Abstract
The most recently identified class of actin nucleators, WASp Homology domain 2 (WH2) – nucleators, use tandem repeats of monomeric actin-binding WH2 domains to facilitate actin nucleation. WH2 domains are involved in a wide variety of actin regulatory activities. Structurally, they are expected to clash with interprotomer contacts within the actin filament. Thus, the discovery of their role in nucleation was surprising. Here we use Drosophila Spire (Spir) as a model system to investigate both how tandem WH2 domains can nucleate actin and what differentiates nucleating WH2-containing proteins from their non-nucleating counterparts. We found that the third WH2 domain in Spir (Spir-C or Sc), plays a unique role. In the context of a short nucleation construct (containing only two WH2 domains), placement of Sc in the N-terminal position was required for the most potent nucleation. We found that the native organization of the WH2 domains with respect to each other is necessary for binding to actin with positive cooperativity. We identified two residues within Sc that are critical for its activity. Using this information we were able to convert a weak synthetic nucleator into one with activity equal to a native Spir construct. Lastly, we found evidence that Sc binds actin filaments, in addition to monomers.
Keywords: WH2, nucleation, actin, cytoskeleton, Spir
Introduction
Actin polymerization is a tightly regulated event, essential to countless fundamental cellular processes. It follows that formation of new filaments (nucleation) and subsequent growth (elongation), as well as disassembly of filaments, are carefully controlled. Many of the proteins that regulate these actin dynamics contain a short (~20 amino acids) actin-binding motif called the WASp Homology domain 2 (WH2) [1,2]. WH2 domains are components of proteins that sequester actin monomers, promote filament elongation, sever filaments, activate nucleators, and even nucleate on their own [3,4]. How can such a short sequence do so many different things? It is easy to imagine that the WH2 domain itself is simply an actin-binding module and that sequences flanking the WH2 domain determine its role. Indeed this is an important part of the story, but the WH2 domains themselves are not all equivalent [5–7].
Do WH2 domains differ because they bind to actin in distinct orientations? A number of WH2 domains have been co-crystalized with actin [8–14]. Despite variability in the short sequence, they bind actin in nearly identical orientations. The core WH2 domain begins with a three-turn amphipathic α-helix that binds actin in the hydrophobic cleft between subdomains 1 and 3. A short linker follows the helix and the core ends with a "LKKT/V" motif, which binds the monomer in an extended conformation reaching towards subdomain 2 of actin. Notably, the sequence beyond the LKKT/V is usually missing in actin co-crystals, making it difficult to assess the structural contribution of residues adjacent to the core WH2 domain.
Recent findings indicate that equilibrium binding affinities help specify the nature of interactions between WH2 domains and actin. Two proteins, Tβ4 and Ciboulot, contain motifs closely related to WH2 domains, which are often referred to as Tβ4 domains. The first Tβ4 domain in Ciboulot (CibD1) does not potently inhibit filament elongation, in contrast with Tβ4 which sequesters actin monomers. It has been shown that sequestration by Tβ4 depends on its N-terminal half binding the barbed face of the actin monomer, thereby blocking interactions with the pointed end of a filament, while the C-terminal half of Tβ4 wraps around the actin monomer, blocking interactions with the barbed end of a filament [6]. However, Didry et al. [15] found that small changes within the N-terminal halves of Tβ4 and CibD1 were sufficient to reverse their interactions with actin [15]. Specifically, replacing one residue within the CibD1 linker (between the α-helix and the LKKT/V) with the corresponding residue from Tβ4 led to 5-fold tighter binding and converted CibD1 from a protein that permitted filament elongation to an actin monomer sequesterer. Likewise, substituting the complementary residue from CibD1 in Tβ4 decreased binding ~20-fold and converted this protein to one which no longer sequesters monomers or inhibits elongation. Thus, binding thermodynamics play a major role in determining the activity of Tβ4 domains and perhaps WH2 domains as well.
Can affinities explain how proteins with multiple WH2 domains function? Many of the proteins that contain tandem repeats of WH2 domains can nucleate actin assembly. These so-called WH2-nucleators include several eukaryotic proteins, such as Cordon bleu (Cobl), JMY and Spire (Spir) [4,7,16,17]. They are implicated in a variety of physiological processes including neural development and polarity establishment [18–22]. The tandem WH2 construction has also been co-opted by some pathogenic bacteria, which use them to hijack the host actin cytoskeleton, playing an important role in pathogenicity [23,24]. WH2-nucleators are commonly considered a single class of actin nucleators although they function by distinct mechanisms. In all cases, regions between or adjacent to the WH2 domains are critical to nucleation [7,16,17,25,26]. How or even whether their constituent WH2 domains define their mechanisms is not known.
Here we focus on Spir, a protein first described as a Drosophila polarity factor and since identified as essential to oogenesis in mammals as well as flies [18,27]. Spir has four tandem WH2 domains and a ~15 amino acid linker sequence (called Linker 3 or the MBL domain) between the last two WH2 domains, all of which contribute to nucleation activity [7,17]. The last two WH2 domains of Spir, flanking Linker 3, are an effective minimal nucleation unit [7,17]. Previously, we used this information to build a synthetic actin nucleator [7]. We started with the tandem WH2 domains of N-WASp, which do not nucleate [7,11]. When we inserted Linker 3 between them, this construct could indeed stimulate actin assembly. However, it was notably weaker than the analogous minimal nucleation unit derived from Spir, suggesting that the sequences of the WH2 domains play an important role. Mutagenesis of each of Spir’s WH2 domains in the context of the N-terminal half of Spir indicated that all four WH2 domains contribute to nucleation to differing degrees [17]. These experiments left an open question regarding the relative importance of WH2 domain positioning with respect to Linker 3 versus the importance of the specific sequence of a given WH2 domain to nucleation activity. To eliminate this complexity, in this study we used the minimal nucleation unit within Spir and variations on this model (Spir–C3D or Sc3d; Figure 1). Together, our data indicate that the third WH2 domain in Spir, Sc, is specialized for a role in nucleation but its affinity for actin does not by itself define its role. Instead, we found that Sc associates with actin in a second mode, filament binding. We also identified key residues within Sc that make it an effective actin nucleator. Finally, our data suggest that a combination of kinetics and cooperative interactions define the roles of Spir’s WH2 domains in nucleation.
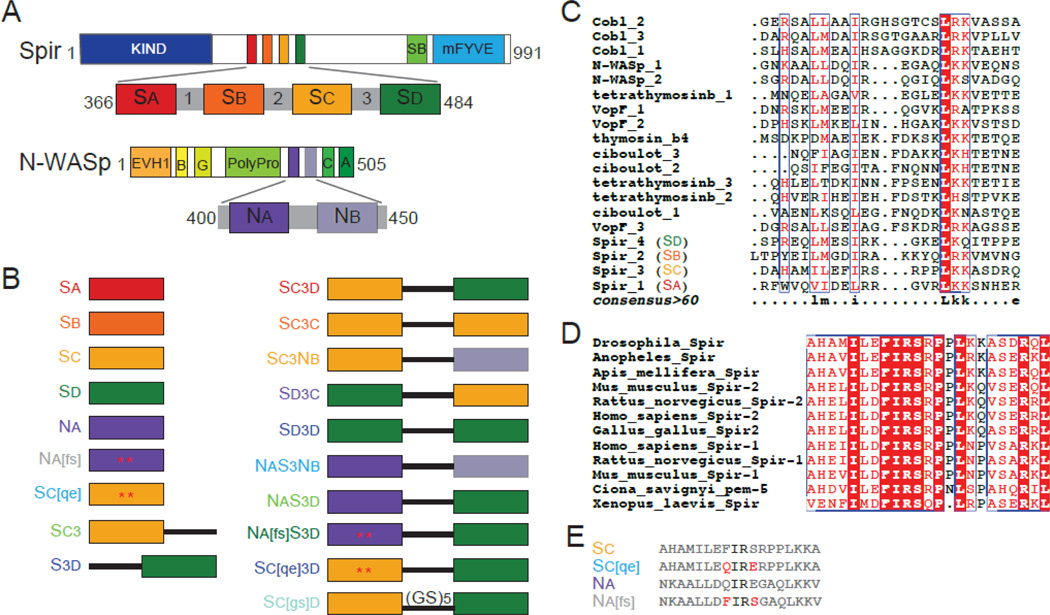
Figure 1.
Spir and N-WASp domains and constructs used. (A) Schematic of Drosophila Spir domain structure: KIND, kinase noncatalytic C-lobe domain (dark blue), , Spir box (light green), mFYVE, modified Fab1/YOTB/Vac1/EEA1 zinc-binding domain (light blue). Expanded: WH2, WASp homology-2 cluster (red, orange, yellow, and dark green) and linker domain regions (grey). Schematic of Rattus N-WASp domain structure: EVH1, Ena/VASP homology-1 domains (orange), B, basic region (light orange), G, GTPase binding domain (yellow), PolyPro, proline rich region (light green), C, central region (green), A, acidic domain (dark green). Expanded: WH2 domains (dark and light purple). (B) Schematic of the constructs used. Colored boxes represent the WH2 domains reported in (A), the black line represents Linker 3 from Spir, and mutated residues are indicated by an asterisk and shown is (E). The color of the construct name is used in future figures. (C) We compared the amino acid sequence of WH2 domains from proteins containing tandem WH2 domains with ClustalW [44]. The bottom line (consensus>60) reflects the consensus as follows: uppercase is identity; lowercase is consensus level great than 60%. (D) Alignment of Sc from nine species of Spir. The entire domain is highly conserved, including the residues flanking the isoleucine and arginine commonly found in the alpha helix of WH2 domains. (E) Sequences of the mutated WH2 domains and their wild type counterparts. Mutated residues in red are represented by asterisks in (B).
