Abstract
Epigenetic changes including genomic imprinting may affect risk of late-onset Alzheimer’s disease (LOAD). There are >100 known imprinted genes and most of them are expressed in human brain. In this study, we examined the association of single nucleotide polymorphisms (SNPs) in 93 imprinted genes with LOAD risk in 1291 LOAD cases and 958 cognitively normal controls. We performed single-site, gene-based and haplotype analyses. Single-site analysis showed 14 significant associations at P<0.01. The most significant SNP (rs11770199; P=0.0003) in single-site analysis was located on chromosome 7 in the GRB10 gene. Gene-based analyses revealed 4 significant associations in the WT1, ZC3H12C, DLGAP2 and GPR1 genes at P <0.05. The haplotype analysis also revealed significant associations with three genes (ZC3H12C, DLGAP2 and GPR1). These findings suggest a possible role of imprinted genes in AD pathogenesis that show specific expression in the brain.
Keywords: Late Onset Alzheimer’s Disease (LOAD), Imprinting, Brain, Gene expression
Introduction
Late-onset Alzheimer’s disease (LOAD) is a major form of dementia. Both genetic and environmental factors determine its etiology [1]. Until recently, APOE was the only established susceptibility gene for LOAD [2]. Since 2009, large scale genome wide association studies (GWAS) have identified more than twenty non-APOE susceptibility loci for LOAD (CR1, BIN1, INPP5D, MEF2C, TREM2, CD2AP, HLA-DRB1/HLA- DRB5, EPHA1, NME8, ZCWPW1, CLU, PTK2B, PICALM, SORL1, CELF1, MS4A4/MS4A6E, SLC24A4/RIN3,FER- MT2, CD33, ABCA7, CASS4) [3-6]. However, common variation in known LOAD genes explain only ~24-33% of total phenotypic variance of AD [7, 8]. This indicates the involvement of additional genes that affect the risk of LOAD. An attractive group of candidate genes that might affect the risk of LOAD include those that participate in genomic imprinting. To date, approximately 150 imprinted genes have been reported in mice and humans, representing less than 1% of the genome [9-11]. Imprinted genes are highly expressed in placenta and the brain [12]. In brain, imprinted genes are associated with neurodevelopment and behavior [13].
Genomic imprinting leads to parental origin specific gene expression, also called parent of origin effect [14]. A number of studies have reported parent of origin effect in LOAD [15-20]. Epigenetic studies in AD have also shown that methylation process involved in imprinting is altered in AD cases compared to controls [21-24]. Given the role of imprinting in neurodevelopment and also in the pathogenesis of LOAD, the present study examined the association of genetic variation in 93 imprinted genes with the risk of LOAD in a large case-control sample. As all of these genes are known for their temporal expression in brain, we hypothesized that genetic variation in these genes may be associated with LOAD risk due to dysregulation of epigenetic mechanisms.
Materials and Methods
Subjects
The case-control sample used in this study is described elsewhere [25]. All subjects were European Americans. The mean age-at-onset (AAO) for LOAD cases (n=1,440) was 72.6±6.4 years (65.6% women and 23.5% autopsy-confirmed). Control subjects (n=1,000) were 60 years or older (mean age 74.07±6.20 years; 59.8% women and 0.2% autopsy-confirmed). LOAD cases were recruited from the University of Pittsburgh Alzheimer’s Disease Research Center (ADRC), all of whom met the National Institute of Neurological and Communication Disorders and Stroke (NINCDS)/Alzheimer’s Disease and Related Disorders Association (ADRDA) criteria for probable or definite AD. The University of Pittsburgh ADRC follows a standard evaluation protocol, including medical history, general medical and neurological examinations, psychiatric interview, neuro-psychological testing and magnetic resonance imaging scan. Controls were cognitively normal subjects that were derived from the same geographical area as the cases. Following standard quality control and exclusion criteria, 2,249 subjects (1,291 LOAD cases and 958 controls) were included in the final analysis. All subjects were recruited with informed consent, and the study was approved by the University of Pittsburgh Institutional Review Board.
Genotyping
A list of 107 human imprinted genes was developed from imprinted gene database (www.geneimprint.com) and catalogue of imprinted genes (http://igc.otago.ac.nz). Only 93 imprinted genes were selected for final association analysis based on their representation in refgene database and our genotyped database. The Illumina Omni1-Quad chip was used for genotyping as described by Kamboh et al. [25]. There were a total of 3,401 single-nucleotide polymorphisms (SNPs) present on the Illumina chip in the examined 93 candidate genes.
