ABSTRACT
Diverse colony morphologies are a hallmark of Burkholderia pseudomallei recovered from infected patients. We observed that stresses that inhibit aerobic respiration shifted populations of B. pseudomallei from the canonical white colony morphotype toward two distinct, reversible, yet relatively stable yellow colony variants (YA and YB). As accumulating evidence supports the importance of B. pseudomallei enteric infection and gastric colonization, we tested the response of yellow variants to hypoxia, acidity, and stomach colonization. Yellow variants exhibited a competitive advantage under hypoxic and acidic conditions and alkalized culture media. The YB variant, although highly attenuated in acute virulence, was the only form capable of colonization and persistence in the murine stomach. The accumulation of extracellular DNA (eDNA) was a characteristic of YB as observed by 4′,6-diamidino-2-phenylindole (DAPI) staining of gastric tissues, as well as in an in vitro stomach model where large amounts of eDNA were produced without cell lysis. Transposon mutagenesis identified a transcriptional regulator (BPSL1887, designated YelR) that when overexpressed produced the yellow phenotype. Deletion of yelR blocked a shift from white to the yellow forms. These data demonstrate that YB is a unique B. pseudomallei pathovariant controlled by YelR that is specifically adapted to the harsh gastric environment and necessary for persistent stomach colonization.
IMPORTANCE
Seemingly uniform populations of bacteria often contain subpopulations that are genetically identical but display unique characteristics which offer advantages when the population is faced with infrequent but predictable stresses. The pathogen Burkholderia pseudomallei is capable of forming several reversible colony types, and it interconverted between one white type and two yellow types under certain environmental stresses. The two yellow forms exhibited distinct advantages in low-oxygen and acidic environments. One yellow colony variant was the only form capable of chronic stomach colonization. Areas of gastric infection were marked by bacteria encased in a DNA matrix, and the yellow forms were able to produce large amounts of extracellular DNA in vitro. We also identified the regulator in control of yellow colony variant formation. These findings demonstrate a role in infection for colony variation and provide a mechanism for chronic stomach colonization—a frequently overlooked niche in melioidosis.
INTRODUCTION
Burkholderia pseudomallei is a Gram-negative soil bacterium and the opportunistic pathogen responsible for the disease melioidosis (1). The resistance of melioidosis to effective public health strategies, its high mortality rate, and its expanding geographical reach make it a serious public health concern and an emerging infectious disease (2). Symptoms of melioidosis are similar to those of protean diseases, and thus, the infection is frequently misdiagnosed. Melioidosis is also highly recalcitrant to antibiotic therapy (3). Vaccine development is complicated, as even B. pseudomallei infection does not confer protection from future infections (1). The combination of these challenges in diagnosis and treatment has convinced many that this bacterium has potential for weaponization, and the Centers for Disease Control and Prevention have categorized B. pseudomallei as a Tier 1 Select Agent (4).
Despite its public health impact, the clinically relevant route of infection has not been clearly established, especially with regard to latent colonization and delayed disease presentation. The natural routes of infection are believed to be inhalation, transcutaneous inoculation, and ingestion (5–7). Although inhalation and inoculation are highly effective at causing acute disease, animal models based on these routes of infection do not recapitulate the features of latent infection observed clinically—specifically, delayed disease presentation and highly variable primary symptoms (8–10). Recently, however, epidemiological investigations of melioidosis outbreaks have identified contaminated water sources as reservoirs of infectious B. pseudomallei observed in clinical outbreaks (11–15), implicating ingestion as the clinically relevant route of infection. In addition to the epidemiological observations, a recent animal model of chronic melioidosis has been described that replicates aspects of chronic melioidosis (16). In chronically colonized animals, the site of persistent infection is the stomach, further suggesting that ingestion is a clinically relevant route of exposure in cases of chronic melioidosis Furthermore, B. pseudomallei was recovered during gastric biopsies in human patients referred for endoscopy examination (17).
Whether patients are infected via inhalation, inoculation, or ingestion, B. pseudomallei recovered from infected hosts is well known for varied and multiple colony morphologies (18–20). While the high prevalence and range of colony morphotypes are hallmarks of B. pseudomallei, colony morphology variation and switching are not unique to B. pseudomallei. Most pathogens presenting with altered colony morphology are variable in a discrete set of attributes: either cell surface proteins or composition of outer membrane lipids (21). This behavior has been proposed to be a strategy for immune evasion by the pathogens (21).
Various B. pseudomallei Malaysian isolates displayed diversity in colony morphologies whose morphotypes were correlated with altered levels of drug resistance and attenuation in an animal model (22), and seven colony morphotypes of B. pseudomallei have been described based on colony characteristics (18). Of these colony types, the most common showed variant-specific pathogenicity in cell culture. They also showed differential expression of virulence factors, including production of elastase, protease, and lipase as well as biofilm formation, swarming motility, and swimming motility. However, the number of mechanistically distinct colony variants and their mechanisms of formation have not been determined, nor have their respective relationships in a natural infection cycle been established.
Here, we describe two yellow colony variants of B. pseudomallei that were routinely isolated under conditions that limited aerobic respiration. Investigation of these variants revealed a unique constellation of characteristics that present a compelling case for their role in establishing persistent gastric colonization following ingestion as a natural route of exposure. This is underscored by the finding that only one of the yellow variants but not the parental form colonized the stomach even though it was significantly less acutely virulent.
RESULTS
Yellow variants emerge under respiration-limiting conditions.
After extended exposure of the parental white form of B. pseudomallei (WHT) to either hypoxia or nitric oxide, we observed that up to 50% of the culture was a mixture of two yellow-pigmented colony morphotypes when plated on Luria-Bertani-Lennox (LB) agar (YA and YB, Fig. 1). Other stresses such as antibiotic exposure, heat shock, nutrient starvation, and iron starvation did not result in observation of yellow-type colony morphotypes (not shown). YA exhibited a smooth, domed, glistening surface and moderate yellow pigmentation. YB was darker in pigmentation with an indurated center and a defined margin (Fig. 1). These starkly different colony types were confirmed to be B. pseudomallei and not contamination by subculture on the highly selective Ashdown medium as well as by whole-genome sequencing. When a population of predominately WHT-form bacteria was placed under hypoxic conditions continuously, yellow colony variants were isolated at increasing frequency over time (Fig. 2). However, the kinetics of variant emergence varied with technical repetition of the experiment. Under hypoxic conditions, it took from 48 to 96 h until at least 50% of each culture had converted into yellow variants. These findings indicate that even though the altered colony morphology provided an extreme advantage under hypoxia, lack of oxygen was not the primary environmental signal or trigger for induction of the phenotype. The initial emergence of yellow variants appeared to be a stochastic event, and the length of time for significant yellow variant emergence was likely determined by the variable number of undetectable yellow variants in the original inoculum.
FIG 1 .
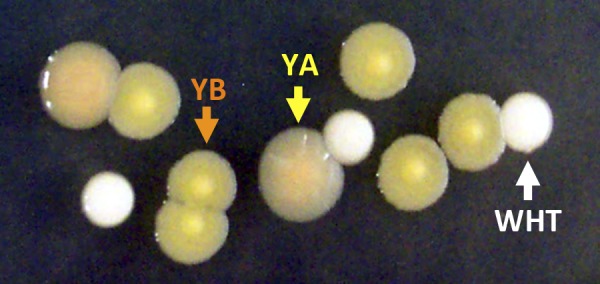
Representative colony morphologies. WHT (white arrow), YA (yellow arrow), and YB (orange arrow) on LB agar.
FIG 2 .

White cultures switch to yellow forms under hypoxic conditions. White-type B. pseudomallei bacteria were inoculated into LB medium at pH 5 (top) or 7 (bottom) at an OD600 of 0.1 in a 0.2% oxygen hypoxia chamber. Colony morphotype distribution was determined at 3, 6, 12, 24, and 48 h. Bars indicate the percentages of culture that were WHT (white), YA (yellow), and YB (orange).
Growth and reversion rates.
YA and YB grew more slowly under aerobic conditions than did WHT. YA had a median doubling time of 70.4 min, YB had a median doubling time of 69.9 min, and WHT had a median doubling time of 63.6 min. Under well-aerated logarithmic growth conditions, YA displayed a relatively high rate of reversion (Table 1). Under well-aerated conditions, YB reverted to YA, not directly to WHT, at a much lower and less predictable rate (Table 1).
TABLE 1 .