Results
Domain order and key residues determine the nucleation activity of Spir
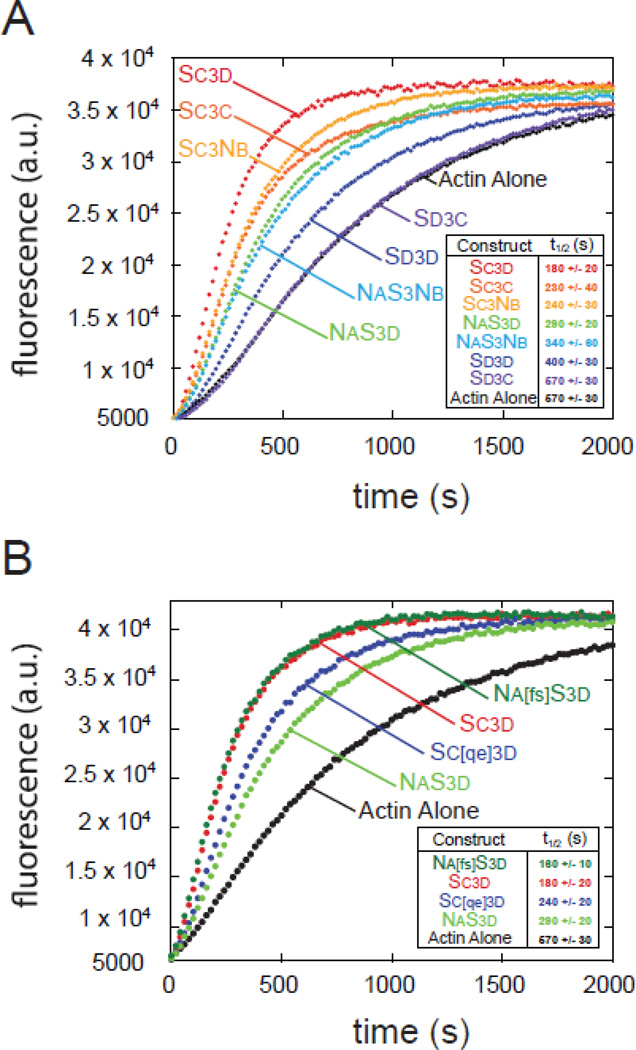
To study the importance of domain order and the specific sequence of a given WH2 domain to nucleation, we used the simplified system of WH2-Linker 3-WH2 constructs. (Figure 1 includes diagrams of all constructs used in this paper and sequences are in Figure S1.) We tested a permutated set of peptides containing the third (Sc) and fourth (Sd) WH2 domains of Spir and the two WH2 domains from the non-nucleating N-WASp, which we refer to as Na and Nb. We tested each construct in a pyrene-actin polymerization assay, using the time until half-maximum polymerization (t1/2) as a metric to compare the nucleation activity between the constructs. This set of constructs displayed a broad range of nucleating activities (t1/2s from ~180 s to 570 s; Figure 2A). The native Spir domain order, Sc3d, to which we refer as wild type, was the most potent nucleator (t1/2 = 180 +/− 20 s). SC3C was somewhat weaker (230 +/− 40 s) and D3D was much weaker (400 +/− 30 s), confirming that the specific sequence of the WH2 domains contributes to activity levels. Strikingly, when SC and SD were reversed (SD3C), nucleation activity was abolished (t1/2 indistinguishable from actin alone, 570 +/− 30 s). That is, two domains that normally nucleate fail to do so when their positions with respect to Linker 3 are changed, demonstrating the importance of domain order to nucleation activity.
Figure 2.
Domain order and key residues affect nucleation. (A) Representative traces of actin polymerization assays monitored by pyrene fluorescence. Minimal nucleation construct variants (0.25 µM) were added to 4 µM actin. (B) Gain of function point mutations convert NaS3d into a construct as potent as Sc3d (Na[fs]S3d). The converse mutations in Sc3d (Sc[qe]3d) result in loss of activity. (A,B) The time until half-maximum polymerization (t1/2) reported is the average of three independent trials +/− standard deviations.
We also observed that the identity of the N-terminal WH2 domain was a major determinant of nucleation activity. The strongest among these nucleators were the constructs with Sc in the first position, Sc3d, Sc3c, and Sc3nb. Likewise, the two constructs with Na in the N-terminal position, NaS3Nb and NaS3d, had equivalent activities. We tested two additional constructs that contained only one WH2 domain and Linker 3, SC3 and S3D. These had no detectable activity (data not shown), confirming that two WH2 domains are necessary for nucleation.
We note that the steady state pyrene signal (plateau) of SC3C was lower than the other constructs, suggesting that this construct sequesters actin more than others, an activity previously reported for Spir [17]. To further examine this effect we compared actin polymerization by three constructs, Sc3c, Sc3d, and NaS3Nb, chosen because they have two, one or zero copies of Sc, respectively. Both constructs with Sc demonstrated slight decreases in plateaus as their concentrations were increased from 250 nM to 1 µM (Figure S2A,B). In contrast, the plateau was constant over this range of concentrations for NaS3Nb (Figure S2C). We note that at all concentrations the plateaus of Sc3c were lower than for the other two constructs. “Dose dependence” is not apparent when comparing the number of Sc domains using these three constructs (Figure S2D), suggesting that potent sequestration is a product of more than just the individual WH2 domains and may be a product of the larger construct.
Because of the contribution Sc makes to nucleation activity, we examined the impact its sequence has on Sc3d. We observed that the residues flanking the conserved isoleucine and arginine within the α-helix of Sc (Phe-438 and Ser-441 in Drosophila Spir) are conserved in all known Spir sequences (Figure 1D). To test what impact these residues have on nucleation activity, we replaced the cognate residues in N-WASp Na, Gln-408 and Glu-411, with those from Sc and then connected it to Linker 3 and Sd (Na[fs]S3d) (Figure 1B,E). We used NaS3d, with no mutations, for a baseline (Figure 2B). NaS3d nucleates with a t1/2 of 290 s, whereas Na[fs]S3d was as potent as Sc3d with a t1/2 of 180 s. Next, we made the converse mutations in Sc3d, replacing Phe-438 with glutamine and Ser-441 with glutamate (Sc[qe]3d). This mutant lost activity, exhibiting a t1/2 similar to NaS3d (t1/2 = 260 s) (Figure 2B). Intriguingly, neither single point mutant in NaS3d showed an effect on nucleation activity (data not shown). Thus, we can dramatically alter the ability of a WH2 domain to nucleate by changing only two residues.
In summary, we found that the order of the WH2 domains influences nucleation activity. In particular, placing Sc N-terminal to Linker 3 creates the strongest nucleator and two residues within its α-helix are sufficient to impart this level of activity.
Cooperative binding by tandem WH2 domains depends on domain order
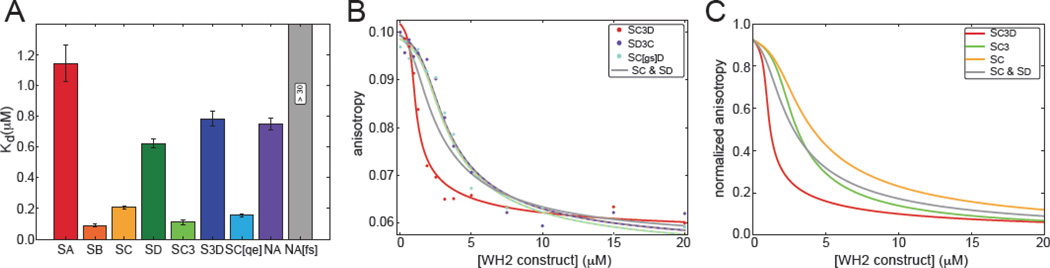
In order to understand how Spir’s WH2 domains contribute to the larger nucleating complexes, we measured equilibrium binding between actin and each individual WH2 domain using competition fluorescence anisotropy (Figures 3A and S3). We measured binding in the same buffer conditions used in our actin assembly assays, using latrunculin B bound actin (latBactin) to prevent polymerization. We first measured the affinity of AlexaFluor488 labeled Sd with Lys-Cys-Lys added to the C-terminus of the domain for labeling (Sd-KCK-AlexaFluor488). We then competed labeled Sd off by titrating in the unlabeled WH2 domains. Despite their short and similar sequences, equilibrium dissociation constants of the Spir WH2 domains vary by an order of magnitude (Kd: 0.09 – 1.13 µM; Figures 3A and S3; Table 1). Spir’s second WH2 domain, Sb, and Sc bind most tightly to monomeric actin (Kds ≤ 0.21 µM) while the first WH2 domain, Sa, and Sd bind with higher equilibrium dissociation constants (Kds ≥ 0.62 µM). We also asked whether the addition of Linker 3 altered the affinity of Sc or Sd. In neither case was the change dramatic: the Kd of Sc3 was 0.11 µM, about twice as tight as that for Sc alone (Kd (Sc) = 0.21 µM); in contrast, S3d is slightly weaker than Sd alone (Kd (S3d) = 0.78 µM vs. Kd (Sd) = 0.62 µM). In our previously published co-crystal of Sc3d with actin, we observed Sd and part of Linker 3 bound to actin [9]. Linker 3 extends away from the actin monomer, suggesting that it would not contribute significantly to equilibrium binding, consistent with what we report here. Because it is C-terminal to the WH2 domain, we imagine that Linker 3 is more likely to contact the actin monomer bound to Sc, and could thereby increase the affinity of this WH2 domain for actin, as we see here.
