Abstract
Small molecules inhibit a mutant enzyme confined to tumors, supporting therapeutic approaches that can reprogram metabolism in cancer.
Many human cancers, particularly gliomas and acute myelogenous leukemia (AML), contain mutations in the genes IDH1 or IDH2, which encode two isoforms of the metabolic enzyme isocitrate dehydrogenase (1, 2). These mutant enzymes produce the (R)-enantiomer of 2-hydroxyglutaric acid [(R)-2HG], a molecule that inhibits histone- and DNA-modifying enzymes, thereby altering gene expression and promoting the acquisition of malignant features (3–5). Reports by Losman et al. (6) as well as by Wang et al. (7) and Rohle et al. (8) on pages XXX and YYY of this issue, respectively, find that inhibitors of the mutant forms of IDH1/2 suppress the growth of (R)-2HG–producing tumor cells (6–8). The findings imply that that curtailing (R)-2HG supply normalizes gene expression and reverses malignancy.
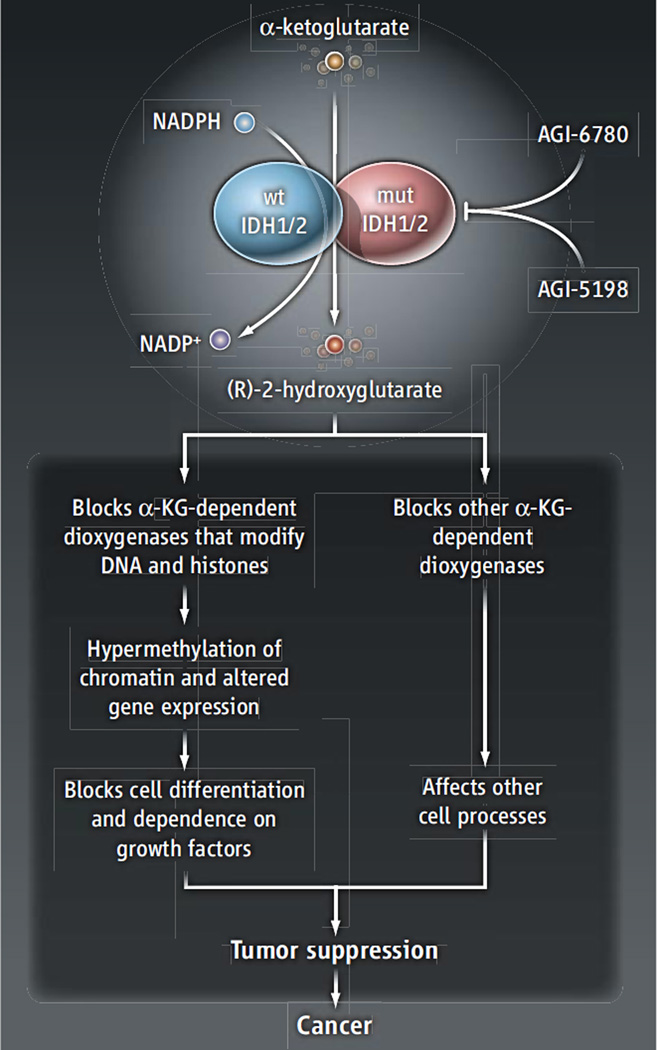
Metabolic reprogramming in cancer has long been considered a potential source of therapeutic targets. However, much of this reprogramming reflects the enhancement of normal metabolic activities already present in nonmalignant tissue, rather than the appearance of novel activities confined to the tumor. This makes it challenging to develop strategies that impair tumor metabolism without disturbing metabolism elsewhere. By contrast, IDH1/2 mutations are somatically acquired and elicit an entirely new function for the enzymes (so-called “gain-of-function” or neomorphic activity) that is absent outside of the tumor (1–3). Wild-type IDH homodimers catalyze the nicotinamide adenine dinucleotide phosphate (NADP+)–dependent conversion of isocitrate to α-ketoglutarate. In tumors with monoallelic mutations in IDH1 or IDH2, heterodimers containing one mutant and one wild-type subunit catalyze the reduction of α-ketoglutarate to (R)-2HG, a reaction that depends on NADPH (the reduced form of NADP+) (see the figure) (3, 9). (R)-2HG accumulates to millimolar concentrations within the tumor (3). The identification of this particular metabolite as the product of mutant IDH1/2 is compelling because its (L)-enantiomer [(L)-2HG] is associated with pediatric brain tumors (10). These observations implicate mutant IDH1/2, and specifically (R)-2HG, as functional drivers of malignancy. More than half of “low-grade” gliomas (slow growing but eventually lethal) and almost 10% of AML cases contain IDH1/2 mutations, and a number of other tumors (including chondrosarcomas and cholangiosarcoma) also harbor mutations in these genes.
Figure 1. Reversing the perfect storm.
Heterodimers of wild-type and mutant subunits of the metabolic enzymes IDH1/2 catalyze the production of (R)-2-hydroxyglutarate. Its accumulation impairs α-ketoglutarate–dependent dioxygenases, including those that modify DNA and histones (including the demethylation of 5-methylcytosine by TET-family hydroxylases and histone demethylases of the JmjC domain-containing family). This alters the epigenetic landscape, thereby blocking cell differentiation and promoting the acquisition of malignant features. Inhibitors (AGI-6780 and AGI-5198) that block (R)-2-hydroxyglutarate–producing IDH isoforms limit the growth of gliomaand AML-derived cancer cells.
