Abstract
Myeloid cells represent a major component of the tumor microenvironment where they play divergent dual roles: they can induce antitumor immune responses but mostly they promote immune evasion, tumor progression and metastases formation. Thus, strategies aiming at reprogramming the tumor microenvironment represent a promising immunotherapy approach. Myeloid cells respond to environmental factors including signals derived from commensal microbes. In this Cancer Immunology at the Crossroads overview we discuss recent advances on the effects of the commensal microbiota on myeloid-cell function and how that impacts the response to cancer therapy.
The microbiota modulates inflammation and immunity by priming myeloid-cell differentiation and functions
Commensal microorganisms are abundant on all our epithelial barrier surfaces where, directly or through released molecules, they interact with innate receptors and cytoplasmic sensors thus regulating the development, tone, and maintenance of local inflammation and immunity (1). The interplay between the host immune system and the microbiota prevents tissue-damaging inflammatory responses to the commensals and controls the growth of indigenous pathobionts while it sets the stage for immune responses against pathogenic infections (2–4). This homeostatic immune regulation may be disrupted by changes in the microbial community that alter the symbiotic relationship with the microbiota and the resultant microbial imbalance is commonly referred to as dysbiosis (5). Many regulatory mechanisms involved in these local interactions have been elucidated (6). In addition to local immunity, the commensal microbiota regulates systemic inflammation, innate resistance and adaptive immunity affecting both resistance to infection and autoimmunity (7–12). Maturation of the immune system is dependent on exposure to the microbiota following birth (13). In germ-free (GF) mice, which are protected from exposure to external microbes, spleens and peripheral lymph nodes are hypoplastic, mesenteric lymph nodes are mostly missing while primary immune organs, thymus and bone marrow, have normal appearance (7). However, GF mice mount normal or heightened responses to nominal purified antigens but defective responses to pathogens due to deficient innate and antigen-presenting cell functions (7, 8, 14). Unlike barrier immunity that can be modulated in a compartmentalized manner by the local microbiota (15), the abundant gut microbiota has been considered primarily responsible for the control of immune homeostasis at the systemic level, however, contributions of microbiota from other anatomic locations (e.g. oral cavity) need to be reevaluated (16).
The mechanism by which the microbiota regulates immunity at distant sterile anatomic sites remains largely unknown. Tight junctions among epithelial cells as well as mechanisms mediated by soluble factors (e.g. antibacterial peptides, antibodies) and innate or adaptive immune cells render the skin/mucosal barrier relatively impermeable to microbes and their products (17). However, some bacterial translocation takes place even under normal physiologic conditions. In addition, increased barrier permeability may be induced by infections, inflammation and immunodeficient states that alter anti-microbial defense mechanisms and epithelial integrity (18–21).
Dysbiosis directly affects immunity and also, by changing the predominance of bacterial species with different effects on host immunoregulation, alters the composition of other colonizing microorganisms. For example, overgrowth of the commensal fungal Candida species is often observed following antibiotics-induced gut dysbiosis and it has been shown to result in increased prostaglandin E2 plasma concentration and M2-macrophage polarization in the lung leading to heightened allergic airways inflammation (22).
Recent studies on the modulation of immunity against infection by microbiota have provided insight into how commensals regulate systemic immunity. GF or antibiotics-treated mice have defective myelopoiesis and impaired neutrophil homeostasis with an increased susceptibility to late-onset sepsis (23). Defective myelopoiesis also makes GF mice unable to resist acute infection with Listeria monocytogenes, however, they have an enhanced adaptive immune response to vaccination with an attenuated L. monocytogenes strain, a result compatible with normal or heightened adaptive response to nominal antigens in GF mice (14, 24, 25).
Mice deprived of commensal microbiota have impaired ability to respond to virus infection or virus-derived products. Antibiotic treatment diminishes the immune response to respiratory influenza virus because of lowered constitutive expression of genes encoding pro-IL1β and pro-IL18, and the inability of immune cells to produce and respond to interferon (IFN) (8, 9). Intranasal or systemic administration of the toll-like receptor 4 (TLR4) ligand lipopolysaccharide (LPS) to antibiotics-treated mice corrected the defective anti-influenza virus immune response (8). Dendritic cells (DC) from GF mice fail to respond to the TLR3-ligand poly(I:C) and to LPS with production of cytokines such as type I IFN, IL12, IL6 and tumor necrosis factor (TNF) and to induce natural killer (NK)-cell activation. LPS-induced recruitment of IRF3 and NF-κB as well as Pol II to the promoter region of inflammatory genes such as Ifnb1, Il6 and Tnf is reduced in DCs from GF or antibiotics-treated mice as compared to DCs from specific pathogen-free (SPF) animals (26). DCs from GF mice also show a reduced association of histone H3K4me3 around the transcriptional start site of the same genes, suggesting that signals from the microbiota are required to epigenetically poise these genes for transcription (26). Thus, the microbiota regulates immune homeostasis both locally and systemically acting primarily although not exclusively at the level of myeloid cells. Microbiota-primed myeloid cells have a heightened response to stimuli derived from pathogens or tissue damage, thus establishing a tonic threshold of inflammation that is required for both innate resistance (e.g. activation of NK cells and other innate lymphocytes) and adaptive immunity (12, 26).
