Abstract
Background
Iron is an essential micronutrient for all living organisms, and virulence and sequestration of iron in pathogenic bacteria are believed to be correlated. As a defence mechanism, potential hosts therefore keep the level of free iron inside the body to a minimum. In general, iron metabolism is well studied for some bacteria (mostly human or animal pathogens). However, this area is still under-investigated for a number of important bacterial pathogens. Aliivibrio salmonicida is a fish pathogen, and previous studies of this bacterium have shown that production of siderophores is temperature regulated and dependent on low iron conditions. In this work we studied the immediate changes in transcription in response to a sudden decrease in iron levels in cultures of A. salmonicida. In addition, we compared our results to studies performed with Vibrio cholerae and Vibrio vulnificus using a pan-genomic approach.
Results
Microarray technology was used to monitor global changes in transcriptional levels. Cultures of A. salmonicida were grown to mid log phase before the iron chelator 2,2’-dipyridyl was added and samples were collected after 15 minutes of growth. Using our statistical cut-off values, we retrieved thirty-two differentially expressed genes where the most up-regulated genes belong to an operon encoding proteins responsible for producing the siderophore bisucaberin. A subsequent pan-transcriptome analysis revealed that nine of the up-regulated genes from our dataset were also up-regulated in datasets from similar experiments using V. cholerae and V. vulnificus, thus indicating that these genes are involved in a shared strategy to mitigate low iron conditions.
Conclusions
The present work highlights the effect of iron limitation on the gene regulatory network of the fish pathogen A. salmonicida, and provides insights into common and unique strategies of Vibrionaceae species to mitigate low iron conditions.
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-015-0342-7) contains supplementary material, which is available to authorized users.
Keywords: Aliivibrio salmonicida, Iron homeostasis, Ferric uptake regulator, Siderophore, Bisucaberin, Microarray
Background
Iron is an essential micronutrient for all living organisms [1-3], and withholding of iron is recognized as a first line of defence against microorganisms (e.g., bacteria) [4,5]. Extremely low iron concentrations create an efficient barrier against potential invading pathogens that may have entered the organism through, for example, an open wound on the skin surface. This defence strategy puts extraordinary pressure on invading pathogens to carry extremely efficient mechanisms to sequester iron from within the host [3,6]. Iron acquisition systems are therefore regarded as important virulence factors. Low iron conditions force pathogens into a stress mode, which results in the down-regulation of genes that encode iron-using and iron-storage proteins, and up-regulation of genes involved in iron acquisition [4,7,8]. Consequently, pathogenic bacteria often express and utilize multiple iron sequestering systems ranging from siderophore based systems, heme uptake systems and systems for uptake of free iron [8].
Although iron is an essential micronutrient, high concentrations of iron in the presence of oxygen are potentially harmful due to formation of oxidative radicals [9]; thus, influx and intracellular processing of iron must be tightly regulated. The ferric uptake regulator (Fur) is the main regulator of genes involved in iron uptake, storage and metabolism, and acts in an iron-dependent manner [10-13]. In E. coli, Fur acts mainly as a transcriptional repressor: at high iron concentrations it binds iron and forms homodimers which suppress the transcription of genes involved in a wide range of metabolic functions. Genes regulated by Fur not only include genes directly involved in iron homeostasis, but also DNA and energy metabolism, redox stress resistance, chemotaxis, bioluminscence and production of toxins and other virulence factors [2,10,14,15]. Fur is therefore regarded as a global regulator. Finally, it is also well established that Fur can indirectly activate gene expression by blocking the expression of the small RNA named RyhB, which typically targets mRNA for degradation. For example, RyhB targets the fur mRNA in a feedback regulation loop, and also targets mRNA that encode iron-using or iron-storing proteins like Bfr, SodB and FumA [16].
Fur recognizes and binds to a site on the DNA known as the Fur-box. Several alternative hypotheses for Fur-boxes have been proposed; for example a palindromic 19 bp site, three 6 bp repeats, and 7–17 motif [17-20]. In 2009, Ahmad and co-workers suggested a Vibrio Fur binding site consensus to be 5′-AATGATAATNATTTCATT-3′ [21]. This Vibrio consensus is similar to the suggested Fur box in other bacteria like Bacillus subtilis, Yersinia pestis, E. coli and Pseudomonas aeruginosa [22-25].
The importance of iron, and the elaborate regulation of iron uptake and homeostasis in bacterial cells in general, has prompted a number of researchers to study the roles of iron with regard to bacterial virulence and pathogenicity. In two recent studies, global responses to low iron conditions in cultures of Vibrio vulnificus and Vibrio cholerae (both human pathogens from the diverse family Vibrionaceae) were studied [11,26]. Here, cultures of V. cholerae and V. vulnificus were grown to mid log phase with iron chelators included in the growth medium from the beginning of the experiments. The results from these two experiments showed up-regulation of genes involved in siderophore biosynthesis and transport: TonB systems, heme transport and utilization, ferrous iron transport, and superoxide dismutase. In addition, the V. vulnificus experiment showed an up-regulation of a Tad-1 cluster.
We are studying the roles of iron in another Vibrionaceae representative, Aliivibrio salmonicida. A. salmonicida is the causative agent of cold-water vibriosis, and possesses several iron acquisition systems that may be important for its pathogenicity [27]. This assumption is based on the observation that the bacterium only produces significant amounts of siderophores when grown at or below 10°C [28,29], which coincides with the observation that outbreaks of cold-water vibriosis are normally associated with temperatures below 10°C [28]. Another intriguing feature of A. salmonicida is that it produces the dihydroxamate siderophore bisucaberin that has not yet been found in other Vibrionaceae representatives [30,31]. The bisucaberin biosynthesis genes (VSAL_I0134-I0136) in A. salmonicida strain LFI1238 are located on a genomic island that has likely been acquired by horizontal gene transfer from an unknown source [27]. The genome of the LFI1238 strain also harbors another siderophore biosynthesis system (VSAL_II0273-VSAL_II0278), which is commonly found in Vibrionaceae. However these latter genes are assumed to be disrupted and are annotated as pseudogenes [27]. Also, the transport of siderophores is carried out through siderophore receptors, and the energy for transport of the iron-siderophore complex across the membrane is provided by TonB systems. Vibrionaceae genomes usually contain 2–3 TonB systems [32-34]. The A. salmonicida genome encodes three TonB systems [27], where the TonB1 gene VSAL_I751 (tonB1) contains a frame-shift mutation and likely produce a non-functional protein.
