Abstract
Background
The molecular alterations that drive tumorigenesis in intrahepatic cholangiocarcinoma (ICC) remain poorly defined. We sought to determine the incidence and prognostic significance of mutations associated with ICC among patients undergoing surgical resection.
Methods
Multiplexed mutational profiling was performed using nucleic acids that were extracted from 200 resected ICC tumor specimens from 7 centers. The frequency of mutations was ascertained and the effect on outcome was determined.
Results
The majority of patients (61.5 %) had no genetic mutation identified. Among the 77 patients (38.5 %) with a genetic mutation, only a small number of gene mutations were identified with a frequency of >5 %: IDH1 (15.5 %) and KRAS (8.6 %). Other genetic mutations were identified in very low frequency: BRAF (4.9 %), IDH2 (4.5 %), PIK3CA (4.3 %), NRAS (3.1 %), TP53 (2.5 %), MAP2K1 (1.9 %), CTNNB1 (0.6 %), and PTEN (0.6 %). Among patients with an IDH1-mutant tumor, approximately 7 % were associated with a concurrent PIK3CA gene mutation or a mutation in MAP2K1 (4 %). No concurrent mutations in IDH1 and KRAS were noted. Compared with ICC tumors that had no identified mutation, IDH1-mutant tumors were more often bilateral (odds ratio 2.75), while KRAS-mutant tumors were more likely to be associated with R1 margin (odds ratio 6.51) (both P < 0.05). Although clinicopathological features such as tumor number and nodal status were associated with survival, no specific mutation was associated with prognosis.
Conclusions
Most somatic mutations in resected ICC tissue are found at low frequency, supporting a need for broad-based mutational profiling in these patients. IDH1 and KRAS were the most common mutations noted. Although certain mutations were associated with ICC clinicopathological features, mutational status did not seemingly affect long-term prognosis.
Biliary tract cancers include a spectrum of invasive carcinomas encompassing cancers arising in the intrahepatic, perihilar, or distal biliary tree (cholangiocarcinoma), as well as carcinomas arising from the gallbladder. Intra-hepatic cholangiocarcinoma (ICC) represents a unique entity with particular clinical challenges. ICC is the second most common form of liver malignancy, with an incidence and mortality that have steadily increased over the last decade.1 Although a subset of individuals with ICC have identifiable risk factors such as primary sclerosing cholangitis or liver fluke infestation, the majority have no underlying risk factors that can be used to develop screening strategies for early detection. Although resection remains the sole curative treatment option, surgery is only feasible in the 10–20 % of patients who present with early-stage disease.1,2 For those patients with advanced disease, treatment typically includes systemic therapy with gemcitabine and cisplatin combination chemotherapy. However, the median survival of patients with locally advanced or metastatic disease continues to be less than 1 year.3
There remains an unmet need to identify novel molecular signatures in cholangiocarcinoma with prognostic and therapeutic implications. Recently, data on the genetic signatures and molecular mechanisms underlying the pathogenesis of ICC have begun to emerge.4,5 For example, some groups have reported somatic alterations in the KRAS, TP53, CDKN2A, and SMAD4 (DPC4) genes in cholangiocarcinoma.6–9 Other investigators have identified mutations in genes encoding for molecules of the phosphatidylinositide 3-kinase (PI3K) cell-signaling pathway (e.g., PIK3CA, PTEN, and AKT1)6,8,9, as well as for isocitrate dehydrogenase (IDH) 1 and 2.6,10,11 Most data on the topic of ICC genetic profiling come from small, single-institution experiences. In addition, some previous reports included data on cholangiocarcinoma from various anatomic locations, including hilar, distal lesions, or gallbladder cancer, with some even including various stages of tumors, making the data heterogeneous and difficult to interpret.6,12 Furthermore, the prognostic significance of newly identified genetic signatures in a well-defined ICC population remains unknown or controversial. Therefore, the aim of the current study was to characterize the genomic profile of ICC among a multi-institutional, international cohort of patients using a broad-based mutational profiling platform. Specifically, we sought to define the frequency of well-established cancer gene mutations, assess the association of these mutations with clinical and morphologic features, and correlate mutations with long-term oncologic outcomes in patients with resected ICCs.
PATIENTS AND METHODS
Patients and Samples
Using an international multi-institutional database, 200 patients with ICC who underwent surgical resection with curative intent between October 1973 and February 2013 at one of seven institutions were identified (Massachusetts General Hospital, Boston, MA; Johns Hopkins School of Medicine, Baltimore, MD; University of Virginia, Charlottesville, VA; Fundeni Clinical Institute of Digestive Disease, Bucharest, Romania; Medical College of Wisconsin, Milwaukee, WI; Cliniques universitaires Saint-Luc, Brussels, Belgium; Queen Mary Hospital at The University of Hong Kong, Hong Kong, China). The institutional review board of each respective institution approved this study. Only patients with histologically confirmed ICC who received their initial treatment for ICC at a study center were included.