The microbiota and cancer
Although cancer-cell proliferation and fitness are linked to genetic mutations and alterations, tumor progression and metastasis formation is highly regulated by the host inflammatory and immune responses (27–31). Inflammation also regulates cancer-predisposing conditions such as obesity (30). Cancer often originates in tissues inflamed due to infection or to physical, chemical, or genetic causes (30). Inflammation may directly induce or facilitate genetic instability and mutations and it always create a tumor-promoting environment that is associated with the production of growth and angiogenic factors, as well as tissue-remodeling enzymes (28, 30). Chemical or infectious carcinogens such as oncogenic viruses and Helicobacter pylori may have a direct mutagenic or cell-transforming ability but they also induce inflammation required for tumor promotion. Established tumors are infiltrated with inflammatory cells and this cancer-associated inflammation maintains the tumor-promoting and immunosuppressive microenvironment (32, 33). Cancer co-morbidities such as anorexia/cachexia, immunodeficiency, pain and depression are known to be modulated by inflammatory unbalance and are likely induced or affected by cancer-associated inflammation (30). The commensal microbiota by setting the inflammatory/immune tone and modulating host response to oncogenic pathogens, cancer-associated inflammation, and tumor-induced tissue damage is a major player affecting the outcome of carcinogenesis, tumor progression, cancer co-morbidities, and response to therapy.
Impact of commensal microbiota on cancer therapy
The gut microbiota influences the response to cancer immunotherapy and chemotherapy by affecting the differentiation and functions of myeloid cells in the tumor microenvironment (Fig. 1). Intratumoral injection of CpG-oligodeoxynucleotide (CpG-ODN) combined with antibody neutralization of IL10 signaling is a very effective treatment of large transplanted subcutaneous tumors in conventional mice but it is largely ineffective in GF or antibiotics-treated mice (34). Within hours following CpG-ODN and anti-IL10R treatment tumors undergo an extensive hemorrhagic necrosis that is dependent on TNF and nitric oxide (NO) production by tumor-infiltrating myeloid cells (35). DCs are then activated, and they migrate to the draining lymph nodes where they induce a CD8 T cell-mediated tumor-specific response required for tumor eradication. In antibiotics-treated or GF mice, tumor-infiltrating myeloid-derived cells fail to produce inflammatory cytokines, including TNF and IL12, in response to CpG-ODN (34). Oral gavage of antibiotics-treated mice with LPS partially rescued the deficient response to CpG-ODN (34). Tumors from antibiotics-treated mice contained a lower number of monocyte-derived Ly6C+ MHC-class II+ macrophage-like cells while the number of Ly6Chigh MHC class II− inflammatory monocytes was equivalent to that of control mice without antibiotics treatment (34). Tumor-associated myeloid-cell subsets have been shown to be mostly derived from circulating inflammatory monocytes that differentiate in situ (36–38). Although inflammatory monocytes appear to infiltrate the tumors in equivalent number, their differentiation after reaching the tumor microenvironment is altered in the absence of gut microbiota and this may affect their response to CpG-ODN (34). Thus, gut commensal bacteria-derived LPS (and possibly other innate receptor ligands) directly or indirectly primes tumor-infiltrating myeloid cells and/or their inflammatory monocyte precursors through TLR4 receptor and enables them to respond to the TLR9 ligand CpG-ODN. The fecal microbiota composition in mice showing high and low TNF responses to CpG-ODN appeared distinct. In particular, the abundance of several individual Gram+ and Gram− bacterial spp positively correlated with the CpG-ODN response, whereas the presence of commensal Lactobacillus spp decreased the response (34). In vivo association experiments confirmed that the Gram− Alistipes shaii enhances the CpG-ODN response while L. fermentum attenuates it (34).
