Abstract
Background and Aims
Increased amyloid deposition in HIV-infected brains may contribute to the pathogenesis of neurocognitive dysfunction in infected patients. We have previously shown that exposure to HIV results in enhanced amyloid β (Aβ) levels in human brain microvascular endothelial cells, suggesting that brain endothelial cells contribute to accumulation of Aβ in HIV-infected brains. Importantly, Aβ not only accumulates in the cytoplasm of HIV-exposed cells but also enters the nuclei of brain endothelial cells.
Methods
cDNA microarray analysis was performed in order to examine changes in the transcriptional profile associated with Aβ nuclear entry in the presence of HIV-1.
Results
Gene network analysis indicated that inhibition of nuclear entry of Aβ resulted in enrichment in gene sets involved in apoptosis and survival, endoplasmic reticulum stress response, immune response, cell cycle, DNA damage, oxidative stress, cytoskeleton remodeling and transforming growth factor b (TGFβ) receptor signaling.
Conclusions
The obtained data indicate that HIV-induced Aβ nuclear uptake affects several cellular stress-related pathways relevant for HIV-induced Aβ pathology.
Keywords: DNA microarray, HIV, Blood-brain barrier, Amyloid beta
Introduction
Strong evidence indicates increased amyloid deposition in the brains of HIV-infected patients (1,2). HIV-associated neurocognitive disorders (HAND) in the older population have been partially linked to early signs of β-amyloidosis, demonstrating the importance of amyloid β (Aβ) deposition for the clinical outcome of HIV infection (3,4). Morphologically, prominent Aβ localization is observed in the perivascular space in HIV brains (3–6), indicating the importance of cerebral microvasculature in amyloid pathology. Indeed, brain vascular dysfunction and the blood-brain barrier (BBB) are critical in Aβ accumulation in the brain (7). Observations from our laboratories have shown that exposure to HIV results in enhanced Aβ levels in human brain microvascular endothelial cells (HBMEC) and increased transendothelial transfer (8,9), supporting the hypothesis that brain endothelial cells may contribute to brain Aβ accumulation in HIV infection.
Recently, we demonstrated that Aβ not only accumulates in the cytoplasm of HIV-exposed cells but also enters the nuclei (10). Inhibition of dynamin by dynasore (Dy), a blocker of GTP-ase activity of dynamin, effectively attenuated HIV-induced Aβ nuclear uptake. To explore the possible mechanisms involved we performed cDNA microarray analysis in order to investigate the transcriptional profile changes in brain endothelial cells associated with the inhibition of HIV-induced Aβ nuclear uptake. Data from this study suggest that blocking HIV-induced Aβ nuclear uptake affects several cellular stress-related pathways that may be relevant for HIV-induced Aβ pathology. These gene signature changes indicate promising new directions for mechanistic studies related to Aβ pathology in the context of HIV brain infection.
Materials and Methods
Cell Cultures, HIV Infection, and Treatment Factors
Human brain microvascular endothelial cells (HBMEC) representing a stable, well-characterized, and differentiated human brain endothelial cell line (11) were cultured as previously reported (10). Briefly, normal human brain endothelial cells were transduced by lentiviral vectors incorporating human telomerase or SV40 T antigen. Among several stable immortalized clones obtained by sequential limiting dilution cloning of the transduced cells, hCMEC/D3 (referred here as HBMEC) was selected as expressing normal endothelial markers and demonstrating blood-brain barrier characteristics (11). Cells were kindly supplied by Dr. Couraud (Institut Cochin, CNRS UMR 8104-INSERM U567, Université René Descartes, Paris, France).
HIV stock was generated using human embryonic kidney (HEK) 293T cells (American Type Culture Collection, Manassas, VA) transfected with pYK-JRCSF plasmid containing full-length proviral DNA as described earlier (12). Throughout the study, HBMEC were exposed to HIV particles for 24 h at the p24 level of 30 ng/mL as previously reported (10). Freshly solubilized Aβ (1–40) HCl (Anaspec, San Jose, CA) solutions without pre-aggregation were used to treat cells at a concentration of 1 μM for 10 min as published earlier (10). The inhibitor dynasore hydrate (Dy) (Sigma Aldrich, St. Louis, MO) was dissolved in DMSO and further diluted in cell culture media. Cells were pre-treated with 50 μM Dy for 30 min prior to Aβ exposure. Treatment was terminated by changing the media to normal growth medium in which cells were incubated for the next 4 h, followed by harvesting the cells and RNA isolation.
RNA Purification
Total RNA was purified using RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Isolated total RNA was assessed by RNA 6000 Nano chips and an Agilent Bioanalyzer with 2100 Expert software V.B.02 (Agilent Technologies, Palo Alto, CA). The quality of total RNA was characterized using the RNA integrity number (RIN) (13). A RIN value of 10 indicates intact RNA, whereas a RIN of 5 represents partially degraded RNA. The RIN value was 10 for all samples (4 for each treatment group).