Figure 3.
Cooperative binding by tandem WH2 domains depends on domain order. (A) The reported Kd values of WH2 constructs bound to latB-actin are the mean of three independent trials of competition fluorescence anisotropy with Sd-AlexaFluor488 (Figure S3). Na[fs] binds too weakly to determine its affinity for actin monomers. Error bars represent one standard deviation. (B) Representative competition fluorescence anisotropy with Sd-AlexaFluor488 and latB-actin as a function of added Sc3d (red circles), Sd3c (purple circles) or Sc[gs]d (aqua circles). Data are fit with a two-site equilibrium binding model. Regressions are in the same color as the data set. The dashed lines represent modeling assuming the Kd for a second actin monomer is infinite for Sd3c or Sc[gs]d. The grey trace is a theoretical plot based on measured affinities for Sc and Sd, with independent binding. (C) Anisotropy curves of Sc3d, Sc3 and Sc based on measured affinities were normalized for comparison. The grey trace is a theoretical plot based on measured affinities for Sc and Sd, with independent binding.
Table 1.
Summary of results.
| WH2 Constructs |
ΔrGd(kJ/mol) | Kd(µM) | Elongation rate (sub/s) |
max rate at 3µM (a.u.) |
ΔrGdf (kJ/mol) |
Kdf(µM) | Inhibition of nucleation |
|---|---|---|---|---|---|---|---|
| Sa | −0.3 ± 0.3 | 1.14 | 1.5 ± 0.2 | 4.1 ± 0.5 | N/D | 28 | +++ |
| Sb | 5.9 ± 0.3 | 0.09 | 6.5 ± 0.7 | 3.5 ± 0.4 | −8 ± 3 | N/D | ++ |
| Sc | 3.9 ± 0.1 | 0.21 (0.01) | 6.6 ± 1.3 | 3.2 ± 0.4 | −1.6 ± 0.3 | 2.0 | − |
| Sd | 1.2 ± 0.1 | 0.62 (0.03) | 6.3 ± 0.5 | 3.1 ± 0.1 | −6.2 ± 0.6 | 13 | +++ |
| Cs3 | 5.4 ± 0.3 | 0.11 | N/D | 4.0 ± 0.3 | −0.9 ± 0.5 | 1.4 | ++ |
| S3d | 0.6 ± 0.2 | 0.78 | N/D | 5.2 ± 0.7 | −15 ± 5 | 500 | + |
| Sc[qe] | 4.6 ± 0.2 | 0.15 | N/D | 2.1 ± 0.2 | −1.6 ± 0.4 | 1.9 | +++ |
| Na | 0.7 ± 0.1 | 0.75 | N/D | 3.0 ± 0.3 | ## | ## | ++ |
| Na[fs] | * | * | N/D | 5.5 ± 0.4 | N/D | N/D | − |
The values in parentheses are the predicted Kd’s for a second actin monomer binding to Sc3d as described for Figure 3B.
Na[fs] affinity is too weak to determine with confidence. It is > 30 µM.
Regression for Na with Kdf free did not converge.
N/D = not determined.
We also measured the affinity of actin for Na and Sc containing the mutations tested in the larger constructs, Na[fs]S3d and Sc[qe]3d, respectively (Figures 3A and S3; Table 1). Surprisingly, the affinities of Sc and Sc[qe] were similar (Kd (Sc) = 0.21 µM vs. Kd (Sc[qe]) = 0.15 µM) despite the impact these mutations had on nucleation activity (t1/2 = 200 s vs. 260 s). In contrast, when the reverse mutations were made in Na, both Kd and the t1/2 changed significantly (Kd (Na) = 0.71 vs. Kd (Na[fs]S3d > 30 µM and t1/2 = 300 vs. 180 s). This result is even more intriguing given that Na[fs]S3d has wild type-like nucleation activity but S3d has no nucleation activity. It follows that equilibrium binding affinity of individual WH2 domains and actin is not predictive of nucleation activity.
Next we examined equilibrium binding of a larger construct, Sc3d. Cooperative binding between tandem WH2 domains and actin was described for a construct of human Spir1 that contains all four WH2 domains [28]. To ask whether Drosophila Spir binds actin cooperatively we again performed competition fluorescence anisotropy. Data were analyzed using a two-site binding model. We assumed that the first binding event was equivalent to actin binding to either individual WH2 domain. Then, we performed regression analysis leaving the value of the second binding event as a free parameter (see Methods). The results for Sc3d yielded dissociation constants of 10 +/− 2 nM or 30 +/− 5 nM for a second actin monomer binding to either Sc or Sd, respectively (Figure 3B, red line, Table 1). These Kd values are much lower than the independently measured Kd values, suggesting that the Sc3d construct exhibits positive cooperative binding.
To ask whether cooperative actin binding contributes to nucleation we compared our results with Sc3d to Sd3c and Sc[gs]d (Linker 3 is replaced by five Gly-Ser repeats as reported in Zuchero et al., 2009). These constructs contain the same WH2 domains as Sc3d but neither nucleates. We performed the same analysis and in both cases found the most probable dissociation constant for binding a second actin monomer is essentially infinite, that is, no actin binding is detected (Figure 3B, dashed lines). Thus these constructs exhibit negative cooperativity. Failure of these two constructs to bind a second actin monomer is consistent with their inability to nucleate.
To analyze these results in greater detail, we compared our competition anisotropy results from three different constructs that contain Sc: Sc, Sc3 and Sc3d (Figure 3C). To do so we pooled all of the data for each case to calculate a representative Kd. We then plotted normalized theoretical anisotropy curves for each construct. There is a progression from Sc to Sc3d, with binding becoming tighter. Cooperative binding by Sc3d is most evident at low concentrations, where the red line dips to low anisotropy values rapidly. For reference, we also plotted a theoretical line representing a construct, which contains Sc and Sd each able to bind actin independently (grey line). The difference between Sc and Sc3 reflects the difference in affinities. Linker 3 enhances binding by Sc.
In sum, the binding data are consistent with our earlier results that domain order influences nucleation activity. We cannot conclude that positive cooperativity is required for nucleation because we do not have a construct that binds two actin monomers without cooperativity. However, the data demonstrate that Linker 3 and domain order are necessary for enhanced binding affinity and suggest that cooperative binding contributes to formation of a nucleus.