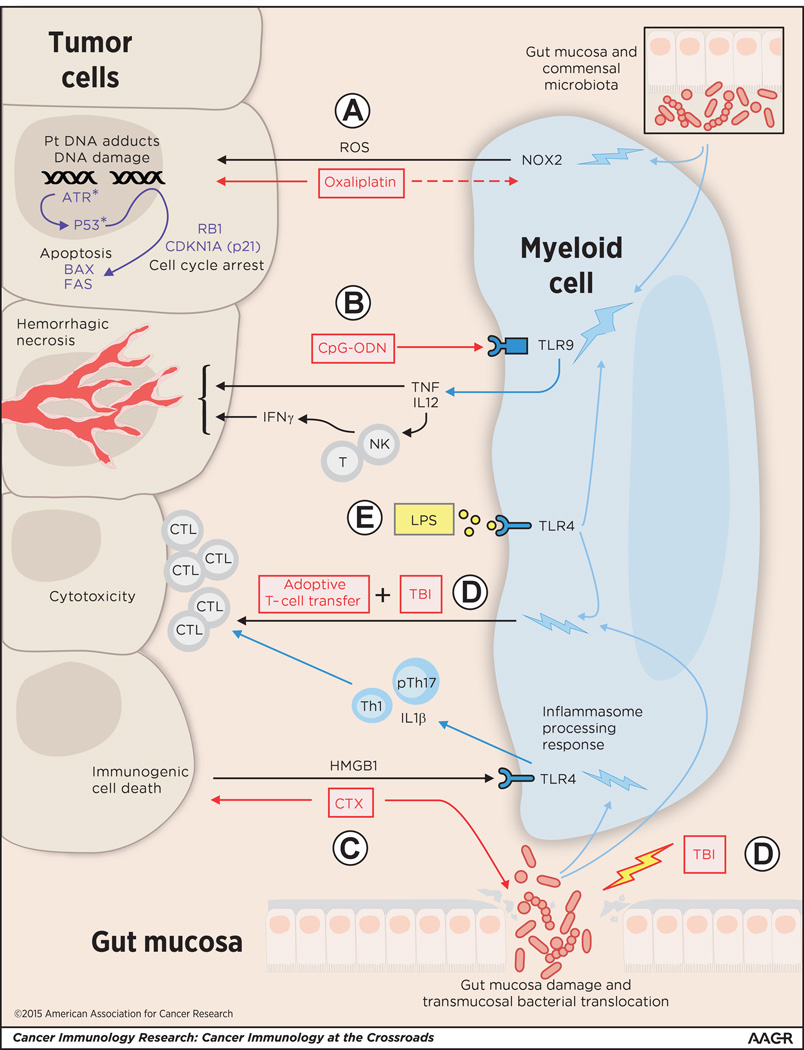
Figure 1. The commensal microbiota conditions tumor-associated myeloid cells to promote cancer therapies.
(A) The genotoxic effects of oxaliplatin comprise DNA damage, cell growth arrest and apoptosis. Oxaliplatin forms platinum (Pt)-DNA adducts, which in the presence of reactive oxygen species (ROS), induces DNA damage and activation of the DNA repair pathway genes (including ataxia telangiectasia and rad3-related [ATR] kinase, p53, BAX, FAS, RB1, CDKN1A) resulting in cell-cycle arrest and apoptosis (34). In the absence of microbiota, myeloid cells fail to produce NADPH oxidase (NOX2)-dependent ROS following oxaliplatin treatment and minimal DNA damage is observed (34). (B) The presence of gut commensal microbiota is required for tumor-associated myeloid cells to respond to intratumoral injection of the TLR9-agonist CpG-ODN with production of TNF and IL12 (which induces IFNγ production primarily from NK and T cells in the tumor microenvironment) and induction of tumor hemorrhagic necrosis (34). Treatments with (C) cyclophosphamide (CTX) and (D) total body irradiation (TBI; used as part of the myeloablative conditioning regimen that augments the effectiveness of adoptive T-cell transfer therapy) induce mucosal damage, dysbiosis, and transmucosal bacterial translocation (43, 44). This altered exposure to intestinal bacteria increases the ability of tumor-associated myeloid cells to sustain CTL generation and/or expansion and antitumor cytotoxicity (44). In CTX therapy, drug-induced immunogenic tumor-cell death releases danger signal HMGB1, which binds to pattern recognition receptor TLR4 activating inflammasome processing and production of IL1β, resulting in the induction of antitumor “pathogenic” Th17 (pTh17) cells and the downstream activation of memory Th1 cells and antitumor effector CTLs. In the absence of microbiota, induction of T effector and memory cells are not observed (44). (E) Treatment of mice with LPS conditions the myeloid cells for response to CpG-ODN and adoptive T-cell transfer even in the absence of the microbiota (34, 43). Blue bolts signify conditioning effect on myeloid cells.