Genotype Analysis
After independent pathologic review (VD) of the tumor samples to confirm the diagnosis of ICC, micro-dissection was performed to obtain only tumor samples for DNA extraction. Total nucleic acids were then extracted from formalin-fixed, paraffin-embedded diagnostic tumor tissue obtained from ICC patients using a custom automated platform based on the Agencourt FormaPure System on a Biomek NXP workstation (Beckman Coulter Genomics, Danvers, MA). Mutational profiling was performed on these nucleic acids, which simultaneously queried for over 150 previously described hotspot mutations across 15 cancer genes, including AKT1, APC, BRAF, CTNNB1, EGFR, ERBB2, IDH1, IDH2, KIT, KRAS, MAP2K1, NOTCH1, NRAS, PIK3CA, PTEN, and TP53. This was performed using a custom modified ABI Prism SNaPshot Multiplex System on an ABI Prism 3730 DNA Analyzer (Life Technologies/Applied Biosystems), as previously described.13 The SNaPshot genotyping assay is a fast, high-throughput, multiplex mutational profiling method that has the advantage over conventional dideoxy-nucleotide (Sanger) sequencing in that mutations can be detected when mutant DNA comprises as little as 5 % of the total DNA. The specific mutations that were assessed using this SNaPshot approach are listed in Supplemental Table 1. Of note, testing of the tumor suppressor genes TP53, APC and PTEN was limited to only the most common mutation sites, where approximately 30, 15, and 15 %, of all known somatic mutations in these genes were covered. Mutational profiling was performed at the Translational Research Laboratory, Massachusetts General Hospital Cancer Center.
Data Collection
Standard demographic and clinicopathologic data were collected, including sex, age, and primary tumor characteristics. Specifically, data were collected on primary tumor location, size, and number as well as morphologic subtype and presence of vascular invasion, defined as minor and/or major. Data on treatment-related variables, such as type of surgery, receipt of lymphadenectomy, and adjuvant therapy, were also obtained. Resection was classified as less than hemi-hepatectomy, hemi-hepatectomy, or extended hepatectomy. Margin and nodal status were ascertained on the basis of final pathologic assessment. Date of last follow-up and vital status were collected on all patients.
Statistical Analysis
Summary statistics were obtained using established methods. Discrete variables were described as medians with interquartile range (IQR). Categorical variables were described as totals and frequencies. Univariate comparisons were assessed using the chi-squared or analysis of variance test as appropriate. Overall survival time was calculated from date of surgery to date of death or date of last follow-up. Cox proportional hazards models were developed using relevant mutations to determine the association of each with overall survival. Cumulative event rates were calculated using the Kaplan–Meier method. Univariate and multivariate logistic regression models were constructed to determine the association of relevant clinicopathologic factors with any identified mutation. Each mutation was tested for any possible association with clinical characteristics or tumor morphology using logistic regression models. Relative risks were expressed as hazard ratios (HR) with 95 % confidence intervals (CI). Significance levels were set at P < 0.05; all tests were two sided. All analyses were carried out with Stata version 12.0 (StataCorp, College Station, TX).
RESULTS
Clinical and Treatment Characteristics
Among the 200 patients, there were 111 men (55.5 %) and 89 women (44.5 %). The median patient age was 63 years (IQR 53–70). Median tumor size was 6.0 cm (IQR 4.5–8.5) and most patients had a solitary tumor (n = 156, 78.0 %). At the time of surgery, the extent of resection was less than a hemi-hepatectomy in 54 patients (29.0 %), a hemi-hepatectomy in 99 patients (53.2 %), and an extended hemi-hepatectomy in 33 patients (17.8 %). Surgical margins were R0 in the majority of patients (n = 179, 90.4 %), while a smaller number of patients had an R1 margin (n = 19, 9.6 %). Lymphadenectomy was performed in 86 patients (43.0 %). On final pathology, a majority of patients had T1 tumors (n = 59, 50.4 %), while smaller subsets had T2 (n = 33, 28.2 %) or T3/T4 (n = 25, 21.4 %) tumors. Among the 86 patients who had at least one lymph node evaluated, 32 patients (16.0 %) had lymph node metastasis. As such, 33 patients (38.8 %) were classified as having stage III disease, while 36 (42.4 %) and 14 (16.5 %) were classified as having stage I or II disease, respectively. Microscopic and major vascular invasion was present in 37 patients (18.5 %) and 33 patients (16.5 %), respectively, while 23 patients (11.5 %) had perineural invasion (Table 1).
TABLE 1.