A key insight into the role of IDH1/2 in cancer was that (R)-2HG interferes with dioxygenases that use α-ketoglutarate as a cosubstrate. These include enzymes that chemically modify histone proteins and DNA to orchestrate gene expression. Tumors with IDH1/2 mutations contain a distinctive profile of DNA and histone hypermethylation and express a suite of genes associated with undifferentiated progenitors (2, 11). Losman et al. found that exogenous expression of mutant enzymes, or treatment with (R)-2HG, is sufficient to reprogram epigenetics, impair differentiation, and promote malignant features in cultured erythroleukemia cells. Thus, the prevailing model is that IDH1/2 mutations provide an oncogenic function by producing a metabolite, (R)-2HG, which arrests differentiation and creates a cellular context conducive to malignancy.
The suitability of these mutant enzymes as therapeutic targets hinges on whether cutting off the (R)-2HG supply is sufficient to induce cell differentiation and/or slow growth. Losman et al. demonstrate that reducing (R)-2HG production in erythroleukemia cells with a chemical inhibitor eliminated growth factor–independent proliferation and restored the expression of cellular differentiation markers, revealing the reversibility of the effects of mutant IDH1 and (R)-2HG (6). Wang et al. developed a small-molecule allosteric inhibitor of IDH2/R140Q (IDH2 containing the common mutation Arg140 →Gln) and demonstrated its selectivity against mutant homodimers and mutant-containing heterodimers of the enzyme. When used to treat leukemia cells, the inhibitor reduced the amount of (R)-2HG produced and activated the expression of genes associated with erythroid differentiation. Furthermore, in primary human AML cells, this inhibitor suppressed cell growth, reduced numbers of immature blast cells, and increased differentiation along macrophage and granulocytic lineages. None of these effects were observed in leukemic cells lacking the IDH2/R140Q mutation.
Rohle et al. observed that an inhibitor against IDH1/R132H (IDH1 containing the mutation Arg132 → His), the most common IDH mutation in gliomas, suppressed colony formation and xenograft growth of cells from a human anaplastic oligodendroglioma, and induced the expression of genes associated with differentiation into mature glial cells. At high oral doses in mice, inhibition of IDH1/R132H reduced some histone methylation marks [whose removal is blocked by (R)-2HG]. Surprisingly, a lower dose of the drug impaired tumor growth but had no effect on differentiation or methylation signatures, which suggests that reversal of the epigenetic program induced by (R)-2HG is unnecessary to suppress glioma growth in this model. Thus, inhibiting enzymes that directly modify histones and DNA may not be equivalent to inhibiting mutant IDH1.
The appearance of IDH1/2 mutations early in the progression of glioma and AML raised concern that (R)-2HG functions in tumor initiation but is dispensable once a more durable transformed state is established by the acquisition of additional mutations. It is therefore noteworthy that inhibitors to IDH1/R132H or IDH2/R140Q were effective against human-derived tumor mutations. The data argue that (R)-2HG functions in tumor maintenance and support IDH1/2 mutants as practical therapeutic targets.
The inhibitors used by Wang et al. and Rohle et al. produced cytostatic rather than cytotoxic effects, in line with the idea that they stimulate cell differentiation rather than cell death. If the inhibitors induce a permanent state of differentiation, perhaps no cytotoxicity is needed to achieve therapeutic efficacy. However, the survival of viable cells still containing a potentially transforming constellation of mutations makes it important to determine whether the therapeutic effects will persist over long time frames.
These studies underscore the complexity of IDH1/2 function in neoplasia. Glioma and AML progenitors are susceptible to IDH1/2 mutations, whereas cells giving rise to many other types of cancer seem to be relatively impervious. Perhaps the basis for this selectivity relates to tissue-specific roles for the panoply of α-ketoglutarate–dependent dioxygenases in tumor suppression. Even in glioma, non-epigenetic effects of IDH1/2 mutations appear to contribute substantially to tumor growth (8). It will undoubtedly be useful to understand the full scope of these effects and to maximize their suppression in cancer therapy.
Acknowledgments
R.J.D. is supported by the National Cancer Institute (grant R01 CA157996), the Robert A. Welch Foundation (grant I-1733), and a Damon Runyon Cancer Research Foundation Clinical Investigator Award.
References
- 1.Yan H, et al. N. Engl. J. Med. 2009;360:765. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Figueroa ME, et al. Cancer Cell. 2010;18:553. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dang L, et al. Nature. 2009;462:739. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu W, et al. Cancer Cell. 2011;19:17. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu C, et al. Nature. 2012;483:474. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Losman JA, et al. Science. 2013;339:1621. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang F, Wang L, Filippenko AV, Zhang T, Zhao X. Science. 2013;340 doi: 10.1126/science.1231502. XXX. [DOI] [PubMed] [Google Scholar]
- 8.Rohle D, et al. Science. 2013;340 YYY 10.1126/science.1236062.10.1126/science.1236062. [Google Scholar]
- 9.Pietrak B, et al. Biochemistry. 2011;50:4804. doi: 10.1021/bi200499m. [DOI] [PubMed] [Google Scholar]
- 10.Kranendijk M, Struys EA, Salomons GS, Van der Knaap MS, Jakobs C. J. Inherit. Metab. Dis. 2012;35:571. doi: 10.1007/s10545-012-9462-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turcan S, et al. Nature. 2012;483:479. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]