DNA Microarray and Processing
The Affymetrix GeneChip Human Gene 2.0 ST microarray platform represents 44,699 protein coding genes. It contains >6.0 million distinct probes covering coding and non-coding transcripts resulting in >285,000 covered full-length transcripts. Samples were first amplified and labeled using Ovation RNA Amplification kit (NuGEN). Converted sense-strand cDNA samples were then fragmented and labeled using Encore Biotin Module (NuGEN) and hybridized to the Affymetrix GeneChip Human Gene 2.0 ST microarray for 17 h at 65°C according to the manufacturer’s recommendation (Affymetrix, Santa Clara, CA).
Statistical and Pathway Analyses
Initial data processing and quality control (QC) were performed using the Affymetrix® Expression Console™ Software. Data were Robust Multi-array Analysis (RMA-Sketch) normalized both at the gene and exon levels and QC checks were performed to identify sample and transcript outliers using multiple metrics and principal Components Analysis (PCA). Post-QC data analyses were carried out using the Affymetrix® Transcriptome Analysis Console (TAC) 2.0 Software, assuming the algorithm option One-Way Between-Subject ANOVA (unpaired) and default filter criteria. Coding transcript and splicing analyses were conducted, and results with an ANOVA p <0.05 and fold change < −2 or fold change >2 were considered significant. Hierarchical clustering was also performed using the TAC software. Further enrichment, pathway analyses, and mapping were conducted using the MetaCore pathway/network analysis (14), which is based on the hypergeometric statistical distribution. As with single transcript analyses, only pathways or networks with false discovery rate (FDR) p <0.05 were selected as significantly enriched.
Quantitative Real-time Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
To validate the cDNA array findings, expression of selected genes was analyzed by quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR). Total RNA was isolated using RNeasy kit (Qiagen) and reverse transcribed into cDNA using the RT2 First Strand Kit (Qiagen). Real-time PCR was conducted using the ABI 7500 standard system (Applied Biosystems, Foster City, CA). PCR amplification was performed on custom 96-well RT2 Profiler PCR array (Qiagen) for ATF4, PAI-1 (SERPINE-1), IL-6, GADD45 B, ANKFY1, MANF, ATP10D, and SLC30A1. The array contained the following controls: GAPDH and β-actin as housekeeping genes, human genomic DNA contamination control, reverse transcription control, and positive PCR control. Thermocycling conditions were 95°C for 10 min followed by 95°C for 15 sec, and 60°C for 60 sec for 40 cycles. Expression of mRNA was calculated and analyzed by the comparative CT method. PCR amplification of human GAPDH was used to normalize expression levels of the target genes.
Results
Gene Expression Signature Associated with Inhibition of HIV-induced Aβ Nuclear Uptake
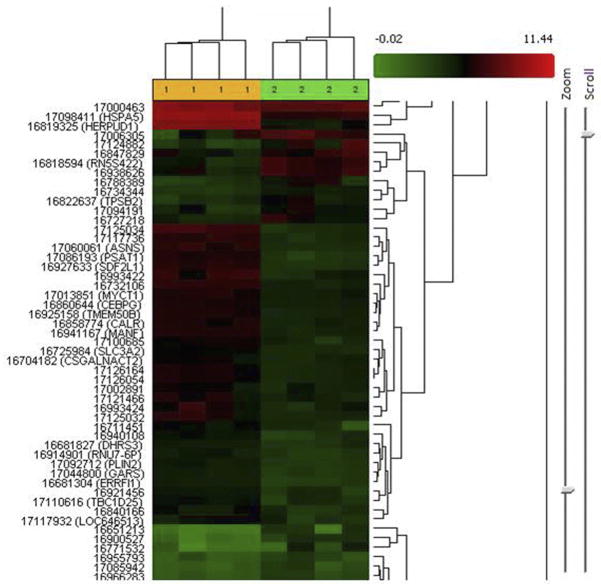
We recently demonstrated that Aβ not only accumulates in the cytoplasm of HIV-exposed cells but also enters the nuclei of brain endothelial cells. Moreover, inhibition of dynamin by Dy (50 μM Dy pretreatment for 30 min prior to Aβ exposure) effectively attenuated HIV-induced Aβ nuclear uptake (10). To explore the possible mechanisms involved, we investigated gene signature changes associated with this event. For this purpose, we compared gene expression profiles of the HIV + Aβ + Dy treatment group to the HIV + Aβ group. Heatmap representation with clustergram analysis of the differentially expressed genes is shown in Figure 1. Comparison of the HIV + Aβ + Dy group to the HIV + Aβ group using the statistical criteria described in Materials and Methods indicated that 281 genes were differentially expressed with 90 genes being downregulated and 191 genes upregulated (ANOVA p <0.05). When applying the more stringent statistical criteria on exon analysis, this list was narrowed down to 13 different transcripts with FDR p <0.05 (Table 1). The list included genes coding for endoplasmic reticulum (ER) stress proteins, transcription factors, cell cycle regulators, ion channels and transporters. The most upregulated transcripts were DDIT3 (DNA-damage-inducible transcript 3) ~19.4 fold, HER-PUD1 (homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1) 11.5-fold, ATP10D (ATPase, class V, type 10D) 13.7-fold, HSPA5 (heat shock 70-kDa protein 5 [glucose-regulated protein, 78 kDa]), 9.65-fold. In addition, MANF (mesencephalic astrocyte-derived neurotrophic factor) was upregulated 3.49-fold, and SLC30A1 (solute carrier family 30 [zinc transporter], member 1), 3.72-fold. Only two transcripts were differentially downregulated with ANKFY1 (ankyrin repeat and FYVE domain containing 1 [inhibits C/EBP DNA binding activity]) being the most affected by −8.58-fold.