Variant doubling time and reversion rate
| Colony type | Doubling time (95% CIc) (min) | Reversion rate |
|---|---|---|
| WHT | 51.0. (44.5–61.5) | |
| YA | 61.7 (49.0–83.3) | 0.15 ± 0.05a |
| YB | 65.0 (61.2–69.2) | 0.005 ± 0.06b |
YA reverted to WHT.
YB reverted to YA.
CI, confidence interval.
Competitive advantage of YA and YB under hypoxia.
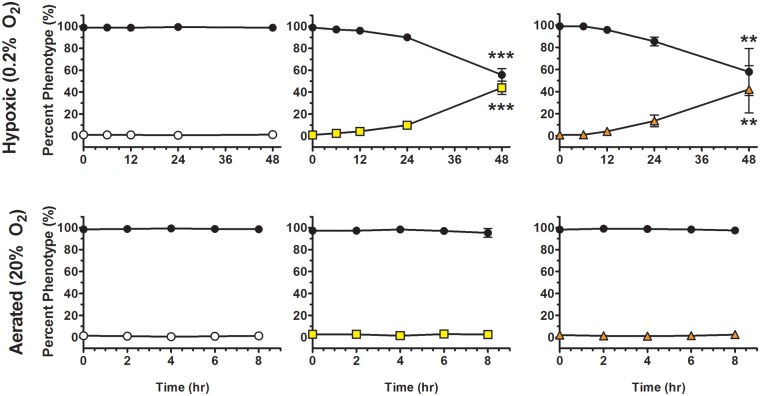
Since yellow variants were initially isolated under conditions that restricted respiration, we sought to determine if these variants possessed a competitive advantage during hypoxia. To control for the rapid reversion of yellow colony variants to the WHT form in competition experiments, we inserted the xylE gene into the genome of the WHT form to be used as a genetic marker to distinguish it from yellow revertants. Competitions were performed with WHT (xylE+), YA, or YB, and the mixtures were placed under hypoxic conditions in acidified growth medium (pH 5) to model a gastric environment. We observed that YA and YB increased from 1% of the population at the beginning of the experiment to over 40% by 48 h under hypoxic conditions (Fig. 3), a statistically significant difference when comparing the measured percent genotype at each time point to the initial percent genotype at the beginning of the experiment. For the YA competition under hypoxia, the increase in percent xylE-negative and decrease in percent xylE-positive colonies became significant at 48 h (Fig. 3). For the YB competition under hypoxia, the increase in percent xylE-negative colonies and the decrease in percent xylE-positive colonies also became significant at 48 h (Fig. 3). No competitive advantage was observed for YA, YB, or the unmarked WHT-type bacteria under well-aerated conditions, nor was there an advantage for the xylE-marked or unmarked WHT-type bacteria under hypoxic and acidic conditions (Fig. 3).
FIG 3 .
Yellow forms have a competitive growth advantage during hypoxia. WHT (white circles), YA (yellow squares), or YB (orange triangles) bacteria were added at 1% of the total population to a 99% xylE-marked WHT culture in LB at pH 5 (black circles). Cultures were grown under hypoxic or aerobic conditions, and the percent composition of each form was determined at 0, 6, 12, 24, and 48 h for the hypoxic cultures and 0, 2, 4, 6, and 8 h for the aerobic cultures. ** denotes P < .01 and *** denotes P < 0.001.
YA and YB alkalize cultures without production of ammonia.
We observed that cultures of YA and YB became alkaline in stationary phase, whereas the parental WHT type remained neutral throughout its growth cycle (not shown). Hypothesizing that this behavior could provide an advantage under extremely acidic environments such as in the stomach, we grew cultures of WHT, YA, and YB forms in moderately acidified rich medium (LB, pH 5.0). YA and YB rapidly neutralized the acidic medium and shifted the pH of the culture from 5.0 to 8.6 within 48 h, whereas the WHT morphotype shifted the pH to approximately 6.5 within the same time frame and grew to a higher density (Fig. 4A and B).
FIG 4 .
Yellow variants alkalize growth medium without production of ammonia. (A and B) WHT (white circles), YA (yellow squares), or YB (orange triangles) bacteria were grown in LB (pH 7) and sampled at 0, 3, 6, 12, 24, and 48 h for OD600 (A) and pH (B). (C) Cultures at experimental endpoints (48 h) were harvested and assayed for ammonia content. ** denotes P < .01 and *** denotes P < 0.001.
Ammonia is a common byproduct of bacterial growth on amino acids, and its production is a mechanism for acid neutralization employed by other stomach-tolerant bacteria (23). The presence of ammonia in culture medium might possibly explain the observed increase in culture pH of yellow variant cultures. Ammonia levels were therefore assayed at experimental endpoints. Only the WHT-morphotype cultures produce large amounts of ammonia as a byproduct of growth on rich medium (Fig. 4C). Unexpectedly, the yellow variants produce significantly less ammonia under the same conditions. YA and YB were capable of producing ammonia to the same degree as the WHT bacteria when grown in defined medium using amino acids as the sole carbon and nitrogen sources (not shown), indicating that yellow variant bacteria were not deficient in the capacity for ammonia production or growth on amino acids.
Colony morphotypes display divergent infection strategies.
Hypoxia and acid stress are key characteristics of the gastric microenvironment, and because the stomach is the one site in healthy individuals with both selective pressures, we chose to determine, using a mouse model of melioidosis, if yellow colony variants of B. pseudomallei had an advantage during gastric infection. WHT, YA, or YB bacteria were used to infect BALB/c mice orally. Mice were monitored for up to 66 days and euthanized at predetermined endpoints. Infection with the WHT morphotype produced severe acute disease and mortality in 50% of mice by day 12 (Fig. 5). Infection with the YA morphotype produced a trend toward more-severe acute disease, resulting in death of 70% of the mice by 12 days postinfection. In contrast, infection with the YB morphotype typically resulted in a latent colonization and delayed presentation of melioidosis, with only 2 in 9 mice developing acute disease relatively late in the model. We considered two courses of infection, acute and chronic, with the acute course from day 0 of infection to day 12 and the chronic infection lasting for the duration of the experiment. Lethality of infection with YB significantly differed from that of infection with WHT or YA over the acute course: WHT and YA are significantly more lethal than YB over the acute course, but WHT and YA are not significantly different in their lethality during acute infection.
FIG 5 .

Yellow variants display altered virulence following oral infection. BALB/c mice (n = 10 per group) were infected with ~104 CFU of B. pseudomallei orally: WHT (white circles), YA (yellow squares), and YB (orange triangles). Mice were monitored for survival and euthanized upon reaching predetermined endpoints. Data were pooled from two independent experiments. *, P < 0.05 between YB and WHT at day 12 (acute infection) and between YA and YB at day 66 (chronic infection); **, P < 0.01 between YA and YB at day 12 in infection.
To identify sites of bacterial persistence following oral infection, bacterial burdens in mice surviving to day 66 or 71 were determined. All tissues from survivors infected with the WHT or YA morphotype were negative for colonies of B. pseudomallei (limit of detection, 1 to 10 CFU/organ) (Fig. 6). Three of the six surviving mice infected with the YB morphotype were positive for colonies of B. pseudomallei in both stomach and feces. One animal also yielded a liver colonization of 20 CFU. If the animals infected with YB (which caused chronic but not acute infection) are compared to those infected with WHT and YA combined (i.e., those that caused acute infection), YB was statistically more likely to achieve chronic gastric carriage and shedding than were WHT and YA combined (P = 0.044). Colonies recovered from YB-infected animals also displayed YB morphology when subcultured on LB agar plates. Thus, the chronically persistent YB morphotype was the only form capable of persistent gastric colonization and colonized with little evidence of disease until late in the model.
FIG 6 .
Only YB variants persist in the gastrointestinal (GI) tract of mice. BALB/c mice were infected with ~104 CFU of B. pseudomallei orally: WHT (white circles; 2.2 × 104 CFU), YA (yellow squares; 8.3 × 103 CFU), and YB (orange triangles; 5.7 × 103 CFU). Bacterial burdens were determined in mice surviving 66 or 71 days postinfection (WHT, n = 5; YA, n = 3; YB, n = 6). Bacterial burdens in stomach, distal GI tract (small intestine, cecum, and colon), feces, lung, liver, and spleen were determined. Data are graphed as individual values with bars representing the mean log10 CFU/organ titer (feces = log10 CFU/gram). The limit of detection for lung, liver, and spleen was 10 CFU/organ; that for stomach and distal GI tract was 1 CFU/organ; and that for feces was 10 to 20 CFU/g. Data were pooled from two independent experiments. *, YB caused chronic infection relative to the combined WHT and YA morphotypes (P = 0.044).
