ABSTRACT
Avian species are reservoirs of influenza A viruses and could harbor viruses with significant pandemic potential. We examined the antibody and cellular immune responses to influenza A viruses in field or laboratory workers with a spectrum of occupational exposure to avian species for evidence of zoonotic infections. We measured the seroprevalence and T cell responses among 95 individuals with various types and degrees of prior field or laboratory occupational exposure to wild North American avian species using whole blood samples collected in 2010. Plasma samples were tested using endpoint enzyme-linked immunosorbent assay (ELISA) and hemagglutination (HA) inhibition (HAI) assays to subtypes H3, H4, H5, H6, H7, H8, and H12 proteins. Detectable antibodies were found against influenza HA antigens in 77% of individuals, while 65% of individuals tested had measurable T cell responses (gamma interferon [IFN-γ] enzyme-linked immunosorbent spot assay [ELISPOT]) to multiple HA antigens of avian origin. To begin defining the observed antibody specificities, Spearman rank correlation analysis showed that ELISA responses, which measure both head- and stalk-binding antibodies, do not predict HAI reactivities, which measure primarily head-binding antibodies. This result suggests that ELISA titers can report cross-reactivity based on the levels of non-head-binding responses. However, the strongest positive correlate of HA-specific ELISA antibody titers was receipt of seasonal influenza virus vaccination. Occupational exposure was largely uncorrelated with serological measures, with the exception of individuals exposed to poultry, who had higher levels of H7-specific antibodies than non-poultry-exposed individuals. While the cohort had antibody and T cell reactivity to a broad range of influenza viruses, only occupational exposure to poultry was associated with a significant difference in antibody levels to a specific subtype (H7). There was no evidence that T cell assays provided greater specificity for the detection of zoonotic infection. However, influenza vaccination appears to promote cross-reactive antibodies and may provide enhanced protection to novel influenza viruses.
IMPORTANCE
Annual vaccinations are necessary to ameliorate influenza disease due to drifted viral variants that emerge in the population. Major shifts in the antigenicity of influenza viruses can result in immunologically distinct viruses that can cause more severe disease in humans. Historically, genetic reassortment between avian, swine, or human influenza viruses has caused influenza pandemics in humans several times in the last century. Therefore, it is important to design vaccines to elicit broad protective responses to influenza infections. Because avian influenza viruses have an important role in emerging infections, we tested whether occupational exposure to birds can elicit immune responses to avian influenza viruses in humans. Instead of a specific occupational exposure, the strongest association of enhanced cross-reactive antibody responses was receipt of seasonal influenza vaccination. Therefore, individuals with preexisting immune responses to seasonal human influenza viruses have substantial cross-reactive antibody and T cell responses that may lead to enhanced protection to novel influenza viruses.
INTRODUCTION
The primary reservoir of influenza A viruses is within waterfowl and shorebirds, and those viruses entering the human population (either directly or through an intermediate host) do not generally have significant pandemic potential, but they can cause severe disease, as is seen with H5N1 infections. While no emergent influenza viruses were as devastating as the 1918 H1N1 pandemic, antigenically different strains have entered the human population on at least four other occasions in the last century: 1957 (H2N2), 1968 (H3N2), 1977 (H1N1), and 2009 (H1N1), causing considerable illness in humans (1–3). The surface hemagglutinin (HA) and neuraminidase (NA) proteins are two of the most immunogenic influenza proteins, and development of neutralizing antibodies directed against the HA or NA is associated with immunological protection, a hallmark of the seasonal influenza vaccination strategy. There are currently eighteen known HA proteins, the most recent being H17 and H18, identified in bats (4, 5). The H1 to H16 proteins can be divided into two major antigenic groups with five distinct clades: group 1 consists of clades H1a (H1, H2, H5, and H6), H1b (H11, H13, and H16), and H9 (H8, H9, and H12), while group 2 consists of clade H3 (H3, H4, and H14) and clade H7 (H7, H10, and H15) (6).
Poultry-specific avian influenza virus (AIV) outbreaks that have caused significant morbidity and mortality in infected humans have involved A/H5N1, A/H7N2, A/H7N3, A/H7N7, A/H7N9, and A/H9N2 viral subtypes (7–10). With the exception of A/H5N1 and A/H7N9, these viruses generally do not have high pathogenicity and are not effectively transmitted from person to person. The immune response in individuals exposed to AIV is relatively understudied, but empirical serological evidence suggests that humans with frequent animal contact, including veterinarians and field workers, have elevated antibody titers against AIV subtypes (11–13). At least in the case of A/H5N1, humans have been documented to be infected directly from birds, with no intermediate host necessary, following direct contact with sick poultry, including butchering and preparing meats for food consumption. In addition to serological evidence of AIV exposure, cross-reactive immune memory responses to A/H5N1 exist, where CD4+ and CD8+ T cells from humans can recognize cross-reactive internal influenza proteins, particularly the matrix and nonstructural proteins (14).
Because observational studies suggest that exposure to birds can elicit detectable antibodies to avian influenza viruses, we hypothesized that individuals with significant occupational exposure to wild bird populations, either in the field or as a laboratory worker, will have increased levels of humoral and cellular immune responses to diverse avian influenza viruses. In this study, we examined the presence of antibodies and T cell responses to HA proteins of avian origin in a human cohort with diverse contact with North American migratory wild bird populations enrolled during an American Ornithologist Union annual meeting in 2010 (described previously in reference 12). We did not observe any enhancement of diverse antibody or T cell responses in individuals reporting greater exposure to birds compared to those of their counterparts with little avian contact, except for H7 responses in those individuals reporting occupational or recreational exposure to poultry. Instead, the strongest correlate of enhanced cross-reactive antibody responses was the receipt of seasonal influenza vaccination. These results provide insights into the diversity of cross-reactive antibody responses in this occupationally exposed population and suggest that measuring T cell responses to the HA does not provide a significantly different impression of antigen exposure than antibody assays.
RESULTS
Individuals are seropositive to multiple HA antigens of avian origin.