Figure 1.
Heatmap representation with clustergram analysis of the differentially expressed genes in the HIV + Aβ + Dy vs. HIV + Aβ treatment groups.
Table 1.
List of the differentially expressed gene transcripts from the exon analysis (FDR p <0.05) in the HIV + Aβ + Dy vs. HIV + Aβ treatment groups
| Gene symbol | Description | Fold change | ANOVA p | FDR-p |
|---|---|---|---|---|
| DDIT3 | DNA-damage-inducible transcript 3 | 19.38 | 7.05E-07 | 0.036865 |
| HERPUD1 | homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 | 11.51 | 2.93E-07 | 0.036865 |
| ANKFY1 | ankyrin repeat and FYVE domain containing-1 | −8.58 | 6.01E-07 | 0.036865 |
| DNAJB9 | DnaJ (Hsp40) homolog, subfamily B, member 9 | 6.7 | 5.62E-07 | 0.036865 |
| HSPA5 | heat shock 70 kDa protein 5 (glucose-regulated protein, 78 kDa) BiP | 9.65 | 7.31E-07 | 0.036865 |
| DNAH9 | dynein, axonemal, heavy chain 9 | −4.5 | 0.000001 | 0.042587 |
| MANF | mesencephalic astrocyte-derived neurotrophic factor | 3.49 | 0.000001 | 0.042587 |
| TFRC | transferrin receptor (p90, CD71) | 3.72 | 0.000001 | 0.042587 |
| SLC30A1 | solute carrier family 30, member 1 | 3.04 | 0.000002 | 0.049175 |
| CARS | cysteinyl-tRNA synthetase | 11.65 | 0.000003 | 0.049175 |
| CCNB1IP1 | cyclin B1 interacting protein 1, | 4.6 | 0.000002 | 0.049175 |
| TNRC6B | trinucleotide repeat containing 6B | 1.65 | 0.000002 | 0.049175 |
| ATP10D | ATPase, class V, type 10D | 13.68 | 0.000002 | 0.049175 |
Gene Network Analysis of the Differentially Expressed Genes Associated with Inhibition of Aβ Nuclear Uptake
Gene network analysis of the differentially expressed genes in the HIV + Aβ + Dy group vs. HIV + Aβ group suggested enrichment in gene sets mainly involved in apoptosis and survival, ER stress response, immune response, cell cycle, DNA damage, oxidative stress, cytoskeleton remodeling and TGFβ receptor signaling (Table 2). These were the top scored pathway maps (i.e., maps with the lowest p value) based on the enrichment distribution sorted by “statistically significant maps” set. All experimental data were linked to and visualized on the maps as thermometer-like figures. Upward thermometers have red color and indicate upregulated signals.
Table 2.
Top scored maps of the pathway gene enrichment analysis of the HIV + Aβ + Dy vs. HIV + Aβ treatment groups
| Maps with genes |
|---|
|
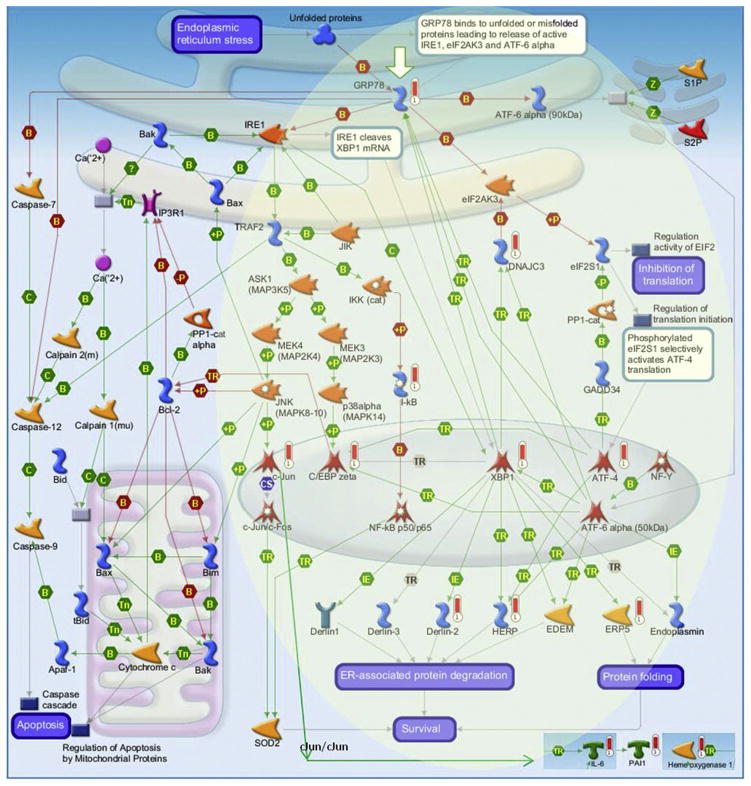
Figure 2 shows the most significant signaling pathway map, namely, “apoptosis and survival—endoplasmic reticulum stress response”—additionally enriched with differentially expressed genes from the “immune response-IL-1 signaling” pathway. We previously showed the involvement of the TGFβ/Smad signaling in the HIV-induced Aβ nuclear uptake and evaluated the impact of dynamin on nuclear levels of pSmad2 and Smad2. Overall, Dy diminished activation of TGFβ/Smad signaling while decreasing Aβ nuclear uptake in cells exposed to HIV (10). Data from this network analysis indicate that several Smad target genes are upregulated as the result of Dy exposure (namely, IL-6, PAI1, HERPUD1, ATF4) (Figure 2), indicating that they may have been repressed by the Aβ nuclear uptake.