Here, we have studied the immediate global responses in cultures of A. salmonicida to low iron conditions using microarray, and compared the results with comparable studies in V. cholerae [11] and V. vulnificus [26] using a pan-genome approach. In the two latter studies long-term responses to low iron was monitored (using microarray). We hypothesize that it is the immediate phase that is most critical for bacterial survival during iron starvation. Hence, we wanted to identify the genes that are first affected by low iron conditions, and avoid secondary effects such as unrelated stress responses. Our results provide new insights into how A. salmonicida responds to low iron conditions.
Methods
Bacterial strains, culture conditions and sampling
A. salmonicida strain LFI1238 [27] was cultured in LB medium containing 1% NaCl (Luria-Bertani broth Miller, Difco) at 8°C with 200 rpm in all experiments. To determine the optimal concentration of iron chelator 2,2'-dipyridyl (Sigma-Aldrich), A. salmonicida was grown to an optical density (600 nm) of 0.4 before the culture was split into 6 separate flasks. One flask was kept as control whereas 10–500 μM 2,2'-dipyridyl was added to the remaining cultures.
For Northern blots and microarray analysis (see below), six individual colonies (i.e. biological replicates) of A. salmonicida LFI1238 were grown until they reached an OD600 of approximately 0.5. The replicates were then split into two sub samples: one of these parallels was kept as the control, while 50 μM of the iron chelator 2,2’-dipyridyl was added to the other half. Samples were harvested after 15 min, spun down and frozen for later use.
Total RNA purifications
For microarray analysis and Northern blotting total RNA was extracted from the cell pellets using IsolRNA (5prime) and DNA was removed using the DNA-free kit (Applied Biosystems). DNase-treated total RNA was subsequently run through RNeasy MinElute Cleanup columns (Qiagen) to remove any remaining contaminants, and to further concentrate the RNA. The RNA was finally dissolved in 16 μl RNase free water.
Microarray analysis
cDNA was made by using the Amino Allyl Labeling cDNA Kit as described by the manufacturer protocol (Applied Biosystems). Reactions contained 18 μg total RNA. cDNA samples were labeled with the CyDye™ Post-Labelling Reactive Dye Pack (GE healthcare). Control samples (i.e. untreated samples) were labeled with Cy3, which produces green fluorescent light when scanned (at 532 nm), and treated samples were labeled with Cy5, which produces red light (at 635 nm). Two of the six slides were labeled in the opposite manner, and were used as dye-swap controls to adjust for unequal labeling efficiencies between the fluorescent dyes.
The labelled samples were hybridized to “Vibrio salmonicida V1.0.1 AROS” slides (Eurofins MWG Operon) at 42°C for 20 hours on a TECAN HS4800 hybridisation station, and microarray slides were subsequently washed, once in 0.1 × SSC/0.1% SDS for 5 min at 42°C, then once in 0.1 × SSC/0.1% SDS for 10 min at room temperature, and finally four times in 0.1 × SSC for 1 min at room temperature. Slides were scanned using a GenePix 4000D scanner (Axon Instruments Inc.) at 532 and 635 nm. Images were explored and initial data analyses were performed by using the GenePix Pro v6.1 software. The final analysis of expression data was done using the R-based Limma software.
Northern blot analysis
Northern blot analysis was used to validate the microarray expression data. Treated and untreated RNA samples from each of the six biological replicates were pooled. Ten μg total RNA was separated on 1.2% denaturing formamide agarose gels, and run at 90 V for four hours in 1× MOPS buffer at 4°C. RNA was next transferred to a Hybond-N+ nylon membrane (Amersham) by capillary transfer. Selected gene specific dsDNA was amplified using PCR and labelled with [α-32P] and was used as probes according to the Amersham Megaprime DNA labeling system kit (GE healthcare). Hybridizations were performed over-night at 42°C using ULTRAhyb hybridization solution (Applied biosystems) and signals were acquired on phosphoimaging screens (Fujifilm) and scanned using a BAS-5000 phosphoimager (Fujifilm). Quantification of signals was done using the ImageGauge software v4.0 (Fujifilm) in profile mode. The intensities of the different bands were normalized to 16S rRNA probes.
Computation of core, accessory and unique transcripts
To compare our microarray results with data from similar experiments using V. vulnificus strain CMCP6 [26] and V. cholerae strain 0395 [11], the protein sequences of differentially expressed genes were downloaded from GenBank (using geneID numbers). For V. cholerae the geneIDs are from the genome sequence of V. cholerae O1 N16961 and not strain O395 that was used in the experiment. Accession numbers for the V. cholerae O1 N16961 are AE003852 and AE003853, accession numbers for V. vulnificus CMCP6 genome are AE016795 and AE016796, and finally accession numbers for the A. salmonicida LFI1238 are FM178379 and FM178380. OrthoMCL [35] was used to identify core genes, i.e. genes present in all three genomes. Genes up-regulated in two datasets were denoted as accessory transcripts and unique transcripts were up-regulated in one dataset. Percent identity cut-off and percent match cut-offs were set to 50%. The inflation parameter was set to 0.
Ethics statement
The research presented in this paper do not involve human subjects, and we see no ethical issues.