YB bacteria develop a focal site of colonization in the stomach.
To verify stomach colonization by YB bacteria, stomachs from two mice infected with YB were collected at 30 and 66 days postinfection and washed. Tissues were fixed, sectioned, stained with fluorescent in situ hybridization (FISH) probes, and inspected by fluorescence microscopy (Fig. 7). Four different sites in the stomach, one in the antrum and three in the corpus, were identified by FISH as positive for B. pseudomallei. Bacteria appeared to group together in a focal, colony-like structure. This community was observed to be lumenal to the gastric mucin layer and associated with the epithelium. Since stomachs were flushed prior to processing, these foci were at least moderately adherent to the mucin layer. These bacterial foci were also observed to be surrounded by 4′,6-diamidino-2-phenylindole (DAPI)-staining material, indicating the association of eDNA with the bacteria.
FIG 7 .
YB variants form gastric colonies. BALB/c mice were infected orally with YB variant B. pseudomallei and euthanized at day 66 for mouse 3 and day 30 for mouse 5. Stomach tissues were fixed and stained with FISH probes. B. pseudomallei was labeled red, enteric bacteria were labeled green, and DNA was stained blue (DAPI). (A) Images at ×200 final magnification. (B) Images at ×1,000 final magnification. (C) Control image stained with the irrelevant FISH probe (green and red) and DAPI (blue) and captured at ×200 final magnification. (D) Sites of B. pseudomallei colonization indicated with green boxes on a representative stomach image.
YB bacteria produce large amounts of eDNA.
Many different extracellular polymers can comprise the matrices of different microbial communities, but two observations indicated that YB established gastric colonies by production of an extracellular DNA (eDNA) matrix. First, the DAPI staining observations revealed eDNA in the microcolony matrix (Fig. 7), and second, static in vitro cultures of YB (and, to a lesser extent, YA) were observed to be highly viscous during manipulations. Viscosity in this system was abolished with DNase I, suggesting that the foundation of the extracellular matrix produced by YB was indeed eDNA.
An in vitro stomach model was established to investigate the behaviors of colony variants under conditions approximating a mouse stomach at the peak of a meal. This model consisted of gastric mucin-coated multiwell plates, a moderately acidic rich growth medium (LB, pH 5), 2.0% oxygen tension, and a nonagitated culture. Production of eDNA by YB in this model was dependent on attachment or presence of mucin, as viscosity was not observed in wells inoculated with YB in the absence of mucin. The supernatant of well contents was analyzed by extracting cell-clarified supernatant from each well with phenol-chloroform and analyzing the extracted aqueous phase by agarose gel electrophoresis and ethidium bromide staining (Fig. 8B). In this analysis, bands from all colony types were observed. These bands were degraded with addition of DNase I, which confirmed the presence of eDNA in the in vitro stomach model (Fig. 8B).
FIG 8 .
YB produces abundant eDNA in vitro. Parental WHT (white circles), YA (yellow squares), and YB (orange triangles) bacteria were grown in an in vitro stomach model. (A and B) At 0, 4, 8, 12, 16, 20, 24, and 48 h, the model growth system was assayed for CFU (A), or supernatants were analyzed by agarose gel electrophoresis for extracellular DNA (B): lane 1, WHT at 12 h; lane 2, YA at 12 h; lane 3, YB at 12 h; lane 4, WHT at 24 h; lane 5, YA at 24 h; lane 6, YB at 24 h; lane 7, WHT at 48 h; lane 8, YA at 48 h; lane 9, YB at 48 h (B, lower panel). The presence of DNA in supernatant fractions was confirmed by addition of DNase I. (C) Quantification of eDNA produced by WHT, YA, and YB was performed in the gel. (D) eDNA levels were converted to theoretical genomes, and the increase in extracellular genomes per CFU per hour was calculated for each time period in the time course. *** denotes statistical significance of P < 0.001.
Extracellular DNA in the settled culture model was observed to increase dramatically in YB-inoculated wells but not in WHT-inoculated wells and to a lesser degree in YA-inoculated wells (Fig. 8C). Accumulation of eDNA inoculated with YB was significantly higher than that for WHT (Fig. 8C). The bulk of eDNA accumulation in YB-inoculated wells occurred between 16 and 24 h (Fig. 8C). This coincided with a period when there was no measurable loss of CFU (Fig. 8A). Comparison of eDNA production levels by all three variants over the time course revealed differences between YB and WHT at 24 and 48 h but no significant difference between WHT and YA at any point.
The source of eDNA in biofilms has been an issue of contention. Our data suggest that the source of eDNA, at least in B. pseudomallei biofilms, is extrusion by live cells and not lysis. Although YB does undergo a loss of CFU between 12 and 16 h of approximately 7.96 107 CFU, which equates to only 6.23 × 102 ng eDNA, yet we observe an increase in eDNA of 1.88 × 103 ng (the equivalent of 2.4 × 108 genomes). This trend of increasing eDNA continues from 12 h until the end of the experiment at 48 h, although the rate at which it is produced drops between 24 and 48 h. If the observed eDNA was due to lysis, then the living bacteria in the system would need to produce genomes and their accompanying cellular biomass purely for lysis at an astoundingly increasing rate as described by the exponential regression equation y = (5 × 108)/x5.87, where y is minutes per extracellular genome and x is minutes under hypoxia (from 12 to 24 h) (r2 = 0.982). Thus, the bacteria in the system would need to be generating approximately 7 genomes per bacterium per hour to produce the observed eDNA and all without an apparent drop in live-cell titers—a feat not matched by B. pseudomallei growing at a maximum growth rate of 63.6 min under optimum growth conditions (Table 1).
YB bacteria survive the cytopathic effects of SGF.
At 24 h in the in vitro model of gastric colonization described above, after addition of synthetic gastric fluid (SGF) at pH 2, YB exhibited only an average 19% loss of viable cells, whereas YA and WHT lost 88% and 97%, respectively (Fig. 9). No protection against acid was observed for any type in agitated cultures. Treatment of agitated cultures with culture medium below pH 3 caused a loss in viable cells immediately after addition, with no viable bacteria measurable after 1 h of exposure (not shown).
FIG 9 .

YB exhibits enhanced protection in synthetic gastric fluid. Survival of WHT, YA, and YB colony variants was determined in an in vitro stomach model at 24 h. Percent survival was calculated relative to known bacterial titers for this time point. ** denotes P < .01 and *** denotes P < 0.001.
The transcriptional regulator BPSL1887 controls yellow phenotypes.
To determine genetic elements responsible for the yellow colony phenotype, we mutagenized the white form of strain K96243 with the mariner transposon pTBurk1. Of approximately 25,000 mutants isolated, 5 were strikingly yellow and rough upon initial observation of the mutant colony (Fig. 10C). No smooth, yellow transposon mutants were identified in the screen. Identification of the site of insertion for the mobile genetic element was determined by arbitrary PCR (ARB PCR) for all 5 of the yellow transposon mutants (see the supplemental material). Of the five mutants obtained, 2 were determined to be sisters to one of 3 unique mutants. All 3 unique insertions occurred in a 50-bp region immediately 5′ to the gene encoding a sigma 54-dependent transcriptional regulator, BPSL1887, and all three occurred in the same orientation—with the kanamycin resistance cassette being transcribed toward the open reading frame of BPSL1887, suggesting a gain-of-function mutation in these mutants. When the coding sequence of BPSL1887 was expressed in a wild-type background from the arabinose-inducible overexpression vector pHERD-Km, yellow colony morphology was observed (Fig. 10A).
FIG 10 .

Deletion of BPSL1887 (yelR) abrogates yellow pigment production. Replicative plasmid containing yelR (pHERD::yelR) electroporated into WHT wild-type B. pseudomallei K96243 (A), yelR deletion mutant (B), yellow transposon mutant (C), and yelR deletion mutant complemented with the pHERD::yelR plasmid (D) were grown on LB agar for 48 h.
When BPSL1887 was deleted from the wild-type genome using an allelic exchange system (24), the resulting mutant strain never developed yellow variant morphology under the same conditions used to switch the parental strain to yellow forms (Fig. 10B). Deletion of BPSL1887 ablated the ability of the bacteria to produce eDNA (see Fig. S1 in the supplemental material). Deletion of BPSL1887 also significantly removed the bacterium’s ability to alkalize growth medium but increased ammonia production as observed in the WHT form (see Fig. S1).