Figure 2.
Differentially expressed gene enrichment of the top scored pathway maps. The most significant signaling pathway map for Apoptosis and survival-Endoplasmic reticulum stress response pathway was additionally enriched with differentially expressed genes from the Immune response-IL-1 signaling pathway. Enrichment and pathway analyses were conducted using the MetaCore pathway/network analysis (14). Experimental data were linked to and visualized on the maps as thermometer-like figures. Upward thermometers have red color and indicate upregulated signals. For additional figure legend go to http://lsresearch.thomsonreuters.com/static/uploads/files/2014-05/MetaCoreQuickReferenceGuide.pdf.
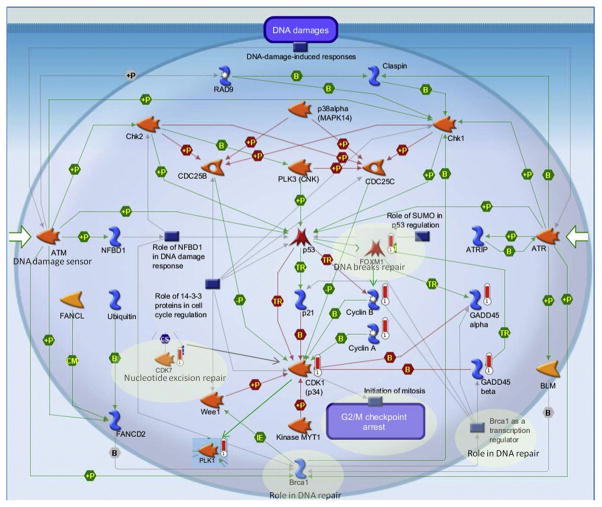
Figure 3 reflects the pathway map for “DNA damage-ATM/ATR regulation of G2/M checkpoint” additionally enriched with differentially expressed genes from the “cell cycle–initiation of mitosis” map. Several genes were differentially upregulated on these maps like PLK1, FOXM1, CDK1 (p34), CDK7, GADD45 alpha, Cyclin A, Cyclin B, GADD45 beta.
Figure 3.
Gene enrichment of the “Cell cycle-Initiation of mitosis” and “DNA damage-ATM/ATR regulation of G2/M checkpoint” combined pathways. The pathway map for “DNA damage-ATM/ATR regulation of G2/M checkpoint” was enriched with differentially expressed genes from the “Cell cycle-Initiation of mitosis” map. Enrichment and pathway analyses were conducted using the MetaCore pathway/network analysis (14). Experimental data were linked to and visualized on the maps as thermometer-like figures. Upward thermometers have red color and indicate upregulated signals. For additional figure legend go to http://lsresearch.thomsonreuters.com/static/uploads/files/2014-05/MetaCoreQuickReferenceGuide.pdf.
In addition to the top scored maps, gene enrichment analysis of the differentially expressed genes pointed to several other pathways including oxidative stress, cytoskeleton remodeling, and TGFβ receptor signaling (Table 2). Several genes (hemoxygenase-1, PAI-1, JUN, GADD45 B) from these maps could also be linked to the previously described top scored maps. Furthermore, finding the SMAD2 gene in the TGFβ receptor signaling gene list is consistent with our previous data showing the involvement of Smad2 and pSmad2 in HIV-induced nuclear Aβ entry (10).