Effects of WH2 domains on actin assembly
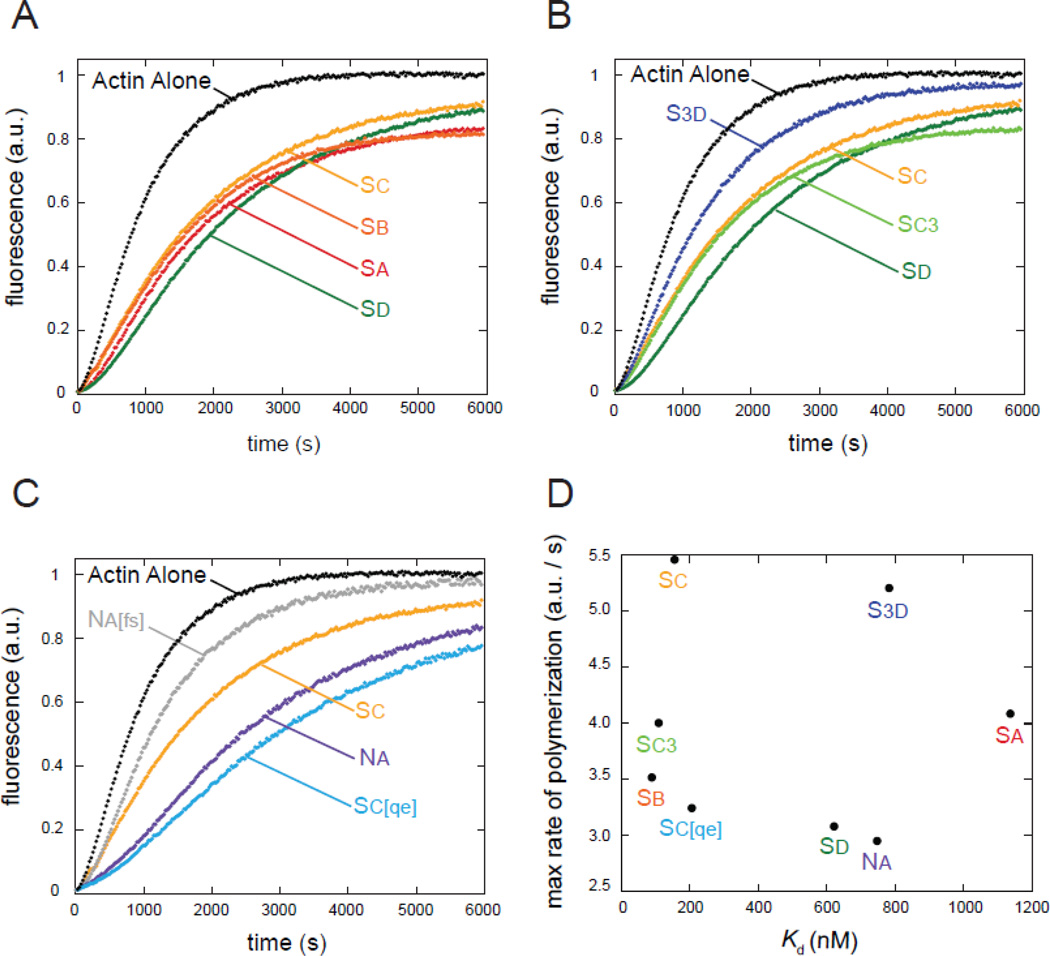
Our binding data suggest that the four Spir WH2 domains interact distinctly with actin at equilibrium. We next asked whether this was also true in a dynamic assay. We tested the effects that each individual WH2 domain has on actin assembly, monitored with a bulk pyrene-actin assembly assay. We performed this assay by adding different concentrations of each WH2 domain (1.5 – 18 µM) to 4 µM actin and initiating polymerization by adding salts. We show 3 µM WH2 as a representative case (Figure 4). Examination of the kinetic traces reveals that the four Spir WH2 domains are slower than actin alone as might be expected for actin monomer binding domains (Figure 4A). However, given the range of affinities for actin, we were surprised to observe that all four traces are quite similar to one another. Specifically, if these proteins were sequestering actin, 3 µM Sa would leave ~2 µM free actin and 3 µM Sb would leave ~1.2 µM free actin at equilibrium. Polymerization of 1.2 versus 2 µM actin alone would be readily distinguishable because nucleation is proportional to the cube of the actin concentration and elongation is directly proportional to monomer concentration [29]. Despite the predicted difference, actin assembly with these two WH2 domains is almost indistinguishable. Subtle differences among the four traces likely reflect differential impact on nucleation or elongation by individual WH2 domains. We consider these contributions below. In order to quantify this observation, we compared the maximum rates of polymerization (Table 1). As our initial inspection suggested, these values did not correlate with affinity for actin (Figure 4D).
Figure 4.
Inhibition of actin polymerization by WH2 domains does not correlate with actin binding affinity. (A) Representative traces of actin polymerization in the presence of the four wild-type Spir-WH2 domains group together despite very different affinities for actin. (B) Adding Linker 3 has little effect on Sc but markedly decreases inhibition by Sd. (C) Mutations in Na decrease inhibition, making it behave more like Sc and mutations in Sc increase inhibition, making it behave more like Na. (A–C) 3 µM WH2 added to 4 µM actin is shown in all cases.
Next, we asked whether addition of Linker 3 to either Sc or Sd changed the behavior of the WH2 domains (Figure 4B). We see a slight increase in actin sequestration by Sc3, shown by the decrease in the plateau compared to Sc, which is consistent with the measured Kds. Surprisingly, S3d is a markedly weaker polymerization inhibitor than Sd alone despite the small difference seen in their affinities for monomeric actin. Although the Sc3d-actin co-crystal reported by Chen et al., does not show an interaction between Sd and Linker 3, these data suggest Linker 3 could alter the kinetics of binding between Sd and actin. We also examined the components of the mutant chimeras we used in the experiments described above (Figure 4C). Of these, we note that Na and Sc[qe], both components of weaker nucleators, inhibit spontaneous polymerization to the greatest degree. In contrast, Na[fs] and Sc, both components of potent nucleators, have only minimal inhibitory effect. Together these data suggest that WH2 domains that are well tuned for nucleation do not individually prevent polymerization; that is, they may readily release actin monomers once incorporated in a filament and/or have binding kinetics that favor filament elongation over sequestration.
Because we found that larger constructs bind actin cooperatively, it may not be surprising that interactions between individual WH2 domains and actin are not predictive of the maximum polymerization rates. While this metric provides useful information about an actin assembly reaction, construction of filaments involves nucleation, elongation, and steady state polymerization. To better understand the consequences of adding WH2 domains to the bulk actin polymerization assay, we conducted a series of experiments to isolate each phase of actin assembly.
Nucleation
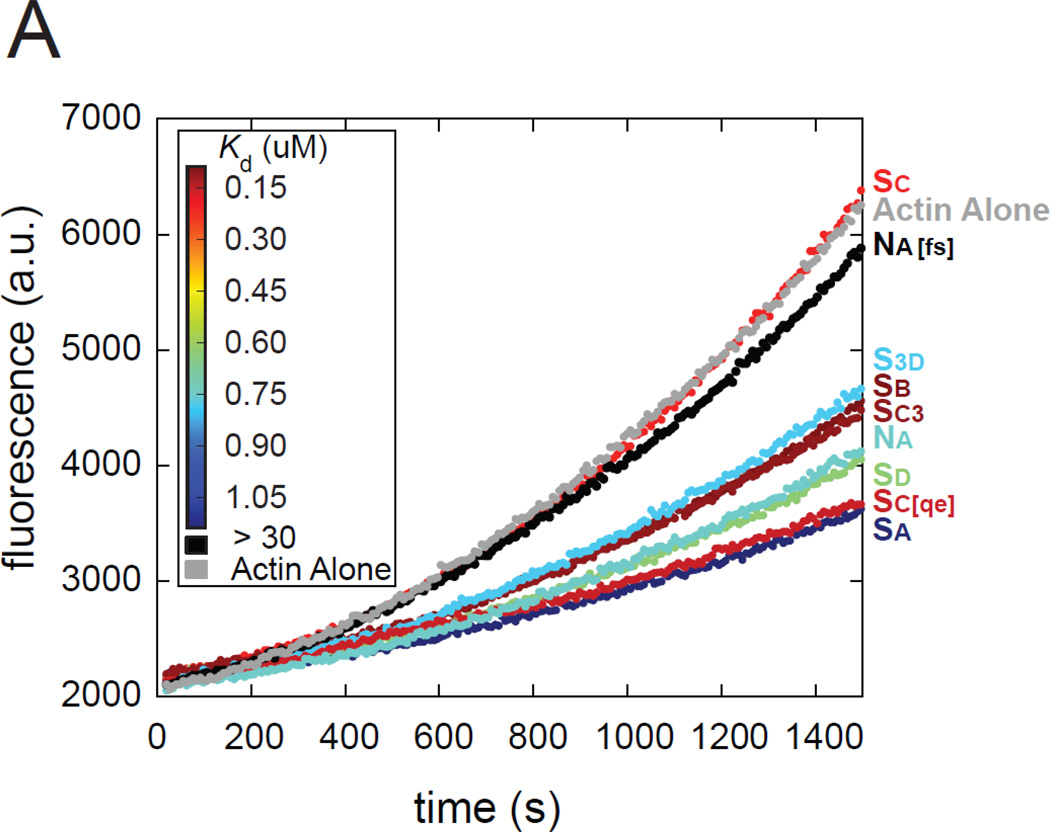
First we examined early time points of the bulk actin polymerization assay, when nucleation is the dominant activity. There was no clear correlation between the rate of these traces and actin binding affinity (Figure 5). Most of the WH2 domains inhibited nucleation to some degree, as expected. Interestingly, addition of Sc or Na[fs], the two WH2 domains in the N-terminal position of the most potent nucleation constructs, had essentially no inhibitory effect.
Figure 5.
Effects of individual WH2 domains on actin nucleation. (A) Representative pyreneactin polymerization assays of different WH2 domain constructs (1.5 µM) added to actin (2 µM) during nucleation. Each trace is colored according to the actin binding affinity of the added WH2 domain according to the inset heat map (red is high affinity). If the effect on nucleation correlated to affinity, the traces would have a rainbow-like trend, but it does not.