Complementation of the mutant with the overexpression vector pHERD::BPSL1887 shifted mutant BPSL1887 bacterial colonies to a yellow morphotype most similar to YA (Fig. 10D). Complementation of the mutant also restored the phenotypes of medium alkalization, diminished ammonia production, and eDNA production (see Fig. S1 in the supplemental material). However, the complemented strains demonstrated intermediate phenotypes, suggesting that loss of endogenous transcriptional control negatively impacted regulation of these phenotypes. Since deletion and complementation of BPSL1887 removed and restored expression of the phenotypes of colony appearance, medium alkalization, ammonia production, and eDNA production, respectively, we propose to name the gene BPSL1887 “yelR” for Yellow program Regulator.
yelR is critical for hypoxic growth.
A random library of 25,000 transposon mutants was collected and subjected to selection by entrance into hypoxia and eventually anaerobiosis using a model described earlier (25). Survivors were then analyzed by high-throughput transposon sequencing (TnSeq) to determine genes important for growth under hypoxia and anaerobiosis. Several gain-of-function mutants were identified that were highly enriched in the final output libraries. The primary genomic region enriched by hypoxic growth was the intergenic region between BPSL1888 and BPSL1887 and the intragenic region within BPSL1888 5′ to BPSL1887 (yelR) (see Table S2 in the supplemental material) covering 9-TA dinucleotide transposon insertion sites. We observed an increase for the region from 6.52% of the total reads to 46.4% of the total reads by 60 h in the hypoxia model (see Table S2). Sixty hours is a point at which the oxygen within the system is fully consumed and bacterial growth stops (25). This enrichment for transposons upstream of yelR further establishes YelR as a critical regulator of hypoxic growth.
DISCUSSION
Environmental bacteria routinely adapt to dramatic changes in their surroundings. Although many of these challenges can be met by transcriptional changes in response to stress, other situations require either more extensive adaptations or a unique physiological structure already in place to survive in the new environment. In these instances, bacteria take advantage of population heterogeneity to survive predictable fluctuations in their environment. Population heterogeneity can be mediated by several mechanisms, including reversible, epigenetic mechanisms that allow genetically identical bacteria to behave in radically divergent manners under the same circumstance (26). In some instances, population heterogeneity is manifested by altered colony morphologies, as is the case for the yellow and white colony variants of B. pseudomallei.
We have established that B. pseudomallei strain K96243 can persist in at least three semistable reversible states, one white and two that are yellow on rich solid medium. The reversibility of each form and the lack of detectable mutations in the yellow variants, including the yelR locus, by whole-genome sequencing (see Fig. S3 in the supplemental material) demonstrate that the B. pseudomallei variants represent a heterogeneous tristable population. Interestingly, the parental white morphotype is most often the form isolated from the environment while colony morphotypes isolated directly from patients are often more variable (18). A large number of different colony morphologies have been described for B. pseudomallei, including 7 general morphotypes (18); thus, it is possible that additional population diversity exists beyond the three types described here. However, the large number of described colony morphotypes is likely a result of a combination of different parental genetic backgrounds combined with a defined number of reversible mechanisms for population heterogeneity.
The finding that expression of yelR mediated the phenotypic switch from the white form to a yellow form is consistent with colony variation being mediated by a bistable switch. The molecular mechanisms underlying bistability are numerous but often rely on the stability of a single master regulatory protein (27). The master regulator often has autoregulatory properties such that it can induce its own expression and is typically degraded or otherwise rendered inert under most circumstances, although its concentration may be influenced by environmental factors. When the regulatory protein achieves a steady-state concentration sufficient to maintain its own expression, due to loss of degradation or a stochastic increase in its own expression, the phenotypes governed by the regulator are then expressed. We have demonstrated that increasing yelR expression shifted the white form to a yellow form. Conversely, eliminating yelR ablated the ability to shift to either yellow form. These observations and the reversible and semistable nature of the variants suggest that YelR is part of a regulatory network controlling bistable or perhaps tristable expression of the yellow variant phenotypes.
While it is evident that the yellow variants have a growth advantage under microaerobic conditions, these conditions did not appear to initiate the original shift to the yellow forms. The time required for yellow forms to be firmly established in a microaerobic culture was highly variable unless the original culture contained measurable yellow forms at the start of the experiment. Thus, it appears that stochastic mechanisms, perhaps influenced by culture age, generated the initial yellow variants, while the proportion of yellow variants in a population was influenced by environmental factors such as hypoxia.
While the harsh conditions of the stomach make bacterial colonization rare, the unique characteristics of the yellow variants are ideal for gastric colonization. The stomach is a severely hypoxic environment with little delivery of oxygen from the circulatory system and small amounts of atmospheric gases swallowed during meals (28). As a reference, the antrum of the mammalian stomach measures at 25 to 30 mm Hg (2.95% atmospheric oxygen) (29). The metabolism of both yellow forms was highly adapted to microaerobic conditions, as they displayed a significant growth advantage during hypoxia, while growth in well-aerated cultures resulted in a shift back to the white parental form. The extreme acidity of the stomach is the primary challenge for bacterial survival, with a resting pH of approximately 1.5 and a postmeal pH of 6 to 7 (30). We have shown that B. pseudomallei yellow variants rapidly alkalized acidic environments and did so by a novel mechanism without production of ammonia. Not only did the YB variant neutralize its local environment, but it was also significantly more tolerant to extreme acidic conditions. The YB acid tolerance was due in part to formation of an organized microbial community, as agitation or mechanical disruption of an extracellular DNA matrix resulted in no enhanced protection for the variant.
B. pseudomallei was recently discovered in the stomach microbiota in a panel of human volunteers (17). To determine if the unique characteristics of the yellow variants provided an advantage during infection, we used the recently described mouse model of gastric colonization (16). Using this model of melioidosis, we found that the YA variant was as virulent as the parental white form; however, the YB variant was severely attenuated during the early stages of infection and developed disease less frequently and much later in infection. However, only the YB form was able to colonize and persist in the stomach. Either the white and YA forms killed the mouse rapidly, or they were rapidly cleared. These findings demonstrate that YB attenuates virulence and allows for persistent gastric colonization similar to what was observed for the strain (1026b) that was used for development of the gastrointestinal model (16). These findings suggest that infections that progress to acute disease are due to the expression of the white or YA morphotype and that an infection that establishes a chronic infection is due to expression of the YB morphotype. Furthermore, the late onset of disease observed in YB-infected mice indicates that regression and development of acute disease symptoms following apparent recovery could be due to a switch away from the YB morphotype to the WHT or YA morphotype. Such a switch may explain the eventual development of systemic disease in some YB-infected mice, with one animal dying at 15 days, another dying at 30 days, and a third showing splenic bacteria and signs of acute disease at the experimental endpoint.
Interestingly, all B. pseudomallei bacteria that had colonized the stomach were found in DNA-encased foci. Hypoxia and acid stress are not the only barriers to stomach colonization. The contents of the organ are mechanically cleared into the duodenum and anaerobic small intestine on a regular basis by contraction of the smooth muscle layer surrounding it. Extracellular biopolymer matrices that provide structure to bacterial communities can help a population resist mechanical clearance due to their adhesive properties. Extracellular DNA has been identified as an essential component of biofilms of Pseudomonas aeruginosa, Bacillus cereus, and a variety of other bacteria (31–34). The extremely long polyanionic nucleotide polymer structure of DNA is ideally suited for use as a structural component for a microbial community in acidic environments. As a covalently linked polymer, often micrometers in length, it can physically retain adhered objects against mechanically dispersive forces. The phosphate backbone of DNA can also act as a buffering agent and help establish a more neutral localized pH. We have shown that YB produced large amounts of eDNA in vitro. The rapid eDNA accumulation in YB cultures, which reached a rate of 7 genomes per hour, was far beyond the maximum growth rate of less than one division per hour for B. pseudomallei under optimum growth conditions. These findings indicated that eDNA accumulation was likely from DNA extrusion from live bacteria, as the growth and death rate necessary to produce the observed eDNA accumulation by cell lysis would have had to be 7 times the maximum growth rate observed for B. pseudomallei. Interestingly, genes within two open reading frames of yelR are annotated as genes encoding components of a type IV pilus system. The type IV pilus system has been implicated as important for biofilm formation and maintenance of biofilm architecture, as well as DNA exchange, in multiple species (35). The type IV pili could potentially be involved in YB attaching to the DNA matrix to enhance gastric colonization.
