Recent studies have started to reveal that protein synthesis is a more highly regulated process than was previously appreciated, although many aspects of translation regulation remain unknown. Two new reports describe the discovery of a bacterial protein factor, energy-dependent translation throttle A (EttA), which can regulate translation in response to changes in cellular energy homeostasis1,2. The ability of EttA to link protein synthesis to changing metabolite levels opens up the exciting possibility that other translation factors might also respond to different physiological cues, as is the case for transcriptional regulation.
EttA (previously known as YjjK) is a member of the ATP-binding cassette (ABC) protein family, best known for their role in membrane transport. However, many ABC proteins are not membrane-associated transporters and have instead been implicated in a wide range of other roles including DNA repair and replication (UvrA), enzyme regulation (GCN20), macrolide-lincosamide - streptogramin antibiotic resistance (MsrA) and translation regulation (ABC50, eEF3 and ABCE1)3,4. EttA is a member of the ABC-F subfamily, whose members all lack membrane-spanning domains. Its similarity to other proteins implicated in translation prompted the authors of the two studies in this issue to probe its role in more detail.
Boël et al.1 report the X-ray crystal structure of nucleotide-free EttA at 2.4-Å resolution ,and Chen et al.2 report the cryo-EM structure of a mutant version of EttA bound to the ribosome. This EttA mutant (E188Q E470Q, termed EttA-EQ2) is a key tool in the present studies because it is able to bind, but not hydrolyze, ATP. In the crystal lattice, EttA forms a domain-swapped dimer, although several observations, including the cryo-EM reconstitution, suggest that the monomer is the biologically active form1,2. The tandem nucleotide-binding domains (NBDs) of EttA resemble those of other ABC proteins except for two insertions termed the ‘toe’ and ‘arm', the latter being a longer extension comprising two α-helices. An ~70-residue ‘linker’ motif, unique to ABC-F proteins lies between the tandem NBDs and contains an extended α-helix. Overall, the protein adopts an ‘open’ conformation, with the subdomains organized loosely with respect to one another, as expected for an ABC protein without a bound nucleotide1.
The extended features of EttA revealed by crystallography allowed EttA-EQ2 to be modeled unambiguously into the cryo-EM map of the ribosome complex. EttA-EQ2 binds the exit (E) site, filling the space between the L1 stalk and P-site tRNA (Fig. 1)2. A model of EttA in the ‘closed’ conformation best fits the cryo-EM density, consistent with the EttA-EQ2 variant being trapped in the ATP-bound state. The arm of EttA-EQ2 interacts with the L1 stalk, a dynamic element of the ribosome responsible for facilitating movement of tRNA from the peptidyl (P) to the E site during translocation. On the other side of the ATPase, the linker motif contacts fMet-tRNA, which is bound to the P site. Because this motif is conserved within the ABC-F subfamily, it was termed the P-site tRNA-interaction motif (PtIM). The terminus of PtIM interacts with the acceptor end of the P-site tRNA, potentially influencing tRNA positioning in the peptidyl transferase center.
Figure 1.
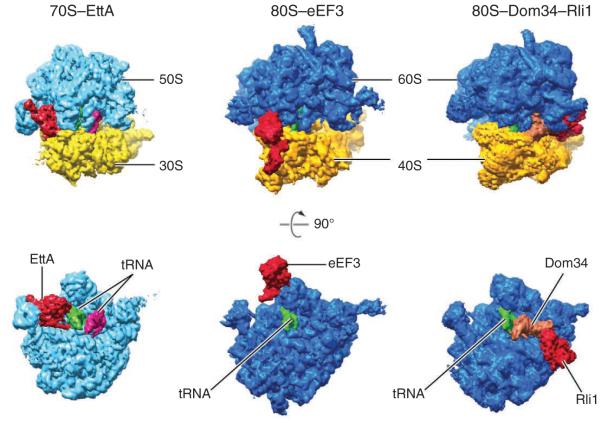
ABC proteins bind to different sites on the ribosome. Comparison of the positions of EttA on the 70S ribosome (left), eEF3 on the 80S ribosome (center) and Dom34-Rli1 on the 80S ribosome (right). Top images show the entire ribosome, viewed down the subunit interface from above; bottom images show the large subunit viewed from the interface. Red, ABC proteins; green, P-site tRNA; magenta, A-site tRNA; orange, release-factor paralog Dom34.
The intriguing observation that EttA binds the ribosomal E site and contacts the tRNA in the P site suggested that the ABC-F protein might regulate translation elongation in response to the ATP/ADP ratio in the cell. Consistent with this possibility, expression of EttA-EQ2 in Escherichia coli conferred dominant lethality and strongly inhibited protein synthesis, effects that depend on the EttA arm motif. To investigate this hypothesis directly, Frank, Hunt and colleagues tested the effects of wild-type EttA and mutant EttA-EQ2 on translation elongation in vitro using purified components. The authors formed 70S initiation complexes, provided them with aminoacyl-tRNAs, elongation factors, and nucleotides in various combinations, and monitored the products of elongation (di-, tri- and tetrapeptides) following addition of EttA or EttA-EQ2. EttA-EQ2 strongly inhibited translation elongation after formation of the first peptide bond.
In light of the cryo-EM reconstruction, the simplest interpretation of these data is that EttA-EQ2 inhibits translocation by sterically occluding the E site, a conclusion further supported by single-molecule fluorescence resonance energy transfer experiments. Under the same conditions, wild-type EttA had no inhibitory effect on elongation. However, when the reaction contained ADP rather than ATP, EttA caused substantial inhibition of translation. Qualitatively, the effects of EttA–ADP and EttA-EQ2–ATP differed, with EttA–ADP uniquely inhibiting formation of the first peptide bond. Interestingly, at high ATP concentration, wild-type EttA slightly stimulated translation elongation. These findings show that EttA can regulate translation in response to variations in the ATP/ADP ratio, at least in vitro.