Statistical analyses
Association of 3,401 SNPs located in 93 imprinted genes was tested using the logistic regression under an additive model adjusting for age, sex, and the first four principal components as covariates using PLINK [25]. Further adjustment was made for APOE genotype following initial association tests. Nominal P-values less than 0.05 were considered suggestive evidence of association in these candidate genes
Versatile Gene-Based Association (VEGA) analysis was performed to determine the association of imprinted gene with LOAD. Tagger analysis was performed in Haploview with the following parameters: r2=0.80, minor-allele frequency (MAF) <0.01 to identify tagSNPs in each gene [26], (www.broadinstitute.org/haploview). This information was used to calculate P-value threshold for significance (Supplementary Table 1).
Haplotype analysis within each gene was done using a sliding-windows approach, including 4 SNPs per window. The global P-value less than 0.05 was considered evidence for association between the trait and the haplotype window. Only SNPs with allele frequencies of 0.01 and higher in the pooled case-control sample were included in the haplotype analysis. Sliding-window haplotype analysis was performed using haplo.glm function in Haplo.Stats R package (version 1.5.5).
Results
Single-locus analysis
We examined the association of 3,401 SNPs in 93 candidate genes with the risk of LOAD. A total of 40 SNPs yielded nominal association (P<0.05) and 14 of them were significant at P<0.01 (Table 1). The most significant SNP, rs11770199 (P=0.0003) was located in the GRB10 gene on chromosome 7 followed by rs11161237 (P=0.0008) in ATP10A and rs6992443 (P=0.0009) in DLGAP2 on chromosome 8. Amongst the 14 significant SNPs at P<0.01, 8 revealed protection against AD risk (odds ratio (OR) range = 0.63-0.83) while the remaining 6 were associated with AD risk (OR range = 1.21-1.68)
Table 1.
Most significant SNPs in 14 genes in single-locus analysis
| Gene | CHR1 | Base Pair Position |
Most Significant SNP |
P-value | OR2 (95%CI)3 |
|---|---|---|---|---|---|
| GRB10 | 7 | 50653096 | rs11770199 | 0.0003 | 0.748 (0.64-0.87) |
| ATP10A | 15 | 23633521 | rs11161237 | 0.0008 | 1.689 (1.24-2.29) |
| DLGAP2 | 8 | 1637345 | rs6992443 | 0.0009 | 1.537 (1.19-1.98) |
| GLIS3 | 9 | 4121657 | rs1340252 | 0.0015 | 1.241 (1.09-1.41) |
| KCNQ1 | 11 | 2504751 | rs2283155 | 0.0017 | 1.218 (1.07-1.37) |
| PHACTR2 | 6 | 144083752 | rs6915880 | 0.0029 | 0.829 (0.73-0.93) |
| ANO1 | 11 | 69711744 | rs3740722 | 0.003 | 0.764 (0.64-0.91) |
| MAGI2 | 7 | 78013532 | rs1978326 | 0.0043 | 0.658 (0.49-0.87) |
| WT1 | 11 | 32366048 | rs5030328 | 0.0054 | 0.632 (0.45-0.87) |
| GPR1 | 2 | 206775748 | rs16838070 | 0.0058 | 1.245 (1.06-1.45) |
| ZNF331 | 19 | 58720819 | rs2708771 | 0.006 | 0.803 (0.69-0.94) |
| NTM | 11 | 131287579 | rs656644 | 0.0067 | 0.812 (0.69-0.94) |
| ZC3H12C | 11 | 109547367 | rs4754425 | 0.0084 | 0.836 (0.73-0.95) |
| SLC22A2 | 6 | 160558735 | rs2450974 | 0.0088 | 1.331 (1.07-1.65) |
Chromosomal location
Odds ratio
Confidence Interval
Gene-based analysis
Gene-based analyses including all SNPs in a given gene, revealed 4 significant associations (WT1, ZC3H12C, DLGAP2, and GPR1) at P <0.05 (Table 2). Interestingly, all of these 4 genes (WT1, ZC3H12C, DLGAP2, and GPR1) also revealed associations at P<0.01 in single-locus analysis. The most significant association was seen with WT1 (P=0.010). However, this association did not remain significant after correcting for gene-based multiple comparisons.
Table 2.