The findings of this study suggest that, if ingested as a mixed population, the WHT-type bacteria would be selected against by the harsh gastric environment and yellow-form B. pseudomallei could adhere to the gastric mucous layer. Once adhered, the yellow variant bacteria could develop an attached microbial community. This community would be protected from acid stress by a thick extracellular matrix, as observed in mouse stomachs. Because gastric mucosa is degraded at the lumenal face by the action of acid on the intermolecular bonds (36), a microbial community that buffers the mucosa from acid exposure could stabilize the mucous layer, thus providing a potentially long-term niche environment for the pathogen to inhabit. A switch to the white or YA form would allow escape from the gastric community and a switch to invasive disease. It is likely that once the yellow forms exit the body via feces, some would switch to the more environmentally suited white form due to increased oxygen availability outside the gastrointestinal tract. However, as yelR homologs exist in related species such as the pathogenic B. mallei, as well as the nonpathogenic Burkholderia thailandensis and Burkholderia oklahomensis, it is likely that the yellow forms evolved in Burkholderia to survive rapid changes in pH and oxygen in the environment. Changes in both pH and oxygen have drastic effects on proton translocation, suggesting a role in proton homeostasis for the yellow forms. These mechanisms seem to have been exploited by B. pseudomallei to establish a chronic infection within mammalian hosts.
Conclusions.
The findings presented here demonstrate that the yellow colony morphotypes of B. pseudomallei are a far-reaching specific physiological form with major implications for persistent colonization and virulence. The yellow variants are governed at least in part by the transcriptional regulator YelR, which is both necessary and sufficient for control of the yellow variant phenotypes. It appears that the different colony morphotypes of B. pseudomallei are a strategy for the pathogen to inhabit the diverse niches encountered during environmental and infectious stages of its life cycle. The yellow variants are ideally suited for acidic and hypoxic environments found during infection, especially in the stomach and perhaps abscesses. The unique ability of the YB form to produce vast amounts of eDNA and persist on the gastric mucosa indicates that it is a form employed specifically for gastric colonization.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All in vitro experiments were performed in a Select Agent-approved biosafety level 3 (BSL-3) facility at the University of Colorado, Anschutz Medical Campus. Except where noted, B. pseudomallei strain K96243 was used (37). White-type bacteria are designated WHT, type A yellow variants are designated YA, and type B yellow variants are designated YB. Unless otherwise noted, bacteria were grown in Luria-Bertani-Lennox (LB) medium prepared at either pH 7 or pH 5 at 37°C and shaken at 275 rpm. Overnight cultures were grown in 15-ml conical-bottom Bio-Reaction tubes. Aerobic cultures and outgrowth cultures were grown in plastic 125-ml vented, screw-cap Erlenmeyer flasks. For in vivo experiments, overnight cultures were grown in LB-Miller (LB-M) broth, pH 7, at 37°C. Stock solutions were stored in 20% (vol/vol) sterile glycerol at −80°C.
Growth rate determination.
Overnight cultures were diluted 1:100 in fresh LB medium and incubated with shaking for 3 h. Cultures were normalized to an optical density at 600 nm (OD600) of 0.1, returned to the shaking incubator, and allowed to grow for 12 h. Cultures were then subcultured to an OD600 of 0.1 and returned to the incubator. At 20-min intervals, cultures were assayed for OD600. Growth rate (G) was calculated according to the standard method.
Reversion rate determination.
A colony of B. pseudomallei was used to inoculate an overnight culture and then normalized to an OD600 of 0.1 in LB broth at pH 5. For hypoxic cultures, 3 ml of normalized culture was placed in a 20-mm by 125-mm glass tube containing a 12- by 7-mm octagonal magnetic stir bar and fitted with a screw cap. The caps were left loose, and cultures were stirred in a hypoxia chamber equilibrated to 0.2% O2 (Coy Laboratories) and were sampled at 6, 12, 24, 48, 72, and 96 h. For aerobic cultures, 25 ml of normalized culture was placed in a 125-ml vented, screw-cap Erlenmeyer flask and shaken at 315 rpm. After entrance into log phase, cultures were diluted to an OD600 of 0.1; sampled at 0, 2, 4, 6, and 8 h; and then assayed for percent composition of colony type. At least 1,000 colonies per replicate per data point were counted and typed. Reversion rates were calculated according to the following formula (38), where ΔNR is the change in number of reverting or white colonies, ΔNT is the change in total number of bacteria, and R is the reversion rate: R = (ΔNR/ΔNT)/ln2. R is reported in the percent chance of the switching event occurring per cell per generation.
Competitive advantage of yellow colony variants during hypoxia.
YA, YB, or WHT cultures were mixed with a white-type K96243 xylE+ culture at 1:100 (test strain to control strain) and incubated under either 0.2% O2 or atmospheric O2. Overnight cultures were diluted to an OD600 of 0.1 in LB (pH 5), and 1% of normalized xylE+ culture was replaced with an equal volume of WHT-, YA-, or YB-type culture at the same OD600. For hypoxic competitions, 3 ml of mixed culture was placed in the stirred tube model (described above) and sampled at 0, 6, 12, 24, and 48 h for percent distribution of both genotype and phenotype. For aerobic competitions, 25 ml of mixed culture was grown at 37°C with shaking at 315 rpm and sampled at 0, 2, 4, 6, and 8 h for percent distribution of both genotype and phenotype by plating an aliquot of dilute culture onto an LB agar plate. Once colonies were counted and typed for phenotype, plates were sprayed with pyrocatechol as described previously (24) and were counted a second time to record genotype.
pH shift and ammonia production.
Overnight cultures were normalized to an OD600 of 0.1 in 25 ml LB (pH 5), and shaking cultures were sampled at 3, 6, 12, 24, and 48 h to determine OD600, CFU/ml, and pH. At 48 h, 10 ml of each culture was clarified by centrifugation and supernatant was filter sterilized twice by passage through a 0.2-µm filter device. Sterile samples were assayed for ammonia content as described previously (25).
Mouse infections.
Female BALB/c mice were purchased from Jackson Laboratories. Mice were 9 to 36 weeks of age at the time of infection. All experiments involving animals were approved by the Institutional Animal Care and Use Committee at Colorado State University (CSU), All in vivo experiments were performed in a BSL-3 facility at CSU. Animal infections were performed as described previously (16). Oral inoculations were performed using a stainless steel 22-gauge gavage needle, and mice were inoculated using a total volume of 100 µl at doses listed in the text.
Quantification of tissue bacterial burden and fecal shedding.
Bacterial burdens were quantified as described previously (39). Briefly, mice were euthanized and organs were placed in 4 ml sterile phosphate-buffered saline (PBS). Stomach and cecum tissues were sectioned. Organs were homogenized using a Stomacher 80 Biomaster (Seward) homogenizer and serially diluted in sterile PBS. Lung, liver, and spleen were plated on LB-M agar plates. Stomach, small intestine, cecum, and colon homogenates were plated on NAP-A agar (16, 40). For quantification of fecal shedding, fecal pellets were collected and processed as described previously (16). Briefly, pellets were collected from individual mice in plastic containers and transferred to sterile PBS at a concentration of ~0.1 g per ml. Fecal pellets were homogenized by mechanical disruption in a Whirl-Pak stomacher bag (Nasco), and serial dilutions were prepared in sterile PBS and plated on NAP-A agar plates.
FISH.
