Abstract
Many fields of biology – including vertebrate Evo-Devo research – are facing an explosion of genomic and transcriptomic sequence information and a multitude of fish species are now swimming in this ‘genomic tsunami’.
Here, we first give an overview of recent developments in sequencing fish genomes and transcriptomes that identify properties of fish genomes requiring particular attention and propose strategies to overcome common challenges in fish genomics. We suggest that the generation of chromosome-level genome assemblies - for which we introduce the term ‘chromonome’ – should be a key component of genomic investigations in fish because they enable large-scale conserved synteny analyses that inform orthology detection, a process critical for connectivity of genomes. Orthology calls in vertebrates, especially in teleost fish, are complicated by divergent evolution of gene repertoires and functions following two rounds of genome duplication in the ancestor of vertebrates and a third round at the base of teleost fish.
Second, using examples of spotted gar, basal teleosts, zebrafish-related cyprinids, cavefish, livebearers, icefish, and lobefin fish, we illustrate how next generation sequencing technologies liberate emerging fish systems from genomic ignorance and transform them into a new model army to answer longstanding questions on the genomic and developmental basis of their biodiversity.
Finally, we discuss recent progress in the genetic toolbox for the major fish models for functional analysis, zebrafish and medaka, that can be transferred to many other fish species to study in vivo the functional effect of evolutionary genomic change as Evo-Devo research enters the postgenomic era.
Keywords: teleost fish, genome duplication, ortholog, paralog, ohnolog, rayfin, spotted gar, lobefin, genome editing, chromonome
1. What fish can teach us about the evolution of vertebrate development
In both a numerical and a phylogenetic sense, asking about the genomic basis of fish Evo-Devo is the better part of asking about the genomic basis of vertebrate Evo-Devo. Not only are more than 50% of all living vertebrate species a ‘fish’ of some sort (Nelson, 2006), but also tetrapods are in fact specialized lobefin fish that happen to cope with life on land (Fig. 1). Casting hyperbole aside, dramatic recent progress in the genomic analysis of bony vertebrate (euteleostome) fish contributes substantially to our understanding of vertebrate Evo-Devo. This progress has been boosted by the revolution in ‘next generation’ sequencing techniques as well as major upgrades to the developmental genetic toolbox enabling the functional analysis of fish gene functions in an evolutionary framework.
Fig. 1. Bony vertebrate fish models for evolutionary developmental genomics (Evo-Devo-Geno).
Phylogenetic relationships are shown to the left, available genomic resources (genome assembly; genetic maps; transcriptomes; BAC libraries) and functional tools (embryology: described and available embryonic developmental stages; transgenic and genome editing methods) to the right. See Table 1 for Evo-Devo-Geno research questions for each model. Genome circles: open circle, ongoing genome sequencing project(s); red circle with ‘C’, chromosome-level reference genome (chromonome) available. Embryology circles: grey circle, spawns occasionally available, but technically highly demanding. VGD1/2: vertebrate genome duplications 1 and 2; TGD: teleost genome duplication. Tree topology is based on Nelson (2006), Near, et al. (2012b), and Betancur, et al. (2013).
Bony vertebrate fish have conquered almost every aquatic habitat on earth (and on earth in the form of tetrapods) and their biodiversity offers unmatched opportunities to study the genomic basis of adaptation, speciation, morphological and physiological change, and behavioral divergence. Among bony vertebrates, teleost fish are the most species-rich group (Nelson, 2006) and they offer unique advantages for the study of gene regulation, morphological evolution, and biodiversity at the functional level, taking advantage of methods developed for the developmental model teleosts zebrafish (Danio rerio) and medaka (Oryzias latipes), (Furutani-Seiki and Wittbrodt, 2004) (Fig. 1).
The following discussion first illustrates recent progress in fish genomics and transcriptomics, highlighting specific challenges that come from applying nucleotide sequencing, assembly and annotation methods to fish genomes, then gives an overview of recent insights obtained from the genomic analysis of selected bony vertebrate fish models and the prospects these models offer for advancing understanding in the future. This overview emphasizes the diversity of fish genomes, which we are just beginning to understand. Finally, this review describes recent methodological advances and new strategies for analyzing the functional evolution of fish genes, mostly based on progress in molecular techniques for zebrafish and medaka, but advances that should be applicable to many other fish systems. Along the way, we will specifically stress the importance of genomic and functional resources that we consider integral to a comprehensive analysis of Evo-Devo genomics (‘Evo-Devo-Geno’) in emerging fish systems. These resources consist of genomic components (chromosome-level genome assemblies which we call ‘chromonomes’, high-resolution genetic maps, reference transcriptomes, and BAC libraries) in combination with the applicability of developmental approaches (access to spawns and developmental staging series, as well as methods for transgenesis and genome editing) (Fig. 1).
2. Sequencing and annotating fish genomes and transcriptomes
2.1 Assembly and annotation of fish gen(om)es: the apples-to-apples principle
Despite the large portion of vertebrate fauna that are rayfin fish, genome-sequencing efforts were until recently focused on a few phylogenetically isolated species. This cohort of five front swimmer species, sequenced mostly with classical Sanger methods, included the major developmental genetic model species zebrafish (Howe et al., 2013b) and medaka (Kasahara et al., 2007), two species of pufferfish that were selected because of their small genome size (Aparicio et al., 2002, Jaillon et al., 2004), and threespine stickleback (Jones et al., 2012), the only species among the ‘first wave’ of fish genomes chosen predominantly to address evolutionary questions. As a result, one or a few of these genomic pioneers have been used as “the fish” in comparative studies with other vertebrate groups, an approach that falls short due to the diversity of rayfin – and especially teleost – genomes.
The recent revolution in sequencing methods has made possible the generation of genomic, transcriptomic, and epigenomic data at a speed and low cost undreamed of a decade ago. As rayfins are now swimming in the ‘genomic tsunami’, it has become almost impossible to keep track of fish genome projects. For recent censuses of ongoing fish genome projects see Bernardi et al. (2012) and Spaink et al. (2013). Many of the more recent fish genome initiatives are motivated by their interest for aquaculture and the fishing industry, such as Atlantic cod (Star et al., 2011), salmonids (rainbow trout: Berthelot et al., 2014; salmon: Davidson et al., 2010), and tongue sole (Chen et al., 2014), or as biomedical models for human disease (Albertson et al., 2009, Schartl, 2013) like platyfish (Schartl et al., 2013). Nevertheless, sequencing a rayfin fish genome just for the sake of tackling evolutionary developmental questions has become achievable for smaller research communities or even single labs, particularly considering the often relatively small size of fish genomes compared to tetrapod genomes.
Sequencing, assembling, and annotating rayfin genomes, and teleost genomes in particular, however, come with a specific set of pitfalls. Some factors such as the diversity of rayfin repetitive elements (Volff et al., 2003, Chalopin et al., 2013) or high rates of polymorphisms often found in fish genomes, even within inbred lab strains (Kasahara, et al., 2007, Brown et al., 2012), may specifically complicate the assembly of rayfin genomes (Bradnam et al., 2013). Another significant factor is that polyploidization, i.e., the duplication of the entire genome, is a common phenomenon, including famous examples such as salmonids, carps, and sturgeons (reviewed in Braasch and Postlethwait, 2012). Polyploidization may be a major obstacle for genome assembly, because the younger the genome duplication event, the more difficult it becomes to distinguish alleles from gene duplicates (paralogs).
A polyploidization event relevant to nearly all teleost genome analyses is the teleost genome duplication (TGD), which occurred in an ancestor of the teleost lineage (Amores et al., 1998, Taylor et al., 2003, Hoegg et al., 2004, Jaillon, et al., 2004, Crow et al., 2006, Hurley et al., 2007, Amores et al., 2011). Even though this polyploidization event is fairly old (~226–350 million years; Braasch and Postlethwait, 2012), it has an important influence on gene annotation, particularly on the determination of orthology, a prerequisite for any evolutionary meaningful analysis, i.e., comparing apples to apples or orthologs to orthologs. The evolutionary significance of the TGD for the diversification of the teleost lineage is a matter of ongoing debate (e.g., Donoghue and Purnell, 2005, Hurley, et al., 2007, Santini et al., 2009, Van de Peer et al., 2009), less controversial, however, is that it had a major impact on the evolution of teleost genome structure and teleost gene functions (Postlethwait et al., 2004, Braasch and Postlethwait, 2012). The rise of teleosts and the TGD have been associated with (i) a phase of major genomic rearrangement (Amores, et al., 2011), (ii) an increase in the molecular evolutionary rate, especially in non-coding regions (Ravi and Venkatesh, 2008, Lee et al., 2011), and (iii) rediploidization of the tetraploid genome after the TGD, i.e., massive loss (nonfunctionalization) of genes and other genomic elements from a duplicated state back to a singleton state. In extant teleost genomes, around 20–25% of protein coding genes are still retained as two TGD paralogs (Braasch and Postlethwait, 2012), with important differences among functional gene classes with regard to the retention and loss of their TGD paralogs (Brunet et al., 2006, Kassahn et al., 2009, Howe, et al., 2013b, Schartl, et al., 2013). Genes of major concern for Evo-Devo research, such as transcription factor and other developmental genes are overrepresented among retained TGD paralogs (Brunet, et al., 2006, Kassahn, et al., 2009). Sequence divergence between TGD paralogs often occurs in an asymmetric manner, with one of the two TGD duplicates evolving faster than the other (Brunet, et al., 2006, Steinke et al., 2006). Furthermore, the processes of non-functionalization, rediploidization, and divergence in sequence and function often occur in a lineage-specific way (Postlethwait, et al., 2004, Semon and Wolfe, 2007, Braasch and Postlethwait, 2012).
Annotation of genomes and transcriptomes of ‘non-model’ fish species usually uses BLAST-based methods against the gene and protein sets of a ‘model’ species as a reference for which orthology assignments have already been generated (e.g., the ‘front swimming five’: zebrafish, medaka, stickleback, two pufferfish). This practice, however, is problematic because of lineage-specific reshuffling of teleost genomes following the TGD, which leads, for example, to the confusion of orthologs and paralogs (apples and oranges), the non-existence of orthologous genes due to reciprocal gene loss, the misassignment of GO terms, etc. This effect is exacerbated because zebrafish, the most detailed annotated teleost genome, as a cyprinid is phylogenetically isolated from most other sequenced teleost genomes (Hiller et al., 2013), including the many rising models belonging to the percomorphs (Fig. 1).
Problems with orthology assignment can often be solved on a gene-by-gene basis using a combination of phylogenetic methods and, if available, data on conserved synteny, i.e., conservation of gene arrangements. Generating phylogenies for every gene family present in entire genomes and transcriptomes [e.g. the EnsemblCompara Gene Trees (Vilella et al., 2009) generated during the Ensembl genome annotation process], remains challenging (Dessimoz et al., 2012). Often, phylogenetic data alone are insufficient to provide clear orthology assignments, especially in the case of fast and asymmetrically diverging teleost paralogs prone to all kinds of tree artifacts. In such cases, conserved synteny information becomes essential for inferring orthologies and the evolutionary origin of teleost genes (e.g., Postlethwait, 2007, Catchen et al., 2009, Catchen et al., 2011b). Although bioinformatic tools for the analysis of conserved syntenies are available [e.g., SyntenyDatabase (Catchen, et al., 2009); Genomicus (Louis et al., 2013)], large-scale analyses of this kind require chromosome-level genome assemblies to perform. Most current second generation genome assemblies, however, remain fragmentary scaffold-level assemblies. To distinguish genome assemblies at the scaffold level from those at the chromosomal level, we introduce here the term ‘chromonome’ for the latter.
2.2 A call for fish ‘chromonomes ‘
Chromonomes are essential for understanding genome evolution in the face of genome duplication with the associated problems of orthology assignment listed above. Fortunately, even for non-model organisms, the advent of second generation sequencing techniques enables the generation of high-density genetic linkage maps containing chromosome-length arrays of sequence-based markers along which the fragmentary scaffolds of a genome assembly can be aligned. An especially good method is restriction site associated DNA (RAD) marker sequencing, single nucleotide polymorphisms (SNPs) occurring in, say, 100 nucleotide long fragments sequenced adjacent to defined restriction sites (Miller et al., 2007, Baird et al., 2008). RAD sequencing (RAD-Seq) methods are now applied in variant forms on numerous types of genetic investigations, including population genomic and genome-wide association studies (GWAS), mapping of quantitative trait loci (QTL), mutant mapping, phylogenomics, and the generation of meiotic maps (see Davey et al., 2011, Hohenlohe et al., 2012 for recent overviews). RAD-Seq can provide thousands of polymorphic markers for the generation of genetic linkage maps (Amores, et al., 2011, Catchen et al., 2011a). The fact that many fish produce hundreds to thousands of embryos from a single cross makes RAD-Seq an ideal method to generate high-density linkage maps for any fish species for which a spawn can be obtained. RAD-seq is so efficient at identifying SNPs that one can usually abandon the former approach of mating widely divergent populations or even different species and making F2 or backcross populations, which was formerly the case, and now just take a single female and a single male from the wild, mate them, and directly obtain a map based on male and on female meiosis from F1 offspring. A meiotic map for the spotted gar was the first RAD-based linkage map of a rayfin (or of any vertebrate) (Amores, et al., 2011), and RAD-based linkage maps have since been generated for numerous other fish species including platyfish (Schartl, et al., 2013, Amores, et al., 2014), salmonids (e.g., Miller et al., 2012, Gagnaire et al., 2013), flatfishes (Palaiokostas et al., 2013a, Chen, et al., 2014), cichlids (Palaiokostas et al., 2013b, Recknagel et al., 2013), and Mexican tetra (O'Quin et al., 2013).
In the case of spotted gar, platyfish, tongue sole, and rainbow trout, RAD marker-based genetic maps were used to anchor draft genome assemblies onto linkage groups, generating chromosome-level genome assemblies, i.e., chromonomes. These chromonomes have enabled unprecedented insight into the evolution of teleost genome structure, sex chromosomes, and gene family histories (Amores et al., 2011, Schartl et al., 2013, Amores et al., 2014, Berthelot et al., 2014, Chen et al., 2014). Because teleosts are, on the one hand, particularly in need of large-scale conserved synteny analyses for orthology detection as result of the TGD and lineage-specific paralog losses, and are, on the other hand, well suited to generate high-density genetic maps due to their fecundity and ease of spawning, we propose that the generation of a RAD-based genetic map as part of a genome assembly project should become a standard in the field of teleost genomics. This ‘call for chromonomes’ will be a recurrent theme throughout the remainder of this article.
2.3 Fish transcriptomics and teleost GO terms
The evolution of RNA-Seq methods spurred a gold rush into sequencing transcriptomes and generating reference transcriptome assemblies for fish models (see Qian et al., 2014). Generating de novo reference transcriptomes is a cost-effective way to obtain insight into the gene content of a non-model species, especially when the aim is to compare multiple species and when a reference genome assembly of a closely related species is unavailable. Nevertheless, with the dropping costs in genome sequencing and assembly, it may be worth considering putting efforts into generating a reference genome assembly first. Generally, reference-based transcriptome assembly strategies have several advantages and tend to be of higher quality compared to de novo assemblies generated without a reference genome (e.g., Martin and Wang, 2011, Vijay et al., 2013).
Numerous paralogs present in teleost genomes as a result of the TGD and the earlier vertebrate genome duplication events (see below) superimposed on lineage-specific gene losses cause particular problems in dealing with the annotation of teleost transcriptomes. The puzzling effect of lineage-specific paralog loss is particularly severe when a transcriptome is the only available source of gene sequence information, because missing content could be due either to insufficient transcript sampling or due to a true absence of a gene from the genome. Reciprocal failure to find paralogs in different lineages will give errors in orthology calling. As described above, taking conserved synteny information into account is often the only way by which orthology of teleost genes can be inferred, and conserved synteny data are by definition unavailable for transcriptomes. Thus, we suggest that a good strategy is to generate a reference genome assembly coupled to a meiotic linkage map of about 10,000 markers or more using about 100 or more map progeny, align the genomic sequencing scaffolds to the genetic map, and to construct a chromonome. This resource will inform the annotation of transcriptomic data, and reciprocally, a transcriptome will inform gene annotations for the genome.
Most large-scale gene expression analyses assign Gene Ontology (GO) terms to differentially expressed genes. GO terms for fish are unfortunately underdeveloped. Zebrafish is the best-curated species thus far, probably with a bias towards early developmental functions. Zebrafish, however, is phylogenetically distant from many teleost models (Fig. 1). Because of lineage-specific subfunctionalization after the TGD (Postlethwait, et al., 2004), one can assume that in some cases, specific subfunctions (potentially representing one or more GO terms) may have been retained in one TGD paralog in zebrafish, while being retained for the other TGD paralog in, for example, a percomorph. We currently don’t understand how often reciprocal subfunctionalization occurs. A conservative strategy to improve putative GO term assignment in a fish species could thus be to use GO terms of both zebrafish paralogs, even if the orthology of the percomorph gene to one of the two zebrafish paralogs is unquestioned.
Besides the analysis protein-coding transcriptomes, efforts using RNA-Seq strategies are also underway in fish to identify and catalogue different types of non- coding RNAs, such as micro RNAs (miRNAs) and long non-coding RNAs (lncRNAs) (e.g., Pauli et al., 2012, Andreassen et al., 2013, Bizuayehu et al., 2013, Kitano et al., 2013). Zebrafish is here the best-documented fish species again, but non-coding RNAs from fish remain to be explored in detail and are currently undercurated in public databases like miRBase (Desvignes et al., 2014).
2.4 Don’t give up on fish BAC libraries
A major leap forward for ‘next-generation’ genomes was the elimination of laborious bacterial cloning steps, making it seem that large insert size libraries such as fosmids and bacterial artificial chromosomes (BACs) had become obsolete. Sound reasons, however, suggest that investing in such types of libraries remains to be worthwhile. First, modified fosmids and BACs can be used as mate-pair libraries for Illumina-based genome assemblies (Williams et al., 2012), which assist with scaffolding efforts. Second, certain genomic regions, including highly repetitive sex chromosomes, are difficult to assemble from genomic DNA, and sequencing BAC clones tiling such regions may be an alternative strategy (Huddleston et al., 2014). Third, current genome assemblies are speckled with assembly gaps, which can be frustrating if such gaps are around your gene of interest, for example when investigating the presence/absence of a conserved non-coding element. Re-sequencing of the region from a BAC clone may help to close these gaps. And finally, BAC transgenesis in zebrafish and medaka has become an important method to study the cis-regulatory circuitry of a genomic region (see section 4). Therefore, we believe that BAC clones should remain an important component of the functional Evo-Devo-Geno toolbox.
3. Emerging fish systems for Evo-Devo-Geno research
In the following sections, we illustrate by the example of seven fish lineages how the new sequencing techniques provide insights into genome evolution of rayfin fish and the development of their diverse phenotypes. For each fish model, we will highlight recent progress as well as common problems that often apply to other fish systems. The narrative presents fish models according to their amenability to functional studies from routine (zebrafish) to impossible (coelacanth). In addition, Figure 1 and Table 1 give a brief overview of the genomic and developmental tools available for these species and other fish models and research questions for which various fish models are exquisitely poised. Detailed discussions of stickleback and cichlids, likely the most studied fish models in Evo-Devo-Geno research, are beyond the scope of the present article, but key articles on these two systems are listed in Table 1.