Results and discussion
Microarray analysis of iron depleted A. salmonicida
Using our model organism A. salmonicida we tested the immediate global changes in gene expression in response to low-iron stress conditions using a microarray approach. The iron chelator 2,2'-dipyridyl was used to create iron-limited conditions, and the appropriate chelator concentration was found by comparing the growth of A. salmonicida in the absence or presence of various 2,2'-dipyridyl concentrations. As shown in Figure 1, the growth of the bacterium was clearly affected when the growth medium contained 100 or 500 μM 2,2'-dipyridyl. Based on this result we decided to use 50 μM 2,2'-dipyridyl because it resulted in only a slight growth reduction, and we assumed that a strong growth inhibition would induce broader and less relevant stress responses.
Figure 1.
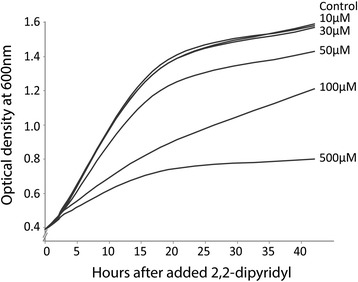
Titration of 2,2’-dipyridyl concentration. A. salmonicida strain LFI1238 was grown in LB containing 1% NaCl to an optical density at 0.4 (600 nm). The culture was split into six individual flasks and supplemented with different concentrations of 2,2'-dipyridyl before growth was monitored for 44 hours. Culture treated with 50 μM 2,2'–dipyridyl showed a slight reduction in growth and this concentration was therefore used in all subsequent experiments.
Samples for microarray analysis were prepared by growing A. salmonicida in LB with 1% NaCl at 8°C. A. salmonicida requires NaCl for growth, and the NaCl concentration of the medium is known to affect growth, motility and other activities [36]. In our experiment we used a NaCl concentration close to the physiological conditions the bacterium would experience inside its natural host (Atlantic salmon) [37], as well as temperature where up-regulation of iron sequestration systems is known to occur, and the bacterium is known to develop cold water vibriosis [28,29]. The cultures were grown to mid log phase (OD600nm 0.5) before 2,2’-dipyridyl was added to a final concentration of 50 μM, and samples were collected after 15 minutes to monitor immediate responses.
Table 1 lists 32 differentially expressed genes that fulfilled our criteria (fold change values ≥ 1.5 and p-values ≤ 0.10). These threshold values were chosen after evaluating alternative combinations of cut-off values, and evaluating the biological soundness of the resulting data (i.e. keep maximum valuable data while minimizing the introduction of noise). Also, biological replicates tend to create more variation between samples compared to technical replicates, and too strict cut-off values can therefore exclude biologically sound data. In our analysis, all differentially expressed genes (Table 1) are up-regulated (in treated sample) and associated with predicted Fur-boxes [21]. Moreover, based on the current annotations, the majority (at least 21 of 32) of genes have predicted functions in iron homeostasis. The operon associated with the highest fold change values contains three genes (bibABC) for biosynthesis of the siderophore bisucaberin. Interestingly, of all sequenced bacteria in the relatively large Vibrionaceae family, A. salmonicida was until recently the only representative with this system. Using the amino acid sequences for the bibABC genes in a Blast search we identified homologous genes (99% identity) in the very closely related Aliivibrio logei [38]. This observation favours that the system was acquired by horizontal gene transfer in the most recent common ancestor of the two Aliivibrio species. This scenario is more parsimonious than the alternative, which is that the system was lost in all other Vibrionaceae representatives. Other genes on the list with functions in iron metabolism include siderophore receptors, heme receptors and the associated ATP-binding cassette (ABC) and TonB transport systems. The gene encoding ferrioxamine B receptor (BfrH) is possibly encoding a siderophore receptor (ferrioxamines are siderohores). The same operon encodes a TonB3 system. The operon encoding FhuC and FhuD (associated with siderophore-iron transport) is also up-regulated under iron limited conditions. The CDSs encoding TolR2, a TonB dependent receptor and a putatively exported protein are located in the same operon and are all up-regulated. A recent publication suggests that TolR is likely TtpC, which is necessary for stabilisation of TonB2 binding in Vibrio anguillarum and V. cholerae [39]. TonB1 is the only TonB, which appears to be up-regulated. Apparently, this TonB is most likely non-functional due to a frameshift in A. salmonicida.
Table 1.