The effect of the microbiota on chemotherapy was analyzed utilizing platinum compounds (e.g. oxaliplatin, cisplatin) (34). These compounds mediate genotoxicity by forming platinum DNA-adducts followed by formation of intrastrand cross-links that inhibit protein synthesis and proliferation, and induce apoptosis in part downstream of the ataxia telangiectasia and rad3-related (ATR) kinase recruitment to DNA damage and p53 activation (39). In addition to their genotoxicity, oxaliplatin but not cisplatin induces immunogenic cell death that activates antitumor T-cell immunity (40). The therapeutic effect of oxaliplatin and cisplatin on mouse sterile subcutaneous transplanted tumors was dramatically reduced in antibiotics-treated or GF mice (34). In antibiotics-treated mice, platinum adducts to tumor-cell DNA were formed at a level comparable to that observed in control mice, however, already at 48 hour post-treatment there was a significant decrease in DNA damage and cytotoxicity. Antibiotics treatment of mice largely suppressed all the gene expression modification induced in the tumor by oxaliplatin. In antibiotics-treated mice tumor-infiltrating myeloid cells failed to produce reactive oxygen species (ROS) via the NADPH oxidase NOX2. ROS are needed for the oxaliplatin antitumor effect. Although the genotoxic effect of platinum compounds was known to require ROS and particularly H2O2 production (41), this requirement was mostly studied in tumor cell lines in vitro. In these conditions ROS was endogenously produced in tumor cells. In the in vivo tumor microenvironment, however, most of the ROS required for oxaliplatin cytotoxicity is produced by tumor-associated myeloid cells. Thus, the microbiota affects oxaliplatin early tumor genotoxicity by systemically priming tumor-associated myeloid cells for ROS production.
The effects of microbiota deprivation in the response to CpG-ODN and platinum compounds were evident at very early times following treatment, suggesting that tumor-associated myeloid cells were primed for responsiveness to therapy by the preexisting microbiota composition. However, chemotherapy and radiation therapy in addition to their antitumor effect also induce damage of the intestinal mucosa affecting mucosal permeability and inducing dysbiosis and bacterial transmucosal translocation. One of the most successful anti-cancer immune therapies is the adoptive transfer of tumor-specific cytotoxic CD8+ T cells (42). For best survival of the T cells and effectiveness of the transfer, lympho- and myelo-ablation are necessary (43). Both in human patients and in mice, total body irradiation (TBI) improves therapy efficacy by increasing DC activation and homeostatic cytokine levels (43). Due to the mucosal damage effect of TBI, commensal gut bacteria were found to infiltrate the mesenteric lymph nodes of irradiated animals and elevated endotoxin levels were observed in their sera (43). The ability of TBI to improve tumor regression induced by adoptive T-cell transfer was reduced in animals treated with broad spectrum antibiotics, by neutralization of serum LPS using polymyxin B, or in mice genetically deficient in CD14 or TLR4 that are unable to respond to LPS. Administration of LPS or LPS-containing serum from irradiated animals to non-irradiated lymphopenic mice was able to enhance the number and function of transferred CD8+ T cells, leading to long-term cure of mice with large transplanted tumors (43).
Similarly to oxaliplatin, cyclophosphamide (CTX) is a member of a group of chemotherapy agents that induce immunogenic cell death. The ability of CTX to induce adaptive antitumor responses is decreased in GF or antibiotics-treated tumor-bearing mice (44). CTX treatment of conventionally raised animals induces dysbiosis and mucositis. Due to the mucosal damage, Gram+ bacteria translocate into the mesenteric draining lymph nodes, prime pathogenic effector Th17 cells and memory Th1 cells, all of which were not observed in microbiota-depleted mice (44). Thus, the activation of antigen-presenting cells and the subsequent induction of antitumor immune responses by chemotherapy-induced immunogenic cell death not only depend on the release of endogenous mediators of inflammation as previously shown (40), but also on the priming and/or activating effects mediated by commensal bacteria and/or by their products.
Clinical perspectives
Inflammation and immunity are hallmarks of cancer, affecting tumor initiation, progression, dissemination and response to therapy (45). The molecular pathways involved in this modulation of the neoplastic disease offer targets of therapeutic intervention for both cancer prevention and therapy. The power of the antitumor immune response present in patients but masked by tumor immunosuppressive mechanisms has been successfully harnessed in immunotherapy approaches (46). The most successful approaches have utilized either adoptive T-cell transfer or inhibition of immune checkpoints, such as CTLA-4 and PD-1/PD-L1/2, which free antitumor T cells from the immunologic brakes expressed by the tumor cells or by other cells in the tumor microenvironment (42, 46, 47). Therapeutic cancer vaccines have not been as successful yet because of the difficulty to permanently overcome the inadequacy of antigen-presenting cells in the tumor and the presence of immune checkpoints restricting both the natural and vaccine-induced immune responses (48, 49). The evidence discussed here adds a layer of complexity indicating that the composition of commensal microbiota and its alteration by neoplastic disease or therapy significantly affect all stages of tumor development and therapy effectiveness via their local or systemic effects on inflammation, immunity and metabolism. These regulatory mechanisms provide new targets and possibilities for therapeutic intervention.
