ABSTRACT
Understanding the structure, functions, activities and dynamics of microbial communities in natural environments is one of the grand challenges of 21st century science. To address this challenge, over the past decade, numerous technologies have been developed for interrogating microbial communities, of which some are amenable to exploratory work (e.g., high-throughput sequencing and phenotypic screening) and others depend on reference genes or genomes (e.g., phylogenetic and functional gene arrays). Here, we provide a critical review and synthesis of the most commonly applied “open-format” and “closed-format” detection technologies. We discuss their characteristics, advantages, and disadvantages within the context of environmental applications and focus on analysis of complex microbial systems, such as those in soils, in which diversity is high and reference genomes are few. In addition, we discuss crucial issues and considerations associated with applying complementary high-throughput molecular technologies to address important ecological questions.
Minireview
Microorganisms inhabit almost every imaginable environment in the biosphere, play integral and unique roles in ecosystems, and are involved in the biogeochemical cycling of essential elements, such as carbon, oxygen, nitrogen, sulfur, phosphorus, and various metals. Their structure, function, interaction, and dynamics are critical to our existence, yet their detection, identification, characterization, and quantification pose several great challenges. First, microbial communities can be extremely diverse, and the majority of microorganisms in natural environments have not yet been cultivated (1, 2). Second, in any ecosystem, various microorganisms interact with each other to form complicated networks whose behavior is hard to predict (3, 4). Establishing mechanistic linkages between microbial diversity and ecosystem functioning adds an additional challenge to understanding the interactions and activities of complex microbial communities (5, 6). Effective high-throughput technologies for analyzing microbial community structure and functions are critical for advancing this mechanistic understanding.
Sequencing and phylogenetic analysis of 16S rRNA genes provided the foundation for modern study of microbial communities. PCR-based 16S rRNA cloning analysis has driven the explosion of information about community memberships and vastly expanded the known diversity of microbial life (7). PCR-based analyses of 16S rRNA genes have three major limitations: (i) PCR limits the information obtained to the sequence between the primers, thereby disregarding functional information; (ii) PCR-based analysis is only somewhat quantitative, with most measurements providing only relative abundance information; and (iii) PCR primer mismatches may result in some lineages being missed entirely (8). All three challenges have been addressed by the development of metagenomic analyses involving direct sequencing or screening of unamplified environmental DNA (9–12). These methods constitute critical “open formats,” which do not require prior knowledge of the community, thereby enabling unprecedented discovery of new taxa and genes and associations between them.
Analysis of cloned DNA has largely been replaced by next-generation sequencing of DNA extracted from environmental sources, which has transformed the field of microbial ecology by increasing the speed and throughput of DNA sequencing by orders of magnitude. Now the metagenomic databases are packed with high-quality sequence information from diverse habitats across the globe, revolutionizing molecular analyses of biological systems (13, 14) and facilitating research on questions that formerly could not be approached. Although functional metagenomics, in which clones containing metagenomic DNA are screened for expressed activities, holds great promise to shape ecological theory and understanding, it has lagged behind shotgun sequencing because of the comparatively slow advances in screening technology. Ecological insights from the massive data sets generated by high-throughput sequencing (open formats) have been facilitated by sophisticated computational methods and by closed-format methods, such as microarrays, which can be used to rapidly query taxa, genes, or transcripts over space and time in complex communities.
High-throughput sequencing and microarray technologies have been applied to diverse communities. The plethora of research using these methods has stimulated several excellent reviews (15–17), particularly as applied to the human microbiome (18–20). Our intent here is to complement previous reviews by focusing primarily on DNA-based metagenomic technologies applied to complex environmental communities, such as those found in soils.
OVERVIEW OF OPEN AND CLOSED MOLECULAR DETECTION APPROACHES
Since 1990, various molecular methods capable of tracking one to hundreds of biomarkers have been widely used to analyze microbial community structure, such as PCR amplification-based gene cloning, sequencing of 16S rRNA genes (21) and functional genes (22), amplified ribosomal DNA restriction analysis (23), denaturing gradient gel electrophoresis (24), terminal restriction fragment length polymorphism (25), phospholipid fatty acid analysis (26), and BioLog EcoPlates for measuring carbon and nitrogen metabolisms (27). Especially in the last decade, high-throughput molecular technologies capable of tracking multiple thousands of biomarkers have been developed for characterizing microbial communities, including high-throughput DNA/RNA sequencing (18, 28–31), PhyloChip (32), GeoChip (33), mass spectrometry-based proteomics for community analysis (34), and metabolite analysis (35).
We can group high-throughput molecular microbial detection technologies into two major categories: open and closed formats (16, 17). “Open format” refers to technologies whose potential experimental results cannot be anticipated prior to performing the analysis, and thus, the experimental outcome is considered open. For instance, when using sequencing to analyze a microbial community, we will not know what types of sequences will be obtained prior to sequencing. The main characteristics of technologies of this type are that they typically do not require a priori sequence information from the community of interest (16, 17) and, overall, they enable discovery of new genes, pathways, and taxa (Table 1). This category includes a variety of molecular techniques, such as high-throughput sequencing technologies, screening for functional expression, fingerprinting methods, and mass spectrometry-based proteomic and metabolomic approaches.
TABLE 1 .
Key differences among open and closed high-throughput platforms for microbial community analysisa
| Step or parameter | Characteristic or consideration | Description of characteristic or consideration in indicated type of analysisb |
Comments | ||||
|---|---|---|---|---|---|---|---|
| Open format |
Closed format |
||||||
| TGS | SMS | MTS | FGAs | PGAs | |||
| Sample preparation and analysis | Sample/target preparation | Complicated | Simple | Very complicated | Simple | Simple | DNA/RNA quality is important for all approaches |
| Analysis of multiplex samples per assay | Large potential | Medium potential | Medium potential | Low (only one or two) | Low (only one or two) | FGAs and PGAs use 1 or 2 dyes for labeling, and it is difficult to multiplex samples in a single assay | |
| PCR amplification or whole-genome analysis | Yes | No | No | No/yes | Yes/no | Amplification introduces major problems for quantification | |
| Potential uneven hybridization | NA | NA | NA | Yes | Yes | Signal normalization is needed within and between arrays to correct signal differences due to systematic errors | |
| Data processing and analysis | Raw data processing | Relatively easy | Difficult | Difficult | Easy | Easy | A major challenge for SMS and MTS with large raw datasets |
| Phylogeny | Yes | Some | Some | No/yes | Yes | GeoChip uses gyrB for phylogeny | |
| Taxonomic resolution | Strain, species, genus | Strain, species | Strain, species | Strain, species | Genus, family | It depends on molecular markers with high resolution for functional genes | |
| Functional features | No/yes | Yes | Yes | Yes | No | TGS can analyze DNA and RNA for functional genes | |
| Signal threshold | Yes | NA | NA | Yes | Yes | Both PGAs and FGAs require a threshold to call positive signals, which is more or less arbitrary. Thus, some ambiguity exists for positive or negative spots. | |
| Requires a priori knowledge | No/yes | No | No | Yes | Yes | Closed-format technologies are designed based on known sequences | |
| Analysis of α diversity | Very good | Good | Very poor | Fair | Fair | Here, α diversity estimation is based on a single gene | |
| Data comparison across samples | Moderate | Difficult | Difficult | Easy | Easy | Random or undersampling is a major issue for open-format approaches | |
| Performance | Coverage/breadth (no. of different genes detected) | Very low | High | High | High | Very low | TGS can analyze phylogenetic or functional genes |
| Sampling depth (no. of sequences or OTUs per gene) | Very high | Low/medium | Low/medium | Medium | High | The sampling depth for closed-format approaches depends on the number of probes used | |
| Detection of rare species/genes | Medium | Difficult | Difficult | Easy | Easy | Easy for closed format as long as the appropriate probes are present | |
| Quantification | Low | Not known | Not known | High | Low/medium | Not rigorously tested for SMS and MTS; for PhyloChip, if RNA is used instead of DNA (no PCR step), quantification is high | |
| Susceptibility to the artifacts associated with random sampling process | Medium | High | High | Low | Medium/low | A major problem for sequencing approaches; PCR amplification may be involved in PhyloChip | |
| Potential discovery of novel genes/species | Yes | Yes | Yes | No | No | ||
| Results skewed by dominant populations | Yes | Yes | Yes | No | No | ||
| Sensitivity to (host) DNA/RNA contamination | No/yes | Yes | Yes | No | No | Difficult to remove host DNA/RNA contamination | |
| Applicability and cost | Most promising applications | In-depth studies of microbial diversity or specific functional groups and discovery of novel genes | Surveys of microbial genetic diversity of unknown communities and discovery of novel genes | Surveys of functional activity of unknown microbial communities and discovery of novel genes | Comparisons of functional diversity and structure of microbial communities across many samples | Comparisons of taxonomic or phylogenetic diversity and structure of microbial communities across many samples | The choice of technology mainly depends on the biological questions and hypotheses to be addressed |
| Relative cost per assay | Medium | High | High | Low | Low | It is challenging to make general statements of cost because they depend on technology platforms, depth of analysis, and approaches used for processing and analyzing data | |
| Cost per sample ($) | 30–150 | 1200–4000 | 1500–4500 | 150–800 | 150–1000 | This is only based on the cost of materials for target gene amplicon preparations and sequencing. | |
| Cost for bioinformatic analysis | Medium | High | High | Low | Low | ||
Since various technologies have different features, it is difficult to make straightforward, point-by-point direct comparison. Thus, our attempt is to highlight the major differences of various technologies in a general sense. We attempt to focus on the issues important to microbial ecology within the context of environmental applications and complex microbial communities like those in soil rather than list the differences of various technologies in a comprehensive manner.
TGS, target gene (e.g., 16S rRNA, amoA, nifH) sequencing; SMS, shotgun metagenome sequencing; MTS, metatranscriptome sequencing; FGAs, functional gene arrays: the listed analysis is mostly based on GeoChip; PGAs, phylogenetic gene arrays: the listed analysis is mostly based on PhyloChip; NA, not applicable.
“Closed format” refers to the detection technologies whose range of potential experimental results is defined prior to performing the analysis, and thus, the experimental outcome is considered closed. For example, when a functional gene array containing 10,000 probes is used for analyzing a microbial community, the experimental results from this sample cannot go beyond the detection capability of the probes (10,000) fabricated on the array. The main features of technologies of this type are that they require a priori sequence information (16, 17) and they do not provide new molecular information because all molecules used for designing the querying devices are known. DNA arrays (32, 33), protein arrays (36), carbohydrate arrays (37), phenotype arrays (38), and BioLog EcoPlates (27), as well as quantitative PCR, are all considered closed-format technologies.
Open- and closed-format technologies typically differ in sample preparation and quality control, data processing and analysis, performance, and application (Table 1), and each presents advantages and limitations. In the following discussion, we compare features important to meaningful applications of each platform by giving special consideration to their usefulness in analyzing complex microbial communities like those in soils. Since next-generation sequencing and microarrays are the best and most widely used representatives of open- and closed-format technologies, respectively, our comparison and discussion are primarily focused on these technologies.
SEQUENCING-BASED HIGH-THROUGHPUT MOLECULAR TECHNOLOGIES FOR MICROBIAL COMMUNITY ANALYSIS.
Sequencing technologies and applications.
