/pathogen interactions in a simple animal host that can inform studies of infectious diseases in humans.
Introduction
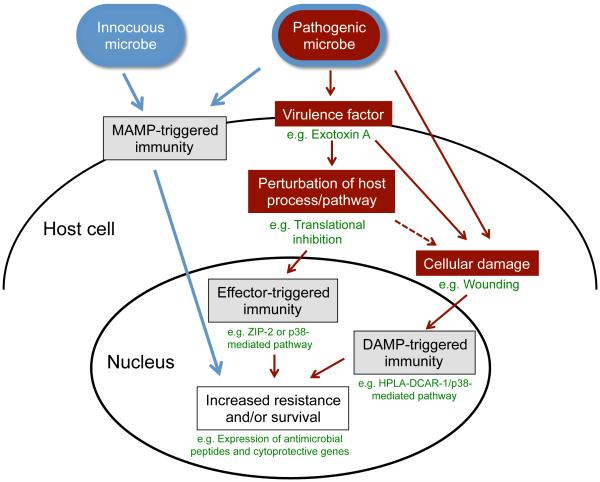
Epithelial cells cover the internal and external body surfaces and thus are often the first responders to pathogenic attack by microbes [1,2]. The significance of epithelial cells for immunity in humans is increasingly appreciated, and epithelial cells are now realized to be key players in defense against infection of the lung, skin and intestine. Furthermore, inappropriate activation of epithelial cell immune pathways can lead to inflammatory diseases, and thus understanding the immune pathways in these cells is of critical importance for human health. Epithelial cells, especially those of the intestine, are in regular contact with a wide array of microbes that include pathogenic microbes that cause disease, as well as innocuous or even beneficial microbial species. Therefore, epithelial cells have the challenge not just of how to discriminate self from non-self (a classic question in immunology), but also how to discriminate pathogen from non-pathogen. Canonical innate immune pathways triggered by pattern-recognition receptors (PRRs) are not sufficient to provide this distinction [3]. PRRs detect molecules associated with broad classes of microbes – these molecules were originally called pathogen-associated molecular patterns, or PAMPs, but they are now commonly called microbe-associated molecular patterns, or MAMPs, in keeping with the idea that they can be found in both pathogenic and non-pathogenic microbes [4,5] (Figure 1). Understanding how hosts distinguish pathogens from other microbes is a rapidly developing field within the area of infection and innate immunity, and is particularly relevant for epithelial cell defense [5,6].
Figure 1. Model for diverse triggers of defense gene expression in response to infection.
C. elegans uses effector-triggered immunity, as well as DAMP-triggered immunity, to upregulate defense gene expression in response to infection. MAMP-triggered immunity is well-described for other hosts, but has not yet been described for C. elegans.
Background on C. elegans infections and host defense
Many animal species lack professional immune cells, and yet thrive in a diverse microbial world by relying on defense by epithelial cells. The nematode Caenorhabditis elegans is one such species, and indeed nematodes are among the most numerous animals on the planet [7]. Many different microbial pathogens have been shown to attack and induce a defense response in the epithelial cells of C. elegans [8-16]. Pseudomonas aeruginosa is an opportunistic bacterial pathogen of humans and the most commonly studied pathogen in C. elegans, where it causes a lethal infection of intestinal epithelial cells [17]. In addition, several other bacterial, fungal and viral pathogens can infect the C. elegans intestine, and penetrating fungal species can infect epithelial cells of the epidermis. C. elegans has no known dedicated, migratory immune cells like macrophages to aid in defense against infection of the intestine or epidermis, and does not appear to have canonical cytokine and chemokine signaling pathways used to recruit those cells. However, C. elegans does use system-wide signaling to respond to stress and infection by upregulating defense pathways in epithelial cells, which is a topic that has been covered in other reviews [9,12,18-21].
C. elegans provides a powerful model system to address questions about innate immune pathways that are independent of classic PRR/MAMP signaling: C. elegans lacks components of some of the PRR pathways used by other metazoans, and it has yet to be shown to respond to MAMPs. In particular, C. elegans does not have an obvious NFκB ortholog, nor does it have Nod-like receptors (NLRs), and its single Toll-like receptor (TLR) does not play a substantial role in defense [14,22]. Interestingly, these signaling components are found in cnidaria, a clade that includes coral, jellyfish and hydra. Like C. elegans, these animals have outer and inner epithelial layers without known dedicated immune cells [23], so the presence of these immune signaling components does not correlate with the presence of a professional immune system. Given the evolutionary relationships among these animals, C. elegans most likely lost these genes during evolution, and presumably other pathways have been able to compensate for their role. Importantly, C. elegans does have a robust inducible defense system. In response to both intestinal and epidermal infection, C. elegans epithelial cells upregulate secreted antimicrobial peptides, detoxifying enzymes and efflux pumps, with distinct responses to distinct pathogens [24]. While some of this transcriptional response might be due to MAMP detection in C. elegans [25-27], it is clear that other signals from pathogens trigger a substantial part of the C. elegans transcriptional response to infection [28-30]. Previous studies of the inducible transcriptional response to infection have indicated that several signaling pathways control these responses, but one central pathway is a p38 MAP kinase (MAPK) pathway that includes a p38 MAPK called PMK-1 [31]. The PMK-1 p38 kinase cascade is an evolutionarily conserved pathway and is important for defense against microbial attack of both the C. elegans intestine and the epidermis. Several transcription factors have been shown to act downstream of PMK-1 in different contexts to control inducible defenses upon infection [32]. Other defense pathways operate in parallel to the p38 kinase cascade, including one regulated by the bZIP transcription factor ZIP-2 [8]. The upstream activators of these pathways, both pathogen-derived and host-derived, are just now being elucidated as described below.