Characteristics of patients with intrahepatic cholangiocarcinoma
| Characteristic | Total (n = 200) | Any mutation (n = 77, 38.5 %)a |
No mutation (n = 123, 61.5 %) |
P |
|---|---|---|---|---|
| Age, median (IQR) | 63 (53–70) | 67 (55–72) | 60 (53–68) | 0.03 |
| Male gender | 111 (55.5) | 35 (45.5) | 76 (61.8) | 0.01 |
| White race | 149 (74.5) | 65 (84.4) | 84 (68.3) | 0.02 |
| Histologic grade (n = 184) | 0.48 | |||
| 1 | 22 (12.0) | 7 (10.3) | 15 (12.9) | |
| 2 | 112 (60.9) | 46 (67.6) | 66 (56.9) | |
| 3 | 49 (26.6) | 15 (22.1) | 34 (29.3) | |
| 4 | 1 (0.5) | 0 | 1 (0.9) | |
| Size, median (IQR) | 6.0 (4.5–8.5) | 6.0 (4.5–8.5) | 6.0 (4.1–8.0) | 0.33 |
| No. of lesions, mean (SD) | 1.7 (1.5) | 1.6 (1.5) | 1.8 (1.5) | 0.41 |
| Solitary lesion | 156 (78.0) | 58 (75.3) | 98 (79.7) | 0.47 |
| Bilobar involvement | 45 (22.5) | 22 (28.6) | 23 (18.7) | 0.09 |
| AJCC stage (n = 85) | 0.46 | |||
| 1 | 36 (42.4) | 19 (41.3) | 17 (43.6) | |
| 2 | 14 (16.5) | 6 (13.0) | 8 (20.5) | |
| 3 | 33 (38.8) | 19 (41.3) | 14 (35.9) | |
| 4 | 2 (2.3) | 2 (4.4) | 0 | |
| AJCC T stage (n = 117) | 0.42 | |||
| 1 | 59 (50.4) | 34 (50.8) | 25 (50.0) | |
| 2 | 33 (28.2) | 21 (31.3) | 12 (24.0) | |
| 3 | 21 (18.0) | 9 (13.4) | 12 (24.0) | |
| 4 | 4 (3.4) | 3 (4.5) | 1 (2.0) | |
| Liver resection (n = 186) | 0.34 | |||
| Less than hemihepatectomy | 54 (29.0) | 18 (23.7) | 36 (32.7) | |
| Hemihepatectomy | 99 (53.2) | 42 (55.3) | 57 (51.8) | |
| Extended hepatectomy | 33 (17.8) | 16 (21.0) | 17 (15.5) | |
| Margin (n = 198) | 0.02 | |||
| R0 | 179 (90.4) | 65 (84.4) | 114 (94.2) | |
| R1 | 19 (9.6) | 12 (15.6) | 7 (5.8) | |
| Lymphadenectomy | 86 (43.0) | 44 (57.1) | 42 (34.1) | 0.04 |
| Lymph node metastases | 32 (16.0) | 16 (20.8) | 16 (13.0) | 0.56 |
| Vascular invasion | ||||
| Microscopic | 37 (18.5) | 13 (16.9) | 24 (19.5) | 0.73 |
| Major | 33 (16.5) | 13 (16.9) | 20 (16.3) | 0.85 |
| Perineural invasion | 23 (11.5) | 10 (13.0) | 13 (10.6) | 0.66 |
| Biliary invasion | 22 (11.0) | 9 (11.7) | 13 (10.6) | 0.76 |
| Satellite lesions | 45 (22.5) | 19 (24.7) | 26 (21.1) | 0.58 |
| Intrahepatic metastases | 20 (10.0) | 9 (11.7) | 11 (8.9) | 0.54 |
| Recurrence | 98 (49.0) | 42 (54.5) | 56 (45.5) | 0.33 |
| Site of recurrence (n = 98) | 0.30 | |||
| Intrahepatic only | 44 (44.9) | 20 (48.8) | 24 (42.1) | |
| Extrahepatic only | 26 (26.5) | 7 (17.1) | 19 (33.3) | |
| Both intra- and extrahepatic | 28 (28.6) | 14 (34.1) | 14 (24.6) | |
| Adjuvant therapy | 60 (30.0) | 25 (32.5) | 35 (28.5) | 0.83 |
| Death | 112 (56.0) | 46 (59.7) | 66 (53.7) | 0.71 |
Mutations: IDH1, IDH2, BRAF, CTNNB1, KRAS, MAP2K1, NRAS, PIK3CA, PTEN, and TP53. Thirty-eight patients were tested for IDH1 or IDH2 only
In the postoperative setting, about one-third of patients (n = 60, 30.0 %) received adjuvant therapy. At a median follow-up of 23.2 months, 1-, 3-, and 5-year survival was 80.8, 46.7, and 34.8 %, respectively; median overall survival was 31.4 months. Several factors were associated with overall survival. Specifically, tumor size ≥5 cm (HR 1.73, 95 % CI 1.11–2.71), nodal status (HR 3.52, 95 % CI 2.14–5.78), microscopic/major vascular invasion (HR 1.71, 95 % CI 1.16–2.52), satellite lesions/intrahepatic metastasis (HR 2.91, 95 % CI 1.95–4.36), and perineural invasion (HR 1.80, 95 % CI 1.06–3.05) were all associated with a worse long-term prognosis (Supplemental Table 2).