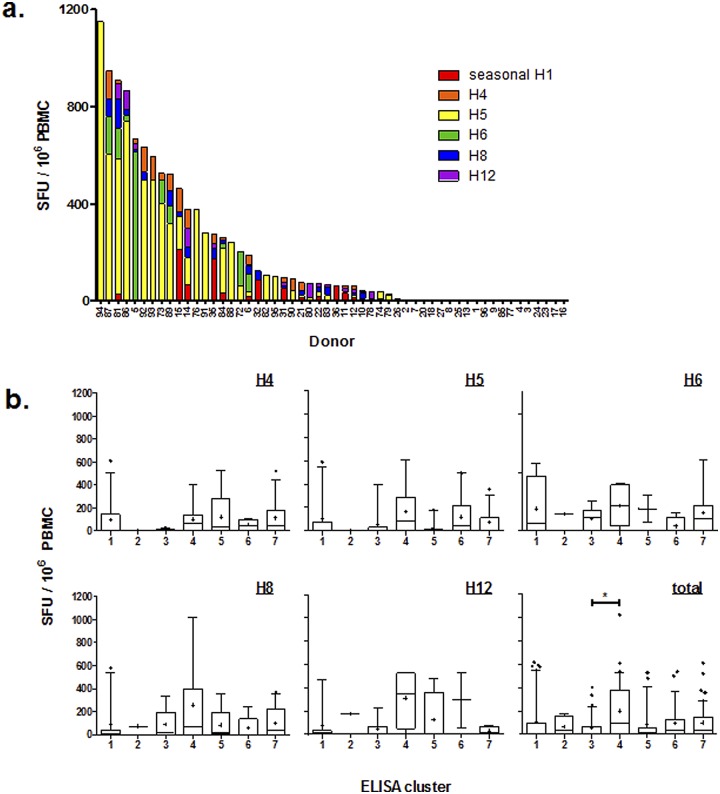
Influenza A viruses are classified into two major groups based on their antigenic and structural similarities (15–18), and since antibodies to HA directly correlate with disease protection (19, 20), endpoint HA-specific antibody titers were first determined for each cohort individual. Detectable antibodies were found against avian influenza HA antigens in 77% of the donors. Not surprisingly, most individuals tested had a strong antibody response to the seasonal H3N2 human virus-derived H3, but many also had strong measurable antibody responses to group 1 HA (avian H5, H6, H8, H12) and group 2 HA (avian H4, human H7) (Fig. 1a), with 66% of donors having some level of detectable seropositivity to four or more HA proteins (data not shown).
FIG 1 .

Individuals exhibit cross-reactive antibody and cellular responses to multiple HA antigens of avian origin. (a) Endpoint ELISA titers were determined for group 1 avian H5, H6, H8, and H12, group 2 avian H4, and group 2 human H7 hemagglutinin antigens. (b) Heat map representing color-coded antibody titers. K-means clustering analysis revealed 7 distinct clusters of individuals based on the endpoint ELISA titers. Gray, samples not tested.
Individuals could be clustered based on their measured endpoint antibody titers to HA antigens (Fig. 1b). Seven distinct clusters of donors were identified, where the clusters could essentially be visualized from least to most seroresponsive. Cluster 1 included primarily individuals with the highest antibody responses to H3, while cluster 7 contained individuals with the highest magnitude of antibody responses to all 6 antigens tested, including the H7 from the newly emerging Asian A/Anhui. There were no apparent defining characteristics, including age or species exposure patterns, among individuals within any of the clusters (characteristics are listed in Table 1); however, the magnitude of response to antigens within a specific HA group (1 or 2) correlated with responses within that group, i.e., individuals were more likely to have strong responses to both H5 and H6 (same clade) but not H8, suggesting that these responses may be more cross-reactive rather than from direct exposure.
TABLE 1 .
Demographic characteristics of study participantsa
| Characteristic | No. | % |
|---|---|---|
| Gender | ||
| Male | 47 | 49 |
| Female | 47 | 49 |
| Unknown | 1 | 1 |
| Age (yrs) | ||
| 20–29 | 24 | 25 |
| 30–39 | 35 | 37 |
| 40–49 | 15 | 16 |
| 50–59 | 10 | 11 |
| 60–69 | 9 | 9 |
| 70–79 | 1 | 1 |
| Unknown | 1 | 1 |
| Race/ethnicity | ||
| White, non-Latino | 87 | 92 |
| Latino | 1 | 1 |
| Asian/Pacific Islander | 2 | 2 |
| Other/multiracial | 5 | 5 |
| Experience (yrs) | ||
| <1 year | 6 | 6 |
| 1–2 years | 8 | 8 |
| 2–5 years | 14 | 15 |
| >5 years | 67 | 71 |
| Work experience | ||
| Banding and measuring | 89 | 94 |
| Bleeding | 60 | 63 |
| Cloacal swabbing | 22 | 23 |
| Sample collection and preparation | 12 | 13 |
| General care | 1 | 1 |
| Surgery | 0 | |
| Otherb | 7 | 7 |
| Avian species handledc | ||
| Passerines | 81 | 85 |
| Raptors | 21 | 22 |
| Waterfowl | 16 | 17 |
| Shorebirds | 16 | 17 |
| Poultryd | 7 | 7.4 |
| Othere | 12 | 13 |
| Handling season (North America) | ||
| Spring | 78 | 82 |
| Summer | 85 | 89 |
| Fall | 55 | 58 |
| Winter | 50 | 53 |
The total number of participants was 95; the values in the table omit missing data (e.g., no answer was given).
Specified work experience included experimental infections, catch and release, and nest monitoring.
Participants could indicate exposure to more than one avian species.
Poultry handling reported included both occupational and recreational exposure.
Specified avian species handled included other sea birds, such as those belonging to the order Pelecaniformes.
Together, these data suggest that, similar to previous studies (11–13), individuals with exposure to wild birds generate functional antibody responses to diverse avian strains, but these responses do not appear strongly correlated with occupational exposure.
Humoral and cellular responses to avian HA antigens do not correlate in the same individual.
Because T cell responses likely have an important role in resolving infection (21–23), we sought to determine if the individuals who had significant antibody responses could also mount meaningful cellular responses to avian HAs. Therefore, to identify influenza HA-specific cellular responses, we determined the frequency of gamma interferon (IFN-γ)-producing cells within bulk peripheral blood mononuclear cells (PBMCs) incubated with antigen-presenting cells pulsed with purified avian HA proteins (Fig. 2). The scope and magnitude of the cellular responses varied considerably between donors, and most individuals exhibited cellular responses to avian influenza virus HA as measured by IFN-γ production (Fig. 2a). The H5 protein appeared to elicit particularly robust responses. No association was revealed between the measured antibody response and IFN-γ response to a particular HA in the same individual (data not shown). We next asked whether T cells from individuals within the defined seroresponsiveness clusters from Fig. 1b reacted similarly to each other (Fig. 2b). However, no differences were observed in T cell IFN-γ responses among the 7 clusters, either in total or when comparing responses to specific HAs. While there were no overt characteristics present that defined each cluster, donors from cluster 3 had higher antibody responses to group 2 HAs (H3, H4, H7) and minimal antibody titers to group 1 HAs (H5, H6, H8, H12). On the other hand, cluster 4 donors had antibody responses to H5 in addition to the group 2 HAs. Together, these data indicate that the individual who mounts a robust antibody response to a particular HA does not necessarily mount an equally robust cellular response to the same HA.