Mechanisms of microbial pathogenesis and host defense in C. elegans have been reviewed previously [8-16]. Here we describe major developments from the last two years with a focus on bacterial infections, but also mention infections by other microbes when relevant. An emerging body of data suggests that nematodes monitor disruptions in cellular homeostasis as a means to detect pathogen infection and mount protective host responses. New data implicate these signals in the activation of conserved immune pathways, including the p38 pathway. In addition, several studies have implicated a conserved role for epithelial autophagy in C. elegans host defense against a broad array of pathogens. Finally, studies of bacterial pathogens have yielded insights both into the strategies employed by microbes to establish infection and the pathogen-encoded factors that lead to C. elegans immune pathway activation.
Surveillance or "effector-triggered" immunity induces host defense by monitoring core processes perturbed by pathogens
One feature that distinguishes pathogens from other microbes is their delivery of toxins and other effector molecules into host cells to disable core processes and pathways that might otherwise aid in defense. The immune responses to these attacks have been termed “effector-triggered” immunity or surveillance immunity, which is a concept that has been pioneered in plant immunity and more recently been appreciated in animal hosts, including C. elegans [4,5,33] (Figure 1).
A common mode of bacterial attack is to disable the process of host mRNA translation, which can prevent production of anti-microbial molecules and therefore improve bacterial survival. An effector-triggered immune pathway activated by inhibition of translation elongation was discovered in C. elegans through analysis of the transcriptional response to P. aeruginosa infection. Previous studies indicated that P. aeruginosa pathogenicity induced protective transcriptional responses mediated by the p38 pathway as well as by the ZIP-2 transcription factor [34]. More recently, it has been shown that a trigger for these pathways is inhibition of translation by the P. aeruginosa secreted Exotoxin A [35]. Studies by McEwan et al demonstrated that C. elegans detects the translation-blocking effects of this toxin, instead of directly detecting Exotoxin A molecular structure [36]. Furthermore, they showed that ZIP-2, as well as the p38 pathway and the G-protein-coupled receptor FSHR-1 provide defense against its toxic effects. In a parallel study, Dunbar et al used an RNAi screen and found that translational inhibition as well as perturbation of several other core processes, such as mitochondrial pathways and transcription-related pathways, could trigger ZIP-2-mediated defense gene expression [37]. They found that host endocytosis of Exotoxin A appeared to block translation specifically in the intestine, which then led to a paradoxical increase in levels of ZIP-2 protein via an upstream open reading frame that controls ZIP-2 expression. In mammals and flies, perturbation of translation has been shown to upregulate cytokine expression, demonstrating a broadly conserved link between surveillance of translation and immune responses [38,39]. In a related study, Melo and Ruvkun knocked down genes in core processes in C. elegans and observed induction of infection response genes and avoidance of normally attractant food, which is a characteristic behavioral response to infection that allows animals to avoid pathogenic microbes [40]. They found that inhibition of many core processes, such as translation, ATP synthesis, proteasome function, in the hypodermis and/or the intestine (common sites of infection) was sufficient to induce avoidance, and expression of defense genes. Altogether, these three studies indicated that C. elegans can sense perturbations of core processes as a method to detect pathogenic attack, and provided insight to a specific mechanism by which blockade of mRNA translation by the pathogen P. aeruginosa can induce host defense gene expression (Table 1, Figure 1).
Table 1.
