ABSTRACT
Ribosomal elongation factor 4 (EF4) is highly conserved among bacteria, mitochondria, and chloroplasts. However, the EF4-encoding gene, lepA, is nonessential and its deficiency shows no growth or fitness defect. In purified systems, EF4 back-translocates stalled, posttranslational ribosomes for efficient protein synthesis; consequently, EF4 has a protective role during moderate stress. We were surprised to find that EF4 also has a detrimental role during severe stress: deletion of lepA increased Escherichia coli survival following treatment with several antimicrobials. EF4 contributed to stress-mediated lethality through reactive oxygen species (ROS) because (i) the protective effect of a ΔlepA mutation against lethal antimicrobials was eliminated by anaerobic growth or by agents that block hydroxyl radical accumulation and (ii) the ΔlepA mutation decreased ROS levels stimulated by antimicrobial stress. Epistasis experiments showed that EF4 functions in the same genetic pathway as the MazF toxin, a stress response factor implicated in ROS-mediated cell death. The detrimental action of EF4 required transfer-messenger RNA (tmRNA, which tags truncated proteins for degradation and is known to be inhibited by EF4) and the ClpP protease. Inhibition of a protective, tmRNA/ClpP-mediated degradative activity would allow truncated proteins to indirectly perturb the respiratory chain and thereby provide a potential link between EF4 and ROS. The connection among EF4, MazF, tmRNA, and ROS expands a pathway leading from harsh stress to bacterial self-destruction. The destructive aspect of EF4 plus the protective properties described previously make EF4 a bifunctional factor in a stress response that promotes survival or death, depending on the severity of stress.
IMPORTANCE
Translation elongation factor 4 (EF4) is one of the most conserved proteins in nature, but it is dispensable. Lack of strong phenotypes for its genetic knockout has made EF4 an enigma. Recent biochemical work has demonstrated that mild stress may stall ribosomes and that EF4 can reposition stalled ribosomes to resume proper translation. Thus, EF4 protects cells from moderate stress. Here we report that EF4 is paradoxically harmful during severe stress, such as that caused by antimicrobial treatment. EF4 acts in a pathway that leads to excessive accumulation of reactive oxygen species (ROS), thereby participating in a bacterial self-destruction that occurs when cells cannot effectively repair stress-mediated damage. Thus, EF4 has two opposing functions—at low-to-moderate levels of stress, the protein is protective by allowing stress-paused translation to resume; at high-levels of stress, EF4 helps bacteria self-destruct. These data support the existence of a bacterial live-or-die response to stress.
INTRODUCTION
Translation elongation factor 4 (EF4) has the intriguing property of being one of the most conserved proteins in nature while also being dispensable for growth (1–4). Biochemical work shows that EF4 back-translocates posttranslational ribosomes for efficient protein synthesis (3), especially during mild stress created by high ionic strength, low pH, or low temperature (2). EF4 is usually stored in the cell membrane; however, during stress, it exits from its storage site (2, 5), binds to the A site of ribosomes, back-translates, and gives stalled ribosomes a chance to resume translation (3). Thus, EF4 provides protein synthesis with an antistalling, error correction mechanism. Since reversing stress-mediated ribosome pausing should limit abortive translational events that deplete resources, EF4 has been thought to protect from stress. Indeed, the effects of several moderate forms of stress are exacerbated by a deficiency of lepA, the gene encoding EF4 in Escherichia coli, or its orthologs in other bacteria (2, 6–9). Thus, EF4 may contribute to survival under unfavorable conditions (9).
EF4 is also found to inhibit the action of transfer-messenger RNA (tmRNA) (8). tmRNA functions as both a tRNA and an mRNA in trans translation (10–12), a process in which tmRNA shifts the translation of nascent, truncated peptides from truncated mRNAs lacking an in-frame translational stop codon to itself. In the process, tmRNA adds a proteolysis tag and a stop codon to the truncated peptide, releases the tagged peptide from the stalled ribosome for degradation, and recycles stalled ribosomes for new translation (12). By reducing tmRNA function, EF4 is expected to elevate the level of untagged, truncated proteins derived from stress-induced mRNA cleavage (8, 13–15). Since some truncated proteins might be toxic, we reasoned that EF4 may have a destructive function when stress is harsh. These observations raise the possibility that EF4 may have both protective and destructive roles in response to stress.
The dual functions of EF4 associated with stress fit our proposal that the response to some forms of stress, in particular, stress caused by lethal antibiotics, can be either protective or destructive, depending on the type and magnitude of the stress (14, 16). An example is a response that involves the MazEF toxin-antitoxin module of E. coli. On the one hand, the MazF endoribonuclease cleaves mRNA and halts translation during stress (17). Such translational pausing is expected to give bacterial cells time to repair damage. On the other hand, cleavage of mRNA can lead to the accumulation of truncated mRNA and subsequently to truncated proteins. Insertion of abnormal, truncated proteins into the cell membrane is proposed to perturb the respiratory chain, cause a subsequent buildup of reactive oxygen species (ROS), and lead ultimately to self-destruction (16, 18–21). Indeed, the MazF orthologue in Bacillus subtilis is protective at low levels of UV irradiation and destructive at high levels (14). Whether EF4, through its inhibition of tmRNA, is part of the MazF-ROS stress response is unknown.
In the present work, we examined how the absence of EF4 in E. coli (ΔlepA) affects the response to a variety of lethal stresses. The lepA deficiency had little effect on growth or the bacteriostatic action of several lethal stressors. However, the mutation increased bacterial survival in a tmRNA/ClpP-dependent manner; the mutation also decreased stress-stimulated intracellular ROS accumulation. Thus, the wild-type form of EF4 has destructive activity that increases the lethal effects of several types of stress. Moreover, lepA and mazF were epistatic, which indicates that EF4 acts through a pathway that involves the MazF toxin. These data indicate that, under stressful conditions, EF4 helps bacteria make a live-or-die decision via a MazF-tmRNA-ROS pathway. The highly conserved nature of lepA raises the possibility that many bacterial species have an EF4 activity that can be either self-destructive or self-protective.
RESULTS
A lepA deficiency diminishes effects of lethal agents.
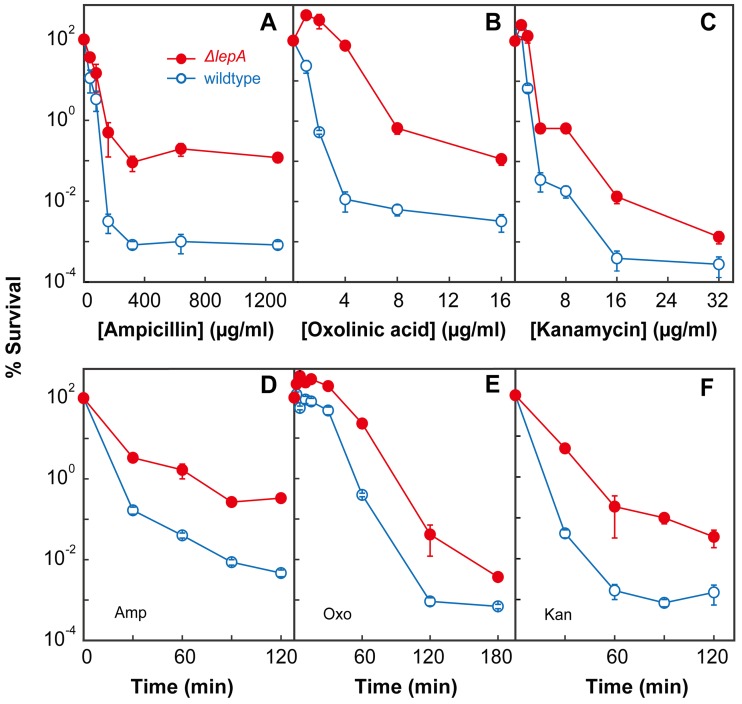
To better understand the role of EF4 when bacteria are faced with harsh stress, we examined the effects of three lethal antimicrobials on relevant bacterial mutants (the strain used are described in Table 1). Deletion of lepA had no effect on growth inhibition by ampicillin, oxolinic acid, or kanamycin, as measured by MIC determination (Table 2). The absence of an effect on the MIC indicates that EF4 has no effect on resistance to these antibiotics, because resistance factors such as reduced drug uptake, efflux, and target affinity generally affect the MIC. However, when the survival of a wild-type strain (3001) and that of an otherwise isogenic ΔlepA mutant (3500) were compared at various concentrations of the three antimicrobials, the survival of the ΔlepA mutant dropped less than that of wild-type cells (Fig. 1A to C). A similar result was obtained when using various treatment times for a fixed concentration of the three compounds (Fig. 1D to F). Thus, EF4 is likely to be part of a poststress cellular response, because its absence decreases stress-mediated lethality without affecting growth inhibition, a surrogate measure for the primary lesions caused by stress.