FIG 2 .
(a) The frequency distributions of participants with IFN-γ-producing cells as measured by ELISPOT assay to individual HA proteins are shown as spot-forming units (SFU) per million PBMCs. (b) Individual PBMC IFN-γ responses for each HA protein or total response to any HA protein are plotted by ELISA cluster. Frequencies have been corrected for background IFN-γ production in unstimulated control cultures.
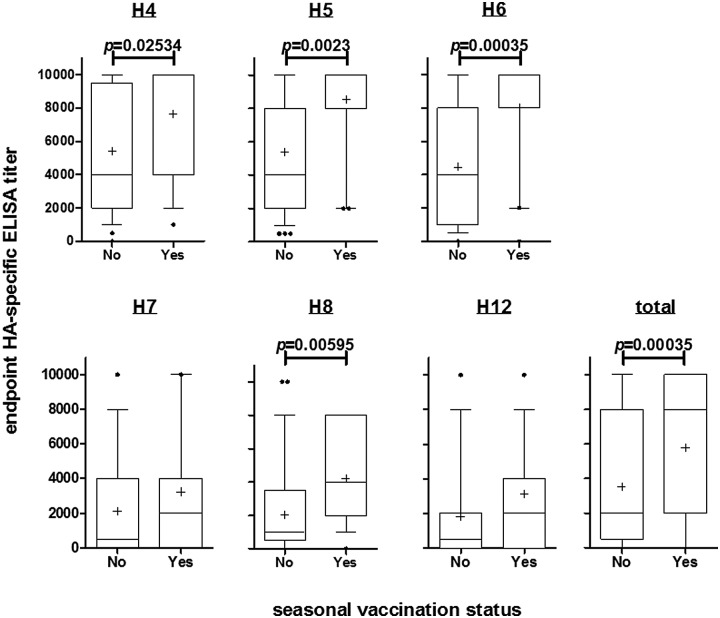
HA-specific antibody titers are enriched in individuals who report receiving seasonal influenza virus vaccination.
Cluster analysis revealed no clear occupational or demographic discriminating factors between individuals with high or low HA antibody titers, but those who had reported receiving seasonal influenza vaccination had significantly higher antibody titers to H4 (false discovery rate [FDR] P = 0.025), H5 (FDR P = 0.002), H6 (FDR P = 0.00035), and H8 (FDR P = 0.00595) proteins than individuals who were not vaccinated (Fig. 3). No differences were found in antibody responses to H7 or H12 in groups that received a seasonal influenza vaccination. There were no differences in the numbers of IFN-γ-producing PBMCs in individuals who reported they had received seasonal vaccination (see Fig. S1 in the supplemental material), implying that seasonal vaccination was not associated with influenza HA-specific cellular responses in this cohort.
FIG 3 .
Reported seasonal influenza virus vaccination is associated with enriched HA-specific antibody titers. Endpoint ELISA titers were determined for avian H4, H5, H6, H8, and H12 and human H7 hemagglutinin antigens. Seasonal vaccination status indicates whether participants had reported receipt of any seasonal (pre-2009) influenza vaccination or the H1N1pdm09 influenza vaccine. Whiskers of box plots represent 10th and 90th percentiles, and the horizontal line and plus sign in each box give the median and mean titers of individuals, respectively. Variability in titer is shown by plotting the first and third quartiles of the titers as the outer limits of the box. Points beyond the whiskers denote outliers. Groups were compared by two-tailed, unpaired t test, and false discovery rate-adjusted P values are indicated.
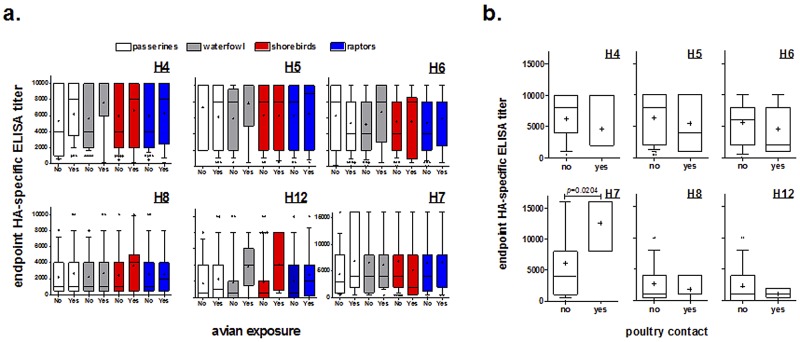
Enriched antibody titers are observed in participants with avian exposure.
Because this cohort reported frequent occupational exposure to wild avian species, we investigated whether subgroups of individuals could be identified by the type of avian species they identified in the questionnaire (12). Participants were asked if they had exposure to passerines (e.g., perching birds, songbirds), waterfowl (e.g., ducks, geese, swans), shorebirds (e.g., waders, gulls), and raptors. There were no differences between antibody responses to avian H4, H5, H6, H8, and H12 and human H7 proteins in groups with and without exposure to particular avian species (Fig. 4a). In contrast, participants with poultry contact, defined as having contact either occupationally or recreationally, had significantly higher antibody responses to A/Anhui H7 protein (FDR P = 0.0204), but to none of the other proteins tested (Fig. 4b), than participants without contact.
FIG 4 .
(a) Exposure to wild avian species is not associated with enriched HA-specific antibody titers. Endpoint ELISA titers were determined for avian H4, H5, H6, H8, and H12 and human H7 hemagglutinin antigens. (b) Contact with poultry is associated with increased levels of anti-H7 antibodies. Endpoint ELISA titers were determined for avian H4, H5, H6, H8, and H12 and human H7 hemagglutinin antigens. Whiskers of box plots represent 10th and 90th percentiles, and the horizontal line and plus sign in each box give the median and mean titers of individuals, respectively. Variability in titer is shown by plotting the first and third quartiles of the titers as the outer limits of the box. Points beyond the whiskers denote outliers. Box color represents the participant-reported avian species exposure (white, passerines; gray, waterfowl; red, shorebirds; blue, raptors). Groups were compared by two-tailed, unpaired t test, and false discovery rate-adjusted P values were determined.
Cross-reactive ELISA responses do not predict HAI reactivity.