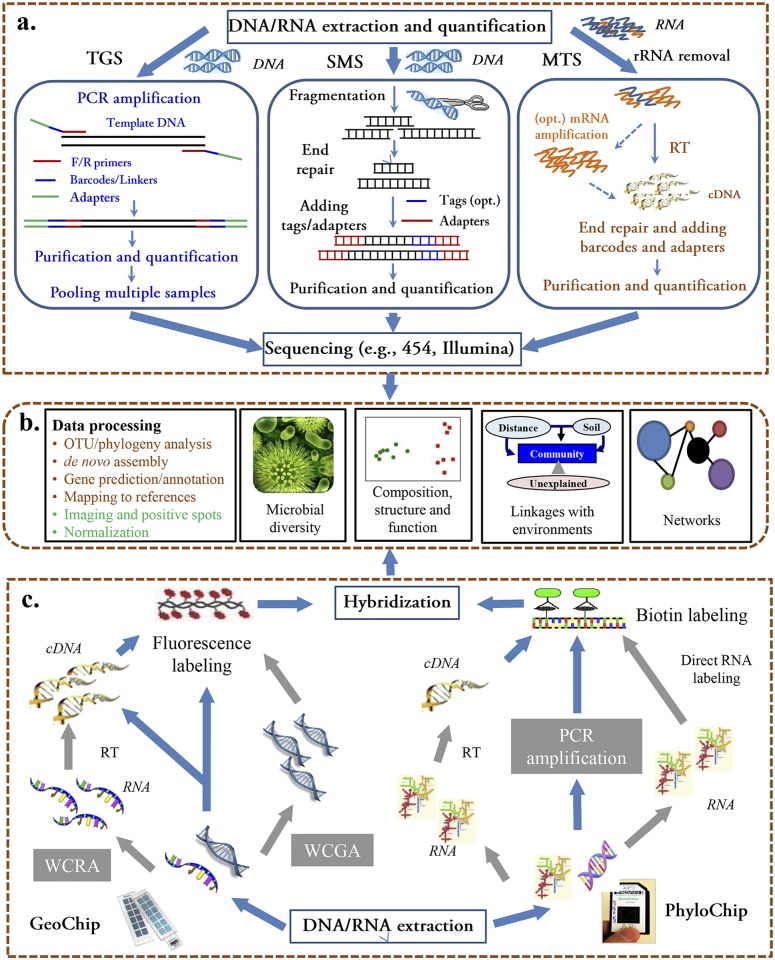
Several high-throughput sequencing platforms have been developed and are widely used, including the Illumina (e.g., HiSeq, MiSeq), Roche 454 GS FLX+, SOLiD 5500 series, and Ion Torrent/Ion Proton platforms. The advantages and limitations of these platforms are detailed elsewhere (18, 29, 30, 39–42). Currently, the majority of microbial ecology studies apply high-throughput sequencing by focusing on either targeted gene sequencing with phylogenetic (e.g., 16S rRNA) (29, 43) or functional (e.g., amoA, nifH) (44, 45) gene targets or on shotgun metagenome sequencing (Fig. 1a). For targeted gene sequencing, community DNA is extracted from environmental samples (e.g., samples from soils, sediments, water, bioreactors, or humans) using various extraction and purification methods (46, 47). After high-quality DNA is obtained, targeted genes can be amplified with conserved primers. Each set of primers is generally barcoded with short oligonucleotide tags (6- to 12-mer), as well as sequencing adapters, so that multiple samples can be pooled and sequenced simultaneously (29, 43). Then, after nontarget DNA fragments are removed by gel electrophoresis, target DNA is quantified, sequenced, and analyzed using bioinformatic approaches, such as operational taxonomic unit (OTU) assignment, sequence assembly, phylogeny, and annotation (Fig. 1a) (41).
FIG 1 .
Key steps of high-throughput metaomic technologies for microbial community analysis. (a) Sequencing-based open-format technologies. Extracted DNA/RNA samples are prepared for sequencing by target gene sequencing (TGS), shotgun metagenome sequencing (SMS), and/or metatranscriptome sequencing (MTS). RT, reverse transcription. (b) Data processing and analysis. Both sequencing- and microarray-based data are processed and then statistically analyzed to address specific microbial ecology questions related to community diversity, composition, structure, function, and network, as well as their linkages with environmental factors. (c) Array-based closed-format technologies. For the GeoChip and PhyloChip, extracted DNA is directly labeled and hybridized, while RNA is first reverse transcribed (RT) to cDNA. DNA and RNA can be amplified by whole-community genome amplification (WCGA) or by whole-community RNA amplification (WCRA), respectively, when there is not enough mass for direct hybridization, but this compromises quantification. Images from both arrays are digitized for further data processing and statistical analysis.
Although targeted gene sequencing is a powerful tool for providing information on specific genes within a microbial community, its suitability for analyzing the whole genetic and functional diversity of communities is limited (18). To query broader characteristics and identify novel genes, shotgun metagenome sequencing has been widely used (10, 28, 48–50). Briefly, community DNA is randomly sheared using various methods, including nebulization, endonucleases, or sonication (Fig. 1a) (40). The sheared fragments are end repaired prior to ligation to platform-specific adaptors, which serve as the priming sites for template amplification (40). A transposon-based approach for simultaneous fragmentation and tagging has also become available (40). Subsequent sequencing produces vast amounts of short reads, which can be assembled and annotated for functional characterization (41, 51). The shotgun metagenomic sequencing approach provides community-level information in complex environments with thousands to millions of different archaeal, bacterial, and eukaryotic species (52, 53), such as soil (49), ocean (10, 28), groundwater (54), cow rumen (50), and human microbiome (48), although short read sequences from complex communities cannot always be assembled and only a fraction may be useful for functional or phylogenetic analyses.
Targeted and shotgun sequencing of DNA provide snapshots of the gene content and genetic diversity of microbial communities but cannot distinguish between expressed and nonexpressed genes in a given environment. In contrast, metatranscriptomic sequencing (i.e., metatranscriptomics) involves random sequencing of expressed microbial community RNA (Fig. 1a) (31, 55–57). Typically, total RNA extracted from microbial communities is dominated by rRNA, which must be removed to obtain high levels of mRNA transcripts (55, 58). Then, the remaining RNAs are reverse transcribed into cDNAs, ligated to adapters, and sequenced (Fig. 1a) (55, 58). Metatranscriptomic studies have provided insight into microbial community functions and activities from diverse habitats, including soil (59), sediment (60), seawater (31, 57), gut microbiomes (61, 62), and activated sludge (63). However, major challenges include the inherent lability of mRNA, requiring proper nucleic acid stabilization and storage procedures to obtain sufficient quantities of high-quality mRNA. Furthermore, mRNA is still one or more steps away from actualized microbial community functions. Therefore, the further development of proteomics and metabolomics is important to understand microbial community functions in the environment.
Key features of sequencing-based open-format detection technologies.
One of the most appealing features of the sequencing-based open-format approaches is that they are ideal for novel discovery (Table 1). Many new genes, phylotypes, regulators, and/or pathways have been discovered using shotgun metagenome sequencing (48–50, 64). For example, in cow rumen samples, 15 uncultured microbial genomes involved in biomass decomposition were reconstructed along with 27,755 putative carbohydrate-active genes, dozens of which were demonstrated to exhibit carbohydrate-degrading activity despite a <55% average amino acid similarity to known proteins (50). Based on mate-paired short-read oceanic metagenomes, the genome of an uncultured member of a novel class of marine photoheterotrophic Euryarchaeota was reconstructed (65). Sequence analyses of this genome also suggested that proteorhodopsin (28, 66, 67) appears to be of euryarchaeal origin.
Metatranscriptomics has also provided new insights into microbial community activities and functions, as well as discovery of novel genes and regulatory elements. For example, the first metatranscriptomic analysis of seawater communities demonstrated that this technique is capable of detecting novel gene- and taxon-specific expression patterns and led to the discovery of novel gene categories undetected in previous DNA-based surveys (31). Subsequently, Shi et al. employed metatranscriptomics to discover well known small RNAs and previously unrecognized putative small RNAs in the ocean’s water column (57). More recently, Haroon et al. (68) used a combination of metagenomics and metatranscriptomics to demonstrate a novel archaeal pathway for anaerobic oxidation of methane coupled with nitrate reduction in an anaerobic bioreactor.
Another distinguishing characteristic of the sequencing-based open-format approaches is in the assessment of α and γ diversity. While α diversity is the diversity within a particular area or ecosystem, which is usually expressed as the number of taxa and abundance of each taxon within a community, γ diversity refers to the overall total diversity of taxa/genes for the different ecosystems within a region. Since new genes and taxa can be detected by sequencing-based open-format technologies, deep sequencing of phylogenetically informative genes (e.g., 16S rRNA) or functional genes (e.g., nifH, amoA) is more suitable for estimating α and γ diversity of microbial communities at the whole-community level or functional-population level. With current high-throughput technologies, it is possible to recover substantial portions of the microbial diversity in complex communities, even if only a few samples are analyzed. For instance, deep pyrosequencing analysis of amoA gene fragments in soil communities identified novel amoA sequences and previously undiscovered phylogenetic lineages (44, 45). In addition, many samples can be multiplexed for analysis in a single assay by targeted gene sequencing (29), and so, the experimental cost per single assay or per sample can be very low for this technique (Table 1).
There are distinct differences between targeted and shotgun metagenome sequencing approaches in terms of sample preparation, sequence output, and data analysis (Fig. 1a), and some of these differences are particularly important for microbial ecology research (Table 1). Targeted sequencing can provide greater depth of coverage for specific gene(s) of interest (e.g., nifH), while shotgun metagenome sequencing captures information about the community as a whole, as well as divergent homologs not captured by the primers employed.
Functional metagenomics can be treated as another open-format approach that does not presuppose or require sequence information, providing the opportunity for novel discovery and representing a powerful complement to shotgun sequencing. This approach involves screening cloned DNA for expressed functional activity in a surrogate host cell (12, 69, 70). Given that the majority of genes in most metagenomic databases do not have homologs with biochemically characterized functions, the opportunity for discovery in the metagenomic sequence space is vast. Although this approach has been used to successfully discover new biosynthetic enzymes (71), degradative enzymes (11, 72), and antibiotics (73, 74), active clones are typically identified at low frequency (typically 1 clone in 10,000 to 100,000 is active). Selective screening, e.g., using antibiotic resistance, can facilitate screening libraries containing 107 or more clones. Functional metagenomic studies of antibiotic resistance in soil (70, 75, 76), water (77), and insect, bird, pig, cow, and human microbiomes (78–80) have yielded a new understanding of the genes encoding antibiotic resistance in natural and managed environments and provide the basis for comparing frequencies of antibiotic resistance among habitats.
Challenges and limitations associated with open-format techniques.
The open-format techniques described above each have their challenges. Some of the major technical challenges for targeted gene sequencing are bias caused by PCR amplification (81–84), sequencing errors, and chimeric sequences (83, 85, 86). In one study, based on 90 identical mock community samples, the average error rate in 16S rRNA pyrotag sequences was 0.6%, and the chimera rate was 8% (83). Sequencing errors have been reduced 30-fold (from 0.6 to 0.02%) by the use of effective sequence analysis pipelines (83, 86). Recently, low-error amplicon sequencing approaches have been developed for human and plant microbiome studies (87, 88). Although the sequencing errors and chimera rates are less problematic for analyses based on assembled sequences, due to sequence overlap and redundancy, they are challenging in studies based on individual sequence reads (83). Sequence errors and chimeras can generate numerous spurious OTUs, which can inflate community diversity estimates by as much as 2 orders of magnitude (16, 82, 86). There is an intense debate regarding how much of the “rare biosphere” is due to sequencing artifacts (43, 82, 83). Thus, great caution and attention to denoising the data are needed when using high-throughput sequencing technologies for estimating microbial community diversity.