TABLE 1 .
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source and/or reference |
|---|---|---|
| Strains | ||
| 2987 | ΔchpA(mazF)781::Kanr | Keio Collection, CGSC 10158a |
| 2989 | ΔchpB773::Kanr | Keio Collection, CGSC 11012 |
| 3001 | Wild type (BW25113) | CGSC 7636 |
| 3308 | ΔclpP723::Kanr | CGSC 8590 |
| 3326 | ΔclpP | This work, by kanamycin marker excision; 64 |
| 3353 | TA 330, W3110 ssrA::FRT-cat-FRT | 70 |
| 3438 | ΔlepA738::Kanr | CGSC strain 10029 |
| 3496 | 3001 ΔmazF::Kanr | This work, by P1-mediated transduction from 2987 into 3001 |
| 3497 | 3001 ΔmazF::Kanr ΔlepA | This work, by P1-mediated transduction from 2987 into 3499 |
| 3499 | 3001 ΔlepA | This work, by P1-mediated transduction from 3438 into 3001, followed by kanamycin marker excision; 64 |
| 3500 | 3001 ΔlepA738::Kanr | This work, by P1-mediated transduction from 3438 into 3001 |
| 3502 | 3001 ΔssrA::FRT-cat-FRT | This work, by P1-mediated transduction from 3353 into 3001 |
| 3503 | 3001 ΔssrA::FRT-cat-FRT ΔlepA | This work, by P1-mediated transduction from 3353 into 3499 |
| 3682 | 3001 transformed with pLEPA | This work |
| 3683 | 3499 transformed with pLEPA | This work |
| 3684 | 3001 transformed with pWSK29 | This work |
| 3685 | 3499 transformed with pWSK29 | This work |
| 3688 | 3001 ΔchpB773::Kanr ΔlepA738 | This work, by P1-mediated transduction from 2989 into 3499 |
| 4114 | ΔlepA::Kanr ΔclpP | This work, by P1-mediated transduction from 3438 into 3326 |
| Plasmids | ||
| pWSK29 | E. coli expression vector | 67 |
| pLEPA | 2.9-kb HindIII DNA fragment encompassing lepA cloned into pWSK29 | 8 |
CGSC, Coli Genetic Stock Center.
TABLE 2 .
Antimicrobial susceptibility
| Strain | Relevant genotype | MIC (μg/ml)a |
|||
|---|---|---|---|---|---|
| Amp | Oxo | Cip | Kan | ||
| 2989 | ΔchpB773::Kanr | 20 | 0.5 | ND | ND |
| 3001 | Wild type | 20 | 0.5 | 0.01 | 2 |
| 3326 | ΔclpP | 20 | ND | ND | ND |
| 3496 | ΔmazF::Kanr | 25 | 0.5 | ND | ND |
| 3497 | ΔmazF::Kanr ΔlepA | 25 | 0.5 | ND | ND |
| 3499 | ΔlepA | 20 | 0.5 | 0.01 | 2 |
| 3500 | ΔlepA-738::Kanr | 20 | 0.5 | 0.01 | ND |
| 3502 | ΔssrA::cat | 25 | 0.5 | ND | ND |
| 3503 | ΔlepA ΔssrA::cat | 25 | 0.5 | ND | ND |
| 3682 | 3001 transformed with pLEPA | ND | 0.5 | ND | 2 |
| 3683 | 3499 transformed with pLEPA | ND | 0.5 | ND | 2 |
| 3684 | 3001 transformed with pWSK29 | ND | 0.5 | ND | 2 |
| 3685 | 3499 transformed with pWSK29 | ND | 0.5 | ND | 2 |
| 3688 | ΔchpB773::Kanr ΔlepA738 | 25 | 0.5 | ND | ND |
| 4114 | ΔlepA::Kanr ΔclpP | 20 | ND | ND | ND |
Abbreviations: Amp, ampicillin; Oxo, oxolinic acid; Cip, ciprofloxacin; Kan, kanamycin; ND, not determined.
FIG 1 .
A lepA deficiency increases bacterial survival following exposure to lethal stress. Exponentially growing cultures of E. coli were treated with ampicillin (A), oxolinic acid (B), or kanamycin (C) at the concentrations indicated for 120, 120, and 30 min, respectively. Cultures were also treated with a fixed concentration of ampicillin at 32 times the MIC (D), oxolinic acid at 32 times the MIC (E), or kanamycin at 4 times the MIC (F) for the times indicated. Symbols: empty circles, wild-type strain 3001; filled circles, ΔlepA mutant strain 3499 (for kanamycin exposure only, strain 3500 for other stressors). Percent survival was measured as described in Materials and Methods. Error bars represent standard errors of the means (some error bars are covered by symbols).
To attribute protection from lethal stress to the lepA deficiency, complementation experiments were performed. A low-copy-number plasmid expressing EF4 was introduced into a lepA-deficient strain that was then examined for survival during lethal stress. The complemented strain (3683) was killed to the same extent as the wild type transformed with the empty vector (strain 3684) when treated with various concentrations of oxolinic acid or kanamycin (see Fig. S1 in the supplemental material), each of which represents a different antimicrobial class. We conclude that ΔlepA mutant cells are better able than lepA+ cells to survive several types of lethal stress; therefore, wild-type EF4 must have a destructive role during lethal stress.
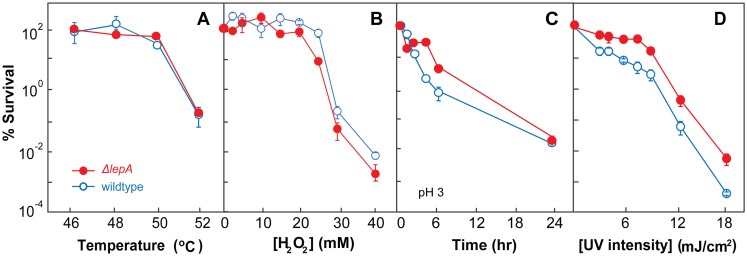
We next examined whether EF4 also affects killing by environmental stress. When cells were treated with heat, hydrogen peroxide, a low pH, or UV irradiation, the protective effect of the lepA deficiency seen with antimicrobials was not observed with heat or exogenous hydrogen peroxide (Fig. 2A and B). The effect on low-pH- and UV-mediated killing was only slightly reduced by the lepA deficiency (Fig. 2C and D). Thus, the lepA mutation protects from killing by some, but not all, types of stress. Possible reasons for the absence of a protective effect against some of these stressors are addressed in the Discussion.
FIG 2 .
A lepA deficiency exhibits little protective effect against environmental stress. Exponentially growing cultures of E. coli were incubated for 10 min at the temperatures indicated (A), incubated for 10 min with hydrogen peroxide at the concentrations indicated (B), incubated for the times indicated in medium at pH 3 (C), or exposed for 30 s to UV irradiation at the intensities indicated (D). After incubation, percent survival was determined as described in Materials and Methods. Symbols: wild type (strain 3001), empty circles; ΔlepA mutant (strain 3500), filled circles. Error bars represent standard errors of the means.
ROS are involved in EF4-mediated enhancement of lethal stress.