Mutation Analyses
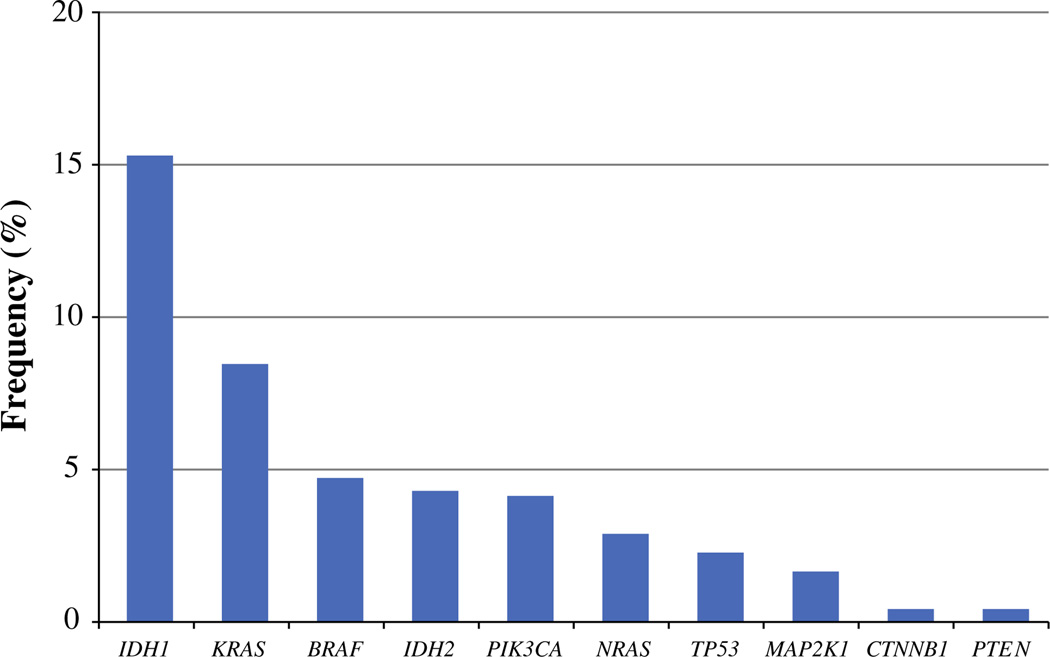
Of the 200 tumor samples evaluated, 162 tumors (81 %) were available for full mutational profiling. The majority (n = 92, 56.8 %) had no genetic mutation identified. Among the 70 patients (43.2 %) who had a tumor with an identified genetic mutation, only a small number of gene mutations were identified with a frequency of greater than approximately 5 % (Fig. 1). Specifically, well-known tumor associated genes such as KRAS (8.6 %) and BRAF (4.9 %) were mutated in roughly 5–10 % of patients. A concurrent KRAS and BRAF mutation was not noted in any patient. Alternations were also identified in the PI3K pathway. Although only one tumor (0.6 %) was found to have a mutation in the most common hotspot regions of PTEN, the incidence of PIK3CA mutations was higher (n = 7, 4.3 %). Genetic mutations in other pathways were identified in very low frequency: NRAS (3.1 %), TP53 (2.5 %), MAP2K1 (1.9 %), and CTNNB1 (0.6 %) (Table 2).
FIG. 1.
A multiplexed mutational profiling platform was used to identify cancer gene mutations in diagnostic cholangiocarcinoma tissue. The frequency of cancer gene mutations identified are expressed as a percentage of all tumors that were tested. Mutational profiling was performed on 162 patient samples. An additional 38 samples were included for IDH1 and IDH2 analysis only (n = 200)
TABLE 2.