Elongation
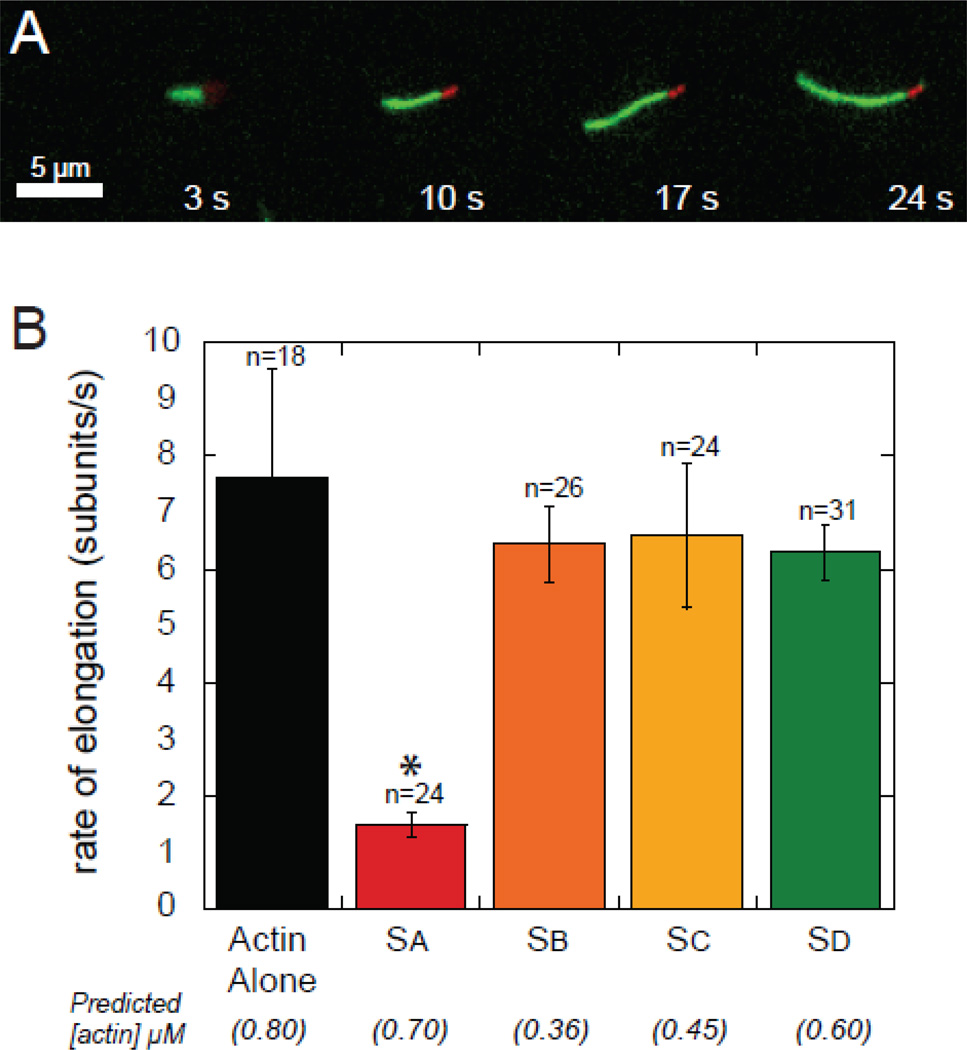
We used Total Internal Reflection Fluorescence (TIRF) microscopy to investigate the effects the individual WH2 domains have on elongation of actin filaments. AlexaFluor647-phalloidin-stabilized actin seeds and 0.8 µM (15% Oregon green) actin were used to measure the rates of elongation in the presence of the WH2 domains (Figure 6; Table 1). We mixed WH2 domains with actin immediately before addition to the slide under polymerizing conditions. Only Sa slowed elongation significantly compared to actin alone (p < 0.01). Based on the affinity we measured, we predict that 0.7 µM actin would be available for elongation in the presence of 1 µM Sa at equilibrium. Surprisingly, the elongation rate is 4-fold lower than actin alone. We interpret this as evidence that Sa binds filament barbed ends reversibly, slowing filament growth. The other Spir-WH2 domains appear to allow elongation as opposed to capping or sequestering under these conditions. In all cases, binding affinity does not correlate with the elongation rate.
Figure 6.
Elongation rates of actin determined by TIRF microscopy. (A) Example of seeded actin (red) and elongation (green) in the presence of Sc observed by TIRF. (B) Actin elongation rates in the presence of 1 µM of the indicated WH2 domains. The values in parentheses are the predicted concentrations of free actin, given the Kds determined in this study. Asterisk indicates statistical significance (p < 0.01) when compared with actin alone (n is the number of filaments analyzed for each case; error bars are one standard deviation).
Steady state
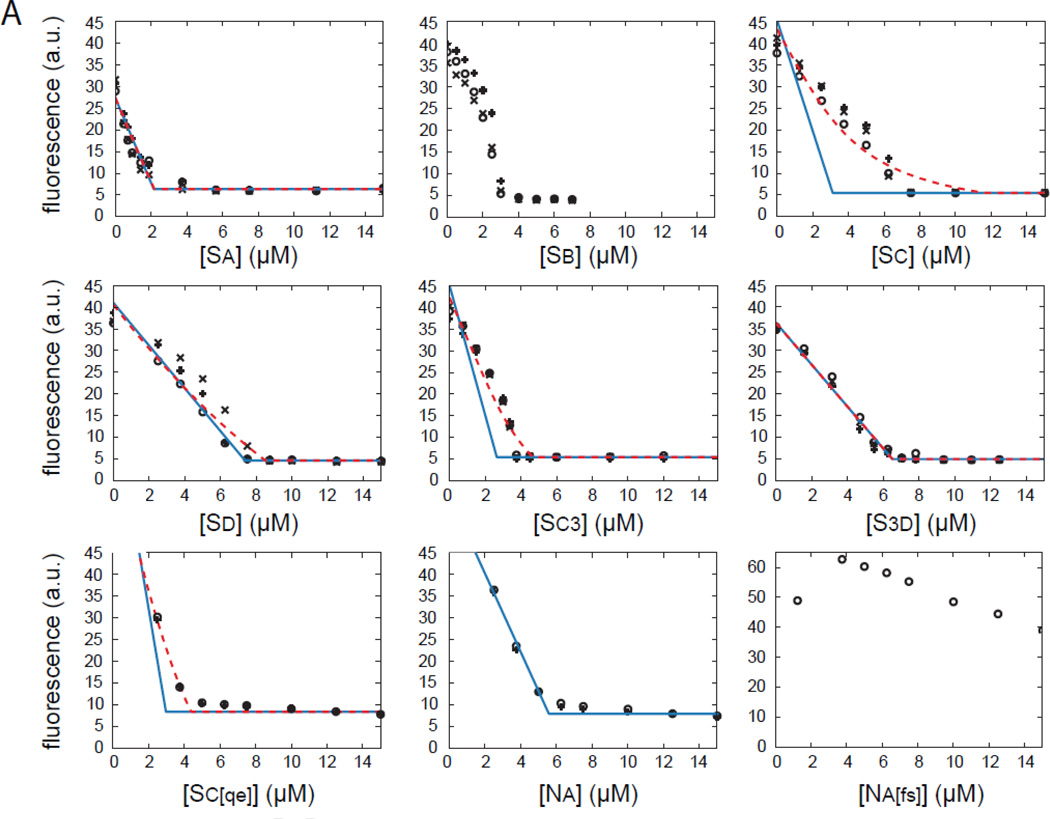
Finally, we performed steady state polymerization titration (SSPT) assays, in which various concentrations of a given WH2 domain were added to pre-polymerized pyrene-actin and total fluorescence was measured after the mixture had come to steady state (Figure 7). As expected, increasing concentrations of WH2 domains caused a decrease in polymer concentration until no remaining polymer could be detected. We note that after 72 hours of incubation with polymerized actin, Sb, and to a lesser extent Sd and Sc3, did not reach steady state. This is evident from the concavity of the titration curve before saturation. Na[fs] does not bind actin tightly enough to cause a significant drop in fluorescence at the tested concentrations; therefore, we do not consider this case or Sb in our analysis.
Figure 7.
Effect of individual WH2 domains on actin at steady state. Raw data (black symbols) and analysis (blue and red traces) of steady state polymerization titration (SSPT) assays for all nine constructs are shown. Each concentration was tested three times, represented by different symbols. Predicted values based on the model described in the appendix, with Kd fixed by our anisotropy measurements, are shown in blue. Extending this model to include filament binding (Kdf) is shown in red. Due to extremely weak binding, we could not analyze the Na[fs] curves. Sb was not analyzed because it did not reached steady state, as evidenced by the concavity of the titration curves. No red trace is shown for Na because the regression did not converge.
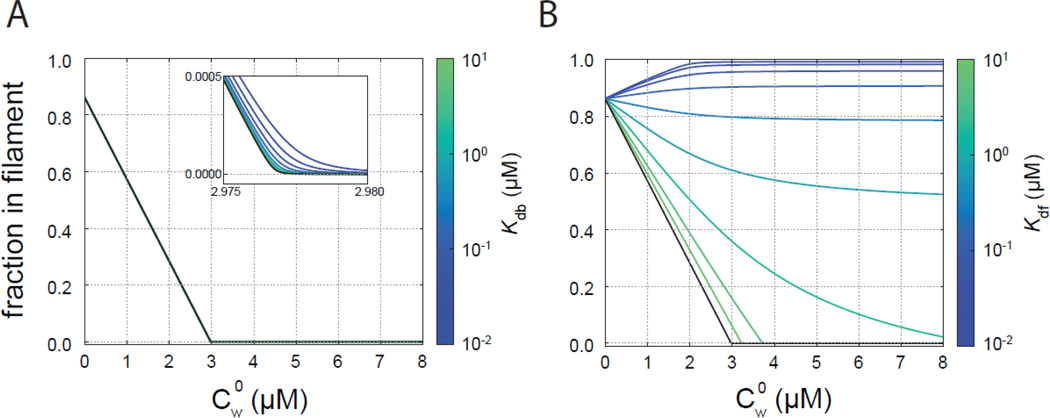
To analyze the SSPT data, we developed a model of steady state polymerization and actin binding by WH2 domains (see Appendix). We first plotted the predicted fluorescence intensity as a function of WH2 concentration, assuming that the WH2 domains act solely by sequestering actin monomers (Figure 7, blue lines). To do so, we fixed the Kd to the value we measured for each WH2 domain. We note that data from constructs containing Sc, in particular, differed from the predicted curves. Therefore, in our theoretical treatment we also considered barbed-end binding and binding to the filaments by WH2 domains. As described in the appendix, the assay is not sensitive to barbed end binding (Figure 8A). It follows that we cannot independently test whether Sa binds the end of actin filaments with this steady state assay. The assay is sensitive to filament binding, as we demonstrate by varying the affinity between WH2 domains and the actin filament (Kdf) while keeping the affinity for actin monomer constant (Figure 8B). As Kdf decreases, the corner concentration moves to the right (increases) and the curve becomes less sharp. This phenomenon occurs because the ability of WH2 domains to bind filaments shifts the equilibrium away from sequestration of actin monomers by WH2 domains.
