Abstract
HERC2 is a large E3 ubiquitin ligase with multiple structural domains that has been implicated in an array of cellular processes. Mutations in HERC2 are linked to developmental delays and impairment caused by nervous system dysfunction, such as Angelman Syndrome and autism-spectrum disorders. However, HERC2 cellular activity and regulation remain poorly understood. We used a broad proteomic approach to survey the landscape of cellular proteins that interact with HERC2. We identified nearly 300 potential interactors, a subset of which we validated binding to HERC2. The potential HERC2 interactors included the eukaryotic translation initiation factor 3 complex, the intracellular transport COPI coatomer complex, the glycogen regulator phosphorylase kinase, beta-catenin, PI3 kinase, and proteins involved in fatty acid transport and iron homeostasis. Through a complex bioinformatic analysis of potential interactors, we linked HERC2 to cellular processes including intracellular protein trafficking and transport, metabolism of cellular energy, and protein translation. Given its size, multidomain structure, and association with various cellular activities, HERC2 may function as a scaffold to integrate protein complexes and bridge critical cellular pathways. This work provides a significant resource with which to interrogate HERC2 function more deeply and evaluate its contributions to mechanisms governing cellular homeostasis and disease.
Keywords: HERC2, UBE3A (E6AP), eIF3, translation, translational initiation, intracellular transport, vesicle-mediated transport, cellular energy metabolism
Introduction
HERC2 is one of the six members of the HERC family of proteins, each of which possesses at least one regulator of chromosome condensation 1 (RCC1)-like domain (RLD) and a homologous to E6AP carboxy terminus (HECT) domain characteristic of a group of ubiquitin ligases.1−3 The HERC2 protein is comprised of 4834 amino acids with a molecular weight exceeding 500 kDa and contains multiple structural domains.2,4 Mutations in the HERC2 gene have been linked to neurodevelopmental delay and dysfunction seen in both Angelman Syndrome and autism-spectrum disorders;5,6 however the role of HERC2 in these disorders is unknown and underlies the need for a greater understanding of HERC2 function.
To date, the most widely studied function of HERC2 involves the cellular response to DNA damage. HERC2 has been reported to interact with RNF8 at double-stranded DNA breaks, and HERC2 expression is required for RNF8-mediated histone ubiquitylation, as well as the recruitment and retention of downstream repair factors.7 HERC2 has also been reported to interact with and regulate the stability of the RING ubiquitin ligase BRCA1 and the nucleotide excision repair factor Xeroderma pigmentosum A (XPA),8−11 potentially through ubiquitin-mediated proteasomal degradation.9,11 BRCA1 also affects HERC2 binding with claspin, an upstream regulator of the checkpoint kinase Chk1 and facilitator of ATR-dependent Chk1 phosphorylation.12 Despite the interactions between HERC2 and proteins important for maintaining genomic integrity and repair, the function of HERC2 in these processes remains largely unknown.
More recently, HERC2 was shown to interact with p53 through the p53 tetramerization domain, thus affecting p53 oligomerization and downstream transcriptional activity.13 HERC2 also interacts with the deubiquitinating enzyme USP33 and the SCF protein FBXL5, regulating their stability through ubiquitin-mediated proteasomal degradation.14,15 HERC2 has also been shown to regulate centrosome morphology and ubiquitin ligase activity through interactions with NEURL4 and UBE3A (also known as E6AP), respectively. NEURL4 and HERC2 were identified as interactors of the centrosomal protein CP110, and their binding is required for maintenance of centrosome integrity.16,17 E6AP was the first mammalian E3 ubiquitin ligase to be identified, and its association with the human papillomavirus (HPV) oncoprotein E6 was shown to redirect the E6AP ligase activity to mediate the ubiquitylation of p53, resulting in p53 proteasomal degradation.18,19 HERC2 complexes with E6AP bound to E6 in HPV16-positive cells20 and regulates E6AP ligase activity in HEK 293T cells.21 Our laboratory showed that HERC2 interacts with NEURL4 and E6AP, along with the serine/threonine kinase MAPK6, in a large molecular weight complex of at least 2 MDa.22 No function has yet been described for this complex, but its size and multiple domains suggest that it may serve as a platform coordinating the activities of several interacting proteins.
To more widely probe the landscape of cellular HERC2 interactions, we undertook a mass spectrometry (MS)-based proteomic approach to identify and validate additional HERC2 interacting proteins. Our analysis identified close to three hundred putative HERC2 interactors that are involved in distinct biological processes including vesicular transport, organelle organization, GTP-associated signal transduction, protein translation, energy and iron metabolism, and DNA repair. We used bioinformatics to identify cellular pathways through which HERC2 may function, and we have validated more than a dozen new HERC2–protein interactions. These interacting partners implicate HERC2 in cellular protein transport, energy metabolism, and protein synthesis. Because of its size, various structural domains, and ability to bind multiple partners, HERC2 is likely involved in a variety of cellular functions and might serve as a platform to integrate different intracellular functions. Our proteomic analysis provides a substantial resource for subsequent in-depth studies of HERC2 cellular function.
Experimental Procedures
Plasmids and Cloning
The HERC2 expression plasmids were obtained from Neils Mailand7 in the pFlag-CMV2 vector (Sigma). To generate vectors containing the amino terminal hemagglutinin tag (N-HA) for expression of the individual HERC2 fragments, the pFlag-CMV2 vectors were amplified by PCR with primers for recombination in the pDONR223 vector (Invitrogen). The forward and reverse primers for each fragment were designed containing the recombination sequence as a leader (forward: 5′-GGGGACAACTTTGTACAAAAAAGTTGG-3′, reverse: 5′-GGGGACAACTTTGTACAAGAAAGTTGGGTC-3′) upstream of the coding sequence for each fragment. The remaining primer sequences for each HERC2 fragment (following the recombination leader sequence) are fragment 1 (F1, amino acids 1–1000) forward 5′-CATGCCCTCTGAATCTTTCTGTTTGG-3′ and reverse 5′-AAGACTCACAGATCCCCGTATTAATAAGG-3′, fragment 2 (F2, amino acids 950–1750) forward 5′-CGCAGCCATTACTGCAGAGATCC-3′ and reverse 5′-AGGCATCCATCAAAACATTTCG-3′, fragment 3 (F3, amino acids 1700–2700) forward 5′-CATTCCTGAGGGAATCG-3′ and reverse 5′-AGGGTACCAACTCCATTTC-3′, fragment 4 (F4, amino acids 2600–3600) forward 5′-CGATGGATTGCATGATCTCAATGTGC-3′ and reverse 5′-ATGTGGCCACATCCTCCAACTCGG-3′, fragment 5 (F5, amino acids 3550–4500) forward 5′-CGTAGACCTGCTCAAGC-3′ and reverse 5′-ACAGGGGCGTGAGTCCG-3′, fragment 6 (F6, amino acids 4421–4834) forward 5′-CAACCGCATCCAGGTCAAACG-3′ and reverse 5′-AGTGTCCTGTTAAATAATCTTG-3′. Following PCR amplification and BP recombination into pDONR223, each HERC2 fragment was then recombined (LR recombination) into the vector pHAGE-P CMVt N-HA GAW, where GAW indicates GATEWAY cassette. A 6.5 kb fragment encoding the carboxy-terminal half of the HERC2 gene (F456, amino acids 2600–4834) was amplified and cloned in the pENTR-D-TOPO vector (Invitrogen) and recombined into the same lentiviral vector as the fragments. This lentiviral vector was derived from an HIV-1-based backbone named “pHAGE”,23 which was modified by standard molecular biology techniques to work as a destination vector for the GATEWAY system (Invitrogen) and to express an N-terminally tagged protein under the control of a tetracycline regulatable promoter (TetON). It also contains a puromycin resistance gene for selection (PHAGE based vectors expressing HA-tagged proteins). The vectors created in the Howley lab for expression of HERC2 fragments are HERC2-F1 (p6325), HERC2-F2 (p6326), HERC2-F3 (p6327), HERC2-F4 (p6328), HERC2-F5 (p6329), HERC2-F6 (p6330), and HERC2-F456 (p6332). The expression vectors for genes to be tested as HERC2 interactors were obtained from the Human ORFeome database (http://horfdb.dfci.harvard.edu/index.php?page=home) in pDONR and recombined into the pHAGE vector described above. These vectors were used to establish stable cell lines for examination of their interaction with the endogenous full-length HERC2. Vectors were created for expression and analysis of HERC2-interacting proteins for the genes AURKB (p7052), BICD2 (p7053), COPE (p7054), COPZ1 (p7055), CPT1A (p7056), EIF3E (p7135), EIF3H (p7136), EIF3M (p7137), EME1 (p7140), FDFT1 (p7058), FTH1 (p7126), FTL (p7127), HLA-A (p7128), HM13 (p7129), KIF20A (p7130), MUS81 (p7141), PHKB (p7138), PHKG2 (p7139), PRKAR2B (p7133), WDR92 (p7134), and ZFYV9 (p7142). All plasmids were generated for this study and have been deposited in Addgene.
Cell Culture and Protein Expression
All cells lines used in this study were maintained in DMEM 10% FBS at 37 °C and 5% CO2. T-REx 293 cells (Invitrogen), which stably express the Tet repressor (TetON), were maintained in 5 μg/mL Blasticidin (InvivoGen). After infection with lentiviruses carrying pHAGE-P CMVt N-HA GAW-derived vectors, transduced cells were selected with 2 μg/mL puromycin (Sigma) and maintained after selection in 1 μg/mL puromycin. To induce the expression of the HA-tagged proteins, the cells were kept 24 h in medium without antibiotics and then induced at roughly 80% confluency with 1 μg/mL doxycycline (Sigma) for 24 h. Alternatively, N-HA tagged protein expression was performed in 293T cells by transient transfection of 2 μg of plasmid DNA in 6 cm culture dishes at approximately 50% confluency using TransIT-293 (Mirus) according to the manufacturer’s instructions. Cells were harvested 48 h post-transfection for expression and immunoprecipitation analysis described below.
