Summary
Small solutes are useful probes of large conformational changes in RNA polymerase (RNAP)-promoter interactions and other biopolymer processes. In general, a large effect of a solute on an equilibrium constant (or rate constant) indicates a large change in water-accessible biopolymer surface area in the corresponding step (or transition state), resulting from conformational changes, interface formation, or both. Here, we describe nitrocellulose filter binding assays from series used to determine the urea dependence of open complex formation and dissociation with Escherichia coli RNAP and λPR promoter DNA. Then, we describe the subsequent data analysis and interpretation of these solute effects.
Keywords: bacterial RNA polymerase, open complex formation, kinetics, mechanism, solute effects, conformational changes
1. Introduction
1.1 Solute Interactions and Solute Effects
Small solutes are emerging as useful probes and perturbants of intermediates and transition states and of overall changes in surface area in transcription initiation and other protein and nucleic acid processes. In general, large amounts of protein and nucleic acid surface are buried or exposed when interfaces are made or disrupted or in coupled conformational changes. The effect of a solute on an equilibrium constant (or rate constant) is determined by its favorable or unfavorable interactions with the functional groups on the surface buried or exposed in the corresponding step (or transition state).
These solute effects vary widely for different solutes and different surfaces. For example, urea interacts favorably with most protein and nucleic acid functional groups. High concentrations (typically more than 4 M) of urea denature proteins and nucleic acids at 25 °C because urea interacts favorably with the amide and hydrocarbon groups exposed in unfolding proteins and with the nucleobase groups exposed in melting nucleic acid helices (1). At lower concentrations, these same interactions cause urea to disfavor steps in transcription initiation that bury RNA polymerase (RNAP) or promoter DNA surface, and to favor the reverse direction of these steps (described below). Glycine betaine (GB) and proline, on the other hand, interact unfavorably with many protein functional groups and with nucleic acid phosphates and therefore stabilize proteins (though not nucleic acids) against denaturation (1, 2) and favor the early and late steps in transcription initiation that bury RNAP and promoter DNA surface ((3) and unpublished data for proline). Replacement of KCl by potassium glutamate (KGlu) has a qualitatively similar effect to replacing urea by GB in that this replacement favors protein folding and assembly (but not DNA helix formation), and therefore favors the early and late steps in transcription initiation (4).
Solute effects are analyzed and interpreted using the slope of a plot of the logarithm of the observed equilibrium constant (lnKobs) or rate constant (lnkobs) versus solute concentration ([solute]). These plots are typically linear up to at least 1 M [solute]. Their slopes, closely related to what are called solute m-values, can be analyzed at several levels, starting with the qualitative level discussed above, where the sign of the effect reports on whether surface is buried or exposed in the step (when lnKobs is analyzed) or the formation of the transition state (when lnkobs is analyzed) and the magnitude of the effect reports on how much surface is involved. As examples of this technique, details of the use of urea and GB to probe conformational changes and interface formation in the steps of forming and stabilizing the open promoter complex by E. coli RNAP are provided in this article (3–6).
1.2 Mechanism of Formation and Stabilization of the Open Promoter Complex in Transcription Initiation
The mechanism of transcription initiation by E. coli and T7 phage RNAP have been extensively investigated and are model systems for transcription initiation. Both polymerases form the open complex by a “bind-bend-melt” mechanism (7, 8). Kinetic studies using E. coli RNAP (R) and the PR promoter (P) of phage λ show that a minimum of four steps and three kinetically-significant intermediates (designated I1, I2, I3) are involved in formation of the transcriptionally-competent open complex (RPo) by the bacterial polymerase: [Mechanism 1 goes here] Immediately after mixing RNAP and λPR promoter DNA under conditions typically used to study transcription initiation, relatively unstable closed complexes designated I1 are formed (9). For λPR, I1 does not appear to be a single species but rather an ensemble (i.e a mixed population) of relatively unstable closed complexes which dissociate rapidly to free RNAP and promoter DNA and therefore equilibrate with each other and with free promoter on the time scale required to convert to I2. Likewise in a λPR dissociation kinetics experiment, the relatively unstable open intermediates I3 and I2 convert rapidly back to the stable open complex (RPo) on the time scale required for I2 to convert to I1. The interconversion I1 ⇄ I2, opening and closing the 13 base pair transcription bubble (9), is the rate-determining step in both directions for λPR, with the highest free energy transition state (TS) in Mechanism 1. While RPo is relatively well-characterized, much less is known about these intermediates and TS (8). This information is needed to determine the mechanism of operation of RNAP as a molecular DNA-opening machine. The use of solutes as probes of large conformational changes and interface formation in these intermediates and transition state, as illustrated in this chapter, provides information that is not readily obtained by other methods.