Recent discoveries about microbial pathogens, their virulence factors, and the immune pathways they activate in C. elegans.
| Infection | Virulence Factor /Attack |
Immunity Triggers |
C. elegans innate immune pathways |
References |
|---|---|---|---|---|
|
P. aeruginosa
(Slow killing model) |
Exotoxin A | Translational inhibition |
ZIP-2, PMK-1, FSHR-1 dependent pathways |
[36,37,40] |
| unknown | Mitochondrial stress |
ATFS-1, ZIP-2, and ceramide/mevalonate pathways |
[37,40-42] | |
|
P. aeruginosa
(Liquid killing model) |
Pyoverdin (iron scavenging) |
Hypoxia | HIF-1 associated defense response |
[62] |
|
P. aeruginosa
(Fast killing model) |
Phenazine-1- carboxylic acid |
Oxidative stress |
AGE-1 PI3K pathway | [60,61] |
| N. parisii | unknown | Proteasomal inhibition |
Ubiquitylation-related pathways |
[44] |
| D. coniospora | Wounding of the cuticle |
HPLA | DCAR-1 receptor activation of PMK-1 pathway |
[47] |
|
S. aureus,
P. aeruginosa (Slow killing model), S. enterica, N. parisii |
unknown | unknown | Autophagy-mediated defense response (TFEB-1 mediated for S. aureus) |
[44,56,57] |
Mitochondrial function is essential for the organismal health, and there are many genera of bacteria found in the C. elegans natural habitat that induce mitochondria stress upon infection. As mentioned above, perturbation of mitochondrial pathways was able to induce aversive behavioral responses, as well as infection response gene expression [37,40]. Two new studies provide mechanistic insight about how mitochondrial surveillance pathways trigger this defense gene expression [41,42]. Pellegrino et al found an immune role for the bZIP transcription factor ATFS-1, as well as ZIP-2, in mitochondrial stress [42]. Previously, these authors had shown that during mitochondrial stress, ATFS-1 is prevented from trafficking to mitochondria, where it normally localizes, and instead moves to the nucleus where it induces the mitochondrial unfolded protein response [43]. More recently, these authors have shown that this ATFS-1 nuclear localization also occurs upon P. aeruginosa infection, and ATFS-1 can induce expression of anti-microbial peptides and secreted lysozyme as part of the innate immune response, acting together with the ZIP-2 transcription factor [42]. Liu et al showed that ceramide and mevalonate biosynthetic pathways are involved in mitochondrial surveillance responses, and ceramide appears to act upstream of ATFS-1. In addition, they demonstrated that certain bacterial species block the induction of defense gene expression triggered by mitochondrial surveillance. Thus, C. elegans uses ATFS-1, ZIP-2, ceramide and mevalonate surveillance pathways to detect disruption of mitochondrial function and induce immune responses.
Another core host system recently shown to be involved in the inducible response to infection in C. elegans is the ubiquitin proteasome system, which targets proteins for degradation. Two intracellular pathogens of C. elegans shown to infect worms in the natural environment are the Orsay virus and the fungal-like microsporidian pathogen Nematocida parisii. Surprisingly, these very distinct pathogens were found to induce very similar transcriptional responses in C. elegans, which were distinct from responses to other pathogens and were characterized by an upregulation of ubiquitin ligase components [44,45]. These ubiquitin ligase components provided defense against infection, and interestingly, their expression could also be induced by inhibition of proteasome function, suggesting that detection of these intracellular pathogens is related to surveillance of the core cellular process of proteasomal degradation [40,44].
Upstream activators of the p38 MAPK immune pathway are diverse and damage-associated
In plant defense, effector-triggered immunity can detect pathogen-induced changes to host cells before there is overt damage, and the same may hold true in animal defense (Figure 1). In addition, prolonged or extensive damage can also trigger defense against infection, through so-called "damage-associated molecular patterns" or DAMPs [46]. DAMPs in mammalian immunity include a diverse range of molecules, including ATP, interleukin 1α, uric acid, S100 cytoplasmic proteins, the nuclear protein HMGB1 and the extracellular matrix molecule hyaluronan. Key insights into DAMP-mediated activation of p38 signaling in C. elegans have come from recent studies of fungal infection, which previously had been shown to trigger a G-protein signaling cascade upstream of p38 in epidermal cells [32,47]. These signaling events can be activated by infection with the fungus Drechmeria coniospora, which penetrates the C. elegans cuticle outside the epidermis, and by sterile wounding of the epidermis, indicating that damage to the epidermis elicits an immune response. Zugasti et al used RNAi screening to identify a G-protein coupled receptor called DCAR-1 to be responsible for activating p38 signaling in both cases [47]. Furthermore, they identified an endogenous ligand called HPLA, which is a tyrosine derivative generated by infection, and likely also by wounding. Thus, HPLA can be considered a DAMP, and together with DCAR-1 provide the first-described ligand/receptor pair that induce immune responses in C. elegans. Future studies will likely investigate the mechanism by which HPLA is induced, and whether a similar signaling pathway exists in mammals.