Table 1. Research questions of bony vertebrate fish model systems.
See Figure 1 for phylogenetic relationships, available genomic resources, and functional tools.
| Lineage | Exemplary Evo-Devo-Geno research questions | Further reading |
|---|---|---|
| Danio complex | pigmentation, dwarfism, novel morphological structures, sex determination, barbels | section 3.3; Howe, et al., 2013b |
| Carps/Goldfish | fin structures, scale patterns, pigmentation, domestication, polyploidy | section 3.3; Henkel, et al., 2012c |
| Goldenline barbels | cave phenotypes, eye loss, albinism, brain structure, sensory organs, polyploidy | section 3.4 |
| Blind cavefish | cave phenotypes, eye loss, albinism, brain structure, craniofacial structures, sensory organs | section 3.4 |
| Catfishes | skeletal structures, pigmentation, mimicry, polyploidy (Corydoras) | Alexandrou et al., 2011, Jiang et al., 2013 |
| Salmonids | hox clusters, polyploidy, skeletal structures, sex determination, evolution of anadromous features | Berthelot et al., 2014; Davidson, et al., 2010 |
| Xiphophorus complex | pigmentation, fin structures, viviparity, sex-linked traits, sex determination, behavior, hybridization | section 3.5; Schartl, et al., 2013 |
| Guppy | pigmentation, fin structures, viviparity, sex-linked traits, sex determination, behavior | section 3.5 |
| Torquoise killifish | aging, pigmentation | Valenzano et al., 2009, Petzold et al., 2013 |
| Oryzias complex | pigmentation, fin structures, sex determination, craniofacial traits | Kasahara, et al., 2007, Takeda and Shimada, 2010, Spivakov et al., 2014 |
| Cichlids | craniofacial adaptations, pigmentation, body shape, sex determination, behavior, adaptive radiation | Kocher, 2004, Salzburger, 2009, Fan et al., 2012, Santos and Salzburger, 2012 |
| Flatfishes | left-right asymmetry, metamorphosis, pigmentation, sex determination/reversal | Cerda and Manchado, 2013, Chen, et al., 2014, Shao et al., 2014 |
| Stickleback | skeletal traits, craniofacial structures, sex determination, pigmentation, behavior | Peichel, 2005, Cresko et al., 2007, Hohenlohe et al., 2010, Jones, et al., 2012 |
| Antarctic icefish | bone development, loss of hemoproteins, cold adaptation, heat shock, adaptive radiation | section 3.6 |
| Pufferfishes | body plan reduction, genome size reduction, sex determination | Aparicio, et al., 2002, Amores et al., 2004, Jaillon, et al., 2004 |
| Sygnathids | male pregnancy, body plan changes, fin structures | Stolting and Wilson, 2007, Mobley et al., 2011 |
| Osteoglosso-morpha | genome evolution, hox clusters, air breathing, electric organs (mormyrids) | section 3.2 |
| Eels | genome evolution, hox clusters, fin loss, body elongation | section 3.2; Henkel, et al., 2012a, Henkel, et al., 2012b |
| Spotted gar | genome evolution, fin structures, air breathing, teleost innovations (outgroup), tetrapod innovations (outgroup) | section 3.1 |
| Paddlefish | tetrapod fin-to-limb transition (outgroup), fin structures, polyploidy, hox clusters, electroreception | section 3.1 |
| Bichirs | fin structures, early embryonic development, hox clusters | section 3.1 |
| Coelacanth | tetrapod fin-to-limb transition and other tetrapod innovations (outgroup), ‘living fossil’ | section 3.7; Amemiya, et al., 2013, Nikaido, et al., 2013 |
| Lungfishes | tetrapod fin-to-limb transition and other tetrapod innovations (outgroup), dentition, genome evolution | section 3.7 |
3.1 Basal rayfin fish and the benefits of the spotted gar model
What makes a teleost a teleost? What distinguishes teleosts from other rayfin fish? What is the secret to teleost evolutionary success? With more than 27,000 species, teleosts populate the majority of the rayfin fish fauna (Nelson, 2006), yet they are genomic outliers among bony vertebrates due to their paleopolyploid origin. To better understand the dynamics of genome evolution and gene function following the TGD, it is essential to define the ancestral, unduplicated state. Until recently, the only available well-studied outgroups diverging before the TGD were the tetrapods, especially mouse and human. The use of mouse as an ersatz unduplicated outgroup for the TGD was clearly just a makeshift solution because teleost and tetrapod lineages have experienced independent genomic and morphological evolution for the last 450 million years. A better outgroup to characterize the pre-TGD rayfin genome would be a ‘model basal rayfin’ species selected from the few extant non-teleost rayfin lineages (Fig. 2A): Polypteriformes (bichirs and reedfish), Acipenseriformes (sturgeons and paddlefish), and the lineage of Holostei consisting of Amiiformes (bowfin) and Lepisosteiformes (gars). Major caveats exist, however, for the use of most of these lineages as genomic and/or functional rayfin basal models. Common to all, and a major challenge for use as laboratory model species, is their long generation times of usually several years.
Fig. 2. Rayfin phylogenetic relationships and the spotted gar model.
A) Phylogeny of major living rayfin lineages. TGD: teleost genome duplication; AGDs: acipenserid (sturgeon) genome duplications; PGD: paddlefish genome duplication. B) Development of the spotted gar (Lepisosteus oculatus) in days post fertilization (dpf). Assignment of developmental stages (st.) follows Long and Ballard (2001). C) Comparison of conserved syntenies between teleosts (stickleback and zebrafish) with respect to gar and human (adapted from Amores et al., 2011). Branch lengths are proportional to conserved syntenies per 100 million years of divergence time. Branch lengths between non-teleosts (gar and Mark Spitz) are much shorter than lengths between either of these two species and teleosts, suggesting massive chromosomal rearrangements in teleosts after their divergence from the gar lineage.
Polypteriforms represent the most basal rayfin lineage and thus, on the one hand, are important models for the early events of rayfin evolution, but because of their very basal position they are, on the other hand, still relatively distant from teleosts and the TGD. In addition, polypteriform genomes tend to be larger than those of human (Takeuchi et al., 2009a). EST sequences for gray bichir (Polypterus senegalus) are available (Takechi et al., 2011), and analyses of bichir hox clusters, sequenced from BAC clones, suggest that they have a composition of hox genes and gene regulatory elements that is intermediate between teleosts and tetrapods (Chiu et al., 2004, Raincrow et al., 2011). Bichirs can be cultured and bred in the lab and are accessible to RNA in situ hybridization and microinjection (Takeuchi, et al., 2009a). Bichirs show many ancestral vertebrate features at the morphological and molecular level, including gene expression during early bichir development that has similarities to lamprey (Takeuchi et al., 2009b). Other morphological features of bichirs, however, such as fin structure, are derived in a polypteriform-specific manner not representative of the ancestral rayfin condition (Metscher and Ahlberg, 2001, Takeuchi, et al., 2009a), a disadvantage for understanding the origin of teleost-specific functions.
Sturgeons and paddlefish are common in aquaculture. Paddlefish is accessible for laboratory studies and has been used as outgroup in comparative studies to investigate, for example, Hox gene evolution during the fin-to-limb transition in lobefins (e.g., Davis et al., 2007, Crow et al., 2012). A problem for genomic analyses, however, is that Acipenseriformes have undergone several rounds of their own lineage-specific polyploidizations (Ludwig et al., 2001, Crow, et al., 2012). Therefore, the dynamics of their genome evolution, while interesting in themselves, make them represent a special case among rayfins and unsuitable as surrogates for the ancestral rayfin genomic condition. The morphological diversity of sturgeons has been underappreciated until recently (Rabosky et al., 2013), and it will be interesting to analyze if there are causal links between the multiple acipenseriform genome amplifications and morphological change in this group.
In the lineage most closely related to teleosts, the holosteans, the only living member of the amiiform lineage is the bowfin (Amia calva), which to the best of our knowledge has not been bred in captivity. Members of the family of Lepisosteiformes (gars) in the holostean lineage, in contrast, are of interest for aquaculture, and they can be consistently bred in captivity (Alfaro et al., 2008), making them available as laboratory animals for developmental studies. Work from our group has identified one of the smaller species, the spotted gar (Lepisosteus oculatus), as the most suitable model gar (Fig. 2B). Embryonic development of gars is relatively well described (Long and Ballard, 2001) and gars are suitable for RNA in situ hybridization, immunohistochemistry, and other developmental methods (e.g., Eames et al., 2012, Braasch et al., 2014). Importantly, spotted gar is well-suited to represent an approximation to the ancestral, pre-teleost and thus pre-TGD, developmental morphology. Ancestral morphological features of gars include their hardy and protective ganoid scales, which share biochemical similarities with tetrapod tooth enamel (Sasagawa et al., 2013), a lung-like gas bladder used for air breathing (Longo et al., 2013), and a dorso-ventrally asymmetrical (heterocercal) tail rather than the pseudosymmetrical (homocercal), evolutionarily innovative tail of teleosts (Metscher and Ahlberg, 2001). Given its advantages, we proposed spotted gar as the best suited non-teleost rayfin fish model species (Amores, et al., 2011).
To investigate whether or not gars are indeed diploid and neither share the genome duplication common to all teleosts nor have their own lineage-specific genome duplication, a high-density genetic map for the spotted gar using RAD-Seq was generated (Amores, et al., 2011). By anchoring transcriptomic sequences derived from two map-cross embryos to the genetic map, it was possible to study patterns of conserved synteny between gar and tetrapods as well as gar and teleosts, even without a reference genome (Amores, et al., 2011). This work has thus provided definitive proof that the TGD happened after the divergence of teleosts from holosteans and is therefore a teleost-specific event. Furthermore, the mapping work showed that genome rearrangements occurred more frequently in teleosts compared to tetrapods and gar (Amores, et al., 2011; Fig. 2C). The genome of the spotted gar has since been sequenced and anchored onto the genetic map and is available in Ensembl [http://www.ensembl.org/Lepisosteus_oculatus].
The spotted gar chromonome is an important resource not only for the study of genome evolution in teleosts, but also for the investigation of the ancestry of bony vertebrate gene functions in general. Following the ‘genomic big bang’ of the earlier rounds of whole genome duplication at the base of the vertebrate lineage, VGD1 and VGD2 (Fig. 1) (reviewed in Canestro, 2012), and subsequent lineage-specific genome reorganization and rediploidizations, the genomes of agnathan, cartilaginous, and bony vertebrates and the gene repertoires they encode have significantly diverged. Gene duplicates that originate in genome duplications are called “ohnologs” (Wolfe, 2001). Following VGD1 and VGD2, many genes were present in four ohnologous copies organized in four ohnologous regions (ohnologons, like the four Hox clusters and many surrounding genes) that are still apparent in extant vertebrate genomes (up to eight in teleosts following the TGD) (reviewed in Canestro, 2012). Gene families with four remaining VGD1/2 ohnologs are, however, rather rare and VGD1/2 ohnologs have frequently gone missing in a lineage-specific manner. Therefore, it often becomes difficult to interpret orthology of genes across vertebrate lineages and to detect instances of ‘hidden paralogy’ among vertebrate genes (Kuraku, 2013). Lineage-specific ‘ohnologs gone missing’ may not only confound interpretations of functional gene evolution across vertebrate lineages (Braasch and Postlethwait, 2012), but can also lead to situations in which orthologous genes simply no longer exist to study the ancestry of gene functions. For example, numerous genes have been retained in teleosts and the lobefin coelacanth but were lost during the water-to-land transition in tetrapods (Amemiya et al., 2013). Likewise, some VGD ohnologs may have been lost in teleosts but retained in tetrapods (e.g., Pou5a1/Oct4, Frankenberg and Renfree, 2013; Prrx2, Braasch, et al., 2014) so that studying their ancestral function in fish cannot be modeled with zebrafish or medaka, requiring a model that has retained many ancestral ohnologs and is amenable to functional studies. While coelacanth is not available for functional studies and lungfishes are only rarely available (see section 3.7), spotted gar can fill this gap (Braasch, et al., 2014).
3.2 Genomic diversity of basally diverging teleost lineages
Shortly after the TGD event, the teleost lineage split into three major branches (Figs. 1, 2A): clupeocephalans (encompassing the majority of teleost diversity, including the radiations of the Ostariophysi and Percomorpha), elopomorpha (eels, tarpons, tenpounders, bonefish), and osteoglossomorpha (bony tongues, goldeye, etc.). Unfortunately, available teleost genome assemblies are almost exclusively from clupeocephalan teleosts. For elopomorphs, draft genome assemblies for two species of eel, European eel (Anguilla anguilla) and Japanese eel (A. japonica), are available (Henkel et al., 2012a, Henkel et al., 2012b). Transcriptome data from the European eel (Coppe et al., 2010, Ager-Wick et al., 2013) and a genetic map for the Japanese eel based on microsatellites and AFLP data (Nomura et al., 2011) are available as well. Developmental studies of eels are hindered by their enigmatic, catadromous life styles that start with spawning in the open ocean, impeding artificial breeding efforts – a major drawback considering the economic importance of eels (Henkel, et al., 2012a, Henkel, et al., 2012b).
Osteoglossomorphs remain a large blind spot for teleost genomics. The superorder Osteoglossomorpha consists of two orders, Osteoglossiformes (bonytongues) and Hiodontiformes (mooneyes) (Nelson, 2006). No genome assembly is currently available for any osteoglossomorph. A low-density genetic map and a first transcriptome have been recently published for the Asian arowana (Scleropages formosus) (Shen et al., 2013a). For Hiodontiformes, a few genes including hox cluster genes of the goldeye (Hiodon alosoides) have been analyzed through PCR screens (Hurley, et al., 2007, Chambers et al., 2009).
Despite the current paucity of elopomorph and osteoglossomorph genomic data, we can deduce a few early trends for the basally diverging teleosts:
Clupeocephalans, elopomorphs, and osteoglossomorphs differ markedly in their retention of ancestral VGD1/2 ohnologs. While many hox paralogy groups are missing from clupeocephalans, eels have kept almost all of them intact (Henkel, et al., 2012a, Henkel, et al., 2012b). Another example, the prrx2 gene (a VGD1 ohnolog of prrx1) is absent from clupeocephalans, but present in elopomorphs (eels) and osteoglossomorphs (butterflyfish), showing that prrx2 is not a pan-teleost ohnolog gone missing (Braasch, et al., 2014) as was initially assumed.
The three major teleost lineages differ in their retention of TGD paralogs. Even clupeocephalans show significant diversity between zebrafish and percomorphs (medaka, stickleback, pufferfishes, etc.) in terms of TGD paralog retention and loss, although the overall level of TGD paralog retention is similar among extant clupeocephalan genomes (e.g., see Braasch et al., 2009, Sato et al., 2009, Schartl, et al., 2013). It can be expected that the diversity in TGD paralogs is even more pronounced when comparing the three early diverging teleost lineages. Indeed, while most clupeocephalan lineages have reduced their hox complement to seven hox clusters, eight hox clusters are present in eels (Henkel, et al., 2012a, Henkel, et al., 2012b) and the osteoglossomorph goldeye (Chambers, et al., 2009). Vice versa, some genes appear to be single copy in elopomorphs and/or osteoglossormorphs, while having been retained in two copies after the TGD in clupeocephalans, e.g., sox9 (Shen, et al., 2013a) or pomc (see below).
According to nomenclature conventions, ‘a’ and ‘b’ suffixes are added to teleost gene names for TGD paralogs. While calling orthologs of the a- vs. b-paralogs is often straightforward among clupeocephalans (e.g., comparing zebrafish to medaka), phylogenetic methods frequently fail in assigning the a/b-orthology of clupeocephalans to elopomorphs and osteoglossomorphs. An example for this phenomenon, the pro-opiomelanocortin (pomc) gene, is shown in Fig. 3. Clupeocephalans have two pomc TGD paralogs, called pomca and pomcb (de Souza et al., 2005). In contrast, eels have a single pomc gene (Ager-Wick, et al., 2013). From pilot genome sequencing, we identified only one pomc gene for goldeye (I. Braasch, P. Batzel, A. Amores, J. H. Postlethwait, unpublished data). This result suggests that eels (elopomorphs) and maybe goldeye (osteoglossomorphs) lost one of the two TGD pomc paralogs at some point during their evolution following the TGD. Phylogenetic reconstructions, however, fail to assign orthology of the single remaining pomc gene of these two basal teleost lineages to one of the two pomc TGD paralogs of clupeocephalans. Likewise, we cannot tell whether the eel pomc gene is the ortholog or a TGD paralog of goldeye pomc (Fig. 3A). Even analysis of small-scale conserved syntenies doesn’t help in this case because, although the direct genomic environment of eel and goldeye pomc genes has been sequenced, these scaffolds are too short to infer whether these gene regions are more closely related to the cluepocephalan a- or b-paralogons by means of conserved syntenies (Fig. 3B). Other examples for unclear orthologies of elopomorph/osteoglossormoph and clupeocephalan genes include many hox TGD paralogs in eel and goldeye (Chambers, et al., 2009, Crow et al., 2009, Henkel, et al., 2012a), sox9 in the osteoglossomorph arowana (Shen, et al., 2013a), and prrx1 paralogs in elopomorph eels and the osteoglossomorph butterflyfish (Braasch, et al., 2014). Additional measures such as splicing patterns and/or long-range conserved syntenies need to be taken into account for orthology detection of elopomorph/osteoglossomorph vs. clueocephalan genes (Braasch, et al., 2014). Even these measures may not necessarily provide certainty because TGD paralogs and paralogons of clupeocephalans and the other two lineages could also evolve convergently following the TGD.
Fig. 3. Evolution of the rayfin proopiomelanocortin (pomc) gene.
A) Maximum likelihood phylogeny (JTT+I+G+F model) of full-length Pomc proteins in rayfins rooted with bichir Pomc. Node values indicate support from 100 bootstrap replications, showing bootstrap values >50% only. Clupeocephalans teleosts retained both TGD pomc paralogs; their assignment to a- and b-paralogs follows de Souza et al. (2005) (monophyly of clupeocephalan Pomca is not supported in the current tree). In eels (elopomorphs) only a single pomc gene has been found and for goldeye (osteoglossomorphs), we have found so far only one pomc gene (but we cannot exclude the presence of the second TGD paralogs yet). The relationship of these pomc singletons among each other as well as to clupeocephalan pomca and pomcb remain unresolved (indicated by ‘?’). Two pomc genes are present in paddlefish, most likely as result of the paddlefish-specific polyploidization; two pomca paralogs are present in platyfish and other percomorphs as result of an additional gene duplication (Harris et al., 2014). B) Conserved syntenies of rayfin pomc gene regions. Neighboring genes of the single pomc gene in goldeye and eel are conserved with spotted gar and both zebrafish pomc paralogons, which makes it impossible to make an a/b-orthology call, which would require more large-scale chromosomal assemblies from the basal teleosts. Tetraodon pomc paralogons are more rearranged than those from zebrafish. Boxed numbers indicate intervening regions that do not contribute to conserved synteny.