Immunoprecipitation for MS
HERC2 HA-tagged fragment proteins were immunopurified, processed, and analyzed by MS as previously described24 with modifications described in Martinez-Noël et al., 2012.22 Briefly, T-REx cells from five 15-cm culture dishes (approximately 80–90% confluent) expressing each HERC2 fragment were washed once with PBS at room temperature and then lysed on the plates with 5 mL of ice cold lysis buffer (50 mM Tris-HC pH 7.5, 150 mM NaCl, 0.5% NP-40, 12.5 mM NaF, 1 mM Na3VO4, 12.5 mM β-glycerphosphate, complete protease inhibitor cocktail (Roche)). Cleared lysates were filtered through a 0.2 μm SFCA-PF syringe filter (Corning) and immunoprecipitated with 50 μL (settled) of anti-HA agarose beads (Sigma) for 2 h at 4 °C. The beads were then washed twice with 10 mL of lysis buffer and twice with 10 mL of PBS, transferred with 1 mL PBS into a microfuge tube, and then eluted with three 50 μL washes of 0.5 N NH4OH, 0.5 mM EDTA. For IP and Western analysis of HERC2 HCIPs, 293T-REx (stable) or 293T (transient) cells expressing HA-tagged proteins were isolated from 6 cm dishes, washed with 1 mL PBS, lysed with 250 μL lysis buffer, and collected in microfuge tubes. Lysates were pelleted for insoluble material at 13K rpm, 4 °C for 10 min and then immunoprecipitated with 20 μL (settled) of anti-HA agarose beads for 2 h at 4 °C. Beads were washed six times with 1 mL of lysis buffer, then twice with 500 μL of PBS, and immunoprecipitated protein was eluted with three 20 μL washes of 0.5 N NH4OH, 0.5 mM EDTA.
MS and Proteomic Analysis
Eluates were TCA-precipitated and subsequently trypsinized in 10% acetonitrile/50 mM ammonium bicarbonate pH 8. Peptides were acidified by addition of formic acid to 2.5%, desalted with Empore C18 extraction media (3 M), and reconstituted in 10 μL of 5% acetonitrile/5% formic acid. A total of 2 × 4 μL of peptide mixture was loaded by a Famos autosampler (LC Packings, San Francisco, CA) onto a 100 μm (i.d.) × 18 cm fused silica microcapillary column in-house-packed with C18 reverse-phase resin (Magic C18AQ; 5-μm particles; 200-Å pore size; Michrom Bioresources, Auburn, CA), and separated with an Agilent 1100 series binary pump with in-line flow splitter across a 50 min 8–26% acetonitrile gradient. Spectra were acquired using an LTQ-Velos linear ion trap mass spectrometer (Thermo Scientific, San Jose, CA) with a data-dependent Top-10 method.25 Each sample was analyzed twice in succession, followed by a wash with 70% acetonitrile, 30% isopropanol to reduce any potential carry-over. The resulting spectra were searched using SEQUEST26 against a target-decoy database of human (UniProt release 2011_05) tryptic peptides.27 Search parameters included: 2 Da precursor ion tolerance; up to two missed cleavages; a static modification of 57.0215 Da (carboxyamidomethylation) on cysteine; a dynamic modification of 15.9949 Da (oxidation) on methionine. Peptides (false discovery rate (FDR) of <1%) were assembled into proteins accounting for protein redundancy using the principles of parsimony and Occam’s razor constraints.28 The resulting list of identifications for each duplicate run was loaded into CompPASS were the data were merged and further analyzed.24
Analysis of MS Data Using CompPASS
The IP-MS data were analyzed by CompPASS as previously described.24,29 We used a stats table with 175 pulldowns from 293 cells generated by the Harper lab. The WD score threshold was calculated in a way that 95% of the data falls below it. The normalized WD (NWD) score results from dividing each WD score by the calculated threshold. We considered HCIPs those proteins with a NWD score equal to or higher than 1 (WD score equal to or higher than the calculated threshold). We did not use a peptide cutoff number for the CompPASS analysis; the number of peptides for each interacting protein is listed in the Supplemental Tables, Supporting Information.
Western Blot Analysis
Protein extracts were prepared using the lysis buffer described above. Immunoprecipitations from 293T-REx or 293T cells analyzed by Western blot were prepared following elution with 0.5 N NH4OH, 0.5 mM EDTA by incubating 20 μL of eluate (one-third of the immunoprecipitated material) in gel loading buffer at 65 °C, followed by SDS-PAGE and electroblotting to PVDF. Blots were blocked with 5% nonfat dried milk in TNET (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 2.5 mM EDTA, and 0.1% Tween 20) and then incubated with the following primary antibodies: HA conjugated to HRP (clone 3F10, Roche), E6AP (Clone E6AP-300, Sigma), HERC2 (BD Transduction Laboratories), MAPK6 (ERK3, Cell Signaling), NEURL4 (ProteinExpress), and actin (Millipore.). Except when the HA-HRP antibody was used, the membranes were washed in TNET and incubated with HRP-conjugated antimouse or antirabbit secondary antibodies (GE Healthcare). Signals were generated using Western Lightning chemiluminescent substrate (PerkinElmer) and recorded on BioMax MR films (Kodak).
Bioinformatic Analysis
The Genomatix Pathway System (GePS, www.genomatix.de) was used to create biological knowledge-based HERC2 networks as previously described.30 Briefly, functional enrichment analysis of the HERC2 gene set was performed with the FuncAssociate algorithm31 as implemented in the Genomatix’ GeneRanker module. GeneRanker assesses functional enrichment with a Fisher’s Exact Test and then estimates an adjusted p-value via 1000 simulated null hypotheses. Gene Ontology and Pathway Data were collected from the Gene Ontology and NCI Pathway Interaction Databases32,33 and the Memorial Sloan-Kettering Cancer Center’s Cancer Cell Map.34 The basal pathway and network data were supplemented with information obtained from Genomatix’ LitInspector35 and MatBase,36 UniProt,37 DBSubLoc,38 STITCH,39 Gene Expression Atlas,40 Reactome,41 and ENCODE.42 Nodes were color-coded according to NWD score with the median score 2.12.
Immunofluorescence and Microscopy
293 T-REx cells grown on 12 mm glass coverslips were washed twice with PBS and fixed in 4% formaldehyde (Sigma) for 15–20 min. Coverslips were then washed twice with PBS and permeabilized for 5 min in buffer containing PBS, 1% BSA, 5% normal goat serum, and 0.5% Triton X-100 (Sigma) followed by incubation in permeabilization buffer containing primary antibody for 1 h. Primary antibodies were used for staining HERC2 (BD Transduction Laboratories, 1 μg/mL, mouse) and HA (Abcam, 2 μg/mL, rabbit). Coverslips were then washed four times in PBS and incubated in permeabilization buffer containing secondary antibody for 1 h. Secondary antibodies used were goat antirabbit or antimouse conjugated to Alexa 594 (Molecular Probes, 1:500), goat antirabbit conjugated to Alexa 488 (Molecular Probes, 1:500), or goat antimouse conjugated to Alexa 488 (Abcam, 1:500). Controls for HA-specificity included cells expressing HA-EIF3E probed with antirabbit IgG, or cells not expressing HA-EIF3E probed with anti-HA antibody. Coverslips were then washed four times in PBS and either mounted onto glass microscope slides with SlowFade Gold +DAPI (Invitrogen) for single protein stain, or for sequential staining for colocalization, incubated in primary and secondary antibodies as described above, and then mounted. Coverslips were secured to slides with nailpolish. All steps were performed at room temperature. For immunofluorescence, coverslips on slides were visualized with an Olympus Fluoview FV1000 IX81 inverted confocal microscope with motorized stage. Images were acquired using a PLAPON 60X O/1.42 NA oil immersion objective and FV10-ASW 3.1 software. Images were generated from nonoverlapping emission spectra at 1024 × 1024 pixel (12 bit/pixel) resolution. The software was used to overlay two images generated from 405, 488, and 594 nm lasers.
Results
Proteomic Identification of HERC2-Interacting Proteins
To gain further insight into the various cellular functions of HERC2, we conducted a proteomic analysis of HERC2-interacting proteins. HERC2 is a large protein with multiple domains, some of which have been shown to interact with specific cellular proteins (Figure 1A). Since the full-length HERC2 protein proved difficult to stably express at a sufficient level for MS analysis, six overlapping fragments comprising the complete HERC2 ORF were expressed. Each fragment, approximately 1000 amino acids in length (F1–F6, Figure 1A), was stably expressed in tetracycline-inducible HEK 293 cells (T-REx 293, Invitrogen) with an amino-terminal HA tag (N-HA) for immunoprecipitation (Figure 1B). Proteins coprecipitating with each HERC2 fragment were identified by LC–MS-MS, and a list of high confidence interacting proteins (HCIPs) was generated by CompPASS analysis.24 HA-tagged GFP served as a negative control. Our analysis identified NEURL4 and UBE3A as HCIPs for F1 and F4, respectively. Each of these proteins has previously been identified as a HERC2-interacting protein, thus providing validation for our studies.17,21
Figure 1.
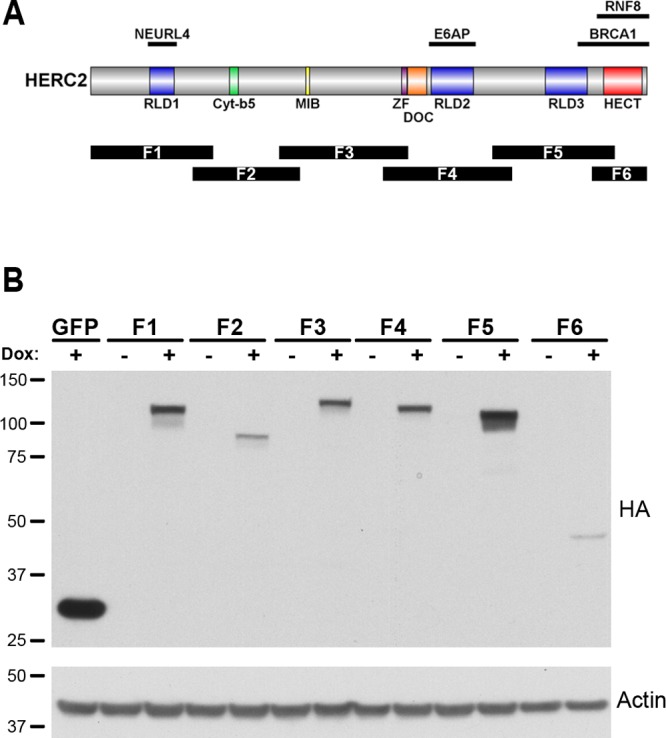
HERC2 schematic and fragment expression. (A) The HERC2 open reading frame encodes a 4834 amino acid polypeptide containing three RCC1-like domains (RLD) and a carboxy terminal HECT domain found in a class of ubiquitin ligases. Additional domains share homology to cytochrome b5 (Cyt-b5), zinc finger (ZF) and two other ubiquitin ligases (MIB and DOC). HERC2 binding sites with known interactors are shown above, and the region of each HERC2 fragment expressed for proteomic analysis is shown below the corresponding HERC2 sequence (F1–F6). (B) The HERC2 open reading frame was expressed as six separate overlapping fragments under control of a tetracyclin-responsive element and stably transfected into 293 T-REx cells (Invitrogen). Treatment with 1 μg/mL doxycycline for 24 h resulted in HERC2 fragment expression of the expected size (major band in “Dox +” lanes). Size markers in kilodaltons are on the left.