Although less is known about the upstream activators of p38 in response to bacterial infection in C. elegans, a recent study indicated that this pathway can be activated by an exogenous compound to promote resistance against infection [48]. An earlier high-throughput screen for anti-infectives yielded several compounds that appeared to act on the host, instead of the pathogen [49]. Pukkila-Worley et al characterized one of these compounds and demonstrated that it provided protection against killing by P. aeruginosa infection. Although its exact molecular target is unknown, they demonstrated that this small molecule could activate genes that are also induced by P. aeruginosa infection and the p38 pathway, and that its effects on pathogen resistance were partially dependent on this pathway. Further studies by these authors identified the conserved Mediator subunit MDT-15/ MED15 to play a role in compound-mediated resistance through the p38 pathway [50]. Given that this compound is toxic to worms, these studies suggest bacterial activation of the intestinal p38 pathway may be due to pathogen-induced damage or perturbation of core physiology. Indeed, a wide variety of bacterial-associated stimuli appear to induce the p38 pathway. Recent discoveries indicate that the translation-blocking Shiga toxin from pathogenic E. coli (which has the same mechanism of action as Exotoxin A from P. aeruginosa) can induce the p38 pathway [51]. Furthermore, non-pathogenic soil-associated bacteria have been shown to induce immune responses via p38 signaling [52]. It is still unclear in these contexts the exact pathogenic triggers and how they are detected by C. elegans.
Autophagy mediates defense against a broad range of pathogenic microbes
Another form of immunity that can be triggered by damage is the cellular process of autophagy, or self-eating, and several recent studies in C. elegans have demonstrated a role for autophagy in defense against infection. Autophagy is the process of de novo membrane formation to engulf large, damaged cellular components for digestion and recycling. This process has gained attention for having a key role in defense against intracellular pathogens in professional immune cells as well as in epithelial cells [53]. Targeting intracellular pathogens for degradation via the autophagy pathway has been deemed 'xenophagy', and can be triggered by damage to intracellular membranes. In C. elegans, autophagy was first shown to have a role in defense against infection by the human bacterial pathogen Salmonella enterica, which is a facultative intracellular pathogen [54]. In these studies, Salmonella was only found intracellular in the C. elegans intestine if autophagy was compromised, indicating that C. elegans autophagy prevents invasion or replication within intestinal cells. However, a subsequent study did not confirm that inhibition of autophagy leads to intracellular Salmonella in the intestine, although it did show that autophagy was required in the intestine to promote survival upon infection [55]. Direct localization of C. elegans autophagy machinery was recently shown for the intracellular pathogen N. parisii, and autophagy played a role in controlling levels of this pathogen [44].
In addition to promoting defense against intracellular microbes, autophagy has recently been shown to promote defense against bacteria that are predominantly extracellular. In particular, Visvikis et al demonstrated that the conserved autophagy transcription factor TFEB-1 activated cytoprotective and anti-microbial gene expression in response to infection with the Gram-positive pathogen Staphylococcus aureus and promoted survival [56]. These authors found a similar role for TFEB-1 in response to S. aureus infections in mammalian cells. An unusual role for autophagy in defense was shown by Zou et al, who demonstrated that autophagy activation prevents necrosis, and provides defense against P. aeruginosa infection [57]. Interestingly, autophagy did not reduce pathogen load (i.e. increase resistance) in S. aureus or P. aeruginosa infections, but rather improved tolerance of pathogen infection [58]. Thus, these new findings indicate that autophagy promotes tolerance against extracellular pathogens by inducing defense gene expression and preventing infection-induced damage, in addition to promoting resistance by directly targeting intracellular pathogens for destruction (xenophagy) and lowering pathogen load.
New insights into strategies of pathogenic attack in C. elegans infections
Pathogens often rely on an extensive arsenal of virulence factors to attack their hosts, and they can deploy different factors under different conditions. One pathogen in particular that has a large number of virulence factors is P. aeruginosa, which can cause a range of different infections in humans. To explore the variety of virulence strategies used by P. aeruginosa, several pathogenesis models have been developed in C. elegans. All the studies mentioned above used the most common pathogenesis model for P. aeruginosa, which is the slow-killing model, where bacteria are grown on minimal media plates and fed to C. elegans. Under these conditions, killing of C. elegans requires the bacteria to be alive and involves accumulation of bacteria in the intestinal lumen of C. elegans. A recently described genome-wide screen with this model identified 170 genes required for virulence, although it did not identify secretion systems or obvious individual effector molecules [59]. This study together with others mentioned above indicate that individual virulence factors such as Exotoxin A are not required for gene induction or for killing, but are sufficient to induce gene expression and cause damage, implying there is extensive redundancy in the toxins deployed by P. aeruginosa under these slow-killing conditions [36,37].