Incidence of mutation and association with OS for individual and combinations of mutations
| Characteristic | n (%) | HR | 95 % CI | P | Median OS, mo |
|---|---|---|---|---|---|
| WT | 92 (56.8) | Ref | – | 31.35 | |
| Any mutations | 70 (43.2) | 1.10 | 0.72–1.68 | 0.66 | 31.38 |
| BRAF | 8 (4.9) | 1.06 | 0.38–2.94 | 0.92 | 25.46 |
| CTNNB1 | 1 (0.6) | 1.17 | 0.16–8.55 | 0.88 | – |
| KRAS | 14 (8.6) | 1.99 | 0.96–4.12 | 0.07 | 20.33 |
| MAP2K1 | 3 (1.9) | 1.74 | 0.54–5.64 | 0.36 | 26.94 |
| NRAS | 5 (3.1) | 1.01 | 0.25–4.19 | 0.98 | 28.62 |
| PIK3CA | 7 (4.3) | 0.55 | 0.17–1.76 | 0.31 | 37.34 |
| PTEN | 1 (0.6) | 1.11 | 0.15–8.09 | 0.92 | – |
| TP53 | 4 (2.5) | 1.26 | 0.39–4.08 | 0.70 | 10.82 |
| KRAS or NRAS | 19 (11.7) | 1.69 | 0.86–3.30 | 0.13 | 28.62 |
| KRAS or NRAS or BRAF | 27 (16.7) | 1.44 | 0.80–2.59 | 0.22 | 25.46 |
| MAP2K1 + PIK3CA | 1 (0.6) | 1.23 | 0.17–8.99 | 0.84 | – |
| KRAS or BRAF | 22 (13.6) | 1.54 | 0.83–2.87 | 0.17 | 20.33 |
| PIK3CA or PTEN | 8 (4.9) | 0.62 | 0.22–1.73 | 0.36 | 43.26 |
Full mutational profiling except for IDH (n = 162)
OS overall survival, HR hazard ratio, CI confidence interval, WT wild type
Regarding IDH mutational analyses, 200 tumors samples were available for mutational profiling. A genetic mutation in IDH1 was identified in 31 samples (15.5 %), compared with only 9 samples (4.5 %) for IDH2. Of note, among patients with an IDH1-mutant tumor, approximately 7 % were associated with a concurrent PIK3CA gene mutation, and to a much lower extent, a mutation in MAP2K1 (4 %). No concurrent mutations in IDH1 and KRAS were noted (Table 3).
TABLE 3.
Incidence of mutation and association with overall survival for individual and combinations of mutations for IDH mutation
| Characteristic | n (%) | HR | 95 % CI | P | Median OS, mo |
|---|---|---|---|---|---|
| WT | 123 (61.5) | Ref | – | – | 30.95 |
| Any mutations | 77 (38.5) | 1.08 | 0.75–1.58 | 0.68 | 31.38 |
| IDH1 | 31 (15.5) | 0.98 | 0.58–1.65 | 0.94 | 39.31 |
| IDH2 | 9 (4.5) | 1.16 | 0.47–2.88 | 0.75 | 25.33 |
| IDH1 or IDH2 | 40 (20.0) | 1.01 | 0.63–1.63 | 0.96 | 31.25 |
| IDH1 + PIK3CA | 2 (1.0) | – | – | – | – |
| IDH1 + MAP2K1 | 1 (0.5) | 1.81 | 0.25–13.15 | 0.56 | – |
| IDH2 + BRAF + PIK3CA | 1 (0.5) | 2.57 | 0.35–18.76 | 0.35 | – |
IDH mutations (n = 200)
CI overall survival, OS overall survival
Association of Mutation Status with Clinicopathological Factors and Survival Outcomes
When patients were stratified according to whether “any” mutation was or was not identified, there were no differences in most clinicopathological and treatment characteristics (Table 1). Certain mutations were, however, associated with specific morphologic and pathologic findings. For example, compared with ICC tumors that had no identified mutation, IDH1-mutant tumors were more often bilateral [odds ratio (OR) 2.75, 95 % CI 1.17–6.44], while KRAS-mutant tumors were more likely to be associated with adjacent organ involvement (OR 10.00, 95 % CI 1.29–77.51), and R1 margin status (OR 6.51, 95 %CI 1.63–26.11) (all P < 0.05). Although other clinicopathological features such as tumor size and number as well as nodal status were associated with survival, no specific mutation was associated with these prognostic factors (Table 4).
TABLE 4.
List of mutations that is significantly associated with tumor morphology (compared to no-mutation group)
| Clinical factor | Odds ratio | 95 % CI | P |
|---|---|---|---|
| IDH1 | |||
| Bilobar invasion | 2.75 | 1.17–6.44 | 0.02 |
| KRAS | |||
| R1 margin | 6.51 | 1.63–26.11 | 0.01 |
| Direct involvement of adjacent organ | 10.00 | 1.29–77.51 | 0.03 |
| NRAS | |||
| Intrahepatic metastasis | 6.73 | 1.01–44.68 | 0.05 |
| IDH1 or IDH2 | |||
| Bilobar invasion | 2.72 | 1.24–5.98 | 0.01 |
| NRAS or KRAS | |||
| R1 margin | 5.82 | 1.63–20.81 | 0.01 |
| NRAS, KRAS, or BRAF | |||
| R1 margin | 5.70 | 1.80–18.01 | 0.003 |
CI confidence interval
Median survival among patients with no identified mutation was the same as survival among patients with “any” mutation (both 31.4 months) (Table 2; Supplemental Fig. 1a). Compared with wild-type tumors, median survival was 20.3 months for KRAS mutant cases (P = 0.07) and 25.5 months for BRAF mutant cases (P = 0.92). Cases with either KRAS or BRAF mutations had a median overall survival of 20.3 months (P = 0.17) (Supplemental Fig. 1b). Mutations in the PI3K pathway had a median survival comparable to wild-type cases (PIK3CA mutation only: 37.3 months vs. PIK3CA or PTEN mutation: 43.3 months) (all P > 0.05) (Supplemental Fig. 1c). Similarly, no association with mutations in IDH was noted (IDH1 mutation only: 39.3 months vs. IDH2 mutation only: 25.3 months vs. IDH1 or IDH2 mutation: 31.3 months) (all P > 0.05) (Supplemental Fig. 1d).