The endpoint enzyme-linked immunosorbent assay (ELISA) provides a good estimate of the total polyclonal antibody response to the specific HA in question, irrespective of the neutralizing capacity, but does not distinguish between regions of specificity (i.e., the head domain which contains the major antigenic sites versus the more conserved stalk region). To determine whether the associations we observed were driven by conventional head-binding antibodies, we took the remaining plasma (n = 59 participants) and performed hemagglutination inhibition (HAI) assays against representative avian influenza viruses circulating in North America prior to 2010, including a mallard/H7 (Table 2). Within the subcohort we tested, 41% of individuals had HAI titers to H4N6 (geometric mean titer [GMT] [95% confidence interval (CI)] of 82.34 [59.98 to 113.1]). Ten percent of individuals had HAI titers to H5N5 (GMT [95% CI] of 89.8 [23.63 to 341.2]), and 29% had detectable HAI titers to H6N1 (GMT [95% CI] of 106.4 [65.27 to 173.5]). Surprisingly, 63% of individuals had HAI antibody titers detectable against a North American H7N3 (GMT [95% CI] of 47.35 [37.47 to 59.82]), while only two participants had HAI titers to the newly identified H7N9 (GMT of 160.0). Thus, the overall pattern we observed by ELISA of a high degree of cross-reactivity among these individuals was maintained.
TABLE 2 .
Hemagglutination inhibition results against alternative circulating North American avian influenza virusesa
| ID | Titer |
|||||||
|---|---|---|---|---|---|---|---|---|
| H3N8 | H4N6 | H5N5 | H6N1 | H7N3 | H7N9b | H8N4 | H12N5 | |
| 1 | ||||||||
| 3 | 40 | |||||||
| 11 | 40 | |||||||
| 16 | 40 | 80 | 40 | 160 | 80 | ND | ND | |
| 17 | ||||||||
| 18 | 40 | |||||||
| 24 | 40 | 80 | 40 | 160 | 80 | ND | ND | |
| 26 | 40 | 40 | 40 | 160 | 80 | ND | ND | |
| 27 | 80 | 40 | ND | ND | ||||
| 28 | 40 | 40 | 80 | 40 | 40 | |||
| 29 | 40 | |||||||
| 35 | ||||||||
| 36 | 320 | 320 | 320 | 640 | 80 | ND | ND | |
| 37 | 40 | 40 | ND | ND | ||||
| 38 | 40 | 40 | 40 | |||||
| 41 | 2,560 | 1,280 | 640 | 1,280 | 2,560 | 320 | ND | ND |
| 44 | 40 | |||||||
| 39 | 40 | |||||||
| 40 | 40 | ND | ND | |||||
| 45 | 40 | 40 | ||||||
| 47 | 40 | |||||||
| 48 | 40 | |||||||
| 49 | 80 | 40 | 40 | ND | ND | |||
| 51 | 80 | 40 | ||||||
| 53 | 40 | |||||||
| 54 | 80 | 80 | 40 | ND | ND | |||
| 61 | 80 | 80 | 40 | ND | ND | |||
| 64 | 80 | 80 | 40 | ND | ND | |||
| 65 | 80 | 80 | 40 | ND | ND | |||
| 68 | 40 | |||||||
| 70 | 80 | 80 | 40 | 40 | ||||
| 71 | 40 | 80 | 40 | 40 | ND | ND | ||
| 72 | 40 | 80 | 40 | 160 | 40 | ND | ND | |
| 73 | 160 | 40 | 40 | ND | ND | |||
| 57 | 40 | |||||||
| 58 | 40 | 40 | 40 | |||||
| 59 | 40 | |||||||
| 60 | ||||||||
| 75 | 40 | 40 | ||||||
| 76 | ||||||||
| 77 | ||||||||
| 78 | ||||||||
| 79 | ||||||||
| 80 | ||||||||
| 81 | 40 | |||||||
| 83 | 40 | |||||||
| 84 | ||||||||
| 85 | 40 | 40 | 40 | |||||
| 86 | 80 | 40 | 40 | 40 | ||||
| 87 | 80 | |||||||
| 88 | ||||||||
| 89 | 80 | |||||||
| 90 | 80 | |||||||
| 91 | ||||||||
| 92 | ||||||||
| 93 | ||||||||
| 94 | ||||||||
| 95 | 40 | 40 | ||||||
| 96 | ||||||||
| Reference serac | 2,560 | >5,120 | 2,560 | 1,280 | >5,120 | 640 | 40 | 80 |
| GMTd (95% CI) | 74.64 (27.35–203.7) | 82.34 (59.98–113.1) | 89.80 (23.63–341.2) | 106.4 (65.27–173.5) | 47.35 (37.47–59.82) | 160.0 | 40 | 40 |
Positive HAI titers are considered ≥1:40, and no value indicates an HAI titer of <40. ND, not determined.
Recently identified A/Anhui/1/2013 (H7N9).
See Materials and Methods for the reference sera used in these assays.
GMT, geometric mean titer.
It was not clear, however, that the individuals that demonstrated cross-reactivity by HAI were the same individuals who demonstrated cross-reactivity by endpoint ELISA. HAI assays measure only head-binding (and not stalk-binding) antibodies, while the ELISA assay captures antibodies binding to both the head and the stalk region of the HA. If individuals mounting strong ELISA responses also mounted strong HAI responses, the ELISA responses may be driven by the strong head-binding antibodies. Conversely, if the ELISA and HAI responses did not correlate, it would suggest that strong responses to the ELISA were driven by non-head-binding (likely stalk-binding) antibodies.
To test this, we performed a Spearman rank correlation between the responses measured to each of the tested antigens, either by ELISA or HAI (Fig. 5). The primary statistically significant associations were observed only among the same type of assay, i.e., ELISA responses correlated largely with other ELISA responses, while HAI responses correlated with other HAI responses. The exception was the ELISA response to the recently emerged H7N9 virus, which did not correlate with any other ELISA response but did correlate with three HAI responses (including the HAI response to itself, H4N6, and H6N1). Among the ELISA response, H3, H8, and H12 responses correlated significantly with all other antigens measured in ELISA. In the HAI, all responses to all antigens were correlated significantly, with the exception of H7N3 and H7N9 with each other. These data indicate that an individual’s ELISA response to a specific antigen is somewhat predictive of their ELISA response to other antigens but is not predictive of their HAI response and vice versa.
FIG 5 .

ELISA responses do not predict HAI reactivity. A Spearman rank correlation was performed on each individual’s ELISA and HAI responses. Above the diagonal, significant associations are marked with a circle, while nonsignificant associations are blank. Similarly, below the diagonal, the correlation coefficient is listed in the square where significant associations were found. ELISA antigens are in green, while HAI antigens are listed in orange following the nomenclature of Table 4. A significance level of less than 0.1 was applied after FDR adjustment.
To support this analysis, we looked at the 10 individuals with the highest average HAI and ELISA titers (Table 3), including only subjects with complete ELISA and HAI results (36 individuals). Only 3 individuals appeared on both lists, reinforcing the impression that strong responses in one type of assay are not predictive of responses in the other.