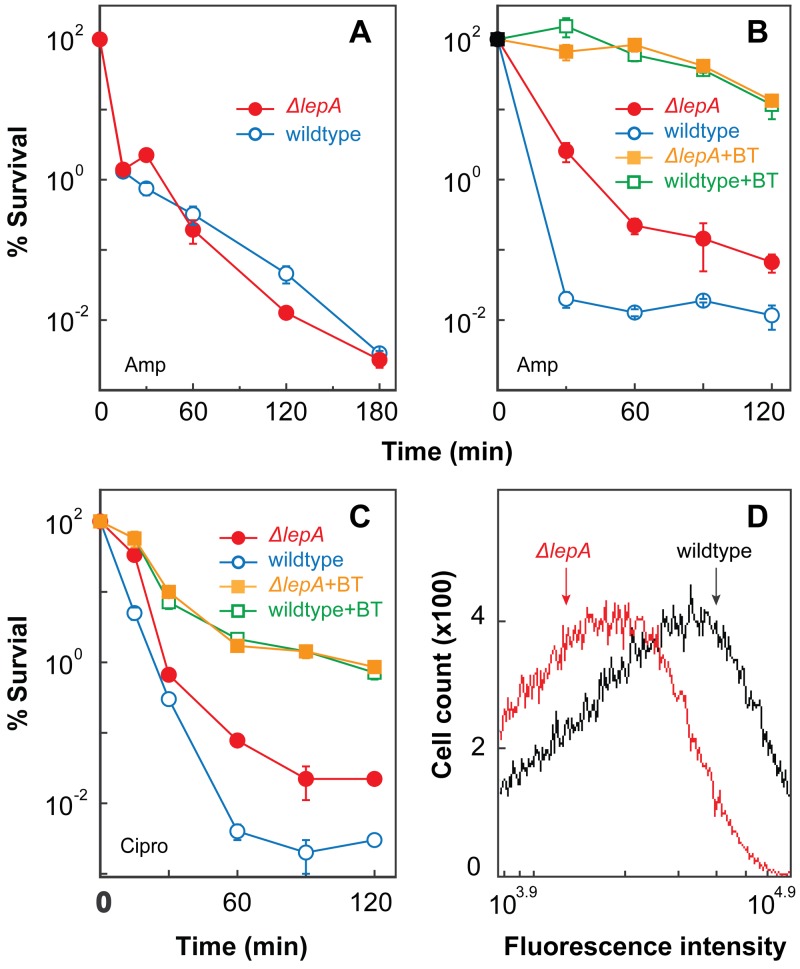
Since several lines of previous work support the idea that ROS contribute to the lethal activity of antimicrobials (19, 21–28), the possibility arose that the destructive action of EF4 might involve ROS. To explore this possibility, we first determined whether anaerobiosis eliminates differences between wild-type cells and a lepA mutant. When the wild-type strain (3001) and a ΔlepA mutant (3500) were compared for survival, the protection from ampicillin conferred by the lepA deficiency under aerobic conditions (Fig. 1A) was absent under anaerobic conditions (Fig. 3A). Protection was also absent during anaerobiosis when an alternate electron acceptor, 100 mM potassium nitrate, was added to the LB-glucose-phosphate medium (29) used for anaerobic bacterial growth (see Fig. S2A in the supplemental material). Similar results were obtained with kanamycin when LB medium supplemented with nitrate was used (see Fig. S2B).
FIG 3 .
ROS are involved in lepA deficiency-mediated protection from lethal stress. Exponentially growing cultures of E. coli were treated for the times indicated with ampicillin at 32 times the MIC under anaerobic conditions (panel A), ampicillin (Amp) at 32 times the MIC following a 5-min pretreatment with or without 0.5 times the MIC of 2,2′-bipyridyl plus thiourea (BT) (panel B), ciprofloxacin at 5 times the MIC following a 5-min pretreatment with or without 0.5 times the MIC of 2,2′-bipyridyl plus thiourea (panel C). After incubation, survival was determined as described in Materials and Methods. Symbols: wild type (strain 3001), empty circles; ΔlepA mutant (strain 3500), filled circles; wild type pretreated with 0.5 times the MIC of 2,2′-bipyridyl plus thiourea for ampicillin or for ciprofloxacin (Cipro), empty squares; ΔlepA mutant pretreated with 0.5 times the MIC of 2,2′-bipyridyl plus thiourea for ampicillin or for ciprofloxacin, filled squares. Error bars represent standard errors of the means. (Panel D) Exponentially growing cultures of E. coli were treated with 10 µM H2DCFDA for 20 min before ampicillin (5 times the MIC) was added. Samples were then taken every 30 min for analysis by flow cytometry. A representative 30-min ampicillin treatment data set is shown (see Fig. S3 in the supplemental material for other time point data sets). Lines: wild type, black; ΔlepA mutant, red. Three replicate experiments gave similar results.
As a second test, we determined whether ROS scavengers eliminate ΔlepA-mediated protection. When wild-type and ΔlepA mutant cells were pretreated with subinhibitory concentrations of thiourea plus 2,2′-bipyridyl to block hydroxyl radical accumulation (23), the lethal action of ampicillin was reduced in both strains by a factor of 100 to several thousand, and the ΔlepA-mediated protection shown in Fig. 1A was eliminated (Fig. 3B).
We next asked whether EF4 participates in both ROS-dependent and ROS-independent killing pathways. For this experiment, we chose ciprofloxacin as a stressor because its ROS-dependent and ROS-independent mechanisms of killing are more evenly partitioned than those of ampicillin (compare Fig. 3B and C for killing in the absence or presence of thiourea plus 2,2′-bipyridyl). The survival of wild-type cells treated only with ciprofloxacin was about 0.01% (Fig. 3C). A deficiency of lepA raised that value by >10-fold. When subinhibitory concentrations of thiourea plus 2,2′-bipyridyl were added with ciprofloxacin to block hydroxyl radical accumulation, a protective effect was observed in both the wild-type and ΔlepA mutant strains. However, the difference between wild-type and lepA mutant cells seen in the absence of thiourea plus 2,2′-bipyridyl was eliminated (Fig. 3C). These are the results expected for ROS involvement in EF4 activity. Substantial killing occurred that was independent of ROS: 99% of the cells were killed by ciprofloxacin even in the presence of thiourea and 2,2′-bipyridyl (Fig. 3C). The absence of EF4 had no effect on this ROS-independent killing. Collectively, the data show that wild-type EF4 stimulates antimicrobial lethality largely through an ROS-dependent pathway.
As a further test of ROS involvement in the action of EF4, we examined wild-type and lepA-deficient cells treated with the fluorogenic dye H2DCFDA during ampicillin exposure. H2DCFDA readily penetrates E. coli cells (30, 31); once it enters cells, H2DCFDA is converted by cellular esterases into a non-membrane-permeating compound that can be oxidized to a fluorescent form by ROS (30). We treated cells with 10 µM H2DCFDA and 5 times the MIC of ampicillin, and then we used flow cytometry to measure intracellular fluorescence. The ΔlepA mutant culture exhibited less fluorescent labeling than wild-type cells following treatment with ampicillin for 0.5 to 2 h (a 0.5-h example is shown in Fig. 3D; additional time points are shown in Fig. S3 in the supplemental material). Thiourea plus 2,2′-bipyridyl, which protects from ampicillin-mediated lethality (Fig. 3B), also reduced ROS accumulation (see Fig. S4 in the supplemental material), as we have observed in previous work (23). Collectively, the data described above are consistent with wild-type EF4 promoting an ROS cascade during lethal stress, possibly through perturbation of the respiratory chain (32).
MazF toxin and EF4 act in the same pathway to enhance effects of lethal stress.
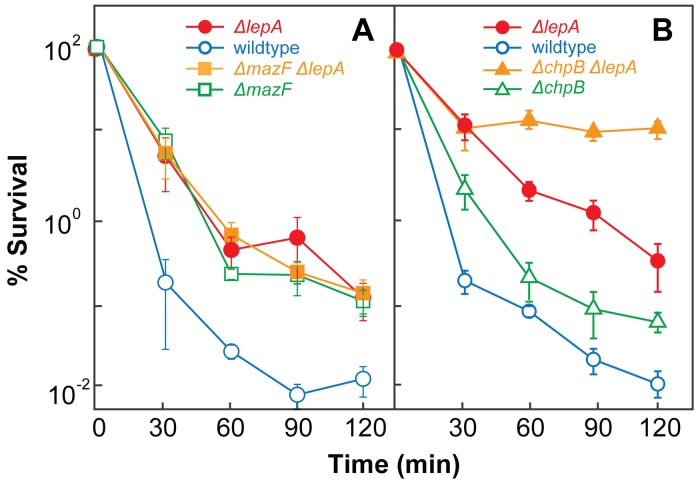
Since toxin-antitoxin modules can increase the fraction of cells that die when stress is extreme (14, 33, 34) and since killing mediated by both EF4 and MazF toxin correlates with ROS accumulation (14, 33, 34), we next determined whether EF4 acts in the same genetic pathway as the MazF toxin. To do so, we measured the lethal activity of ampicillin with a set of isogenic strains. When the wild-type strain (3001), the ΔmazF mutant (3496), the ΔlepA mutant (3500), and the ΔmazF ΔlepA double mutant (3497) were compared for survival, the wild-type strain was more readily killed by ampicillin than were the three other strains (Fig. 4A). The double mutant exhibited a loss of survival that was similar to that observed with each single mutant. Oxolinic acid treatment showed results similar to those obtained with ampicillin (see Fig. S5A in the supplemental material). Thus, the toxin-encoding mazF gene and lepA appear to function in the same pathway during lethal stress.
FIG 4 .