Two other models for P. aeruginosa pathogenesis have been developed in C. elegans. One model is the "fast-killing" model, where growth of P. aeruginosa on nutrient-rich, high osmolarity plates leads to rapid killing of C. elegans. This killing does not require P. aeruginosa to be alive during exposure, does not involve accumulation of P. aeruginosa cells within the intestine, and appears to be caused by diffusible toxins. Previous work had implicated host oxidative stress responses and bacterial-derived phenazines as key factors in this infection model [60], and Cezairliyan et al used a combination of mutant, biochemical and metabolomic studies to demonstrate that the predominant toxin in this model is phenazine-1-carboxylic acid [61]. A new model for P. aeruginosa pathogenicity in C. elegans is the "liquid-killing" format, which identified a separate set of factors important for pathogenicity [62]. Kirienko et al showed that iron-scavenging by P. aeruginosa-derived pyoverdin is a virulence strategy, and can lead to a hypoxic crisis in C. elegans. The C. elegans ortholog of hypoxia-inducible factor (HIF-1) promoted survival upon infection in this model; HIF-1 has also been shown to regulate host resistance in other infection models [63,64]. Given that pyoverdin and phenazines have been implicated in P. aeruginosa killing in mammals, further analysis of these pathogenesis strategies in medically relevant infections is warranted.
In addition to toxin-based killing, a visually dramatic form of pathogenic attack was described by Hodgkin et al, who showed how Leucobacter bacteria attack the epidermal surface of C. elegans and use a trapping strategy to create "worm-stars", which are structures of dozens of worms captured by their tails radiating outward, preventing their mobility and leading to their early demise [65]. These studies focused on natural epidermal pathogens and found that C. elegans resistance to one strain caused by changes in surface glycosylation led to susceptibility to a second strain, and a sub-lethal infection with the second strain induced resistance to the first strain. Thus, they demonstrated the trade-offs that are often found in host defense against microbial infection, and also identified a novel mechanism by which bacterial pathogens can attack their hosts.
Conclusion
Discoveries in recent years have shown how C. elegans can detect pathogens not simply through the identification of their physical presence, but through detecting the common perturbations they cause in core processes to upregulate defense. Studies have also indicated that host autophagy serves to increase pathogen tolerance against extracellular pathogens, as well as pathogen resistance against intracellular pathogens. Recent findings have also highlighted the diversity of pathogenic strategies that bacteria use to attack C. elegans. This animal host provides a powerful model for further investigating pathogen attack and damage-triggered or effector-triggered immunity in epithelial cells, because it appears to be the foundation of defense for C. elegans.
Highlights.
C. elegans detects infection with surveillance or "effector-triggered" immunity
Multiple upstream inputs activate the p38 MAPK pathway
Autophagy provides defense against intracellular and extracellular pathogens
Pathogen attack involves toxins, as well as physical trapping of the host
Acknowledgements
We thank Read Pukkila-Worley, Suzannah Szumowski and Keir Balla for helpful comments on the manuscript. This work was supported by NIH predoctoral training grant T32 GM07240 to L.B.C., and NIAID R01 AI087528, the Searle Scholars Program, Packard Foundation and Burroughs Wellcome Fund fellowship to E.R.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nature reviews. Immunology. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benjamin JL, Sumpter R, Jr., Levine B, Hooper LV. Intestinal epithelial autophagy is essential for host defense against invasive bacteria. Cell Host Microbe. 2013;13:723–734. doi: 10.1016/j.chom.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 4.Rajamuthiah R, Mylonakis E. Effector triggered immunity: Activation of innate immunity in metazoans by bacterial effectors. Virulence. 2014;5 doi: 10.4161/viru.29091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stuart LM, Paquette N, Boyer L. Effector-triggered versus pattern-triggered immunity: how animals sense pathogens. Nat Rev Immunol. 2013;13:199–206. doi: 10.1038/nri3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murfin KE, Dillman AR, Foster JM, Bulgheresi S, Slatko BE, Sternberg PW, Goodrich-Blair H. Nematode-bacterium symbioses--cooperation and conflict revealed in the "omics" age. Biol Bull. 2012;223:85–102. doi: 10.1086/BBLv223n1p85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pukkila-Worley R, Ausubel FM. Immune defense mechanisms in the Caenorhabditis elegans intestinal epithelium. Curr Opin Immunol. 2012;24:3–9. doi: 10.1016/j.coi.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ermolaeva MA, Schumacher B. Insights from the worm: The C. elegans model for innate immunity. Semin Immunol. 2014 doi: 10.1016/j.smim.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Partridge FA, Gravato-Nobre MJ, Hodgkin J. Signal transduction pathways that function in both development and innate immunity. Dev Dyn. 2010;239:1330–1336. doi: 10.1002/dvdy.22232. [DOI] [PubMed] [Google Scholar]