Figure 8.
Effects of Kdb and Kdf on steady state actin polymerization. For these illustrative curves, Kda is that determined for 10% pyrene labeled actin and Kd = 0.2 µM. The black curves have Kdb or Kdf → ∞. (A) SSPT curves for various values of Kdb with Kdf → ∞. (B) SSPT curves for various values of Kdf.
We tested whether the discrepancies between our model and experimental data were due to binding of the WH2 domains to filaments by performing a regression with Kd constrained to the value we measured and with Kdf as a free parameter. The deviations are recovered as shown with red lines (Figure 7). For Sa, Sd and S3d the Kdf values are greater than 10 µM and unlikely to be physiologically relevant (Table 1). For constructs containing Sc, the resulting Kdf values are less than or equal to 2 µM, weaker than their respective affinities for actin monomers but tight enough to be consequential (Table 1). In light of this result, we asked whether Sc cosediments with actin filaments. We did not detect significantly more protein pelleting with actin than in our negative control (Figure S4A). Controls with actin monomers and polymers confirm that Sc does not alter pyrene fluorescence directly (Figure S4B). We also examined whether Sc binds actin monomers with a different stoichiometry (see Supplementary Materials). When we considered a two-site binding model, we found that this stoichiometry actually shifted the curve to the left, the opposite direction from that which we observed (Figure S4D). Only if two molecules of Sc were required to sequester actin would the curve be shifted to the right. Given that this case is unlikely, we favor the interpretation of the SSPT data in which Sc binds actin filaments in addition to monomers. We discuss this in more detail below.
In summary, equilibrium binding does not predict behavior of WH2 domains in nucleation, elongation or steady state assays. None of the Spir-WH2 domains inhibit elongation, but Sa does have measurable capping activity. Finally, Sc is distinct in several ways. Our steady state measurements indicate that it binds monomer and filaments, though we were unable to directly observe filament binding.
Discussion
Sc is unique in its contribution to the nucleation activity of Spir
We set out to determine how the WH2 domains of Spir contribute to actin nucleation and how they differ from WH2 domains in non-nucleating proteins. Our data suggest that Sc is specialized for nucleation. In the context of a minimal nucleation complex, WH2-Linker 3-WH2, we found that the most potent nucleating activity depended on having Sc N-terminal to Linker 3, as it is in Spir. Closer examination of Sc’s sequence led us to two residues—Phe-438 and Ser-441—that are conserved in Sc. Supporting our claim that these residues are important, introducing them into a weaker construct (NaS3d) was sufficient to increase the activity to wild type Sc3d levels. When modeled in complex with an actin monomer based on the Sd/actin co-crystal by Chen et al., these two residues are found near the end of the three-turn α-helix in the hydrophobic cleft of actin (Figure S5). We speculate that the large, hydrophobic Phe-438 can stack with Phe-352 of actin with minimal rearrangements. The stacking would cause a shift in the α-helix of Sc, which would be unimpeded by the small, uncharged Ser-441, allowing it to be embedded deeper within the actin hydrophobic cleft. The modulated interaction between Sc and the actin hydrophobic cleft may alter the stability of the typical interaction. Alternatively, it could change the path the adjacent sequence would take when extending out of the cleft, thereby allowing a unique interaction to occur. To date, several WH2/actin co-crystals have been obtained and they are all very similar to one another [8–14]. They provide little insight into how this domain can function in such distinct ways. We, and others, have tried to co-crystallize Sc with actin without success. In both cases, we were working with constructs that contained multiple WH2 domains. Ducka et al., [10] may not have seen actin bound to Sc because of the incompatibility of fully occupied tandem WH2 domains in crystal structures as reported by Rebowski et al. [11]. Our attempts were with Sc3d and we were surprised to recover a crystal with Sd occupied but not Sc despite the fact that Sc alone binds actin more tightly [2]. Hopefully, future work will produce this crystal.
Consistent with our speculation that Sc’s interaction is distinct from the other WH2 domains, it was the only one that exhibited significant filament binding activity. While we do not know if this alternate binding is physiological, the affinity we measured, ≤ 2 µM, suggests that it may be. The fact that this binding is approximately 10-fold weaker than for actin monomers indicates that more “traditional” monomer binding still dominates. However, because WH2 binding to the hydrophobic cleft of actin monomers is thought to be incompatible with longitudinal actin filament contacts [11,30], an alternate interaction between Sc and actin could contribute to nucleation activity, by allowing this region to “get out of the way” of the newly forming filament or perhaps by stabilizing the nucleus. Why don’t we observe Sc in the pellet of the cosedimentation assay? Of course, our interpretation of the SSPT results may be incorrect. Two ways to reconcile the SSPT results and the cosedimenation data are as follow: The off rate of Sc could be too high to detect in cosedimentation assays, or Sc could bind only to small actin assemblies, large enough to enhance pyrene fluorescence but too small to sediment. The later possibility still lends itself to an interesting role in nucleation. Finally, we note that Spir WH2 domains sever filaments [9,28], which presumably required filament binding. Perhaps Sc mediates this interaction.
Previously, we carried out nucleation studies using the N-terminal half of Spir (SpirNT), which included the entire cluster of four WH2 domains [17]. Mutations to remove actin binding were introduced into each WH2 domain, one at a time. Mutating Sd had the greatest effect, followed by mutations in Sc. In contrast with our findings, these data suggested that Sd contributes the most to nucleation. We believe the difference in observations depends on the construct. That is, two functioning WH2 domains must flank Linker 3 to form a functional nucleator. In the context of SpirNT, Linker 3 still has functional WH2 domains on either side of it when Sc is mutated but not when Sd is mutated. Thus the minimal nucleation construct was instrumental in revealing the important role of Sc.
Spir has emergent properties
We found little to no correlation between affinity and how a WH2 domain affects actin assembly. This lack of correspondence was emphasized by studying each of the three phases of actin polymerization in isolation—nucleation, elongation, and steady state. For example, Spir’s four WH2 domains behaved similarly in bulk actin assembly assays despite affinities for actin that range over an order of magnitude. In addition, single filament elongation assays indicate that Sa has some barbed end capping activity and SSPT assays suggest that Sc binds to filaments. These data show that interaction with actin of different WH2 domains can differ greatly and that Spir’s nucleation activity cannot be predicted by the sum of its parts. Of course, actin assembly is a dynamic process and detailed kinetics assays may prove useful for predictions.
We also observed that domain order is a significant determinant of nucleation activity: Sc3d is a relatively strong nucleator, while the reversed construct, Sd3c, has no detectable nucleation activity. Here binding assays were able to provide insight. Sc3d bound actin with positive cooperativity, while Sd3c and Sc[gs]d bound actin with negative cooperativity. Linker 3 is necessary but not sufficient for positive cooperativity. We were intrigued to find that the negative cooperativity of Sd3c and Sc[gs]d binding to actin is so extreme as to only support binding to a single actin monomer, despite the presence of two WH2 domains. This binding behavior explains why these constructs fail to nucleate and emphasizes that Spir activity is greater than the sum of its parts.
How does Spir work?
Experimental data demonstrate that all four WH2 domains can bind actin but none functions as a tight sequesterer, like Tβ4. Comparison of our experimental observations to a theoretical framework revealed that Sc may interact with actin filaments or very short polymers. Given the evidence that Linker 3 plays an important role in nucleation, it is perhaps fitting that Sc, one of the domains adjacent to Linker 3, has an alternate binding modality. Our results also suggest that an ability to release actin is important for nucleation by these actin-binding motifs. Together these data support a model where each WH2 domain binds an actin monomer, brings them into close proximity with one another, and then subsequently adjusts without completely dissociating to allow for filament formation. Our data, and that of Bosch et al., [28] suggest that at least two structures can be formed – a nucleus and a stable sequestration complex. Future work will address when/how one or the other is built and what happens when Spir is dimerized, which accelerates nucleation, as is the case when it binds to Cappuccino [31,32].
Methods
DNA constructs
D. melanogaster Spir and R. rattus N-WASp constructs were generated by PCR amplification from full-length templates, CG10076 and D88461.1, respectively. Truncations were subcloned into the pGEX-6P-2 vector (GE Healthcare, Piscataway NJ) or pET20b(+) (Novagen, Hornsby Australia). Constructs used are depicted in Figures 1 and S1. Point mutations were introduced using QuikChange Site Directed Mutagenesis (Stratagene, Santa Clara, CA). Chimeras were constructed by SOEing [33].