DISCUSSION
There has been an emerging interest in the molecular mechanisms of many different gastrointestinal malignancies. For example, data have suggested an important role in the mutation of the two proto-oncogenes, KRAS and BRAF, among many patients with colon cancer.14–16 For hepatocellular carcinoma, genetic events such as gene mutation (e.g., TP53, CTNNB1, KRAS), DNA methylation, and other gene expressions (e.g., IGF, VEGFR, CD24) have been implicated in the multi-step process of hepatocarcinogenesis.17–19 Fewer data are available regarding the molecular underpinnings of ICC. Cholangiocarcinoma is a heterogeneous malignancy with probable varied gene signatures for intrahepatic, proximal, and distal cancers.20 Although previous reports on the genetic profiling of ICC have been published, these data were based on small, single-institution cohorts. The current study is important because we utilized a broad, multi-institutional cohort of patients who underwent surgery for ICC. Genetic profiling was performed at a single center (MGH) and revealed that only a small number of gene mutations were identified with a frequency of approximately 5 %. Specifically, genes such as KRAS and BRAF, as well as those such as PIK3CA, IDH1, and IDH2 were mutated in about 5–15 % of patients with ICC. Interestingly, although we found that certain mutations (e.g., KRAS and IDH1) were associated with specific clinicopathologic and pathologic tumor characteristics, no mutation was a strong predictor of long-term survival.
Mutations in KRAS and BRAF have been noted to be important drivers of tumorigenesis in colon cancer and, to a lesser extent, ICC.10,14–16,21 Although mutations in KRAS and BRAF have been reported in ICC, the frequency of these mutations has varied considerably, ranging from 5 to 50 %.21–23 The reasons for the reported wide-ranging incidence of KRAS and BRAF mutations in ICC are likely multifactorial, including possible large variations that can result from deriving proportions from low sample sizes. In addition, some previous studies reported combined data on both ICC and extrahepatic cholangiocarcinoma, which may have a different incidence of KRAS mutations—thereby further confounding these reports.6,12 In the current study we identified mutations in KRAS in 8.6 % of cases and BRAF mutations in an additional 4.9 % of cases. The identification of KRAS and BRAF mutations in a small subset of patients is consistent with several previous studies.21–23 Interestingly, although it did not reach significance perhaps as a result of lack of statistical power, KRAS mutation tended to be associated with a worse outcome (Supplemental Fig. 1b). Specifically, the median survival of patients with ICC characterized by a KRAS mutation (20.3 months) was about 50 % shorter than the survival of patients with no identified mutation (31.4 months) (P = 0.07) (Table 2). Interestingly, in a separate smaller study, Robertson et al.21 previously reported a comparable difference in survival among patients with KRAS (13.5 months) and wild-type (37.3 months) ICC cases. The identification of KRAS and BRAF mutated tumors may help inform future targeted therapy for ICC. For example, agents such as vemurafenib have antitumor activity in patients with BRAF mutations, whereas patients with KRAS or BRAF mutations are unlikely to be good candidates for EGFR inhibitor therapy.15,16,21,24 In another study with an integrated genetic and genomic analysis, Andersen et al.12 also identified genetic alterations of key signaling molecules and the relevance of EGFR and HER2 targeting in ICC.
IDH1 and IDH2 are genes that have gained considerable interest in patients with ICC.6,8,11 IDH1 and IDH2 (IDH1/2) normally function to catalyze the oxidative carboxylation of isocitrate to α-ketoglutarate. The recurrent cancer mutations in these enzymes confer neomorphic activity through the reduction of α-ketoglutarate to the metabolite R(−)-2-hydroxyglutarate (2HG), resulting in 2HG accumulation in the tumor tissue.25,26 High intracellular levels of 2HG are sufficient for promoting the tumorigenic effects of mutant IDH activity that are associated with enhanced proliferation and impaired differentiation.27 Several previous studies found these mutations to be a significant molecular feature present in approximately 20 % of ICC cases.6,11,28 In the current study, we found a comparable mutation rate for IDH1 (15.5 %), but a lower incidence in the mutation of IDH2 (4.5 %). For IDH mutations, earlier reports suggested that mutations in IDH1 or IDH2 were associated with a longer overall survival and time to tumor recurrence after resection.11 However, a recent study showed the opposite trend; subjects with IDH1 or IDH2 mutations had 3-year survival of 33 % compared with 3-year survival of 81 % for subjects with wild-type IDH genes (P = 0.0034).10 In the current study, we failed to find any association of IDH1/2 mutation with survival (Supplemental Fig. 1d). Collectively, these data suggest that although IDH1 and IDH2 may be one of the more commonly identified genetic mutations in ICC tumors, its effect on patient prognosis remains uncertain.