Imprinted gene loci associated with Alzheimer Disease in VEGA analysis
| Gene | CHR1 | SNP | Base-Pair Location | Total SNPs |
No. of Tagging SNPs |
P- Value Threshold for Significance |
P-value |
|---|---|---|---|---|---|---|---|
| WT1 | 11 | rs5030328 | 32365897-32413657 | 29 | 17 | 0.002941 | 0.0100 |
| ZC3H12C | 11 | rs4754425 | 109469296-109547776 | 30 | 11 | 0.004545 | 0.0227 |
| DLGAP2 | 8 | rs6992443 | 1436975-1644049 | 95 | 60 | 0.000833 | 0.0364 |
| GPR1 | 2 | rs16838070 | 206748284-206791016 | 17 | 14 | 0.003571 | 0.0371 |
Chromosomal location
Haplotype Analysis
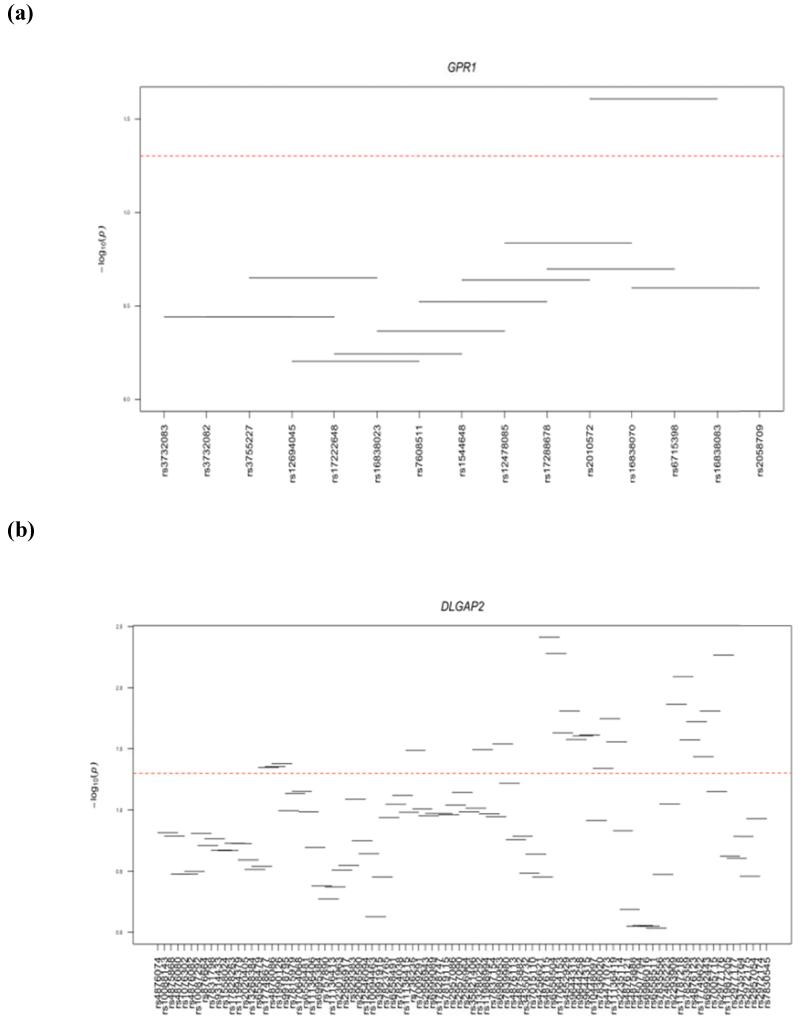
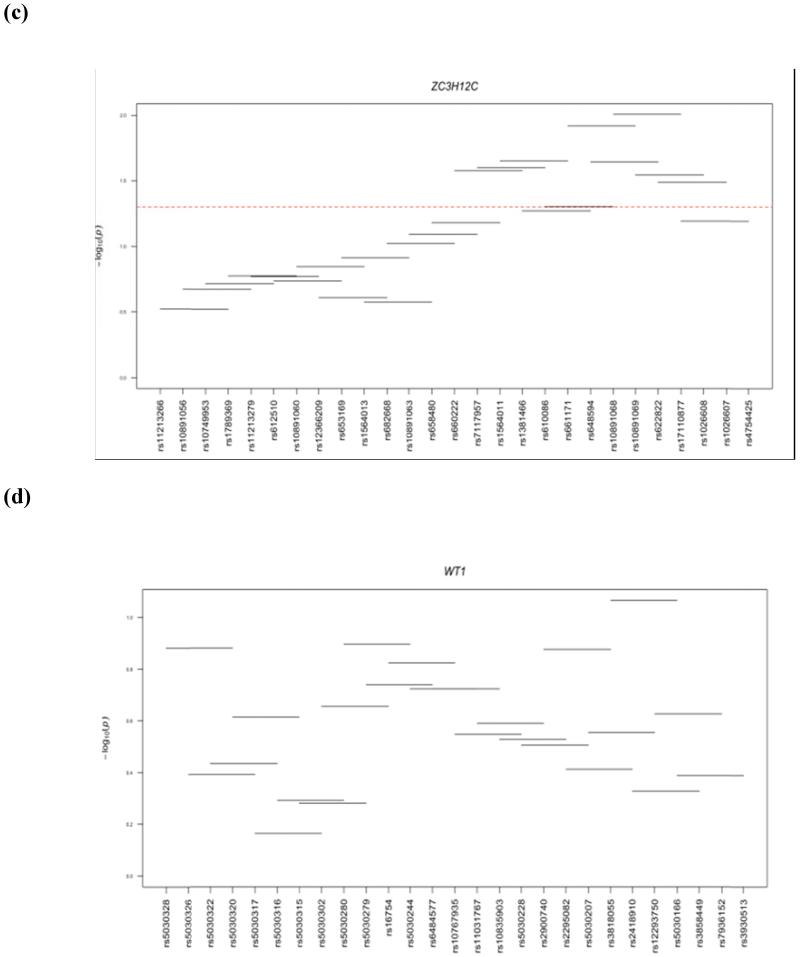
Haplotype analysis was performed within 4 genes (WT1, ZC3H12C, DLGAP2, and GPR1) that showed significant gene-based associations. Three of the 4 genes examined (ZC3H12C, DLGAP2, and GPR1) showed significant haplotype window associations with LOAD (Fig 1). ZC3H12C had 8 significant windows and the best association was observed for window 21 (SNPs rs10891068-rs10891069-rs622822-rs17110877; P=0.009801). DLGAP2 showed 23 significant windows and the best association was observed for window 58 (SNPs rs4256621-rs4876115-rs6558504-rs17064153; P=0.003875). Only one window was significant in GPR1 (SNPs rs2010572-rs16838070-rs6715398-rs16838083; P=0.024761). No significant haplotype window was observed in the WT1 gene (Fig 1).
Figure 1.
Haplotye windows showing association with AD. Horizontal lines show the windows tested, with the corresponding SNP rs number along the horizontal axis and global p-value on the vertical axis. Significant association fall above the red dotted line at –log10 (0.05) = 1.3.
Discussion