Table 1 displays a list of proteins identified as HCIPs for each fragment. Proteins analyzed by CompPASS were considered HCIPs by meeting the statistical threshold cutoff of 0.95 and a calculated normalized weighted D-score (NWD) of 1.0 or greater. The top 25 HCIPs for each fragment are listed in Table 1 (complete results are in Supplementary Table S1, Supporting Information). Overall, 283 potential interactors were identified, 239 of which were identified in the analysis of only a single fragment. The remaining 44 HCIPs were identified in the analysis of two (30 HCIPs), three (10 HCIPs), four (3 HCIPs), or five (1 HCIP) different fragments. HCIP identification with more than one fragment could potentially be due to fragment overlap with the site of HCIP interaction, a complex interaction between two or more proteins interacting with different regions of HERC2, or from a stable but nonspecific interaction. Overall, more than 80% of the HCIPs identified in our analysis were found to interact with a single fragment of HERC2.
Table 1. HERC2 Fragment MS Analysis and HCIP Identificationa.
| F1 1–1000 |
F2 950–1750 |
F3 1700–2700 |
F4 2600–3600 |
F5 3550–4500 |
F6 4421–4834 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | NWD | Gene | NWD | Gene | NWD | Gene | NWD | Gene | NWD | Gene | NWD |
| MUS81 | 4.24 | ASAH1 | 6 | ITPR2 | 8.49 | BICD1 | 14.7 | PHKA2 | 7.75 | LCP1 | 18 |
| CCDC22 | 4.24 | MAVS | 3.5 | PTPN1 | 8.49 | BICD2 | 14 | ENPP1 | 6.93 | SELO | 15 |
| AGAP3 | 4.24 | CST4 | 3 | GNA13 | 8.49 | ZFYVE9 | 12.37 | EIF3H | 6.12 | SPECC1L | 10.39 |
| ARHGEF5 | 3.97 | SPTLC2 | 2.42 | FNDC3A | 6 | CEP170 | 11.64 | MIOS | 5.66 | SNX6 | 10.39 |
| EME1 | 3.35 | ZG16B | 2.27 | EPCAM | 5.2 | SNAPIN | 8.49 | PHKB | 5.41 | ARAF | 10.39 |
| HERC1 | 3.35 | NCCRP1 | 2 | NCOA4 | 4.08 | UBE3A | 6.39 | EIF3A | 5.33 | TUBB3 | 8.16 |
| KIF20A | 3.18 | HLA-A | 1.98 | MAVS | 3 | SLC30A9 | 6 | PHKG2 | 5.29 | RAF1 | 6.89 |
| HPS3 | 3 | HM13 | 1.75 | LACTB | 3 | PCM1 | 5.29 | OCRL | 5.2 | ABR | 6.09 |
| HPS5 | 2.12 | VDAC2 | 1.75 | NEK11 | 2.83 | FHOD3 | 5.2 | MTMR3 | 5.2 | RER1 | 6 |
| TUBA1C | 2.1 | KRT80 | 1.6 | HLA-E | 2.83 | AKAP9 | 4.9 | EIF3E | 4.78 | TXLNG | 6 |
| MLL4 | 2 | CSTA | 1.5 | LMNB2 | 2 | NIN | 4.41 | PHKA1 | 4.74 | CBX2 | 6 |
| RAB34 | 2 | BLMH | 1.21 | TUBA1C | 1.77 | NUDCD2 | 4.24 | WDR59 | 4.58 | PRR14L | 3 |
| NEURL4 | 1.98 | PSMB4 | 1.03 | FTH1 | 1.73 | SLC27A4 | 3.67 | EIF3B | 4.38 | SENP1 | 3 |
| ZDHHC17 | 1.73 | PSMD1 | 1.11 | MLEC | 1.7 | ASPM | 3.46 | EIF3L | 4.04 | ATP4A | 3 |
| EPHA7 | 1.73 | PSMD11 | 1.02 | PSMD1 | 1.66 | PEX1 | 3.46 | EIF3M | 3.96 | EXD2 | 2.83 |
| ZBTB5 | 1.7 | PELP1 | 1 | LMNA | 1.56 | USP20 | 3.32 | EIF4G1 | 3.81 | SSX2IP | 2.83 |
| NUP153 | 1.7 | TEX10 | 1 | HM13 | 1.5 | ETAA1 | 3 | EIF3G | 3.75 | TUBA1C | 2.83 |
| GTF3C2 | 1.7 | BIRC6 | 1.5 | PRR14L | 3 | EIF3C | 3.74 | RNGTT | 2.64 | ||
| MLF2 | 1.7 | C19orf2 | 1.5 | RIC8A | 3 | EIF3K | 3.47 | CDC25C | 2.12 | ||
| ARNT | 1.67 | PPM1B | 1.5 | PHKG2 | 2.83 | EIF3D | 3.19 | MYO9B | 2.12 | ||
| RNGTT | 1.5 | COPB2 | 1.37 | SCO2 | 2.83 | CTNNA1 | 3 | SLC27A4 | 2.12 | ||
| C10orf118 | 1.5 | OBSL1 | 1.33 | WDR62 | 2.6 | CTNNB1 | 2.83 | CXorf57 | 2.12 | ||
| TRMT2A | 1.5 | COPG2 | 1.31 | GNAS | 2.6 | WDR24 | 2.3 | KIAA0889 | 2.12 | ||
| VSIG7 | 1.5 | COPG | 1.29 | TUBA1C | 2.19 | EIF3I | 2.26 | FLII | 2.08 | ||
| FDFT1 | 1.34 | COPA | 1.25 | PHKB | 2.12 | GCC2 | 2.22 | FASTKD5 | 2 | ||
The 25 highest scoring HERC2 interactors for each fragment following immunoprecipitation, mass spectrometry, and CompPASS. The scores for each HCIP were generated at a 0.95 confidence threshold. The genes in bold indicate HCIPs with NWD ≥ 1 when using a 0.98 threshold. Genes in italics were identified as an HCIP with more than one HERC2 fragment.
F1 of HERC2 contains a single RLD domain and yielded 133 HCIPs, 103 of which were unique to F1. The RLD1 of HERC2 is defined by high homology to RCC1, a protein that functions as a guanine nucleotide-exchange factor (GEF) promoting the release of GDP and uptake of GTP by the G protein Ran, and is important in cell division and mitosis.43,44 The Rho GEF ARHGEF5 and the GTPase-activating protein AGAP3 were among the HCIPs identified with F1, in addition to KIF20A, a kinesin known to interact with GTP-bound Ras family members RAB6A and RAB6B.45,46 These findings are significant as multiple proteins serve to link HERC2 with G protein activity and regulation.
HERC2 has been previously implicated in genomic repair,7,10 and among the HCIPs identified with F1 were EME1 and MUS81, components of an endonuclease heterodimer active in the repair of structure-specific DNA nicks at Holliday junctions, 3′-flaps, and stalled replication forks.47,48 HERC2 has been found at stalled replication forks;12 however, EME1 and MUS81 have not previously been identified as HERC2 interactors. Another interactor, the coiled-coil domain containing 22 protein (CCDC22), is involved in NF-κB regulation through its interaction with COMMD proteins.49 COMMD proteins contribute to the ubiquitylation of the NF-κB subunit RELA, leading to its degradation, and thereby down-regulating NF-κB activity.50 COMMD1, COMMD3, COMMD4, and COMMD8 were also identified as HCIPs in the F1 analysis, suggesting a complex interaction with HERC2 and CCDC22.
Fifteen HCIPs were identified in the analysis of F2, eight of which were unique to F2. Although this is a low number of HCIPs relative to other fragments, several of these proteins share important cellular functions. SPTLC2 and ASAH1 are enzymes critical to sphingolipid biosynthesis and degradation, respectively.51,52 ASAH1 is a lysosomal enzyme and cathepsin A (CTSA), another HCIP, is a serine protease found in the lysosome.53 In general, cathepsins are responsible for several physiological processes, the most well-known being protein turnover in the lysosome.54 Three different proteins involved in immune function were identified as HCIPs. The mitochondrial antiviral signaling protein (MAVS) is necessary for transcription factor activation regulating beta interferon expression in the innate immune response.55 The histocompatibility minor protein 13 (HM13) and major complex class I A protein (HLA-A) are involved in signal peptide processing and antigen presentation, respectively.56,57 HLA-A was also found as an HCIP of F1, along with MHC class I proteins HLA-C and HLA-E, which were also identified with HM13 as HCIPs in the analysis of F3. The identification of these HLA proteins by separate fragments with different functional domains all within the amino terminal half of HERC2 suggests this region of HERC2 may have some role in immune functions through interaction with multiple HLA proteins.