MazF and ChpB toxin deficiencies have different effects on ΔlepA-mediated protection from lethal stress. Exponentially growing cultures of E. coli were treated with ampicillin at 32 times the MIC for the times indicated. Percent survival was measured as described in Materials and Methods. (A) Effect of mazF deficiency. Symbols: wild type (strain 3001), empty circles; ΔlepA mutant (strain 3500), filled circles; ΔmazF mutant (strain 3496), empty squares; ΔmazF ΔlepA double mutant (strain 3497), filled squares. (B) Effect of chpB deficiency. Symbols: wild type, empty circles; ΔlepA mutant, filled circles; ΔchpB mutant (strain 2989), empty triangles; ΔchpB ΔlepA double mutant (strain 3688), filled triangles. Error bars represent standard errors of the means.
To determine whether the observed epistatic effect of lepA and mazF applies to other toxin-antitoxin modules, we also examined the ChpB toxin. When the wild-type strain (3001), the ΔchpB mutant (2989), the ΔlepA mutant (3500), and the ΔchpB ΔlepA double mutant (3688) were compared for survival following treatment with ampicillin, the wild type was more readily killed than the other three strains. The ΔchpB ΔlepA double mutant was the least susceptible (Fig. 4B), and the ΔchpB mutant exhibited a loss of survival that was intermediate between those of ΔlepA single mutant and wild-type cells. This additive effect, which was also observed with oxolinic acid treatment (see Fig. S5B in the supplemental material), indicates that EF4 acts in a pathway different from that involving the ChpB toxin. Thus, the epistatic relationship between mazF and lepA is not common to all toxin-antitoxin modules.
tmRNA is required for EF4-mediated enhancement of stress-mediated lethality.
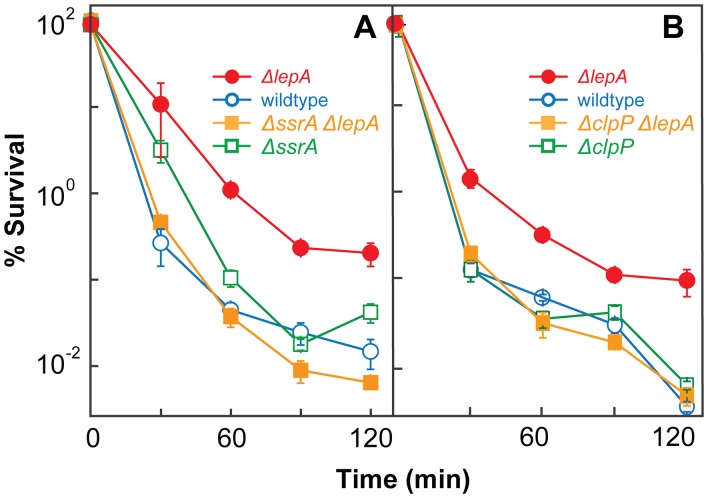
To better understand how EF4 is connected to the lethal cascade of ROS, we examined the effects of tmRNA on the protective action of the ΔlepA mutation. As pointed out in the introduction, interference with tmRNA tagging by EF4 could facilitate the accumulation of truncated proteins, some of which may indirectly stimulate ROS production. When the wild-type strain (3001), a ΔssrA (tmRNA) mutant (3502), a ΔlepA mutant (3500), and a ΔssrA ΔlepA double mutant (3503) were compared for survival following treatment with ampicillin, the survival of the ΔssrA mutant was similar to that of the wild-type strain (Fig. 5A). As expected, the survival of the ΔlepA mutant was greater than that of wild-type cells, as also shown in Fig. 1. Addition of an ssrA deficiency to the ΔlepA strain eliminated both the protective effect of the lepA deletion on killing (Fig. 5A) and its reduction of ampicillin-mediated ROS accumulation (see Fig. S6 in the supplemental material). To further test whether truncated protein accumulation is involved in ΔlepA-mediated protection, we combined ΔlepA with a deficiency in clpP, since ClpP is the major protease responsible for degradation of tmRNA-tagged proteins (12, 35). As expected, addition of a clpP deficiency to the ΔlepA mutant strain eliminated the ΔlepA-mediated protective effect (Fig. 5B). Collectively, these data indicate that the protective effect of an EF4 deficiency involves tmRNA and truncated-protein degradation: the destructive effect of wild-type EF4 requires functional tmRNA and ClpP protease.
FIG 5 .
Functional tmRNA and ClpP are required for lepA deficiency-mediated protection from lethal stress. Exponentially growing cultures of E. coli were treated with ampicillin (32 times the MIC) for the times indicated. After incubation, percent survival was determined as described in Materials and Methods. (A) Effect of ssrA deficiency. Symbols: wild type (strain 3001), empty circles; ΔlepA mutant (strain 3500), filled circles; ΔssrA mutant (strain 3502), empty squares; ΔssrA ΔlepA double mutant (strain 3503), filled squares. (B) Effect of clpP deficiency. Symbols: wild type (strain 3001), empty circles; ΔlepA mutant (strain 3500), filled circles; ΔclpP mutant (strain 3226), empty squares; ΔclpP ΔlepA double mutant (strain 4114), filled squares. Error bars represent standard errors of the means.
DISCUSSION
Harsh stress, such as that created by lethal antimicrobials, causes a bacterial response that now includes translation elongation factor 4 (EF4). The present work provides three insights into the action of EF4. First, it solves the mystery of why few strong phenotypes have been reported previously for defects in lepA, the highly conserved gene that encodes EF4: phenotypes are often related to growth, not the response to lethal stress. Second, this work, in conjugation with earlier biochemical studies, demonstrates that EF4 has both protective and destructive functions that depend on the severity of the stress encountered. Third, we tie EF4 to a MazF-tmRNA-ROS pathway involved in stress-mediated self-destruction. Each of these points is discussed below.
The lack of a phenotype of EF4-deficient strains derives from traditional phenotype assays usually seeking effects on bacterial growth, such as a change in the MIC of an antimicrobial. Stress response genes such as lepA act after the primary stress-mediated damage occurs; thus, they function largely on survival rather than on growth. Indeed, we found that a ΔlepA mutation has no effect on the MIC (Table 2), while the mutation confers protection against killing by several antimicrobials (Fig. 1). Thus, the absence of a phenotype was likely due to the nature of the assay. The conservation of the EF4 amino acid sequence may reflect the importance of self-destruction to bacterial population fitness.
Support for the dual function of EF4 comes from two directions. First, EF4 allows translation to restart when EF-G malfunctions (3). Such activity is expected to contribute to the recovery of cells from moderate perturbations (2). Second, more recent biochemical studies show that EF4 not only back-translates posttranslocation (POST) ribosomes toward a pretranslocation (PRE) state (predominantly to an intermediate state [I3] before PRE) (36) but can also compete with EF-G for binding to the PRE complex (37). That competition effectively sequesters a catalytically active ribosome and transiently inhibits elongation. New crystal structure work indicates that binding of EF4 to ribosomes causes backward rotation of the 30S subunit (38), supporting a role for EF4 as a back-translocase that can reverse the process performed by either EF-G or EF-Tu. Interfering with EF-G and/or EF-Tu function by EF4 may suppress tmRNA from rescuing ribosomes stalled by truncated mRNAs, because loading of tmRNA to the A site of ribosomes requires EF-Tu and translocating tmRNA from the A site to the P site, needed to shift translation from the truncated mRNA to tmRNA itself, involves EF-G (39, 40). Indeed, EF4 inhibits the activity of tmRNA (8), a protective system that releases truncated proteins from stalled ribosomes and tags the peptides for proteolytic degradation (11, 41). Since excessive untagged, truncated proteins produced during severe stress resist proteolysis and thus may be more harmful to cells than their tagged cognates, EF4 interference with tmRNA under harsh stress may be destructive. Taken together, the biochemical evidence suggests that EF4 might have both protective and destructive effects following stress. The present work established the destructive effects of EF4 when living cells are challenged with harsh stress.