Differentially expressed genes in A. salmonicida LFI1238 stimulated with 50 μM 2,2'-dipyridyl
| CDS | Gene | Product 1 | Fold change 2 | p-value 2 |
|---|---|---|---|---|
| Transport/binding proteins | ||||
| VSAL_I1734 | heme receptor (pseudogene) | 1.5 | 0.03 | |
| VSAL_I1751 | tonB1 | TonB protein (pseudogene) | 5.0 | 0 |
| VSAL_I1752 | exbB1 | TonB system transport protein | 2.4 | 0.01 |
| VSAL_I1754 | hmuT | heme transporter protein, putative periplasmic binding protein | 4.3 | 0 |
| VSAL_I2257 | feoA | ferrous iron transport protein FeoA | 1.8 | 0.06 |
| VSAL_I2258 | feoB | ferrous iron transport protein FeoB | 1.8 | 0.07 |
| VSAL_I2259 | feoC | ferrous iron transport protein FeoC | 1.8 | 0 |
| VSAL_I2588 | fbpA | iron(III) ABC transporter, periplasmic iron-compound-binding protein | 2.1 | 0.08 |
| VSAL_II0110 | TonB dependent receptor | 3.4 | 0 | |
| VSAL_II0112 | tolR2 | biopolymer transport protein | 2.0 | 0 |
| VSAL_II0150 | fhuC | ferrichrome transport ATP-binding protein | 3.2 | 0 |
| VSAL_II0151 | fhuD | ferrichrome-binding periplasmic protein | 3.2 | 0.01 |
| VSAL_II0909 | bfrH | ferrioxamine B receptor | 3.3 | 0 |
| VSAL_p320_27 | iron ion ABC transporter, periplasmic component | 2.4 | 0.01 | |
| VSAL_p320_29 | iron ion ABC transporter ATP-binding protein | 1.7 | 0.07 | |
| Adaptation | ||||
| VSAL_I1749 | huvX | heme uptake and utilization protein | 1.7 | 0 |
| Biosynthesis of cofactors, carriers | ||||
| VSAL_I0134 | bibA* | Bisucaberin siderophore biosynthesis protein A | 7.6 | 0 |
| VSAL_I0135 | bibB* | Bisucaberin siderophore biosynthesis protein B | 5.8 | 0.01 |
| VSAL_I0136 | bibC* | Bisucaberin siderophore biosynthesis protein C | 1.9 | 0.06 |
| VSAL_I1750 | phuW | putative coproporphyrinogen oxidase | 2.2 | 0 |
| Cell envelope | ||||
| VSAL_I1248 | membrane protein | 2.9 | 0 | |
| VSAL_I1785 | putative exported protein | 2.2 | 0 | |
| VSAL_I1786 | putative iron-regulated protein | 2.8 | 0 | |
| VSAL_I1864 | putative outer membrane protein | 4.2 | 0 | |
| VSAL_II0074 | membrane protein | 3.4 | 0 | |
| VSAL_II0111 | putative exported protein | 2.3 | 0 | |
| VSAL_II0717 | putative membrane protein | 1.6 | 0.02 | |
| VSAL_II0868 | putative lipoprotein | 3.4 | 0 | |
| sRNA | ||||
| VSAL_I3102s | VSsrna22 small RNA RyhB | 4.6 | 0 | |
| Unknown function, no known homologues | ||||
| VSAL_I2980 | hypothetical protein | 1.5 | 0.1 | |
| VSAL_I2892 | hypothetical protein | 3.7 | 0 | |
| VSAL_II0148 | hypothetical protein | 3.8 | 0 | |
1Annotated product of CDS 2Fold change values are shown for ≥ 1.5 differentially expressed genes with p-values ≤ 0.1. Positive fold change value indicate up-regulation compared to untreated control. * bibA is annotated as L-2,4-diaminobutyrate decarboxylase in the A. salmonicida genome annotations, and bibB and bibC are annotated as iucD and iucC, respectively [26,30].
None of the differentially expressed genes on our lists were down-regulated (i.e. did not fulfil the cut-off criteria). This is surprising since ryhB is moderately up-regulated (4.6 fold) under low iron conditions, and down-regulation of known RyhB targets is expected based on evidence from other species. There are two possible explanations for this finding: the data is valid and all significantly differentially expressed genes are up-regulated, however we cannot rule out that unknown technical issues have affected our data leading to this result. Although we have not validated any potential RyhB targets by Northern blot analysis, the overall agreement between fold change values in our microarray and Northern blot data (see below) support the conclusion that the microarray data is valid and we have no reason to suspect serious technical issues. Known RyhB targets that are identified in both E. coli and V. cholerae include sodB, sdhC, fumA and gltB1 [40,41]. All these are present in our dataset, but are not differentially expressed (fold change −1.03, −1.16, −1.10 and −1.27, respectively). In V. cholerae, 31 genes are up-regulated in a ryhB null mutant; however, the fold change values for these potential RyhB targets are very moderate (majority varies between 1.6 − 3.3 fold) [41]. Similarly, when RyhB is over-expressed in E. coli, fold change values for the majority of down-regulated genes vary between 1.5 − 6 [40]. Based on this information, it may not be surprising that secondary effects such as for example RyhB regulation is not detected in our experiment considering that; i) RyhB is only moderately up-regulated, ii) A. salmonicida has a relatively long doubling time (6–8 hours) at 8°C, and iii) we measured effects after a very short exposure time (15 min) to the iron chelator.
Northern blot analysis
Northern blot analysis was used to validate the microarray expression data of 5 up-regulated genes; VSAL_I0134, VSAL_I0135, VSAL_I0448, VSAL_II0148, VSAL_II0110. The intensities of the different bands were normalized to 16S rRNA. Autoradiogram pictures are shown in Figure 2, and show that the Northern blot data are in good overall agreement with the microarray analysis. For example, for VSAL_I0135 (bibB) the microarray and Northern blot analyses show fold change values of 5.8 and 5.2, respectively, and for VSAL_II0110 the respectively fold change values were 4.5 and 3.2. The microarray fold change value that differs most in magnitude from the Northern Blot result is for VSAL_I0134 (bibA). Here, the microarray and Northern Blot values were 7.6 and 14.7, respectively.
Figure 2.
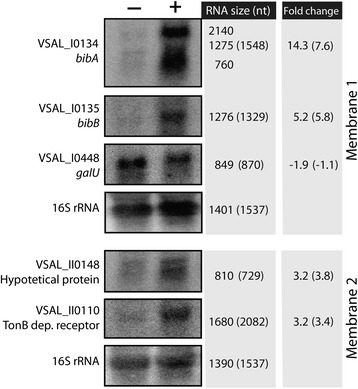
Validation of selected microarray results using Northern blot analysis. RNAs from six replicates of A. salmonicida were pooled and separated on denaturating 1.2% formamide agarose gels, transferred to two membranes and tested for presence of specific RNAs using radio-labeled probes. Plus (+) indicates addition of 50 μM of the iron chelator 2,2'-dipyridyl to the cultures 15 min prior to sampling, whereas minus (−) indicates the untreated control. Numbers in the left column indicate the size of the RNA as measured from the gel, and numbers in parentheses indicate the theoretical size. The right column indicate normalized fold change values calculated from the Northern blot autoradiograms, while numbers in parentheses show the corresponding microarray fold change values.