TABLE 3 .
Top 10 average ELISA and HAI responses
| Assay | IDa | Avg titer |
|---|---|---|
| ELISA | 47 | 10,857 |
| 49 | 9,143 | |
| 18 | 8,857 | |
| 41 | 8,571 | |
| 77 | 8,571 | |
| 1 | 8,286 | |
| 40 | 7,714 | |
| 61 | 7,714 | |
| 71 | 7,429 | |
| 72 | 7,429 | |
| HAI | 41 | 60.0 |
| 36 | 40.0 | |
| 16 | 33.3 | |
| 24 | 33.3 | |
| 26 | 33.3 | |
| 72 | 33.3 | |
| 73 | 33.3 | |
| 28 | 33.3 | |
| 54 | 33.3 | |
| 61 | 26.7 |
IDs appearing in both lists are underlined.
DISCUSSION
In this study, we had access to a subset of individuals described by Shafir et al. (12), where individuals who handled migratory birds were sero-surveilled for exposure to avian influenza viruses. Shafir et al. reported that transmission of avian influenza viruses from migratory birds to humans was relatively rare, measured by microneutralization assay. We found, in a portion of this same cohort, that individuals with repeated and diverse (field or laboratory) exposure to wild birds indeed have measurable levels of humoral and cellular immunity cross-reactive toward the influenza HA protein alone. However, there was no clear association between humoral and cellular immunity and the type of occupational exposure, with the exception of poultry-exposed individuals having higher levels of H7 antibodies than those of individuals not exposed. Furthermore, those individuals receiving seasonal influenza vaccination had increased levels of cross-reactive responses to multiple avian influenza subtypes compared to those of unvaccinated individuals. Finally, a subset of our cohort had substantial levels of head-binding HA antibodies, indicating that these cross-reactive responses may elicit functional immune protection.
While the number of virologically confirmed cases of avian influenza in humans reported by the WHO continues to rise, the frequency of subclinical avian influenza virus infections of humans remains understudied, and it is unclear the role cross-reactive responses have in immunity elicited against diverse influenza antigens, particularly the distinct surface proteins. Evidence suggests that humans in frequent occupational or recreational contact with poultry or wild birds have some level of seropositivity to avian influenza viruses (11, 24–29).
The HA stalk domain contains multiple conserved antigenic sites that may result in cross-protection against novel viruses (30–32), and development of a universal vaccine targeting conserved HA stem regions, particularly those that span group 1 and group 2 HAs, is an appealing approach (30–33). Broadly neutralizing antibodies are produced in many individuals receiving seasonal influenza vaccinations that cross-react with H1, H2, H5, H6, and H9 antigens (34–36), and evidence suggests that HA stalk antibodies increase in individuals naturally exposed to influenza viruses over a 20-year time period (37). Regarding newly emerging influenza viruses, recombinant Newcastle disease virus expressing HA of a North American linage H7 virus elicited cross-reactive immune responses in mice, resulting in protection from challenge with a broad range of heterologous H7 viruses, including the novel H7, suggesting that conserved antigenic sites do exist, even in phylogenetically divergent strains (38). Glycosylation sites on the HA head domain can lead to immune escape, but these sites can also result in polyclonal cross-reactive antibodies against drifted antigenic variants, at least in the case of A/H1N1(2009) (39). Many of the HA responses in our cohort were found to be nonneutralizing HA-specific antibody titers, measured by ELISA or HAI, and despite their poor hemagglutination inhibition properties, previous studies have found that even these antibodies can provide protection in murine models of influenza infection since HA stalk-binding antibodies, as confirmed by competition assays, are capable of neutralizing influenza in a complement-mediated enhancement manner (35, 40) but do not function in the HAI assay (41–43). Our data showing that having high reactivity in ELISA assays did not predict reactivity in HAI assays is consistent with this result and suggests that the high ELISA-reactive individuals may have cross-protective antistalk antibodies. In contrast, those individuals with cross-reactive HAI responses may be targeting conserved head epitopes. Intriguingly, a recent study suggested that inhibition of germinal center formation and immunoglobulin class switching by sirolimus treatment can enhance heterosubtypic immunity in mice (44).
Currently circulating North American AIV may not pose a significant health threat, but our data suggest that individuals with preexisting immune responses to seasonal human influenza viruses have substantial and measurable cross-reactive antibody and T cell responses that may lead to enhanced protection to novel influenza viruses. However, the antibody and T cell responses were not well correlated with each other. Together, our data indicate that the individual who mounts a robust antibody response does not necessarily mount an equally robust cellular response, and therefore vaccination strategies that elicit both cellular and humoral responses would be advantageous. New vaccination modalities, including adjuvants and attenuated viral vectors, may support this aim of eliciting broad population immunity by utilizing multiple immune effector mechanisms.
MATERIALS AND METHODS
Participants.
Samples were obtained from participants attending the American Ornithologist Union annual meeting in 2010. A detailed description of the cohort was previously published (12). Inclusion criteria required that participants handled wild birds and were at least 18 years of age. The Institutional Review Boards of the University of California, Los Angeles, and St. Jude Children’s Research Hospital approved the study. Written informed consent was obtained from participants at the time of enrollment, and only those participants who gave consent for participation in the substudy were included. Demographic characteristics are listed in Table 1.
PBMC isolation.
PBMCs were isolated from 5 to 8 ml of peripheral blood collected by venipuncture. Plasma was collected by centrifuging whole blood for 10 min at 200 × g, and PBMCs were collected by density gradient centrifugation using lymphocyte separation medium (MP Biomedicals, Santa Ana, CA) and stored in liquid nitrogen for future use.
IFN-γ ELISPOT assay.
MultiScreen-IP 96-well plates (Millipore, Bedford, MA) were precoated with anti-human IFN-γ monoclonal antibody (clone 1-D1K; MABTECH, Mariemont, OH) overnight at 4°C. PBMCs were incubated with 1 µg of purified HA protein from virus strains per 1 × 105 cells (Table 4) (eEnzyme, Gaithersburg, MD) for 4 h at 37°C. Stimulated cells were washed with RPMI 1640 with 10% fetal bovine serum (FBS) and plated at 1 × 105 cells per well in duplicate or triplicate per condition depending on cell availability. Responder PBMCs were added at 1 × 105 cells per well, for a total of 2 × 105 cells per well. Following 48 h of incubation, IFN-γ spots were developed by incubating with biotinylated anti-human IFN-γ monoclonal antibody (MAb) (clone 7-B6-1; MABTECH). IFN-γ spot counts were enumerated using a Zeiss Axioplan 2 microscope and KS enzyme-linked immunosorbent spot assay (ELISPOT) software (Munich-Hallbergmoos, Germany). Positive control wells contained whole PBMCs stimulated with 5 µg/ml concanavalin A (Sigma-Aldrich, St. Louis, MO). Negative control wells contained whole PBMC incubated with medium alone. The number of specific spot-forming cells (SFC) was calculated by subtracting the number of spots in the negative-control wells from the number of spots in each experimental well. The magnitude of the HA-specific response was reported as the number of SFC per million cells.