The pulldown and CompPASS analysis of F3 of HERC2 resulted in the identification of 52 proteins, 35 of which were only recognized with F3. Among the unique F3 HCIPs were seven subunits of the COPI coatomer protein complex (COPA, COPB1, COPB2, COPE, COPG1, COPG2, and COPZ1) involved in vesicle-mediated Golgi to ER protein transport.58 Five of these COPI proteins are known to interact with TMED10, a transmembrane protein important for COPI vesicle formation, also found as an HCIP in the analysis of F3. Other HCIPs for this fragment include the ferritin heavy and light chains (FTH1, FTL) and the nuclear receptor coactivator 4 (NCOA4), the latter of which was recently reported to bind and regulate ferritin transport during autophagy.59
F4 of HERC2 contains the sequence previously shown to interact with the ubiquitin ligase UBE3A.21 This was confirmed in our analysis as UBE3A was one of 30 unique HCIPs (48 total) found to interact with F4. Multiple HCIPs found with F4 are also reported to bind UBE3A, including glutamate dehydrogenase (GLUD1), a metallochaperone involved in cytochrome c oxidase biogenesis, SCO2, and the centrosomal protein CEP170. Additional F4 HCIPs include BICD1 and BICD2, as well as the zinc finger and FYVE domain-containing protein ZFYVE9 (also known as SARA). BICD1 and BICD2 are proteins involved in the motility of membrane-bound organelles regulating their transport.60 ZFYVE9 was originally identified as an anchor for SMAD2 and SMAD3 activation, and general regulator of TGF-β signaling.61 More recently, ZFYVE9 has been implicated in cellular trafficking due to its localization to the endosome62 and reported interactions with the GEF Rab 563 and vesicle mediator syntaxin-3.64
Thirty-seven proteins were identified as HCIPs in the analysis of F5, 27 of which were unique to this fragment. Most significant among the 27 unique F5 HCIPs were 12 components of the eukaryotic translation initiation factor 3 complex (eIF3). The eIF3 scaffold is the largest initiation complex formed during cellular protein translation and serves multiple distinct functions including the facilitation of mRNA and ribosomal binding, large and small ribosomal subunit dissociation, and stabilization of the eIF2-GTP-Met-tRNAi complex, as well as interacting with several different initiation factors.65 Also identified in the F5 analysis were all three regulatory subunits (PHKA1, PHKA2, and PHKB) and one catalytic subunit (PHKG2) of phosphorylase kinase (PhK), a serine/threonine kinase involved in glycogen metabolism.66
F6 is the shortest of all the HERC2 fragments and contains the carboxy-terminal HECT domain responsible for ubiquitin ligase activity. Forty-eight proteins were identified as HCIPs, 32 of which were identified only with F6. There is an important diversity among the function of the F6 HCIPs, but most proteins possess known roles in microtubule-based binding and vesicle-mediated trafficking (LCP1, SPECC1L, MYO9B, FLII, TUBA1C, TUBB3, SNX6, RER1, TXLNG, IQGAP1, LRPPRC), cellular growth signaling (ARAF, RAF1, CDC25C), and various mechanisms governing cellular transcription and translation (VBP1, PFDN4, PFDN5, PFDN6, RPS27L, RNGTT, CBX2, GCN1L1, XPOT). The interaction of these proteins with F6 of HERC2 is also important due to the nature of the HERC2 HECT domain, contained within this fragment, and the possibility that these proteins may serve to regulate HERC2 ubiquitin ligase activity or may be substrates for HERC2-mediated ubiquitylation. Our analysis greatly expands the HERC2–protein interactome, providing numerous potential HERC2 regulators and substrates, and implicating HERC2 in several important cellular processes.
HCIP Analysis Links HERC2 to Intracellular Trafficking and Energy Metabolism
Our proteomic analysis identified 283 distinct HERC2 HCIPs, the majority of which have not been previously reported. To probe the potential cellular functions of HERC2 more deeply, functional enrichment of gene ontology (GO) biological process (BP) terms and subsequent biological network analyses of the HERC2 HCIP gene list were performed using Genomatix Pathway System (GePS, http://mygga.genomatix.de/). The highest NWD score for each HCIP (if identified in multiple experiments) was used to color code the corresponding gene node in the interaction networks generated for each GO BP term. Two hundred seventy-one significantly enriched terms were identified in our analysis. Given the high degree of gene redundancy among GO definitions, functionally enriched GO BP terms were grouped by distinct function using a combination of term similarity and percentage of input genes representing the total number of genes associated with a term. This proved useful in distilling the large number of GO terms and identifying potential cellular pathways in which HERC2 may be involved.
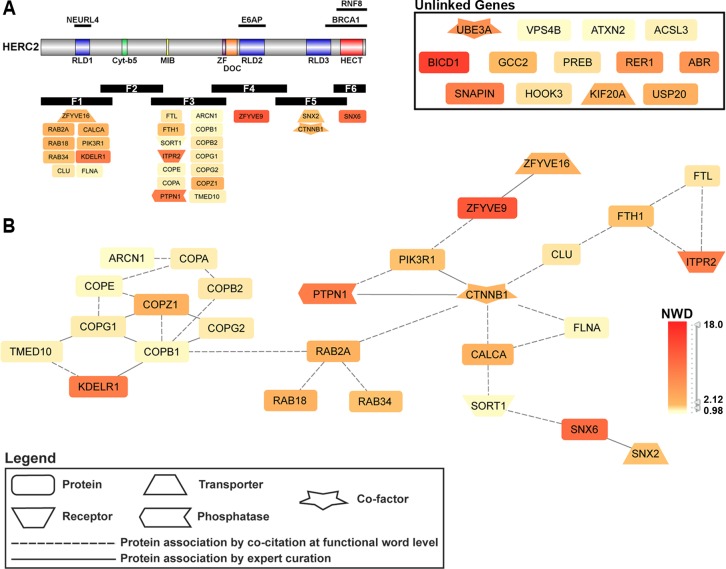
HERC2 HCIP GePS analysis revealed 15 functionally enriched GO BP terms involving intracellular vesicle-mediated organization and transport. A nonredundant gene interaction network was generated encompassing all input genes associated with vesicle-mediated transport-related GO BP terms (Figure 2). Of the 40 HERC2 HCIPs associated with the 15 enriched GO BP terms, 27 HCIPs have a known biological relationship, which is depicted in the network in Figure 2. The GePS builds interaction networks from known biological data, which include expertly curated and experimentally validated associations, combined with natural language processing of PubMed abstracts. Consequently, the edges between nodes represents the association between two or more genes as defined by one of the methods described above. The 13 HCIPs not displayed in the interaction network had no known association with any of the genes involved in the network or with each other (unlinked genes).
Figure 2.
Intracellular transport pathways involving HERC2. GePS analysis of HERC2 HCIPs yielded 15 distinct GO biological process terms related to various intracellular protein transport and trafficking pathways (Table 2). Forty HERC2-interacting genes were represented, 27 of which comprise the major network shown in (B). Network HCIPs and the corresponding HERC2 fragment used for identification are displayed in (A). The GTPase RAB2A is linked to the COPI coatomer complex and beta-catenin, the latter of which integrates PI3K signaling with other subnetworks involving vesicle-mediated transport and localization. Nodes are colored according to the gene’s NWD score from the HERC2 CompPASS analysis.
Table 2. GO Biological Process Terms Associated with HERC2 HCIPs Related to Intracellular Vesicle-Mediated Organization and Transporta.
| GO-Term | GO-Term ID | p-value | Adj p-value | # input genes | # total genes |
|---|---|---|---|---|---|
| retrograde vesicle-mediated transport, Golgi to ER | GO: 0006890 | 3.41 × 10–12 | 0.00 × 100 | 10 | 25 |
| COPI-coated vesicle budding | GO: 0035964 | 6.07 × 10–12 | 0.00 × 100 | 8 | 13 |
| Golgi transport vesicle coating | GO: 0048200 | 6.07 × 10–12 | 0.00 × 100 | 8 | 13 |
| COPI coating of Golgi vesicle | GO: 0048205 | 6.07 × 10–12 | 0.00 × 100 | 8 | 13 |
| Golgi vesicle budding | GO: 0048194 | 1.40 × 10–11 | 0.00 × 100 | 8 | 14 |
| vesicle targeting to, from, or within Golgi | GO: 0048199 | 2.94 × 10–10 | 0.00 × 100 | 9 | 27 |
| vesicle targeting | GO: 0006903 | 8.71 × 10–9 | 0.00 × 100 | 9 | 38 |
| vesicle coating | GO: 0006901 | 1.12 × 10–8 | 0.00 × 100 | 9 | 39 |
| Golgi vesicle transport | GO: 0048193 | 2.07 × 10–8 | 0.00 × 100 | 17 | 192 |
| vesicle organization | GO: 0016050 | 3.29 × 10–8 | 0.00 × 100 | 13 | 110 |
| vesicle-mediated transport | GO: 0016192 | 2.19 × 10–7 | 0.00 × 100 | 40 | 1010 |
| establishment of vesicle localization | GO: 0051650 | 4.15 × 10–6 | 0.00 × 100 | 11 | 118 |
| intra-Golgi vesicle-mediated transport | GO: 0006891 | 1.01 × 10–5 | 0.00 × 100 | 6 | 31 |
| vesicle localization | GO: 0051648 | 2.96 × 10–5 | 0.00 × 100 | 11 | 145 |
| Golgi localization | GO: 0051645 | 7.17 × 10–3 | 1.00 × 10–2 | 2 | 8 |
Fifteen gene ontology biological process terms with a p-value ≤ 0.05 used to generate the network shown in Figure 2. The number of HERC2 HCIPs (Input Genes) associated with each GO term and total number of genes associated with each term are listed.
The major network shown in Figure 2 contains all subunits of the COPI coatomer complex involved in retrograde Golgi-ER transport (ARCN1, COPA, COPB1, COPB2, COPE, COPG1, COPG2, COPZ1). This complex is linked to the Rab GTPase family member RAB2A. The COPI complex and Rab signaling represent vesicular trafficking and transport pathways that are integrated in this network through beta-catenin (CTNNB1), a component of adherens junctions necessary for anchoring the actin cytoskeleton, as well as being a core member of the Wnt signaling pathway.67 Beta-catenin additionally links lipid kinase signaling (PI3KR1), and calcium and iron regulation (CALCA and FTH1/FTL, respectively), with these transport pathways and implicates HERC2 in the coordination of intracellular protein transport.
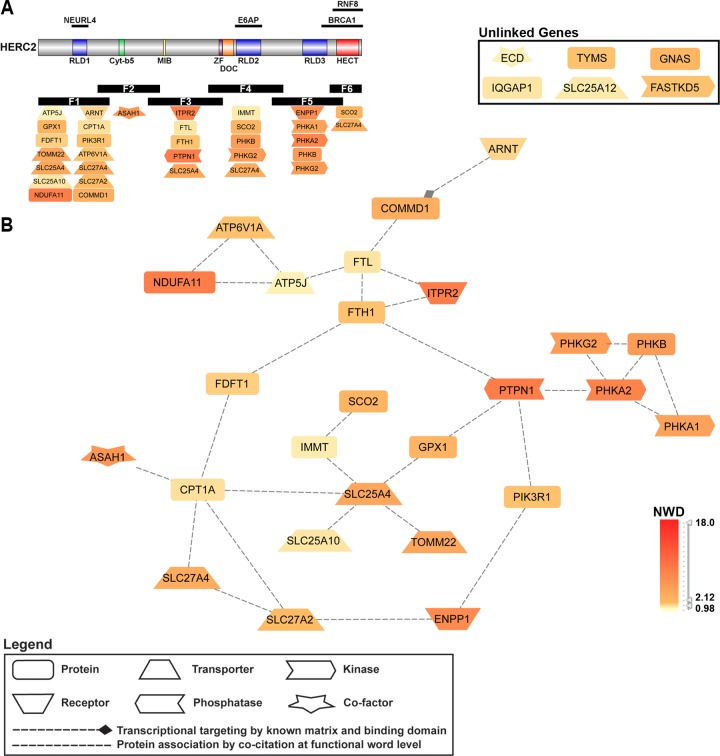
Several proteins identified as potential HERC2 interactors were associated with cellular processes involved in the production and metabolism of cellular energy. Using GePS, 14 functionally enriched GO BP terms were identified containing 19 HERC2 HCIPs. Of these 19 HCIPs, nine were linked in the original combined network (original network not shown). We then used GePS to extend all 19 HCIPs by known associations with other HCIPs not produced in the enrichment analysis. This extension added 13 HCIPs through known associations with the nine HCIPs from the original network and four of the previously unlinked genes, totaling 26 HCIPs. All 26 linked HCIPs were reconfigured to generate the interaction network displayed in Figure 3.