1.3 Examples of Solute Effects on Steps of Transcription Initiation and their Interpretation
Solutes can be used as probes for the change in surface area in a step or the formation of a transition state. As an example from transcription initiation, the equilibrium constant and both rate constants for the interconversion of I1 and I2, the step which includes opening/closing the 13 base pair initiation bubble, are independent of urea and of the choice of anion (Cl− versus Glu−) and only weakly dependent on salt concentration (3–5). This contrasts greatly with the situation for DNA melting in solution, where the equilibrium constant for opening DNA oligomers of this length is a strong function of urea and salt concentration, and provides one of several lines of evidence that DNA opening occurs in the active site cleft of RNA polymerase in an environment very different from that of the solution.
As a second example of the use of solutes as probes, the dissociation rate constant kd is a strong function of urea, GB, and salt concentration, and is profoundly affected by replacing Cl− by Glu− (3–5). Effects of urea and KCl are entirely in the equilibrium constant K3 for the conversion of the initial open complex I2 to the stable open complex RPo and provide strong evidence for folding and assembly of approximately120 residues of downstream mobile elements (DME) (6) of WT RNAP on duplex DNA as part of stabilizing the λPR open complex by a factor of about 105 (4, 5)
In addition to their use as probes, solutes can also be used as selective perturbants. As one example, because urea and KCl are found to destabilize RPo greatly with respect to I2, but do not affect the rate of conversion of I2 to I1, rapid increases in concentration of these solutes create a transient, majority population (called a burst) of the otherwise-undetectable intermediate I2 for characterization by DNA footprinting (9). In another study, GB was used to increase the I1 binding constant and hence increase the size of the burst population of I1, and to reduce the rate of its conversion to open complexes, to footprint I1 in manual mixing experiments (10). The original papers should be consulted for details of these experiments.
In what follows, we describe the methods used to perform nitrocellulose filter binding assays to study the urea dependence of the kinetics of RPo formation and of RPo dissociation into free RNAP and promoter DNA. We then explain the data analysis required to determine the solute concentration dependences of the individual steps of Mechanism I, and describe the conclusions we have reached using such analyses in our studies of the mechanism with λPR.
2. Materials
2.1. Reagents
RNAP: Prepare using standard protocols, such as described in (reference chapter in this volume describing this prep). Dialyze into RNAP Storage Buffer (see below), divide into 100 µL aliquots, and store at −80°C. Store a 100 µL aliquot at −20°C while in use.
Radiolabeled DNA: Use T4 Polynucleotide Kinase (New England Biolabs) and γ 32P-labeled ATP (Perkin Elmer) to 32P-label one primer, then use this primer to amplify the fragment containing the promoter via PCR. Purify PCR fragment from an agarose gel. Store at 4°C (see Note 1).
8× Transcription Buffer Salts: 320 mM Tris (pH 8.5 at room temperature), 960 mM KCl, 80 mM MgCl2. Dissolve 3.876 g Tris HCl, 7.157 g KCl, and 1.626 g MgCl2 into about 90 mL deionized H2O. Adjust pH to 8.5 using HCl. Bring to 100 mL with deionized H2O in a volumetric flask. Store at 4°C.
4× Transcription Buffer Salts: 160 mM Tris (pH 8 at assay temperature), 480 mM KCl, 40 mM MgCl2. Combine 25 mL 8× Transcription Buffer Salts with about 20 mL deionized H2O. Equilibrate at intended assay temperature and adjust the pH to 8.0 using HCl. Bring to 50 mL in a volumetric flask using deionized H2O. Store at 4°C.
1 M dithiothreitol (DTT). Mix 0.154 g DTT with 1 mL deionized H2O. Vortex to dissolve. Filter using a 0.45 µm syringe filter. Store at −20°C in 50 µL aliquots.
50 mg/mL bovine serum albumin (BSA). Mix 100 mg with 2 mL deionized H2O. Filter using a 0.45 µm syringe filter. Store at −20°C in 50 µL aliquots.
50 mg/mL heparin. Mix 100 mg with 2 mL deionized H2O. Filter using a 0.45 µm syringe filter. Store at −20°C in 50 µL aliquots. (See Note 2).
RNAP Storage Buffer: 50% v/v glycerol, 10 mM Tris (pH 7.5 at 4°C), 100 mM NaCl, 0.1 mM DTT, 0.1 mM Na2EDTA. Dissolve 0.061 g Tris HCl and 0.292 g NaCl in about 20 mL deionized H2O. Add 10 µL 0.5 M Na2EDTA (pH 8.0 at room temperature) and 5 µL 1 M DTT. Equilibrate at 4°C in a water bath and adjust pH to 7.5 using HCl. Bring to 25 mL with deionized H2O in a volumetric flask. Bring to 50 mL with glycerol in a volumetric flask. Store at −20 °C.