Comparison of results with other global expression profiling studies from Vibrionaceae
Next, we wanted to compare our result to similar global expression profiling studies (microarray) where the response of other representatives of the Vibrionaceae family to low-iron conditions was studied. By uncovering responses that are shared between bacteria belonging to the Vibrionaceae family, or that are unique to one species, we may eventually provide a deeper understanding of mechanisms involved in virulence. Two such datasets are currently available: Crosa and co-workers [26] tested responses of V. vulnificus strain CMCP6 to iron-limiting conditions by adding 50 μM ethylenediamine-di-(o-hydroxyphenylacetic) acid (EDDA) (iron-depleted conditions) to TSBS medium cultures from the beginning, in addition to untreated controls, and harvested cells at mid-log phase (i.e. OD600nm 0.6-0.8). Three biological replicates were pooled before cDNA synthesis to avoid culture variations in microarray analysis. In their analysis they were able to identify 49 genes that are differentially up-regulated during iron-depleted conditions. In another study by Mey et al. [11], V. cholerae strain 0395 was grown in EZ RDM defined medium with no added iron (i.e. iron-depleted conditions), or EZ RDM with 40 μM of ferrous sulfate (iron-replete conditions) to OD650nm 0.3. In their study, they identified 84 differentially expressed genes during iron-depleted conditions.
In our comparative analysis we adopted the Pan genome concept and organized the differentially-expressed transcripts into core, accessory and unique transcripts. A Venn-diagram representation of the comparative analysis are shown in Figure 3. More detailed information about the comparative analysis results are found in Additional file 1. Core transcripts are differentially expressed in all datasets, unique transcripts are differentially expressed in one dataset, whereas accessory transcripts are differentially expressed in two datasets. Although the three experiments were performed differently (e.g., different growth media, different iron chelators/iron-deplete medium, different sampling time/cell densities, etc.), we believe they could identify potentially interesting common or unique responses to low iron conditions among the three bacteria.
Figure 3.
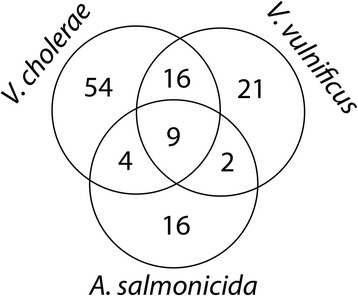
Venn diagram summarizing numbers of transcripts that are differentially expressed in A. salmonicida , V. cholerae and V. vulnificus under low-iron conditions. Numbers are based on this study, and the microarray studies using V. cholerae [11] and V. vulnificus [26].
We used the software OrthoMCL [35] with percent identity and percent match cut-off set to 50 and the inflation value set to 0 to identify potential homologs. Subsequently, we curated the generated homology clusters manually and identified 9 core transcripts (10 in V. vulnificus as VV1_1660 and VV1_1661 are paralogs, and both cluster together with VC0608 in V. cholerae and VSAL_I2588 in A. salmonicida). Three of the core transcripts belonging to a ferrous iron transport system (feoA, feoB and feoC), one transcript belonging to a TonB system (exbB1), two transcripts encode proteins that are potentially involved in heme uptake/utilization, two transcripts which may belong to a TonB2 system, and finally one transcript encoding a ferric iron ABC transporter periplasmic iron-compound-binding protein. Therefore, all differentially expressed core transcripts encode proteins involved in iron homeostasis.
Sixteen accessory transcripts are shared between V. vulnificus and V. cholerae. These encode products involved in siderophore biosynthesis, siderophore and heme transport and utilization, iron storage (bfd and bfr), and oxidative stress response (sodA). Of the 16 accessory genes shared between V. cholerae and V. vulnificus, ten are not present in the A. salmonicida genome (e.g. bfr, bfd and the different vtc component genes). Moreover, the finding that three siderophore biosynthesis sequences (VC0771/VV2_0838, VC0773/VV2_0835 and VC0774/VV2_0834) are shared only between V. vulnificus and V. cholerae does not seem reasonable, and may reflect the fact that some siderophore biosynthesis proteins are more related between V. vulnificus and V. cholerae than they are to the bisucaberin biosynthesis system in A. salmonicida. Four accessory transcripts are shared between V. cholerae and A. salmonicida (VSAL_I2892/VC0091, VSAL_I1786/VC1264, VSAL_I1785/VC1265 and VSAL_II0074/VC1588), and finally two accessory transcripts are shared between A. salmonicida and V. vulnificus (i.e. TonB1 VSAL_I1751 and VV21614, and a periplasmic heme binding protein encoded by VSAL_I1754 and VV21611). The fact that V. cholerae and V. vulnificus share the highest number (i.e. sixteen) of common up-regulated transcripts is reasonable since they are more closely related to each other than to A. salmonicida. In addition, the experimental conditions used for V. cholerae and V. vulnificus are more similar.
Fifty-four, twenty-one and sixteen transcripts are unique to V. cholerae, V. vulnificus and A. salmonicida, respectively, and of these at least eighteen, eight and ten transcripts are directly associated with iron homeostasis. In V. cholerae the unique transcripts encode proteins with functions in vibriobactin biosynthesis (vibD-H), siderophore transport (viuA, viuC, viuD, viuG, viuP, irgA, vctA, fhuA and fhuC), heme transport (hutA and hutD), iron transport (tonB1, tonB2 and exbD2), transcription regulation (irgB and vctR), various enzymatic catalysis (e.g. ligA-2, fumC, ptrB, napA-D, napF, menB), and finally hypothetical functions. In V. vulnificus half of the eight unique transcripts encode proteins that are involved in vulnibactin biosynthesis and transport (VV20839, VV20840, VV20841 and VV20844), and the remaining four have functions in iron transport, i.e. TonB systems (VV21618, VV20359 and VV20360) and an ABC-type Fe3+ transport protein (VV11662). In A. salmonicida unique transcripts are directly associated with bisucaberin biosynthesis genes (bibA, bibB and bibC), ferrioxamine B receptor (bfrH), ferrichrome binding (fhuC and fhuD), iron transport, i.e. an ABC transport system (VSAL_p320_27 and VSAL_p320_29), a TonB2 dependent receptor (VSAL_II0110), and a heme receptor (VSAL_I1734).