Validation of Differentially Expressed Genes by Real-time RT-PCR
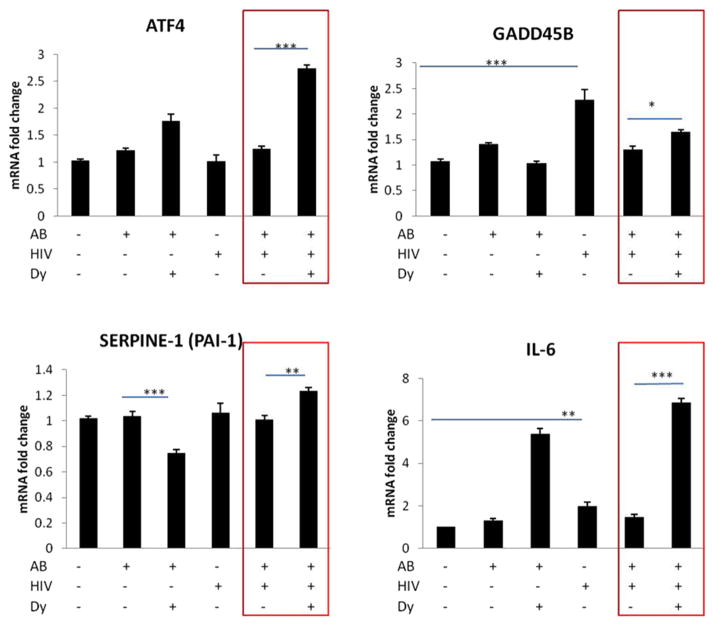
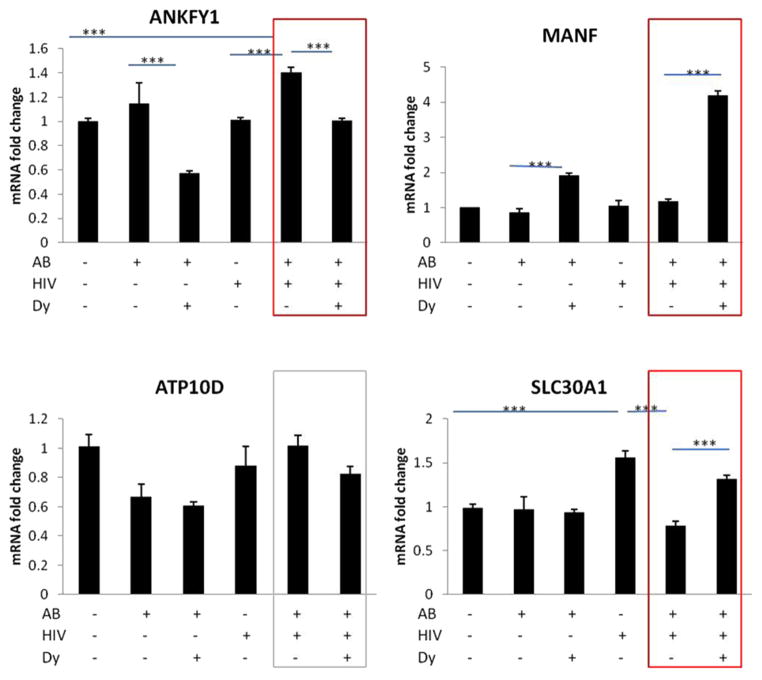
We validated several Smad target genes with real-time RT-PCR including ATF4, IL-6, PAI-1, GADD45 B (Figure 4). In addition, we selected gene transcripts from Table 1, up-or downregulated in the DNA microarray (with FDR < 0.05) and analyzed the results by real-time RT-PCR for MANF, SLC30A1, ANKFY1, ATP10D (Figure 5). We validated the transcript expressional change trend in the HIV + Aβ + Dy group vs. HIV + Aβ group for ATF4, IL-6, PAI-1, GADD45 B, MANF, SLC30A1, ANKFY1 (red boxes) but not for ATP10D (grey box) (Figure 5). Differential gene expression was similar between the results of the DNA microarray and real-time RT-PCR, meaning that the level of expression of these genes changed in the same direction. However, there were quantitative differences between these methods (Figures 4 and 5).
Figure 4.
Validation of Smad target gene changes by qRT-PCR. HBMEC were exposed to HIV particles at p24 levels of 30 ng/mL for 24 h followed by co-treatment with 1 μM Aβ (1–40) for 10 min. Selected cultures were pretreated with dynamin inhibitor dynasore (Dy, 50 μM) for 30 min followed by co-treatment with 1 μM Aβ (1–40) for 10 min. Real-time RT-PCR was conducted on custom 96-well RT2 Profiler PCR array (Qiagen) using the SYBRgreen technique. Expression of mRNA was calculated and analyzed by the comparative CT method and normalized to human GAPDH. Values are mean ± SEM, n = 5. *Statistically significant at p <0.05, ***Statistically significant at p <0.001.
Figure 5.
Validation of selected gene changes from the top scored list (FDR p <0.05) by qRT-PCR. HBMEC were treated and qRT-PCR was performed as in Figure 4. Expression of mRNA was calculated and analyzed by the comparative CT method and normalized to human GAPDH. Values are mean ± SEM, n = 5. *Statistically significant at p <0.05, ***Statistically significant at p <0.001.
Discussion
There is an increasing need for integrative molecular approaches on the entire human transcriptome instead of just focusing on one gene or on a set of target genes in order to better understand biological processes. Microarray technology is a constantly expanding field allowing an integrated insight into transcriptional changes. This approach is frequently used as a useful tool to “fingerprint” different diseases (15) or to create new hypotheses and/or potential novel mechanisms. Thus, this approach was used in the present study to identify a transcriptional profile associated with HIV-induced Aβ accumulation in the nuclei in brain endothelial cells. To this extent, we analyzed gene signatures in cells in which Aβ entry into the nuclei was blocked by inhibition of dynamin by Dy. Although Dy is very effective in attenuation of Aβ nuclear entry in cells exposed to HIV and Aβ, one of the pitfalls is a potential influence of blocking GTP-ase activity of dynamin, which may have an effect beyond Aβ trafficking into the nuclei.