6 M Urea in 1× Transcription Buffer Salts (see Note 3): 43 mm (millimolal) Tris (pH 8 at assay temperature), 129 mm KCl, 10.7 mm MgCl2, 6 M urea. Combine 12.5 mL 4× Transcription Buffer Salts and 6.25 mL RNAP Storage Buffer. Bring to 50 mL with deionized H2O in a volumetric flask. Dissolve 18.018 g dry urea into a small amount of this solution of 1× Transcription Buffer Salts. Transfer this solution into a 50 mL volumetric flask and add additional 1× Transcription Buffer Salts to bring the final volume to 50 mL (see Note 4 and Section 3.1). Store at 4 °C.
Filter Binding Wash Buffer: 10 mM Tris (pH 8.0 at room temperature), 100 mM NaCl, 0.1 mM Na2EDTA. Dissolve 5.844 g NaCl and 1.211 g Tris in about 950 mL deionized H2O. Add 200 µL 0.5 M Na2EDTA stock (pH 8.0 at room temperature). Adjust pH using HCl. Bring to 1 L with deionized H2O. Store at 4°C.
2.2 Supplies and equipment
0.45 µm nitrocellulose filters (Schleich and Schuell). Soak in deionized H2O at least 30 minutes prior to assay. Store at 4°C in deionized H2O.
10-place filter-holding vacuum manifold capable of achieving pressures of 15–30 inches of Hg (Hoefer Scientific, San Francisco, CA).
Three-syringe Rapid Quench-Flow Fast Mixer (Model RQF-3; KinTek Co. Austin, TX) with temperature-regulated water bath for rapid-mix, rapid-quench experiments.
Approximately 20 1.5 mL microcentrifuge tubes with holes made in the top. (see Note 5).
Scintillation counter (Hewlett-Packard Tri-Carb 2100 TR).
Laboratory timer for manual mixing experiments.
3. Methods
This section provides details of nitrocellulose filter binding kinetics experiments that detect long-lived competitor-resistant (CR) open complexes and serve to determine the dependences of equilibrium and rate constants of individual steps in Mechanism I on solute concentration. For λPR under the wide range of solution conditions investigated, the kinetics of open-complex dissociation are single exponential, as are the kinetics of association when one reactant (typically RNAP) is in excess. Under such conditions, overall rate constants for dissociation (kd) and association (βCR or αCR; see Equations 2–4 below) are directly determined from filter binding experiments. From the dependence of αCR on RNAP concentration, the equilibrium constant K1 and rate constant k2 in Mechanism 1 are determined. A different assay, capable of detecting I1 directly, is necessary to obtain individual rate constants k1 and k−1 in addition to K1. Rapid destabilization of RPo using high concentrations of salt or urea yields a burst of I2, allowing one to determine k−2 directly and therefore dissect kd to obtain K3 (and in favorable cases k3 and k−3 (5)).
Filter binding studies of the effects of different solutes and solute concentration on the observable rate constants kd and βCR or αCR are analyzed similarly to determine which individual rate or equilibrium constants are affected by that solute, and by how much. Methods described below are for the destabilizing solute urea, which favors the exposure of RNAP and DNA surface and hence destabilizes both binding and folding steps. In most cases these methods are easily adapted to study protein-stabilizing solutes like GB and proline, effects of which on protein folding are comparable in magnitude but opposite in direction to urea. (See Note 6.)
It is important to note that solute-as-probe experiments typically are performed in the solute concentration range 0 – 1 M. Higher concentrations of protein-stabilizing solutes can be used if needed to detect small effects, but higher concentrations of urea and other destabilizing solutes should generally be avoided. In experiments using urea as a perturbant to rapidly destabilize RPo (5), concentrations of urea in the molar range can be used because RNAP in promoter complexes is stabilized against subunit dissociation and unfolding by binding free energy.
Below, we describe assays utilizing both manual mixing and rapid quench-flow fast mixing with a three syringe mixer. For best results, fast mixing should be used for reactions with half-lives shorter than about 100 seconds. For λPR these include studies of the kinetics of RPo formation at temperatures above 20 °C and of RPo dissociation at high urea or salt concentrations. However, slower reactions can be easily (and more efficiently) assayed using manual mixing (see (3–6, 11)). In fast mixing experiments, each time point is an individual reaction that proceeds for a defined amount of time and then is stopped with a quench solution. In manual mixing experiments, individual aliquots are taken from the same large continuous reaction at different times and quenched.