Integration of EF4 into a pathway that involves ROS, MazF toxin, and tmRNA is supported by several lines of evidence. First, connections between EF4 and ROS were indicated by the absence of a ΔlepA effect on antimicrobial lethality with cells (i) growing under anaerobic conditions (Fig. 3A) or (ii) pretreated with thiourea plus 2,2′-bipyridyl, agents that block hydroxyl radical accumulation (16, 19, 21) (Fig. 3B and C). Second, measurement of ROS by H2DCFDA-mediated fluorescence using flow cytometry revealed that ampicillin treatment raises the fraction of ROS-positive cells more with a wild-type strain than with a ΔlepA mutant (Fig. 3D). A link between EF4 and MazF was established by the epistatic relationship between strains deficient in these functions when the lethal activity of ampicillin was measured (Fig. 4A). Such is not the case for all toxin-antitoxin systems, since the ChpB module showed an additive, rather than an epistatic, effect with EF4 (Fig. 4B). Third, evidence of the involvement of tmRNA/ClpP derives from the similar lethal effects of ampicillin on a single mutant (ΔssrA [tmRNA deficient] or ΔclpP [ClpP protease deficient]) and a double mutant (ΔlepA ΔssrA or ΔlepA ΔclpP), each of which exhibited lower survival than a ΔlepA single mutant (Fig. 1 and 5); the absence of tmRNA/ClpP eliminated the protective effect of an EF4 deficiency.
The experiments described above and previous work suggest a plausible stress response scenario that involves MazF, EF4, tmRNA, and ROS. When bacteria encounter low-to-moderate stress, translational elongation may be stalled because of ribosome pausing (2). That triggers the movement of EF4 from its membrane storage site to the cytosol (2, 5). EF4 then binds to stalled ribosomes, back-translates, and helps restore translation (3). When E. coli experiences harsh stress, toxin- and/or ribosome pause-mediated mRNA cleavage becomes extensive (17), thereby producing many truncated mRNAs lacking translation stop codons. Such truncated mRNAs halt ribosome elongation. As a countermeasure, tmRNA shifts the translation of nascent, truncated peptides from the truncated mRNA to tmRNA itself and releases stalled ribosomes for new translation (12). In that process, tmRNA adds a proteolysis tag and an in-frame stop codon to the truncated protein (12). EF4 inhibits shifting translation from truncated mRNA to tmRNA (8), which interferes with tmRNA-mediated tagging of abnormal proteins for degradation (35, 42, 43). That elevates the accumulation of truncated proteins. Since truncated proteins are often misfolded, insertion of those proteins into cell membranes is expected to perturb the respiratory chain, activate an ROS cascade (16, 18, 20), and promote cell death. The absence of EF4 leads to loss of tmRNA inhibition, reduced abundance of truncated proteins, and lower ROS-mediated lethality.
Previous work showed that some lethal stressors are insensitive to the effects of the MazF-ROS pathway. Examples are a highly active fluoroquinolone (44) and high temperature (16). While the ROS cascade generally contributes to the lethal action of quinolones with E. coli (19, 21, 44), an exception exists if a fluoroquinolone fragments bacterial chromosomes so rapidly and extensively that an ROS cascade would make only a small additional contribution (16, 44, 45). The present work shows that much of the lethal action of ciprofloxacin falls into this category and that only the ROS-dependent portion of killing by ciprofloxacin is protected by a lepA deficiency (Fig. 3C). High temperature (Fig. 2A) is expected to produce such large amounts of misfolded protein that the amount arising from the action of EF4 may be insignificant. Likewise, the action of exogenous hydrogen peroxide (Fig. 2B) is expected to overshadow endogenous-ROS-mediated effects. Although we observed only slight protection from low pH-mediated killing by a lepA deficiency (Fig. 2C), the result is surprising, since previous studies have shown that a lepA deficiency confers hypersusceptibility to acidic conditions (7, 9). We attribute this discrepancy to the different natures of killing and growth inhibition assays (for a discussion, see reference 46). The much smaller difference between the wild-type and lepA mutant strains in survival at a low pH or during UV exposure than during antimicrobial treatment may be similar to the comparison of high temperature and antibiotics: a low pH causes massive protein denaturation, making the effect of EF4-mediated release of misfolded peptide fragments trivial. UV irradiation itself causes massive DNA damage such that EF4-mediated ROS reduction is insufficient to protect massively damaged cells. Clearly, the lethal action of some stressors is independent of an ROS cascade; such ROS-independent stressors would be insensitive to a lepA deficiency.
Several recent reports (47–49) have challenged the general idea that an ROS cascade is associated with the lethal action of antimicrobials. The central issue is whether ROS serve as an ancillary killing mechanism, as repeatedly stressed (19, 50), or whether ROS replace other known killing mechanisms, as challenged (47). No evidence supports the later scenario, but many independent lines of work (21, 23–28, 44) and a recent rebuttal (22) establish the role of ROS as a common lethal mechanism that adds to well-studied antimicrobial-specific mechanisms. For example, ROS involvement in antimicrobial activity remains the best explanation for several situations, such as anaerobiosis selectively blocking killing by some quinolones (29, 51), defects in peroxide-detoxifying enzymes increasing antimicrobial lethality (21), and overexpression of peroxide-detoxifying enzymes decreasing lethality and ROS-mediated protein aggregation (22, 52). These issues have been discussed in recent reviews (46, 53, 54). Nevertheless, there remains a need to establish specificity for perturbations and assays of ROS accumulation, and the protective effect of bipyridyl and thiourea in the absence of oxygen is far from understood.
The response to lethal stress is complex; studies need to consider that ROS increase the rate of killing rather than the extent (23), that safety valves (peroxidases [21, 22] and the YihE protein kinase [16]) must sometimes be overcome to observe killing, and that experiments must use multiple drug concentrations for various times to ensure that a window in which the ROS effect can be observed is not missed. Moreover, the choice of stressor can be critical (46, 53).
In conclusion, the finding that EF4 has both protective and destructive roles connected to MazF and ROS, which are also both protective and destructive (14, 55–60), supports the general idea that bacteria respond to stress by either repairing the damage or self-destructing, a decision that is based on the level of stress (14, 16). Discovering ways to manipulate the lethal stress response could reveal novel approaches for enhancing antimicrobial therapy both by making antimicrobials more lethal and by enhancing stressful host defense systems. Of immediate medical relevance is the inhibitory effect that antioxidants may have on antimicrobial therapy. Consumption of antioxidants as nutritional supplements, which occurs in more than half of U.S. households (61), could counter the lethal action of many antimicrobials (62). Effects of antioxidants on antimicrobial efficacy may merit clinical investigation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The E. coli K-12 strains listed in Table 1 were grown in LB liquid medium or on LB agar plates (63). Construction of E. coli mutant strains was performed by sequential antibiotic marker excision (64) and by bacteriophage P1-mediated transduction (65), the latter using phage prepared from E. coli deletion mutants in which the gene of interest was replaced by the in-frame insertion of a kanamycin resistance marker (66). Anaerobic growth was achieved by passing a Bioblend gas mixture (85% nitrogen, 5% carbon dioxide, and 10% hydrogen; obtained from GTS-Welco Gas Corp., Newark, NJ) through the cultures grown in LB broth supplemented with 1% glucose and 100 mM equimolar sodium phosphate buffer (pH 6.8) for 3 h as previously described (29). The Bioblend gas was also passed through antimicrobial solutions before being added to anaerobically growing cultures. For complementation experiments, pLEPA and its vector pWSK29 (67) were introduced into E. coli strains by bacterial transformation (68).
Chemicals and reagents.
Oxolinic acid, kanamycin, ampicillin, thiourea, hydrogen peroxide, and 2,2′-bipyridyl were obtained from Sigma Chemical Co. (St. Louis, MO). Ciprofloxacin hydrochloride monohydrate was obtained from Bayer AG (Wuppertal, Germany). Antibiotics were dissolved in distilled water (ampicillin to 100 mg/ml, kanamycin to 50 mg/ml, ciprofloxacin to 10 mg/ml) or 100 mM sodium hydroxide (oxolinic acid to 10 mg/ml). Thiourea (1 M) was dissolved in distilled water, and 2, 2′-bipyridyl (50 mM) was dissolved in dimethyl sulfoxide. Stock solutions of reagent were stored at −20°C for several weeks during experimentation.
Susceptibility measurements.
Growth inhibition, measured as the MIC, was determined by broth dilution in which two sets of staggered 2-fold drug concentration increments, offset by 50%, were used; each assay tube contained ~105 CFU. Growth was determined visually by the presence of turbidity.