Most of the studies examining the influence of commensal microbiota on cancer therapy have been performed in experimental animals, and to what extent the microbiota regulates human myeloid-cell function and response to cancer therapy remains to be determined. While some of the molecular mechanisms by which the microbiota locally affects inflammation, immunity, and cancer have been elucidated, our understanding of the systemic effects is still rudimental. Nevertheless, it is tempting to speculate that the variable response to cancer therapy observed in patients may be in part due to different microbiota composition affecting the inflammatory tone and myeloid-cell functions in the tumor microenvironment. This would offer the possibility to improve the effectiveness of immune, chemical, and radiation therapy by altering the microbiota, targeting the pathways by which the microbiota communicates with inflammatory cells, or directly targeting the molecular mechanisms in myeloid cells and antigen-presenting cells that restrict their functions.
Since Hippocrates’ recognition that “all disease begins in the gut” there has been many attempts, most often not scientifically controlled, to modify the gut microbiota to correct dysbiosis and to promote health. Clinical therapeutic procedures including the use of probiotics, diet modification and prebiotics, fecal or defined microbiota transfer have been utilized to enhance patients’ response to cancer therapy (50). The major difficulties include our incomplete understanding of what constitutes a healthy microbiota, our current knowledge of which bacterial species affect which type of cancer therapy needs to be extended, and the little that we know is based on experimental animal data which may not reflect conditions in humans. Furthermore, the same bacterial species may have opposite effects in different types or at different stages of therapy. For example, in the experimental model of CpG-ODN tumor therapy the abundance in the gut or the oral administration of Lactobacillus spp decreased the ability of the tumor-associated myeloid cells to produce effector cytokines whereas bacteria of the same genus increased the ability of antigen-presenting cells to induce an effective antitumor immune response following CTX-induced immunogenic cell death (34, 44, 51).
In the absence of precise indications of which specific bacterial species or their combination would offer the best possibility to prevent or cure disease, one approach has been to replace the disease-associated dysbiotic microbiota in patients with that from healthy subjects using fecal microbiota transplantation. However, fecal transplant may have both anti-inflammatory and pro-inflammatory effects on mucosal inflammation as well as on systemic metabolism and immunity (52). Fecal transplants have been very successful in the treatment of Clostridium difficile infections most likely by altering the dysbiosis permissive for colonization by this pathobiont (53). Although the composition of the commensal microbiota is defined by our genetics, nutrition and environmental exposure, fecal transplants change the composition of the recipient gut microbiota to resemble that of the donor for at least several weeks (54). Fecal transplants have also been proposed as a treatment for inflammatory bowel diseases and metabolic disorders; transfer of intestinal microbiota from lean donors has been shown to increase sensitivity to insulin in patients with metabolic syndrome (55). However, the use of not well characterized microbial mixture could increase or induce bowel inflammation elicited by the transplanted microbiota, resulting in bacteremia (56, 57). In theory, fecal transplants could treat dysbiosis that are associated with cancer-comorbidities (30) and could contribute to optimal response to cancer therapy. There is, however, controversy on how and if fecal transplant should be regulated and the FDA presently is considering feces as a drug that requires IND approval (58). In any case, several safety and consistency concerns remain that suggest the usefulness of developing better defined and safer microbial replacement therapeutic procedures (52, 53, 55, 59).
An alternative to the use of bacterial preparations is to administer bacterial-derived or bacterial-induced products that modulate the immune system. Ligands for TLRs or other innate receptors are being developed for clinical use and could be used in combination cancer therapies (60). LPS for example has been shown to reestablish the resistance to influenza virus infection, the response to CpG-ODN therapy and T-cell adoptive transfer in mice depleted of commensal microbiota (8, 34, 43). Short-chain fatty acids (SCFA) are bacterial products derived by fermentation of dietary fibers. SCFAs modulate mucosal immunity via G-protein-coupled receptors by inducing IL18 production by enterocytes and directly acting on the IL10-producing T-regulatory pool and regulating its size and functions (61–63). In ApcMin/+ Msh2−/− mice antibiotics treatment or a diet reduced in carbohydrates, which changes the microbiota composition and results in lower SCFA production, decreased colon polyps incidence (64). While these data show that it is possible to affect cancer by modifying the microbiota, it also cautions that a clear understanding of the roles and functions of different microbial species in cancer is necessary because SCFAs have been shown in other experimental conditions to be protective against colon and mammary cancer (61, 65, 66).