Figure 3.
Cellular energy and metabolism pathways involving HERC2. GePS analysis of HERC2 HCIPs yielded 14 distinct GO biological process terms regarding processes involved in energy and cellular metabolism (Table 3). Nineteen HERC2-interacting genes were represented, 10 of which were not linked in any network. Four of the unlinked genes (ENPP1, ITPR2, ARNT, SCO2) and all other linked genes were extended by known interactions to other input genes and reorganized to generate the major network shown in B. Network HCIPs and the corresponding HERC2 fragment used for identification are displayed in A (five HCIPs were identified with more than one fragment). Nodes are colored according to the gene’s NWD score from the HERC2 CompPASS analysis.
Table 3. GO Biological Process Terms Associated with HERC2 HCIPs Related to Cellular Energy and Metabolisma.
| GO-Term | GO-Term ID | p-value | Adj p-value | # input genes | # total genes |
|---|---|---|---|---|---|
| cellular carbohydrate catabolic process | GO: 0044275 | 2.86 × 10–4 | 0.00 × 100 | 6 | 55 |
| glycogen catabolic process | GO: 0005980 | 6.02 × 10–4 | 1.00 × 10–3 | 4 | 24 |
| glucan catabolic process | GO: 0009251 | 7.07 × 10–4 | 1.00 × 10–3 | 4 | 25 |
| cellular polysaccharide catabolic process | GO: 0044247 | 7.07 × 10–4 | 1.00 × 10–3 | 4 | 25 |
| energy derivation by oxidation of organic compounds | GO: 0015980 | 8.20 × 10–4 | 1.00 × 10–3 | 14 | 316 |
| polysaccharide catabolic process | GO: 0000272 | 9.56 × 10–4 | 1.00 × 10–03 | 4 | 27 |
| energy reserve metabolic process | GO: 0006112 | 1.59 × 10–3 | 1.00 × 10–3 | 9 | 163 |
| generation of precursor metabolites and energy | GO: 0006091 | 2.72 × 10–3 | 5.00 × 10–3 | 16 | 439 |
| glucose metabolic process | GO: 0006006 | 3.27 × 10–3 | 4.00 × 10–3 | 10 | 216 |
| polysaccharide metabolic process | GO: 0005976 | 4.96 × 10–3 | 4.00 × 10–3 | 6 | 95 |
| glycogen metabolic process | GO: 0005977 | 5.02 × 10–3 | 2.00 × 10–3 | 5 | 67 |
| glucan metabolic process | GO: 0044042 | 5.35 × 10–3 | 3.00 × 10–3 | 5 | 68 |
| cellular glucan metabolic process | GO: 0006073 | 5.35 × 10–3 | 3.00 × 10–3 | 5 | 68 |
| hexose metabolic process | GO: 0019318 | 9.43 × 10–3 | 8.00 × 10–3 | 10 | 252 |
Fourteen gene ontology biological process terms with a p-value ≤ 0.05 used to generate the network shown in Figure 3. The number of HERC2 HCIPs (Input Genes) associated with each GO term and total number of genes associated with each term are listed.
The central nodes represent proteins involved in energy and metabolite production in the mitochondria (SLC25A4, SLC25A10, TOM22, IMMT, SCO2). These are integrated into the network by the outer ring through known associations to proteins involved in fatty acid metabolism (CPT1A), and the regulation of antioxidants (via GPX1) and insulin (via PTPN1). Other HCIPs composing the outer ring of the network are involved in the generation and transport of fatty acids (SLC27A2, SLC27A4, FDFT1), iron homeostasis (FTH1 and FTL), and insulin metabolism (PIK3R1 and ENPP1). Additional subnetworks include the components of the glycogen regulator phosphorylase kinase (PHKA1, PHKA2, PHKB, PHKG2) and the establishment of proton gradients and electron transport in the cell (ATP5J, NDUFA11, ATP6 V1A). Overall, the association of multiple HERC2 HCIPs important to the regulation of insulin, glycogen, and fatty acid generation and transport, support a role for HERC2 as a component of pathways governing the metabolism and maintenance of cellular energy.
Validation of HERC2-Interacting Proteins
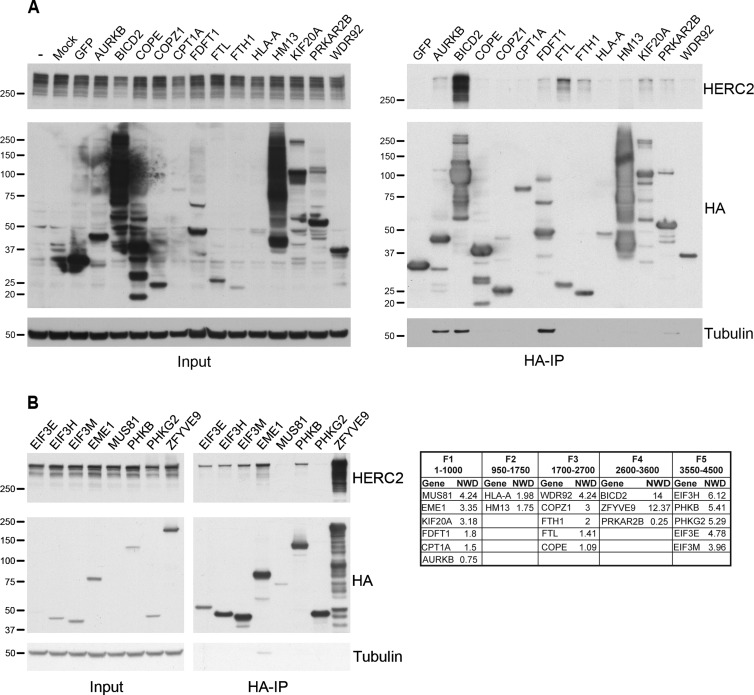
We next attempted to validate the interaction of endogenous HERC2 with HCIPs identified in our MS/CompPASS analysis. To do so, potential interactors were expressed as N-HA proteins in 293T-REx cells and analyzed for their interaction with full-length endogenous HERC2 using an antibody to HERC2 (Figure 4). Available expression vectors were selected from the Human ORFeome Database and cloned into vectors containing an N-HA tag. Variable levels of expression were observed following transient transfection, but full-length protein could generally be detected following HA-immunoprecipitation (Figure 4). Binding to endogenous HERC2 was analyzed by Western blot with HA-GFP serving as a negative control. We examined a total of 21 cellular proteins identified in our proteomic analysis above as potential HERC2 interactors (19 HCIPs) and were able to confirm binding of 13 to endogenous HERC2. This validation provides a level of confidence for the many other HERC2 HCIPs revealed by the CompPASS analysis above.
Figure 4.
Validation of HERC2 HCIPs. HERC2 HCIPs were transiently expressed as N-HA tagged proteins in 293T cells (A) or stably transfected in 293T-REx cells and expressed following induction with 1 μg/mL doxycycline (B). Expressed proteins were immunoprecipitated (HA-IP) from equivalent amounts of total cellular protein. Variable levels of expression were seen for each protein (Input, middle panel) and the expressed protein was appropriately purified (HA-IP, middle panel). Protein interaction with HERC2 was examined by Western blot for endogenous full-length HERC2 (HA-IP, top panel). Size markers in kilodaltons are on the left. Gene symbol and NWD score of proteins examined for binding endogenous HERC2 are displayed in the table.
Of the proteins examined, the cellular transport adaptor BICD2 and the endosomal protein ZFYVE9 coprecipitated the greatest amount of HERC2, though each bait protein was also expressed at a very high level (Figure 4). We corroborated the HERC2-ZFYVE9 interaction by a reciprocal MS-CompPASS analysis and identified HERC2 as an HCIP of ZFYVE9 (data not shown). The mitotic kinesin KIF20A also coprecipitated a modest amount of HERC2. When analyzed by NWD score, BICD2 and ZFYVE9 were among the top three HCIPs for F4, and KIF20A was among the highest for F1 (Table 1). ZFYVE9 and KIF20A were among the HCIPs linking HERC2 with intracellular transport (Figure 2). BICD2 is known to regulate Golgi-ER transport, and its paralogue BICD1 was also associated with HERC2-linked intracellular transport. The strong interaction between HERC2 and BICD2, ZYFVE9 and KIF20A, provides additional support for a potential role of HERC2 in intracellular protein trafficking and transport.
Intracellular iron storage is mediated by the protein ferritin, which is composed of heavy (FTH1) and light (FTL) subunit chains. Both the FTH1 and FTL subunits were identified as HCIPs with F3, and each displayed an interaction with endogenous HERC2 (Figure 4A). Responsible for regulating iron storage and transport, ferritin is a component in the major networks linking HERC2 with intracellular protein transport and cellular energy metabolism (Figures 2 and 3). NCOA4 is important for mediating the autophagic turnover of ferritin,59 and while our results show an interaction between HERC2 and FTH1/FTL, we cannot rule out that the interaction is a consequence of overexpression and is indirect via the known interaction of NCOA4 with ferritin.
Among the proteins linking HERC2 with cellular energy metabolism (Figure 3), the PHKB and PHKG2 subunits of PhK were examined for binding endogenous HERC2. Despite good protein expression, only the PHKB subunit bound an appreciable amount of full-length HERC2 (Figure 4B). FDFT1, an important enzyme for the synthesis of cholesterol, was identified as an HCIP associated with F3 and is a component of the cellular energy metabolism pathways linked to HERC2 (Figure 3). FDFT1 was found to interact with endogenous HERC2 (Figure 4A), and along with PHKB, supports a role for HERC2 in the regulation of cellular energy.