- 11.Clark LC, Hodgkin J. Commensals, probiotics and pathogens in the Caenorhabditis elegans model. Cell Microbiol. 2014;16:27–38. doi: 10.1111/cmi.12234. [DOI] [PubMed] [Google Scholar]
- 12.Kim DH. Bacteria and the aging and longevity of Caenorhabditis elegans. Annu Rev Genet. 2013;47:233–246. doi: 10.1146/annurev-genet-111212-133352. [DOI] [PubMed] [Google Scholar]
- 13.Balla KM, Troemel ER. Caenorhabditis elegans as a model for intracellular pathogen infection. Cell Microbiol. 2013;15:1313–1322. doi: 10.1111/cmi.12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irazoqui JE, Urbach JM, Ausubel FM. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat Rev Immunol. 2010;10:47–58. doi: 10.1038/nri2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mylonakis E, Aballay A. Worms and flies as genetically tractable animal models to study host-pathogen interactions. Infect Immun. 2005;73:3833–3841. doi: 10.1128/IAI.73.7.3833-3841.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sifri CD, Begun J, Ausubel FM. The worm has turned--microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 2005;13:119–127. doi: 10.1016/j.tim.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 17.Kirienko NV, Cezairliyan BO, Ausubel FM, Powell JR. Pseudomonas aeruginosa PA14 Pathogenesis in Caenorhabditis elegans. Methods Mol Biol. 2014;1149:653–669. doi: 10.1007/978-1-4939-0473-0_50. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Zhang Y. Neural-immune communication in Caenorhabditis elegans. Cell Host Microbe. 2009;5:425–429. doi: 10.1016/j.chom.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 19.Tan MW, Shapira M. Genetic and molecular analysis of nematode-microbe interactions. Cell Microbiol. 2011;13:497–507. doi: 10.1111/j.1462-5822.2011.01570.x. [DOI] [PubMed] [Google Scholar]
- 20.Aballay A. Role of the nervous system in the control of proteostasis during innate immune activation: insights from C. elegans. PLoS Pathog. 2013;9:e1003433. doi: 10.1371/journal.ppat.1003433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Oosten-Hawle P, Morimoto RI. Transcellular chaperone signaling: an organismal strategy for integrated cell stress responses. J Exp Biol. 2014;217:129–136. doi: 10.1242/jeb.091249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, Tan MW, Ray KP, Solari R, Johnson CD, Ewbank JJ. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. 2001;11:809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- 23.Bosch TC. Cnidarian-microbe interactions and the origin of innate immunity in metazoans. Annu Rev Microbiol. 2013;67:499–518. doi: 10.1146/annurev-micro-092412-155626. [DOI] [PubMed] [Google Scholar]
- 24.Shivers RP, Youngman MJ, Kim DH. Transcriptional responses to pathogens in Caenorhabditis elegans. Curr Opin Microbiol. 2008;11:251–256. doi: 10.1016/j.mib.2008.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pukkila-Worley R, Ausubel FM, Mylonakis E. Candida albicans infection of Caenorhabditis elegans induces antifungal immune defenses. PLoS Pathog. 2011;7:e1002074. doi: 10.1371/journal.ppat.1002074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irazoqui JE, Troemel ER, Feinbaum RL, Luhachack LG, Cezairliyan BO, Ausubel FM. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 2010;6:e1000982. doi: 10.1371/journal.ppat.1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Twumasi-Boateng K, Shapira M. Dissociation of immune responses from pathogen colonization supports pattern recognition in C. elegans. PLoS One. 2012;7:e35400. doi: 10.1371/journal.pone.0035400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Rourke D, Baban D, Demidova M, Mott R, Hodgkin J. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 2006 doi: 10.1101/gr.50823006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pujol N, Cypowyj S, Ziegler K, Millet A, Astrain A, Goncharov A, Jin Y, Chisholm AD, Ewbank JJ. Distinct innate immune responses to infection and wounding in the C. elegans epidermis. Curr Biol. 2008;18:481–489. doi: 10.1016/j.cub.2008.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, Tanaka-Hino M, Hisamoto N, Matsumoto K, Tan MW, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- 32.Luallen RJ, Troemel ER. Breaking barriers: a GPCR triggers immunity in nematodes. Nat Immunol. 2014;15:826–828. doi: 10.1038/ni.2963. [DOI] [PubMed] [Google Scholar]
- 33.Spoel SH, Dong X. How do plants achieve immunity? Defence without specialized immune cells. Nature reviews. Immunology. 2012;12:89–100. doi: 10.1038/nri3141. [DOI] [PubMed] [Google Scholar]
- 34.Estes KA, Dunbar TL, Powell JR, Ausubel FM, Troemel ER. bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2010;107:2153–2158. doi: 10.1073/pnas.0914643107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleino A, Silverman N. UnZIPping mechanisms of effector-triggered immunity in animals. Cell Host Microbe. 2012;11:320–322. doi: 10.1016/j.chom.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McEwan DL, Kirienko NV, Ausubel FM. Host translational inhibition by Pseudomonas aeruginosa Exotoxin A Triggers an immune response in Caenorhabditis elegans. Cell Host Microbe. 2012;11:364–374. doi: 10.1016/j.chom.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• The authors show that P. aeruginosa-derived Exotoxin A is suficient to produce an immune response in C. elegans, and the ZIP-2, p38 and FSHR-1 pathways provide defense against this toxin.