Lethal action of antimicrobials was measured with cultured cells grown aerobically or anaerobically to mid-log phase (~5 × 108 CFU/ml) with shaking at about 250 rpm. Treated cultures were exposed to antimicrobials or hydrogen peroxide for various times and at various concentrations, diluted in drug-free medium, plated on drug-free agar, and incubated overnight at 37°C. Percent survival was estimated by colony formation relative to that of an untreated control sampled at the time of antimicrobial or peroxide addition. For UV-mediated killing, serial dilutions of E. coli cultures were applied as 10-µl aliquots to LB agar plates and exposed to UV light at 254 nm at the intensities and for the times indicated in the figure legends. To measure lethal effects of thermal treatment, serial dilutions of E. coli cultures were placed in a PCR Thermal Cycler (GeneAmp PCR system 9700; Applied Biosystems, Carlsbad, CA) as 50-µl aliquots and incubated at various temperatures for 10 min, after which samples were processed as in the antimicrobial killing assay described above. To measure the lethal activity of a low pH, cultured cells were grown in LB medium to about 2 × 108 CFU/ml, concentrated by centrifugation, resuspended in LB medium adjusted to pH 3 with HCl, and sampled for colony formation at various times. The effect of hydroxyl radical on antibiotic-mediated lethality was assessed by treating cultured cells grown to mid-log phase with a subinhibitory combination of 75 mM thiourea plus 0.6 mM 2,2′-bipyridyl (0.5 times the MIC for both agents) for 5 min and then with ampicillin (32 times the MIC) or ciprofloxacin (5 times the MIC) for various times. All experiments were repeated at least twice with similar results.
Determination of ROS accumulation.
E. coli cells were stained with H2DCFDA (Life Technologies, Grand Island, NY), a compound that is oxidized to green fluorescent dichlorofluorescein by ROS (69). For these experiments, E. coli strains were grown in liquid medium at 37°C to early exponential phase (~2 × 108 CFU/ml) and then H2DCFDA was added to 10 µM. Ampicillin was added to 5 times the MIC 20 min after H2DCFDA addition. Samples were then taken at 30-min intervals, and the mean fluorescence intensity was determined with a BD Accuri C6 flow cytometer (BD Biosciences, San Jose, CA). Quantification of fluorescence was taken as a measure of intracellular ROS accumulation.
SUPPLEMENTAL MATERIAL
Complementation of lepA deficiency-mediated protection from oxolinic acid and kanamycin lethality. Exponentially growing cultures of E. coli were treated with oxolinic acid for 2 h (A) or kanamycin for 45 min (B) at the concentrations indicated. Symbols: wild-type cells transformed with pWSK29 (strain 3684), empty circles; ΔlepA mutant cells transformed with pWSK29 (strain 3685), filled circles; wild-type cells transformed with pLEPA (strain 3682), empty squares; ΔlepA mutant cells transformed with pLEPA (strain 3683), filled squares. Error bars represent standard errors of the means. Download
Alternate electron acceptor has no effect on loss of protection from anaerobic antimicrobial killing by ΔlepA. (A) Ampicillin-mediated killing. E. coli cultures were grown in LB-glucose-phosphate medium (29) supplemented with 100 mM potassium nitrate anaerobically to exponential phase before they were treated with ampicillin at 32 times the MIC under anaerobic conditions for the times indicated. Samples were then taken, diluted, and applied to drug-free LB agar for recovery growth. Percent survival was calculated from the number of colonies recovered divided by the colony count of an untreated control sampled at the time of ampicillin addition. (B) Kanamycin-mediated killing. The conditions were the same as those described for panel A, except that (i) glucose and phosphate were omitted from the medium (glucose supplementation increases the kanamycin MIC by 128-fold) and (ii) kanamycin at 4 times the MIC (64 µg/ml) was used. Symbols: wild-type, empty circles; ΔlepA mutant, filled circles. Error bars represent standard errors of the means. Download
A deficiency in lepA suppresses ampicillin-mediated intracellular accumulation of ROS. Exponentially growing cultures of E. coli were treated with 10 µM H2DCFDA for 20 min before ampicillin (5 times the MIC) was added. Samples were then taken at 1, 1.5, and 2 h after ampicillin addition for analysis by flow cytometry. Lines: wild type, black; lepA mutant, red. Three replicate experiments gave similar results. The loss of ΔlepA-mediated ROS reduction after a long incubation time may be due to ampicillin-mediated cell lysis. Download
Bipyridyl plus thiourea suppresses ampicillin-mediated intracellular accumulation of ROS. Exponentially growing cultures of wild-type E. coli were treated with 10 µM H2DCFDA for 15 min. The culture was then split into two samples, one of which was treated with 0.5 times the MIC of 2,2′-bipyridyl plus thiourea for another 5 min before ampicillin at 5 times the MIC was added to both samples. Aliquots were then taken at 0.5 h after ampicillin addition for analysis by flow cytometry. Lines: wild type, black; wild type pretreated with 2,2′-bipyridyl plus thiourea, red. Three replicate experiments gave similar results. Download
Effects of MazF and ChpB toxins on ΔlepA-mediated protection from oxolinic acid-mediated killing. Exponentially growing cultures of E. coli were treated with various concentrations of oxolinic acid for 2 h. Percent survival was measured as described in Materials and Methods. (A) Effect of mazF deficiency. Symbols: wild type (strain 3001), empty circles; ΔlepA mutant (strain 3500), filled circles; ΔmazF mutant (strain 3496), empty squares; ΔmazF ΔlepA double mutant (strain 3497), filled squares. (B) Effect of chpB deficiency. Symbols: wild type, empty circles; ΔlepA mutant, filled circles; ΔchpB mutant (strain 2989), empty triangles; ΔchpB ΔlepA double mutant (strain 3688), filled triangles. Error bars represent standard errors of the means. Download
Combination of an ssrA deficiency with a ΔlepA mutation eliminates the ΔlepA-mediated reduction of ROS accumulation during exposure to ampicillin. Exponentially growing cultures of E. coli were treated with 10 µM H2DCFDA for 20 min before ampicillin (5 times the MIC) was added. Samples were then taken at 0.5 h after ampicillin addition for analysis by flow cytometry. Lines: wild type, black; ΔlepA mutant, red; ΔssrA mutant, blue line; ΔssrA ΔlepA double mutant, yellow line. Three replicate experiments gave similar results. Download
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (AI103781, AI073491, and 1DP2OD007423) and the National Natural Science Foundation of China (grant 81473251).
We thank Kurt Fredrick for the pWSK29 vector and plasmid pLEPA. We also thank Marila Gennaro, Richard Pine, and Issar Smith for critical comments on the manuscript.
Footnotes
Citation Li L, Hong Y, Luan G, Mosel M, Malik M, Drlica K, Zhao X. 2014. Ribosomal elongation factor 4 promotes cell death associated with lethal stress. mBio 5(6):e01708-14. doi:10.1128/mBio.01708-14.