Fluorescent in situ hybridization (FISH) was performed as described previously, with slight modifications (16). Briefly, antisense single-stranded DNA (ssDNA) probes were purchased from Integrated DNA Technologies. Probes used in this study included the B. pseudomallei-specific probes Bpm427 and Bpm975, as well as the Eub338 probe, which recognizes a sequence present in all bacteria, and the irrelevant control probe Non338 (see Table S1 in the supplemental material). Bpm427 and Bpm975 were labeled with Cy3, Eub338 was labeled with Alexa Fluor 488, and Non338 was labeled with Cy3 or Alexa Fluor 488. Stomach tissue sections were permeabilized by heating in 10 mM citrate buffer, pH 6.1, at 90°C for 12 min and allowed to cool gradually at room temperature. After a water wash, tissues were treated with 0.3% Triton X-100 in PBS for 6 min at room temperature and washed in PBS. Next, tissue sections were hybridized with ssDNA probes diluted in hybridization buffer (16). Tissues were hybridized with a cocktail of Bpm427, Bpm975, and Eub338 probes each used at a final concentration of 1 µg/ml, or the Non338 probe conjugated to Cy3 or Alexa 488 at a final concentration of 2 or 1 µg/ml, respectively. Probes were hybridized in a humidified chamber at 37°C for 3 h. Sections were washed for 15 min in 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate) at 37°C, for 15 min in 1× SSC at 54°C, for 15 min in 0.5× SSC at 54°C, and for 10 min in 0.5× SSC at room temperature. Slides were stained with 100 ng/ml DAPI for 10 min at room temperature, washed in 2× SSC, air dried, and mounted with Pro-Long Gold (Life Technologies). Fluorescence microscopy was performed on a Leica DM4500 fluorescence microscope (Leica Microsystems Inc.). Images were captured with a Retiga-2000R camera (Q-Imaging) utilizing QCapture Pro software version 5.1 (Q-Imaging).
eDNA quantification and CFU determination in settled culture model.
Five hundred microliters each of an overnight culture of WHT, YA, or YB type diluted to an OD600 of 0.1 in LB at pH 5 was placed in a single well of a mucin-coated 24-well plate. At 4, 8, 12, 16, 20, and 24 h, one well was used to determine both CFU and eDNA per well. One milliliter of PBS was added to each well and was aggressively agitated by pipetting. The 1.5-ml volume of each well was transferred to a 2-ml screw-cap tube and vortexed for 30 s. A 20-µl aliquot of the resulting suspension was then plated for CFU, and the remainder was clarified by centrifugation at maximum speed for 5 min. Seven hundred fifty microliters of supernatant was removed to a 1.5-ml tube, and 750 µl of Tris-saturated phenol-chloroform-isoamyl alcohol (25:24:1) was added. Tubes were mixed aggressively for 10 s and then intermittently for 2 min. Phases were then separated by centrifugation at maximum speed for 15 min. The aqueous phase was removed to a new tube. Fifteen-microliter aliquots of the aqueous phase from each sample were loaded into an agarose gel, run, and then stained with ethidium bromide. Gels were visualized on a Bio-Rad ChemiDoc XRS gel imaging station, and bands were quantified with ImageLab software (Bio-Rad) using 0.3 µg of GeneRuler 1KB Plus quantitative DNA ladder (Fermentas Life Sciences) as a standard (r2 minimum of 0.90). Mucin-coated plates were prepared as described earlier (41) with the modification that the mucin was sterilized before use as described previously (42), and 500 µl of solubilized mucin was added per well to tissue-culture-treated 24-well plates.
Percent survival in presence of synthetic gastric fluid.
Settled cultures of WHT, YA, and YB were prepared as described above. Briefly, mucin-coated 24-well plates were inoculated with 0.5 ml of culture at an OD600 of 0.10 and grown in the hypoxic chamber at 2% O2 for 24 h. At 24 h of growth, 0.5 ml prewarmed synthetic gastric fluid (SGF) at pH 2 was added to each well. Multiwell plates were returned to the hypoxia chamber and incubated at 37°C for 2.5 h. Multiwell plates were removed, and 0.5 ml sterile-filtered 0.5% (wt/vol) sodium bicarbonate was carefully added to each well. The bicarbonate was allowed to neutralize the acid in SGF for 15 min at room temperature without agitation. The contents of each well were homogenized, and 20 µl of each well was assayed for CFU/ml. SGF was prepared as described previously (43) with the following minor modifications: the final pH of SGF was adjusted to pH 2, and nutrient medium was sterilized by vacuum filtration. Lysozyme and pepsin were reconstituted and added to SGF immediately before use.
Plasmids, primers, and strain construction.
Escherichia coli strains were grown in LB broth at 30°C, and antibiotics were added as needed at the indicated concentrations: ampicillin, 100 µg/ml; kanamycin, 50 µg/ml; and gentamicin, 30 µg/ml. For selection in B. pseudomallei, LB plates contained kanamycin at 200 µg/ml and zeocin at 50 µg/ml. The overexpression vector pHERD was generated from pHERD30T (44) and pMO130 (24) using the kanamycin resistance marker from pMO130 and the restriction sites BsrGI and AclI. The overexpression vector pHERD::BPSL1887 was generated for this study using primers listed in Table S1 in the supplemental material. The BPSL1887 sequence was then cloned into the pGEM-T Easy (Invitrogen) vector using the manufacturer’s protocol. The xylE-integrating vector pCA001 was generated from pMO130 by inserting two adjacent DNA sequences with chromosomal homology into the two restriction sites flanking the xylE marker (see Text S1 in the supplemental material). This vector was transformed into electrocompetent E. coli S17-1, and positively selected colonies were mated with wild-type B. pseudomallei as previously described (24). XylE-positive cointegrants were plated on LB selection agar and identified by screening with pyrocatechol. Single colonies were resolved of the plasmid as described earlier (24). Clones retaining the xylE marker were identified by screening with pyrocatechol and verified for loss of the plasmid by demonstrating sensitivity to kanamycin. The BPSL1887 knockout vector pMO130::BPSL1887KO was generated from pMO130 using the strategy described in Text S1 and primers listed in Table S1 in the supplemental material. Plasmids with correct inserts were transformed into chemically competent E. coli S17-1, and the resulting strain was used to mate the knockout construct into wild-type, WHT B. pseudomallei K96243. The knockout was derived as described previously (24). The transposon pTBurk1, which contains T7 promoters behind both inverted repeats of the transposable element, was generated from pMar2xT7 (45) and pHBurk1 (46) as described in Text S1 in the supplemental material.
Transposon mutagenesis.
Transposition was performed as described for pHBurk1 (46) with minor modifications, including that transposon delivery into B. pseudomallei was achieved using electroporation as described previously (47). Briefly, an overnight culture of wild-type B. pseudomallei was diluted 1:10 and grown for 3 h at 42°C. Ten milliliters of culture was pelleted by centrifugation and washed 3 times with 0.3 M sterile sucrose. Cells were suspended in 0.3 M sucrose to which was added 10 ng pTBurk1. The suspension was mixed and electroporated. Nine hundred microliters of LB was added, and cells were allowed to recover for 30 min at 30°C before they were then diluted, spread onto LB selection agar, and incubated at 37°C. Transposons that were starkly yellow after initial selection were struck for isolation on LB selection agar and stocked. The site of transposon insertion for isolated yellow transposons was determined by arbitrary PCR (ARB PCR) (see Text S1 in the supplemental material). ARB PCR products were Sanger sequenced by the University of Colorado Cancer Center DNA Sequencing Center at UCD Anschutz using the ARBseq primer. Twenty-five thousand transposon mutants were collected for TnSeq.
TnSeq.
High-throughput transposon sequencing was performed as previously described (48). Briefly, total genomic DNA (gDNA) from input and output libraries was collected using the Qiagen DNeasy blood and tissue kit. Sequencing libraries were prepared by shearing total DNA. Fragment ends were repaired using the NEBNExt end repair module (New England Biolabs), cleaned using the Qiagen PCR purification kit, and then 3′ polyadenylated using Klenow fragment Exo (New England Biolabs). Polyadenylated products were cleaned using the Qiagen PCR purification kit and then ligated to adapter oligonucleotides (see Table S1 in the supplemental material) using T4 DNA ligase. Gel extraction was performed on adapter-ligated fragments of 200 to 300 bp in size. Sites of transposon insertion were enriched by PCR using primers PE PCR 1.0 and HITS PCR 2.0-Indexx 1, 2, 3, 4, or 5 (see Table S1).
Final sequencing libraries were purified by and quantified using the Agilent Bioanalyzer DNA7500 chip, multiplexed, cluster amplified, and sequenced on the Illumina MiSeq platform. High-quality sequencing reads containing the Himar1 internal repeat (IR) sequence and the adjacent TA were selected from the raw fastq file for analysis. The internal repeat sequence was removed, and the processed reads were mapped onto the B. pseudomallei K96243 reference genome using the program Bowtie 2 (49). Annotation of TA sites was accomplished using Seqanno (https://github.com/brwnj/seqanno). Seqanno scripts were used to quantify reads over a given sequence, compile those counts at the gene level, compare results between samples, and add functional annotation using a UniProt flat file for B. pseudomallei.