More recently, exome sequencing has also identified frequent inactivating mutations in multiple chromatin-remodeling genes including BAP1 and ARID1A.10,29,30 Interestingly, comparisons between fluke-related and non-fluke-related ICCs demonstrated statistically significant differences in some mutation patterns including BAP1, which was more frequently mutated in non-fluke-related ICCs.29 In addition to the mutations, FGFR gene fusions have also emerged as a frequent molecular event in ICC.31 In the current study, we did identify low frequency mutations in some other genes including NRAS (3.1 %), TP53 (2.5 %), MAP2K1 (1.9 %), and CTNNB1 (0.6 %), as well as genes in the PI3K pathway. Activation of EGFR can activate downstream pathways such as PI3K.32 Previous reports on mutations in the PI3K pathway among patients with ICC are rare. Compared with the 4 % incidence of PIK3CA mutation noted in the current study, two previous smaller studies reported mutations of 9 and 32 %.9,33
The current study had several limitations. Despite being one of largest series of ICC patients undergoing molecular profiling reported in the literature, the current study still had a relatively small sample size. Given the overall low frequency of genetic mutations found in the cohort, statistical tests assessing differences in the subgroups may have been underpowered. Because only 162 out of 200 patients in our cohort underwent complete mutation analyses, the assessment of concurrent mutations with IDH mutations may be underestimated. Although the multi-institutional study design did offer the benefits of increased generalizability, collaborating with multiple institutions limited the ability to easily standardize all diagnostic and treatment criteria. Furthermore, given that all patients included in the study had undergone surgical resection, these data may not be representative of other patients with more advanced, unresectable ICC. Last, the inherent limitations of multiplexed mutational profiling platform exclude the assessment of other mutations or gene fusions that are potentially relevant in ICC. SNaPshot genotyping platform has inherent limitations and this may explain, at least in part, why only 57 % of tumors had mutations identified in our study compared with a recent report by Ross et al.34 that noted an average of 2.9 alterations per ICC tumor. We are planning to expand our initial findings from this study and use more comprehensive genomic technology [i.e., Next-Generation Sequencing (NGS)] for a future study to assess the prevalence and prognostic significance of other mutations including those in the ARID family and others that have been recently reported.
In conclusion, most patients with resected ICC had no somatic mutation identified on multiplexed mutational profiling. KRAS and IDH1 were the most common mutations noted. Although certain mutations were associated with ICC clinicopathological features, mutational status did not seemingly affect long-term prognosis. Future studies should strive to enhance our understanding of the molecular underpinnings of ICC with more advanced genomic testing platforms in order to refine the prognosis, as well as identify potential therapeutic targets, for patients with this disease.
Supplementary Material
ACKNOWLEDGMENT
We thank Kenneth C. Fan, Hector U. Lopez, and Christina R. Matulis for their technical assistance and Daniel J. Harris for data collection. Supported in part by Agios.
Footnotes
Presented in part at the Annual Society of Surgical Oncology Meeting, Phoenix, AZ, 2014. A video of the presentation of the data in this article at the 67th Annual Society of Surgical Oncology Cancer Symposium is available online (http://www.surgonc.org/vm/).
Electronic supplementary material The online version of this article (doi:10.1245/s10434-014-3828-x) contains supplementary material, which is available to authorized users.
DISCLOSURE The authors declare no conflict of interest.
REFERENCES
- 1.Shaib YH, Davila JA, McGlynn K, et al. Rising incidence of intrahepatic cholangiocarcinoma in the United States: a true increase? J Hepatol. 2004;40:472–477. doi: 10.1016/j.jhep.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 2.Poultsides GA, Zhu AX, Choti MA, et al. Intrahepatic cholangiocarcinoma. Surg Clin North Am. 2010;90:817–837. doi: 10.1016/j.suc.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 3.Valle J, Wasan H, Palmer DH, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med. 2010;362:1273–1281. doi: 10.1056/NEJMoa0908721. [DOI] [PubMed] [Google Scholar]
- 4.Hezel AF, Deshpande V, Zhu AX. Genetics of biliary tract cancers and emerging targeted therapies. J Clin Oncol. 2010;28:3531–3540. doi: 10.1200/JCO.2009.27.4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sia D, Tovar V, Moeini A, et al. Intrahepatic cholangiocarcinoma: pathogenesis and rationale for molecular therapies. Oncogene. 2013;32:4861–4870. doi: 10.1038/onc.2012.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borger DR, Tanabe KK, Fan KC, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17:72–79. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tannapfel A, Sommerer F, Benicke M, et al. Genetic and epigenetic alterations of the INK4a-ARF pathway in cholangiocarcinoma. J Pathol. 2002;197:624–631. doi: 10.1002/path.1139. [DOI] [PubMed] [Google Scholar]