TABLE 4 .
Influenza virus strains tested by assay
| Strain | Antigena | Source | Assayb |
|---|---|---|---|
| A/Brisbane/59/2007 (H1N1) | H1 | Whole virus; SJCRH | ELISPOT |
| A/Brisbane/10/2007 (H3N2) | H3 | Purified HA; eEnzyme IA-0042W-005P | ELISA |
| A/mallard/Alberta/160/2010 (H3N8) | H3N8 | Whole virus; SJCRH | HAI |
| A/mallard/Ohio/657/2002 (H4N6) | H4 | Purified HA; eEnzyme IA-006-005P | ELISPOT, ELISA |
| A/mallard/Alberta/297/2010 (H4N6) | H4N6 | Whole virus; SJCRH | HAI |
| A/American green-winged teal/California/HKWF609/2007 (H5N2) | H5 | Purified HA; eEnzyme IA-018W-005P | ELISPOT, ELISA |
| A/mallard/Alberta/383/2009 (H5N5) | H5N5 | Whole virus; SJCRH | HAI |
| A/northern shoveler/California/HKWF115/2007 (H6N1) | H6 | Purified HA; eEnzyme IA-021W-005P | ELISPOT, ELISA |
| A/northern pintail/Alberta/3/2009 (H6N1) | H6N1 | Whole virus; SJCRH | HAI |
| A/mallard/Alberta/167/2010 (H7N3) | H7N3 | Whole virus; SJCRH | HAI |
| A/Anhui/1/2013 (H7N9) | H7 | Purified HA; Mt. Sinai Hospital | ELISA |
| H7N9 | Whole virus; SJCRH | HAI | |
| A/pintail duck/Alberta/114/1979 (H8N4) | H8 | Purified HA; eEnzyme IA-010-005P | ELISPOT, ELISA |
| A/mallard/Alberta/242/2003 (H8N4) | H8N4 | Whole virus; SJCRH | HAI |
| A/green-winged teal/Alberta/199/1991 (H12N5) | H12 | Purified HA; eEnzyme IA-013-005P | ELISPOT, ELISA |
| A/ruddy turnstone/Delaware/119/2007 (H12N5) | H12N5 | Whole virus; SJCRH | HAI |
Depending on the assay used, the antigen is either whole virus or purified HA protein.
The assays included hemagglutination inhibition (HAI) assay, ELISPOT, or ELISA.
Enzyme-linked immunosorbent assays.
Enzyme immunoassay (EIA)/radioimmunoassay (RIA) plates (Costar, Corning, NY) were coated with 250 ng of purified HA protein (Table 4) in phosphate-buffered saline (PBS) overnight at 4°C. The wells were then blocked with PBS containing 5% nonfat skim milk for 2 h at room temperature. Plasma samples were diluted serially and 100 µl was added per well. After incubating for 3 h at room temperature, wells were washed with PBS containing 0.05% Tween 20 and incubated with horseradish peroxidase (HRP)-conjugated goat anti-human IgG antibody (Invitrogen, Frederick, MD) for 1 h at room temperature. The wells were washed again and developed with TMB substrate (ThermoScientific, Rockford, IL). After 15 min, the enzyme reaction was stopped by adding 0.18 M H2S04, and the absorbance was read at 450 nm using a microplate reader. Responses were regarded as positive if they had at least four times the mean background reading in the negative-control wells.
Hemagglutination inhibition assay.
Influenza HA-specific HAI titers were measured in plasma from 59 (out of 95 total) individuals. Reference sera and donor plasma were treated with receptor-destroying enzyme (RDE; Denka Seiken Co., Ltd., United Kingdom), and serial 2-fold dilutions of the pretreated and inactivated plasma (1:40) were incubated with a standardized amount, or 4 agglutinating doses (AD4 HA units), of the designated influenza virus (Table 1) for 1 h at room temperature to allow antigen-antibody binding. An equal volume of 0.5% chicken red blood cell suspension was then added to the mixture. HAI titers were determined after incubation for 30 min at room temperature as the reciprocal of the highest plasma dilution that completely inhibited hemagglutination. Reference sera used as controls included H3 (A/duck/Shantou/1283/2001), H4N9 (A/duck/Shantou/461/2000), H5 (A/tern/South Africa/1961), H6 (A/turkey/Massachusetts/3740/1965), H7 (A/fowl plague virus/Rostock/1934), H7N9 (A/Anhui/1/2013), H8 (A/turkey/Ontario/6118/1968), and H12N5 (A/ruddy turnstone/Delaware/119/2007).
Statistical analysis.
K-means clustering analysis was used to classify individuals by endpoint HA-specific ELISA titer. Data were grouped into a predetermined number of nonhierarchical clusters based on their similarity using TIBCO Spotfire Decision Site software (Boston, MA). Geometric mean HAI antibody titers (GMTs) were determined, where a value of 1 was substituted for individual HAI antibody titers of <40 (undetectable titer). Differences in T cell responses between ELISA clusters were evaluated by one-way analysis of variance (ANOVA) with Tukey’s multiple comparison posttest. To control the error rates among differences in vaccination and avian exposure groups, P values evaluated by Student’s t test were false discovery rate (FDR) adjusted using the method of Benjamini and Hochberg described in reference 45. FDR-adjusted P values of <0.05 were considered significant. Whiskers of box plots represent 10th and 90th percentiles, and the horizontal line and plus sign in each box give the median and mean titer of individuals, respectively. Variability in antibody titer is shown by plotting the first and third quartiles of the titers as the outer limits of the box. Statistical analyses were performed using GraphPad Prism (La Jolla, CA) and R Statistical Software (46) version 3.1.1 (Vienna, Austria). Spearman rank correlation and FDR-adjusted significance (<0.1) was calculated using the psych package and plotted with the corrplot package (47, 48).
SUPPLEMENTAL MATERIAL
The frequencies of IFN-γ-producing cells in individuals receiving seasonal influenza vaccination as measured by ELISPOT assay to individual HA proteins are shown as spot-forming units (SFU) per million PBMC. Mean values for each donor are denoted as a red line. Groups were compared by two-tailed, unpaired t test, and false discovery rate-adjusted P values were determined. Download
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health Center of Excellence for Influenza Research and Surveillance (HHSN266200700005C), 1R56AI091938-01, and ALSAC.