Siderophore biosynthesis in Vibrionaceae
The approximately 150 different Vibrionaceae species (157 species in the NCBI taxonomy database when excluding unclassified sp. and subspecies) [42] have the potential to synthesize various siderophore iron chelators. For example, V. cholerae encodes the system VibABCDEFH that is responsible for the production of vibriobactin. Similarly, A. salmonicida contains the bibABC genes, which encode enzymes involved in production of the bisucaberin siderophore [30]. V. vulnificus produces the species specific siderophore vulnibactin [43]. Vulnibactin is structurally similar to vibriobactin, but its biosynthesis pathway is not fully understood [44]. The genes venB, vvsA and vvsB are involved in the biosynthesis, but their roles are unclear. V. vulnificus also synthesizes a hydroxamate-type siderophore. Unfortunately, neither its structure nor its biosynthetic pathway have been identified [45].
In our analysis the three genes involved in bisucaberin biosynthesis in A. salmonicida top our list of differentially up-regulated genes/operons. The result resembles expression data from both V. cholerae [11] and V. vulnificus [26] where siderophore biosynthesis genes were highly up-regulated (although they did not top the list of up-regulated genes) after being grown in low iron medium. Together these results strongly support the idea that siderophore production and utilization represent one of the first and probably most important responses to mitigate low iron conditions. It is however still unclear why different Vibrio/Aliivibrio species use different siderophores. One possible explanation is that the utilization of multiple siderophores represents an advantage in the competition for scarce resources. However, some vibrios can partly mitigate such strategies by utilizing siderophores produced by other bacteria.
TonB systems
In Gram negative bacteria the transport of ferri-siderophores and heme across the membrane requires energy. The energy is provided by TonB systems, which consist of the TonB, ExbB and ExbD proteins. In vibrios TonB2 systems also include the TtpC protein [1,39]. Vibrio genomes typically contain two or three TonB systems [32,46]. Interestingly, in our analysis tonB1, tonB2 and exbD2 from V. cholerae, and tonB2 from V. vulnificus, are considered unique, whereas the remaining TonB transcripts (exbB1, exbB2 and exbD1) are either core or accessory transcripts. tonB1 is shared between A. salmonicida and V. vulnificus even though the A. salmonicida tonB1 gene is a pseudogene (contains one frameshift mutation). tonB2 transcripts were not identified in the A. salmonicida microarray dataset. Intriguingly the V. cholerae tonB1 transcript was considered unique, but after further examinations we realized that this transcript was excluded from the results because it was just below the cut-off settings for identities, while the TonB1 transcripts from V. vulnificus and A. salmonicida were just above the cut-off setting. This show the weaknesses of a small dataset and the problems of setting specific cut-offs.
Conclusions
We studied the immediate effect of low iron conditions, and compared this to similar studies where effects were examined after prolonged growth in low iron conditions. We identified 32 up-regulated genes, whereas no genes were found to be down-regulated. Although caution should be taken in extrapolating in vitro results to what may occur in vivo, it is our belief that studies such as those performed here will provide a better understanding of iron uptake and metabolism in bacteria, and eventually provide us with some insights into their virulence and survival mechanisms, their ability to adapt to changing environmental conditions, and finally their evolution. We have studied expression of genes that are essential for iron homeostasis in a single species, but by studying a collection of species from a broader spectrum of bacteria e.g., from the same family (i.e. Vibrionaceae), unique and common strategies for mitigating low iron conditions can be identified. A future goal for us is to use such knowledge to compare environmental isolates with known pathogens to better understand the relevance of iron homeostasis in virulence. Finally, increased knowledge on iron uptake systems and regulation is highly relevant to on-going efforts where such systems are used as targets for potential drugs with the goal to control pathogenic bacteria.
Availability of supporting data
Microarray data are available in the ArrayExpress database under accession number GSE57996.
Acknowledgements
The authors would like to acknowledge Ph.D. Geir Å. Hansen for assistance with Northern blot analysis, and The Microarray Resource Center in Tromsø (MRCT) for offering facilities and equipment. We also thank Adele Kim Williamson (UiT-The Arctic University of Norway) for final proofreading of the manuscript. The project was funded by the The Norwegian Research Council through the The National graduate school in structural biology (BioStruct), and UiT-The Arctic University of Norway.
Abbreviations
- PCR
Polymerase chain reaction
- LB
Luria Bertani broth
- Fur
Ferric uptake regulator
- dsDNA
Double-stranded DNA
- rpm
Rounds per minute
- OD
Optical density
- min
minutes
- MOPS
3-(N-morpholino)propanesulfonic acid
- SSC
Saline-sodium citrate
- SDS
Sodium dodecyl sulfate
- CDS
Coding sequence
- rRNA
Ribosomal RNA
- mRNA
Messenger RNA
- sRNA
Small RNA
- AS
Aliivibrio salmonicida
- ABC
ATP-binding cassette
Additional file
The table lists differentially expressed (up-regulated) core, accessory and unique transcripts in A. salmonicida, V. vulnificus and V. cholerae. Amino acid sequences of corresponding genes were retrieved from ENA, and used as input for clustering of orthologs using OrthoMCL with amino acid percent match and percent identity cut-offs set to 50%.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
PH and HH conceived of the study and designed experiments. HH supervised the microarray analysis, and PH supervised the study and helped drafting the manuscript. SKT participated in the design of experiments, performed the wet-lab experiments, initial microarray data analysis and Northern data analysis, and drafted the manuscript. TK performed the pan-genome analysis, and EMR performed the Limma based microarray data analysis. All authors read and approved the final manuscript.