The ideal therapeutic intervention would be to target directly the tumor-infiltrating myeloid cells and change their properties so that they could enhance and sustain therapeutic responses and antitumor immunity. In the tumor microenvironment many factors are present that prevent anti-cancer immunity or response to therapy, including products of both classically- and alternatively-activated macrophages and other myeloid-cell subsets, T-regulatory cells and the presence of anti-inflammatory factors such as TGF-β and IL10 (67–70). These mechanisms provide targets of intervention and many of which have been tested experimentally and some clinically (71, 72). The ability of IL10 antagonism to enhance the response of tumor-associated myeloid cells to CpG-ODN indicates that it is possible to revert at least partially the unresponsive phenotype of those cells in vivo. However, even when IL10 signaling is blocked myeloid cells still require commensal microbiota for their response indicating that the unresponsive phenotype of these cells is controlled by multiple signals (34, 35). In order to identify new possibilities of precise targeting of myeloid cells in the treatment of cancer it is essential to define the genetic and epigenetic pathways affected by the microbiota and their relevance in response to therapy (9, 26, 34).
Acknowledgments
Funding Support: The work of RSG, AD, NPR, and GT were supported by the intramural research program of the NCI. The work of LZ and SV were supported by Institut National du Cancer (INCa), la Ligue contre le cancer (LIGUE labellisée), SIRIC Socrate, LABEX and PACRI Onco-Immunology.
Footnotes
Disclosure of potential conflicts of interest: The authors disclosed no potential conflicts of interest.
References
- 1.Belkaid Y, Naik S. Compartmentalized and systemic control of tissue immunity by commensals. Nat Immunol. 2013;14:646–653. doi: 10.1038/ni.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chow J, Mazmanian SK. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe. 2010;7:265–276. doi: 10.1016/j.chom.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Underwood MA. Intestinal dysbiosis: Novel mechanisms by which gut microbes trigger and prevent disease. Prev Med. 2014;65:133–137. doi: 10.1016/j.ypmed.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 6.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhn KA, Stappenbeck TS. Peripheral education of the immune system by the colonic microbiota. Semin Immunol. 2013;25:364–369. doi: 10.1016/j.smim.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chervonsky AV. Microbiota and autoimmunity. Cold Spring Harb Perspect Biol. 2013;5:a007294. doi: 10.1101/cshperspect.a007294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011;10:311–323. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hooper DC, Molowitz EH, Bos NA, Ploplis VA, Cebra JJ. Spleen cells from antigen-minimized mice are superior to spleen cells from germ-free and conventional mice in the stimulation of primary in vitro proliferative responses to nominal antigens. Eur J Immunol. 1995;25:212–217. doi: 10.1002/eji.1830250135. [DOI] [PubMed] [Google Scholar]
- 15.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brusca SB, Abramson SB, Scher JU. Microbiome and mucosal inflammation as extra-articular triggers for rheumatoid arthritis and autoimmunity. Curr Opin Rheumatol. 2014;26:101–107. doi: 10.1097/BOR.0000000000000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menard S, Cerf-Bensussan N, Heyman M. Multiple facets of intestinal permeability and epithelial handling of dietary antigens. Mucosal Immunol. 2010;3:247–259. doi: 10.1038/mi.2010.5. [DOI] [PubMed] [Google Scholar]
- 18.Wanke I, Skabytska Y, Kraft B, Peschel A, Biedermann T, Schittek B. Staphylococcus aureus skin colonization is promoted by barrier disruption and leads to local inflammation. Exp Dermatol. 2013;22:153–155. doi: 10.1111/exd.12083. [DOI] [PubMed] [Google Scholar]
- 19.Hahn BL, Sohnle PG. Direct translocation of staphylococci from the skin surface to deep organs. Microb Pathog. 2013;63:24–29. doi: 10.1016/j.micpath.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci USA. 2008;105:20858. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP, et al. Symbiotic Bacterial Metabolites Regulate Gastrointestinal Barrier Function via the Xenobiotic Sensor PXR and Toll-like Receptor 4. Immunity. 2014 doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim YG, Udayanga KG, Totsuka N, Weinberg JB, Nunez G, Shibuya A. Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE(2) Cell Host Microbe. 2014;15:95–102. doi: 10.1016/j.chom.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deshmukh HS, Liu Y, Menkiti OR, Mei J, Dai N, O'Leary CE, et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med. 2014;20:524–530. doi: 10.1038/nm.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khosravi A, Yanez A, Price JG, Chow A, Merad M, Goodridge HS, et al. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. 2014;15:374–381. doi: 10.1016/j.chom.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mittrucker HW, Seidel D, Bland PW, Zarzycka A, Kaufmann SH, Visekruna A, et al. Lack of microbiota reduces innate responses and enhances adaptive immunity against Listeria monocytogenes infection. Eur J Immunol. 2014 doi: 10.1002/eji.201343927. [DOI] [PubMed] [Google Scholar]