Among different treatments analyzed in the present study, only the HIV + Aβ + Dy vs. HIV + Aβ comparison resulted in differentially expressed genes with the highest significance (FDR p <0.05) (Table 1). Several of the genes that play a role in ER stress were upregulated, e.g., DDIT3 (C/EBP-zeta, CHOP), HERPUD1, DNAJB9, HSPA5, MANF. The top scored pathway map (i.e., map with the lowest p value) from the gene network analysis also indicated enrichment in gene sets involved in apoptosis and survival-ER stress response. The third scored map on immune response-IL-1 signaling showed several upregulated genes playing a role in inflammatory responses. These genes could also be linked into the top scored map, demonstrating that ER stress and immune/inflammatory responses work in concert and the outcome can be either cell apoptosis or survival. However, the enrichment analysis depicted on these maps suggests that the differentially upregulated genes tip the balance towards cell survival (shaded area on the map). These observations are consistent with the evidence that ER stress plays a role in Aβ pathology (16).
A novel and interesting observation is the association of ER stress response with the inhibition of nuclear Aβ uptake. Although the mechanisms of this phenomenon are unknown, we observed that nuclear uptake is a substantial part of the overall HIV-induced Aβ accumulation in HBMEC (10). Thus, inhibition of this process is likely to lead to higher levels of Aβ in the cytoplasm, resulting in ER stress. Indeed, the unfolding protein responses are known inducers of ER stress with the subsequent pathways leading to autophagy and/or apoptosis. In this context, an interesting observation was increased (FDR p <0.05) expression of MANF (mesencephalic astrocyte-derived neurotrophic factor) associated with the inhibition of HIV-induced Aβ nuclear uptake (Figure 5). MANF was previously shown to be upregulated and secreted in ER stress (17), protecting cells from ER stress-induced apoptosis (18). Furthermore, MANF was reported to be protective to neurons in cerebral ischemia (19,20). To our knowledge, there are no previous reports on endothelial MANF; thus, it may be interesting to investigate the novel role of this protein in HIV-associated vascular pathology of Aβ or in other neurodegenerative conditions.
Evaluation of the pathway map for “DNA damage-ATM/ATR regulation of G2/M checkpoint” additionally enriched with differentially expressed genes from the “cell cycle–initiation of mitosis” map (PLK1, FOXM1, CDK7) also produced several interesting observations (Figure 3). PLK1 and CDK7 are serine/threonine kinases and could be linked to the “DNA damage-ATM/ATR regulation of G2/M checkpoint” map through cyclin-dependent kinase 1, CDK1 (p34). FOXM1, on the other hand, is a transcription factor and is linked to this pathway by regulating expression of cell cyclins (Figure 3). ATR (ataxia telangiectasia and Rad3 related) and ATM (ataxia telangiectasia mutated) are closely related kinases that are activated by DNA damage. ATM/ATR initiates the DNA damage checkpoint by phosphorylating a number of key proteins. Once activated, the checkpoint leads to cell cycle arrest and either DNA repair or apoptosis (20). CCNA2 (cyclin A2) and CCNB1 (cyclin B1) belong to the highly conserved cyclin family whose members are characterized by a dramatic periodicity in protein abundance through the cell cycle. Cyclins function as regulators of CDK kinases. Different cyclins exhibit distinct expression and degradation patterns that contribute to the temporal coordination of each mitotic event (21). Several genes were differentially upregulated on these maps like PLK1, FOXM1, CDK1 (p34), CDK7, GADD45 alpha, Cyclin A, Cyclin B, GADD45 beta. The shaded areas on the map show different genes having a role in DNA repair mechanisms. Although it appears that inhibition of nuclear entry of Aβ stimulates several DNA repair mechanisms, these results may be caused by an unspecific effect of the inhibitor used in the present study. Dy itself may affect several nuclear import/export mechanisms leading to DNA damage needing DNA repair.
Analysis of the maps presented above indicated that several Smad target genes including ATF4, IL-6, PAI-1 (SERPINE-1) and GADD45 B were differentially expressed upon inhibition of HIV-induced Aβ nuclear uptake. All these proteins may play an important role in HIV and/or Aβ pathology. ATF4 is a transcription factor and an important player in ER stress responses (16) that was shown to be involved in the TGFβ/Smad signaling pathway (22). We have demonstrated before that HIV Tat protein induced IL-6 (23) and this effect was amplified by Aβ. Interestingly, blocking HIV-induced nuclear Aβ uptake also induced IL-6 expression. PAI-1 was shown to have a role in HIV-associated cardiovascular and Aβ pathology. In fact, PAI-1 expression level predicted myocardial infarction in HIV-infected patients (24) and knocking out PAI-1 gene in a mouse model of Alzheimer’s disease reduced Aβ burden (25). Thus, PAI-1 may be a potential target for therapy to protect against Aβ-related pathology (26). GADD45 B (growth arrest and DNA-damage-inducible, beta) involvement in TGFβ/Smad signaling was demonstrated by several laboratories (27,28). However, there are no literature data on endothelial GADD45 B gene expression changes in the context of HIV or Aβ pathology.