In this study we have tested the hypothesis that genetic variation in imprinted genes is associated with LOAD risk. We performed single-locus, gene-based and haplotype analyses on 93 imprinted genes. Single-locus analysis demonstrated 14 significant loci at P<0.01 and four of them also revealed significant associations in gene-based analysis. Four genes that showed significant association in gene-based and single locus analysis include WT1, ZC3H12C, DLGAP2, and GPR1. Although the association signals in these genes do not survive after multiple testings, they deserve attention as they are expressed in the brain and thus are potential candidates for AD. Our single-locus analysis showed that SNPs in two of these genes (DLGAP2/rs6992443; GPR1/rs16838070) are associated with increased risk for LOAD (OR=1.24-1.53). Also the significant haplotype windows within these genes contained best SNPs from single-locus analysis. These two genes have been reported as maternally imprinted. DLGAP2 (disks large-associated protein 2) is highly expressed in the brain [27] and it encodes membrane-associated guanylate kinases that plays a role in molecular organization of synapses and signaling of neuronal cells. GPR1 (G Protein-Coupled Receptor 1) encodes for a receptor that has a role in regulation of inflammation and is expressed in hippocampus [28].
The other two genes that showed decreased risk for AD were ZC3H12C/ rs4754425 (OR=0.84) and WT1/ rs5030328 (OR=0.63). ZC3H12C showed 8 significant haplotype windows; however, we did not find any significant haplotype window for WT1. The WT1 (Wilms Tumor 1) gene encodes a transcription factor. It plays an important role in cellular development and cell survival. WT1 has been reported as paternally imprinted in placenta and fetal brain but maternally imprinted in fibroblasts and lymphocytes. It is suggested that neuronal apoptosis plays an important role in the pathogenesis of AD and relevant to this is the observation that levels of Wilms tumor 1 protein are increased during neuronal apoptosis [29]. Zinc Finger CCCH-Type containing 12C (ZC3H12C) functions as RNase and it regulates the levels of target RNA species [30].