- 26.Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171–186. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 27.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galon J, Angell HK, Bedognetti D, Marincola FM. The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity. 2013;39:11–26. doi: 10.1016/j.immuni.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 30.Trinchieri G. Cancer and inflammation: an old intuition with rapidly evolving new concepts. Annu Rev Immunol. 2012;30:677–706. doi: 10.1146/annurev-immunol-020711-075008. [DOI] [PubMed] [Google Scholar]
- 31.Bronte V, Chappell DB, Apolloni E, Cabrelle A, Wang M, Hwu P, et al. Unopposed production of granulocyte-macrophage colony-stimulating factor by tumors inhibits CD8+ T cell responses by dysregulating antigen-presenting cell maturation. J Immunol. 1999;162:5728–5737. [PMC free article] [PubMed] [Google Scholar]
- 32.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bronte V, Apolloni E, Cabrelle A, Ronca R, Serafini P, Zamboni P, et al. Identification of a CD11b(+)/Gr-1(+)/CD31(+) myeloid progenitor capable of activating or suppressing CD8(+) T cells. Blood. 2000;96:3838–3846. [PMC free article] [PubMed] [Google Scholar]
- 34.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guiducci C, Vicari AP, Sangaletti S, Trinchieri G, Colombo MP. Redirecting in vivo elicited tumor infiltrating macrophages and dendritic cells towards tumor rejection. Cancer Res. 2005;65:3437–3446. doi: 10.1158/0008-5472.CAN-04-4262. [DOI] [PubMed] [Google Scholar]
- 36.Franklin RA, Liao W, Sarkar A, Kim MV, Bivona MR, Liu K, et al. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344:921–925. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Movahedi K, Laoui D, Gysemans C, Baeten M, Stange G, Van den Bossche J, et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 2010;70:5728–5739. doi: 10.1158/0008-5472.CAN-09-4672. [DOI] [PubMed] [Google Scholar]
- 38.Shand FH, Ueha S, Otsuji M, Koid SS, Shichino S, Tsukui T, et al. Tracking of intertissue migration reveals the origins of tumor-infiltrating monocytes. Proc Natl Acad Sci U S A. 2014;111:7771–7776. doi: 10.1073/pnas.1402914111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 40.Kroemer G, Galluzzi L, Kepp O, Zitvogel L. Immunogenic cell death in cancer therapy. Annu Rev Immunol. 2013;31:51–72. doi: 10.1146/annurev-immunol-032712-100008. [DOI] [PubMed] [Google Scholar]
- 41.Ozben T. Oxidative stress and apoptosis: impact on cancer therapy. J Pharm Sci. 2007;96:2181–2196. doi: 10.1002/jps.20874. [DOI] [PubMed] [Google Scholar]
- 42.Restifo NP, Dudley ME, Rosenberg SA. Adoptive immunotherapy for cancer: harnessing the T cell response. Nat Rev Immunol. 2012;12:269–281. doi: 10.1038/nri3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paulos CM, Wrzesinski C, Kaiser A, Hinrichs CS, Chieppa M, Cassard L, et al. Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. J Clin Invest. 2007;117:2197–2204. doi: 10.1172/JCI32205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 46.Couzin-Frankel J. Breakthrough of the year 2013. Cancer immunotherapy. Science. 2013;342:1432–1433. doi: 10.1126/science.342.6165.1432. [DOI] [PubMed] [Google Scholar]
- 47.Brahmer JR, Pardoll DM. Immune checkpoint inhibitors: making immunotherapy a reality for the treatment of lung cancer. Cancer Immunol Res. 2013;1:85–91. doi: 10.1158/2326-6066.CIR-13-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eggermont AM. Therapeutic vaccines in solid tumours: can they be harmful? Eur J Cancer. 2009;45:2087–2090. doi: 10.1016/j.ejca.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 49.Schlom J, Hodge JW, Palena C, Tsang KY, Jochems C, Greiner JW, et al. Therapeutic cancer vaccines. Adv Cancer Res. 2014;121:67–124. doi: 10.1016/B978-0-12-800249-0.00002-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ursell LK, Van Treuren W, Metcalf JL, Pirrung M, Gewirtz A, Knight R. Replenishing our defensive microbes. Bioessays. 2013;35:810–817. doi: 10.1002/bies.201300018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Viaud S, Daillere R, Boneca IG, Lepage P, Pittet MJ, Ghiringhelli F, et al. Harnessing the Intestinal Microbiome for Optimal Therapeutic Immunomodulation. Cancer Res. 2014;74:4217–4221. doi: 10.1158/0008-5472.CAN-14-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pamer EG. Fecal microbiota transplantation: effectiveness, complexities, and lingering concerns. Mucosal Immunol. 2014;7:210–214. doi: 10.1038/mi.2013.117. [DOI] [PubMed] [Google Scholar]