These early trends have important implications for teleost genomics. The example of pomc genes illustrates the need for long-range, chromosome length synteny information for elopomorphs and osteoglossomorphs in the form of chromonomes. Only with chromonomes will we be able to fully appreciate the diversity of teleost genomes and infer the early events of teleost genome evolution shortly after the TGD. Chromonomes will enable tests of the ‘genomic big bang hypothesis’ that the three teleost ‘genome galaxies’ (elopomorphs, osteglossomorphs, and clupeocephalans) have been drifting apart in genome evolutionary space since the TGD, potentially leaving few genomic commonalities behind. Such analysis is important because it will illuminate general principles that also characterized the aftermath of the more ancient VGD1/2 genome duplications at the base of the vertebrate radiation that shape the genomes of all vertebrates (including our own), which due to their even deeper ancestry and paucity of surviving, anciently diverging lineages are even more difficult to study.
The unfolding diversity of teleost genomes has additional relevance for phylogenomic analyses of basal teleost relationships. Based on morphological and molecular data, three hypothesis have been discussed (Li et al., 2008): (1) Osteoglossomorphs are the sister lineage of all other teleosts: (osteoglosso (elopo+ clupeo)); (2) Elopomorphs are the sister lineage of all other teleosts: (elopo (osteoglosso + clupeo)); (3) Clupeocephalans are the sister lineage of a clade combining osteoglosso- and elopomorphs: (clupeo (osteoglosso + elopo)). While the basal position of osteoglossormorphs (hypothesis 1) is supported by phylogenies of mitochondrial genomes (Inoue et al., 2003, Setiamarga et al., 2009), several recent multi-locus phylogenomic studies based on 10–20 nuclear marker genes support the basal divergence of elopomorphs (hypothesis 2) (Near et al., 2012b, Betancur et al., 2013, Broughton et al., 2013). Nuclear markers used in these studies were designed from comparative genomic screens for singleton genes of zebrafish and pufferfish and other percomorphs (Li et al., 2007). In using these markers, the studies assume that the markers are singletons in all teleosts, that the loss of their TGD paralogs occurred shortly after the TGD and predate the divergence of living teleost lineages, and that these markers are thus strictly orthologous. Reciprocal loss of TGD paralogs, i.e., loss of the a-paralog in one lineage and loss of the b-paralog in another lineage leaving paralogous and not orthologous singletons behind, appears to be rather rare within clupeocephalans (<10% between zebrafish and percomorphs; Semon and Wolfe, 2007, Kassahn, et al., 2009). The unfolding diversity of basally diverging teleost lineages described above, however, suggest that singletons in one lineage (clupeocephalans in this case) may not necessarily be singletons in the other two major lineages (elopomorphs, osteoglossormophs). Furthermore, even if a marker gene is present in only one copy in all three teleost lineages, these singletons may not necessarily be true orthologs and could be – at an unknown frequency – instances of ‘hidden paralogy’ (Kuraku, 2013). Hidden paralogy becomes more likely the shorter the delay between the TGD event and the divergence of the three teleost lineages, and current studies suggest that these incidents happened within a short time window of a few tens of millions of years (Near, et al., 2012b, Broughton, et al., 2013). Future studies therefore will need to address whether or not the assumptions made in the use of singleton nuclear markers developed for clupeocephalans are met in the other two teleost lineages and assess how potential violations of these assumptions may have influenced recent conclusions about the relationships of basally diverging teleost lineages. Such assessment, again, will require large-scale conserved synteny data and thus chromonomes from elopomorphs and osteoglossomorphs to improve orthology calls among the three teleost lineages.
3.3 The Danio flotilla and the cyprinid armada
Cyprinidae is the most diverse family in all of Vertebrata with more than 2,400 species and 200 genera. This group includes some of the best-known fish in the world such as carps, goldfish, and zebrafish, the most widely used teleost developmental model organism. The wealth of cyprinid biodiversity has spawned projects such as the Cypriniformes Tree of Life [http://bio.slu.edu/mayden/cypriniformes/overview.html].
A number of cyprinids, including many of the seventeen currently recognized Danio species, display characteristics that make the group of zebrafish and related species, the danionins, interesting as an Evo-Devo model species complex. Phenotypic differences among danionins include both discrete changes (e.g. loss or fusion of craniofacial elements, reduction and loss of barbels and breeding tubercles), and more complex changes (e.g., reduction of adult size, mineralization, and diverse pigmentation patterns) (Fang, 2003, Parichy, 2007). Danionins and other related cyprinids further allow the investigation of repeated evolution of miniaturization in this group (Ruber et al., 2007). Two zebrafish-related species of note are Paedocypris progenetica, which holds the record as the smallest known fish (Kottelat et al., 2006), and Danionella dracula, which grows evolutionarily novel “fangs” on its jaws as its name suggests (Britz et al., 2009). The most obvious and well-studied differences between Danio species and related genera, however, are found in their diversity of adult coloration and striking pigment patterns, some of which resemble zebrafish pigmentation mutants (Parichy, 2007). Clever complementation tests in interspecies hybrids derived from these zebrafish mutants and other Danio species have helped to elucidate the signaling pathways involved in the differentiation and evolution of pigment patterns across the genus (Parichy and Johnson, 2001, Quigley et al., 2005). Follow up studies in zebrafish have subsequently explored cellular interactions of different types of pigment cells leading to stripe formation (Inaba et al., 2012, Hamada et al., 2014) and the genetic mechanisms causing pigment cell differentiation (Patterson and Parichy, 2013).
These studies illustrate three major strengths of the danionins for studies in evolution and development. First, these species are able to hybridize, bringing together genomes separated by millions of years in a nested series of evolutionary time scales and allowing access to the wealth of naturally occurring genetic diversity that has arisen as the species diversified. Interspecies hybrids will allow one to investigate the genetic and developmental basis of this variation as the genomes of two different species interact sharing the same cell nucleus within single individuals. Second, the unmatched infrastructure for genetic analysis and imaging of zebrafish development as well as ease of laboratory culture allows for investigation of the genomic and cellular mechanisms underlying the evolution of development of the danionins. Third, the zebrafish reference chromonome (Howe, et al., 2013b), along with accessibility of zebrafish mutants for a multitude of genes (Kettleborough et al., 2013), transgenic lines, and the curation of zebrafish research data by ZFIN, the Zebrafish Information Network (Howe et al., 2013a) allows for follow-up experiments to functionally disentangle the genetic and cellular basis of evolutionary change in the Danio species complex (see also section 4). The danionins are therefore ripe to become the ‘drosophilids’ of vertebrate Evo-Devo-Geno research.
Beyond the Danio system, the species-richness of the cyprinid family provides a wealth of natural genetic and phenotypic variation for study in an Evo-Devo framework. To this end, the Phenoscape project aims to extend the genetic and developmental knowledge gained from zebrafish to cyprinids and Ostariophysi (Fig.1) to develop explicit Evo-Devo hypotheses (Mabee et al., 2012).
Cyprinidae also embraces the subfamily Cyprininae, which includes common carp (Cyprinus carpio) and goldfish (Carassius auratus), the oldest domestication experiments of the fish world. In addition to the TGD, this group underwent an additional whole genome duplication event within the last few million years (Ohno et al., 1967, Wang et al., 2012). This recent carp genome duplication allows for the study of post-duplication genome evolution on a different timescale than the basal teleost models mentioned previously. Striking phenotypes in goldfish and carps are the result of centuries of artificial selection and represent another potential avenue of interest for Evo-Devo research, similar to domestication in dogs or pigeons. Historical relationships among various goldfish strains have only recently been determined and the genomic basis of color patterns and morphological monstrosities like bubble eyes, pearlscale, or lionhead remains to be uncovered (Komiyama et al., 2009, Wang et al., 2013).
The importance of carps in aquaculture has resulted in numerous studies of gene expression and physiology, genetic maps, and a recent first genome assembly for common carp (Henkel et al., 2012c). Carp is the second available cyprinid genome draft after zebrafish, now allowing, for example, the inference that the high repeat content in zebrafish is not common to cyprinids in general, because the genome of carp – despite of its tetraploid origin – is similar in size to zebrafish thanks to a much lower repeat content (Henkel, et al., 2012c). Syntenies appear to be almost perfectly conserved between carp and zebrafish (Henkel, et al., 2012c), as well as between the zebrafish and a RAD-based genetic map of gudgeon (Gnathopogon), another cyprinid (Kakioka et al., 2013). A second cyprinid chromonome, however, will be necessary to evaluate in detail whether the increase in interchromosomal rearrangements found for zebrafish compared to percomorphs (Braasch and Postlethwait, 2012, Howe, et al., 2013b) is a pervasive feature of cyprinids in general or a specific phenomenon of the zebrafish lineage.
3.4 Cavefishes: convergent evolution in the dark
Cavefish populations of the Mexican tetra (Astyanax mexicanus) are paradigmatic examples for evolutionary change in an extreme environment: constant darkness. Astyanax cavefish (also known as ‘blind cavefish’) are characterized by a combination of ‘regressive’ and ‘constructive’ changes including the reduction of eyes and pigmentation (albinism), changes to their sensory organs, central nervous system, and craniofacial structures, modified feeding behavior, loss of sleep and schooling behaviors and many more. The blind cavefish is thus an ideal fish system to study the genomic basis of vertebrate Evo-Devo at the microevolutionary scale (reviewed in Jeffery, 2008).
Fertile hybrids between cave and ancestral surface populations allow extensive QTL mapping analyses to identify genetic loci that differentiate cave from surface forms. These studies revealed a complex genomic architecture of cavefish evolution, with multiple loci underlying regressive and constructive traits (e.g., Protas et al., 2006, Protas et al., 2007, Yoshizawa et al., 2012, Kowalko et al., 2013, Gross et al., 2014). Population genomic analysis based on RAD-seq data further suggests that standing genetic variation significantly contributed to the evolutionary changes observed in cave populations (Bradic et al., 2013). This hypothesis gets further support by the observation that experimental inhibition of stress response as well as water conditions found in caves can unmask cryptic eye variation in surface fish (Rohner et al., 2013).
The question of whether regressive phenotypes in cavefish, like loss of eyes and pigmentation, are based on neutral processes and drift or whether they are adaptive and under positive selection, possibly as an indirect effect of selection for pleiotropic genes, is a hotly debated matter (see e.g., Jeffery, 2009, Wilkens, 2010, Retaux and Casane, 2013). For example, it has been suggested that cavefish eye loss could be an indirect effect of selection for increased sensory neuron numbers enabling adaptive feeding behavior in the dark using vibration response (Yoshizawa, et al., 2012). Inhibition of pigment synthesis on the other hand has been proposed to increase tyrosine levels in albinistic cavefish. Because tyrosine is a precursor of both melanin pigments and neurotransmitters, cavefish albinism may thus be adaptive by making additional tyrosine available for neurotransmitter synthesis, thereby contributing to complex cavefish behaviors (Bilandzija et al., 2013).
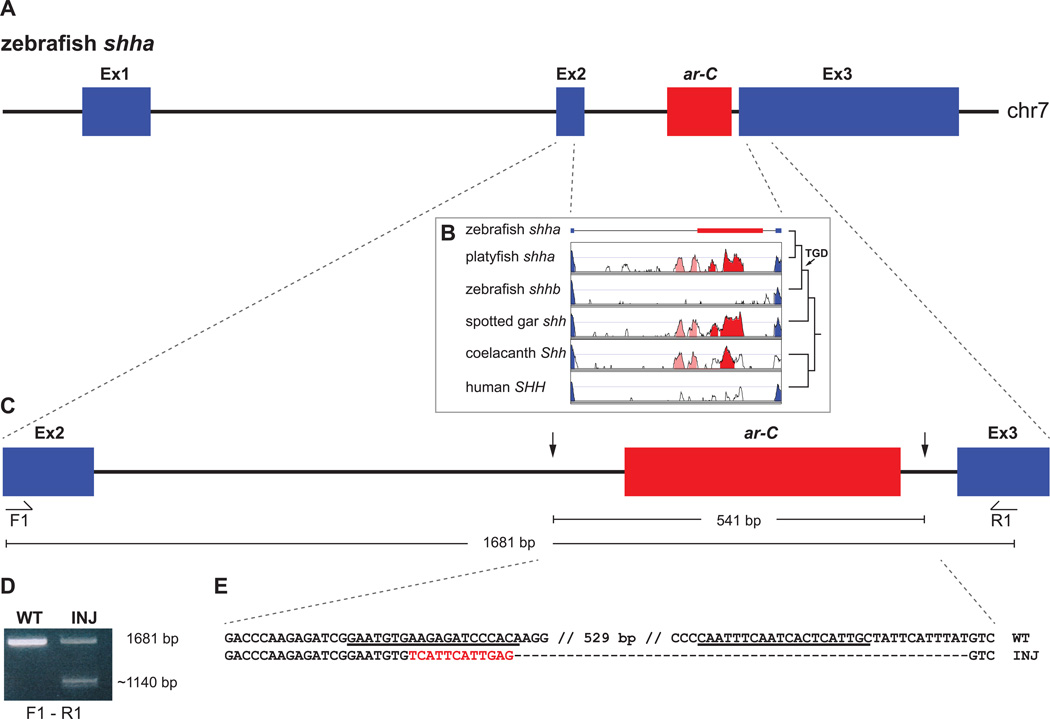
Astyanax cavefish is well suited for functional studies due to the availability of embryos from different populations and its relatively close phylogenetic relationship with zebrafish (Fig. 1), which has helped in the cloning of candidate genes for functional tests and adopting much of the zebrafish methodological toolkit to Astyanax research (Jeffery, 2008). As described in section 4, functional studies on gene candidates underlying cavefish QTLs have led to the identification of the genomic basis of cavefish pigmentation phenotypes. While albinism is caused by mutations in oca2 (Protas, et al., 2006), mutations in the mc1r gene are responsible for the ‘brown’ phenotype (Gross et al., 2009). Candidate gene expression studies furthermore identified the hedgehog signaling pathway as a key player for eye regression in cavefish. Expanded expression of sonic hedgehog along the anterior midline was observed in cavefish compared to surface fish, inducing cavefish eye regression through lens apoptosis, but also leading to advantageous increases in jaw size and taste bud numbers (Yamamoto et al., 2004, Yamamoto et al., 2009) – further indication for the potential linkage of regressive and constructive traits. Surprisingly, shh or other components of the hedgehog pathway do not locate to the QTL regions involved in eye regression suggesting that these loci may contain yet unidentified upstream regulators of hedgehog signaling (Protas, et al., 2007, Jeffery, 2009, Yoshizawa, et al., 2012).
Transcriptomic analyses of Astyanax cave vs. surface fish further highlight cavefish-specific mutations and expression differences in the visual system and metabolic pathways (Gross et al., 2013, Hinaux et al., 2013). The Astyanax genome assembly (not yet available as a chromonome) became available recently [http://www.ensembl.org/Astyanax_mexicanus] and represents an important step for teleost genomics in general because it is the first non-cyprinid genome available from the superorder Ostariophysi, which accounts for almost one-third of known fish species (Fig. 1) (Nelson, 2006). The impressive recent advances in detailed genetic analyses, genomic resource availability, and its functional accessibility establish the Astyanax system as a fully equipped ‘model system’ to study vertebrate Evo-Devo.
In addition to Astyanax about 150 other fish species live in cave environments (Retaux and Casane, 2013), offering avenues for asking about the convergent evolution of cave-dwelling fish traits. Current evidence points towards different molecular evolutionary trajectories that can lead to convergence in cave phenotypes in fish. For example, important differences in the molecular evolution of the circardian system have been found between Astyanax cavefish and the Somalian cavefish (Phreatichthys andruzzii), a cyprinid (Beale et al., 2013). Furthermore, many cave species are found among goldenline barbels, cyprinids of the genus Sinocyclocheilus from China, that are further characterized by conspicuous horns and humps of mostly unknown function (Zhao et al., 2011) (Fig. 1). Goldenline barbels display important differences in the cellular mechanism for eye regression compared to Astyanax. While shh-induced apopotosis of the lens causes eye regression in Astyanax (Yamamoto, et al., 2004, Jeffery, 2009), such lens degeneration is not observed in the Sinocyclocheilus cavefish, which instead show regressive changes to the retina of their small, internalized eyes (Meng et al., 2013a). Transcriptomic comparisons of cave and surface eyes in Sinocyclocheilus further support the hypothesis of retinal degeneration as photoreceptor-specific genes such as opsins and their common upstream regulator Crx are downregulated in Sinocyclocheilus cavefish (Meng, et al., 2013a, Meng et al., 2013b). The molecular genetic mechanism leading to lower crx expression in cavefish eyes remains to be elucidated. Comparing gene expression of cave vs. surface forms of Sinocyclocheilus and Astyanax revealed few expressional changes shared between the two lineages. This finding suggests that different molecular mechanisms lead to convergent evolution of eye regression in Sinocyclocheilus and Astyanax (Meng, et al., 2013a).
3.5 Life in Technicolor: poeciliid livebearers
The family of Poeciliidae, also known as ‘livebearers’, has been studied for more than a century for their amazing morphological adaptations and phenotypic variation (see Evans et al., 2011 for an overview). The family contains some of the most common aquarium species: guppy, mollies, platyfish, and swordtails. The first sequenced poeciliid genome is that of the platyfish (Xiphophorus maculatus), a chromonome build by the inclusion of a RAD-based high-density genetic map (Schartl, et al., 2013, Amores et al., 2014). The platyfish chromonome highlights the stability of percomorph karyotypes because only few interchromosomal rearrangements were found between platyfish, medaka, stickleback, and pufferfish (Schartl, et al., 2013). Transcriptomes of other poeciliid species, such as additional species of the genus Xiphophorus (Cui et al., 2013, Schartl, et al., 2013, Shen et al., 2013b) or the guppy (Poecilia reticulata) (Fraser et al., 2011), have become available as well and multiple poeciliid genome assemblies are in progress.
As implied by their common name, all but one of the more than 260 species of livebearers are characterized by their viviparous reproduction mode; species across the group show a spectrum of continuous complexity in maternal provisioning, including the evolution of placentas (Pollux et al., 2009). A first avenue towards understanding the evolution of viviparity in poeciliids comes from sequence comparison of viviparity candidate genes from the platyfish genome and Xiphophorus transcriptomes with those of other teleost species leading to the identification of several viviparity candidate genes under positive selection in poeciliids (Schartl, et al., 2013).
Poeciliid sexual dimorphism is seen in many morphological structures, most prominently in the male gonopodium, a transformed anal fin used as intromittent organ for internal fertilization, as well as in the ventral sword-like caudal fin extension in swordtails of the genus Xiphophorus. Considering swordtails as a model for the evolution by sexual selection actually dates back to Darwin’s Descent of Man, and Selection in Relation to Sex (1871) (Darwin, 1871). Recent large-scale phylogenomic analysis based on RAD markers (Jones et al., 2013) and transcriptomes from multiple Xiphophorus species (Cui, et al., 2013) support the hypothesis that the development of a sword in Xiphophorus males is the ancestral state in the genus and that the sword has been lost secondarily most likely more than once in the genus.
Another striking phenotype of poeciliids – and of platyfish and guppy in particular – that has fascinated scientists for a century is their diversity in color patterns, which are often sexually dimorphic and to a large degree based on sex- linked loci (Winge, 1927, Basolo, 2006). While the genetic basis of these sex-linked color loci remains largely unresolved, the platyfish chromonome excluded the vast majority of classic pigmentation genes as candidates for these loci because they were found not to locate to the platyfish sex chromosomes (Schartl, et al., 2013). In contrast, the receptor tyrosines kinase genes kit and csf1r, well known from zebrafish pigment mutants and their involvement in pigment pattern divergence among Danio species (Parichy, 2007), were identified as the genomic bases of two autosomal color loci in guppy (Kottler et al., 2013).