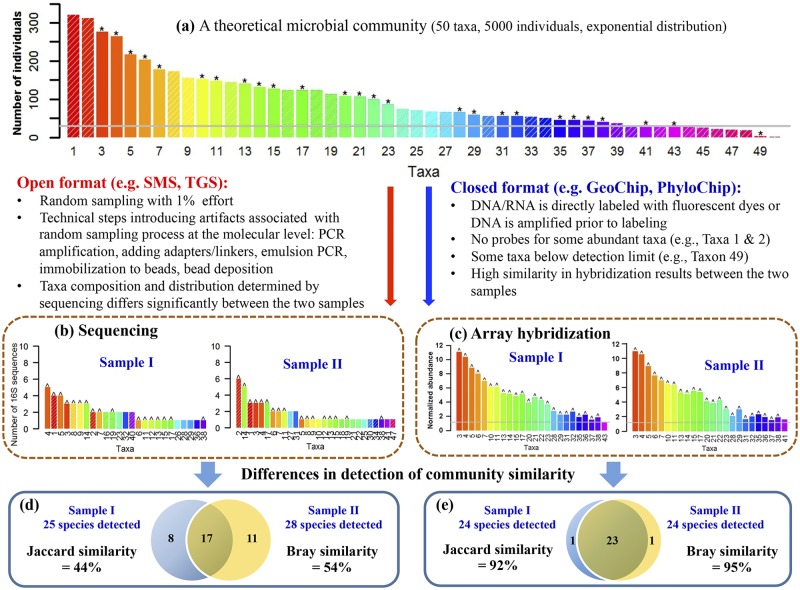
Another technical challenge for amplicon-based sequencing approaches can be low reproducibility (84, 85, 89–93, 163–165) and poor quantitation (89) due to the artifacts associated with inadequate random sampling (89, 94, 95), amplification biases (82, 83), and/or sequencing errors (83). For example, the subset of 16S molecules that are amplified and the subset of tagged amplified fragments that are attached to the surface of the flow cell (e.g., Illumina) or allocated to beads (e.g., 454) for sequencing is totally random and follows a Poisson random sampling distribution (95). How such artifacts associated with inadequate molecular-level sampling can lead to low technical reproducibility was described with an analogy to reading random words in a book (96) and explicitly demonstrated by recent mathematical modeling and simulations (95). To better visualize the potential differential effects of inadequate random sampling on open- and closed-format detection, it is useful to consider a hypothetical community. We assume that such a microbial community has 50 exponentially distributed taxa with 5,000 individuals (or 16S rRNA molecules) (Fig. 2a), and the community is sampled twice with 1% effort (i.e., 50 individuals) as technical replicates (Fig. 2b). Due to the molecular-level random sampling artifacts generated by insufficient sequences to represent all taxa, the taxon membership and abundance distribution are quite different between these two samples even though they are from the same community (Fig. 2b). Based on mathematical simulation, the overlap between these two samples is approximately 50% (Fig. 2d), which is consistent with experimental observations (84, 85, 89–93). However, as the sampling effort increases, the overlap between samples increases, achieving 95% overlap between two samples with ~20% of the community sampled. If all individuals are effectively sampled, erasing all the random sampling artifacts, 100% overlap is theoretically expected. For one real soil microbial community, on average, more than 60,000 16S rRNA sequences per sample were needed to achieve 90% OTU overlap among three technical replicates (95). Due to artifacts associated with inadequate random sampling, PCR amplification biases, chimeras, and/or sequencing errors, amplicon-based target sequencing is not considered quantitative (85, 89). This is consistent with the results of previous pyrotag sequencing studies (81) and with a general consensus that conventional PCR amplification of the template can introduce significant biases and artifacts (97).
FIG 2 .
Illustration of random sampling processes and their impacts on the analysis of microbial communities using open- and closed-format metagenomic technologies. (a) A theoretical community contains 50 taxa with 5,000 individuals and follows exponential distribution, λe−λx (λ = 0.01 in this case). The taxa are ranked based on their abundance. Two technical replicates of this community are taken for analysis (sample I and sample II). Also, assume that a microarray is constructed, covering about 50% of the taxa, as indicated by asterisks (*). (b) For sequencing, 1% sampling effort is performed. Overlapping taxa detected in the two samples are indicated by carets (^). (c) The community DNA is directly labeled and hybridized with the microarrays. Because some populations are below the detection limit, only certain portions are detected. Overlapping taxa detected in the two samples are also indicated by carets (^). In both cases (b and c), similar numbers of taxa were detected. (d and e) Jaccard and Bray-Curtis overlaps for the open- and closed-format technologies.
Targeted sequencing of functional genes can provide important functional gene information from microbial communities (45, 98); however, there are several challenges associated with this approach. First, widespread lack of sequence conservation across functionally homologous genes can make PCR primer design difficult, leading to lack of detection of relevant functional genes in the environment. Second, even though fairly conserved primers can be designed for some functional genes of interest (e.g., amoA, nifH, nirS, nirK), the success of amplification is habitat/ecosystem dependent, most likely due to variations in the quality of extracted DNA, community complexity, sequence divergence, and target gene abundance. As a result, comparative studies can be compromised or impossible (99). In addition, preparing high-quality libraries of amplified PCR products for various functional genes from multiple samples is often difficult. Nonspecific amplification requires the tedious and time-consuming step of additional gel purification of PCR products prior to sequencing, which could substantially slow down the sequencing process as a whole.
Shotgun metagenomic sequencing avoids many of the biases encountered in amplicon sequencing because it does not require amplification prior to sequencing. While it often fails to provide sufficient sequence depth to assemble and model the genomes of individual species (41, 100), especially in complex microbial communities like those found in soils, whole-genome recovery from ever more complex communities is now possible (50, 64, 101). Another obstacle to adequate sequence coverage is contaminant DNA, particularly in host-associated microbiome studies, where sequence data may be predominantly from the host (41, 102). Sequence-based open-format approaches can also be impaired by dominant populations in the sample, which may be excessively oversampled. In metatranscriptomic studies, this issue can be compounded by high rRNA abundance (55).
Data analysis can be challenging for the open-format sequencing technologies, particularly shotgun sequencing data, as the assembly and analysis of large sequencing data sets are computationally demanding and often require specialized computing hardware (50, 51, 64). Many genome-oriented analyses of interest are still impractical with short reads alone (102). Also, although many studies are focused on single-read-based analysis, statistical analysis of large short read datasets is time consuming and sometimes only a fraction of reads are usable for biological inference (103), depending on the length of the reads and the availability of representative reference genomes. With frequent changes in technology, there may be little consensus on appropriate procedures for quality filtering and statistical validation. However, with recent rapid advances in both hardware and software for data analysis, plus an ever-growing genome database, sequence data analysis is constantly improving. In addition, for MiSeq-based target gene sequencing data, considerable variations (up to 10-fold) of the estimated OTU numbers can be obtained from the same data set with different computational software tools (e.g., UCLUST versus UPARSE) (86), which presents a challenge for microbial diversity assessments; however, an increasing number of controlled benchmarking experiments are addressing these issues.
The challenges of functional metagenomics are largely associated with barriers to heterologous gene expression. Transcription and translation machinery of the surrogate host must recognize cues in the foreign DNA, and authentic posttranslational modification, protein secretion, and/or availability of precursors for synthesis of active small molecules may not be sufficiently coordinated to enable detection of the active product (104, 105). These challenges have been addressed using phylogenetically diverse hosts (106) and promoters tailored to the host species (107, 108). Functional metagenomics is also laborious and time consuming, providing deep information about a small collection of clones that is in sharp contrast with the expansive views provided by high-throughput sequencing or microarrays. Yet the functional analysis of novel gene products that lack sequence similarity to genes of known function is necessary to illuminate the contents of the vast collection of genes with no known functions that are now in metagenomic databases.
CLOSED-FORMAT MICROARRAY-BASED HIGH-THROUGHPUT DETECTION APPROACHES FOR MICROBIAL COMMUNITY ANALYSIS
Array-based detection technologies.
Various types of DNA microarrays have been developed for microbial detection and community analyses (109), including phylogenetic and functional gene arrays as two main categories. Phylogenetic gene arrays often target rRNA genes, which are useful for identifying specific taxa within microbial communities and studying phylogenetic relationships among different microorganisms. Different types of phylogenetic gene arrays have been developed for microbial ecology applications, such as the PhyloChip (32) that broadly targets known taxa, a microbiota microarray (110) targeting human gut microbiomes, COMPOCHIP targeting compost-degrading microbial communities (111), and SRP-PhyloChip for detecting sulfate-reducing microorganisms (112).
PhyloChip is the most comprehensive and widely used phylogenetic gene array. It is a photolithographic Affymetrix-based technology with 25-mer oligonucleotide probes to discriminate 16S rRNA gene sequences in microbial communities. The most recent version of the PhyloChip (G3) has probes targeting ~60,000 operational taxonomic units (OTUs), representing 2 domains (Archaea and Bacteria), 147 phyla, 1,123 classes, 1,219 orders, 1,464 families, and 10,993 subfamilies (32). Generally, 16S rRNA genes are extracted and PCR amplified from microbial community DNA and then biotin labeled for PhyloChip hybridization and digital image detection (Fig. 1) (32, 113, 114).
Functional gene arrays contain probes targeting genes involved in various biogeochemical cycling processes or specific genomes (115), pangenomes (116), or metagenomes (117), which are useful for monitoring the functional composition and structure of microbial communities (Fig. 1c). Over the past decade, different types of functional gene arrays have been developed, including GeoChip, a generic functional array targeting hundreds of functional gene categories for biogeochemical, ecological, and environmental analyses (33, 118), as well as arrays for detecting specific functional processes, such as nitrogen cycling (119, 120), methanotrophy (121), virulence (122, 123), stress responses (124), hydrogen production and consumption (125), marine microbial communities (117), and bioleaching potential (Fig. 1c) (126).
The most recent GeoChip (version 5.0) contains about 167,000 50-mer oligonucleotide probes covering ~395,000 coding sequences from >1,590 functional genes related to microbial (archaea, bacteria, fungi, and protists) carbon, nitrogen, sulfur, and phosphorus cycling, energy metabolism, antibiotic resistance, metal homeostasis and resistance, secondary metabolism, organic remediation, stress responses, bacteriophages, and virulence. GeoChip also uses phylogenetic markers like gyrB rather than 16S rRNA genes for fine-level phylogenetic analysis (33, 127). To fabricate the GeoChip, it is designed using sequences retrieved from public databases and the CommOligo program (128). Once probes are selected, microarrays are spotted or photolithographically manufactured (e.g., Roche NimbleGen and Agilent). In general, community nucleic acids are extracted, directly labeled with fluorescent dyes, hybridized with GeoChip, and digitally imaged (Fig. 1).
Specificity, sensitivity, and quantitation are critical parameters for any technique used to detect and monitor microorganisms in natural environments, due to the presence of numerous orthologous sequences for each gene in a sample (33). Extremely stringent conditions can improve microarray hybridization specificity, generating results that can be species/strain specific (33, 129). Also, only moderate amounts of total community DNA are needed for PhyloChip and GeoChip analyses. For instance, generally, 0.5 to 2.0 µg of PCR amplicons or ~2.0 µg of total RNA are needed for PhyloChip hybridization (113, 114), and the PhyloChip exhibits a detection limit of 107 copies or 0.01% of nucleotides hybridized to the array (114, 130). For GeoChip hybridization, samples comprising 0.2 to 2.0 µg of DNA or 2 to 5 µg of total RNA (33, 118) are needed, depending on the array format. If the amount of community DNA or RNA is not sufficient, it can be amplified using whole-community genome amplification (131) or whole-community RNA amplification (132), with initial DNA concentrations as low as 10 fg (~2 bacterial cells) resulting in positive detection but not accurate quantification (131). With appropriate amounts of unamplified material, reliable quantitation can be obtained with microarrays like the GeoChip (33) and PhyloChip (130). For example, GeoChip-based studies have shown good correlations between target DNA or RNA concentrations and hybridization signal intensities using pure cultures, mixed cultures, and environmental samples without amplification (33, 129–132) over DNA input amounts varying by 5 orders of magnitude (0.01 to 500 ng) (131). Good correlations have also been reported between PhyloChip signal intensities and quantitative PCR copy numbers of over 5 orders of magnitude (130, 133). It should be noted that PCR amplification biases also occur with the PhyloChip-based detection approach if the 16S rRNA genes are PCR amplified for detection prior to hybridization. Recently, two PCR-independent methods have been developed as viable alternatives to PCR-amplified microbial community analysis for PhyloChip analysis (113, 114).
Key features of array-based detection.
Technical reproducibility in array-based closed-format technologies is less affected by inadequate random sampling than open-format sequencing technologies. To better illustrate this point, we return to the hypothetical community described above (Fig. 2a) to analyze it with a microarray-based technology. Even if the arrays only have probes covering half of the taxa in the community, the simulated overlap between two replicate samples is expected to be above 90% (Fig. 2e). However, taxa with no probes or taxa whose abundance is below the array detection limit will remain undetected by the microarrays (Fig. 2c). The number of taxa detected is defined by the probe sets on the array, and the overlap between samples is less dependent on the level of sampling effort. Furthermore, because hybridization is reasonably quantitative, the taxon identities and abundance distribution are very similar between replicates.