To test the idea that EttA regulates translation in energy-depleted cells, Boël et al.1 assessed the fitness of ΔettA cells grown in cultures that were subjected to intervals of stationary phase conditions. Whereas ΔettA cells grew as well as did control cells in mixed cultures under growth-permissive conditions, control cells took over the population when cultures were subjected to increased time in stationary phase. These data imply that EttA contributes either to survival in stationary phase or to the transition from stationary to logarithmic phase after dilution into fresh media.
The authors propose a model, based on their collective findings, for how EttA might regulate translation: as cells enter stationary phase, the increased concentration of ADP causes EttA to inhibit translation initiation by binding the vacant E site of ribosomes positioned at the start codon. This inhibitory function may be part of a larger cellular mechanism, involving pathways such as the stringent response, to place certain ribosomal complexes in a state of hibernation. Upon nutrient replenishment, exchange of ADP for ATP on EttA enables EttA to promote dipeptide formation, hydrolyze ATP and dissociate from the E site. Although this model provides an initial framework for understanding EttA function, many open questions remain. One is how ATP hydrolysis can promote the release of EttA from the ribosome, whereas ADP can stabilize EttA and cause translation inhibition. Another is whether EttA acts in a general manner or targets a particular subset of ribosomal complexes. Future studies to address these questions are undoubtedly on the docket.
EttA is the third soluble ABC protein whose binding site on the ribosome has been determined, as cryo-EM structures reported by Beckmann and co-workers previously revealed the positions of eEF3 and ABCE1 (Rli1) on the 80S ribosome5,6. Notably, these three ABC proteins bind the ribosome at completely different sites (Fig. 1). This arrangement contrasts with that of the translational GTPases (e.g., IF2, EF-Tu, EF-G, RF3 and LepA), which all bind an overlapping site on the aminoacyl (A)-site side of the ribosome7–11. Eukaryotic elongation factor 3 (eEF3) is an ABC-F protein that has important roles in both translation elongation and recycling in fungi12–14. This ATPase binds along the ‘top’ of the ribosome, bridging the central protuberance of the large subunit and the head of the small subunit (Fig. 1)6. ABCE1 (Rli1 in yeast) is highly conserved in Eukarya and Archaea and contributes to both termination and ribosome recycling15–17. Rli1 acts in concert with eRF1 and eRF3 when the ribosome encounters a stop codon and additionally acts with release-factor paralogs Dom 34 and Hbs1 when the ribosome becomes stalled inadvertently (for example, in the ‘no-go decay’ and ‘non-stop decay’ pathways). The binding site for Rli1 overlaps that of the translational GTPases (Fig. 1)5, consistent with biochemical evidence that Rli1 action follows GTP hydrolysis and release of eRF3 or Hbs1 in recycling path-ways16. That EttA, eEF3 and ABCE1 act on distinct regions of the ribosome underscores the functional diversity of this family of molecular motors, and it seems likely that more roles in regulating translation will be assigned to ABC family members in the future.
ACKNOWLEDGMENTS
We thank J. Frank, B. Chen and M. Thomas for providing the figure. Work on translation in the authors’ labs is supported by grants from the US National Institutes of Health (GM 072528 to K.F. and GM065183 to M.I.).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Boël G, et al. Nat. Struct. Mol. Biol. 21:143–151. doi: 10.1038/nsmb.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen B, et al. Nat. Struct. Mol. Biol. 21:152–159. doi: 10.1038/nsmb.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davidson AL, Dassa E, Orelle C. Chen, J. Microbiol. Mol. Biol. Rev. 2008;72:317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kerr ID. Biochem. Biophys. Res. Commun. 2004;315:166–173. doi: 10.1016/j.bbrc.2004.01.044. [DOI] [PubMed] [Google Scholar]
- 5.Becker T, et al. Nature. 2012;482:501–506. doi: 10.1038/nature10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersen CB, et al. Nature. 2006;443:663–668. doi: 10.1038/nature05126. [DOI] [PubMed] [Google Scholar]
- 7.Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. Cell. 2005;121:703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 8.Schmeing TM, et al. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao YG, et al. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J, Lancaster L, Trakhanov S, Noller HF. RNA. 2012;18:230–240. doi: 10.1261/rna.031187.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connell SR, et al. Nat. Struct. Mol. Biol. 2008;15:910–915. doi: 10.1038/nsmb.1469. [DOI] [PubMed] [Google Scholar]
- 12.Triana-Alonso FJ, Chakraburtty K, Nierhaus KH. J. Biol. Chem. 1995;270:20473–20478. doi: 10.1074/jbc.270.35.20473. [DOI] [PubMed] [Google Scholar]
- 13.Kurata S, et al. Proc. Natl. Acad. Sci. USA. 2010;107:10854–10859. doi: 10.1073/pnas.1006247107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurata S, et al. Nucleic Acids Res. 2013;41:264–276. doi: 10.1093/nar/gks958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pisarev AV, et al. Mol. Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoemaker CJ, Green R. Proc. Natl. Acad. Sci. USA. 2011;108:E1392–E1398. doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franckenberg S, Becker T, Beckmann R. Curr. Opin. Struct. Biol. 2012;22:786–796. doi: 10.1016/j.sbi.2012.08.002. [DOI] [PubMed] [Google Scholar]