Sex chromosomes are generally genomic hotspots for accumulating loci contributing to poeciliid diversity. While poeciliid sex determination genes remain to be identified, the genetic basis of some sex-linked loci has been elucidated. The classic example is a tumor locus that causes the development of melanoma in interspecific Xiphophorus hybrids. These skin cancers are caused by Xmrk, an oncogenic, sex-linked, local gene duplicate of the receptor tyrosinase gene egfrb (Meierjohann and Schartl, 2006); Xmrk has been proposed to represent a Xiphophorus ‘speciation gene’ (see Schartl, 2008 for a critical discussion). The sex-linked ‘puberty locus’ determining the onset of male maturity and mating behavior in Xiphophorus appears to be caused by copy number variation of multiple melanocortin 4 receptor (mc4r) gene duplicates that were identified by sequencing BAC clones from the highly repetitive platyfish sex chromosomes (Lampert et al., 2010, Volff et al., 2013).
Poeciliids are popular models to study teleost behaviors. Coupled with complex behavior, genes implicated in cognition are retained at a particularly high rate, around 45% compared to only around 15% for liver genes, as became apparent from comparisons of the platyfish gene content to that of other teleosts and mammals (Schartl, et al., 2013). Likewise, the zebrafish genome project showed a high TGD duplicate retention for neuronal genes (Howe, et al., 2013b). These findings suggest that highly complex cognitive abilities and behaviors found in teleosts may be based on functional divergence of gene duplicates retained from the TGD.
Functional studies in poeciliids are hampered by their livebearing lifestyle that inhibits genetic manipulations and transgenesis. The recent progress in poeciliid genomics, in combination with functional approaches such as testing poeciliid gene functions in medaka (section 4) thereby serves as a paradigm for other fish systems that are challenged by internal development such as sygnathids (pipefishes, sea horse, sea dragons) or mouthbrooding cichlids.
3.6 Adaptation to the cold: Antarctic rockcods and icefish
Antarctic fish constitute an Evo-Devo model of many extremes because of their polar habitats and the technical challenges that accompany this extreme and remote environment. In the freezing cold waters of the Southern Ocean (−1.9°C), Antarctic fish diversity is largely dominated by a single suborder, the notothenioids, which form a remarkable example of adaptive radiation in an extreme cold environment (Eastman, 2005). As Antarctic waters cooled, which reaching their current frigid conditions about 10–14 Myr ago (Kennett, 1977), many taxa became locally extinct, liberating various ecological niches (Eastman, 1993, Eastman and McCune, 2000) that notothenioids were able to conquer based on dramatic adaptive changes towards cold tolerance and the use of new habitats.
The most striking example of adaptive evolution to frigid environment is the gain of antifreeze glycoproteins (AFGPs) that evolved from pancreatic trypsinogen and allowed notothenioids to survive, adapt and diversify in freezing waters in which other species would freeze to death (Cheng and Chen, 1999, Kock, 2005b, Kock, 2005a, Cheng and Detrich, 2007). Several other molecular and cellular changes accompanied the adaptation to a constant icy-cold environment including an apparent loss of inducible heat shock response in steady cold ambient temperature (Hofmann et al., 2000, Buckley et al., 2004, Place and Hofmann, 2005, Huth and Place, 2013).
With the development of transcriptomic techniques, recent studies have looked at the effects of acclimation temperature and heat-shocks on gene expression in several species of cold-adapted notothenioids and suggested (i) numerous gene duplications (Chen et al., 2008, Coppe et al., 2013) consistent with large genome size (Detrich et al., 2010), (ii) the involvement of the ubiquitin-conjugated protein cytosolic protein-degradation pathway of misfolded or damaged protein as putative factor of cold adaptation (Shin et al., 2012, Bilyk and Cheng, 2013, Huth and Place, 2013), and (iii) increased mitochondrial function (Coppe, et al., 2013).
Being able to survive in icy waters, notothenioids diversified from a benthic scavenger common ancestor to species inhabiting all parts of the water column spanning from benthic to pelagic habitats (Eastman, 1993). Although all notothenioids lack a swim bladder, the diversification along the vertical water column and evolution to become ambush feeders was accompanied by buoyancy modifications (Kock, 2005a) (Fig. 4). Among changes, the most striking ones are the delay and reduction of skeletal ossification through the alteration of collagen gene expression (Albertson, et al., 2009), the loss or reduction of mineralized elements such as scales and dermal bones (Kock, 2005a, Albertson, et al., 2009), and the evolution of extensive unique lipid deposits (Friedrich and Hagen, 1994, Hagen et al., 2000). These evolutionary alterations reflect paedomorphic changes by the retention of larval characteristics in the adult.
Fig. 4. Diversity of ecological and physiological features in notothenioids.
Phylogenetic relationships and the buoyancy data are from Near et al. (2012a) and information on adult lifestyles is reviewed in Rutschmann et al. (2011). A dissected heart is shown for each species and presence (+) and absence (−) of hemoglobin and myoglobin (Hb/Mb) is indicated as well.
One synapomorphy of the family of icefishes, the Channichthyidae (Kock, 2005b, Kock, 2005a), has baffled biologists from the first report by J. T. Ruud in 1954 of “Vertebrates without erythrocytes and blood pigment” (Ruud, 1954). This puzzling condition, lethal in any other vertebrate species, was shown to be a disadvantageous but non-lethal (‘disaptive’) phenotype (Baum and Larson, 1991, Montgomery and Clements, 2000) at low temperature because oxygen concentration is inversely correlated with water temperature. Furthermore, globin-independent loss of cardiac myoglobin has occurred in six of sixteen species of Channichthyidae (Sidell et al., 1997, Cheng and Detrich, 2007) (Fig. 4). The loss of ability to express globin and myoglobin genes was compensated by molecular and morphological adaptations such as diffusion of oxygen through the scale-less skin of icefishes and modest decrease in metabolic oxygen demand (Hemmingsen, 1991), an increased heart size and pumping volume in relation to body size (Hemmingsen, 1991),a more developed vascular system (Egginton et al., 2002), and quantitative changes in mitochondrial density and morphology (Sidell, et al., 1997, O'Brien et al., 2000).
Interestingly, these oxygen-carrier-lacking vertebrates could help understand molecular mechanisms of erythropoiesis and anemia (Albertson, et al., 2009). BAC libraries from erythrocytes of the red-blooded Yellow belly rock-cod Notothenia coriiceps and from leukocytes of the clear-blooded Blackfin Icefish Chaenocephalus aceratus have been generated (Detrich and Amemiya, 2010, Detrich, et al., 2010). Representational Difference Analysis of cDNA libraries from pronephric kidney of the same two species identified a novel gene, bloodthirsty, that was further shown to play a role in differentiation of the committed red cell progenitor by functional analysis in zebrafish (Yergeau et al., 2005).
The Antarctic notothenioids represent a compelling opportunity to study morphological, physiological and molecular evolutionary development adaptations of a marine species-flock to freezing cold environment. Breeding and raising notothenoids, however, is technically difficult and is currently only possible at Antarctic research stations. Therefore, a combination of genomic and transcriptomic characterization of notothenoids in combination with functional tests within other fish species - as exemplified by the bloodthirsty case (Yergeau et al., 2005; see also section 4) - appears to be the method of choice to unravel the extraordinary morphological and physiological adaptations of these element-braving fish.
3.7 Coelacanth and lungfish: how lobefin fish left the water
Probably the most iconic fish genome sequenced so far is that of legendary ‘Old Fourlegs’, the coelacanth (Latimeria) (Amemiya, et al., 2013, Nikaido et al., 2013). Coelacanths were thought to be extinct until their rediscovery in 1938 and since then have been considered ‘living fossils’ (Smith, 1939). Coelacanths and lungfishes (Dipnoi) represent one of the only two living lobefin (sarcopterygian) lineages that are fish but not tetrapods (Fig. 1). Therefore, coelacanths and lungfishes are essential to understand the conquest of land and the morphological changes that occurred during the water-to-land transition, especially the fin-to-limb transition. The question of whether coelacanths or lungfishes represent the sister lineage tetrapods has been long debated. Lungfish have the largest genomes of the animal kingdom – estimates range from 40 to 130 Gb – presumably due to massive amplification of repetitive elements early in the lungfish lineage that are now ‘fossilized’, escaping current repeat detection methods (Metcalfe et al., 2012). These gigantic genomes make it impractical to sequence a lungfish genome and transcriptomic characterization of the lungfish gene content is currently the only practical option. Phylogenomic analysis of the coelacanth genome content in combination with gene sequences obtained from lungfish transcriptomes finally settled the tetrapod relation: lungfish are the sister group of tetrapods (Amemiya, al., 2013).
The coelacanth genome sequence enabled testing the status of coelacanths as ‘living fossil’: The molecular evolutionary rate of protein coding genes is indeed significantly slower in coelacanth than that for most other vertebrate lineages (Amemiya, et al., 2013, Nikaido, et al., 2013). As inferred from the scaffold-level coelacanth genome assembly, karyotype evolution also seems to be relatively slow in coelacanth (Amemiya, et al., 2013), but this conclusion would need to be confirmed with a coelacanth chromonome. Transposable elements, in contrast, are abundant and have been recently active in coelacanth (Amemiya, et al., 2013, Chalopin, et al., 2013, Forconi et al., 2013, Nikaido, et al., 2013), which contrasts with the coelacanth’s morphological stasis and slow rate of protein evolution.
The analysis of coelacanth genomes revealed their transitionary position among sarcopterygian vertebrates, showing genomic features of both ‘fish’ and tetrapods (Amemiya, et al., 2013, Nikaido, et al., 2013; coelacanth special issue of this journal). The coelacanth retained ancestral vertebrate genes that were lost in tetrapods and are thus present only in fish. Interestingly, genes lost in tetrapods but present coelacanth and teleost fish are involved in those structures that most dramatically changed during the water-to-land transition, such as paired appendages, which have different functions in water and on land, ears and eyes, which sense the environment different in water and in air, and kidneys, which have different stresses in dry air and water, suggesting that gene losses were an important component for the remodeling of the emerging tetrapod genome (Amemiya, et al., 2013). The most prominent examples for genes ‘left in the water’ are the actinodin genes (Amemiya, et al., 2013, Nikaido, et al., 2013) that encode structural components of fin folds in fish (Zhang et al., 2010).
In other aspects, however, the coelacanth genome shows similarities to tetrapods, for example in the constitution of its immune system (Boudinot et al., 2014, Saha et al., 2014) and olfactory gene repertoires (Nikaido, et al., 2013, Picone et al., 2013). Furthermore, several cis-regulatory elements involved in gene regulation during limb and lung development are unexpectedly conserved between tetrapods and coelacanth showing that important components of limb and lung gene regulatory networks predate the rise of tetrapods and evolved before the fin-to-limb transition and the evolution of terrestrial lungs (Amemiya, et al., 2013, Nikaido, et al., 2013).
Though a lot of additional analyses lie ahead, current studies of coelacanth genomes highlight several tetrapod genomic innovations that may have been involved in the adaptation to life on land. These include, for example, the emergence of putative gene regulatory elements around important developmental genes, including known limb development genes, and positive selection on components of the urea cycle (Amemiya, et al., 2013).
As critically endangered, livebearing, marine deep-water inhabitants, coelacanths are probably the worst imaginable fish to pursue functional investigations. Lungfishes have been informative, e.g., to study the evolution of walking mechanics (King et al., 2011) and of the pelvic apparatus (Boisvert et al., 2013). Furthermore, although mRNA in situ hybridization gene expression studies, e.g., during the development of lungfish digits homologs (Johanson et al., 2007) and lungfish dentition (Smith et al., 2009), have been performed, lungfish genome sizes and the challenges to obtain lungfish embryos make molecular studies very difficult. Functional studies of lobefin fish genes will thus have to rely on tests in other vertebrate species, as have been done for coelacanth conserved non-coding elements (CNEs) and coelacanth BACs in transgenic mice (Amemiya, et al., 2013).
4. Recent advances to study piscine gene functions in the postgenomic era
Regardless of the specific questions addressed using a particular fish model, any Evo-Devo study at some point will inevitably aim to turn insights from genetic, genomic, and transcriptomic analyses into functional hypotheses that ideally would be tested within the species under investigation and during the development of the trait. This requires the analysis of developmental processes in vivo and the study of spatio-temporal gene expression patterns with RNA in situ hybridization and/or immunohistochemistry. As described by examples above, these experiments may or may not be feasible depending on the specifics of the fish species, the regular availability of embryos (Fig. 1), feasibility of microinjections, working antibodies, etc. Functional analysis of gene evolution will require even more investment into developing species-specific tools. Chemical treatments with inhibitors and agonists of major developmental signaling pathways have been applied to a wide range of fish species because they can simply be added to the culture water. For example, mosquitofish (Ogino et al., 2004), bichir (Cuervo et al., 2012), and lungfish (Smith et al., 2009) have been treated in experiments with cyclopamine, an inhibitor of the hedgehog signaling pathway, to understand cell-cell signaling.
Functional assays, however, do not necessarily require doing functional experiments in the endogenous species itself. A seminal example is a study on cavefish albinism, showing that transfecting an Oca2 protein deficient, and therefore unpigmented murine pigment cell line with alleles of the oca2 gene from normally pigmented surface Mexican tetra restores melanin synthesis, but transfection with alleles from albino cavefish populations fails to rescue pigmentation. This analysis provided functional evidence that sequence variation in the oca2 gene, identified by QTL mapping as a major candidate, is indeed causing cavefish albinism (Protas, et al., 2006).
Functional tests will always be challenging in non-model fish species, even with sequenced genomes and other types of genomic resources. Thus, analogous to the study of the genetic basis of human evolution in ‘humanized mice’ (e.g., Prabhakar et al., 2008, Kamberov et al., 2013), we envision that in the future, zebrafish and medaka will be used as ‘functional hubs’ for Ostariophysi and Percomorpha (Fig. 1), respectively, to study the evolution of gene functions from a broad variety of fish. An example for such an interspecies approach is a platyfish-medaka tumor model, enabling functional tests that would not have been possible in platyfish itself because of its livebearing reproduction mode: the Xmrk tumor locus gene from platyfish was integrated into the medaka genome to generate a stable transgenic medaka melanoma model (Schartl et al., 2010), which then enabled the in piscine study of tumorigenic events downstream of the constitutively active receptor tyrosine kinase encoded by platyfish’s oncogene (Schartl et al., 2012, Liedtke et al., 2013). An instance for the use of zebrafish as a functional hub is the functional analysis of a candidate for the ‘brown’ locus in cavefish: Rescuing pigmentation in zebrafish embryos, in which Mc1r function was knocked-down with morpholino antisense oligo nucleotides, was successful with co-injection of surface but not cave mc1r RNAs, supporting that mc1r variation is responsible for the ‘brown’ phenotype (Gross, et al., 2009).
Functional tools for the analysis of gene actions have dramatically evolved for zebrafish and medaka within the past few years. Particularly, the toolbox for the analysis of cis-regulatory elements, which are at the heart of Evo-Devo research (Wray, 2007, Carroll, 2008), is rapidly transitioning. Putative cis-regulatory elements are often identified as conserved non-coding elements (CNEs) (e.g., Woolfe et al., 2005, Lee, et al., 2011, Lowe et al., 2011, Hiller, et al., 2013). The particularly high evolutionary turn-over rate of non-coding sequences in teleosts (Ravi and Venkatesh, 2008, Lee, et al., 2011), and the still insufficient genomic coverage of the rayfin lineage (Hiller, et al., 2013) makes the detection of CNEs in fish challenging. One solution to this problem comes with the application of ChIP-Seq, i.e., chromatin immunoprecipitation combined with high-throughput sequencing. Probing for DNA-protein interactions, ChIP-Seq can identify genome-wide transcription factor binding events or epigenetic marks indicative of enhancers and other types of gene regulatory elements. ChIP-Seq has assayed the activity of regulatory elements during zebrafish development (Aday et al., 2011, Bogdanovic et al., 2013). Because ChIP-Seq works independent of (although correlated with) sequence conservation, it enables to identification of newly evolved, lineage-specific cis-elements that would not turn up as conserved elements by comparing distant taxa.
The function of putative cis-elements are classically tested in transient or stable reporter construct assays in which the sequence of interest is cloned in front of a minimal promoter to drive a fluorescent protein in a spatio-temporal manner. These reporter constructs are then co-injected into 1–2 cell stage embryos of zebrafish or medaka with Tol2 transposon RNA or meganuclease protein, respectively, which mediate integration of the construct into the host genome (e.g., Fisher et al., 2006, Grabher and Wittbrodt, 2007, Bessa et al., 2009). Similar transgenic methods should in principle also work in other fish species for which injectable, fertilized eggs in sufficient numbers are available, as shown for stickleback (Chan et al., 2010), some cichlids (Nakamura et al., 2008, Juntti et al., 2013), killifish (Valenzano et al., 2011), and others.
To study interactions of long-range enhancers and their target genes as well as to test more than just short, isolated, individual non-coding fragments, cross-species transgenesis in zebrafish or medaka with BAC clones from the species under investigation, usually covering ~150kb around the locus of interest, may be a method of choice. BAC transgenesis is now well established in zebrafish (Suster et al., 2011) and medaka (Nakamura, et al., 2008). An extreme example for the application of cross-vertebrate BAC transgenesis is a recent study in which pufferfish BACs from the hoxA and hoxD clusters were transfected into mouse to investigate the regulatory basis of tetrapod digit origins (Woltering et al., 2014).
A common problem of these transgenic methods, however, is that the randomness of integration into the host genome makes results quite variable. The genomic environment of the integration site and its epigenetic state may influence expression of the reporter construct, a phenomenon known as ‘positional effects’ (Roberts et al., 2014). To overcome this problem, a new solution is on the horizon that will likely become a standard method in the fish field soon: PhiC31 integrase systems use landing sites integrated at well characterized spots of the host genome with low positional effects. These genomic attP landing sites recombine with the reporter construct’s attB to accomplish site-directed transgenesis of the construct at a predefined genomic position in a repeatable manner (Hu et al., 2011). Various PhiC31 lines have been developed for medaka (Kirchmaier et al., 2013) and zebrafish (Mosimann et al., 2013, Roberts, et al., 2014), and the method should in principle even enable the integration of large, BAC-sized transgenes in fish (Kirchmaier, et al., 2013).
The breadth of mutants available for zebrafish and medaka further elevates their use as functional hubs for fish Evo-Devo. New methods for directed mutagenesis and genomic manipulation established for zebrafish, medaka, and a few other species (Fig. 1) that should be applicable to more fish systems come through the use of zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and most recently, the regularly interspaced short palindromic repeats (CRISPR)/Cas9 nuclease system (Ansai et al., 2013, Blackburn et al., 2013). The highly efficient CRISPR/Cas9 system uses a bacterial derived endonuclease directed by a short guide RNA to cleave DNA upstream of a protospacer adjacent motif. By inducing site-specific double-strand DNA breaks, the system can create indel mutations, targeted insertions, or substitutions, or, when multiple sites are targeted, larger scale deletions or inversions (Chang et al., 2013, Jao et al., 2013, Xiao et al., 2013). The flexibility and efficiency of these systems allow for the easy creation of loss-of-function mutations or the study of the loss of non-coding regulatory elements and the removal of enhancers. An example of such enhancer deletion comes from our own work (Fig. 5), the deletion of the well-studied sonic hedgehog a (shha) gene enhancer ar-C in zebrafish (Gehrig et al., 2009). Figure 5 shows that the CRISPR/Cas9 genome editing system ushers in a new phase in the analysis of gene regulatory networks in fish to study in vivo the effects of knocking out regulatory elements (and potentially also knocking in or switching alleles) on gene expression of the affected gene(s) and downstream effects in the network beyond just characterizing putative roles of cis-elements with reporter assay approaches.