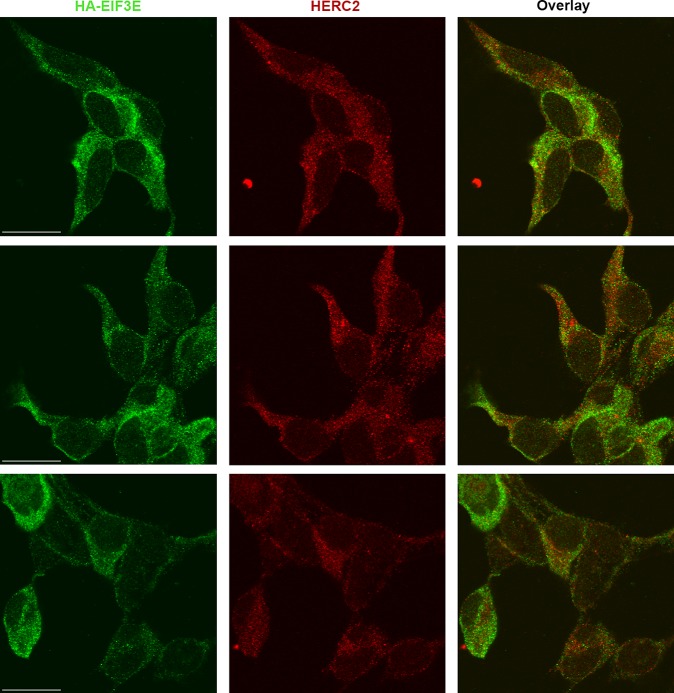
Twelve subunits of eIF3 were identified as F5 HCIPs (Table 1), and we examined the interaction of three different components of this complex (EIF3E, EIF3H, EIF3M) with HERC2. Each eIF3 subunit coprecipitated endogenous HERC2 to a similar degree suggesting a highly stable complex interaction (Figure 4B). We corroborated the eIF3–HERC2 interaction by a reciprocal MS-CompPASS analysis of EIF3E, and identified HERC2 as an HCIP of EIF3E (data not shown). We further validated the interaction between HERC2 and eIF3 by confocal miscroscopy in cells expressing HA-tagged EIF3E. Figure 5 depicts three different fields that demonstrated endogenous HERC2 and EIF3E to be cytoplasmic; partial colocalization is revealed by the yellow and orange cytoplasmic dots evident in the image overlays. The validation of HERC2 interaction with multiple members of the eIF3 complex also suggests a role for HERC2 in the regulation of cellular protein expression through translational initiation.
Figure 5.
Colocalization of HERC2 and EIF3E. 293 T-REx cells stably expressing inducible N-HA EIF3E were probed for HA to visualize EIF3E (green), followed by probing for HERC2 (red). Overlay of HA and HERC2 shows colocalization of EIF3E and HERC2 primarily in the cytoplasm. Similar colocalization and staining patterns were seen regardless of the order of staining for HA and HERC2, or the fluorophore for each secondary antibody. Scale bar in the EIF3E panels, 20 μM.
HERC2 is known to be important for maintenance of genomic integrity and the response to DNA damage through interactions with several DNA repair proteins. In the CompPASS analysis of HERC2 HCIPs, we identified EME1 and MUS81, a heterodimeric endonuclease complex that resolves structure-specific DNA lesions.47 We validated the interaction of HERC2 with EME1 (Figure 4B), but not MUS81, suggesting that HERC2 interacts with this complex directly through EME1 (analysis of nonbinders is provided in the Discussion). Nonetheless, this finding supports the role of HERC2 in DNA damage and repair, potentially placing it in a new context.
The NWD score utilized for identification of HCIPs is a metric combining multiple factors that provides a level of confidence to potential bait–prey interactions. The capture and identification of proteins is impacted by, among other things, the amount of prey protein and the stability of the interaction. Since transient interactions and/or those with proteins of low abundance could be extremely important but produce a low NWD score, we also reviewed NWD scores for each fragment at less stringent confidence thresholds and examined AURKB and PRKAR2B for interaction with endogenous HERC2. AURKB, a serine/threonine kinase involved in regulation of chromosome alignment and mobility,68 displayed modest binding to endogenous HERC2 (Figure 4A). Identified in multiple analyses of F1, AURKB interacts with the same region of HERC2 as NEURL4, the latter of which cooperates with HERC2 in centrosome maintenance, a process that may also involve AURKB. Among several proteins involved in lipid and glucose metabolism, we validated HERC2 binding with the regulatory subunit of protein kinase A PRKAR2B (Figure 4A). PRKAR2B was identified in the analysis of F4, along with two of the PhK subunits, implicating HERC2 in the regulation of cellular energy metabolism through multiple pathways and complexes.
Overall, our analysis has provided a wealth of new information regarding HERC2 protein interactions in the cell. We’ve identified numerous potential HERC2 binders that participate in diverse and functionally important cellular processes. We have confirmed the in vivo interaction between HERC2 and over a dozen HCIPs, implicating HERC2 in previously unlinked pathways and imparted a source of study that promises to enhance our understanding of several mechanisms vital to the cell.
Discussion
HERC2 is a large ubiquitin ligase with known roles in enzyme regulation, the maintenance of centrosome architecture, and the cellular response to DNA damage. Despite its size and the diversity of cellular processes in which HERC2 is reportedly involved, relatively few proteins have been shown to interact directly with HERC2, while the mechanism of HERC2 function in the cell remains largely unknown. In this study, we utilized a systematic proteomic approach to identify HERC2-interacting proteins and utilized bioinformatics tools to provide context for the interactions, yielding greater insight into HERC2 cellular functions.
Our proteomic analysis of stable HERC2 protein interactions utilizing CompPASS identified over 280 HCIPs. Among these interactors were components of cellular protein complexes involved in vesicle-mediated transport (COPI coatomer complex), glycogen metabolism (phosphorylase kinase), and the regulation of cellular protein translation (eIF3 complex). The large number of HCIPs and the heterogeneity of their known cellular functions led us to analyze HERC2 HCIPs using the Genomatix Pathway System. Over 270 functionally enriched GO BP terms were grouped to generate potential HERC2 interaction networks of distinct cellular functions, identifying pathways regulating intracellular protein transport and trafficking (Figure 2), metabolism of cellular energy (Figure 3), and cellular protein translation. A dozen or more HERC2 HCIPs were involved in each of these processes, which provide links to potential HERC2 functions that have not been previously described.
To support the involvement of HERC2 in these pathways, we validated the interaction of various HCIPs with the full-length endogenous HERC2 protein (Figure 4). We were able to confirm binding of HERC2 with genes linked in each network, including FDFT1 and PHKB (energy), ZFYVE9 (trafficking), and FTH1 and FTL (both networks). HCIPs not found to bind HERC2 in our coprecipitation studies often represented one subunit of a complex whose other members displayed stronger HERC2 binding. For example, the PhK catalytic subunit PHKG2 and the DNA repair endonuclease component MUS81 did not coprecipitate with HERC2 and thus may bind HERC2 indirectly through other complex members, such as PHKB and EME1, respectively. Alternatively, the inability of a protein to coprecipitate HERC2 may be because the N-HA tag, placed on the HCIP instead of a HERC2 fragment, may interfere with the HERC2-HCIP interaction. The method of detecting an interaction may also be an issue as mass spectrometry is much more sensitive than Western blot, offering the possibility that the amount of HCIP bound to HERC2 was a limiting factor in the validation.
Among the most interesting findings, several HCIPs involved in cellular protein translation were identified, including the eIF3 complex, the eIF4 complex subunit G1, and four members of the prefoldin family of proteins. Prefoldins are known for regulating chaperone-mediated folding of filament proteins, such as actin and tubulin, and were recently reported to be involved in transcriptional elongation.69,70 GePS analysis did not reveal any significant association among eIF3 and prefoldin members; however the large size of HERC2 and identification of eIF3 and prefoldins by adjacent HERC2 fragments with 80 amino acids of overlap suggest that HERC2 could serve to structurally bridge translation initiation and protein folding by these complexes. As prefoldins assist in the folding of proteins bound for the cytoskeleton, their association with HERC2 also strengthens the potential for HERC2 function in intracellular trafficking and transport pathways.
Regarding an alternative potential role for HERC2 in translation, the HPV E6 oncoprotein has been reported to stimulate the mammalian target of rapamycin complex 1 (mTORC1) and cap-dependent translation, a phenomenon enhanced by E6 interaction with E6AP.71,72 Since HERC2 interacts with E6AP-bound to E6, it is possible that HERC2 and E6AP contribute to cellular protein translation by cooperative interactions with eIF3, eIF4, prefoldin and mTORC1, and this activity may be enhanced in HPV-positive cells. Additionally, the HPV genome encodes several proteins that impact the maintenance of genomic integrity, and recent studies have shown a strong link between HPV genome replication and cellular DNA damage.73−75 Given the involvement of HERC2 in the maintenance of genomic integrity and repair, high risk HPV E6 proteins may possibly exert some effect on viral genome replication by establishing and/or sustaining a cellular DNA repair response through HERC2.
Iron homeostasis is critical to cellular function and is dependent upon the iron carrier ferritin. Both ferritin subunits (FTH1 and FTL) were identified as HCIPs, linked in both major HERC2-interaction networks involving intracellular transport and energy, and bind endogenous HERC2; however, whether the ferritin–HERC2 interaction is direct remains to be established. A recent report has suggested a role for HERC2 in iron metabolism through stability control of FBXL5,15 and another study provided evidence for NCOA4 as an autophagic cargo receptor for ferritin, regulating ferritin stability and turnover upon depletion of intracellular iron.59 In the latter study, HERC2 was identified as an interactor of NCOA4 by mass spectrometry, and here we have corroborated that finding by the reciprocal MS analysis. Further studies are necessary to define the role of HERC2 in NCOA4 and ferritin biology as well as intracellular iron homeostasis.
Our analysis also identified 10 proteasome subunits as HERC2 HCIPs (33 total proteasomal subunits were identified by MS). Given that HERC2 is an ubiquitin ligase and regulator of protein stability, identification of HERC2 association with the proteasome was not unexpected. The binding of HERC2 to the proteasome may be direct, to facilitate degradation of ubiquitylated substrates, or indirect, occurring through E6AP, as E6AP forms a stable interaction with HERC2 and is known to bind the proteasome.22,76−80
No reported HERC2 interactors involved in DNA damage and repair (i.e., RNF8, BRCA1, XPA)7,9,11 were identified as HCIPs in our study. This may be a consequence of the cells used in the analysis, which were not treated to induce DNA damage, or differences in the extraction procedure that we used, or the possibility that adenovirus E1A and/or SV40 large T antigen, which are expressed in the 293T cells, might perturb interactions with HERC2. Additional proteomic analyses of HERC2 under DNA damage conditions may be helpful to determine whether HERC2–protein interactions are altered during DNA damage repair.
In summary, we have utilized a novel proteomic approach to identify potential cellular interacting partners of the giant ubiquitin ligase HERC2. Our analysis generated well over 200 HCIPs that have not been previously reported to interact with HERC2. An in-depth bioinformatic analysis placed HERC2 in new pathways governing critical cellular processes including protein translation, vesicular trafficking, and energy metabolism. These pathways contribute to the functional versatility of HERC2 and will undoubtedly lead to new insights in the dynamics of these processes. This analysis provides a platform to study HERC2 in important disease contexts including HPV infection and cancer, as well as Angelman Syndrome and autism-spectrum disorders. The identification of new pathways involved in those conditions will improve our comprehension of their etiology and could result in new therapeutic targets for their treatment.