Statistical analyses.
Data reported here represent the averages of three biological replicates plus and minus the standard deviation. Statistical significances of doubling time (G) and reversion rate were determined by the two-tailed Student t test under nonparametric assumptions. Statistical significance of growth rate constants was determined using one-way analysis of variance (ANOVA). Statistical differences in animal survival were determined by a Mantel-Cox log rank analysis. Differences in tissue colonization frequency were determined by a two-tailed Fisher exact test.
SUPPLEMENTAL MATERIAL
Supplemental methods. Download
Deletion of yelR ablates eDNA production and medium alkalization. (A and B) The yelR deletion strain, complemented strain, wild-type complemented strain, and one of the yellow transposon strains were assayed for pH (A) and ammonia production (B). (C) Determination of eDNA production by deletion and complemented strains was accomplished by growth in the settled culture model. Download
Fold enrichment of transposon mutants 5′ to BPSL1887 after hypoxic conditions. A 25,000-member random transposon library was placed under hypoxic conditions, and bulk DNA was collected before and after hypoxia and then analyzed by TnSeq. Transposon mutants with insertions at TA dinucleotides 5′ to BPSL1887 were observed to dominate the output population as indicated by the increase in relative abundance of their reads observed by TnSeq. The normalized relative read number was used to calculate fold increase of reads at each site. The fold increase was then transformed by log2 and aligned with the appropriate TA dinucleotide—indicated by TA number and red font coloring above the cartoon of the open reading frames for BPSL1888 and BPSL1887 (yelR). Download
Sequence analysis of the yelR (BPSL1887) locus in YA and YB and revertants of YA. Total DNAs from YA, YB, and two white revertants of YA, YAr3 and YAr4, were sequenced on the 454 Roche deep-sequencing platform, trimmed for quality, and aligned with the published consensus sequence for B. pseudomallei K96243 using the -mpileup command in SAMTools. The output was visualized using the Integrative Genome Viewer, with blue indicating mixed parent and nonparent sequence reads at a given site and gray indicating only parent sequence reads at a given site. No nonparent sequences were observed within the coding sequence region for BPSL1887 or BPSL1888. Potential mutations identified throughout the genomes of YA or YB forms were also present in the revertants, indicating that they were not likely involved in the yellow forms. Download
Oligonucleotides used in this study
TnSeq enrichment sites
ACKNOWLEDGMENTS
This work was funded by a grant from the Rocky Mountain Regional Center for Excellence (RMRCE), U54 AI065357, and an internal TransRMRCE grant. C. R. Austin was funded in part by the institutional training grant T32 AI052066.
We thank Ken Jones and Joe Brown of the University of Colorado High-Throughput Next-Generation Sequencing Core for their assistance with the TnSeq.
Footnotes
Citation Austin CR, Goodyear AW, Bartek IL, Stewart A, Sutherland MD, Silva EB, Zweifel A, Vitko NP, Tuanyok A, Highnam G, Mittelman D, Keim P, Schweizer HP, Vázquez-Torres A, Dow SWC, Voskuil MI. 2015. A Burkholderia pseudomallei colony variant necessary for gastric colonization. mBio 6(1):e02462-14. doi:10.1128/mBio.02462-14.
REFERENCES
- 1.Cheng AC, Currie BJ. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin Microbiol Rev 18:383–416. doi: 10.1128/CMR.18.2.383-416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Currie BJ, Dance DA, Cheng AC. 2008. The global distribution of Burkholderia pseudomallei and melioidosis: an update. Trans R Soc Trop Med Hyg 102(Suppl 1):S1–S4. doi: 10.1016/S0035-9203(08)70002-6. [DOI] [PubMed] [Google Scholar]
- 3.Limmathurotsakul D, Peacock SJ. 2011. Melioidosis: a clinical overview. Br Med Bull 99:125–139. doi: 10.1093/bmb/ldr007. [DOI] [PubMed] [Google Scholar]
- 4.Rotz LD, Khan AS, Lillibridge SR, Ostroff SM, Hughes JM. 2002. Public health assessment of potential biological terrorism agents. Emerg Infect Dis 8:225–230. doi: 10.3201/eid0802.010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White N. 2003. Seminar: melioidosis. Lancet 361:1715–1722. doi: 10.1016/S0140-6736(03)13374-0. [DOI] [PubMed] [Google Scholar]
- 6.Vietri NJ, DeShazer D. 2008. Melioidosis, p 147–166. In Dembeck ZF (ed), Medical aspects of biological warfare. Borden Institute, Walter Reed Army Medical Center, Washington, DC. [Google Scholar]
- 7.Leelarasamee A. 1986. Epidemiology of melioidosis. J Infect Dis Antimicrob Agents 3:84–93. [Google Scholar]
- 8.Easton A, Haque A, Chu K, Lukaszewski R, Bancroft GJ. 2007. A critical role for neutrophils in resistance to experimental infection with Burkholderia pseudomallei. J Infect Dis 195:99–107. doi: 10.1086/509810. [DOI] [PubMed] [Google Scholar]
- 9.Leakey AK, Ulett GC, Hirst RG. 1998. BALB/c and C57BL/6 mice infected with virulent Burkholderia pseudomallei provide contrasting animal models for the acute and chronic forms of human melioidosis. Microb Pathog 24:269–275. doi: 10.1006/mpat.1997.0179. [DOI] [PubMed] [Google Scholar]
- 10.Titball RW, Russell P, Cuccui J, Easton A, Haque A, Atkins T, Sarkar-Tyson M, Harley V, Wren B, Bancroft GJ. 2008. Burkholderia pseudomallei: animal models of infection. Trans R Soc Trop Med Hyg 102(Suppl 1):S111–S116. doi: 10.1016/S0035-9203(08)70026-9. [DOI] [PubMed] [Google Scholar]
- 11.Mayo M, Kaesti M, Harrington G, Cheng AC, Ward L, Karp D, Jolly P, Godoy D, Spratt BG, Currie BJ. 2011. Burkholderia pseudomallei in unchlorinated domestic bore water, tropical Northern Australia. Emerg Infect Dis 17:1283–1285. doi: 10.3201/eid1707.100614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker A, Tahani D, Gardiner C, Bristow KL, Greenhill AR, Warner J. 2011. Groundwater seeps facilitate exposure to Burkholderia pseudomallei. Appl Environ Microbiol 77:7243–7246. doi: 10.1128/AEM.05048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inglis TJ, Garrow SC, Henderson M, Clair A, Sampson J, O’Reilly L, Cameron B. 2000. Burkholderia pseudomallei traced to water treatment plant in Australia. Emerg Infect Dis 6:56–59. doi: 10.3201/eid0601.000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Currie BJ, Mayo M, Anstey NM, Donohoe P, Haase A, Kemp DJ. 2001. A cluster of melioidosis cases from an endemic region is clonal and is linked to the water supply using molecular typing of Burkholderia pseudomallei isolates. Am J Trop Med Hyg 65:177–179. [DOI] [PubMed] [Google Scholar]
- 15.Dai D, Chen Y-S, Chen P-S, Chen Y-L. 2012. Case cluster shifting and contaminant source as determinants of melioidosis in Taiwan. Trop Med Int Health 17:1005–1013. doi: 10.1111/j.1365-3156.2012.03036.x. [DOI] [PubMed] [Google Scholar]
- 16.Goodyear A, Bielefeldt-Ohmann H, Schweizer H, Dow S. 2012. Persistent gastric colonization with Burkholderia pseudomallei and dissemination from the gastrointestinal tract following mucosal inoculation of mice. PLoS One 7:e37324. doi: 10.1371/journal.pone.0037324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khosravi Y, Dieye Y, Poh BH, Ng CG, Loke MF, Goh KL, Vadivelu J. 2014. Culturable bacterial microbiota of the stomach of Helicobacter pylori positive and negative gastric disease patients. Sci World J 2014:610421. doi: 10.1155/2014/610421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chantratita N, Wuthiekanun V, Boonbumrung K, Tiyawisutsri R, Vesaratchavest M, Limmathurotsakul D, Chierakul W, Wongratanacheewin S, Pukritiyakamee S, White NJ, Day NP, Peacock SJ. 2007. Biological relevance of colony morphology and phenotypic switching by Burkholderia pseudomallei. J Bacteriol 189:807–817. doi: 10.1128/JB.01258-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nigg C, Ruch J, Scott E, Noble K. 1956. Enhancement of virulence of Malleomyces pseudomallei. J Bacteriol 71:530–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanton A, Fletcher W. 1925. Melioidosis, a disease of rodents communicable to man. Lancet 205:10–13. doi: 10.1016/S0140-6736(01)04724-9. [DOI] [Google Scholar]