Protein purification and labeling
A. castellani actin was purified and labeled according to published protocols [32,34]. WH2-containing constructs were expressed in E. coli BL21(DE3) cells and purified by standard protocols as described in Supplemental Methods. Briefly, proteins were purified on glutathione- Sepharose 4b. The GST-tag was removed with PreScission protease and a second exposure to glutathione resin. Single WH2-containing proteins were further purified by filtering over an anion exchange column (MonoQ, GE Healthcare) and collecting the flow through. The multiple WH2-containing constructs were purified on a cation exchange column (MonoS, GE Life Sciences). The final products were dialyzed against 10 mM Tris pH 8, and 1 mM DTT overnight at 4°C. Protein aliquots were flash-frozen in liquid nitrogen and stored at −80°C. Protein concentrations were calculated by quantitative SYPRO Red (Life Technologies, Grand Island, NY) stained gels using Amino Acid Analyzed (UCLA Biopolymer Laboratory) Sd as a standard, due to their lack of absorbance at 280nm.
For actin assembly assays, actin was labeled with pyrene iodoacetamide as described [35]. For TIRF assays, actin was labeled with Oregon Green 488 iodoacetamide (Life Technologies, Carlsbad, CA) on cysteine-374 as described [36].
For anisotropy experiments, unlabeled Sd-KCK was incubated for 20 min at 42 °C with 2 mM TCEP and then dialyzed twice for 2 h each against 10mM HEPES pH 7, 50 mM KCl. Protein was then rocked at 25 °C for 30 min with a 2–4 molar excess of AlexaFluor488-C5-maleimide. The reaction was quenched by addition of 10 mM DTT. Unconjugated dye was removed using a PD-10 desalting column (GE Life Sciences) equilibrated with 10 mM Tris-HCl pH 8.0, 1 mM DTT, 100 mM KCl. Protein concentration was determined by quantitative SYPRO Red staining. The concentration of incorporated dye was determined by absorbance at 496 nm using an extinction coefficient of 71,000 cm−1. Labeling efficiency was calculated to be 58%.
Pyrene-actin polymerization assays
Pyrene-actin assembly assays were carried out essentially as described [37]. Briefly, 4 µM actin (5% pyrene labeled) was incubated for 2 min at 25°C with ME buffer (final concentration, 200 µM ethylene glycol tetraacetic acid [EGTA] and 50 µM MgCl2) to convert Ca-G-actin to Mg-Gactin. Polymerization was initiated by adding KMEH polymerization buffer (final concentration, 10 mM HEPES, pH 7.0, 1 mM EGTA, 50 mM KCl, 1 mM MgCl2) to the Mg-G-actin. WH2-containing proteins were combined in the polymerization buffer before addition to Mg-G-actin. Fluorescence was monitored using a TECAN F200 (Tecan Group Ltd., Männedorf, Switzerland) with λexcitation = 365 nm and λemission = 407 nm.
Steady state polymerization titration
To test most of the WH2 domains, 2 µM actin (10% pyrene labeled) was polymerized in KMEH plus 0.5 mM Thesit for 1 h in a 96-well plate at 25°C. Single WH2-containing proteins were titrated in and stored for 72 h at 4°C. The plate was allowed to warm to room temperature for 30 min before fluorescence was measured in a TECAN F200. For Sa, the same procedure was followed except we used 0.5 µM actin (40% labeled). Controls to test for altered pyrene fluorescence as a result of WH2 binding to monomers (latrunculin-actin) or filaments (phalloidinactin) showed less than 1% change (Figure S4A).
The SSPT curves were fit with the model described in the appendix with Kdb → ∞. The dissociation constants, Kda, for addition of an actin monomer to the end of a filament were determined from a critical concentration assays with 10% and 40% pyrene labeled actin (data not shown). The dissociation constants, Kd, for respective WH2 domains with actin monomers were constrained to the values determined from our competition anisotropy assays. The background pyrene fluorescence, pbg was given by the mean of the last four points of the SSPT curve. For the blue curves, Kdf was set to infinite, so ˜p (a scaling factor) was the only free parameter in the regression. For the red curves, Kdf was also a free parameter.
Polarization anisotropy
All assays were carried out with latrunculin bound actin (LatB-actin) in 10 mM HEPES, pH 7.0, 1 mM EGTA, 1 mM TCEP, 0.5 mM Thesit, 50 mM KCl, and 1 mM MgCl2 at 25°C. LatB-actin was made by mixing a 2-fold molar excess of latrunculin B with actin for 1 h at 25°C.
Fluorescence polarization anisotropy of 5 nM Sd-KCK–AlexaFluor488 (58% labeled) mixed with 2 µM LatB-actin was measured with increasing concentrations of actin. The fluorophore was excited by plane-polarized light at 488 nm, and emission was measured at 520 nm at angles parallel and perpendicular to the angle of incidence using a TECAN F200 (Tecan Group Ltd., Männedorf, Switzerland). We performed a regression to determine the equilibrium dissociation constant, Kd*, using a quadratic binding model as previously described [37].
Next, we added increasing amounts of competing individual WH2 domains to 5 nM Sd-KCK-AlexaFluor488 and 2.0 µM LatB-actin. Given the previously measured value of Kd*, the dissociation constants of the respective constructs were obtained using regression tools in EQTK (eqtk.org), a forthcoming analysis tool for coupled equilibria using algorithms from [38,39]. Specifically, we performed a regression corresponding to the reactions, for example for Sc,
| A. Sd* ⬄ A + Sd* | Kd* |
| A. Sc ⬄ A + Sc | Kd, |
where A is LatB-actin and Sd* is Sd-KCK-AlexaFluor488, with similar analysis for Sa, Sb, Sd, C3, S3d, Na, Sc[qe] and Na[fs].
To investigate cooperative binding of actin by Sc3d, the reactions and dissociation constants considered in the regression analysis using EQTK were
| A. Sd* ⬄ A + Sd* | Kd* |
| A. Sc3d ⬄ A + Sc3d | Kd,1 |
| Sc3d.A ⬄ Sc3d + A | Kd,2 |
| A. Sc3d.A ⬄ A + Sc3d.A | Kd,3 |
| A. Sc3d.A ⬄ A. Sc3d + A | Kd,2Kd,3/ Kd,1, |
where the dissociation constant for the last reaction is determined by the previous three as required by the path independence of equilibrium. An analogous set of reactions was used for Sd3c.
TIRF microscopy assays
Coverslips for TIRF elongation assays were prepared as described [9]. Briefly, they were silanized with 5% 3-aminopropyl-triethoxysilane (Sigma-Aldrich, St. Louis, MO) and PEGylated with N-hydroxylsuccinimide–functionalized polyethylene glycol (PEG-NHS; 97% methoxy-PEG-NHS and 3% biotin-PEG-NHS (JenKem Technology, Allen, TX).
Actin was biotinylated using maleimide-PEG11-biotin (Thermo Scientific, Waltham, MA) and mixed with AlexaFluor647-phalloidin at a 1:1 ratio to create 5% biotinylated phalloidin-stabilized actin seeds. Immediately before imaging, a blocking solution (PBS, 1% Pluronic, 0.1 mg/ml casein) was applied to the flow cells for 2 minutes followed by a wash with TIRF buffer (final concentrations: 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10 mM HEPES, pH 7, 0.2 mM ATP, 50 mM DTT, 0.4% methylcellulose). Next 40 nM streptavidin (VWR, Radnor, PA) was applied to the flow cell for 1 min, followed by a wash with TIRF buffer, 30 s of incubation with 10–30 nM actin seeds, and a wash with TIRF buffer. Oregon green-actin (0.8 µM, 15% Oregon green-labeled final concentration) was incubated with ME buffer for 2 min at room temperature. A solution containing 2× TIRF buffer, glucose oxidase (final concentration 0.25 mg/ml), catalase (final concentration 0.05 mg/ml), and any test proteins (1 µM single WH2-containing proteins final concentration) was mixed with the Mg-G-actin solution and added to the flow cell. Filament elongation was visualized on a DMI6000 TIRF microscope (Leica, Wetzlar, Germany) for at least 20 min, capturing images at 10 s intervals. Filament lengths and elongation rates were analyzed with JFilament [40] incorporated into Fiji [41].
Spir Sc model
Using SWISS-MODEL, the sequence of Sc (residues 421 through 428 of Drosophila melanogaster SpirB) was threaded through the Sd crystal structure bound to A. castellanii actin (PDB ID: 4EFH) [9,42]. Sd was then removed from 4EFH, replaced by the generated Sc model, and the co-crystal model was visualized using UCSF Chimera [43].
Supplementary Material
Highlights.
Domain order plays a critical role in nucleation activity by Spir.
Cooperative actin binding may contribute to nucleation.
Linker 3 is necessary but not sufficient for cooperative actin binding.
Two residues in WH2C are necessary for strong nucleation and sufficient to convert a weak nucleator into a potent one.