All the four significant genes in our gene-based analysis are maternally imprinted and thus may provide a mechanism responsible for increased maternal inheritance in AD patients [19]. In this regard, maternally expressed imprinted genes in mice generally, but not always, show their expression in cerebral cortex/hippocampal regions that are associated with cognition and executive functioning [31]. So, it is quite possible that silencing or imprinting of maternal allele disrupt the cognitive functioning of the brain and it could lead to neurodegenerative disorders like AD. This is in line with many imprinting disorders where imprinting status of a gene is associated with abnormal brain development and function [32].
Although the imprinted genes in the brain are candidate for AD, the current study does not tell us about the parental origin of single-base changes. However, finding significant associations with selected number of imprinted genes implies that imprinting may be an important mechanism for AD. Additional genetic studies in independent samples are needed to confirm our findings and to comprehensively examine the role of genetic variation in imprinted genes in relation to LOAD risk.
Supplementary Material
Acknowledgements
This project was supported by Higher Education Commission (HEC) of Pakistan and the U.S. National Institutes of Health through grant numbers AG030653, AG041718, AG005133, AG07562 and AG023651.
References
- [1].Karch CM, Jeng AT, Nowotny P, Cady J, Cruchaga C, Goate AM. Expression of novel Alzheimer’s disease risk genes in control and Alzheimer’s disease brains. PloS One. 2012;7:e50976. doi: 10.1371/journal.pone.0050976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Genin E, Hannequin D, Wallon D, Sleegers K, Hiltunen M, Combarros O, Bullido MJ, Engelborghs S, De Deyn P, Berr C. APOE and Alzheimer disease: a major gene with semi-dominant inheritance. Mol Psychiatr. 2011;16:903–907. doi: 10.1038/mp.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hollingworth P, Harold D, Sims R, Gerrish A, Lambert J-C, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lambert J, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido M, Tavernier B. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- [5].Lambert J-C, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, Jun G, DeStefano AL, Bis JC, Beecham GW. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carrasquillo MM, Lambert JC. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lee SH, Harold D, Nyholt DR, Goddard ME, Zondervan KT, Williams J, Montgomery GW, Wray NR, Visscher PM. Estimation and partitioning of polygenic variation captured by common SNPs for Alzheimer’s disease, multiple sclerosis and endometriosis. Hum Mol Genet. 2013;22:832–841. doi: 10.1093/hmg/dds491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ridge PG, Mukherjee S, Crane PK, Kauwe JS. Alzheimer’s disease: analyzing the missing heritability. PLoS One. 2013;8:e79771. doi: 10.1371/journal.pone.0079771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Barbaux S, Gascoin-Lachambre G, Buffat C, Monnier P, Mondon F, Tonanny M-B, Pinard A, Auer J, Bessières B, Barlier A. A genome-wide approach reveals novel imprinted genes expressed in the human placenta. Epigenetics. 2012;7:1079–1090. doi: 10.4161/epi.21495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Luedi PP, Dietrich FS, Weidman JR, Bosko JM, Jirtle RL, Hartemink AJ. Computational and experimental identification of novel human imprinted genes. Genome Res. 2007;17:1723–1730. doi: 10.1101/gr.6584707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21:457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- [12].Bartolomei MS, Ferguson-Smith AC. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol. 2011;3:a002592. doi: 10.1101/cshperspect.a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kernohan KD, Bérubé NG. Genetic and epigenetic dysregulation of imprinted genes in the brain. Epigenomics. 2010;2:743–763. doi: 10.2217/epi.10.61. [DOI] [PubMed] [Google Scholar]
- [14].Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- [15].Bassett SS, Avramopoulos D, Fallin D. Evidence for parent of origin effect in late onset Alzheimer disease. Am J Med Genet. 2002;114:679–686. doi: 10.1002/ajmg.10648. [DOI] [PubMed] [Google Scholar]