In addition to the previously discussed genes, two other genes from the highly significant gene list (FDR <0.05) (Table 1) and validated by real-time RT-PCR (Figure 5) should be mentioned, specifically, ANKFY and SLC30A1. ANKFY1 (Rabankyrin-5) is a Rab5 effector regulating endocytic trafficking (29). Although endocytosis was linked to HIV-induced Aβ pathology (30), it may be interesting to explore exactly why this member of the endocytotic machinery is highly associated with the inhibition of Aβ nuclear uptake. SLC30A1 gene encodes for a Zn transporter (31), which is widely expressed in different cells and is responsible for pumping Zn out of the cell. Disturbances in Zn homeostasis play a role in Aβ pathology (for review see Reference (32); however, to our knowledge, there are no reports on the role of SLC30A1 (SLC30) in HIV infection.
In conclusion, data from this study indicate that blocking HIV-induced Aβ nuclear uptake may involve several cellular stress related pathways that are relevant for HIV-induced Aβ pathology. One of the promising avenues to explore is the induction of ER stress responses. Data indicate a strong association between induction of ER stress and upregulation of mediators involved in induction of inflammatory pathways. These interlinked pathways may be detrimental to decide the fate and/or functional changes of the cells.
Acknowledgments
pYK-JRCSF was obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIH/NIAID. Supported by MH063022, MH098891, NS39254, DA027569. We acknowledge support from the Miami Center for AIDS Research (CFAR) at the University of Miami Miller School of Medicine funded by a grant (P30AI073961) from the National Institutes of Health (NIH).
References
- 1.Achim CL, Adame A, Dumaop W, et al. Increased accumulation of intraneuronal amyloid beta in HIV-infected patients. J Neuroimmune Pharmacol. 2009;4:190–199. doi: 10.1007/s11481-009-9152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brew BJ, Crowe SM, Landay A, et al. Neurodegeneration and ageing in the HAART era. J Neuroimmune Pharmacol. 2009;4:163–174. doi: 10.1007/s11481-008-9143-1. [DOI] [PubMed] [Google Scholar]
- 3.Xu J, Ikezu T. The comorbidity of HIV-associated neurocognitive disorders and Alzheimer’s disease: a foreseeable medical challenge in post-HAART era. J Neuroimmune Pharmacol. 2009;4:200–212. doi: 10.1007/s11481-008-9136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soontornniyomkij V, Moore DJ, Gouaux B, et al. Cerebral beta-amyloid deposition predicts HIV-associated neurocognitive disorders in APOE epsilon4 carriers. AIDS. 2012;26:2327–2335. doi: 10.1097/QAD.0b013e32835a117c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green DA, Masliah E, Vinters HV, et al. Brain deposition of beta-amyloid is a common pathologic feature in HIV positive patients. AIDS. 2005;19:407–411. doi: 10.1097/01.aids.0000161770.06158.5c. [DOI] [PubMed] [Google Scholar]
- 6.Steinbrink F, Evers S, Buerke B, et al. Cognitive impairment in HIV infection is associated with MRI and CSF pattern of neurodegeneration. Eur J Neurol. 2013;20:420–428. doi: 10.1111/ene.12006. [DOI] [PubMed] [Google Scholar]
- 7.Deane R, Zlokovic BV. Role of the blood-brain barrier in the pathogenesis of Alzheimer’s disease. Curr Alzheimer Res. 2007;4:191–197. doi: 10.2174/156720507780362245. [DOI] [PubMed] [Google Scholar]
- 8.Andras IE, Eum SY, Huang W, et al. HIV-1-induced amyloid beta accumulation in brain endothelial cells is attenuated by simvastatin. Mol Cell Neurosci. 2010;43:232–243. doi: 10.1016/j.mcn.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andras IE, Eum SY, Toborek M. Lipid rafts and functional caveolae regulate HIV-induced amyloid beta accumulation in brain endothelial cells. Biochem Biophys Res Commun. 2012;421:177–183. doi: 10.1016/j.bbrc.2012.03.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andras IE, Toborek M. HIV-1 stimulates nuclear entry of amyloid beta via dynamin dependent EEA1 and TGF-beta/Smad signaling. Exp Cell Res. 2014;323:66–76. doi: 10.1016/j.yexcr.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weksler BB, Subileau EA, Perriere N, et al. Blood-brain barrier-specific properties of a human adult brain endothelial cell line. FASEB J. 2005;19:1872–1874. doi: 10.1096/fj.04-3458fje. [DOI] [PubMed] [Google Scholar]
- 12.Andras IE, Toborek M. HIV-1-induced alterations of claudin-5 expression at the blood-brain barrier level. Methods Mol Biol. 2011;762:355–370. doi: 10.1007/978-1-61779-185-7_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schroeder A, Mueller O, Stocker S, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ekins S, Nikolsky Y, Bugrim A, et al. Pathway mapping tools for analysis of high content data. Methods Mol Biol. 2007;356:319–350. doi: 10.1385/1-59745-217-3:319. [DOI] [PubMed] [Google Scholar]