- 37.Dunbar TL, Yan Z, Balla KM, Smelkinson MG, Troemel ER. C. elegans detects pathogen-induced translational inhibition to activate immune signaling. Cell Host Microbe. 2012;11:375–386. doi: 10.1016/j.chom.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• The authors find that C. elegans senses translational block in the intestine induced by P. aeruginosa-derived Exotoxin A, to induce an innate immune response via increased levels of the ZIP-2 transcription factor.
- 38.Mohr I, Sonenberg N. Host translation at the nexus of infection and immunity. Cell Host Microbe. 2012;12:470–483. doi: 10.1016/j.chom.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemaitre B, Girardin SE. Translation inhibition and metabolic stress pathways in the host response to bacterial pathogens. Nat Rev Microbiol. 2013;11:365–369. doi: 10.1038/nrmicro3029. [DOI] [PubMed] [Google Scholar]
- 40.Melo JA, Ruvkun G. Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell. 2012;149:452–466. doi: 10.1016/j.cell.2012.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • The authors demonstrate that inhibition of core processes induces responses similar to those induced by pathogens, including food avoidance and detoxification gene activation.
- 41.Liu Y, Samuel BS, Breen PC, Ruvkun G. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature. 2014;508:406–410. doi: 10.1038/nature13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• The authors used a genome wide RNAi screen to identify the ceramide biosynthetic pathway as a key component of mitochondrial surveillance, which is used to detect infection from a wide range of bacteria genera.
- 42.Pellegrino MW, Nargund AM, Kirienko NV, Gillis R, Fiorese CJ, Haynes CM. Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature. doi: 10.1038/nature13818. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• In this study, the authors found that after mitochondrial stress, the transcription factor ATFS-1 traffics from the mitochondria to the nucleus, where it upregulates the innate immune response to defend against P. aeruginosa infection.
- 43.Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bakowski MA, Desjardins CA, Smelkinson MG, Dunbar TA, Lopez-Moyado IF, Rifkin SA, Cuomo CA, Troemel ER. Ubiquitin-mediated response to microsporidia and virus infection in C. elegans. PLoS Pathog. 2014;10:e1004200. doi: 10.1371/journal.ppat.1004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • The authors show ubiquitin and autophagy machinery localize to an intracellular pathogen, as well as surveillance of the proteasome as a mechanism to induce responses to intracellular infection.
- 45.Sarkies P, Ashe A, Le Pen J, McKie MA, Miska EA. Competition between virus-derived and endogenous small RNAs regulates gene expression in Caenorhabditis elegans. Genome Res. 2013;23:1258–1270. doi: 10.1101/gr.153296.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Crombruggen K, Jacob F, Zhang N, Bachert C. Damage-associated molecular patterns and their receptors in upper airway pathologies. Cell Mol Life Sci. 2013;70:4307–4321. doi: 10.1007/s00018-013-1356-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zugasti O, Bose N, Squiban B, Belougne J, Kurz CL, Schroeder FC, Pujol N, Ewbank JJ. Activation of a G protein-coupled receptor by its endogenous ligand triggers the innate immune response of Caenorhabditis elegans. Nat Immunol. 2014;15:833–838. doi: 10.1038/ni.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• These authors identify the first receptor/ligand pair to activate the p38 MAPK pathway in C. elegans. The endogenous ligand HPLA is a DAMP produced in reponse to damage to the host cuticle and activates a GPCR.
- 48.Pukkila-Worley R, Feinbaum R, Kirienko NV, Larkins-Ford J, Conery AL, Ausubel FM. Stimulation of host immune defenses by a small molecule protects C. elegans from bacterial infection. PLoS Genet. 2012;8:e1002733. doi: 10.1371/journal.pgen.1002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • The authors show that an anti-infective compound promotes resistance against P. aeruginosa iinfection in part through upregulation of the p38 pathway.
- 49.Moy TI, Ball AR, Anklesaria Z, Casadei G, Lewis K, Ausubel FM. Identification of novel antimicrobials using a live-animal infection model. Proc Natl Acad Sci U S A. 2006;103:10414–10419. doi: 10.1073/pnas.0604055103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pukkila-Worley R, Feinbaum RL, McEwan DL, Conery AL, Ausubel FM. The evolutionarily conserved mediator subunit MDT-15/MED15 links protective innate immune responses and xenobiotic detoxification. PLoS Pathog. 2014;10:e1004143. doi: 10.1371/journal.ppat.1004143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • In this study, the authors use an RNAi screen to identify MDT-15/MED15 as a regulator of the p38 MAPK pathway in response to the xenobiotic toxin RPW-24.