We thank the participants who contributed to our study, Thomas Smith of the UCLA Department of Ecology and Evolutionary Biology, Scott Krauss of SJCRH, and Ali H. Ellebedy of Emory University School of Medicine for valuable discussion. We also are grateful for the technical assistance provided by J. L. McClaren, M. M. Morris, T. H. Oguin, III, and D. Wang of SJCRH. The purified HA protein from A/Anhui/1/2013 (H7N9) was generously provided by Peter Palese of the Icahn School of Medicine at Mt. Sinai Hospital, New York, NY.
Footnotes
Citation Oshansky CM, Wong S-S, Jeevan T, Smallwood HS, Webby RJ, Shafir SC, Thomas PG. 2014. Seasonal influenza vaccination is the strongest correlate of cross-reactive antibody responses in migratory bird handlers. mBio 5(6):e02107-14. doi:10.1128/mBio.02107-14.
REFERENCES
- 1. Palese P. 2004. Influenza: old and new threats. Nat. Med. 10:S82–S87. 10.1038/nm1141. [DOI] [PubMed] [Google Scholar]
- 2. Kilbourne ED. 2006. Influenza pandemics of the 20th century. Emerg. Infect. Dis. 12:9–14. 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sorrell EM, Schrauwen EJ, Linster M, De Graaf M, Herfst S, Fouchier RA. 2011. Predicting “airborne” influenza viruses: (trans-) mission impossible? Curr. Opin. Virol. 1:635–642. 10.1016/j.coviro.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. 2012. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. U. S. A. 109:4269–4274. 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. 2013. New world bats harbor diverse influenza A viruses. PLoS Pathog. 9:e1003657. 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Russell RJ, Kerry PS, Stevens DJ, Steinhauer DA, Martin SR, Gamblin SJ, Skehel JJ. 2008. Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 105:17736–17741. 10.1073/pnas.0807142105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, Lai RW, Orr WK, Shortridge KF. 1999. Human infection with influenza H9N2. Lancet 354:916–917. 10.1016/S0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 8. Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 368:1888–1897. 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 9. de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, Qui PT, Cam BV, Ha doQ, Guan Y, Peiris JS, Chinh NT, Hien TT, Farrar J. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 12:1203–1207. 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rimmelzwaan GF, Schutten M, Van Doornum GJ, Koch G, Bosman A, Koopmans M, Osterhaus AD. 2004. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl. Acad. Sci. U. S. A. 101:1356–1361. 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Myers KP, Setterquist SF, Capuano AW, Gray GC. 2007. Infection due to 3 avian influenza subtypes in United States veterinarians. Clin. Infect. Dis. 45:4–9. 10.1086/518579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shafir SC, Fuller T, Smith TB, Rimoin AW. 2012. A national study of individuals who handle migratory birds for evidence of avian and swine-origin influenza virus infections. J. Clin. Virol. 54:364–367. 10.1016/j.jcv.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Puzelli S, Di Trani L, Fabiani C, Campitelli L, De Marco MA, Capua I, Aguilera JF, Zambon M, Donatelli I. 2005. Serological analysis of serum samples from humans exposed to avian H7 influenza viruses in Italy between 1999 and 2003. J. Infect. Dis. 192:1318–1322. 10.1086/444390. [DOI] [PubMed] [Google Scholar]
- 14. Lee LY, Ha doLA, Simmons C, de Jong MD, Chau NV, Schumacher R, Peng YC, McMichael AJ, Farrar JJ, Smith GL, Townsend AR, Askonas BA, Rowland-Jones S, Dong T. 2008. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J. Clin. Invest. 118:3478–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Russell RJ, Gamblin SJ, Haire LF, Stevens DJ, Xiao B, Ha Y, Skehel JJ. 2004. H1 and H7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes. Virology 325:287–296. 10.1016/j.virol.2004.04.040. [DOI] [PubMed] [Google Scholar]
- 16. Nobusawa E, Aoyama T, Kato H, Suzuki Y, Tateno Y, Nakajima K. 1991. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology 182:475–485. 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- 17. Air GM. 1981. Sequence relationships among the hemagglutinin genes of 12 subtypes of influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 78:7639–7643. 10.1073/pnas.78.12.7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Medina RA, García-Sastre A. 2011. Influenza A viruses: new research developments. Nat. Rev. Microbiol. 9:590–603. 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Couch RB. 1975. Assessment of immunity to influenza using artificial challenge of normal volunteers with influenza virus. Dev. Biol. Stand. 28:295–306. [PubMed] [Google Scholar]
- 20. Clements ML, Betts RF, Tierney EL, Murphy BR. 1986. Serum and nasal wash antibodies associated with resistance to experimental challenge with influenza A wild-type virus. J. Clin. Microbiol. 24:157–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de Bree GJ, van Leeuwen EM, Out TA, Jansen HM, Jonkers RE, van Lier RA. 2005. Selective accumulation of differentiated CD8+ T cells specific for respiratory viruses in the human lung. J. Exp. Med. 202:1433–1442. 10.1084/jem.20051365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Piet B, de Bree GJ, Smids-Dierdorp BS, van der Loos CM, Remmerswaal EB, von der Thüsen JH, van Haarst JM, Eerenberg JP, ten Brinke A, van der Bij W, Timens W, van Lier RA, Jonkers RE. 2011. CD8(+) T cells with an intraepithelial phenotype upregulate cytotoxic function upon influenza infection in human lung. J. Clin. Invest. 121:2254–2263. 10.1172/JCI44675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hillaire ML, van Trierum SE, Bodewes R, van Baalen CA, van Binnendijk RS, Koopmans MP, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. 2011. Characterization of the human CD8(+) T cell response following infection with 2009 pandemic influenza H1N1 virus. J. Virol. 85:12057–12061. 10.1128/JVI.05204-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Khurelbaatar N, Krueger WS, Heil GL, Darmaa B, Ulziimaa D, Tserennorov D, Baterdene A, Anderson BD, Gray GC. 2013. Sparse evidence for equine or avian influenza virus infections among Mongolian adults with animal exposures. Influenza Other Respir. Viruses 7:1246–1250. 10.1111/irv.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Coman A, Maftei DN, Krueger WS, Heil GL, Friary JA, Chereches RM, Sirlincan E, Bria P, Dragnea C, Kasler I, Gray GC. 2013. Serological evidence for avian H9N2 influenza virus infections among Romanian agriculture workers. J Infect. Public Health 6:438–447. 10.1016/j.jiph.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 26. Bridges CB, Lim W, Hu-Primmer J, Sims L, Fukuda K, Mak KH, Rowe T, Thompson WW, Conn L, Lu X, Cox NJ, Katz JM. 2002. Risk of influenza A (H5N1) infection among poultry workers, Hong Kong, 1997-1998. J. Infect. Dis. 185:1005–1010. 10.1086/340044. [DOI] [PubMed] [Google Scholar]