Acknowledgments
We thank members of the Howley and Harper laboratories for helpful discussions and suggestions. We also thank Ed Huttlin of the Gygi Lab (Cell Biology, Harvard Medical School) for the percent sequence coverage of all protein identification data.
Glossary
Abbreviations
- RING
really interesting new gene
- SCF
Skp, Cullin, F-box
- CMV2
cytomegalovirus 2
- MS
mass spectrometry
- SFCA-PF
surfactant free cellulose acetate – pre filter
- TCA
trichloroacetic acid
- LTQ
linear trap quadropole
- FDR
false discovery rate
- CompPASS
Comparative Proteomic Analysis Software Suite
- NWD
normalized weighted D score
- HA
hemagglutinin
- IP
immunoprecipitation
- HRP
horseradish peroxidase
- TNET
Tris, NaCl, EDTA, Tween 20
- NCI
National Cancer Institute
- DBSubLoc
Database of protein Subcellular Localization
- STITCH
Search Tool for Interactions of Chemicals
- ENCODE
Encyclopedia of DNA Elements
- RLD
RCC1-like domain
- GEF
guanine nucleotide exchange factor
- HCIP
high confidence interacting protein
- eIF
eukaryotic initiation factor
- PhK
phosphorylase kinase
- GO
gene ontology
- BP
biological process
- ER
endoplasmic reticulum
- COPI
coat protein I
- HPV
human papillomavirus
- SV40
simian vacuolating virus 40
- ZF
zinc finger
- MIB
mind bomb
- DOC
deleted in oral cancer also known as APC10 - anaphase promoting complex subunit 10
- Cyt-b5
cytochrome-b5
- Dox
doxycycline
Supporting Information Available
Full HERC2 fragment MS and CompPASS proteomic analysis: HERC2_Fragment_Proteomic_Analysis Protein and peptide identification data sets for individual HERC2 fragments: HERC2_Fragment_1_MS_Protein_Peptide_ID HERC2_Fragment_2_MS_Protein_Peptide_ID HERC2_Fragment_3_MS_Protein_Peptide_ID HERC2_Fragment_4_MS_Protein_Peptide_ID HERC2_Fragment_5_MS_Protein_Peptide_ID HERC2_Fragment_6_MS_Protein_Peptide_ID This material is available free of charge via the Internet at http://pubs.acs.org.
This work has been supported by National Institutes of Health Grants T32AI007245 to J.T.G., P01CA50661 to P.M.H., and R01AG011085 to J.W.H.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Huibregtse J. M.; Scheffner M.; Beaudenon S.; Howley P. M. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. U. S. A. 1995, 92115249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y.; Rebert N. A.; Joslin J. M.; Higgins M. J.; Schultz R. A.; Nicholls R. D. Structure of the highly conserved HERC2 gene and of multiple partially duplicated paralogs in human. Genome Res. 2000, 103319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo F. R.; Rosa J. L. The HERC proteins: functional and evolutionary insights. Cell. Mol. Life Sci. 2005, 62161826–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochrainer K.; Mayer H.; Baranyi U.; Binder B.; Lipp J.; Kroismayr R. The human HERC family of ubiquitin ligases: novel members, genomic organization, expression profiling, and evolutionary aspects. Genomics 2005, 852153–164. [DOI] [PubMed] [Google Scholar]
- Puffenberger E. G.; Jinks R. N.; Wang H.; Xin B.; Fiorentini C.; Sherman E. A.; Degrazio D.; Shaw C.; Sougnez C.; Cibulskis K.; Gabriel S.; Kelley R. I.; Morton D. H.; Strauss K. A. A homozygous missense mutation in HERC2 associated with global developmental delay and autism spectrum disorder. Hum. Mutat. 2012, 33121639–1646. [DOI] [PubMed] [Google Scholar]
- Harlalka G. V.; Baple E. L.; Cross H.; Kuhnle S.; Cubillos-Rojas M.; Matentzoglu K.; Patton M. A.; Wagner K.; Coblentz R.; Ford D. L.; Mackay D. J.; Chioza B. A.; Scheffner M.; Rosa J. L.; Crosby A. H. Mutation of HERC2 causes developmental delay with Angelman-like features. J. Med. Genet 2013, 50265–73. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S.; Rendtlew Danielsen J.; Fugger K.; Gromova I.; Nerstedt A.; Lukas C.; Bartek J.; Lukas J.; Mailand N. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat. Cell Biol. 2010, 12180–6sup pp 1–12. [DOI] [PubMed] [Google Scholar]
- Kang T. H.; Lindsey-Boltz L. A.; Reardon J. T.; Sancar A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc. Natl. Acad. Sci. U. S. A. 2010, 107114890–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T. H.; Reardon J. T.; Sancar A. Regulation of nucleotide excision repair activity by transcriptional and post-transcriptional control of the XPA protein. Nucleic Acids Res. 2011, 3983176–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. H.; Park J. M.; Leem S. H.; Kang T. H. Coordinated regulation of XPA stability by ATR and HERC2 during nucleotide excision repair. Oncogene 2012, 33119–25. [DOI] [PubMed] [Google Scholar]
- Wu W.; Sato K.; Koike A.; Nishikawa H.; Koizumi H.; Venkitaraman A. R.; Ohta T. HERC2 is an E3 ligase that targets BRCA1 for degradation. Cancer Res. 2010, 70156384–92. [DOI] [PubMed] [Google Scholar]
- Izawa N.; Wu W.; Sato K.; Nishikawa H.; Kato A.; Boku N.; Itoh F.; Ohta T. HERC2 Interacts with Claspin and regulates DNA origin firing and replication fork progression. Cancer Res. 2011, 71175621–5. [DOI] [PubMed] [Google Scholar]
- Cubillos-Rojas M.; Amair-Pinedo F.; Peiro-Jordan R.; Bartrons R.; Ventura F.; Rosa J. L. The E3 Ubiquitin Protein Ligase HERC2Modulates the Activity of Tumor Protein p53 by Regulating Its Oligomerization. J. Biol. Chem. 2014, 2892114782–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan N. C.; den Besten W.; Sweredoski M. J.; Hess S.; Deshaies R. J.; Chan D. C. Degradation of the Deubiquitinating Enzyme USP33 Is Mediated by p97 and the Ubiquitin Ligase HERC2. J. Biol. Chem. 2014, 2892819789–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroishi T.; Yamauchi T.; Nishiyama M.; Nakayama K. I. HERC2 Targets the Iron Regulator FBXL5 for Degradation and Modulates Iron Metabolism. J. Biol. Chem. 2014, 2892316430–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Kim S.; Kobayashi T.; Liang F. X.; Korzeniewski N.; Duensing S.; Dynlacht B. D. Neurl4, a novel daughter centriole protein, prevents formation of ectopic microtubule organizing centres. EMBO Rep. 2012, 13, 547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hakim A. K.; Bashkurov M.; Gingras A. C.; Durocher D.; Pelletier L. Interaction proteomics identify NEURL4 and the HECT E3 ligase HERC2 as novel modulators of centrosome architecture. Mol. Cell Proteomics 2012, 116M111 014233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse J. M.; Scheffner M.; Howley P. M. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991, 10134129–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M.; Huibregtse J. M.; Vierstra R. D.; Howley P. M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 1993, 753495–505. [DOI] [PubMed] [Google Scholar]
- Vos R. M.; Altreuter J.; White E. A.; Howley P. M. The ubiquitin-specific peptidase USP15 regulates human papillomavirus type 16 E6 protein stability. J. Virol 2009, 83178885–92.19553310 [Google Scholar]
- Kuhnle S.; Kogel U.; Glockzin S.; Marquardt A.; Ciechanover A.; Matentzoglu K.; Scheffner M. Physical and functional interaction of the HECT ubiquitin-protein ligases E6AP and HERC2. J. Biol. Chem. 2011, 2862219410–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Noel G.; Galligan J. T.; Sowa M. E.; Arndt V.; Overton T. M.; Harper J. W.; Howley P. M. Identification and proteomic analysis of distinct UBE3A/E6AP protein complexes. Mol. Cell. Biol. 2012, 32153095–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy G. J.; Mostoslavsky G.; Kotton D. N.; Mulligan R. C. Exogenous control of mammalian gene expression via modulation of translational termination. Nat. Med. 2006, 1291093–9. [DOI] [PubMed] [Google Scholar]
- Sowa M. E.; Bennett E. J.; Gygi S. P.; Harper J. W. Defining the human deubiquitinating enzyme interaction landscape. Cell 2009, 1382389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas W.; Faherty B. K.; Gerber S. A.; Elias J. E.; Beausoleil S. A.; Bakalarski C. E.; Li X.; Villen J.; Gygi S. P. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol. Cell Proteomics 2006, 571326–37. [DOI] [PubMed] [Google Scholar]
- Eng J. K.; McCormack A. L.; Yates J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994, 511976–89. [DOI] [PubMed] [Google Scholar]
- Elias J. E.; Gygi S. P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 2007, 43207–14. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii A. I.; Keller A.; Kolker E.; Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003, 75174646–58. [DOI] [PubMed] [Google Scholar]
- Behrends C.; Sowa M. E.; Gygi S. P.; Harper J. W. Network organization of the human autophagy system. Nature 2010, 466730268–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethunaickan R.; Berthier C. C.; Zhang W.; Kretzler M.; Davidson A. Comparative transcriptional profiling of 3 murine models of SLE nephritis reveals both unique and shared regulatory networks. PLoS One 2013, 810e77489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriz G. F.; King O. D.; Bryant B.; Sander C.; Roth F. P. Characterizing gene sets with FuncAssociate. Bioinformatics 2003, 19182502–4. [DOI] [PubMed] [Google Scholar]
- Ashburner M.; Ball C. A.; Blake J. A.; Botstein D.; Butler H.; Cherry J. M.; Davis A. P.; Dolinski K.; Dwight S. S.; Eppig J. T.; Harris M. A.; Hill D. P.; Issel-Tarver L.; Kasarskis A.; Lewis S.; Matese J. C.; Richardson J. E.; Ringwald M.; Rubin G. M.; Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25125–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer C. F.; Anthony K.; Krupa S.; Buchoff J.; Day M.; Hannay T.; Buetow K. H. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009, 37Database issueD674–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E. G.; Gross B. E.; Demir E.; Rodchenkov I.; Babur O.; Anwar N.; Schultz N.; Bader G. D.; Sander C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011, 39Database issueD685–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch M.; Klocke B.; Haltmeier M.; Frech K. LitInspector: literature and signal transduction pathway mining in PubMed abstracts. Nucleic Acids Res. 2009, 37Web Server issueW135–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K.; Frech K.; Grote K.; Klocke B.; Haltmeier M.; Klingenhoff A.; Frisch M.; Bayerlein M.; Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 2005, 21132933–42. [DOI] [PubMed] [Google Scholar]