Contributor Information
Sunniva Katharina Thode, Email: sunniva.k.thode@uit.no.
Tim Kahlke, Email: kah023@csiro.au.
Espen Mikal Robertsen, Email: espen.m.robertsen@uit.no.
Hilde Hansen, Email: hilde.hansen@uit.no.
Peik Haugen, Email: peik.haugen@uit.no.
References
- 1.Andrews SC, Robinson AK, Rodríguez-Quiñones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–37. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 2.McHugh JP, Rodriguez-Quinones F, Abdul-Tehrani H, Svistunenko DA, Poole RK, Cooper CE, et al. Global iron-dependent gene regulation in Escherichia coli. J Biological Chem. 2003;278:29478–86. doi: 10.1074/jbc.M303381200. [DOI] [PubMed] [Google Scholar]
- 3.Miethke M. Molecular strategies of microbial iron assimilation: from high-affinity complexes to cofactor assembly systems. Metallomics. 2013;5:15–28. doi: 10.1039/c2mt20193c. [DOI] [PubMed] [Google Scholar]
- 4.Collins HL. Withholding iron as a cellular defence mechanism – friend or foe? Eur J Immunol. 2008;38:1803–6. doi: 10.1002/eji.200838505. [DOI] [PubMed] [Google Scholar]
- 5.Ellis AE. Innate host defense mechanisms of fish against viruses and bacteria. Dev Comp Immunol. 2001;25:827–39. doi: 10.1016/S0145-305X(01)00038-6. [DOI] [PubMed] [Google Scholar]
- 6.Thompson FL, Iida T, Swings J. Biodiversity of Vibrios. Microbiol Mol Biol Rev. 2004;68:403–31. doi: 10.1128/MMBR.68.3.403-431.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaible UE, Kaufmann SHE. Iron and microbial infection. Nat Rev Microbiol. 2004;2:946–53. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 8.Wooldridge KG, Williams PH. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol Rev. 1993;12:325–48. doi: 10.1111/j.1574-6976.1993.tb00026.x. [DOI] [PubMed] [Google Scholar]
- 9.Touati D. Iron and oxidative stress in bacteria. Arch of Biochem Biophys. 2000;373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- 10.Hantke K. Iron and metal regulation in bacteria. Curr Opin Microbiol. 2001;4:172–7. doi: 10.1016/S1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- 11.Mey AR, Wyckoff EE, Kanukurthy V, Fisher CR, Payne SM. Iron and Fur regulation in Vibrio cholerae and the role of Fur in virulence. Infect Immun. 2005;73:8167–78. doi: 10.1128/IAI.73.12.8167-8178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Troxell B, Hassan HM. Transcriptional regulation by ferric uptake regulator (Fur) in pathogenic bacteria. Front Cel Infect Microbiol. 2013;3:59. doi: 10.3389/fcimb.2013.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheikh MA, Taylor GL. Crystal structure of the Vibrio cholerae ferric uptake regulator (Fur) reveals insights into metal co-ordination. Mol Microbiol. 2009;72:1208–20. doi: 10.1111/j.1365-2958.2009.06718.x. [DOI] [PubMed] [Google Scholar]
- 14.Escolar L, Perez-Martin J, de Lorenzo V. Opening the iron Box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–9. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyckoff EE, Mey AR, Payne SM. Iron acquisition in Vibrio cholerae. BioMetals. 2007;20:405–16. doi: 10.1007/s10534-006-9073-4. [DOI] [PubMed] [Google Scholar]
- 16.Hoe C-H, Raabe CA, Rozhdestvensky TS, Tang T-H. Bacterial sRNAs: regulation in stress. Int J Med Microbiol. 2013;303:217–29. doi: 10.1016/j.ijmm.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 17.De Lorenzo V, Giovannini F, Herrero M, Neilands JB. Metal ion regulation of gene expression: Fur repressor-operator interaction at the promoter region of the aerobactin system of pCoIV-K30. J Mol Biol. 1988;203:875–84. doi: 10.1016/0022-2836(88)90113-1. [DOI] [PubMed] [Google Scholar]
- 18.Davies BW, Bogard RW, Mekalanos JJ. Mapping the regulon of Vibrio cholerae ferric uptake regulator expands its known network of gene regulation. Proc Natl Acad Sci U S A. 2011;108:12467–72. doi: 10.1073/pnas.1107894108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Escolar L, Perez-Martin J, de Lorenzo V. Binding of the fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J Mol Biol. 1998;283:537–47. doi: 10.1006/jmbi.1998.2119. [DOI] [PubMed] [Google Scholar]
- 20.Baichoo N, Helmann JD. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J Bacteriol. 2002;184:5826–32. doi: 10.1128/JB.184.21.5826-5832.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmad R, Hjerde E, Hansen GA, Haugen P, Willassen NP. Prediction and experimental testing of ferric uptake regulator regulons in vibrios. J Mol Microbiol Biotechnol. 2009;16:159–68. doi: 10.1159/000128322. [DOI] [PubMed] [Google Scholar]
- 22.Fuangthong M, Helmann JD. Recognition of DNA by three ferric uptake regulator (Fur) homologs in Bacillus subtilis. J Bacteriol. 2003;185:6348–57. doi: 10.1128/JB.185.21.6348-6357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou D, Qin L, Han Y, Qiu J, Chen Z, Li B, et al. Global analysis of iron assimilation and fur regulation in Yersinia pestis. FEMS Microbiol Lett. 2006;258:9–17. doi: 10.1111/j.1574-6968.2006.00208.x. [DOI] [PubMed] [Google Scholar]