Consequently, depending on the sampling coverage of microbial communities, technical reproducibility can be a significant issue in open-format approaches, while it is minimized in closed-format approaches (94, 134). As a consequence, open- and closed-format detection can yield different results when they are used for comparing microbial community structure. This could be particularly important in examining microbial taxa-area relationships (TARs), one of the best studied and documented patterns in biogeography (94, 134), because taxon richness data are used. The lower susceptibility to random sampling artifacts associated with closed-format-based detection approaches renders them better suited for assessing β diversity, which describes the site-to-site variability in taxon/gene/population composition among communities (89, 94, 95, 117), as well as for detecting low-abundance organisms (117, 135)
Another feature of the array-based closed-format detection is that it is less affected by dominant genes/populations because, although detection is confined to the defined probe set (Table 1), even low-abundance populations present at numbers above the detection limit will be detected (135). Unlike the sequencing-based open-format detections, the array-based closed-format detections are also less susceptible to contaminant DNAs or rRNAs because only targeted nucleic acids generate signals and, hence, interference from the contaminating nucleic acids is minimal (133).
Compared to other high-throughput technologies that target a single gene, such as targeted sequencing and phylogenetic gene arrays, functional gene arrays have several unique features (Table 1). First, they are capable of simultaneously identifying and quantifying many microbial functional genes/pathways that are important for biogeochemical, environmental, and ecological processes, which is critical for ecosystem-level studies, functional biodiversity (136), and trait-based microbial biogeography (137). In contrast, 16S rRNA gene-based techniques do not provide functional information. Second, functional gene arrays can have higher taxonomic resolution than the 16S rRNA gene-based approaches because functional gene markers are generally more divergent than phylogenetic gene markers (129). High taxonomic resolution is important for differentiating treatment effects and examining fine-scale biogeographical patterns. Moreover, technologies that do not require PCR amplification can provide reliable quantitative information on the genes detected (8, 89, 129), across space, time and environmental gradients. However, unlike the 16S rRNA gene-based technologies, functional arrays may not be suitable for providing phylogenetic information at high taxonomic levels (e.g., family and above), due to faster molecular evolution (i.e., rapid mutation saturation), lack of representation on the array, and complications associated with horizontal gene transfer for some functional genes, especially for the genes involved in metal resistance, antibiotic resistance, and contaminant degradation. Rapid mutational saturation of the functional genes could make them less suitable for broad-scale (e.g., continental) microbial biogeographical investigations because the functional genes among various communities could diverge too quickly to preserve signals that would be reliable for resolving broad-scale biogeographical patterns.
The beneficial characteristics of closed-format technologies, including high throughput, low detection limits, high reproducibility, and/or potential for quantification enables them to provide novel insights into specific ecosystems of interest. For instance, surprisingly rich and diverse metabolic reservoirs of microbial communities were revealed using these technologies in a hydrothermal vent chimney (135), Antarctic dry valleys (138), and urban aerosols (130). The importance of stochasticity in controlling ecological diversity and succession was also recently demonstrated by GeoChip-based functional community structure data (139, 140).
Challenges and limitations of array-based closed-format detection technologies.
Unlike the sequencing-based open-format detection technologies, one of the main drawbacks of the closed-format technologies is that they do not enable novel discoveries, such as new genes, taxa, and/or regulatory elements. This is because the input required for array construction must be based upon known sequence information. Thus, the closed-format approaches are not suitable for novel explorations.
Another major limitation of the array-based closed format is that all of the probes on the arrays are derived from a chosen set of genes/sequences that do not necessarily represent the known diversity of the microbial communities of interest. As a result, closed-format technologies will fail to detect potentially important taxa not represented on the microarrays, potentially underestimating the diversity of microbial communities. Thus, it is necessary to continuously update closed-format technologies to reflect the expanding knowledge generated by open-format technologies. Since high-throughput sequencing is ideal for characterizing diversity and discovering new genes, while functional metagenomics assigns function to genes of previously unknown function, coupling high-throughput sequencing approaches, functional expression, and array hybridization is desirable for describing microbial community structure, function, and activity in a comprehensive manner that includes both depth and breadth, as well as quantitative and qualitative surveys.
Although many technical challenges regarding environmental applications of microarrays have been solved over the last decade, several critical bottlenecks still limit the technology. One critical issue is the designing of oligonucleotide probes specific to the target genes/microorganisms of interest when sequences of a particular phylogenetic/functional gene are highly homologous and/or incomplete. This is especially challenging when using arrays for analyzing complex natural systems, since the majority of microorganisms (1, 2) are not yet cultivated and, even among cultured organisms, the biochemical functions of many genes have not been assigned, dramatically compounding this issue.
In addition, due to the variability of reagents (e.g., dyes) and hybridization dynamics, large variations within or between technical microarray replicates are sometimes observed, so that normalization within and between replicates (32, 33, 127, 141, 142) is generally needed. Such variations could affect the probe numbers detected and their quantitation if they are not well controlled experimentally. Various types of controls and skilled personnel with extensive experience are important to minimize such variations.
Finally, due to sequence conservation and the complicated nature of surface hybridization, there can be low-level cross-hybridization to nontarget genes/strains. The challenge is to distinguish true hybridization signals from nonspecific background noise. Also, differentiating genes/populations with low abundance/expression from those not present or not expressed can be a challenge. Generally, subjective thresholds of signal intensity based on signal-to-noise ratio are applied to call-positive signals (33). Thus, great caution is needed in interpreting the gene numbers detected when estimating microbial diversity.
CRITICAL ISSUES IN THE USE OF HIGH-THROUGHPUT METAGENOMIC TECHNOLOGIES TO ADDRESS ECOLOGICAL QUESTIONS
Quality of community DNA/RNA.
Variations in DNA extraction methods can have dramatic impacts on the results of metagenomic studies, especially in high-diversity communities like those in soil (91, 143). Obtaining representative high-quality DNA and RNA from environmental samples is challenging since different populations within the community may require different lysis conditions and diverse, sometimes unidentified contaminants must be removed (144, 145). Thus, any comparisons between studies, whether the analysis is an open or closed format, must be undertaken with caution if nucleic acid extraction methods vary among the studies compared. High-molecular-weight DNA is required to produce representative, quantitative, and efficient amplification of whole-community DNA for microarray analysis (131), to build long-range mate-pair libraries for effective scaffolding of metagenome sequence, and to perform functional metagenomic studies in which entire genes or gene clusters linked to their regulatory sequences need to be maintained intact. But the gentle extraction methods that produce large DNA fragments may underrepresent cells that are harder to lyse, such as those of Gram-positive bacteria and archaea (131). In addition, metagenomic DNA should be sufficiently pure (e.g., A260/A230 ratios of >1.7) for subsequent experimental analyses, such as template amplifications, tagging, or dye labeling. Although PCR amplification can occasionally be obtained with lower-quality DNA, such amplifications might be unreliable and carry the risk of biases, errors, and artifacts. Since community DNA extracted and processed using many commercial kits is often of low purity or low molecular weight, well established custom-optimized DNA extraction protocols are preferred for certain applications (46, 47), ensuring that reliable experimental data are generated for subsequent resource- and effort-intensive analyses and interpretation.
Biological and technical replicates.
The composition, structure, activities, and dynamics of microbial communities in natural settings are shaped by a variety of biological (e.g., competition, predation, mutualistic interactions) and environmental (e.g., temperature, pH, and moisture) factors, which are generally characterized by high spatial and temporal variability. Quantifying the scale at which variation is of interest (between sites, samples, subsamples, nucleic acid extractions, or PCR amplifications) is necessary to determine the nature and degree of replication and to design proper statistical analysis and interpretation of results. That is, it is not possible to determine whether communities in different environments differ significantly if the within-site variability in sampling and analysis is not known. Technical replicates (splitting one sample into two or more aliquots prior to parallel processing and analysis) are useful for estimating the variability associated with the multiple steps of sample processing and analytical methods. On the other hand, biological replicates (e.g., multiple samples taken from soil plots or microcosms that have been manipulated identically) are necessary for estimating spatial and temporal variability associated with experimental conditions so that proper statistical analysis can lead to appropriate interpretation of data (89, 146). This is especially important for highly heterogeneous soil samples (114, 147).
Having a priori knowledge of the expected ranges of variability allows the experimental design to integrate appropriate numbers and types of replicates. For example, while technical replicates are often not performed with photolithographic microarrays due to the known analytical reproducibility of those platforms, biological replicates are essential for proper statistical analysis (114). In contrast, both technical and biological replicates could be important for PCR amplicon-based sequencing approaches (85, 89). In the early years of molecular microbial ecology, many studies were performed without sufficient biological replicates for valid quantitative comparisons and statistical analysis (146). In particular, targeted gene sequencing data may have higher technical variation, which could make comparative studies challenging, particularly with inadequate sampling and replication (89). Increasing the biological replicates, even at the cost of sampling depth, can be an effective way to improve the comparability of data (4, 89, 146). Based on past experience with soils, 3 to 12 biological replicates are needed in typical microbial ecology studies, and more replicates are needed for proper network analysis (4, 148).
Sampling, replication, and sequencing depth.
The site-to-site variability in species/taxon composition, known as β diversity, is crucial to understanding spatiotemporal patterns of species diversity and the mechanisms controlling community composition and structure, which is a central but poorly understood issue in ecology, especially in microbial ecology. However, quantifying β diversity in microbial ecology by using sequencing-based metagenomic technologies requires proper experimental design, including suitable replication, minimal amplification, adequate depth, and stringent quality control (89, 95).
With recent advances in sequencing technologies and associated reductions in cost, appropriate replication can be attained with greater sequencing coverage (29). Balancing sequencing depth with the number of samples per sequencing run is dependent on the biological question and the complexity of the community (8, 32, 89). If the objective is to differentiate the impacts of various conditions (e.g., warming versus nonwarming or high versus low CO2 exposure) on microbial community structure, sampling only dominant microorganisms could be sufficient, necessitating less sequencing coverage per sample (149). However, if the objective is to focus on microbial diversity, distribution, and biogeography, sampling rare taxa could be more important, and thus, deep sequencing with up to millions of reads per sample may be preferred (29, 150). Increasing the sequencing depth will reduce the chance of artifacts associated with random sampling (95). In addition, the sampling effort generally depends on the variations between microbial communities to be compared. For communities that share great similarity, deeper sequencing is needed to distinguish treatment effects on microbial communities (29).
Relative comparisons.
In analyses of microbial communities with high-throughput molecular technologies, relative comparisons are often valid when absolute measurements are not possible. Making relative comparisons mitigates the possible effects of technical variations associated with both open and closed detection formats, such as incomplete cell lysis in DNA extraction, PCR amplification biases, chimerism, sequencing errors, molecular-level random sampling artifacts, variability in bioinformatics analysis, specificity, sensitivity, and/or quantification issues. In general, relative changes in microbial communities can be reliably measured by sequence abundance or treatment sample/control sample hybridization signal ratios. When ratios are used under the assumption that technical variations are similar between the treatment and control samples, such a relative comparison could cancel out the effects of technical variations on the final experimental outcomes and, hence, increase quantitative accuracy (142). Describing relative changes between treatment and control samples is usually defensible, whereas describing absolute changes is much more complicated (32, 147).
One drawback of applying a strictly relative approach is that changes in relative abundances can easily mask large changes in actual abundances. In some cases, the change in absolute abundance can be more informative in describing the dynamics of a population in a community. For example, a 10-fold increase in the expression of a gene with extremely low abundance may simply be an artifact, whereas a 10-fold increase in a moderately abundant gene is more likely to be biologically meaningful. Ideally, both relative and absolute abundances should be used, but caution is needed in the interpretation of data, including assessments of statistical significance.