- 15.Simunovic F, Yi M, Wang Y, et al. Gene expression profiling of substantia nigra dopamine neurons: further insights into Parkinson’s disease pathology. Brain. 2009;132:1795–1809. doi: 10.1093/brain/awn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Endres K, Reinhardt S. ER-stress in Alzheimer’s disease: turning the scale? Am J Neurodegener Dis. 2013;2:247–265. [PMC free article] [PubMed] [Google Scholar]
- 17.Apostolou A, Shen Y, Liang Y, et al. Armet, a UPR-upregulated protein, inhibits cell proliferation and ER stress-induced cell death. Exp Cell Res. 2008;314:2454–2467. doi: 10.1016/j.yexcr.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glembotski CC, Thuerauf DJ, Huang C, et al. Mesencephalic astrocyte-derived neurotrophic factor protects the heart from ischemic damage and is selectively secreted upon sarco/endoplasmic reticulum calcium depletion. J Biol Chem. 2012;287:25893–25904. doi: 10.1074/jbc.M112.356345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu YQ, Liu LC, Wang FC, et al. Induction profile of MANF/ARMET by cerebral ischemia and its implication for neuron protection. J Cereb Blood Flow Metab. 2010;30:79–91. doi: 10.1038/jcbfm.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan S, Sorrell M, Berman Z. Functional interplay between ATM/ATR-mediated DNA damage response and DNA repair pathways in oxidative stress. Cell Mol Life Sci. 2014;71:3951–3967. doi: 10.1007/s00018-014-1666-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaldis P, Aleem E. Cell cycle sibling rivalry: Cdc2 vs. Cdk2 Cell Cycle. 2005;4:1491–1494. doi: 10.4161/cc.4.11.2124. [DOI] [PubMed] [Google Scholar]
- 22.Lian N, Lin T, Liu W, et al. Transforming growth factor beta suppresses osteoblast differentiation via the vimentin activating transcription factor 4 (ATF4) axis. J Biol Chem. 2012;287:35975–35984. doi: 10.1074/jbc.M112.372458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andras IE, Rha G, Huang W, et al. Simvastatin protects against amyloid beta and HIV-1 Tat-induced promoter activities of inflammatory genes in brain endothelial cells. Mol Pharmacol. 2008;73:1424–1433. doi: 10.1124/mol.107.042028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knudsen A, Katzenstein TL, Benfield T, et al. Plasma plasminogen activator inhibitor-1 predicts myocardial infarction in HIV-1-infected individuals. AIDS. 2014;28:1171–1179. doi: 10.1097/QAD.0000000000000247. [DOI] [PubMed] [Google Scholar]
- 25.Liu RM, van Groen T, Katre A, et al. Knockout of plasminogen activator inhibitor 1 gene reduces amyloid beta peptide burden in a mouse model of Alzheimer’s disease. Neurobiol Aging. 2011;32:1079–1089. doi: 10.1016/j.neurobiolaging.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kutz SM, Higgins CE, Higgins PJ. Novel combinatorial therapeutic targeting of PAI-1 (SERPINE1) gene expression in Alzheimer’s disease. Mol Med Ther. 2012;1:106. doi: 10.4172/2324-8769.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoo J, Ghiassi M, Jirmanova L, et al. Transforming growth factor-beta-induced apoptosis is mediated by Smad-dependent expression of GADD45b through p38 activation. J Biol Chem. 2003;278:43001–43007. doi: 10.1074/jbc.M307869200. [DOI] [PubMed] [Google Scholar]
- 28.Takekawa M, Tatebayashi K, Itoh F, et al. Smad-dependent GADD45-beta expression mediates delayed activation of p38 MAP kinase by TGF-beta. EMBO J. 2002;21:6473–6482. doi: 10.1093/emboj/cdf643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Reiling C, Reinecke JB, et al. Rabankyrin-5 interacts with EHD1 and Vps26 to regulate endocytic trafficking and retromer function. Traffic. 2012;13:745–757. doi: 10.1111/j.1600-0854.2012.01334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X, Hui L, Geiger NH, et al. Endolysosome involvement in HIV-1 transactivator protein-induced neuronal amyloid beta production. Neurobiol Aging. 2013;34:2370–2378. doi: 10.1016/j.neurobiolaging.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang L, Tepaamorndech S. The SLC30 family of zinc transporters—a review of current understanding of their biological and pathophysiological roles. Mol Aspects Med. 2013;34:548–560. doi: 10.1016/j.mam.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 32.Nuttall JR, Oteiza PI. Zinc and the aging brain. Genes Nutr. 2014;9:379. doi: 10.1007/s12263-013-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]