- 51.Chou TC, Chiu HC, Kuo CJ, Wu CM, Syu WJ, Chiu WT, Chen CS. Enterohaemorrhagic Escherichia coli O157:H7 Shiga-like toxin 1 is required for full pathogenicity and activation of the p38 mitogen-activated protein kinase pathway in Caenorhabditis elegans. Cell Microbiol. 2013;15:82–97. doi: 10.1111/cmi.12030. [DOI] [PubMed] [Google Scholar]
- 52.Montalvo-Katz S, Huang H, Appel MD, Berg M, Shapira M. Association with soil bacteria enhances p38-dependent infection resistance in Caenorhabditis elegans. Infect Immun. 2013;81:514–520. doi: 10.1128/IAI.00653-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Randow F, Youle RJ. Self and nonself: how autophagy targets mitochondria and bacteria. Cell Host Microbe. 2014;15:403–411. doi: 10.1016/j.chom.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jia K, Thomas C, Akbar M, Sun Q, Adams-Huet B, Gilpin C, Levine B. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc Natl Acad Sci U S A. 2009;106:14564–14569. doi: 10.1073/pnas.0813319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Curt A, Zhang J, Minnerly J, Jia K. Intestinal autophagy activity is essential for host defense against Salmonella typhimurium infection in Caenorhabditis elegans. Dev Comp Immunol. 2014;45:214–218. doi: 10.1016/j.dci.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 56.Visvikis O, Ihuegbu N, Labed SA, Luhachack LG, Alves AM, Wollenberg AC, Stuart LM, Stormo GD, Irazoqui JE. Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity. 2014;40:896–909. doi: 10.1016/j.immuni.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • In this study, the authors find that HLH-30, the C. elegans ortholog to TFEB-1, activates 80% of the transcriptional reponse to S. aureus, including antimicrobial and autophagy genes. This transcriptional response provides defense and tolerance to infection.
- 57.Zou CG, Ma YC, Dai LL, Zhang KQ. Autophagy protects C. elegans against necrosis during Pseudomonas aeruginosa infection. Proc Natl Acad Sci U S A. 2014;111:12480–12485. doi: 10.1073/pnas.1405032111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Autophagy promotes tolerance of P. aeruginosa infections through inhibition of necrosis.
- 58.Medzhitov R, Schneider DS, Soares MP. Disease tolerance as a defense strategy. Science. 2012;335:936–941. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feinbaum RL, Urbach JM, Liberati NT, Djonovic S, Adonizio A, Carvunis AR, Ausubel FM. Genome-wide identification of Pseudomonas aeruginosa virulence-related genes using a Caenorhabditis elegans infection model. PLoS Pathog. 2012;8:e1002813. doi: 10.1371/journal.ppat.1002813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • The authors perform a genome-wide screen for P. aeruginosa genes that contribute to virulence under the slow killing model. Most of the virulence factors identified are ancient genes that are part of the core P. aeruginosa genome.
- 60.Mahajan-Miklos S, Tan MW, Rahme LG, Ausubel FM. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 61.Cezairliyan B, Vinayavekhin N, Grenfell-Lee D, Yuen GJ, Saghatelian A, Ausubel FM. Identification of Pseudomonas aeruginosa phenazines that kill Caenorhabditis elegans. PLoS Pathog. 2013;9:e1003101. doi: 10.1371/journal.ppat.1003101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • This study identifys phenazine-1-carboxylic acid is the primary toxin produced by P. aeruginosa during fast killing of worms on rich media.
- 62.Kirienko NV, Kirienko DR, Larkins-Ford J, Wahlby C, Ruvkun G, Ausubel FM. Pseudomonas aeruginosa disrupts Caenorhabditis elegans iron homeostasis, causing a hypoxic response and death. Cell Host Microbe. 2013;13:406–416. doi: 10.1016/j.chom.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • A high throughput screen of small molecules revealed liquid killing of C. elegans by P. aeruginosa to be mediated by pyoverdin, an iron chelator, which causes hyoxia.
- 63.Bellier A, Chen CS, Kao CY, Cinar HN, Aroian RV. Hypoxia and the hypoxic response pathway protect against pore-forming toxins in C. elegans. PLoS Pathog. 2009;5:e1000689. doi: 10.1371/journal.ppat.1000689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luhachack LG, Visvikis O, Wollenberg AC, Lacy-Hulbert A, Stuart LM, Irazoqui JE. EGL-9 controls C. elegans host defense specificity through prolyl hydroxylation-dependent and -independent HIF-1 pathways. PLoS Pathog. 2012;8:e1002798. doi: 10.1371/journal.ppat.1002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hodgkin J, Felix MA, Clark LC, Stroud D, Gravato-Nobre MJ. Two Leucobacter strains exert complementary virulence on Caenorhabditis including death by worm-star formation. Curr Biol. 2013;23:2157–2161. doi: 10.1016/j.cub.2013.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • The authors show novel form of pathogenic attack by bacteria that infect the epidermis of C. elegans, and trade-off in defense against different strains.