INTEGRATED FRAMEWORK FOR ANALYZING COMPLEX MICROBIAL COMMUNITIES
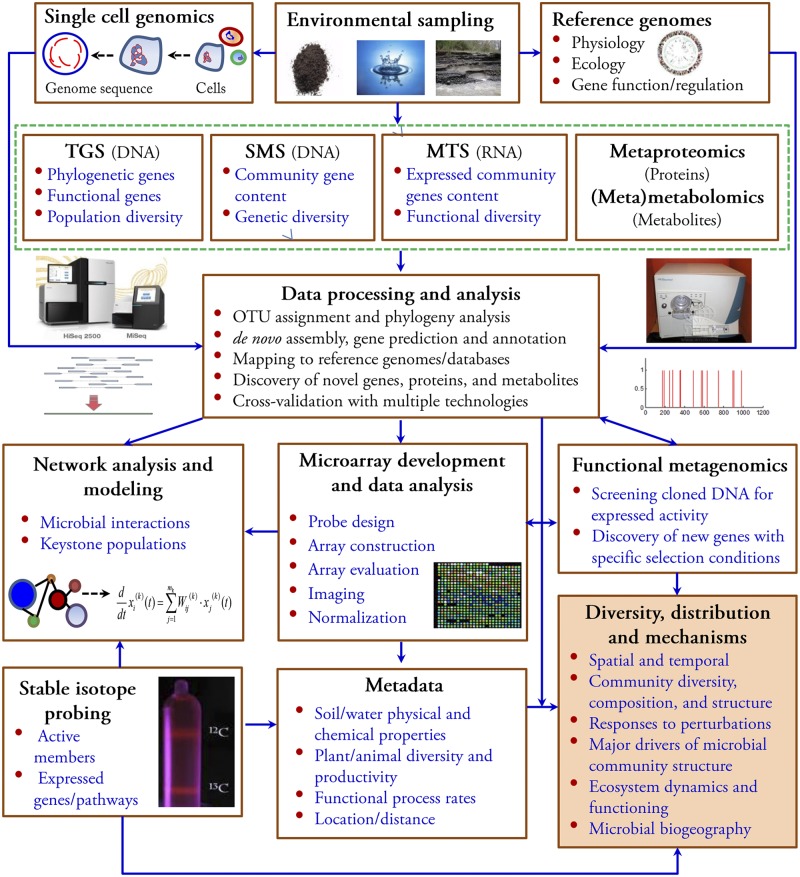
A wide variety of open- and closed-format technologies have been developed, each having distinct features and advantages suitable for different applications in microbial ecology, and thus, they provide complementary approaches for addressing microbial ecology questions (Table 1). Here, we describe an integrated workflow for analyzing microbial communities from different environments using high-throughput metaomic technologies (Fig. 3). Cultivated microorganisms are isolated and sequenced to study their physiology, ecology, gene functions, and regulation. For not-yet cultivated microorganisms, single-cell genomics (151) may provide similar information. To study microbial communities, extracted nucleic acids (DNA/RNA) are analyzed by high-throughput sequencing, including targeted gene sequencing, metagenome, and/or metatranscriptome sequencing. The resulting sequence data are assembled, annotated, and analyzed with information from reference isolates or single-cell genomes (Fig. 3) (152). Functional metagenomics or stable isotope probing (153) can be integrated into the workflow to assign functions to hypothetical genes and uncharacterized populations. Metaomic data and functional information can then be used to develop more comprehensive microarray technologies that complement sequencing. Subsequently, both sequencing and microarray data might be used to link the microbial community structure to ecosystem metadata (e.g., biogeochemical variables) with deeper sampling. In this manner, open- and closed-format technologies can be used as complementary tools for examining microbial community diversity and distribution and to address fundamental questions in microbial ecology. In addition, the data can be used for studying microbial network interactions, identifying keystone species/populations, examining the effects of environmental perturbations, and simulating and modeling community dynamics for predictive microbial ecology (148, 154).
FIG 3 .
An integrated workflow for analyzing microbial communities from different environments using high-throughput metaomic technologies. DNA, RNA, proteins, and/or metabolites are extracted from environmental samples for sequencing and protein/metabolite identification. At the same time, physiological, ecological, and functional information can be obtained via reference genomes and single-cell genomics, which helps with sequencing data analysis and functional annotation, generating useful information for microarray development, especially with novel genes. Microarray-based technologies can be used as a routine tool to address various microbial ecology questions in a rapid and cost-effective manner. Furthermore, metagenomic, metaproteomic, metametabolomic, stable isotope probing, and microarray data can be used alone or coupled with metadata for network analysis and modeling, understanding of microbial diversity, distribution and assembly mechanisms, and linking the microbial community structure with both environmental factors and ecosystem functioning.
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
Significant progress has been made in the development and application of high-throughput molecular technologies for microbial community analysis, but many challenges still remain, especially in the context of environmental applications. For instance, metagenomic sequence assembly, especially from complex communities like those in soil, is one of the grand challenges in bioinformatics (51, 155) although metagenome-specific assembly algorithms (155) and methods for “binning” genomes from metagenome data (64, 156) have led to numerous successes. Single-cell genomics technologies are also proving to be a powerful complement to metagenome studies (Fig. 3) (50, 151, 152).
Another grand challenge for the application of high-throughput molecular tools for microbial community research is the analysis, visualization, and interpretation of massive amounts of both sequencing and array data, especially shotgun metagenome sequencing data (16, 18, 41, 100). For instance, it is difficult to annotate abundant short read sequences to be tabulated and compared in an intuitive manner. This limits our ability to address ecological questions related to microbial biodiversity (e.g., taxonomic, phylogenetic, genetic, functional diversity), functional trait-based microbial biogeography (94, 134, 137), and ecosystem functioning, stability, and succession (157–159). Many excellent bioinformatics tools have been developed for processing, mining, visualizing, and comparing molecular data (41), but they are not optimized for dealing with the vast amounts of experimental data from complex communities like those in soil. Network tools to delineate the interactions among different microbial populations based on high-throughput metagenomics datasets are a promising new development, since understanding the interactions among different species is a central but poorly understood issue in microbial ecology (Fig. 3) (4, 99, 160).
Each omics technology has its strengths and weaknesses and must be selected based on the biological questions and objectives of the study (Fig. 3). In general, open-format technologies are most suitable for exploratory discovery studies, whereas the closed-format technologies can be advantageous for more narrowly defined, hypothesis-driven, quantitative, and comparative studies (117). As sequencing technologies improve and costs decrease, high-throughput sequencing may replace microarrays as the method of choice for many applications (40), but for now, microarray-based closed-format approaches play a valuable role in microbial community analysis, especially for complex microbial communities whose comprehensive sampling remains infeasible (16). Functional metagenomics will continue to identify functions of previously unknown genes. As more functional gene sequences of interest become available, functional arrays that are both more comprehensive (e.g., the next generation of GeoChip, with up to 1 million probes) and more specific (e.g., PathoChip and StressChip) (123, 124) will be developed for addressing different ecological questions and applications. Also, high-throughput molecular technologies should be integrated with other approaches, such as single-cell genomics, metaproteomics (161), and metametabolomics (35, 162), as well as targeted techniques like stable isotope probing (Fig. 3), to address ecological questions and hypotheses within the context of environmental and medical applications. Only in this way will their power for microbial community analysis be realized.
The ultimate goal of microbial ecology is to understand who is where, with whom, doing what, why, and when (159). To answer such questions, reliable, reproducible, quantitative, and statistically valid (146) experimental information on community-wide spatial and temporal dynamics is needed. Also, to achieve this predictive goal, it is essential to model microbial community dynamics and their behaviors at both structural and functional levels (Fig. 3). With the rapid and continuous advances of molecular high-throughput technologies and high-performance computational tools, it is anticipated that in the not-too-distant future, microbiologists will be able to model and predict the behaviors of microbial communities. A new era of quantitative predictive microbial ecology is coming.
ACKNOWLEDGMENTS
We thank Jo Handelsman for her valuable suggestions and editing of this review.
The efforts in preparing this review were supported, through contract DE-AC02-05CH11231 (as part of ENIGMA, a Scientific Focus Area), by funding from the U.S. Department of Energy (DOE), Office of Science, Office of Biological and Environmental Research’s (OBER) Genomics Science Program to Lawrence Berkeley National Laboratory (LBNL), by the OBER Biological Systems Research on the Role of Microbial Communities in Carbon Cycling program (DE-SC0004601), by the U.S. National Science Foundation (NSF) MacroSystems Biology program through grant EF-1065844 to J.Z. and Z.H. and NSF CBET-1336709 to L.A.-C., by Strategic Environmental Research and Development Program grant ER-1587 and National Institute of Environmental Health Sciences Superfund grant P42ES004705 to L.A.-C., by DOE Early Career Research Program grant no. KP/CH57/1 to S.G.T., and by the DOE Office of Science under contract no. DE-AC02-05CH11231 to DOE-Joint Genome Institute (JGI)/LBNL. This work was also supported by funding from the Collaborative Innovation Center for Regional Environmental Quality and State Key Joint Laboratory of Environment Simulation and Pollution Control at Tsinghua University to J.Z.
Footnotes
Citation Zhou J, He Z, Yang Y, Deng Y, Tringe SG, Alvarez-Cohen L. 2015. High-throughput metagenomic technologies for complex microbial community analysis: open and closed formats. mBio 6(1):e02288-14. doi:10.1128/mBio.02288-14.