Fig. 5. Deletion of the shha ar-C enhancer using the CRISPR/Cas9 system in zebrafish.
A) The zebrafish ar-C enhancer is located between exon (Ex) 2 and 3 of the sonic hedgehog a (shha) gene on chromosome (chr) 7. B) VISTA plot showing sequence conservation of zebrafish shha intron 2 (reference sequence) to other species. TGD: teleost genome duplication. The location of the ar-C region was defined by Gehrig et al. (2009). C) The CRISPR/Cas9 system was employed to remove the enhancer by targeting two sites flanking the region. The locations of the targeted regions are indicated with arrows; primers (F1 and R1) were used for PCR detection of induced deletions. D) Deletions of the expected size in injected embryos (INJ) were confirmed by PCR, and were not present in uninjected embryos (WT). E) A representative example of a sequenced allele in an injected embryo verified the removal of the ar-C enhancer with the sequence of the shha coding sequence remaining intact. The target region is underlined and the protospacer adjacent motif site is indicated in bold. Deletions are indicated with dashes while inserted nucleotides are indicated in red. The experiment shows that the targeted deletion of cis-regulatory elements using the CRISPR/Cas9 system is a useful new tool for the Evo-Devo-Geno toolbox in fish.
5. Conclusions
With the current revolution in sequencing techniques, the genomic net covering fish evolution is becoming more and more fine-meshed, but is still leaky in view of the tremendous amount of biodiversity of rayfins and of teleost fish in particular. Orthology detection in teleost fish is more error-prone than in other vertebrate lineages because it needs to account for divergent evolution of teleost genomes and gene functions, which are aftermaths of the TGD, the genomic big bang at the base of the teleost lineage. Enabling conserved synteny analyses, chromosome-level genome assemblies will be essential to help inform teleost gene orthologies. So far, genomic and transcriptomic methods have liberated multiple fish models from genomic ignorance, transforming them into a new model army for a comprehensive analysis of the genomic basis of evolution and development.
In combination with the general advantages of numerous fish systems to study the genetic underpinnings of vertebrate development, such as ease of husbandry, external development, high fertility, natural or chemically inducible embryo transparency, technical advances in the molecular and developmental analysis of fish gene functions suggest an auspicious future in which many fish model systems can enter a phase of functional investigations following the genetic, genomic and/or transcriptomic characterization of their evolutionary diversity. For species in which direct functional testing in vivo is not practical, testing their gene functions in zebrafish or medaka may become the method of choice.
Finally, as outreach and societal impact become increasingly important criteria for research funding, we believe that – because everybody knows (and likes!) fish from fishing, fishkeeping, diving, and the dinner table – the ‘Fish World’ is also an excellent motivating educational device to teach systematics, evolution, developmental biology, morphology, physiology, and ecology to the general public.
Acknowledgments
We would like to thank Marc Robinson-Rechavi and Gunter Wagner for giving us the opportunity to contribute to this special issue. We are grateful to the many peer fish researchers that have contributed to our vision of fish genomics. In particular, we would like to acknowledge fruitful discussions on the many aspects of fish Evo-Devo-Geno with Tom Titus, Angel Amores, Yilin Yan, Catherine Wilson, and the entire Postlethwait lab, Jennifer Anderson, Cristian Cañestro, Fangwei Meng, Jamie Nichols, Chuck Kimmel, Brian Eames, Bill Cresko, Julian Catchen, Clay Small, Susie Bassham, Julia Ganz, the zebrafish community at the University of Oregon, Manfred Schartl, Wes Warren, Ron Walter, Chris Amemiya, Allyse Ferrara, Markus Hecker, Walter Salzburger, Astrid Böhne, Emilia Santos, Neil Shubin, Tetsuya Nakamura, Yann Guiguen, Julien Bobe, Bill Detrich, and our collaborators from the platyfish, coelacanth, and spotted gar genome consortia. We apologize for not being able to cover more fish Evo-Devo-Geno work in the current article.
Supporting grant information: This work was supported by grant I/84 815 of the “Initiative Evolutionary Biology” from the Volkswagen Foundation, Germany (to IB) and NIH grants R01OD011116 (alias R01 RR020833), R01AG031922, R24OD011199, and R24RR032670 (to JHP).
References
- Aday AW, Zhu LJ, Lakshmanan A, Wang J, Lawson ND. Identification of cis regulatory features in the embryonic zebrafish genome through large-scale profiling of H3K4me1 and H3K4me3 binding sites. Dev Biol. 2011;357:450–462. doi: 10.1016/j.ydbio.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ager-Wick E, Dirks RP, Burgerhout E, Nourizadeh-Lillabadi R, de Wijze DL, Spaink HP, van den Thillart GE, Tsukamoto K, Dufour S, Weltzien FA, Henkel CV. The pituitary gland of the European eel reveals massive expression of genes involved in the melanocortin system. PLoS One. 2013;8:e77396. doi: 10.1371/journal.pone.0077396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson RC, Cresko W, Detrich HW, Postlethwait JH. Evolutionary mutant models for human disease. Trends in Genetics. 2009;25:74–81. doi: 10.1016/j.tig.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrou MA, Oliveira C, Maillard M, McGill RA, Newton J, Creer S, Taylor MI. Competition and phylogeny determine community structure in Mullerian co-mimics. Nature. 2011;469:84–88. doi: 10.1038/nature09660. [DOI] [PubMed] [Google Scholar]
- Alfaro RM, Gonzalez CA, Ferrara AM. Gar biology and culture: status and prospects. Aquaculture Research. 2008;39:748–763. [Google Scholar]
- Amemiya CT, Alfoldi J, Lee AP, Fan S, Philippe H, Maccallum I, Braasch I, Manousaki T, Schneider I, Rohner N, Organ C, Chalopin D, Smith JJ, Robinson M, Dorrington RA, Gerdol M, Aken B, Biscotti MA, Barucca M, Baurain D, Berlin AM, Blatch GL, Buonocore F, Burmester T, Campbell MS, Canapa A, Cannon JP, Christoffels A, De Moro G, Edkins AL, Fan L, Fausto AM, Feiner N, Forconi M, Gamieldien J, Gnerre S, Gnirke A, Goldstone JV, Haerty W, Hahn ME, Hesse U, Hoffmann S, Johnson J, Karchner SI, Kuraku S, Lara M, Levin JZ, Litman GW, Mauceli E, Miyake T, Mueller MG, Nelson DR, Nitsche A, Olmo E, Ota T, Pallavicini A, Panji S, Picone B, Ponting CP, Prohaska SJ, Przybylski D, Saha NR, Ravi V, Ribeiro FJ, Sauka-Spengler T, Scapigliati G, Searle SM, Sharpe T, Simakov O, Stadler PF, Stegeman JJ, Sumiyama K, Tabbaa D, Tafer H, Turner-Maier J, van Heusden P, White S, Williams L, Yandell M, Brinkmann H, Volff JN, Tabin CJ, Shubin N, Schartl M, Jaffe DB, Postlethwait JH, Venkatesh B, Di Palma F, Lander ES, Meyer A, Lindblad-Toh K. The African coelacanth genome provides insights into tetrapod evolution. Nature. 2013;496:311–316. doi: 10.1038/nature12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A, Catchen J, Ferrara A, Fontenot Q, Postlethwait JH. Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication. Genetics. 2011;188:799–808. doi: 10.1534/genetics.111.127324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A, Catchen J, Nanda I, Warren W, Walter R, Schartl M, Postlethwait JH. A RAD-tag Genetic Map for the Platyfish (Xiphophorus maculatus) Reveals Mechanisms of Karyotype Evolution Among Teleost Fish. Genetics. 2014 doi: 10.1534/genetics.114.164293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Amores A, Suzuki T, Yan YL, Pomeroy J, Singer A, Amemiya C, Postlethwait JH. Developmental roles of pufferfish Hox clusters and genome evolution in ray-fin fish. Genome Res. 2004;14:1–10. doi: 10.1101/gr.1717804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen R, Worren MM, Hoyheim B. Discovery and characterization of miRNA genes in Atlantic salmon (Salmo salar) by use of a deep sequencing approach. BMC Genomics. 2013;14:482. doi: 10.1186/1471-2164-14-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansai S, Sakuma T, Yamamoto T, Ariga H, Uemura N, Takahashi R, Kinoshita M. Efficient targeted mutagenesis in medaka using custom-designed transcription activator-like effector nucleases. Genetics. 2013;193:739–749. doi: 10.1534/genetics.112.147645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal P, Christoffels A, Rash Hoon S, Smit A, Gelpke MD, Roach J, Oh T, Ho IY, Wong M, Detter C, Verhoef F, Predki P, Tay A, Lucas S, Richardson P, Smith SF, Clark MS, Edwards YJ, Doggett N, Zharkikh A, Tavtigian SV, Pruss D, Barnstead M, Evans C, Baden H, Powell Glusman G, Rowen L, Hood L, Tan YH, Elgar G, Hawkins T, Venkatesh B, Rokhsar D, Brenner S. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One. 2008;3:e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basolo AL. Genetic Linkage and Color Polymorphism in the Southern Platyfish (Xiphophorus maculatus): A Model System for Studies of Color Pattern Evolution. Zebrafish. 2006;3:65–83. doi: 10.1089/zeb.2006.3.65. [DOI] [PubMed] [Google Scholar]
- Baum DA, Larson A. Adaptation reviewed: a phylogenetic methodology for studying character macroevolution. Systematic Zoology. 1991;40:1–18. [Google Scholar]
- Beale A, Guibal C, Tamai TK, Klotz L, Cowen S, Peyric E, Reynoso VH, Yamamoto Y, Whitmore D. Circadian rhythms in Mexican blind cavefish Astyanax mexicanus in the lab and in the field. Nat Commun. 2013;4:2769. doi: 10.1038/ncomms3769. [DOI] [PubMed] [Google Scholar]
- Bernardi G, Wiley EO, Mansour H, Miller MR, Orti G, Haussler D, O'Brien SJ, Ryder OA, Venkatesh B. The fishes of Genome 10K. Marine Genomics. 2012;7:3–6. doi: 10.1016/j.margen.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Berthelot C, Brunet F, Chalopin D, Juanchich A, Bernard M, Noel B, Bento P, Da Silva C, Labadie K, Alberti A, Aury JM, Louis A, Dehais P, Bardou P, Montfort J, Klopp C, Cabau C, Gaspin C, Thorgaard GH, Boussaha M, Quillet E, Guyomard R, Galiana D, Bobe J, Volff JN, Genet C, Wincker P, Jaillon O, Roest Crollius H, Guiguen Y. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun. 2014;5:3657. doi: 10.1038/ncomms4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessa J, Tena JJ, de la Calle-Mustienes E, Fernandez-Minan A, Naranjo S, Fernandez A, Montoliu L, Akalin A, Lenhard B, Casares F, Gomez-Skarmeta JL. Zebrafish enhancer detection (ZED) vector: a new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev Dyn. 2009;238:2409–2417. doi: 10.1002/dvdy.22051. [DOI] [PubMed] [Google Scholar]
- Betancur RR, Broughton RE, Wiley EO, Carpenter K, Lopez JA, Li C, Holcroft NI, Arcila D, Sanciangco M, Cureton Ii JC, Zhang F, Buser T, Campbell MA, Ballesteros JA, Roa-Varon A, Willis S, Borden WC, Rowley T, Reneau PC, Hough DJ, Lu G, Grande T, Arratia G, Orti G. The tree of life and a new classification of bony fishes. PLoS Curr. 2013;5 doi: 10.1371/currents.tol.53ba26640df0ccaee75bb165c8c26288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilandzija H, Ma L, Parkhurst A, Jeffery WR. A potential benefit of albinism in Astyanax cavefish: downregulation of the oca2 gene increases tyrosine and catecholamine levels as an alternative to melanin synthesis. PLoS One. 2013;8:e80823. doi: 10.1371/journal.pone.0080823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilyk KT, Cheng CHC. Model of gene expression in extreme cold - reference transcriptome for the high-Antarctic cryopelagic notothenioid fish Pagothenia borchgrevinki. Bmc Genomics. 2013;14 doi: 10.1186/1471-2164-14-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizuayehu TT, Fernandes JM, Johansen SD, Babiak I. Characterization of novel precursor miRNAs using next generation sequencing and prediction of miRNA targets in Atlantic halibut. PLoS One. 2013;8:e61378. doi: 10.1371/journal.pone.0061378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn PR, Campbell JM, Clark KJ, Ekker SC. The CRISPR system--keeping zebrafish gene targeting fresh. Zebrafish. 2013;10:116–118. doi: 10.1089/zeb.2013.9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanovic O, Fernandez-Minan A, Tena JJ, de la Calle-Mustienes E, Gomez-Skarmeta JL. The developmental epigenomics toolbox: ChIP-seq and MethylCap-seq profiling of early zebrafish embryos. Methods. 2013;62:207–215. doi: 10.1016/j.ymeth.2013.04.011. [DOI] [PubMed] [Google Scholar]
- Boisvert CA, Joss JM, Ahlberg PE. Comparative pelvic development of the axolotl (Ambystoma mexicanum) and the Australian lungfish (Neoceratodus forsteri): conservation and innovation across the fish-tetrapod transition. Evodevo. 2013;4:3. doi: 10.1186/2041-9139-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudinot P, Zou J, Ota T, Buonocore F, Scapigliati G, Canapa A, Cannon J, Litman G, Hansen JD. A tetrapod-like repertoire of innate immune receptors and effectors for coelacanths. J Exp Zool B Mol Dev Evol. 2014 doi: 10.1002/jez.b.22559. [DOI] [PubMed] [Google Scholar]
- Braasch I, Brunet F, Volff JN, Schartl M. Pigmentation pathway evolution after whole-genome duplication in fish. Genome Biol Evol. 2009;1:479–493. doi: 10.1093/gbe/evp050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I, Guiguen Y, Loker R, Letaw JH, Ferrara A, Bobe J, Postlethwait JH. Connectivity of vertebrate genomes: Paired-related homeobox (Prrx) genes in spotted gar, basal teleosts, and tetrapods. Comp Biochem Physiol C Toxicol Pharmacol. 2014 doi: 10.1016/j.cbpc.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I, Postlethwait JH. Polyploidy in Fish and the Teleost Genome Duplication. In: Soltis PS, Soltis DE, editors. Polyploidy and Genome Evolution. Berlin Heidelberg: Springer; 2012. pp. 341–383. [Google Scholar]
- Bradic M, Teotonio H, Borowsky RL. The population genomics of repeated evolution in the blind cavefish Astyanax mexicanus. Mol Biol Evol. 2013;30:2383–2400. doi: 10.1093/molbev/mst136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradnam KR, Fass JN, Alexandrov A, Baranay P, Bechner M, Birol I, Boisvert S, Chapman JA, Chapuis G, Chikhi R, Chitsaz H, Chou WC, Corbeil J, Del Fabbro C, Docking TR, Durbin R, Earl D, Emrich S, Fedotov P, Fonseca NA, Ganapathy G, Gibbs RA, Gnerre S, Godzaridis E, Goldstein S, Haimel M, Hall G, Haussler D, Hiatt JB, Ho IY, Howard J, Hunt M, Jackman SD, Jaffe DB, Jarvis ED, Jiang H, Kazakov S, Kersey PJ, Kitzman JO, Knight JR, Koren S, Lam TW, Lavenier D, Laviolette F, Li Y, Li Z, Liu B, Liu Y, Luo R, Maccallum I, Macmanes MD, Maillet N, Melnikov S, Naquin D, Ning Z, Otto TD, Paten B, Paulo OS, Phillippy AM, Pina-Martins F, Place M, Przybylski D, Qin X, Qu C, Ribeiro FJ, Richards S, Rokhsar DS, Ruby JG, Scalabrin S, Schatz MC, Schwartz DC, Sergushichev A, Sharpe T, Shaw TI, Shendure J, Shi Y, Simpson JT, Song H, Tsarev F, Vezzi F, Vicedomini R, Vieira BM, Wang J, Worley KC, Yin S, Yiu SM, Yuan J, Zhang G, Zhang H, Zhou S, Korf IF. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. Gigascience. 2013;2:10. doi: 10.1186/2047-217X-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britz R, Conway KW, Ruber L. Spectacular morphological novelty in a miniature cyprinid fish, Danionella dracula n. sp. Proceedings of the Royal Society B-Biological Sciences. 2009;276:2179–2186. doi: 10.1098/rspb.2009.0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton RE, Betancur RR, Li C, Arratia G, Orti G. Multi-locus phylogenetic analysis reveals the pattern and tempo of bony fish evolution. PLoS Curr. 2013;5 doi: 10.1371/currents.tol.2ca8041495ffafd0c92756e75247483e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KH, Dobrinski KP, Lee AS, Gokcumen O, Mills RE, Shi X, Chong WW, Chen JY, Yoo P, David S, Peterson SM, Raj T, Choy KW, Stranger BE, Williamson RE, Zon LI, Freeman JL, Lee C. Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. Proc Natl Acad Sci U S A. 2012;109:529–534. doi: 10.1073/pnas.1112163109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet FG, Roest Crollius H, Paris M, Aury JM, Gibert P, Jaillon O, Laudet V, Robinson-Rechavi M. Gene loss and evolutionary rates following whole-genome duplication in teleost fishes. Mol Biol Evol. 2006;23:1808–1816. doi: 10.1093/molbev/msl049. [DOI] [PubMed] [Google Scholar]
- Buckley BA, Place SP, Hofmann GE. Regulation of heat shock genes in isolated hepatocytes from an Antarctic fish, Trematomus bernacchii. J Exp Biol. 2004;207:3649–3656. doi: 10.1242/jeb.01219. [DOI] [PubMed] [Google Scholar]
- Canestro C. Two rounds of whole-genome duplication: evidence and impact on the evolution of vertebrate innovations. In: Soltis PS, Soltis DE, editors. Polyploidy and Genome Evolution. Berlin Heidelberg: Springer; 2012. pp. 309–339. [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH. Stacks: building and genotyping Loci de novo from short-read sequences. G3 (Bethesda) 2011a;1:171–182. doi: 10.1534/g3.111.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen JM, Braasch I, Postlethwait JH. Conserved synteny and the zebrafish genome. Methods Cell Biol. 2011b;104:259–285. doi: 10.1016/B978-0-12-374814-0.00015-X. [DOI] [PubMed] [Google Scholar]
- Catchen JM, Conery JS, Postlethwait JH. Automated identification of conserved synteny after whole-genome duplication. Genome Res. 2009;19:1497–1505. doi: 10.1101/gr.090480.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda J, Manchado M. Advances in genomics for flatfish aquaculture. Genes and Nutrition. 2013;8:5–17. doi: 10.1007/s12263-012-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalopin D, Fan S, Simakov O, Meyer A, Schartl M, Volff JN. Evolutionary active transposable elements in the genome of the coelacanth. J Exp Zool B Mol Dev Evol. 2013 doi: 10.1002/jez.b.22521. [DOI] [PubMed] [Google Scholar]