- UniProt C. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010, 38Database issueD142–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T.; Hua S.; Ji X.; Sun Z. DBSubLoc: database of protein subcellular localization. Nucleic Acids Res. 2004, 32Database issueD122–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M.; von Mering C.; Campillos M.; Jensen L. J.; Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2008, 36Database issueD684–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson H.; Kapushesky M.; Kolesnikov N.; Rustici G.; Shojatalab M.; Abeygunawardena N.; Berube H.; Dylag M.; Emam I.; Farne A.; Holloway E.; Lukk M.; Malone J.; Mani R.; Pilicheva E.; Rayner T. F.; Rezwan F.; Sharma A.; Williams E.; Bradley X. Z.; Adamusiak T.; Brandizi M.; Burdett T.; Coulson R.; Krestyaninova M.; Kurnosov P.; Maguire E.; Neogi S. G.; Rocca-Serra P.; Sansone S. A.; Sklyar N.; Zhao M.; Sarkans U.; Brazma A. ArrayExpress update--from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009, 37Database issueD868–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D.; Mundo A. F.; Haw R.; Milacic M.; Weiser J.; Wu G.; Caudy M.; Garapati P.; Gillespie M.; Kamdar M. R.; Jassal B.; Jupe S.; Matthews L.; May B.; Palatnik S.; Rothfels K.; Shamovsky V.; Song H.; Williams M.; Birney E.; Hermjakob H.; Stein L.; D’Eustachio P. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014, 42Database issueD472–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium E. P. A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 2011, 94e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F. R.; Ponstingl H. Mitotic regulator protein RCC1 is complexed with a nuclear ras-related polypeptide. Proc. Natl. Acad. Sci. U. S. A. 1991, 882310830–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F. R.; Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 1991, 354634880–2. [DOI] [PubMed] [Google Scholar]
- Echard A.; Jollivet F.; Martinez O.; Lacapere J. J.; Rousselet A.; Janoueix-Lerosey I.; Goud B. Interaction of a Golgi-associated kinesin-like protein with Rab6. Science 1998, 2795350580–5. [DOI] [PubMed] [Google Scholar]
- Opdam F. J.; Echard A.; Croes H. J.; van den Hurk J. A.; van de Vorstenbosch R. A.; Ginsel L. A.; Goud B.; Fransen J. A. The small GTPase Rab6B, a novel Rab6 subfamily member, is cell-type specifically expressed and localised to the Golgi apparatus. J. Cell Sci. 2000, 113Pt 152725–35. [DOI] [PubMed] [Google Scholar]
- Ciccia A.; Constantinou A.; West S. C. Identification and characterization of the human mus81-eme1 endonuclease. J. Biol. Chem. 2003, 2782725172–8. [DOI] [PubMed] [Google Scholar]
- Ogrunc M.; Sancar A. Identification and characterization of human MUS81-MMS4 structure-specific endonuclease. J. Biol. Chem. 2003, 2782421715–20. [DOI] [PubMed] [Google Scholar]
- Starokadomskyy P.; Gluck N.; Li H.; Chen B.; Wallis M.; Maine G. N.; Mao X.; Zaidi I. W.; Hein M. Y.; McDonald F. J.; Lenzner S.; Zecha A.; Ropers H. H.; Kuss A. W.; McGaughran J.; Gecz J.; Burstein E. CCDC22 deficiency in humans blunts activation of proinflammatory NF-kappaB signaling. J. Clin Invest 2013, 12352244–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bie P.; van de Sluis B.; Burstein E.; Duran K. J.; Berger R.; Duckett C. S.; Wijmenga C.; Klomp L. W. Characterization of COMMD protein-protein interactions in NF-kappaB signalling. Biochem. J. 2006, 398163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B.; Stoffel W. Human and murine serine-palmitoyl-CoA transferase--cloning, expression and characterization of the key enzyme in sphingolipid synthesis. Eur. J. Biochem. 1997, 2491239–47. [DOI] [PubMed] [Google Scholar]
- Ferlinz K.; Kopal G.; Bernardo K.; Linke T.; Bar J.; Breiden B.; Neumann U.; Lang F.; Schuchman E. H.; Sandhoff K. Human acid ceramidase: processing, glycosylation, and lysosomal targeting. J. Biol. Chem. 2001, 2763835352–60. [DOI] [PubMed] [Google Scholar]
- Galjart N. J.; Morreau H.; Willemsen R.; Gillemans N.; Bonten E. J.; d’Azzo A. Human lysosomal protective protein has cathepsin A-like activity distinct from its protective function. J. Biol. Chem. 1991, 2662214754–62. [PubMed] [Google Scholar]
- Turk V.; Stoka V.; Vasiljeva O.; Renko M.; Sun T.; Turk B.; Turk D. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim. Biophys. Acta 2012, 1824168–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth R. B.; Sun L.; Ea C. K.; Chen Z. J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 2005, 1225669–82. [DOI] [PubMed] [Google Scholar]
- Weihofen A.; Binns K.; Lemberg M. K.; Ashman K.; Martoglio B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science 2002, 29655762215–2218. [DOI] [PubMed] [Google Scholar]
- Li X. C.; Raghavan M. Structure and function of major histocompatibility complex class I antigens. Curr. Opin Organ Transplant 2010, 154499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis T. E.; Lowe M.; Pepperkok R. COPs regulating membrane traffic. Annu. Rev. Cell Dev Biol. 1995, 11, 677–706. [DOI] [PubMed] [Google Scholar]
- Mancias J. D.; Wang X.; Gygi S. P.; Harper J. W.; Kimmelman A. C. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 2014, 5097498105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matanis T.; Akhmanova A.; Wulf P.; Del Nery E.; Weide T.; Stepanova T.; Galjart N.; Grosveld F.; Goud B.; De Zeeuw C. I.; Barnekow A.; Hoogenraad C. C. Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat. Cell Biol. 2002, 412986–992. [DOI] [PubMed] [Google Scholar]
- Tsukazaki T.; Chiang T. A.; Davison A. F.; Attisano L.; Wrana J. L. SARA, a FYVE domain protein that recruits Smad2 to the TGFbeta receptor. Cell 1998, 956779–91. [DOI] [PubMed] [Google Scholar]
- Panopoulou E.; Gillooly D. J.; Wrana J. L.; Zerial M.; Stenmark H.; Murphy C.; Fotsis T. Early endosomal regulation of Smad-dependent signaling in endothelial cells. J. Biol. Chem. 2002, 2772018046–18052. [DOI] [PubMed] [Google Scholar]
- Hu Y.; Chuang J. Z.; Xu K.; McGraw T. G.; Sung C. H. SARA, a FYVE domain protein, affects Rab5-mediated endocytosis. J. Cell Sci. 2002, 115Pt 244755–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang J. Z.; Zhao Y.; Sung C. H. SARA-regulated vesicular targeting underlies formation of the light-sensing organelle in mammalian rods. Cell 2007, 1303535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey J. W. Regulation of protein synthesis and the role of eIF3 in cancer. Braz. J. Med. Biol. Res. 2010, 4310920–30. [DOI] [PubMed] [Google Scholar]
- Brushia R. J.; Walsh D. A. Phosphorylase kinase: the complexity of its regulation is reflected in the complexity of its structure. Front Biosci 1999, 4, D618–D641. [DOI] [PubMed] [Google Scholar]
- Brembeck F. H.; Rosario M.; Birchmeier W. Balancing cell adhesion and Wnt signaling, the key role of beta-catenin. Curr. Opin Genet Dev 2006, 16151–59. [DOI] [PubMed] [Google Scholar]
- Vader G.; Medema R. H.; Lens S. M. The chromosomal passenger complex: guiding Aurora-B through mitosis. J. Cell Biol. 2006, 1736833–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainberg I. E.; Lewis S. A.; Rommelaere H.; Ampe C.; Vandekerckhove J.; Klein H. L.; Cowan N. J. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell 1998, 935863–873. [DOI] [PubMed] [Google Scholar]
- Millan-Zambrano G.; Rodriguez-Gil A.; Penate X.; de Miguel-Jimenez L.; Morillo-Huesca M.; Krogan N.; Chavez S. The prefoldin complex regulates chromatin dynamics during transcription elongation. PLoS Genet 2013, 99e1003776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangle J. M.; Ghosh-Choudhury N.; Munger K. Activation of cap-dependent translation by mucosal human papillomavirus E6 proteins is dependent on the integrity of the LXXLL binding motif. J. Virol 2012, 86147466–7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangle J. M.; Munger K. The human papillomavirus type 16 E6 oncoprotein activates mTORC1 signaling and increases protein synthesis. J. Virol 2010, 84189398–9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody C. A.; Laimins L. A. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog 2009, 510e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara N.; Mitra R.; McBride A. A. The papillomavirus E1 helicase activates a cellular DNA damage response in viral replication foci. J. Virol 2011, 85178981–8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie K. A.; Mehta K. P.; Laimins L. A.; Moody C. A. Human papillomaviruses recruit cellular DNA repair and homologous recombination factors to viral replication centers. J. Virol 2012, 86179520–9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleijnen M. F.; Shih A. H.; Zhou P.; Kumar S.; Soccio R. E.; Kedersha N. L.; Gill G.; Howley P. M. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell 2000, 62409–419. [DOI] [PubMed] [Google Scholar]
- Wang X.; Chen C. F.; Baker P. R.; Chen P. L.; Kaiser P.; Huang L. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry 2007, 46113553–3565. [DOI] [PubMed] [Google Scholar]
- Besche H. C.; Haas W.; Gygi S. P.; Goldberg A. L. Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry 2009, 48112538–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon T. C.; Gottlieb B.; Durcan T. M.; Fon E. A.; Beitel L. K.; Trifiro M. A. Isolation of human proteasomes and putative proteasome-interacting proteins using a novel affinity chromatography method. Exp. Cell Res. 2009, 3152176–189. [DOI] [PubMed] [Google Scholar]
- Tai H. C.; Besche H.; Goldberg A. L.; Schuman E. M. Characterization of the Brain 26S Proteasome and its Interacting Proteins. Front. Mol. Neurosci. 2010, 3, pii: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.