REFERENCES
- 1.Quince C, Curtis TP, Sloan WT. 2008. The rational exploration of microbial diversity. ISME J 2:997–1006. doi: 10.1038/ismej.2008.69. [DOI] [PubMed] [Google Scholar]
- 2.Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D’Hondt S. 2012. Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci U S A 109:16213–16216. doi: 10.1073/pnas.1203849109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuhrman JA. 2009. Microbial community structure and its functional implications. Nature 459:193–199. doi: 10.1038/nature08058. [DOI] [PubMed] [Google Scholar]
- 4.Zhou J, Deng Y, Luo F, He Z, Tu Q, Zhi X. 2010. Functional molecular ecological networks. mBio 1(4):e00169-10. doi: 10.1128/mBio.00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitter AH, Gilligan CA, Hollingworth K, Kleczkowski A, Twyman RM, Pitchford JW, The Members of the Nerc Soil Biodiversity Programme . 2005. Biodiversity and ecosystem function in soil. Funct Ecol 19:369-377. [Google Scholar]
- 6.Levin SA. 2006. Fundamental questions in biology. PLoS Biol 4:e300. doi: 10.1371/journal.pbio.0040300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pace NR. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 8.Zhou J, Deng Y, He Z, Wu L, Van Nostrand JD. 2010. Applying GeoChip analysis to disparate microbial communities. Microbe Magazine 5:60–65. doi: 10.1128/microbe.5.60.1. [DOI] [Google Scholar]
- 9.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 10.Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, Bork P, Hugenholtz P, Rubin EM. 2005. Comparative metagenomics of microbial communities. Science 308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 11.Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR, Loiacono KA, Lynch BA, MacNeil IA, Minor C, Tiong CL, Gilman M, Osburne MS, Clardy J, Handelsman J, Goodman RM. 2000. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol 66:2541–2547. doi: 10.1128/AEM.66.6.2541-2547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Handelsman J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev 68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schena M, Shalon D, Davis RW, Brown PO. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 14.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suenaga H. 2012. Targeted metagenomics: a high-resolution metagenomics approach for specific gene clusters in complex microbial communities. Environ Microbiol 14:13–22. doi: 10.1111/j.1462-2920.2011.02438.x. [DOI] [PubMed] [Google Scholar]
- 16.Roh SW, Abell GC, Kim K-H, Nam Y-D, Bae J-W. 2010. Comparing microarrays and next-generation sequencing technologies for microbial ecology research. Trends Biotechnol 28:291–299. doi: 10.1016/j.tibtech.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Vieites JM, Guazzaroni M-E, Beloqui A, Golyshin PN, Ferrer M. 2009. Metagenomics approaches in systems microbiology. FEMS Microbiol Rev 33:236–255. doi: 10.1111/j.1574-6976.2008.00152.x. [DOI] [PubMed] [Google Scholar]
- 18.Weinstock GM. 2012. Genomic approaches to studying the human microbiota. Nature 489:250–256. doi: 10.1038/nature11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Human Microbione Project Consortium 2012. A framework for human microbiome research. Nature 486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Human Microbione Project Consortium 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt TM, DeLong EF, Pace NR. 1991. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol 173:4371–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Bagwell CE, Wu L, Devol AH, Zhou J. 2003. Molecular diversity of sulfate-reducing bacteria from two different continental margin habitats. Appl Environ Microbiol 69:6073–6081. doi: 10.1128/AEM.69.10.6073-6081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Massol-Deyá A, Weller R, Ríos-Hernández L, Zhou JZ, Hickey RF, Tiedje JM. 1997. Succession and convergence of biofilm communities in fixed-film reactors treating aromatic hydrocarbons in groundwater. Appl Environ Microbiol 63:270–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muyzer G, de Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu WT, Marsh TL, Cheng H, Forney LJ. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol 63:4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frostegård Å, Tunlid A, Bååth E. 2011. Use and misuse of PLFA measurements in soils. Soil Biol Biochem 43:1621–1625. doi: 10.1016/j.soilbio.2010.11.021. [DOI] [Google Scholar]
- 27.Hadwin AM, Del Rio LF, Pinto LJ, Painter M, Routledge R, Moore MM. 2006. Microbial communities in wetlands of the Athabasca oil sands: genetic and metabolic characterization. FEMS Microbiol Ecol 55:68–78. doi: 10.1111/j.1574-6941.2005.00009.x. [DOI] [PubMed] [Google Scholar]
- 28.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers Y-H, Smith HO. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 29.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ. 2012. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 31.Frias-Lopez J, Shi Y, Tyson GW, Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, Delong EF. 2008. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA 105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, Jansson JK, Probst A, Borglin SE, Fortney JL, Stringfellow WT, Bill M, Conrad ME, Tom LM, Chavarria KL, Alusi TR, Lamendella R, Joyner DC, Spier C, Baelum J, Auer M, Zemla ML, Chakraborty R, Sonnenthal EL, D’haeseleer P, Holman H-YN, Osman S, Lu Z, Van Nostrand JD, Deng Y, Zhou J, Mason OU. 2010. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330:204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- 33.He Z, Deng Y, Van Nostrand JD, Tu Q, Xu M, Hemme CL, Li X, Wu L, Gentry TJ, Yin Y, Liebich J, Hazen TC, Zhou J. 2010. GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J 4:1167–1179. doi: 10.1038/ismej.2010.46. [DOI] [PubMed] [Google Scholar]
- 34.Ram RJ, VerBerkmoes NC, Thelen MP, Tyson GW, Ram RJ, Verberkmoes NC, Thelen MP, Tyson GW, Baker BJ, Blake RC, Shah M, Hettich RL, Banfield JF. 2005. Community proteomics of a natural microbial biofilm. Science 308:1915–1920. doi: 10.1126/science.1109070. [DOI] [PubMed] [Google Scholar]
- 35.Cui Q, Lewis IA, Hegeman AD, Anderson ME, Li J, Schulte CF, Westler WM, Eghbalnia HR, Sussman MR, Markley JL. 2008. Metabolite identification via the Madison Metabolomics Consortium Database. Nat Biotechnol 26:162–164. doi: 10.1038/nbt0208-162. [DOI] [PubMed] [Google Scholar]
- 36.Melton L. 2004. Protein arrays: proteomics in multiplex. Nature 429:101–107. doi: 10.1038/429101a. [DOI] [PubMed] [Google Scholar]
- 37.Houseman BT, Mrksich M. 2002. Carbohydrate arrays for the evaluation of protein binding and enzymatic modification. Chem Biol 9:443–454. doi: 10.1016/S1074-5521(02)00124-2. [DOI] [PubMed] [Google Scholar]
- 38.Borglin S, Joyner D, DeAngelis KM, Khudyakov J, D’haeseleer P, Joachimiak MP, Hazen T. 2012. Application of phenotypic microarrays to environmental microbiology. Curr Opin Biotechnol 23:41–48. doi: 10.1016/j.copbio.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 39.Metzker ML. 2010. Sequencing technologies-the next generation. Nat Rev Genet 11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 40.Loman NJ, Constantinidou C, Chan JZ, Halachev M, Sergeant M, Penn CW, Robinson ER, Pallen MJ. 2012. High-throughput bacterial genome sequencing: an embarrassment of choice, a world of opportunity. Nat Rev Microbiol 10:599–606. doi: 10.1038/nrmicro2850. [DOI] [PubMed] [Google Scholar]
- 41.Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, Knight R. 2012. Experimental and analytical tools for studying the human microbiome. Nat Rev Genet 13:47–58. doi: 10.1038/nrg3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartram AK, Lynch MD, Stearns JC, Moreno-Hagelsieb G, Neufeld JD. 2011. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end Illumina reads. Appl Environ Microbiol 77:3846–3852. doi: 10.1128/AEM.02772-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc Natl Acad Sci U S A 103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pester M, Rattei T, Flechl S, Gröngröft A, Richter A, Overmann J, Reinhold-Hurek B, Loy A, Wagner M. 2012. amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ Microbiol 14:525–539. doi: 10.1111/j.1462-2920.2011.02666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gubry-Rangin C, Hai B, Quince C, Engel M, Thomson BC, James P, Schloter M, Griffiths RI, Prosser JI, Nicol GW. 2011. Niche specialization of terrestrial archaeal ammonia oxidizers. Proc Natl Acad Sci U S A 108:21206–21211. doi: 10.1073/pnas.1109000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou J, Bruns MA, Tiedje JM. 1996. DNA recovery from soils of diverse composition. Appl Environ Microbiol 62:316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hurt RA, Qiu X, Wu L, Roh Y, Palumbo AV, Tiedje JM, Zhou J. 2001. Simultaneous recovery of RNA and DNA from soils and sediments. Appl Environ Microbiol 67:4495–4503. doi: 10.1128/AEM.67.10.4495-4503.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto J-M, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K, Wang J. 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 49.Mackelprang R, Waldrop MP, DeAngelis KM, David MM, Chavarria KL, Blazewicz SJ, Rubin EM, Jansson JK. 2011. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 480:368–371. doi: 10.1038/nature10576. [DOI] [PubMed] [Google Scholar]
- 50.Hess M, Sczyrba A, Egan R, Kim T-W, Chokhawala H, Schroth G, Luo S, Clark DS, Chen F, Zhang T, Mackie RI, Pennacchio LA, Tringe SG, Visel A, Woyke T, Wang Z, Rubin EM. 2011. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331:463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 51.Nagarajan N, Pop M. 2013. Sequence assembly demystified. Nat Rev Genet 14:157–167. doi: 10.1038/nrg3367. [DOI] [PubMed] [Google Scholar]
- 52.Torsvik V, Øvreås L, Thingstad TF. 2002. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science 296:1064–1066. doi: 10.1126/science.1071698. [DOI] [PubMed] [Google Scholar]
- 53.Gans J, Wolinsky M, Dunbar J. 2005. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309:1387–1390. doi: 10.1126/science.1112665. [DOI] [PubMed] [Google Scholar]
- 54.Hemme CL, Deng Y, Gentry TJ, Fields MW, Wu L, Barua S, Barry K, Tringe SG, Watson DB, He Z, Hazen TC, Tiedje JM, Rubin EM, Zhou J. 2010. Metagenomic insights into evolution of a heavy metal-contaminated groundwater microbial community. ISME J 4:660–672. doi: 10.1038/ismej.2009.154. [DOI] [PubMed] [Google Scholar]
- 55.Sorek R, Cossart P. 2010. Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat Rev Genet 11:9–16. doi: 10.1038/nrg2695. [DOI] [PubMed] [Google Scholar]
- 56.Moran MA, Satinsky B, Gifford SM, Luo H, Rivers A, Chan L-K, Meng J, Durham BP, Shen C, Varaljay VA, Smith CB, Yager PL, Hopkinson BM. 2013. Sizing up metatranscriptomics. ISME J 7:237–243. doi: 10.1038/ismej.2012.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi Y, Tyson GW, DeLong EF. 2009. Metatranscriptomics reveals unique microbial small RNAs in the ocean's water column. Nature 459:266–269. doi: 10.1038/nature08055. [DOI] [PubMed] [Google Scholar]
- 58.He S, Wurtzel O, Singh K, Froula JL, Yilmaz S, Tringe SG, Wang Z, Chen F, Lindquist EA, Sorek R, Hugenholtz P. 2010. Validation of two ribosomal RNA removal methods for microbial metatranscriptomics. Nat Methods 7:807–812. doi: 10.1038/nmeth.1507. [DOI] [PubMed] [Google Scholar]
- 59.Urich T, Lanzén A, Qi J, Huson DH, Schleper C, Schuster SC. 2008. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One 3:e2527. doi: 10.1371/journal.pone.0002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dumont MG, Pommerenke B, Casper P. 2013. Using stable isotope probing to obtain a targeted metatranscriptome of aerobic methanotrophs in lake sediment. Environ Microbiol Rep 5:757–764. doi: 10.1111/1758-2229.12078. [DOI] [PubMed] [Google Scholar]
- 61.Xiong X, Frank DN, Robertson CE, Hung SS, Markle J, Canty AJ, McCoy KD, Macpherson AJ, Poussier P, Danska JS, Parkinson J. 2012. Generation and analysis of a mouse intestinal metatranscriptome through Illumina based RNA-sequencing. PLoS One 7:e36009. doi: 10.1371/journal.pone.0036009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giannoukos G, Ciulla DM, Huang K, Haas BJ, Izard J, Levin JZ, Livny J, Earl AM, Gevers D, Ward DV, Nusbaum C, Birren BW, Gnirke A. 2012. Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes. Genome Biol 13:r23. doi: 10.1186/gb-2012-13-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu K, Zhang T. 2012. Metagenomic and metatranscriptomic analysis of microbial community structure and gene expression of activated sludge. PLoS One 7:e38183. doi: 10.1371/journal.pone.0038183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wrighton KC, Thomas BC, Sharon I, Miller CS, Castelle CJ, VerBerkmoes NC, Wilkins MJ, Hettich RL, Lipton MS, Williams KH, Long PE, Banfield JF. 2012. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337:1661–1665. doi: 10.1126/science.1224041. [DOI] [PubMed] [Google Scholar]