REFERENCES
- 1. Dibb NJ, Wolfe PB. 1986. Lep operon proximal gene is not required for growth or secretion by Escherichia coli. J. Bacteriol. 166:83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pech M, Karim Z, Yamamoto H, Kitakawa M, Qin Y, Nierhaus KH. 2011. Elongation factor 4 (EF4/LepA) accelerates protein synthesis at increased Mg2+ concentrations. Proc. Natl. Acad. Sci. U. S. A. 108:3199–3203. 10.1073/pnas.1012994108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Qin Y, Polacek N, Vesper O, Staub E, Einfeldt E, Wilson DN, Nierhaus KH. 2006. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell 127:721–733. 10.1016/j.cell.2006.09.037. [DOI] [PubMed] [Google Scholar]
- 4. Zhang D, Qin Y. 2013. The paradox of elongation factor 4: highly conserved, yet of no physiological significance? Biochem. J. 452:173–181. 10.1042/BJ20121792. [DOI] [PubMed] [Google Scholar]
- 5. March PE, Inouye M. 1985. GTP-binding membrane protein of Escherichia coli with sequence homology to initiation factor 2 and elongation factors Tu and G. Proc. Natl. Acad. Sci. U. S. A. 82:7500–7504. 10.1073/pnas.82.22.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bauerschmitt H, Funes S, Herrmann JM. 2008. The membrane-bound GTPase Guf1 promotes mitochondrial protein synthesis under suboptimal conditions. J. Biol. Chem. 283:17139–17146. 10.1074/jbc.M710037200. [DOI] [PubMed] [Google Scholar]
- 7. Bijlsma JJ, Lie-A-Ling M, Nootenboom IC, Vandenbroucke-Grauls CM, Kusters JG. 2000. Identification of loci essential for the growth of Helicobacter pylori under acidic conditions. J. Infect. Dis. 182:1566–1569. 10.1086/315855. [DOI] [PubMed] [Google Scholar]
- 8. Shoji S, Janssen BD, Hayes CS, Fredrick K. 2010. Translation factor LepA contributes to tellurite resistance in Escherichia coli but plays no apparent role in the fidelity of protein synthesis. Biochimie 92:157–163. 10.1016/j.biochi.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang F, Li Z, Hao J, Qin Y. 2014. EF4 knockout E. coli cells exhibit lower levels of cellular biosynthesis under acidic stress. Protein Cell 5:563–567. 10.1007/s13238-014-0050-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garza-Sánchez F, Schaub RE, Janssen BD, Hayes CS. 2011. tmRNA regulates synthesis of the ArfA ribosome rescue factor. Mol. Microbiol. 80:1204–1219. 10.1111/j.1365-2958.2011.07638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Keiler KC, Waller PR, Sauer RT. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990–993. 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 12. Withey JH, Friedman DI. 2003. A salvage pathway for protein structures: tmRNA and trans-translation. Annu. Rev. Microbiol. 57:101–123. 10.1146/annurev.micro.57.030502.090945. [DOI] [PubMed] [Google Scholar]
- 13. Van Melderen L, Saavedra De Bast M. 2009. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 5:e1000437. 10.1371/journal.pgen.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu X, Wang X, Drlica K, Zhao X. 2011. A toxin-antitoxin module in Bacillus subtilis can both mitigate and amplify effects of lethal stress. PLoS One 6:e23909. 10.1371/journal.pone.0023909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamaguchi Y, Park JH, Inouye M. 2011. Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 45:61–79. 10.1146/annurev-genet-110410-132412. [DOI] [PubMed] [Google Scholar]
- 16. Dorsey-Oresto A, Lu T, Mosel M, Wang X, Salz T, Drlica K, Zhao X. 2013. YihE kinase is a central regulator of programmed cell death in bacteria. Cell Rep 3:528–537. 10.1016/j.celrep.2013.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, Inouye M. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12:913–923. 10.1016/S1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 18. Davies BW, Kohanski MA, Simmons LA, Winkler JA, Collins JJ, Walker GC. 2009. Hydroxyurea induces hydroxyl radical-mediated cell death in Escherichia coli. Mol. Cell 36:845–860. 10.1016/j.molcel.2009.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 20. Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. 2008. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135:679–690. 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang X, Zhao X. 2009. Contribution of oxidative damage to antimicrobial lethality. Antimicrob. Agents Chemother. 53:1395–1402. 10.1128/AAC.01087-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dwyer DJ, Belenky PA, Yang JH, MacDonald IC, Martell JD, Takahashi N, Chan CT, Lobritz MA, Braff D, Schwarz EG, Ye JD, Pati M, Vercruysse M, Ralifo PS, Allison KR, Khalil AS, Ting AY, Walker GC, Collins JJ. 2014. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. U. S. A. 111:E2100–E2109. 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu Y, Liu X, Qu Y, Wang X, Li L, Zhao X. 2012. Inhibitors of reactive oxygen species accumulation delay and/or reduce the lethality of several antistaphylococcal agents. Antimicrob. Agents Chemother. 56:6048–6050. 10.1128/AAC.00754-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gusarov I, Shatalin K, Starodubtseva M, Nudler E. 2009. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science 325:1380–1384. 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shatalin K, Shatalina E, Mironov A, Nudler E. 2011. H2S: a universal defense against antibiotics in bacteria. Science 334:986–990. 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- 26. Sampson TR, Liu X, Schroeder MR, Kraft CS, Burd EM, Weiss DS. 2012. Rapid killing of Acinetobacter baumannii by polymyxins is mediated by a hydroxyl radical death pathway. Antimicrob. Agents Chemother. 56:5642–5649. 10.1128/AAC.00756-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grant SS, Kaufmann BB, Chand NS, Haseley N, Hung DT. 2012. Eradication of bacterial persisters with antibiotic-generated hydroxyl radicals. Proc. Natl. Acad. Sci. U. S. A. 109:12147–12152. 10.1073/pnas.1203735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pandey R, Rodriguez GM. 2012. A ferritin mutant of Mycobacterium tuberculosis is highly susceptible to killing by antibiotics and is unable to establish a chronic infection in mice. Infect. Immun. 80:3650–3659. 10.1128/IAI.00229-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Malik M, Hussain S, Drlica K. 2007. Effect of anaerobic growth on quinolone lethality with Escherichia coli. Antimicrob. Agents Chemother. 51:28–34. 10.1128/AAC.00739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bass DA, Parce JW, Dechatelet LR, Szejda P, Seeds MC, Thomas M. 1983. Flow cytometric studies of oxidative product formation by neutrophils: a graded response to membrane stimulation. J. Immunol. 130:1910–1917. [PubMed] [Google Scholar]
- 31. Brandt R, Keston AS. 1965. Synthesis of diacetyldichlorofluorescin: a stable reagent for fluorometric analysis. Anal. Biochem. 11:6–9. 10.1016/0003-2697(65)90035-7. [DOI] [PubMed] [Google Scholar]
- 32. Yang F, Gao Y, Li Z, Chen L, Xia Z, Xu T, Qin Y. 2014. Mitochondrial EF4 links respiratory dysfunction and cytoplasmic translation in Caenorhabditis elegans. Biochim. Biophys. Acta 1837:1674–1683. 10.1016/j.bbabio.2014.05.353. [DOI] [PubMed] [Google Scholar]
- 33. Kolodkin-Gal I, Engelberg-Kulka H. 2009. The stationary-phase sigma factor sigma(s) is responsible for the resistance of Escherichia coli stationary-phase cells to mazEF-mediated cell death. J. Bacteriol. 191:3177–3182. 10.1128/JB.00011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kolodkin-Gal I, Sat B, Keshet A, Engelberg-Kulka H. 2008. The communication factor EDF and the toxin-antitoxin module mazEF determine the mode of action of antibiotics. PLOS Biol. 6:e319. 10.1371/journal.pbio.0060319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gottesman S, Roche E, Zhou Y, Sauer RT. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12:1338–1347. 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu H, Pan D, Pech M, Cooperman BS. 2010. Interrupted catalysis: the EF4 (LepA) effect on back-translocation. J. Mol. Biol. 396:1043–1052. 10.1016/j.jmb.2009.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu H, Chen C, Zhang H, Kaur J, Goldman YE, Cooperman BS. 2011. The conserved protein EF4 (LepA) modulates the elongation cycle of protein synthesis. Proc. Natl. Acad. Sci. U. S. A. 108:16223–16228. 10.1073/pnas.1103820108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gagnon MG, Lin J, Bulkley D, Steitz TA. 2014. Crystal structure of elongation factor 4 bound to a clockwise ratcheted ribosome. Science 345:684–687. 10.1126/science.1253525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ramrath DJ, Yamamoto H, Rother K, Wittek D, Pech M, Mielke T, Loerke J, Scheerer P, Ivanov P, Teraoka Y, Shpanchenko O, Nierhaus KH, Spahn CM. 2012. The complex of tmRNA-SmpB and EF-G on translocating ribosomes. Nature 485:526–529. 10.1038/nature11006. [DOI] [PubMed] [Google Scholar]
- 40. Yamamoto H, Qin Y, Achenbach J, Li C, Kijek J, Spahn CM, Nierhaus KH. 2014. EF-G and EF4: translocation and back-translocation on the bacterial ribosome. Nat. Rev. Microbiol. 12:89–100. 10.1038/nrmicro3176. [DOI] [PubMed] [Google Scholar]
- 41. Karzai AW, Roche ED, Sauer RT. 2000. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat. Struct. Biol. 7:449–455. 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- 42. Choy JS, Aung LL, Karzai AW. 2007. Lon protease degrades transfer-messenger RNA-tagged proteins. J. Bacteriol. 189:6564–6571. 10.1128/JB.00860-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Herman C, Thévenet D, Bouloc P, Walker GC, D’Ari R. 1998. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH). Genes Dev. 12:1348–1355. 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang X, Zhao X, Malik M, Drlica K. 2010. Contribution of reactive oxygen species to pathways of quinolone-mediated bacterial cell death. J. Antimicrob. Chemother. 65:520–524. 10.1093/jac/dkp486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Malik M, Zhao X, Drlica K. 2006. Lethal fragmentation of bacterial chromosomes mediated by DNA gyrase and quinolones. Mol. Microbiol. 61:810–825. 10.1111/j.1365-2958.2006.05275.x. [DOI] [PubMed] [Google Scholar]