- 8.Voss JS, Holtegaard LM, Kerr SE, et al. Molecular profiling of cholangiocarcinoma shows potential for targeted therapy treatment decisions. Hum Pathol. 2013;44:1216–1222. doi: 10.1016/j.humpath.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Xu RF, Sun JP, Zhang SR, et al. KRAS and PIK3CA but not BRAF genes are frequently mutated in Chinese cholangiocarcinoma patients. Biomed Pharmacother. 2011;65:22–26. doi: 10.1016/j.biopha.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Jiao Y, Pawlik TM, Anders RA, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45:1470–1473. doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang P, Dong Q, Zhang C, et al. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene. 2013;32:3091–3100. doi: 10.1038/onc.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersen JB, Spee B, Blechacz BR, et al. Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors. Gastroenterology. 2012;142:1021.e15–1031.e15. doi: 10.1053/j.gastro.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dias-Santagata D, Akhavanfard S, David SS, et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2:146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karagkounis G, Torbenson MS, Daniel HD, et al. Incidence and prognostic impact of KRAS and BRAF mutation in patients undergoing liver surgery for colorectal metastases. Cancer. 2013;119:4137–4144. doi: 10.1002/cncr.28347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bazan V, Agnese V, Corsale S, et al. Specific TP53 and/or Ki-ras mutations as independent predictors of clinical outcome in sporadic colorectal adenocarcinomas: results of a 5-year Gruppo Oncologico dell’Italia Meridionale (GOIM) prospective study. Ann Oncol. 2005;16(Suppl 4):iv50–iv55. doi: 10.1093/annonc/mdi908. [DOI] [PubMed] [Google Scholar]
- 16.Andreyev HJ, Norman AR, Cunningham D, et al. Kirsten ras mutations in patients with colorectal cancer: the “RASCAL II” study. Br J Cancer. 2001;85:692–696. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woo HG, Park ES, Thorgeirsson SS, et al. Exploring genomic profiles of hepatocellular carcinoma. Mol Carcinog. 2011;50:235–243. doi: 10.1002/mc.20691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S, Lee HJ, Kim JH, et al. Aberrant CpG island hypermethylation along multistep hepatocarcinogenesis. Am J Pathol. 2003;163:1371–1378. doi: 10.1016/S0002-9440(10)63495-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minguez B, Tovar V, Chiang D, et al. Pathogenesis of hepatocellular carcinoma and molecular therapies. Curr Opin Gastroenterol. 2009;25:186–194. doi: 10.1097/MOG.0b013e32832962a1. [DOI] [PubMed] [Google Scholar]
- 20.Khan SA, Toledano MB, Taylor-Robinson SD. Epidemiology, risk factors, and pathogenesis of cholangiocarcinoma. HPB (Oxford) 2008;10:77–82. doi: 10.1080/13651820801992641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robertson S, Hyder O, Dodson R, et al. The frequency of KRAS and BRAF mutations in intrahepatic cholangiocarcinomas and their correlation with clinical outcome. Hum Pathol. 2013;44:2768–2773. doi: 10.1016/j.humpath.2013.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tannapfel A, Benicke M, Katalinic A, et al. Frequency of p16(INK4A) alterations and K-ras mutations in intrahepatic cholangiocarcinoma of the liver. Gut. 2000;47:721–727. doi: 10.1136/gut.47.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sia D, Hoshida Y, Villanueva A, et al. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology. 2013;144:829–840. doi: 10.1053/j.gastro.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sosman JA, Kim KB, Schuchter L, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Losman JA, Looper RE, Koivunen P, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339:1621–1625. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kipp BR, Voss JS, Kerr SE, et al. Isocitrate dehydrogenase 1 and 2 mutations in cholangiocarcinoma. Hum Pathol. 2012;43:1552–1558. doi: 10.1016/j.humpath.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 29.Chan-On W, Nairismagi ML, Ong CK, et al. Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nat Genet. 2013;45:1474–1478. doi: 10.1038/ng.2806. [DOI] [PubMed] [Google Scholar]
- 30.Ong CK, Subimerb C, Pairojkul C, et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat Genet. 2012;44:690–693. doi: 10.1038/ng.2273. [DOI] [PubMed] [Google Scholar]
- 31.Wu YM, Su F, Kalyana-Sundaram S, et al. Identification of tar-getable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636–647. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersen JB, Thorgeirsson SS. Genetic profiling of intrahepatic cholangiocarcinoma. Curr Opin Gastroenterol. 2012;28:266–272. doi: 10.1097/MOG.0b013e3283523c7e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riener MO, Bawohl M, Clavien PA, et al. Rare PIK3CA hotspot mutations in carcinomas of the biliary tract. Genes Chromosomes Cancer. 2008;47:363–367. doi: 10.1002/gcc.20540. [DOI] [PubMed] [Google Scholar]
- 34.Ross JS, Wang J, Gay L, et al. New routes to targeted therapy of intrahepatic cholangiocarcinoma revealed by next-generation sequencing. Oncologist. 2014;19:235–242. doi: 10.1634/theoncologist.2013-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.