- Chambers KE, McDaniell R, Raincrow JD, Deshmukh M, Stadler PF, Chiu CH. Hox cluster duplication in the basal teleost Hiodon alosoides (Osteoglossomorpha) Theory Biosci. 2009;128:109–120. doi: 10.1007/s12064-009-0056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal G, Jr, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, Myers RM, Petrov D, Jonsson B, Schluter D, Bell MA, Kingsley DM. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhang G, Shao C, Huang Q, Liu G, Zhang P, Song W, An N, Chalopin D, Volff JN, Hong Y, Li Q, Sha Z, Zhou H, Xie M, Yu Q, Liu Y, Xiang H, Wang N, Wu K, Yang C, Zhou Q, Liao X, Yang L, Hu Q, Zhang J, Meng L, Jin L, Tian Y, Lian J, Yang J, Miao G, Liu S, Liang Z, Yan F, Li Y, Sun B, Zhang H, Zhang J, Zhu Y, Du M, Zhao Y, Schartl M, Tang Q, Wang J. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet. 2014 doi: 10.1038/ng.2890. [DOI] [PubMed] [Google Scholar]
- Chen Z, Cheng CH, Zhang J, Cao L, Chen L, Zhou L, Jin Y, Ye H, Deng C, Dai Z, Xu Q, Hu P, Sun S, Shen Y, Chen L. Transcriptomic and genomic evolution under constant cold in Antarctic notothenioid fish. Proc Natl Acad Sci U S A. 2008;105:12944–12949. doi: 10.1073/pnas.0802432105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CH, Chen L. Evolution of an antifreeze glycoprotein. Nature. 1999;401:443–444. doi: 10.1038/46721. [DOI] [PubMed] [Google Scholar]
- Cheng CHC, Detrich HW. Molecular ecophysiology of Antarctic notothenioid fishes. Philosophical Transactions of the Royal Society B-Biological Sciences. 2007;362:2215–2232. doi: 10.1098/rstb.2006.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CH, Dewar K, Wagner GP, Takahashi K, Ruddle F, Ledje C, Bartsch P, Scemama JL, Stellwag E, Fried C, Prohaska SJ, Stadler PF, Amemiya CT. Bichir HoxA cluster sequence reveals surprising trends in ray-finned fish genomic evolution. Genome Res. 2004;14:11–17. doi: 10.1101/gr.1712904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppe A, Agostini C, Marino IA, Zane L, Bargelloni L, Bortoluzzi S, Patarnello T. Genome evolution in the cold: Antarctic icefish muscle transcriptome reveals selective duplications increasing mitochondrial function. Genome Biol Evol. 2013;5:45–60. doi: 10.1093/gbe/evs108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppe A, Pujolar JM, Maes GE, Larsen PF, Hansen MM, Bernatchez L, Zane L, Bortoluzzi S. Sequencing, de novo annotation and analysis of the first Anguilla anguilla transcriptome: EeelBase opens new perspectives for the study of the critically endangered European eel. BMC Genomics. 2010;11:635. doi: 10.1186/1471-2164-11-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresko WA, McGuigan KL, Phillips PC, Postlethwait JH. Studies of threespine stickleback developmental evolution: progress and promise. Genetica. 2007;129:105–126. doi: 10.1007/s10709-006-0036-z. [DOI] [PubMed] [Google Scholar]
- Crow KD, Amemiya CT, Roth J, Wagner GP. Hypermutability of HoxA13A and functional divergence from its paralog are associated with the origin of a novel developmental feature in zebrafish and related taxa (Cypriniformes) Evolution. 2009;63:1574–1592. doi: 10.1111/j.1558-5646.2009.00657.x. [DOI] [PubMed] [Google Scholar]
- Crow KD, Smith CD, Cheng JF, Wagner GP, Amemiya CT. An independent genome duplication inferred from Hox paralogs in the American paddlefish--a representative basal ray-finned fish and important comparative reference. Genome Biol Evol. 2012;4:937–953. doi: 10.1093/gbe/evs067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow KD, Stadler PF, Lynch VJ, Amemiya C, Wagner GP. The "fish-specific" Hox cluster duplication is coincident with the origin of teleosts. Mol Biol Evol. 2006;23:121–136. doi: 10.1093/molbev/msj020. [DOI] [PubMed] [Google Scholar]
- Cuervo R, Hernandez-Martinez R, Chimal-Monroy J, Merchant-Larios H, Covarrubias L. Full regeneration of the tribasal Polypterus fin. Proc Natl Acad Sci U S A. 2012;109:3838–3843. doi: 10.1073/pnas.1006619109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui R, Schumer M, Kruesi K, Walter R, Andolfatto P, Rosenthal GG. Phylogenomics reveals extensive reticulate evolution in Xiphophorus fishes. Evolution. 2013;67:2166–2179. doi: 10.1111/evo.12099. [DOI] [PubMed] [Google Scholar]
- Darwin C. The descent of man, selection in relation to sex. London: John Murray; 1871. [Google Scholar]
- Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nature Reviews Genetics. 2011;12:499–510. doi: 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]
- Davidson WS, Koop BF, Jones SJ, Iturra P, Vidal R, Maass A, Jonassen I, Lien S, Omholt SW. Sequencing the genome of the Atlantic salmon (Salmo salar) Genome Biol. 2010;11:403. doi: 10.1186/gb-2010-11-9-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MC, Dahn RD, Shubin NH. An autopodial-like pattern of Hox expression in the fins of a basal actinopterygian fish. Nature. 2007;447:473-U473. doi: 10.1038/nature05838. [DOI] [PubMed] [Google Scholar]
- de Souza FS, Bumaschny VF, Low MJ, Rubinstein M. Subfunctionalization of expression and peptide domains following the ancient duplication of the proopiomelanocortin gene in teleost fishes. Mol Biol Evol. 2005;22:2417–2427. doi: 10.1093/molbev/msi236. [DOI] [PubMed] [Google Scholar]
- Dessimoz C, Gabaldon T, Roos DS, Sonnhammer EL, Herrero J. Toward community standards in the quest for orthologs. Bioinformatics. 2012;28:900–904. doi: 10.1093/bioinformatics/bts050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvignes T, Beam MJ, Batzel P, Sydes J, Postlethwait JH. Expanding the annotation of zebrafish microRNAs based on small RNA sequencing. Gene. 2014 doi: 10.1016/j.gene.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detrich HW, 3rd, Amemiya CT. Antarctic notothenioid fishes: genomic resources and strategies for analyzing an adaptive radiation. Integr Comp Biol. 2010;50:1009–1017. doi: 10.1093/icb/icq071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detrich HW, Stuart A, Schoenborn M, Parker SK, Methe BA, Amemiya CT. Genome Enablement of the Notothenioidei: Genome Size Estimates from 11 Species and BAC Libraries from 2 Representative Taxa. Journal of Experimental Zoology Part B-Molecular and Developmental Evolution 314B. 2010:369–381. doi: 10.1002/jez.b.21341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue PC, Purnell MA. Genome duplication, extinction and vertebrate evolution. Trends Ecol Evol. 2005;20:312–319. doi: 10.1016/j.tree.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Eames BF, Amores A, Yan YL, Postlethwait JH. Evolution of the osteoblast: skeletogenesis in gar and zebrafish. BMC Evol Biol. 2012;12:27. doi: 10.1186/1471-2148-12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman JT. Antarctic Fish Biology: Evolution in a Unique Environment. San Diego: Academic Press; 1993. [Google Scholar]
- Eastman JT. The nature of the diversity of Antarctic fishes. Polar Biology. 2005;28:93–107. [Google Scholar]
- Eastman JT, McCune AR. Fishes on the Antarctic continental shelf: evolution of a marine species flock? Journal of Fish Biology. 2000;57:84–102. [Google Scholar]
- Egginton S, Skilbeck C, Hoofd L, Calvo J, Johnston IA. Peripheral oxygen transport in skeletal muscle of Antarctic and sub-Antarctic notothenioid fish. Journal of Experimental Biology. 2002;205:769–779. doi: 10.1242/jeb.205.6.769. [DOI] [PubMed] [Google Scholar]
- Evans JP, Pilastro A, Schlupp I. Ecology and Evolution of Poeciliid Fish. Chicago: The University of Chicago Press; 2011. [Google Scholar]
- Fan S, Elmer KR, Meyer A. Genomics of adaptation and speciation in cichlid fishes: recent advances and analyses in African and Neotropical lineages. Philos Trans R Soc Lond B Biol Sci. 2012;367:385–394. doi: 10.1098/rstb.2011.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F. Phylogenetic analysis of the Asian cyprinid genus Danio (Teleostei, Cyprinidae) Copeia. 2003:714–728. [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, McCallion AS. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nature Protocols. 2006;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- Forconi M, Chalopin D, Barucca M, Biscotti MA, De Moro G, Galiana D, Gerdol M, Pallavicini A, Canapa A, Olmo E, Volff JN. Transcriptional activity of transposable elements in coelacanth. J Exp Zool B Mol Dev Evol. 2013 doi: 10.1002/jez.b.22527. = [DOI] [PubMed] [Google Scholar]
- Frankenberg S, Renfree MB. On the origin of POU5F1. BMC Biol. 2013;11:56. doi: 10.1186/1741-7007-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser BA, Weadick CJ, Janowitz I, Rodd FH, Hughes KA. Sequencing and characterization of the guppy (Poecilia reticulata) transcriptome. BMC Genomics. 2011;12:202. doi: 10.1186/1471-2164-12-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich C, Hagen W. Lipid contents of five species of notothenioid fish from high-Antarctic waters and ecological implications. Polar Biology. 1994;14:359–369. [Google Scholar]
- Furutani-Seiki M, Wittbrodt J. Medaka and zebrafish, an evolutionary twin study. Mech Dev. 2004;121:629–637. doi: 10.1016/j.mod.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Gagnaire PA, Normandeau E, Pavey SA, Bernatchez L. Mapping phenotypic, expression and transmission ratio distortion QTL using RAD markers in the Lake Whitefish (Coregonus clupeaformis) Molecular Ecology. 2013;22:3036–3048. doi: 10.1111/mec.12127. [DOI] [PubMed] [Google Scholar]
- Gehrig J, Reischl M, Kalmar E, Ferg M, Hadzhiev Y, Zaucker A, Song CY, Schindler S, Liebel U, Muller F. Automated high-throughput mapping of promoter-enhancer interactions in zebrafish embryos. Nature Methods. 2009;6:911-U971. doi: 10.1038/nmeth.1396. [DOI] [PubMed] [Google Scholar]
- Grabher C, Wittbrodt J. Meganuclease and transposon mediated transgenesis in medaka. Genome Biology. 2007;8 doi: 10.1186/gb-2007-8-s1-s10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, Borowsky R, Tabin CJ. A Novel Role for Mc1r in the Parallel Evolution of Depigmentation in Independent Populations of the Cavefish Astyanax mexicanus. Plos Genetics. 2009;5 doi: 10.1371/journal.pgen.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, Furterer A, Carlson BM, Stahl BA. An Integrated Transcriptome-Wide Analysis of Cave and Surface Dwelling Astyanax mexicanus. Plos One. 2013;8 doi: 10.1371/journal.pone.0055659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, Krutzler AJ, Carlson BM. Complex Craniofacial Changes in Blind Cave-Dwelling Fish Are Mediated by Genetically Symmetric and Asymmetric Loci. Genetics. 2014 doi: 10.1534/genetics.114.161661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen W, Kattner G, Friedrich C. The lipid compositions of high-Antarctic notothenioid fish species with different life strategies. Polar Biology. 2000;23:785–791. [Google Scholar]
- Hamada H, Watanabe M, Lau HE, Nishida T, Hasegawa T, Parichy DM, Kondo S. Involvement of Delta/Notch signaling in zebrafish adult pigment stripe patterning. Development. 2014;141:318–324. doi: 10.1242/dev.099804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RM, Dijkstra PD, Hofmann HA. Complex structural and regulatory evolution of the pro-opiomelanocortin gene family. Gen Comp Endocrinol. 2014;195:107–115. doi: 10.1016/j.ygcen.2013.10.007. [DOI] [PubMed] [Google Scholar]
- Hemmingsen EA. Respiratory and Cardiovascular Adaptations in Hemoglobin-Free Fish: Resolved and Unresolved Problems. In: di Prisco G, Maresca B, Tota B, editors. Biology of Antarctic Fish. Berlin Heidelberg: Springer; 1991. pp. 191–203. [Google Scholar]
- Henkel CV, Burgerhout E, de Wijze DL, Dirks RP, Minegishi Y, Jansen HJ, Spaink HP, Dufour S, Weltzien FA, Tsukamoto K, van den Thillart GE. Primitive duplicate Hox clusters in the European eel's genome. PLoS One. 2012a;7:e32231. doi: 10.1371/journal.pone.0032231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel CV, Dirks RP, de Wijze DL, Minegishi Y, Aoyama J, Jansen HJ, Turner B, Knudsen B, Bundgaard M, Hvam KL, Boetzer M, Pirovano W, Weltzien FA, Dufour S, Tsukamoto K, Spaink HP, van den Thillart GE. First draft genome sequence of the Japanese eel, Anguilla japonica. Gene. 2012b;511:195–201. doi: 10.1016/j.gene.2012.09.064. [DOI] [PubMed] [Google Scholar]
- Henkel CV, Dirks RP, Jansen HJ, Forlenza M, Wiegertjes GF, Howe K, van den Thillan GEEJM, Spaink HP. Comparison of the Exomes of Common Carp (Cyprinus carpio) and Zebrafish (Danio rerio) Zebrafish. 2012c;9:59–67. doi: 10.1089/zeb.2012.0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Agarwal S, Notwell JH, Parikh R, Guturu H, Wenger AM, Bejerano G. Computational methods to detect conserved non-genic elements in phylogenetically isolated genomes: application to zebrafish. Nucleic Acids Res. 2013;41:e151. doi: 10.1093/nar/gkt557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinaux H, Poulain J, Da Silva C, Noirot C, Jeffery WR, Casane D, Retaux S. De Novo Sequencing of Astyanax mexicanus Surface Fish and Pachon Cavefish Transcriptomes Reveals Enrichment of Mutations in Cavefish Putative Eye Genes. Plos One. 2013;8 doi: 10.1371/journal.pone.0053553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegg S, Brinkmann H, Taylor JS, Meyer A. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J Mol Evol. 2004;59:190–203. doi: 10.1007/s00239-004-2613-z. [DOI] [PubMed] [Google Scholar]
- Hofmann GE, Buckley BA, Airaksinen S, Keen JE, Somero GN. Heat-shock protein expression is absent in the antarctic fish Trematomus bernacchii (family Nototheniidae) J Exp Biol. 2000;203:2331–2339. doi: 10.1242/jeb.203.15.2331. [DOI] [PubMed] [Google Scholar]
- Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA, Cresko WA. Population Genomics of Parallel Adaptation in Threespine Stickleback using Sequenced RAD Tags. Plos Genetics. 2010;6 doi: 10.1371/journal.pgen.1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenlohe PA, Catchen J, Cresko WA. Population genomic analysis of model and nonmodel organisms using sequenced RAD tags. Methods Mol Biol. 2012;888:235–260. doi: 10.1007/978-1-61779-870-2_14. [DOI] [PubMed] [Google Scholar]
- Howe DG, Bradford YM, Conlin T, Eagle AE, Fashena D, Frazer K, Knight J, Mani P, Martin R, Moxon SA, Paddock H, Pich C, Ramachandran S, Ruef BJ, Ruzicka L, Schaper K, Shao X, Singer A, Sprunger B, Van Slyke CE, Westerfield M. ZFIN, the Zebrafish Model Organism Database: increased support for mutants and transgenics. Nucleic Acids Res. 2013a;41:D854–D860. doi: 10.1093/nar/gks938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, McLaren S, Sealy I, Caccamo M, Churcher C, Scott C, Barrett JC, Koch R, Rauch GJ, White S, Chow W, Kilian B, Quintais LT, Guerra-Assuncao JA, Zhou Y, Gu Y, Yen J, Vogel JH, Eyre T, Redmond S, Banerjee R, Chi J, Fu B, Langley E, Maguire SF, Laird GK, Lloyd D, Kenyon E, Donaldson S, Sehra H, Almeida-King J, Loveland J, Trevanion S, Jones M, Quail M, Willey D, Hunt A, Burton J, Sims S, McLay K, Plumb B, Davis J, Clee C, Oliver K, Clark R, Riddle C, Eliott D, Threadgold G, Harden G, Ware D, Mortimer B, Kerry G, Heath P, Phillimore B, Tracey A, Corby N, Dunn M, Johnson C, Wood J, Clark S, Pelan S, Griffiths G, Smith M, Glithero R, Howden P, Barker N, Stevens C, Harley J, Holt K, Panagiotidis G, Lovell J, Beasley H, Henderson C, Gordon D, Auger K, Wright D, Collins J, Raisen C, Dyer L, Leung K, Robertson L, Ambridge K, Leongamornlert D, McGuire Gilderthorp R, Griffiths C, Manthravadi D, Nichol S, Barker G, Whitehead S, Kay M, Brown J, Murnane C, Gray E, Humphries M, Sycamore N, Barker D, Saunders D, Wallis J, Babbage A, Hammond S, Mashreghi-Mohammadi M, Barr L, Martin S, Wray P, Ellington A, Matthews N, Ellwood M, Woodmansey R, Clark G, Cooper Tromans A, Grafham D, Skuce C, Pandian R, Andrews R, Harrison E, Kimberley A, Garnett J, Fosker N, Hall R, Garner P, Kelly D, Bird C, Palmer S, Gehring I, Berger A, Dooley CM, Ersan-Urun Z, Eser C, Geiger H, Geisler M, Karotki L, Kirn A, Konantz J, Konantz M, Oberlander M, Rudolph-Geiger S, Teucke M, Osoegawa K, Zhu B, Rapp A, Widaa S, Langford C, Yang F, Carter NP, Harrow J, Ning Z, Herrero J, Searle SM, Enright A, Geisler R, Plasterk RH, Lee C, Westerfield M, de Jong PJ, Zon LI, Postlethwait JH, Nusslein-Volhard C, Hubbard TJ, Roest Crollius H, Rogers J, Stemple DL. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013b;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Goll MG, Fisher S. PhiC31 integrase mediates efficient cassette exchange in the zebrafish germline. Dev Dyn. 2011;240:2101–2107. doi: 10.1002/dvdy.22699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huddleston J, Ranade S, Malig M, Antonacci F, Chaisson M, Hon L, Sudmant PH, Graves TA, Alkan C, Dennis MY, Wilson RK, Turner SW, Korlach K, Eichler EE. Reconstructing complex regions of genomes using long-read sequencing technology. Genome Research. 2014 doi: 10.1101/gr.168450.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley IA, Mueller RL, Dunn KA, Schmidt EJ, Friedman M, Ho RK, Prince VE, Yang Z, Thomas MG, Coates MI. A new time-scale for ray-finned fish evolution. Proc Biol Sci. 2007;274:489–498. doi: 10.1098/rspb.2006.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huth TJ, Place SP. De novo assembly and characterization of tissue specific transcriptomes in the emerald notothen, Trematomus bernacchii. Bmc Genomics. 2013;14 doi: 10.1186/1471-2164-14-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba M, Yamanaka H, Kondo S. Pigment Pattern Formation by Contact-Dependent Depolarization. Science. 2012;335:677–677. doi: 10.1126/science.1212821. [DOI] [PubMed] [Google Scholar]