The WH2 domains of Spir interact with both actin monomers and filaments in distinct ways.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Paunola E, Mattila PK, Lappalainen P. WH2 domain: a small, versatile adapter for actin monomers. FEBS Lett. 2002;513:92–97. doi: 10.1016/s0014-5793(01)03242-2. [DOI] [PubMed] [Google Scholar]
- 2.Carlier MF, Hertzog M, Didry D, Renault L, Cantrelle FX, van Heijenoort C, et al. Structure, function, and evolution of the beta-thymosin/WH2 (WASP-Homology2) actin-binding module. Ann N Acad Sci. 2007;1112:67–75. doi: 10.1196/annals.1415.037. [DOI] [PubMed] [Google Scholar]
- 3.Husson C, Cantrelle FX, Roblin P, Didry D, Le KH, Perez J, et al. Multifunctionality of the beta-thymosin/WH2 module: G-actin sequestration, actin filament growth, nucleation, and severing. Ann N Acad Sci. 2010;1194:44–52. doi: 10.1111/j.1749-6632.2010.05473.x. [DOI] [PubMed] [Google Scholar]
- 4.Qualmann B, Kessels MM. New players in actin polymerization--WH2-domain-containing actin nucleators. Trends Cell Biol. 2009;19:276–285. doi: 10.1016/j.tcb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Kelly AE, Kranitz H, Dotsch V, Mullins RD. Actin binding to the central domain of WASP/Scar proteins plays a critical role in the activation of the Arp2/3 complex. J Biol Chem. 2006;281:10589–10597. doi: 10.1074/jbc.M507470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nachmias VT. Small actin-binding proteins: the beta-thymosin family. Curr Opin Cell Biol. 1993;5:56–62. doi: 10.1016/s0955-0674(05)80008-0. [DOI] [PubMed] [Google Scholar]
- 7.Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol. 2009;11:451–459. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetmo K, Dominguez R. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci U A. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen CK, Sawaya MR, Phillips ML, Reisler E, Quinlan ME. Multiple forms of Spire-actin complexes and their functional consequences. J Biol Chem. 2012;287:10684–10692. doi: 10.1074/jbc.M111.317792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ducka AM, Joel P, Popowicz GM, Trybus KM, Schleicher M, Noegel AA, et al. Structures of actin-bound Wiskott-Aldrich syndrome protein homology 2 (WH2) domains of Spire and the implication for filament nucleation. Proc Natl Acad Sci U A. 2010;107:11757–11762. doi: 10.1073/pnas.1005347107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rebowski G, Namgoong S, Boczkowska M, Leavis PC, Navaza J, Dominguez R. Structure of a longitudinal actin dimer assembled by tandem w domains: implications for actin filament nucleation. J Mol Biol. 2010;403:11–23. doi: 10.1016/j.jmb.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aguda AH, Xue B, Irobi E, Préat T, Robinson RC. The structural basis of actin interaction with multiple WH2/beta-thymosin motif-containing proteins. Struct Lond Engl 1993. 2006;14:469–476. doi: 10.1016/j.str.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 13.Hertzog M, van Heijenoort C, Didry D, Gaudier M, Coutant J, Gigant B, et al. The beta-thymosin/ WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell. 2004;117:611–623. doi: 10.1016/s0092-8674(04)00403-9. [DOI] [PubMed] [Google Scholar]
- 14.Lee SH, Kerff F, Chereau D, Ferron F, Klug A, Dominguez R. Structural basis for the actin-binding function of missing-in-metastasis. Struct Lond Engl 1993. 2007;15:145–155. doi: 10.1016/j.str.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Didry D, Cantrelle F-X, Husson C, Roblin P, Moorthy AME, Perez J, et al. How a single residue in individual β-thymosin/WH2 domains controls their functions in actin assembly. EMBO J. 2012;31:1000–1013. doi: 10.1038/emboj.2011.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahuja R, Pinyol R, Reichenbach N, Custer L, Klingensmith J, Kessels MM, et al. Cordonbleu is an actin nucleation factor and controls neuronal morphology. Cell. 2007;131:337–350. doi: 10.1016/j.cell.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila Spire is an actin nucleation factor. Nature. 2005;433:382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- 18.Manseau LJ, Schupbach T. cappuccino and spire: two unique maternal-effect loci required for both the anteroposterior and dorsoventral patterns of the Drosophila embryo. Genes Dev. 1989;3:1437–1452. doi: 10.1101/gad.3.9.1437. [DOI] [PubMed] [Google Scholar]
- 19.Ravanelli AM, Klingensmith J. The actin nucleator Cordon-bleu is required for development of motile cilia in zebrafish. Dev Biol. 2011;350:101–111. doi: 10.1016/j.ydbio.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q-C, Liu J, Wang F, Duan X, Dai X-X, Wang T, et al. Role of nucleation-promoting factors in mouse early embryo development. Microsc Microanal Off J Microsc Soc Am Microbeam Anal Soc Microsc Soc Can. 2013;19:559–564. doi: 10.1017/S1431927613000032. [DOI] [PubMed] [Google Scholar]
- 21.Carroll EA, Gerrelli D, Gasca S, Berg E, Beier DR, Copp AJ, et al. Cordon-bleu is a conserved gene involved in neural tube formation. Dev Biol. 2003;262:16–31. doi: 10.1016/s0012-1606(03)00323-3. [DOI] [PubMed] [Google Scholar]
- 22.Ferreira T, Ou Y, Li S, Giniger E, van Meyel DJ. Dendrite architecture organized by transcriptional control of the F-actin nucleator Spire. Dev Camb Engl. 2014;141:650–660. doi: 10.1242/dev.099655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liverman AD, Cheng HC, Trosky JE, Leung DW, Yarbrough ML, Burdette DL, et al. Arp2/3-independent assembly of actin by Vibrio type III effector VopL. Proc Natl Acad Sci U A. 2007;104:17117–17122. doi: 10.1073/pnas.0703196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tam VC, Suzuki M, Coughlin M, Saslowsky D, Biswas K, Lencer WI, et al. Functional analysis of VopF activity required for colonization in Vibrio cholerae. MBio. 2010;1 doi: 10.1128/mBio.00289-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Namgoong S, Boczkowska M, Glista MJ, Winkelman JD, Rebowski G, Kovar DR, et al. Mechanism of actin filament nucleation by Vibrio VopL and implications for tandem W domain nucleation. Nat Struct Mol Biol. 2011;18:1060–1067. doi: 10.1038/nsmb.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu B, Cheng HC, Brautigam CA, Tomchick DR, Rosen MK. Mechanism of actin filament nucleation by the bacterial effector VopL. Nat Struct Mol Biol. 2011;18:1068–1074. doi: 10.1038/nsmb.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfender S, Kuznetsov V, Pleiser S, Kerkhoff E, Schuh M. Spire-type actin nucleators cooperate with Formin-2 to drive asymmetric oocyte division. Curr Biol. 2011;21:955–960. doi: 10.1016/j.cub.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bosch M, Le KH, Bugyi B, Correia JJ, Renault L, Carlier MF. Analysis of the function of Spire in actin assembly and its synergy with formin and profilin. Mol Cell. 2007;28:555–568. doi: 10.1016/j.molcel.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 29.Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophys J. 2001;81:667–674. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zahm JA, Padrick SB, Chen Z, Pak CW, Yunus AA, Henry L, et al. The bacterial effector VopL organizes actin into filament-like structures. Cell. 2013;155:423–434. doi: 10.1016/j.cell.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, Kerkhoff E. Regulatory interactions between two actin nucleators, Spire and Cappuccino. J Cell Biol. 2007;179:117–128. doi: 10.1083/jcb.200706196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vizcarra CL, Kreutz B, Rodal AA, Toms AV, Lu J, Zheng W, et al. Structure and function of the interacting domains of Spire and Fmn-family formins. Proc Natl Acad Sci U A. 2011;108:11884–11889. doi: 10.1073/pnas.1105703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warrens AN, Jones MD, Lechler RI. Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene. 1997;186:29–35. doi: 10.1016/s0378-1119(96)00674-9. [DOI] [PubMed] [Google Scholar]
- 34.MacLean-Fletcher S, Pollard TD. Identification of a factor in conventional muscle actin preparations which inhibits actin filament self-association. Biochem Biophys Res Commun. 1980;96:18–27. doi: 10.1016/0006-291x(80)91175-4. [DOI] [PubMed] [Google Scholar]
- 35.Cooper JA, Walker SB, Pollard TD. Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J Muscle Res Cell Motil. 1983;4:253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]
- 36.Bor B, Vizcarra CL, Phillips ML, Quinlan ME. Autoinhibition of the formin Cappuccino in the absence of canonical autoinhibitory domains. Mol Biol Cell. 2012;23:3801–3813. doi: 10.1091/mbc.E12-04-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zalevsky J, Grigorova I, Mullins RD. Activation of the Arp2/3 complex by the Listeria acta protein. Acta binds two actin monomers and three subunits of the Arp2/3 complex. J Biol Chem. 2001;276:3468–3475. doi: 10.1074/jbc.M006407200. [DOI] [PubMed] [Google Scholar]
- 38.Bois JS. PhD. thesis. California Institute of Technology; 2007. Analysis of Interacting Nucleic Acids in Dilute Solutions. [Google Scholar]
- 39.Dirks RM, Bois JS, Schaeffer JM, Winfree W. Thermodynamic analysis of interacting nucleic acid strands. SIAM Rev. 2007;49:65–88. [Google Scholar]
- 40.Smith MB, Li H, Shen T, Huang X, Yusuf E, Vavylonis D. Segmentation and tracking of cytoskeletal filaments using open active contours. Cytoskelet Hoboken NJ. 2010;67:693–705. doi: 10.1002/cm.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 43.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 44.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinforma Oxf Engl. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.