- 27. Gill JS, Webby R, Gilchrist MJ, Gray GC. 2006. Avian influenza among waterfowl hunters and wildlife professionals. Emerg. Infect. Dis. 12:1284–1286. 10.3201/eid1708.060492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gray GC, McCarthy T, Capuano AW, Setterquist SF, Alavanja MC, Lynch CF. 2008. Evidence for avian influenza A infections among Iowa’s agricultural workers. Influenza Other Respir. Viruses 2:61–69. 10.1111/j.1750-2659.2008.00041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Khuntirat BP, Yoon IK, Blair PJ, Krueger WS, Chittaganpitch M, Putnam SD, Supawat K, Gibbons RV, Pattamadilok S, Sawanpanyalert P, Heil GL, Friary JA, Capuano AW, Gray GC. 2011. Evidence for subclinical avian influenza virus infections among rural Thai villagers. Clin. Infect. Dis. 53:e107–e116. 10.1093/cid/cir525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, Vogels R, Brakenhoff JP, Kompier R, Koldijk MH, Cornelissen LA, Poon LL, Peiris M, Koudstaal W, Wilson IA, Goudsmit J. 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333:843–850. 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JP, Skehel JJ, Lanzavecchia A. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333:850–856. 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 32. Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, Kepler TB, Liao HX, Harrison SC. 2011. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. U. S. A. 108:14216–14221. 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pica N, Palese P. 2013. Toward a universal influenza virus vaccine: prospects and challenges. Annu. Rev. Med. 64:189–202. 10.1146/annurev-med-120611-145115. [DOI] [PubMed] [Google Scholar]
- 34. Corti D, Suguitan AL, Jr, Pinna D, Silacci C, Fernandez-Rodriguez BM, Vanzetta F, Santos C, Luke CJ, Torres-Velez FJ, Temperton NJ, Weiss RA, Sallusto F, Subbarao K, Lanzavecchia A. 2010. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J. Clin. Invest. 120:1663–1673. 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, Mehta A, Razavi B, Del Rio C, Zheng NY, Lee JH, Huang M, Ali Z, Kaur K, Andrews S, Amara RR, Wang Y, Das SR, O’Donnell CD, Yewdell JW, Subbarao K, Marasco WA, Mulligan MJ, Compans R, Ahmed R, Wilson PC. 2011. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J. Exp. Med. 208:181–193. 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ellebedy AH, Krammer F, Li GM, Miller MS, Chiu C, Wrammert J, Chang CY, Davis CW, McCausland M, Elbein R, Edupuganti S, Spearman P, Andrews SF, Wilson PC, García-Sastre A, Mulligan MJ, Mehta AK, Palese P, Ahmed R. 2014. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc. Natl. Acad. Sci. U. S. A. 111:13133–13138. 10.1073/pnas.1414070111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miller MS, Gardner TJ, Krammer F, Aguado LC, Tortorella D, Basler CF, Palese P. 2013. Neutralizing antibodies against previously encountered influenza virus strains increase over time: a longitudinal analysis. Sci. Transl. Med. 5:198ra107. 10.1126/scitranslmed.3006637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Goff PH, Krammer F, Hai R, Seibert CW, Margine I, García-Sastre A, Palese P. 2013. Induction of cross-reactive antibodies to novel H7N9 influenza virus by recombinant Newcastle disease virus expressing a North American lineage H7 subtype hemagglutinin. J. Virol. 87:8235–8240. 10.1128/JVI.01085-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Medina RA, Stertz S, Manicassamy B, Zimmermann P, Sun X, Albrecht RA, Uusi-Kerttula H, Zagordi O, Belshe RB, Frey SE, Tumpey TM, García-Sastre A. 2013. Glycosylations in the globular head of the hemagglutinin protein modulate the virulence and antigenic properties of the H1N1 influenza viruses. Sci. Transl. Med. 5:187ra170. 10.1126/scitranslmed.3005996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li GM, Chiu C, Wrammert J, McCausland M, Andrews SF, Zheng NY, Lee JH, Huang M, Qu X, Edupuganti S, Mulligan M, Das SR, Yewdell JW, Mehta AK, Wilson PC, Ahmed R. 2012. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc. Natl. Acad. Sci. U. S. A. 109:9047–9052. 10.1073/pnas.1118979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mozdzanowska K, Furchner M, Washko G, Mozdzanowski J, Gerhard W. 1997. A pulmonary influenza virus infection in SCID mice can be cured by treatment with hemagglutinin-specific antibodies that display very low virus-neutralizing activity in vitro. J. Virol. 71:4347–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mozdzanowska K, Feng J, Eid M, Zharikova D, Gerhard W. 2006. Enhancement of neutralizing activity of influenza virus-specific antibodies by serum components. Virology 352:418–426. 10.1016/j.virol.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 43. Feng JQ, Mozdzanowska K, Gerhard W. 2002. Complement component C1q enhances the biological activity of influenza virus hemagglutinin-specific antibodies depending on their fine antigen specificity and heavy-chain isotype. J. Virol. 76:1369–1378. 10.1128/JVI.76.3.1369-1378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Keating R, Hertz T, Wehenkel M, Harris TL, Edwards BA, McClaren JL, Brown SA, Surman S, Wilson ZS, Bradley P, Hurwitz J, Chi H, Doherty PC, Thomas PG, McGargill MA. 2013. The kinase mTOR modulates the antibody response to provide cross-protective immunity to lethal infection with influenza virus. Nat. Immunol. 14:1266–1276. 10.1038/ni.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yayh B. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Stat. Methodol. 57:289–300. [Google Scholar]
- 46. Core R. Team; . 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 47. Revelle W. 2014. Psych: procedures for personality and psychological research, 1.4.8. Northwestern University, Evanston, IL: http://CRAN.R-project.org/package=psych. [Google Scholar]
- 48. Wei T. 2013. corrplot: visualization of a correlation matrix. R package version 0.73. http://CRAN.R-project.org/package=corrplot.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The frequencies of IFN-γ-producing cells in individuals receiving seasonal influenza vaccination as measured by ELISPOT assay to individual HA proteins are shown as spot-forming units (SFU) per million PBMC. Mean values for each donor are denoted as a red line. Groups were compared by two-tailed, unpaired t test, and false discovery rate-adjusted P values were determined. Download