- Inoue JG, Miya M, Tsukamoto K, Nishida M. Basal actinopterygian relationships: a mitogenomic perspective on the phylogeny of the "ancient fish". Mol Phylogenet Evol. 2003;26:110–120. doi: 10.1016/s1055-7903(02)00331-7. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, Nicaud S, Jaffe D, Fisher S, Lutfalla G, Dossat C, Segurens B, Dasilva C, Salanoubat M, Levy M, Boudet N, Castellano S, Anthouard V, Jubin C, Castelli V, Katinka M, Vacherie B, Biemont C, Skalli Z, Cattolico L, Poulain J, De Berardinis V, Cruaud C, Duprat S, Brottier P, Coutanceau JP, Gouzy J, Parra G, Lardier G, Chapple C, McKernan KJ, McEwan P, Bosak S, Kellis M, Volff JN, Guigo R, Zody MC, Mesirov J, Lindblad-Toh K, Birren B, Nusbaum C, Kahn D, Robinson- Rechavi M, Laudet V, Schachter V, Quetier F, Saurin W, Scarpelli C, Wincker P, Lander ES, Weissenbach J, Roest Crollius H. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery WR. Emerging model systems in evo-devo: cavefish and microevolution of development. Evol Dev. 2008;10:265–272. doi: 10.1111/j.1525-142X.2008.00235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery WR. Regressive Evolution in Astyanax Cavefish. Annual Review of Genetics. 2009;43:25–47. doi: 10.1146/annurev-genet-102108-134216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Gao X, Liu S, Zhang Y, Liu H, Sun F, Bao L, Waldbieser G, Liu Z. Whole genome comparative analysis of channel catfish (Ictalurus punctatus) with four model fish species. BMC Genomics. 2013;14:780. doi: 10.1186/1471-2164-14-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson Z, Joss J, Boisvert CA, Ericsson R, Sutija M, Ahlberg PE. Fish fingers: digit homologues in sarcopterygian fish fins. J Exp Zool B Mol Dev Evol. 2007;308:757–768. doi: 10.1002/jez.b.21197. [DOI] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC, White S, Birney E, Searle S, Schmutz J, Grimwood J, Dickson MC, Myers RM, Miller CT, Summers BR, Knecht AK, Brady SD, Zhang H, Pollen AA, Howes T, Amemiya C, Baldwin J, Bloom T, Jaffe DB, Nicol R, Wilkinson J, Lander ES, Di Palma F, Lindblad-Toh K, Kingsley DM. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Fan SH, Franchini P, Schartl M, Meyer A. The evolutionary history of Xiphophorus fish and their sexually selected sword: a genome-wide approach using restriction site-associated DNA sequencing. Molecular Ecology. 2013;22:2986–3001. doi: 10.1111/mec.12269. [DOI] [PubMed] [Google Scholar]
- Juntti SA, Hu CK, Fernald RD. Tol2-Mediated Generation of a Transgenic Haplochromine Cichlid, Astatotilapia burtoni. PLoS One. 2013;8:e77647. doi: 10.1371/journal.pone.0077647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakioka R, Kokita T, Kumada H, Watanabe K, Okuda N. A RAD-based linkage map and comparative genomics in the gudgeons (genus Gnathopogon, Cyprinidae) BMC Genomics. 2013;14:32. doi: 10.1186/1471-2164-14-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamberov YG, Wang S, Tan J, Gerbault P, Wark A, Tan L, Yang Y, Li S, Tang K, Chen H, Powell A, Itan Y, Fuller D, Lohmueller J, Mao J, Schachar A, Paymer M, Hostetter E, Byrne E, Burnett M, McMahon AP, Thomas MG, Lieberman DE, Jin L, Tabin CJ, Morgan BA, Sabeti PC. Modeling recent human evolution in mice by expression of a selected EDAR variant. Cell. 2013;152:691–702. doi: 10.1016/j.cell.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M, Naruse K, Sasaki S, Nakatani Y, Qu W, Ahsan B, Yamada T, Nagayasu Y, Doi K, Kasai Y, Jindo T, Kobayashi D, Shimada A, Toyoda A, Kuroki Y, Fujiyama A, Sasaki T, Shimizu A, Asakawa S, Shimizu N, Hashimoto SI, Yang J, Lee Y, Matsushima K, Sugano S, Sakaizumi M, Narita T, Ohishi K, Haga S, Ohta F, Nomoto H, Nogata K, Morishita T, Endo T, Shin-I T, Takeda H, Morishita S, Kohara Y. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- Kassahn KS, Dang VT, Wilkins SJ, Perkins AC, Ragan MA. Evolution of gene function and regulatory control after whole-genome duplication: comparative analyses in vertebrates. Genome Res. 2009;19:1404–1418. doi: 10.1101/gr.086827.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennett JP. Cenozoic Evolution of Antarctic Glaciation, Circum-Antarctic Ocean, and Their Impact on Global Paleoceanography. Journal of Geophysical Research-Oceans and Atmospheres. 1977;82:3843–3860. [Google Scholar]
- Kettleborough RNW, Busch-Nentwich EM, Harvey SA, Dooley CM, de Bruijn E, van Eeden F, Sealy I, White RJ, Herd C, Nijman IJ, Fenyes F, Mehroke S, Scahill C, Gibbons R, Wali N, Carruthers S, Hall A, Yen J, Cuppen E, Stemple DL. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature. 2013;496:494. doi: 10.1038/nature11992. -+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- King HM, Shubin NH, Coates MI, Hale ME. Behavioral evidence for the evolution of walking and bounding before terrestriality in sarcopterygian fishes. Proc Natl Acad Sci U S A. 2011;108:21146–21151. doi: 10.1073/pnas.1118669109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchmaier S, Hockendorf B, Moller EK, Bornhorst D, Spitz F, Wittbrodt J. Efficient site-specific transgenesis and enhancer activity tests in medaka using PhiC31 integrase. Development. 2013;140:4287–4295. doi: 10.1242/dev.096081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano J, Yoshida K, Suzuki Y. RNA sequencing reveals small RNAs differentially expressed between incipient Japanese threespine sticklebacks. BMC Genomics. 2013;14:214. doi: 10.1186/1471-2164-14-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher TD. Adaptive evolution and explosive speciation: the cichlid fish model. Nat Rev Genet. 2004;5:288–298. doi: 10.1038/nrg1316. [DOI] [PubMed] [Google Scholar]
- Kock KH. Antarctic icefishes (Channichthyidae): a unique family of fishes. A review, Part I. Polar Biology. 2005a;28:862–895. [Google Scholar]
- Kock KH. Antarctic icefishes (Channichthyidae): a unique family of fishes. A review, Part II. Polar Biology. 2005b;28:897–909. [Google Scholar]
- Komiyama T, Kobayashi H, Tateno Y, Inoko H, Gojobori T, Ikeo K. An evolutionary origin and selection process of goldfish. Gene. 2009;430:5–11. doi: 10.1016/j.gene.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Kottelat M, Britz R, Hui TH, Witte KE. Paedocypris, a new genus of Southeast Asian cyprinid fish with a remarkable sexual dimorphism, comprises the world's smallest vertebrate. Proceedings of the Royal Society B-Biological Sciences. 2006;273:895–899. doi: 10.1098/rspb.2005.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottler VA, Fadeev A, Weigel D, Dreyer C. Pigment Pattern Formation in the Guppy, Poecilia reticulata, Involves the Kita and Csf1ra Receptor Tyrosine Kinases. Genetics. 2013;194:631. doi: 10.1534/genetics.113.151738. -+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalko JE, Rohner N, Linden TA, Rompani SB, Warren WC, Borowsky R, Tabin CJ, Jeffery WR, Yoshizawa M. Convergence in feeding posture occurs through different genetic loci in independently evolved cave populations of Astyanax mexicanus. Proc Natl Acad Sci U S A. 2013;110:16933–16938. doi: 10.1073/pnas.1317192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku S. Impact of asymmetric gene repertoire between cyclostomes and gnathostomes. Seminars in Cell & Developmental Biology. 2013;24:119–127. doi: 10.1016/j.semcdb.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Lampert KP, Schmidt C, Fischer P, Volff JN, Hoffmann C, Muck J, Lohse MJ, Ryan MJ, Schartl M. Determination of onset of sexual maturation and mating behavior by melanocortin receptor 4 polymorphisms. Curr Biol. 2010;20:1729–1734. doi: 10.1016/j.cub.2010.08.029. [DOI] [PubMed] [Google Scholar]
- Lee AP, Kerk SY, Tan YY, Brenner S, Venkatesh B. Ancient vertebrate conserved noncoding elements have been evolving rapidly in teleost fishes. Mol Biol Evol. 2011;28:1205–1215. doi: 10.1093/molbev/msq304. [DOI] [PubMed] [Google Scholar]
- Li C, Orti G, Zhang G, Lu G. A practical approach to phylogenomics: the phylogeny of ray-finned fish (Actinopterygii) as a case study. BMC Evol Biol. 2007;7:44. doi: 10.1186/1471-2148-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CH, Lu GQ, Orti G. Optimal data partitioning and a test case for ray-finned fishes (Actinopterygii) based on ten nuclear loci. Systematic Biology. 2008;57:519–539. doi: 10.1080/10635150802206883. [DOI] [PubMed] [Google Scholar]
- Liedtke D, Erhard I, Abe K, Furutani-Seiki M, Kondoh M, Schartl M. Xmrk-induced melanoma progression is affected by Sdf1 signals through Cxcr7. Pigment Cell and Melanoma Research. 2013 doi: 10.1111/pcmr.12188. [DOI] [PubMed] [Google Scholar]
- Long WL, Ballard WW. Normal embryonic stages of the longnose gar, Lepisosteus osseus. BMC Dev Biol. 2001;1:6. doi: 10.1186/1471-213X-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo S, Riccio M, McCune AR. Homology of lungs and gas bladders: Insights from arterial vasculature. Journal of Morphology. 2013;274:687–703. doi: 10.1002/jmor.20128. [DOI] [PubMed] [Google Scholar]
- Louis A, Muffato M, Roest Crollius H. Genomicus: five genome browsers for comparative genomics in eukaryota. Nucleic Acids Res. 2013;41:D700–D705. doi: 10.1093/nar/gks1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CB, Kellis M, Siepel A, Raney BJ, Clamp M, Salama SR, Kingsley DM, Lindblad-Toh K, Haussler D. Three periods of regulatory innovation during vertebrate evolution. Science. 2011;333:1019–1024. doi: 10.1126/science.1202702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A, Belfiore NM, Pitra C, Svirsky V, Jenneckens I. Genome duplication events and functional reduction of ploidy levels in sturgeon (Acipenser, Huso and Scaphirhynchus) Genetics. 2001;158:1203–1215. doi: 10.1093/genetics/158.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabee P, Balhoff JP, Dahdul WM, Lapp H, Midford PE, Vision TJ, Westerfield M. 500,000 fish phenotypes: The new informatics landscape for evolutionary and developmental biology of the vertebrate skeleton. Journal of Applied Ichthyology. 2012;28:300–305. doi: 10.1111/j.1439-0426.2012.01985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JA, Wang Z. Next-generation transcriptome assembly. Nat Rev Genet. 2011;12:671–682. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- Meierjohann S, Schartl M. From Mendelian to molecular genetics: the Xiphophorus melanoma model. Trends Genet. 2006;22:654–661. doi: 10.1016/j.tig.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Meng F, Braasch I, Phillips JB, Lin X, Titus T, Zhang C, Postlethwait JH. Evolution of the eye transcriptome under constant darkness in Sinocyclocheilus cavefish. Mol Biol Evol. 2013a;30:1527–1543. doi: 10.1093/molbev/mst079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Zhao Y, Postlethwait JH, Zhang C. Differentially-expressed genes identified in cavefish endemic to China. Curr Zool. 2013b;59:170–174. doi: 10.1093/czoolo/59.2.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe CJ, Filee J, Germon I, Joss J, Casane D. Evolution of the Australian lungfish (Neoceratodus forsteri) genome: a major role for CR1 and L2 LINE elements. Mol Biol Evol. 2012;29:3529–3539. doi: 10.1093/molbev/mss159. [DOI] [PubMed] [Google Scholar]
- Metscher D, Ahlberg PE. Origin of the teleost tail: phylogenetic frameworks for developmental studies. In: Ahlberg PE, editor. Major Events in Early Vertebrate Evolution: Taylor & Francis. 2001. pp. 333–349. [Google Scholar]
- Miller MR, Brunelli JP, Wheeler PA, Liu S, Rexroad CE, 3rd, Palti Y, Doe CQ, Thorgaard GH. A conserved haplotype controls parallel adaptation in geographically distant salmonid populations. Mol Ecol. 2012;21:237–249. doi: 10.1111/j.1365-294X.2011.05305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MR, Dunham JP, Amores A, Cresko WA, Johnson EA. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res. 2007;17:240–248. doi: 10.1101/gr.5681207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley KB, Small CM, Jones AG. The genetics and genomics of Syngnathidae: pipefishes, seahorses and seadragons. Journal of Fish Biology. 2011;78:1624–1646. doi: 10.1111/j.1095-8649.2011.02967.x. [DOI] [PubMed] [Google Scholar]
- Montgomery J, Clements K. Disaptation and recovery in the evolution of Antarctic fishes. Trends in Ecology & Evolution. 2000;15:267–271. doi: 10.1016/s0169-5347(00)01896-6. [DOI] [PubMed] [Google Scholar]
- Mosimann C, Puller AC, Lawson KL, Tschopp P, Amsterdam A, Zon LI. Site-directed zebrafish transgenesis into single landing sites with the phiC31 integrase system. Dev Dyn. 2013;242:949–963. doi: 10.1002/dvdy.23989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Saito D, Tanaka M. Generation of transgenic medaka using modified bacterial artificial chromosome. Dev Growth Differ. 2008;50:415–419. doi: 10.1111/j.1440-169X.2008.01027.x. [DOI] [PubMed] [Google Scholar]
- Near TJ, Dornburg A, Kuhn KL, Eastman JT, Pennington JN, Patarnello T, Zane L, Fernandez DA, Jones CD. Ancient climate change, antifreeze, and the evolutionary diversification of Antarctic fishes. Proceedings of the National Academy of Sciences of the United States of America. 2012a;109:3434–3439. doi: 10.1073/pnas.1115169109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, Davis MP, Wainwright PC, Friedman M, Smith WL. Resolution of ray-finned fish phylogeny and timing of diversification. Proc Natl Acad Sci U S A. 2012b;109:13698–13703. doi: 10.1073/pnas.1206625109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JS. Fishes of the World. Hoboken, New Jersey: John Wiley & Sons, Inc; 2006. [Google Scholar]
- Nikaido M, Noguchi H, Nishihara H, Toyoda A, Suzuki Y, Kajitani R, Suzuki H, Okuno M, Aibara M, Ngatunga BP, Mzighani SI, Kalombo HW, Masengi KW, Tuda J, Nogami S, Maeda R, Iwata M, Abe Y, Fujimura K, Okabe M, Amano T, Maeno A, Shiroishi T, Itoh T, Sugano S, Kohara Y, Fujiyama A, Okada N. Coelacanth genomes reveal signatures for evolutionary transition from water to land. Genome Res. 2013;23:1740–1748. doi: 10.1101/gr.158105.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K, Ozaki A, Morishima K, Yoshikawa Y, Tanaka H, Unuma T, Ohta H, Arai K. A genetic linkage map of the Japanese eel (Anguilla japonica) based on AFLP and microsatellite markers. Aquaculture. 2011;310:329–342. [Google Scholar]
- O'Brien KM, Xue HJ, Sidell BD. Quantification of diffusion distance within the spongy myocardium of hearts from antarctic fishes. Respiration Physiology. 2000;122:71–80. doi: 10.1016/s0034-5687(00)00139-0. [DOI] [PubMed] [Google Scholar]
- O'Quin KE, Yoshizawa M, Doshi P, Jeffery WR. Quantitative genetic analysis of retinal degeneration in the blind cavefish Astyanax mexicanus. PLoS One. 2013;8:e57281. doi: 10.1371/journal.pone.0057281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino Y, Katoh H, Yamada G. Androgen dependent development of a modified anal fin, gonopodium, as a model to understand the mechanism of secondary sexual character expression in vertebrates. FEBS Lett. 2004;575:119–126. doi: 10.1016/j.febslet.2004.08.046. [DOI] [PubMed] [Google Scholar]
- Ohno S, Muramoto J, Christia L, Atkin NB. Diploid-Tetraploid Relationship among Old-World Members of Fish Family Cyprinidae. Chromosoma. 1967;23:1. -&. [Google Scholar]
- Palaiokostas C, Bekaert M, Davie A, Cowan ME, Oral M, Taggart JB, Gharbi K, McAndrew BJ, Penman DJ, Migaud H. Mapping the sex determination locus in the Atlantic halibut (Hippoglossus hippoglossus) using RAD sequencing. BMC Genomics. 2013a;14:566. doi: 10.1186/1471-2164-14-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaiokostas C, Bekaert M, Khan MG, Taggart JB, Gharbi K, McAndrew BJ, Penman DJ. Mapping and validation of the major sex-determining region in Nile tilapia (Oreochromis niloticus L.) Using RAD sequencing. PLoS One. 2013b;8:e68389. doi: 10.1371/journal.pone.0068389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parichy DM. Homology and the evolution of novelty during Danio adult pigment pattern development. Journal of Experimental Zoology Part B-Molecular and Developmental Evolution. 2007;308B:578–590. doi: 10.1002/jez.b.21141. [DOI] [PubMed] [Google Scholar]
- Parichy DM, Johnson SL. Zebrafish hybrids suggest genetic mechanisms for pigment pattern diversification in Danio. Development Genes and Evolution. 2001;211:319–328. doi: 10.1007/s004270100155. [DOI] [PubMed] [Google Scholar]
- Patterson LB, Parichy DM. Interactions with Iridophores and the Tissue Environment Required for Patterning Melanophores and Xanthophores during Zebrafish Adult Pigment Stripe Formation. Plos Genetics. 2013;9 doi: 10.1371/journal.pgen.1003561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, Schier AF. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22:577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peichel CL. Fishing for the secrets of vertebrate evolution in threespine sticklebacks. Dev Dyn. 2005;234:815–823. doi: 10.1002/dvdy.20564. [DOI] [PubMed] [Google Scholar]
- Petzold A, Reichwald K, Groth M, Taudien S, Hartmann N, Priebe S, Shagin D, Englert C, Platzer M. The transcript catalogue of the short-lived fish Nothobranchius furzeri provides insights into age-dependent changes of mRNA levels. Bmc Genomics. 2013;14 doi: 10.1186/1471-2164-14-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picone B, Hesse U, Panji S, Van Heusden P, Jonas M, Christoffels A. Taste and odorant receptors of the coelacanth-A gene repertoire in transition. J Exp Zool B Mol Dev Evol. 2013 doi: 10.1002/jez.b.22531. [DOI] [PubMed] [Google Scholar]