- 65.Iverson V, Morris RM, Frazar CD, Berthiaume CT, Morales RL, Armbrust EV. 2012. Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science 335:587–590. doi: 10.1126/science.1212665. [DOI] [PubMed] [Google Scholar]
- 66.DeLong EF, Béjà O. 2010. The light-driven proton pump proteorhodopsin enhances bacterial survival during tough times. PLoS Biol. 8:e1000359. doi: 10.1371/journal.pbio.1000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frigaard N-U, Martinez A, Mincer TJ, DeLong EF. 2006. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature 439:847–850. doi: 10.1038/nature04435. [DOI] [PubMed] [Google Scholar]
- 68.Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW. 2013. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500:567–570. doi: 10.1038/nature12375. [DOI] [PubMed] [Google Scholar]
- 69.Uchiyama T, Miyazaki K. 2009. Functional metagenomics for enzyme discovery: challenges to efficient screening. Curr Opin Biotechnol 20:616–622. doi: 10.1016/j.copbio.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 70.Allen HK, Moe LA, Rodbumrer J, Gaarder A, Handelsman J. 2009. Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME J 3:243–251. doi: 10.1038/ismej.2008.86. [DOI] [PubMed] [Google Scholar]
- 71.Knietsch A, Bowien S, Whited G, Gottschalk G, Daniel R. 2003. Identification and characterization of coenzyme B12-dependent glycerol dehydratase- and diol dehydratase-encoding genes from metagenomic DNA libraries derived from enrichment cultures. Appl Environ Microbiol 69:3048–3060. doi: 10.1128/AEM.69.6.3048-3060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lorenz P, Schleper C. 2002. Metagenome—a challenging source of enzyme discovery. J Mol Catal B Enzymatic 19–20:13–19. doi: 10.1016/S1381-1177(02)00147-9. [DOI] [Google Scholar]
- 73.Wang GY, Graziani E, Waters B, Pan W, Li X, McDermott J, Meurer G, Saxena G, Andersen RJ, Davies J. 2000. Novel natural products from soil DNA libraries in a streptomycete host. Org Lett 2:2401–2404. doi: 10.1021/ol005860z. [DOI] [PubMed] [Google Scholar]
- 74.Brady SF, Clardy J. 2004. Palmitoylputrescine, an antibiotic isolated from the heterologous expression of DNA extracted from bromeliad tank water. J Nat Prod 67:1283–1286. doi: 10.1021/np0499766. [DOI] [PubMed] [Google Scholar]
- 75.McGarvey KM, Queitsch K, Fields S. 2012. Wide variation in antibiotic resistance proteins identified by functional metagenomic screening of a soil DNA library. Appl Environ Microbiol 78:1708–1714. doi: 10.1128/AEM.06759-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Riesenfeld CS, Goodman RM, Handelsman J. 2004. Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ Microbiol 6:981–989. doi: 10.1111/j.1462-2920.2004.00664.x. [DOI] [PubMed] [Google Scholar]
- 77.Xi C, Zhang Y, Marrs CF, Ye W, Simon C, Foxman B, Nriagu J. 2009. Prevalence of antibiotic resistance in drinking water treatment and distribution systems. Appl Environ Microbiol 75:5714–5718. doi: 10.1128/AEM.00382-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hu Y, Yang X, Qin J, Lu N, Cheng G, Wu N, Pan Y, Li J, Zhu L, Wang X, Meng Z, Zhao F, Liu D, Ma J, Qin N, Xiang C, Xiao Y, Li L, Yang H, Wang J, Yang R, Gao GF, Wang J, Zhu B. 2013. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat J Commun 4:2151. [DOI] [PubMed] [Google Scholar]
- 79.Sommer MO, Dantas G, Church GM. 2009. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325:1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cheng G, Hu Y, Yin Y, Yang X, Xiang C, Wang B, Chen Y, Yang F, Lei F, Wu N, Lu N, Li J, Chen Q, Li L, Zhu B. 2012. Functional screening of antibiotic resistance genes from human gut microbiota reveals a novel gene fusion. FEMS Microbiol Lett 336:11–16. doi: 10.1111/j.1574-6968.2012.02647.x. [DOI] [PubMed] [Google Scholar]
- 81.Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, Ochman H, Hugenholtz P. 2010. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J 4:642–647. doi: 10.1038/ismej.2009.153. [DOI] [PubMed] [Google Scholar]
- 82.Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. 2010. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- 83.Schloss PD, Gevers D, Westcott SL. 2011. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lemos LN, Fulthorpe RR, Roesch LF. 2012. Low sequencing efforts bias analyses of shared taxa in microbial communities. Folia Microbiol (Praha) 57:409–413. doi: 10.1007/s12223-012-0155-0. [DOI] [PubMed] [Google Scholar]
- 85.Pinto AJ, Raskin L. 2012. PCR biases distort bacterial and archaeal community structure in pyrosequencing datasets. PLoS One 7:e43093. doi: 10.1371/journal.pone.0043093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 87.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. 2013. The long-term stability of the human gut microbiota. Science 341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lundberg DS, Yourstone S, Mieczkowski P, Jones CD, Dangl JL. 2013. Practical innovations for high-throughput amplicon sequencing. Nat Methods 10:999–1002. doi: 10.1038/nmeth.2634. [DOI] [PubMed] [Google Scholar]
- 89.Zhou J, Wu L, Deng Y, Zhi X, Jiang Y-H, Tu Q, Xie J, Van Nostrand JD, He Z, Yang Y. 2011. Reproducibility and quantitation of amplicon sequencing-based detection. ISME J 5:1303–1313. doi: 10.1038/ismej.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Flores R, Shi J, Gail MH, Gajer P, Ravel J, Goedert JJ. 2012. Assessment of the human faecal microbiota. II. Reproducibility and associations of 16S rRNA pyrosequences. Eur J Clin Invest 42:855–863. doi: 10.1111/j.1365-2362.2012.02659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vishnivetskaya TA, Layton AC, Lau MC, Chauhan A, Cheng KR, Meyers AJ, Murphy JR, Rogers AW, Saarunya GS, Williams DE, Pfiffner SM, Biggerstaff JP, Stackhouse BT, Phelps TJ, Whyte L, Sayler GS, Onstott TC. 2014. Commercial DNA extraction kits impact observed microbial community composition in permafrost samples. FEMS Microbiol Ecol 87:217–230. doi: 10.1111/1574-6941.12219. [DOI] [PubMed] [Google Scholar]
- 92.Peng X, Yu K-Q, Deng G-H, Jiang Y-X, Wang Y, Zhang G-X, Zhou H-W. 2013. Comparison of direct boiling method with commercial kits for extracting fecal microbiome DNA by Illumina sequencing of 16S rRNA tags. J Microbiol Methods 95:455–462. doi: 10.1016/j.mimet.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 93.Ge Y, Schimel JP, Holden PA. 2014. Analysis of run-to-run variation of bar-coded pyrosequencing for evaluating bacterial community shifts and individual taxa dynamics. PLoS One 9:e99414. doi: 10.1371/journal.pone.0099414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou J, Kang S, Schadt CW, Garten CT. 2008. Spatial scaling of functional gene diversity across various microbial taxa. Proc Natl Acad Sci U S A 105:7768–7773. doi: 10.1073/pnas.0709016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhou J, Jiang Y-H, Deng Y, Shi Z, Zhou BY, Xue K, Wu L, He Z, Yang Y. 2013. Random sampling process leads to overestimation of β-diversity of microbial communities. mBio 4( 3):e00324-13. doi: 10.1128/mBio.00324-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schloss PD, Handelsman J. 2007. The last word: books as a statistical metaphor for microbial communities. Annu Rev Microbiol 61:23–24. doi: 10.1146/annurev.micro.61.011507.151712. [DOI] [PubMed] [Google Scholar]
- 97.Qiu X, Wu L, Huang H, McDonel PE, Palumbo AV, Tiedje JM, Zhou J. 2001. Evaluation of PCR-generated chimeras, mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl Environ Microbiol 67:880–887. doi: 10.1128/AEM.67.2.880-887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Palmer K, Biasi C, Horn MA. 2012. Contrasting denitrifier communities relate to contrasting N2O emission patterns from acidic peat soils in Arctic tundra. ISME J 6:1058–1077. doi: 10.1038/ismej.2011.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Faust K, Raes J. 2012. Microbial interactions: from networks to models. Nat Rev Microbiol 10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 100.Zengler K, Palsson BO. 2012. A road map for the development of community systems (CoSy) biology. Nat Rev Microbiol 10:366–372. doi: 10.1038/nrmicro2763. [DOI] [PubMed] [Google Scholar]
- 101.Castelle CJ, Hug LA, Wrighton KC, Thomas BC, Williams KH, Wu D, Tringe SG, Singer SW, Eisen JA, Banfield JF. 2013. Extraordinary phylogenetic diversity and metabolic versatility in aquifer sediment. Nat Commun 4:2120. doi: 10.1038/ncomms3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gevers D, Pop M, Schloss PD, Huttenhower C. 2012. Bioinformatics for the Human Microbiome Project. PLoS Comput Biol 8:e1002779. doi: 10.1371/journal.pcbi.1002779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, Owens S, Gilbert JA, Wall DH, Caporaso JG. 2012. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci U S A 109:21390–21395. doi: 10.1073/pnas.1215210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Feng Z, Kim JH, Brady SF. 2010. Fluostatins produced by the heterologous expression of a TAR reassembled environmental DNA derived type II PKS gene cluster. J Am Chem Soc 132:11902–11903. doi: 10.1021/ja104550p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Evans TC Jr., Xu M-Q. 2010. Heterologous gene expression in E. coli: methods and protocols, vol 705 Humana Press, New York, NY. [Google Scholar]
- 106.McMahon MD, Guan C, Handelsman J, Thomas MG. 2012. Metagenomic analysis of Streptomyces lividans reveals host-dependent functional expression. Appl Environ Microbiol 78:3622–3629. doi: 10.1128/AEM.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kallifidas D, Kang H-S, Brady SF. 2012. Tetarimycin A, an MRSA-active antibiotic identified through induced expression of environmental DNA gene clusters. J Am Chem Soc 134:19552–19555. doi: 10.1021/ja3093828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hosokawa-Okamoto R, Miyazaki K. 2011. Escherichia coli host engineering for efficient metagenomic enzyme discovery. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 109.He Z, Van Nostrand J, Deng Y, Zhou J. 2011. Development and applications of functional gene microarrays in the analysis of the functional diversity, composition, and structure of microbial communities. Front Environ Sci Eng China 5:1–20. doi: 10.1007/s11783-011-0301-y. [DOI] [Google Scholar]
- 110.Paliy O, Kenche H, Abernathy F, Michail S. 2009. High-throughput quantitative analysis of human intestinal microbiota with phylogenetic microarray. Appl Environ Microbiol 75:3572–3579. doi: 10.1128/AEM.02764-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Franke-Whittle IH, Knapp BA, Fuchs J, Kaufmann R, Insam H. 2009. Application of COMPOCHIP microarray to investigate the bacterial communities of different composts. Microb Ecol 57:510–521. doi: 10.1007/s00248-008-9435-2. [DOI] [PubMed] [Google Scholar]