- 46. Zhao X, Hong Y, Drlica K. Moving forward with reactive oxygen species involvement in antimicrobial lethality. J. Antimicrob. Chemother., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Keren I, Wu Y, Inocencio J, Mulcahy LR, Lewis K. 2013. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339:1213–1216. 10.1126/science.1232688. [DOI] [PubMed] [Google Scholar]
- 48. Liu Y, Imlay JA. 2013. Cell death from antibiotics without the involvement of reactive oxygen species. Science 339:1210–1213. 10.1126/science.1232751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ezraty B, Vergnes A, Banzhaf M, Duverger Y, Huguenot A, Brochado AR, Su SY, Espinosa L, Loiseau L, Py B, Typas A, Barras F. 2013. Fe-S cluster biosynthesis controls uptake of aminoglycosides in a ROS-less death pathway. Science 340:1583–1587. 10.1126/science.1238328. [DOI] [PubMed] [Google Scholar]
- 50. Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat. Rev. Microbiol. 8:423–435. 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lewin CS, Morrissey I, Smith JT. 1991. The mode of action of quinolones: the paradox in activity of low and high concentrations and activity in the anaerobic environment. Eur. J. Clin. Microbiol. Infect. Dis. 10:240–248. [DOI] [PubMed] [Google Scholar]
- 52. Ling J, Cho C, Guo LT, Aerni HR, Rinehart J, Söll D. 2012. Protein aggregation caused by aminoglycoside action is prevented by a hydrogen peroxide scavenger. Mol. Cell 48:713–722. 10.1016/j.molcel.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhao X, Drlica K. 2014. Reactive oxygen species and the bacterial response to lethal stress. Curr. Opin. Microbiol. 21C:1–6. 10.1016/j.mib.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dwyer DJ, Collins JJ, Walker GC. 10 September 2014. Unraveling the physiological complexities of antibiotic lethality. Annu. Rev. Pharmacol. Toxicol. 10.1146/annurev-pharmtox-010814-124712. [DOI] [PubMed] [Google Scholar]
- 55. Burger RM, Drlica K. 2009. Superoxide protects Escherichia coli from bleomycin mediated lethality. J. Inorg. Biochem. 103:1273–1277. 10.1016/j.jinorgbio.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mosel M, Li L, Drlica K, Zhao X. 2013. Superoxide-mediated protection of Escherichia coli from antimicrobials. Antimicrob. Agents Chemother. 57:5755–5759. 10.1128/AAC.00754-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wu Y, Vulić M, Keren I, Lewis K. 2012. Role of oxidative stress in persister tolerance. Antimicrob. Agents Chemother. 56:4922–4926. 10.1128/AAC.00921-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Christensen SK, Pedersen K, Hansen FG, Gerdes K. 2003. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332:809–819. 10.1016/S0022-2836(03)00922-7. [DOI] [PubMed] [Google Scholar]
- 59. Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. 2006. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2:e135. 10.1371/journal.pgen.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tsilibaris V, Maenhaut-Michel G, Mine N, Van Melderen L. 2007. What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J. Bacteriol. 189:6101–6108. 10.1128/JB.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Radimer K, Bindewald B, Hughes J, Ervin B, Swanson C, Picciano MF. 2004. Dietary supplement use by US adults: data from the National Health and Nutrition Examination Survey, 1999-2000. Am. J. Epidemiol. 160:339–349. 10.1093/aje/kwh207. [DOI] [PubMed] [Google Scholar]
- 62. Marathe SA, Kumar R, Ajitkumar P, Nagaraja V, Chakravortty D. 2013. Curcumin reduces the antimicrobial activity of ciprofloxacin against Salmonella typhimurium and Salmonella typhi. J. Antimicrob. Chemother. 68:139–152. 10.1093/jac/dks375. [DOI] [PubMed] [Google Scholar]
- 63. Miller J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 64. Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. 10.1016/0378-1119(95)00193-A. [DOI] [PubMed] [Google Scholar]
- 65. Wall JD, Harriman PD. 1974. Phage P1 mutants with altered transducing abilities for Escherichia coli. Virology 59:532–544. 10.1016/0042-6822(74)90463-2. [DOI] [PubMed] [Google Scholar]
- 66. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio Collection. Mol. Syst. Biol. 2:0008. 10.1038/msb41000502006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene-expression in Escherichia coli. Gene 100:195–199. 10.1016/0378-1119(91)90366-J. [DOI] [PubMed] [Google Scholar]
- 68. Cohen SN, Chang AC, Hsu L. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. U. S. A. 69:2110–2114. 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Eruslanov E, Kusmartsev S. 2010. Identification of ROS using oxidized DCFDA and flow-cytometry. Methods Mol. Biol. 594:57–72. 10.1007/978-1-60761-411-1_4. [DOI] [PubMed] [Google Scholar]
- 70. Abo T, Ueda K, Sunohara T, Ogawa K, Aiba H. 2002. SsrA-mediated protein tagging in the presence of miscoding drugs and its physiological role in Escherichia coli. Genes Cells 7:629–638. 10.1046/j.1365-2443.2002.00549.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complementation of lepA deficiency-mediated protection from oxolinic acid and kanamycin lethality. Exponentially growing cultures of E. coli were treated with oxolinic acid for 2 h (A) or kanamycin for 45 min (B) at the concentrations indicated. Symbols: wild-type cells transformed with pWSK29 (strain 3684), empty circles; ΔlepA mutant cells transformed with pWSK29 (strain 3685), filled circles; wild-type cells transformed with pLEPA (strain 3682), empty squares; ΔlepA mutant cells transformed with pLEPA (strain 3683), filled squares. Error bars represent standard errors of the means. Download
Alternate electron acceptor has no effect on loss of protection from anaerobic antimicrobial killing by ΔlepA. (A) Ampicillin-mediated killing. E. coli cultures were grown in LB-glucose-phosphate medium (29) supplemented with 100 mM potassium nitrate anaerobically to exponential phase before they were treated with ampicillin at 32 times the MIC under anaerobic conditions for the times indicated. Samples were then taken, diluted, and applied to drug-free LB agar for recovery growth. Percent survival was calculated from the number of colonies recovered divided by the colony count of an untreated control sampled at the time of ampicillin addition. (B) Kanamycin-mediated killing. The conditions were the same as those described for panel A, except that (i) glucose and phosphate were omitted from the medium (glucose supplementation increases the kanamycin MIC by 128-fold) and (ii) kanamycin at 4 times the MIC (64 µg/ml) was used. Symbols: wild-type, empty circles; ΔlepA mutant, filled circles. Error bars represent standard errors of the means. Download
A deficiency in lepA suppresses ampicillin-mediated intracellular accumulation of ROS. Exponentially growing cultures of E. coli were treated with 10 µM H2DCFDA for 20 min before ampicillin (5 times the MIC) was added. Samples were then taken at 1, 1.5, and 2 h after ampicillin addition for analysis by flow cytometry. Lines: wild type, black; lepA mutant, red. Three replicate experiments gave similar results. The loss of ΔlepA-mediated ROS reduction after a long incubation time may be due to ampicillin-mediated cell lysis. Download
Bipyridyl plus thiourea suppresses ampicillin-mediated intracellular accumulation of ROS. Exponentially growing cultures of wild-type E. coli were treated with 10 µM H2DCFDA for 15 min. The culture was then split into two samples, one of which was treated with 0.5 times the MIC of 2,2′-bipyridyl plus thiourea for another 5 min before ampicillin at 5 times the MIC was added to both samples. Aliquots were then taken at 0.5 h after ampicillin addition for analysis by flow cytometry. Lines: wild type, black; wild type pretreated with 2,2′-bipyridyl plus thiourea, red. Three replicate experiments gave similar results. Download
Effects of MazF and ChpB toxins on ΔlepA-mediated protection from oxolinic acid-mediated killing. Exponentially growing cultures of E. coli were treated with various concentrations of oxolinic acid for 2 h. Percent survival was measured as described in Materials and Methods. (A) Effect of mazF deficiency. Symbols: wild type (strain 3001), empty circles; ΔlepA mutant (strain 3500), filled circles; ΔmazF mutant (strain 3496), empty squares; ΔmazF ΔlepA double mutant (strain 3497), filled squares. (B) Effect of chpB deficiency. Symbols: wild type, empty circles; ΔlepA mutant, filled circles; ΔchpB mutant (strain 2989), empty triangles; ΔchpB ΔlepA double mutant (strain 3688), filled triangles. Error bars represent standard errors of the means. Download
Combination of an ssrA deficiency with a ΔlepA mutation eliminates the ΔlepA-mediated reduction of ROS accumulation during exposure to ampicillin. Exponentially growing cultures of E. coli were treated with 10 µM H2DCFDA for 20 min before ampicillin (5 times the MIC) was added. Samples were then taken at 0.5 h after ampicillin addition for analysis by flow cytometry. Lines: wild type, black; ΔlepA mutant, red; ΔssrA mutant, blue line; ΔssrA ΔlepA double mutant, yellow line. Three replicate experiments gave similar results. Download