- [16].Berti V, Mosconi L, Glodzik L, Li Y, Murray J, De Santi S, Pupi A, Tsui W, De Leon MJ. Structural brain changes in normal individuals with a maternal history of Alzheimer’s. Neurobiol Aging. 2011;32:e17–e26. doi: 10.1016/j.neurobiolaging.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fallin MD, Szymanski M, Wang R, Gherman A, Bassett SS, Avramopoulos D. Fine mapping of the chromosome 10q11-q21 linkage region in Alzheimer’s disease cases and controls. Neurogenetics. 2010;11:335–348. doi: 10.1007/s10048-010-0234-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Liu Z, Chen HH, Li TL, Xu L, Du HQ. A cross sectional study on cerebrospinal fluid biomarker levels in cognitively normal elderly subjects with or without a family history of Alzheimer’s disease. CNS Neurosci Ther. 2013;19:38–42. doi: 10.1111/cns.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mosconi L, Mistur R, Switalski R, Brys M, Glodzik L, Rich K, Pirraglia E, Tsui W, De Santi S, De Leon M. Declining brain glucose metabolism in normal individuals with a maternal history of Alzheimer disease. Neurology. 2009;72:513–520. doi: 10.1212/01.wnl.0000333247.51383.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bassett SS, Avramopoulos D, Perry RT, Wiener H, Watson B, Go RC, Fallin MD. Further evidence of a maternal parent of origin effect on chromosome 10 in late onset Alzheimer’s disease. Am J Med Genet B Neuropsychiatr Genet. 2006;141:537–540. doi: 10.1002/ajmg.b.30350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mastroeni D, McKee A, Grover A, Rogers J, Coleman PD. Epigenetic differences in cortical neurons from a pair of monozygotic twins discordant for Alzheimer’s disease. PloS One. 2009;4:e6617. doi: 10.1371/journal.pone.0006617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, Jaenisch R, Laird PW, Akbarian S. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PloS One. 2007;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tohgi H, Utsugisawa K, Nagane Y, Yoshimura M, Genda Y, Ukitsu M. Reduction with age in methylcytosine in the promoter region-224 approximately −101 of the amyloid precursor protein gene in autopsy human cortex. Mol Brain Res. 1999;70:288–292. doi: 10.1016/s0169-328x(99)00163-1. [DOI] [PubMed] [Google Scholar]
- [24].Wang H-X, Wahlberg M, Karp A, Winblad B, Fratiglioni L. Psychosocial stress at work is associated with increased dementia risk in late life. Alzheimers Dement. 2012;8:114–120. doi: 10.1016/j.jalz.2011.03.001. [DOI] [PubMed] [Google Scholar]
- [25].Kamboh M, Demirci F, Wang X, Minster R, Carrasquillo M, Pankratz V, Younkin S, Saykin A, Jun G, Baldwin C. Genome-wide association study of Alzheimer’s disease. Transl Psychiatry. 2012;2:e117. doi: 10.1038/tp.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Barrett JC, Fry B, Maller J, Daly M. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- [27].Ranta S, Zhang Y, Ross B, Takkunen E, Hirvasniemi A, de la Chapelle A, Gilliam TC, Lehesjoki A-E. Positional cloning and characterisation of the human DLGAP2 gene and its exclusion in progressive epilepsy with mental retardation. Eur J Hum Genet. 2000;8:381–384. doi: 10.1038/sj.ejhg.5200440. [DOI] [PubMed] [Google Scholar]
- [28].Marchese A, Docherty JM, Nguyen T, Heiber M, Cheng R, Heng HH, Tsui L-C, Shi X, George SR, O’Dowd BF. Cloning of human genes encoding novel G protein-coupled receptors. Genomics. 1994;23:609–618. doi: 10.1006/geno.1994.1549. [DOI] [PubMed] [Google Scholar]
- [29].Lovell MA, Xie C, Xiong S, Markesbery WR. Wilms’ tumor suppressor (WT1) is a mediator of neuronal degeneration associated with the pathogenesis of Alzheimer’s disease. Brain Res. 2003;983:84–96. doi: 10.1016/s0006-8993(03)03032-4. [DOI] [PubMed] [Google Scholar]
- [30].Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: integrating information about genes, proteins and diseases. Trends Genet. 1997;13:163. doi: 10.1016/s0168-9525(97)01103-7. [DOI] [PubMed] [Google Scholar]
- [31].Davies W, Isles AR, Wilkinson LS. Imprinted gene expression in the brain. Neurosci Biobehav Rev. 2005;29:421–430. doi: 10.1016/j.neubiorev.2004.11.007. [DOI] [PubMed] [Google Scholar]
- [32].Falls JG, Pulford DJ, Wylie AA, Jirtle RL. Genomic imprinting: implications for human disease. Am J Pathol. 1999;154:635–647. doi: 10.1016/S0002-9440(10)65309-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.