- 21.Van Der Woude MW, Bäumler AJ. 2004. Phase and antigenic variation in bacteria. Clin Microbiol Rev 17:581–611. doi: 10.1128/CMR.17.3.581-611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S-H, Chong C-E, Lim B-S, Chai S-J, Sam K-K, Mohamed R, Nathan S. 2007. Burkholderia pseudomallei animal and human isolates from Malaysia exhibit different phenotypic characteristics. Diagn Microbiol Infect Dis 58:263–270. doi: 10.1016/j.diagmicrobio.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 23.Cotter PD, Hill C. 2003. Surviving the acid test: responses of Gram-positive bacteria to low pH. Microbiol Mol Biol Rev 67:429–453. doi: 10.1128/MMBR.67.3.429-453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamad MA, Zajdowicz SL, Holmes RK, Voskuil MI. 2009. An allelic exchange system for compliant genetic manipulation of the select agents Burkholderia pseudomallei and Burkholderia mallei. Gene 430:123–131. doi: 10.1016/j.gene.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamad MA, Austin CR, Stewart AL, Higgins M, Vázquez-Torres A, Voskuil MI. 2011. Adaptation and antibiotic tolerance of anaerobic Burkholderia pseudomallei. Antimicrob Agents Chemother 55:3313–3323. doi: 10.1128/AAC.00953-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veening J-W, Smits WK, Kuipers OP. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol 62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- 27.Dubnau D, Losick R. 2006. Bistability in bacteria. Mol Microbiol 61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- 28.Nestle N, Wunderlich A, Nüssle-Kügele K. 2004. In vivo observation of oxygen-supersaturated water in the human mouth and stomach. Magn Reson Imaging 22:551–556. doi: 10.1016/j.mri.2004.01.040. [DOI] [PubMed] [Google Scholar]
- 29.Kivilaakso E, Ahonen J, Aronsen K-F, Höckerstedt K, Kalima T, Lempinen M, Suoranta H, Vernerson E. 1982. Gastric blood flow, tissue gas tension and microvascular changes during hemorrhage-induced stress ulceration in the pig. Am J Surg 143:322–330. doi: 10.1016/0002-9610(82)90101-5. [DOI] [PubMed] [Google Scholar]
- 30.Fordtran JS, Walsh JH. 1973. Gastric acid secretion rate and buffer content of the stomach after eating. J Clin Invest 52:645–657. doi: 10.1172/JCI107226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steinberger RE, Holden PA. 2005. Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl Environ Microbiol 71:5404–5410. doi: 10.1128/AEM.71.9.5404-5410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vilain S, Pretorius JM, Theron J, Brözel VS. 2009. DNA as an adhesin: Bacillus cereus requires extracellular DNA to form biofilms. Appl Environ Microbiol 75:2861–2868. doi: 10.1128/AEM.01317-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 34.Dominiak DM, Nielsen JL, Nielsen PH. 2011. Extracellular DNA is abundant and important for microcolony strength in mixed microbial biofilms. Environ Microbiol 13:710–721. doi: 10.1111/j.1462-2920.2010.02375.x. [DOI] [PubMed] [Google Scholar]
- 35.Giltner CL, Nguyen Y, Burrows LL. 2012. Type IV pilin proteins: versatile molecular modules. Microbiol Mol Biol Rev 76:740–772. doi: 10.1128/MMBR.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen A, Garner A. 1980. Mucus and bicarbonate secretion in the stomach and their possible role in mucosal protection. Gut 21:249–262. doi: 10.1136/gut.21.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holden MT, Titball RW, Peacock SJ, Cerdeño-Tárraga AM, Atkins T, Crossman LC, Pitt T, Churcher C, Mungall K, Bentley SD, Sebaihia M, Thomson NR, Bason N, Beacham IR, Brooks K, Brown KA, Brown NF, Challis GL, Cherevach I, Chillingworth T, Cronin A, Crossett B, Davis P, DeShazer D, Feltwell T, Fraser A, Hance Z, Hauser H, Holroyd S, Jagels K, Keith KE, Maddison M, Moule S, Price C, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Simmonds M, Songsivilai S, Stevens K, Tumapa S, Vesaratchavest M, Whitehead S, Yeats C, Barrell BG, Oyston PCF, Parkhill J. 2004. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci USA 101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Birge EA. 2006. Bacterial and bacteriophage genetics, 5th ed. Birkhäuser Verlag, Basel, Switzerland. [Google Scholar]
- 39.Goodyear A, Kellihan L, Bielefeldt-Ohmann H, Troyer R, Propst K, Dow S. 2009. Protection from pneumonic infection with Burkholderia species by inhalational immunotherapy. Infect Immun 77:1579–1588. doi: 10.1128/IAI.01384-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodyear A, Strange L, Rholl DA, Silisouk J, Dance DA, Schweizer HP, Dow S. 2013. An improved selective culture medium enhances the isolation of Burkholderia pseudomallei from contaminated specimens. Am J Trop Med Hyg 89:973–982. doi: 10.4269/ajtmh.13-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rose H, Baldwin A, Dowson CG, Mahenthiralingam E. 2009. Biocide susceptibility of the Burkholderia cepacia complex. J Antimicrob Chemother 63:502–510. doi: 10.1093/jac/dkn540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitsui Y, Matsumura K, Kondo C, Takashima R. 1976. The role of mucin on experimental Pseudomonas keratitis in rabbits. Invest Ophthalmol 15:208–210. [PubMed] [Google Scholar]
- 43.Beumer RR, de Vries J, Rombouts FM. 1992. Campylobacter jejuni non-culturable coccoid cells. Int J Food Microbiol 15:153–163. doi: 10.1016/0168-1605(92)90144-R. [DOI] [PubMed] [Google Scholar]
- 44.Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. 2008. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl Environ Microbiol 74:7422–7426. doi: 10.1128/AEM.01369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rholl DA, Trunck LA, Schweizer HP. 2008. In vivo Himar1 transposon mutagenesis of Burkholderia pseudomallei. Appl Environ Microbiol 74:7529–7535. doi: 10.1128/AEM.01973-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi K-H, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 48.Wong SMS, Gawronski JD, Lapointe D, Akerley BJ. 2011. High-throughput insertion tracking by deep sequencing for the analysis of bacterial pathogens. Methods Mol Biol 733:209–222. doi: 10.1007/978-1-61779-089-8_15. [DOI] [PubMed] [Google Scholar]
- 49.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods. Download
Deletion of yelR ablates eDNA production and medium alkalization. (A and B) The yelR deletion strain, complemented strain, wild-type complemented strain, and one of the yellow transposon strains were assayed for pH (A) and ammonia production (B). (C) Determination of eDNA production by deletion and complemented strains was accomplished by growth in the settled culture model. Download
Fold enrichment of transposon mutants 5′ to BPSL1887 after hypoxic conditions. A 25,000-member random transposon library was placed under hypoxic conditions, and bulk DNA was collected before and after hypoxia and then analyzed by TnSeq. Transposon mutants with insertions at TA dinucleotides 5′ to BPSL1887 were observed to dominate the output population as indicated by the increase in relative abundance of their reads observed by TnSeq. The normalized relative read number was used to calculate fold increase of reads at each site. The fold increase was then transformed by log2 and aligned with the appropriate TA dinucleotide—indicated by TA number and red font coloring above the cartoon of the open reading frames for BPSL1888 and BPSL1887 (yelR). Download
Sequence analysis of the yelR (BPSL1887) locus in YA and YB and revertants of YA. Total DNAs from YA, YB, and two white revertants of YA, YAr3 and YAr4, were sequenced on the 454 Roche deep-sequencing platform, trimmed for quality, and aligned with the published consensus sequence for B. pseudomallei K96243 using the -mpileup command in SAMTools. The output was visualized using the Integrative Genome Viewer, with blue indicating mixed parent and nonparent sequence reads at a given site and gray indicating only parent sequence reads at a given site. No nonparent sequences were observed within the coding sequence region for BPSL1887 or BPSL1888. Potential mutations identified throughout the genomes of YA or YB forms were also present in the revertants, indicating that they were not likely involved in the yellow forms. Download
Oligonucleotides used in this study
TnSeq enrichment sites