- 112.Loy A, Lehner A, Lee N, Adamczyk J, Meier H, Ernst J, Schleifer K-H, Wagner M. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl Environ Microbiol 68:5064–5081. doi: 10.1128/AEM.68.10.5064-5081.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee PK, Warnecke F, Brodie EL, Macbeth TW, Conrad ME, Andersen GL, Alvarez-Cohen L. 2012. Phylogenetic microarray analysis of a microbial community performing reductive dechlorination at a TCE-contaminated site. Environ Sci Technol 46:1044–1054. doi: 10.1021/es203005k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.DeAngelis KM, Wu CH, Beller HR, Brodie EL, Chakraborty R, DeSantis TZ, Fortney JL, Hazen TC, Osman SR, Singer ME, Tom LM, Andersen GL. 2011. PCR amplification-independent methods for detection of microbial communities by the high-density microarray PhyloChip. Appl Environ Microbiol 77:6313–6322. doi: 10.1128/AEM.05262-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee PK, Cheng D, Hu P, West KA, Dick GJ, Brodie EL, Andersen GL, Zinder SH, He J, Alvarez-Cohen L. 2011. Comparative genomics of two newly isolated Dehalococcoides strains and an enrichment using a genus microarray. ISME J 5:1014–1024. doi: 10.1038/ismej.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hug LA, Salehi M, Nuin P, Tillier ER, Edwards EA. 2011. Design and verification of a pangenome microarray oligonucleotide probe set for Dehalococcoides spp. Appl Environ Microbiol 77:5361–5369. doi: 10.1128/AEM.00063-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shilova IN, Robidart JC, James Tripp H, Turk-Kubo K, Wawrik B, Post AF, Thompson AW, Ward B, Hollibaugh JT, Millard A, Ostrowski M, Scanlan DJ, Paerl RW, Stuart R, Zehr JP. 2014. A microarray for assessing transcription from pelagic marine microbial taxa. ISME J 8:1476–1491. doi: 10.1038/ismej.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lu Z, Deng Y, Van Nostrand JD, He Z, Voordeckers J, Zhou A, Lee Y-J, Mason OU, Dubinsky EA, Chavarria KL, Tom LM, Fortney JL, Lamendella R, Jansson JK, D’Haeseleer P, Hazen TC, Zhou J. 2012. Microbial gene functions enriched in the Deepwater Horizon deep-sea oil plume. ISME J 6:451–460. doi: 10.1038/ismej.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Taroncher-Oldenburg G, Griner EM, Francis CA, Ward BB. 2003. Oligonucleotide microarray for the study of functional gene diversity in the nitrogen cycle in the environment. Appl Environ Microbiol 69:1159–1171. doi: 10.1128/AEM.69.2.1159-1171.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Abell GCJ, Robert SS, Frampton DMF, Volkman JK, Rizwi F, Csontos J, Bodrossy L, 2012. High-throughput analysis of ammonia oxidiser community composition, via a novel, amoA-based functional gene array. PLoS One 7:e51542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bodrossy L, Stralis-Pavese N, Konrad-Köszler M, Weilharter A, Reichenauer TG, Schöfer D, Sessitsch A. 2006. mRNA-based parallel detection of active methanotroph populations by use of a diagnostic microarray. Appl Environ Microbiol 72:1672–1676. doi: 10.1128/AEM.72.2.1672-1676.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Miller SM, Tourlousse DM, Stedtfeld RD, Baushke SW, Herzog AB, Wick LM, Rouillard JM, Gulari E, Tiedje JM, Hashsham SA. 2008. In situ-synthesized virulence and marker gene biochip for detection of bacterial pathogens in water. Appl Environ Microbiol 74:2200–2209. doi: 10.1128/AEM.01962-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lee Y-J, van Nostrand JD, Tu Q, Lu Z, Cheng L, Yuan T, Deng Y, Carter MQ, He Z, Wu L, Yang F, Xu J, Zhou J. 2013. The PathoChip, a functional gene array for assessing pathogenic properties of diverse microbial communities. ISME J 7:1974–1984. doi: 10.1038/ismej.2013.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhou A, He Z, Qin Y, Lu Z, Deng Y, Tu Q, Hemme CL, Van Nostrand JD, Wu L, Hazen TC, Arkin AP, Zhou J. 2013. StressChip as a high-throughput tool for assessing microbial community responses to environmental stresses. Environ Sci Technol 47:9841–9849. doi: 10.1021/es4018656. [DOI] [PubMed] [Google Scholar]
- 125.Marshall IP, Berggren DR, Azizian MF, Burow LC, Semprini L, Spormann AM. 2012. The Hydrogenase Chip: a tiling oligonucleotide DNA microarray technique for characterizing hydrogen-producing and -consuming microbes in microbial communities. ISME J 6:814–826. doi: 10.1038/ismej.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yin H, Cao L, Qiu G, Wang D, Kellogg L, Zhou J, Dai Z, Liu X. 2007. Development and evaluation of 50-mer oligonucleotide arrays for detecting microbial populations in acid mine drainages and bioleaching systems. J Microbiol Methods 70:165–178. doi: 10.1016/j.mimet.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 127.Tu Q, Yu H, He Z, Deng Y, Wu L, Van Nostrand JD, Zhou A, Voordeckers J, Lee YJ, Qin Y, Hemme CL, Shi Z, Xue K, Yuan T, Wang A, Zhou J. 2014. GeoChip 4: a functional gene-arrays-based high-throughput environmental technology for microbial community analysis. Mol Ecol Resour 14:914–928. doi: 10.1111/1755-0998.12239. [DOI] [PubMed] [Google Scholar]
- 128.Li X, He Z, Zhou J. 2005. Selection of optimal oligonucleotide probes for microarrays using multiple criteria, global alignment and parameter estimation. Nucleic Acids Res 33:6114–6123. doi: 10.1093/nar/gki914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tiquia SM, Wu L, Chong SC, Passovets S, Xu D, Xu Y, Zhou J. 2004. Evaluation of 50-mer oligonucleotide arrays for detecting microbial populations in environmental samples. Biotechniques 36:664–670. [DOI] [PubMed] [Google Scholar]
- 130.Brodie EL, DeSantis TZ, Parker JP, Zubietta IX, Piceno YM, Andersen GL. 2007. Urban aerosols harbor diverse and dynamic bacterial populations. Proc Natl Acad Sci U S A 104:299–304. doi: 10.1073/pnas.0608255104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wu L, Liu X, Schadt CW, Zhou J. 2006. Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole-community genome amplification. Appl Environ Microbiol 72:4931–4941. doi: 10.1128/AEM.02738-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gao H, Yang ZK, Gentry TJ, Wu L, Schadt CW, Zhou J. 2007. Microarray-based analysis of microbial community RNAs by whole-community RNA amplification. Appl Environ Microbiol 73:563–571. doi: 10.1128/AEM.01771-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lemon KP, Klepac-Ceraj V, Schiffer HK, Brodie EL, Lynch SV, Kolter R. 2010. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. mBio 1(3):e00129-10. doi: 10.1128/mBio.00129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Martiny JB, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, Ovreås L, Reysenbach A-L, Smith VH, Staley JT. 2006. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol. 4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 135.Wang F, Zhou H, Meng J, Peng X, Jiang L, Sun P, Zhang C, Van Nostrand JD, Deng Y, He Z, Wu L, Zhou J, Xiao X. 2009. GeoChip-based analysis of metabolic diversity of microbial communities at the Juan de Fuca ridge hydrothermal vent. Proc Natl Acad Sci U S A 106:4840–4845. doi: 10.1073/pnas.0810418106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hillebrand H, Matthiessen B. 2009. Biodiversity in a complex world: consolidation and progress in functional biodiversity research. Ecol Lett 12:1405–1419. doi: 10.1111/j.1461-0248.2009.01388.x. [DOI] [PubMed] [Google Scholar]
- 137.Green JL, Bohannan BJ, Whitaker RJ. 2008. Microbial biogeography: from taxonomy to traits. Science 320:1039–1043. doi: 10.1126/science.1153475. [DOI] [PubMed] [Google Scholar]
- 138.Chan Y, Van Nostrand JD, Zhou J, Pointing SB, Farrell RL. 2013. Functional ecology of an Antarctic dry valley. Proc Natl Acad Sci U S A 110:8990–8995. doi: 10.1073/pnas.1300643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhou J, Deng Y, Zhang P, Xue K, Liang Y, Van Nostrand JD, Yang Y, He Z, Wu L, Stahl DA, Hazen TC, Tiedje JM, Arkin AP. 2014. Stochasticity, succession, and environmental perturbations in a fluidic ecosystem. Proc Natl Acad Sci U S A 111:E836–E845. doi: 10.1073/pnas.1324044111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhou J, Liu W, Deng Y, Jiang Y-H, Xue K, He Z, Van Nostrand JD, Wu L, Yang Y, Wang A. 2013. Stochastic assembly leads to alternative communities with distinct functions in a bioreactor microbial community. mBio 4(2):e00584-12. doi: 10.1128/mBio.00584-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wu L, Thompson DK, Li G, Hurt RA, Tiedje JM, Zhou J. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl Environ Microbiol 67:5780–5790. doi: 10.1128/AEM.67.12.5780-5790.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.He Z, Gentry TJ, Schadt CW, Wu L, Liebich J, Chong SC, Huang Z, Wu W, Gu B, Jardine P, Criddle C, Zhou J. 2007. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J 1:67–77. doi: 10.1038/ismej.2007.2. [DOI] [PubMed] [Google Scholar]
- 143.Bintrim SB, Donohue TJ, Handelsman J, Roberts GP, Goodman RM. 1997. Molecular phylogeny of Archaea from soil. Proc Natl Acad Sci U S A 94:277–282. doi: 10.1073/pnas.94.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liles MR, Williamson LL, Rodbumrer J, Torsvik V, Goodman RM, Handelsman J. 2008. Recovery, purification, and cloning of high-molecular-weight DNA from soil microorganisms. Appl Environ Microbiol 74:3302–3305. doi: 10.1128/AEM.02630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Desai C, Madamwar D. 2007. Extraction of inhibitor-free metagenomic DNA from polluted sediments, compatible with molecular diversity analysis using adsorption and ion-exchange treatments. Bioresour Technol 98:761–768. doi: 10.1016/j.biortech.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 146.Prosser JI. 2010. Replicate or lie. Environ Microbiol 12:1806–1810. doi: 10.1111/j.1462-2920.2010.02201.x. [DOI] [PubMed] [Google Scholar]
- 147.Zhou J, Xue K, Xie J, Deng Y, Wu L, Cheng X, Fei S, Deng S, He Z, Van Nostrand JD, Luo Y. 2012. Microbial mediation of carbon-cycle feedbacks to climate warming. Nat Clim Change 2:106–110. doi: 10.1038/nclimate1331. [DOI] [Google Scholar]
- 148.Zhou J, Deng Y, Luo F, He Z, Yang Y. 2011. Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2. mBio 2(4):2:e00122-11. doi: 10.1128/mBio.00122-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Knight R, Jansson J, Field D, Fierer N, Desai N, Fuhrman JA, Hugenholtz P, van der Lelie D, Meyer F, Stevens R, Bailey MJ, Gordon JI, Kowalchuk GA, Gilbert JA. 2012. Unlocking the potential of metagenomics through replicated experimental design. Nat Biotechnol 30:513–520. doi: 10.1038/nbt.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hazen TC, Rocha AM, Techtmann SM. 2013. Advances in monitoring environmental microbes. Curr Opin Biotechnol 24:526–533. doi: 10.1016/j.copbio.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 151.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu W-T, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 152.Lasken RS. 2012. Genomic sequencing of uncultured microorganisms from single cells. Nat Rev Microbiol 10:631–640. doi: 10.1038/nrmicro2857. [DOI] [PubMed] [Google Scholar]
- 153.Dumont MG, Murrell JC. 2005. Stable isotope probing—linking microbial identity to function. Nat Rev Microbiol 3:499–504. doi: 10.1038/nrmicro1162. [DOI] [PubMed] [Google Scholar]
- 154.Barberán A, Bates ST, Casamayor EO, Fierer N. 2012. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J 6:343–351. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pell J, Hintze A, Canino-Koning R, Howe A, Tiedje JM, Brown CT. 2012. Scaling metagenome sequence assembly with probabilistic de Bruijn graphs. Proc Natl Acad Sci U S A 109:13272–13277. doi: 10.1073/pnas.1121464109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sharon I, Morowitz MJ, Thomas BC, Costello EK, Relman DA, Banfield JF. 2013. Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res 23:111–120. doi: 10.1101/gr.142315.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Prosser JI, Bohannan BJ, Curtis TP, Ellis RJ, Firestone MK, Freckleton RP, Green JL, Green LE, Killham K, Lennon JJ, Osborn AM, Solan M, van der Gast CJ, Young JP. 2007. The role of ecological theory in microbial ecology. Nat Rev Microbiol 5:384–392. doi: 10.1038/nrmicro1643. [DOI] [PubMed] [Google Scholar]
- 158.Sutherland WJ, Armstrong-Brown S, Armsworth PR, Tom B, Brickland J, Campbell CD, Chamberlain DE, Cooke AI, Dulvy NK, Dusic NR, Fitton M, Freckleton RP, Godfray HCJ, Grout N, Harvey HJ, Hedley C, Hopkins JJ, Kift NB, Kirby J, Kunin WE, Macdonald DW, Marker B, Naura M, Neale AR, Oliver TOM, Osborn DAN, Pullin AS, Shardlow MEA, Showler DA, Smith PL, Smithers RJ, Solandt J-L, Spencer J, Spray CJ, Thomas CD, Thompson JIM, Webb SE, Yalden DW, Watkinson AR. 2006. The identification of 100 ecological questions of high policy relevance in the UK. J Appl Ecol 43:617–627. doi: 10.1111/j.1365-2664.2006.01188.x. [DOI] [Google Scholar]
- 159.Zhou J. 2009. Predictive microbial ecology. Microb Biotechnol 2:154–156. doi: 10.1111/j.1751-7915.2009.00090_21.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shade A, McManus PS, Handelsman J. 2013. Unexpected diversity during community succession in the apple flower Microbiome. mBio 4(2):e00602-12. doi: 10.1128/mBio.00602-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kleiner M, Wentrup C, Lott C, Teeling H, Wetzel S, Young J, Chang Y-J, Shah M, VerBerkmoes NC, Zarzycki J, Fuchs G, Markert S, Hempel K, Voigt B, Becher D, Liebeke M, Lalk M, Albrecht D, Hecker M, Schweder T, Dubilier N. 2012. Metaproteomics of a gutless marine worm and its symbiotic microbial community reveal unusual pathways for carbon and energy use. Proc Natl Acad Sci U S A 109:E1173–E1182. doi: 10.1073/pnas.1121198109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kell DB. 2004. Metabolomics and systems biology: making sense of the soup. Curr Opin Microbiol 7:296–307. doi: 10.1016/j.mib.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 163.Ushio M, Makoto K, Klaminder J, Takasu H, Nakano S. 2014. High-throughput sequencing shows inconsistent results with a microscope-based analysis of the soil prokaryotic community. Soil Biol Biochem 76:53–56. doi: 10.1016/j.soilbio.2014.05.010. [DOI] [Google Scholar]
- 164.Decelle J, Romac S, Sasaki E, Not F, Mahé F. Intracellular diversity of the V4 and V9 regions of the 18S rRNA in marine protists (Radiolarians) assessed by high-throughput sequencing. PLoS ONE 9:e104297. doi: 10.1371/journal.pone.0104297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhan A, He S, Brown EA, Chain FJJ, Therriault TW, Abbott CL, Heath DD, Cristescu ME, MacIsaac HJ. 2014. Reproducibility of pyrosequencing data for biodiversity assessment in complex communities. Meth Ecol Evol 5:881–890. doi: 10.1111/2041-210X.12230. [DOI] [Google Scholar]