- 24.Gao H, Zhou D, Li Y, Guo Z, Han Y, Song Y, et al. The iron-responsive Fur regulon in Yersinia pestis. J Bacteriol. 2008;190:3063–75. doi: 10.1128/JB.01910-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z, Lewis KA, Shultzaberger RK, Lyakhov IG, Zheng M, Doan B, et al. Discovery of Fur binding site clusters in Escherichia coli by information theory models. Nucleic Acids Re. 2007;35:6762–77. doi: 10.1093/nar/gkm631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alice AF, Naka H, Crosa JH. Global gene expression as a function of the iron status of the bacterial cell: influence of differentially expressed genes in the virulence of the human pathogen Vibrio vulnificus. Infect Immun. 2008;76:4019–37. doi: 10.1128/IAI.00208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hjerde E, Lorentzen M, Holden M, Seeger K, Paulsen S, Bason N, et al. The genome sequence of the fish pathogen Aliivibrio salmonicida strain LFI1238 shows extensive evidence of gene decay. BMC Genomics. 2008;9:616. doi: 10.1186/1471-2164-9-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Enger O, Husevag B, Goksoyr J. Seasonal variations in precense of Vibrio salmonicida and total bacterial counts in Norwegian fish-farm water. Can J Microbiol. 1991;37:618–23. doi: 10.1139/m91-105. [DOI] [PubMed] [Google Scholar]
- 29.Colquhoun DJ, Sørum H. Temperature dependent siderophore production in Vibrio salmonicida. Microb Pathog. 2001;31:213–9. doi: 10.1006/mpat.2001.0464. [DOI] [PubMed] [Google Scholar]
- 30.Winkelmann G, Schmid DG, Nicholson G, Jung G, Colquhoun DJ. Bisucaberin–a dihydroxamate siderophore isolated from Vibrio salmonicida, an important pathogen of farmed Atlantic salmon (Salmo salar) BioMetals. 2002;15:153–60. doi: 10.1023/A:1015206419613. [DOI] [PubMed] [Google Scholar]
- 31.Kadi N, Song L, Challis GL. Bisucaberin biosynthesis: an adenylating domain of the BibC multi-enzyme catalyzes cyclodimerization of N-hydroxy-N-succinylcadaverine. Chem Commun (Camb) 2008;41:5119–21. doi: 10.1039/b813029a. [DOI] [PubMed] [Google Scholar]
- 32.Kustusch RJ, Kuehl CJ, Crosa JH. The ttpC gene is contained in two of three TonB systems in the human pathogen Vibrio vulnificus, but only one is active in iron transport and virulence. J Bacteriol. 2012;194:3250–9. doi: 10.1128/JB.00155-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seliger SS, Mey AR, Valle AM, Payne SM. The two TonB systems of Vibrio cholerae: redundant and specific functions. Mol Microbiol. 2001;39:801–12. doi: 10.1046/j.1365-2958.2001.02273.x. [DOI] [PubMed] [Google Scholar]
- 34.Stork M, Di Lorenzo M, Mourino S, Osorio CR, Lemos ML, Crosa JH. Two tonB systems function in iron transport in Vibrio anguillarum, but only one is essential for virulence. Infect Immun. 2004;72:7326–9. doi: 10.1128/IAI.72.12.7326-7329.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–89. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karlsen C, Paulsen SM, Tunsjø HS, Krinner S, Sørum H, Haugen P, et al. Motility and flagellin gene expression in the fish pathogen Vibrio salmonicida: effects of salinity and temperature. Microb Pathog. 2008;45:258–64. doi: 10.1016/j.micpath.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 37.Bergheim A, Kroglund F, Vatne DF, Rosseland BO. Blood plasma parameters in farmed Atlantic salmon (Salmo salar L.) transferred to sea cages at age eight to ten months. Aquaculture. 1990;84:159–65. doi: 10.1016/0044-8486(90)90345-N. [DOI] [Google Scholar]
- 38.Dikow RB, Smith WL. Genome-level homology and phylogeny of Vibrionaceae (Gammaproteobacteria: Vibrionales) with three new complete genome sequences. BMC Microbiol. 2013;13:80. doi: 10.1186/1471-2180-13-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stork M, Otto BR, Crosa JH. A novel protein, TtpC, is a required component of the TonB2 complex for specific iron transport in the pathogens Vibrio anguillarum and Vibrio cholerae. J Bacteriol. 2007;189:1803–15. doi: 10.1128/JB.00451-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Massé E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol. 2005;187:6962–71. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davis BM, Quinones M, Pratt J, Ding Y, Waldor MK. Characterization of the small untranslated RNA RyhB and its regulon in Vibrio cholerae. J Bacteriol. 2005;187:4005–14. doi: 10.1128/JB.187.12.4005-4014.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.NCBI Vibrionaceae taxonomy database http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=641
- 43.Okujo N, Saito M, Yamamoto S, Yoshida T, Miyoshi S, Shinoda S. Structure of vulnibactin, a new polyamine-containing siderophore from Vibrio vulnificus. Biometals. 1994;7:109–16. doi: 10.1007/BF00140480. [DOI] [PubMed] [Google Scholar]
- 44.Lemos M, Osorio CR. Iron uptake in Vibrio and Aeromonas. In: Cornelis P, Andrews SC, editors. Iron uptake and homeostasis in microorganisms. Great Britain: Caister Academic Press; 2010. pp. 117–41. [Google Scholar]
- 45.Simpson LM, Oliver JD. Siderophore production by Vibrio vulnificus. Infect Immun. 1983;41:644–9. doi: 10.1128/iai.41.2.644-649.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuehl CJ, Crosa JH. The TonB energy transduction systems in Vibrio species. Future Microbiol. 2010;5:1403–12. doi: 10.2217/fmb.10.90. [DOI] [PMC free article] [PubMed] [Google Scholar]