- Place S, Hofmann G. Constitutive expression of a stress-inducible heat shock protein gene, hsp70, in phylogenetically distant Antarctic fish. Polar Biology. 2005;28:261–267. [Google Scholar]
- Pollux BJA, Pires MN, Banet AI, Reznick DN. Evolution of Placentas in the Fish Family Poeciliidae: An Empirical Study of Macroevolution. Annual Review of Ecology Evolution and Systematics. 2009;40:271–289. [Google Scholar]
- Postlethwait J, Amores A, Cresko W, Singer A, Yan YL. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20:481–490. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH. The zebrafish genome in context: ohnologs gone missing. J Exp Zool B Mol Dev Evol. 2007;308:563–577. doi: 10.1002/jez.b.21137. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Visel A, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Morrison H, Fitzpatrick DR, Afzal V, Pennacchio LA, Rubin EM, Noonan JP. Human-specific gain of function in a developmental enhancer. Science. 2008;321:1346–1350. doi: 10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protas M, Conrad M, Gross JB, Tabin C, Borowsky R. Regressive evolution in the Mexican cave tetra, Astyanax mexicanus. Curr Biol. 2007;17:452–454. doi: 10.1016/j.cub.2007.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, Jeffery WR, Zon LI, Borowsky R, Tabin CJ. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 2006;38:107–111. doi: 10.1038/ng1700. [DOI] [PubMed] [Google Scholar]
- Qian X, Ba Y, Zhuang Q, Zhong G. RNA-Seq Technology and Its Application in Fish Transcriptomics. OMICS. 2014;18:98–110. doi: 10.1089/omi.2013.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley IK, Manuel JL, Roberts RA, Nuckels RJ, Herrington ER, MacDonald EL, Parichy DM. Evolutionary diversification of pigment pattern in Danio fishes: differential fms dependence and stripe loss in D. albolineatus. Development. 2005;132:89–104. doi: 10.1242/dev.01547. [DOI] [PubMed] [Google Scholar]
- Rabosky DL, Santini F, Eastman J, Smith SA, Sidlauskas B, Chang J, Alfaro ME. Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation. Nature Communications. 2013;4 doi: 10.1038/ncomms2958. [DOI] [PubMed] [Google Scholar]
- Raincrow JD, Dewar K, Stocsits C, Prohaska SJ, Amemiya CT, Stadler PF, Chiu CH. Hox clusters of the bichir (Actinopterygii, Polypterus senegalus) highlight unique patterns of sequence evolution in gnathostome phylogeny. J Exp Zool B Mol Dev Evol. 2011;316:451–464. doi: 10.1002/jez.b.21420. [DOI] [PubMed] [Google Scholar]
- Ravi V, Venkatesh B. Rapidly evolving fish genomes and teleost diversity. Curr Opin Genet Dev. 2008;18:544–550. doi: 10.1016/j.gde.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Recknagel H, Elmer KR, Meyer A. A hybrid genetic linkage map of two ecologically and morphologically divergent Midas cichlid fishes (Amphilophus spp.) obtained by massively parallel DNA sequencing (ddRADSeq) G3 (Bethesda) 2013;3:65–74. doi: 10.1534/g3.112.003897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retaux S, Casane D. Evolution of eye development in the darkness of caves: adaptation, drift, or both? Evodevo. 2013;4:26. doi: 10.1186/2041-9139-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JA, Miguel-Escalada I, Slovik KJ, Walsh KT, Hadzhiev Y, Sanges R, Stupka E, Marsh EK, Balciuniene J, Balciunas D, Müller F. Targeted transgene integration overcomes variability of position effects in zebrafish. Development. 2014;141:715–724. doi: 10.1242/dev.100347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohner N, Jarosz DF, Kowalko JE, Yoshizawa M, Jeffery WR, Borowsky RL, Lindquist S, Tabin CJ. Cryptic Variation in Morphological Evolution: HSP90 as a Capacitor for Loss of Eyes in Cavefish. Science. 2013;342:1372–1375. doi: 10.1126/science.1240276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruber L, Kottelat M, Tan HH, Ng PKL, Britz R. Evolution of miniaturization and the phylogenetic position of Paedocypris, comprising the world's smallest vertebrate. Bmc Evolutionary Biology. 2007;7 doi: 10.1186/1471-2148-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutschmann S, Matschiner M, Damerau M, Muschick M, Lehmann MF, Hanel R, Salzburger W. Parallel ecological diversification in Antarctic notothenioid fishes as evidence for adaptive radiation. Molecular Ecology. 2011;20:4707–4721. doi: 10.1111/j.1365-294X.2011.05279.x. [DOI] [PubMed] [Google Scholar]
- Ruud JT. Vertebrates without erythrocytes and blood pigment. Nature London. 1954;173:848–850. doi: 10.1038/173848a0. [DOI] [PubMed] [Google Scholar]
- Saha NR, Ota T, Litman GW, Hansen J, Parra Z, Hsu E, Buonocore F, Canapa A, Cheng JF, Amemiya CT. Genome complexity in the coelacanth is reflected in its adaptive immune system. J Exp Zool B Mol Dev Evol. 2014 doi: 10.1002/jez.b.22558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger W. The interaction of sexually and naturally selected traits in the adaptive radiations of cichlid fishes. Mol Ecol. 2009;18:169–185. doi: 10.1111/j.1365-294X.2008.03981.x. [DOI] [PubMed] [Google Scholar]
- Santini F, Harmon LJ, Carnevale G, Alfaro ME. Did genome duplication drive the origin of teleosts? A comparative study of diversification in ray-finned fishes. BMC Evol Biol. 2009;9:194. doi: 10.1186/1471-2148-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos ME, Salzburger W. How Cichlids Diversify. Science. 2012;338:619–621. doi: 10.1126/science.1224818. [DOI] [PubMed] [Google Scholar]
- Sasagawa I, Ishiyama M, Yokosuka H, Mikami M. Teeth and ganoid scales in Polypterus and Lepisosteus, the basic actinopterygian fish: An approach to understand the origin of the tooth enamel. Journal of Oral Biosciences. 2013;55:76–84. [Google Scholar]
- Sato Y, Hashiguchi Y, Nishida M. Temporal pattern of loss/persistence of duplicate genes involved in signal transduction and metabolic pathways after teleost-specific genome duplication. BMC Evol Biol. 2009;9:127. doi: 10.1186/1471-2148-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl M. Evolution of Xmrk: an oncogene, but also a speciation gene? Bioessays. 2008;30:822–832. doi: 10.1002/bies.20807. [DOI] [PubMed] [Google Scholar]
- Schartl M. Beyond the zebrafish: diverse fish species for modeling human disease. Dis Model Mech. 2013 doi: 10.1242/dmm.012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl M, Kneitz S, Wilde B, Wagner T, Henkel CV, Spaink HP, Meierjohann S. Conserved Expression Signatures between Medaka and Human Pigment Cell Tumors. Plos One. 2012;7 doi: 10.1371/journal.pone.0037880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl M, Walter RB, Shen Y, Garcia T, Catchen J, Amores A, Braasch I, Chalopin D, Volff JN, Lesch KP, Bisazza A, Minx P, Hillier L, Wilson RK, Fuerstenberg S, Boore J, Searle S, Postlethwait JH, Warren WC. The genome of the platyfish, Xiphophorus maculatus, provides insights into evolutionary adaptation and several complex traits. Nat Genet. 2013;45:567–572. doi: 10.1038/ng.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl M, Wilde B, Laisney JAGC, Taniguchi Y, Takeda S, Meierjohann S. A Mutated EGFR Is Sufficient to Induce Malignant Melanoma with Genetic Background-Dependent Histopathologies. Journal of Investigative Dermatology. 2010;130:249–258. doi: 10.1038/jid.2009.213. [DOI] [PubMed] [Google Scholar]
- Semon M, Wolfe KH. Reciprocal gene loss between Tetraodon and zebrafish after whole genome duplication in their ancestor. Trends Genet. 2007;23:108–112. doi: 10.1016/j.tig.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Setiamarga DH, Miya M, Yamanoue Y, Azuma Y, Inoue JG, Ishiguro NB, Mabuchi K, Nishida M. Divergence time of the two regional medaka populations in Japan as a new time scale for comparative genomics of vertebrates. Biol Lett. 2009;5:812–816. doi: 10.1098/rsbl.2009.0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Li Q, Chen S, Zhang P, Lian J, Hu Q, Sun B, Jin L, Liu S, Wang Z, Zhao H, Jin Z, Liang Z, Li Y, Zheng Q, Zhang Y, Wang J, Zhang G. Epigenetic modification and inheritance in sexual reversal of fish. Genome Res. 2014 doi: 10.1101/gr.162172.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen XY, Kwan HY, Thevasagayam NM, Prakki SR, Kuznetsova IS, Ngoh SY, Lim Z, Feng F, Chang A, Orban L. The first transcriptome and genetic linkage map for Asian arowana. Mol Ecol Resour. 2013a doi: 10.1111/1755-0998.12212. [DOI] [PubMed] [Google Scholar]
- Shen Y, Garcia T, Pabuwal V, Boswell M, Pasquali A, Beldorth I, Warren W, Schartl M, Cresko WA, Walter RB. Alternative strategies for development of a reference transcriptome for quantification of allele specific expression in organisms having sparse genomic resources. Comp Biochem Physiol Part D Genomics Proteomics. 2013b;8:11–16. doi: 10.1016/j.cbd.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SC, Kim SJ, Lee JK, Ahn do H, Kim MG, Lee H, Lee J, Kim BK, Park H. Transcriptomics and comparative analysis of three antarctic notothenioid fishes. PLoS One. 2012;7:e43762. doi: 10.1371/journal.pone.0043762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidell BD, Vayda ME, Small DJ, Moylan TJ, Londraville RL, Yuan ML, Rodnick KJ, Eppley ZA, Costello L. Variable expression of myoglobin among the hemoglobinless Antarctic icefishes. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:3420–3424. doi: 10.1073/pnas.94.7.3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JLB. A living fish of Mesozoic type. Nature London. 1939;143:455–456. [Google Scholar]
- Smith MM, Okabe M, Joss J. Spatial and temporal pattern for the dentition in the Australian lungfish revealed with sonic hedgehog expression profile. Proceedings of the Royal Society B-Biological Sciences. 2009;276:623–631. doi: 10.1098/rspb.2008.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaink HP, Jansen HJ, Dirks RP. Advances in genomics of bony fish. Brief Funct Genomics. 2013 doi: 10.1093/bfgp/elt046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivakov M, Auer TO, Peravali R, Dunham I, Dolle D, Fujiyama A, Toyoda A, Aizu T, Minakuchi Y, Loosli F, Naruse K, Birney E, Wittbrodt J. Genomic and Phenotypic Characterization of a Wild Medaka Population: Towards the Establishment of an Isogenic Population Genetic Resource in Fish. G3 (Bethesda) 2014 doi: 10.1534/g3.113.008722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star B, Nederbragt AJ, Jentoft S, Grimholt U, Malmstrom M, Gregers TF, Rounge TB, Paulsen J, Solbakken MH, Sharma A, Wetten OF, Lanzen A, Winer R, Knight J, Vogel JH, Aken B, Andersen O, Lagesen K, Tooming-Klunderud A, Edvardsen RB, Tina KG, Espelund M, Nepal C, Previti C, Karlsen BO, Moum T, Skage M, Berg PR, Gjoen T, Kuhl H, Thorsen J, Malde K, Reinhardt R, Du L, Johansen SD, Searle S, Lien S, Nilsen F, Jonassen I, Omholt SW, Stenseth NC, Jakobsen KS. The genome sequence of Atlantic cod reveals a unique immune system. Nature. 2011;477:207–210. doi: 10.1038/nature10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinke D, Salzburger W, Braasch I, Meyer A. Many genes in fish have species specific asymmetric rates of molecular evolution. BMC Genomics. 2006;7:20. doi: 10.1186/1471-2164-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolting KN, Wilson AB. Male pregnancy in seahorses and pipefish: beyond the mammalian model. Bioessays. 2007;29:884–896. doi: 10.1002/bies.20626. [DOI] [PubMed] [Google Scholar]
- Suster ML, Abe G, Schouw A, Kawakami K. Transposon-mediated BAC transgenesis in zebrafish. Nat Protoc. 2011;6:1998–2021. doi: 10.1038/nprot.2011.416. [DOI] [PubMed] [Google Scholar]
- Takechi M, Takeuchi M, Ota KG, Nishimura O, Mochii M, Itomi K, Adachi N, Takahashi M, Fujimoto S, Tarui H, Okabe M, Aizawa S, Kuratani S. Overview of the transcriptome profiles identified in hagfish, shark, and bichir: current issues arising from some nonmodel vertebrate taxa. J Exp Zool B Mol Dev Evol. 2011;316:526–546. doi: 10.1002/jez.b.21427. [DOI] [PubMed] [Google Scholar]
- Takeda H, Shimada A. The art of medaka genetics and genomics: what makes them so unique? Annu Rev Genet. 2010;44:217–241. doi: 10.1146/annurev-genet-051710-151001. [DOI] [PubMed] [Google Scholar]
- Takeuchi M, Okabe M, Aizawa S. The genus Polypterus (bichirs): a fish group diverged at the stem of ray-finned fishes (Actinopterygii) Cold Spring Harb Protoc. 2009a doi: 10.1101/pdb.emo117. 2009:pdb emo117. [DOI] [PubMed] [Google Scholar]
- Takeuchi M, Takahashi M, Okabe M, Aizawa S. Germ layer patterning in bichir and lamprey; an insight into its evolution in vertebrates. Dev Biol. 2009b;332:90–102. doi: 10.1016/j.ydbio.2009.05.543. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, trait shared by 22000 species of ray-finned fish. Genome Res. 2003;13:382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzano DR, Kirschner J, Kamber RA, Zhang E, Weber D, Cellerino A, Englert C, Platzer M, Reichwald K, Brunet A. Mapping Loci Associated With Tail Color and Sex Determination in the Short-Lived Fish Nothobranchius furzeri. Genetics. 2009;183:1385–1395. doi: 10.1534/genetics.109.108670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzano DR, Sharp S, Brunet A. Transposon-Mediated Transgenesis in the Short-Lived African Killifish Nothobranchius furzeri, a Vertebrate Model for Aging. G3 (Bethesda) 2011;1:531–538. doi: 10.1534/g3.111.001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 2009;10:725–732. doi: 10.1038/nrg2600. [DOI] [PubMed] [Google Scholar]
- Vijay N, Poelstra JW, Kunstner A, Wolf JB. Challenges and strategies in transcriptome assembly and differential gene expression quantification. comprehensive in silico assessment of RNA-seq experiments. Mol Ecol. 2013;22:620–634. doi: 10.1111/mec.12014. [DOI] [PubMed] [Google Scholar]
- Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff JN, Bouneau L, Ozouf-Costaz C, Fischer C. Diversity of retrotransposable elements in compact pufferfish genomes. Trends Genet. 2003;19:674–678. doi: 10.1016/j.tig.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Volff JN, Selz Y, Hoffmann C, Froschauer A, Schultheis C, Schmidt C, Zhou Q, Bernhardt W, Hanel R, Bohne A, Brunet F, Segurens B, Couloux A, Bernard-Samain S, Barbe V, Ozouf-Costaz C, Galiana D, Lohse MJ, Schartl M. Gene amplification and functional diversification of melanocortin 4 receptor at an extremely polymorphic locus controlling sexual maturation in the platyfish. Genetics. 2013;195:1337–1352. doi: 10.1534/genetics.113.155952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JT, Li JT, Zhang XF, Sun XW. Transcriptome analysis reveals the time of the fourth round of genome duplication in common carp (Cyprinus carpio) Bmc Genomics. 2012;13 doi: 10.1186/1471-2164-13-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SY, Luo J, Murphy RW, Wu SF, Zhu CL, Gao Y, Zhang YP. Origin of Chinese goldfish and sequential loss of genetic diversity accompanies new breeds. PLoS One. 2013;8:e59571. doi: 10.1371/journal.pone.0059571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkens H. Genes, modules and the evolution of cave fish. Heredity (Edinb) 2010;105:413–422. doi: 10.1038/hdy.2009.184. [DOI] [PubMed] [Google Scholar]
- Williams LJ, Tabbaa DG, Li N, Berlin AM, Shea TP, Maccallum I, Lawrence MS, Drier Y, Getz G, Young SK, Jaffe DB, Nusbaum C, Gnirke A. Paired-end sequencing of Fosmid libraries by Illumina. Genome Res. 2012;22:2241–2249. doi: 10.1101/gr.138925.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winge O. The location of eighteen genes in Lebistes reticulatus. Journal of Genetics. 1927;18:1–43. [Google Scholar]
- Wolfe KH. Yesterday's polyploids and the mystery of diploidization. Nat Rev Genet. 2001;2:333–341. doi: 10.1038/35072009. [DOI] [PubMed] [Google Scholar]
- Woltering JM, Noordermeer D, Leleu M, Duboule D. Conservation and Divergence of Regulatory Strategies at Hox Loci and the Origin of Tetrapod Digits. PLoS Biology. 2014;12:e1001773. doi: 10.1371/journal.pbio.1001773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, Walter K, Abnizova I, Gilks W, Edwards YJ, Cooke JE, Elgar G. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Xiao A, Wang ZX, Hu YY, Wu YD, Luo Z, Yang ZP, Zu Y, Li WY, Huang P, Tong XJ, Zhu ZY, Lin S, Zhang B. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Research. 2013;41 doi: 10.1093/nar/gkt464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Byerly MS, Jackman WR, Jeffery WR. Pleiotropic functions of embryonic sonic hedgehog expression link jaw and taste bud amplification with eye loss during cavefish evolution. Dev Biol. 2009;330:200–211. doi: 10.1016/j.ydbio.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Stock DW, Jeffery WR. Hedgehog signalling controls eye degeneration in blind cavefish. Nature. 2004;431:844–847. doi: 10.1038/nature02864. [DOI] [PubMed] [Google Scholar]
- Yergeau DA, Cornell CN, Parker SK, Zhou Y, Detrich HW. bloodthirsty, an RBCC/TRIM gene required for erythropoiesis in zebrafish. Developmental Biology. 2005;283:97–112. doi: 10.1016/j.ydbio.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Yoshizawa M, Yamamoto Y, O'Quin KE, Jeffery WR. Evolution of an adaptive behavior and its sensory receptors promotes eye regression in blind cavefish. BMC Biol. 2012;10:108. doi: 10.1186/1741-7007-10-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wagh P, Guay D, Sanchez-Pulido L, Padhi BK, Korzh V, Andrade-Navarro MA, Akimenko MA. Loss of fish actinotrichia proteins and the fin-to-limb transition. Nature. 2010;466:234–237. doi: 10.1038/nature09137. [DOI] [PubMed] [Google Scholar]
- Zhao YH, Gozlan RE, Zhang CG. Out of sight out of mind: current knowledge of Chinese cave fishes. Journal of Fish Biology. 2011;79:1545–1562. doi: 10.1111/j.1095-8649.2011.03066.x. [DOI] [PubMed] [Google Scholar]