- 53.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 54.Backhed F, Fraser CM, Ringel Y, Sanders ME, Sartor RB, Sherman PM, et al. Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe. 2012;12:611–622. doi: 10.1016/j.chom.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 55.Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913–916. e7. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 56.Konstantinov SR, Peppelenbosch MP. Fecal microbiota transfer may increase irritable bowel syndrome and inflammatory bowel diseases-associated bacteria. Gastroenterology. 2013;144:e19–e20. doi: 10.1053/j.gastro.2012.12.040. [DOI] [PubMed] [Google Scholar]
- 57.Quera R, Espinoza R, Estay C, Rivera D. Bacteremia as an adverse event of fecal microbiota transplantation in a patient with Crohn's disease and recurrent Clostridium difficile infection. J Crohns Colitis. 2014;8:252–253. doi: 10.1016/j.crohns.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 58.Smith MB, Kelly C, Alm EJ. Policy: How to regulate faecal transplants. Nature. 2014;506:290–291. doi: 10.1038/506290a. [DOI] [PubMed] [Google Scholar]
- 59.Allegretti JR, Hamilton MJ. Restoring the gut microbiome for the treatment of inflammatory bowel diseases. World J Gastroenterol. 2014;20:3468–3474. doi: 10.3748/wjg.v20.i13.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ridnour LA, Cheng RY, Switzer CH, Heinecke JL, Ambs S, Glynn S, et al. Molecular pathways: toll-like receptors in the tumor microenvironment--poor prognosis or new therapeutic opportunity. Clin Cancer Res. 2013;19:1340–1346. doi: 10.1158/1078-0432.CCR-12-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalina U, Koyama N, Hosoda T, Nuernberger H, Sato K, Hoelzer D, et al. Enhanced production of IL-18 in butyrate-treated intestinal epithelium by stimulation of the proximal promoter region. Eur J Immunol. 2002;32:2635–2643. doi: 10.1002/1521-4141(200209)32:9<2635::AID-IMMU2635>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 63.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Belcheva A, Irrazabal T, Robertson SJ, Streutker C, Maughan H, Rubino S, et al. Gut microbial metabolism drives transformation of msh2-deficient colon epithelial cells. Cell. 2014;158:288–299. doi: 10.1016/j.cell.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 65.Elangovan S, Pathania R, Ramachandran S, Ananth S, Padia RN, Lan L, et al. The niacin/butyrate receptor GPR109A suppresses mammary tumorigenesis by inhibiting cell survival. Cancer Res. 2014;74:1166–1178. doi: 10.1158/0008-5472.CAN-13-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bultman SJ, Jobin C. Microbial-derived butyrate: an oncometabolite or tumor-suppressive metabolite? Cell Host Microbe. 2014;16:143–145. doi: 10.1016/j.chom.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sica A, Bronte V. Altered macrophage differentiation and immune dysfunction in tumor development. J Clin Invest. 2007;117:1155–1166. doi: 10.1172/JCI31422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stewart CA, Metheny H, Iida N, Smith L, Hanson M, Steinhagen F, et al. Interferon-dependent IL-10 production by Tregs limits tumor Th17 inflammation. J Clin Invest. 2013;123:4859–4874. doi: 10.1172/JCI65180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pang Y, Gara SK, Achyut BR, Li Z, Yan HH, Day CP, et al. TGF-beta Signaling in Myeloid Cells Is Required for Tumor Metastasis. Cancer Discov. 2013;3:936–951. doi: 10.1158/2159-8290.CD-12-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morris JC, Tan AR, Olencki TE, Shapiro GI, Dezube BJ, Reiss M, et al. Phase I study of GC1008 (fresolimumab): a human anti-transforming growth factor-beta (TGFbeta) monoclonal antibody in patients with advanced malignant melanoma or renal cell carcinoma. PLoS ONE. 2014;9:e90353. doi: 10.1371/journal.pone.0090353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Llorente L, Richaud-Patin Y, Garcia-Padilla C, Claret E, Jakez-Ocampo J, Cardiel MH, et al. Clinical and biologic effects of anti-interleukin-10 monoclonal antibody administration in systemic lupus erythematosus. Arthritis Rheum. 2000;43:1790–1800. doi: 10.1002/1529-0131(200008)43:8<1790::AID-ANR15>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]