In general, for fast mixer reactions, approximately 19 µL of each reactant in sample ports A and B are combined at time t = 0 in the instrument’s reaction loop. At a later time t, approximately 150 µL (consisting of the combined reaction mixtures and the buffers from push syringes A and B used to drive the reaction mixtures together) are combined with half this volume of the quenching reagent loaded into push syringe C and immediately expelled through the exit line (total volume: approximately 220 µL). Only minimal instructions are given here for use of the RQF-3 Rapid Quench-Flow Fast Mixer. For more comprehensive instructions, the instrument manual should be consulted.
3.1 Preparation of Reaction Mixtures
Filter binding assays are performed in 1× Transcription Buffer, which consists of the following concentrations in the absence of urea: 120 mM KCl (129 mm (millimolal)), 10 mM (10.7 mm) MgCl2, 40 mM (43mm) Tris pH 8.0 at assay temperature, 1 mM DTT, 100 µg/mL bovine serum albumin (BSA), and (from storage buffer) 12.5 mM (13.3 mm) NaCl and 6.25% v/v (855 mM, 913 mm) glycerol. This solution can be prepared by mixing one-fourth the final volume 4× Transcription Buffer Salts, one eighth the final volume RNAP Storage Buffer, 1/1000th the volume 1M DTT stock, and 1/500th the volume 50 mg/mL BSA stock with deionized H2O.
As described in Section 2.1, we add dry urea to 1× Transcription Buffer to make our urea stock solutions. This procedure maintains the molal concentrations of the other components constant, and does not significantly change the molar concentrations of other species at final urea concentrations less than 1 M, though the favorable interaction of urea with KCl reduces the effective salt concentration (i.e., the salt activity (1)).
In general, reaction volumes should be large enough to allow for the production of 12–18 time points, which should be sufficient to determine a time course of competitor-resistant open complex formation or dissociation (see Note 7). All volumes described below have been chosen to achieve this number of data points.
All solutions used should be equilibrated at the assay temperature 5–15 minutes prior to initiation of an experiment.
3.2 Nitrocellulose Filter Binding (see Note 8)
Use a pair of tweezers to place a nitrocellulose filter over a manifold port under vacuum. Place one of the provided stainless steel chimney weights over the filter (leaving the center of the filter accessible via the hole in the weight). Open the port to remove water from the filter, close the port, and cover filter with 200–500 µL Filter Binding Wash Buffer.
To bind an experimental sample, open the port to pull through the Wash Buffer. Apply the sample to the center of the filter. Wash with 200–500 µL Wash Buffer. Close the port, and remove the filter from the manifold.
Dry the filter completely. Insert the filter into a scintillation vial and determine its radioactive counts per minute (cpm) using a scintillation counter (see Note 9).
3.3 Examples of Assays to Determine Solute Effects on Kinetics of Open Complex Formation
3.3.1 Manual Mixing Association Experiment at 0.5 M urea
- Prepare Solutions:
- DNA Mix (800µL total): Combine 66.7 µL 6 M Urea in 1× Transcription Buffer Salts, 183 µL 4× Transcription Buffer Salts, 0.80 µL 1 M DTT, 1.60 µL 50 mg/mL BSA, 91.6 µL RNAP Storage Buffer, X µL of radiolabeled DNA (final concentration 50–500 nM; see Note 10), and (456 – X) µL of deionized H2O.
- RNAP Mix (750 µL total): Combine 62.5 µL 6 M Urea in 1× Transcription Buffer Salts, 172 µL 4× Transcription Buffer Salts, 0.75 µL 1 M DTT, 1.50 µL 50 mg/mL BSA, Y µL RNAP in Storage Buffer (to obtain a final concentration twice as large as that desired in the association reaction), (85.9 – Y) µL Storage Buffer, and 427 µL deionized H2O (see Note 11).
- Competitor Mix (500 µL, for 50 µg/mL heparin in assay): Combine 41.7 µL 6 M Urea in 1× Transcription Buffer Salts, 115 µL 4× Transcription Buffer Salts, 0.5 µL 1 M DTT, 1.0 µL 50 mg/mL BSA, 57.3 µL RNAP Storage Buffer, 3 µL 50 mg/mL heparin, and 285 µL dH2O. Aliquot 20 µL of this mix into one tube per filter for the experiment.
Before initiating the reaction, remove 50 µL of DNA Mix and combine it with 50 µL of 1× Transcription Buffer and 20 µL Competitor Mix. Filter 100 µL of this to determine the background filter retention of DNA (cpmbkgd; see section 4.1).
At time 0, combine 750 µL DNA Mix and 750 µL RNAP Mix. Vortex gently or pipet to mix.
At various time points, remove a 100 µL aliquot of the DNA-RNAP reaction mixture, quench by combining it with 20 µL of Competitor Mix in a separate tube for 10 s, and filter 100 µL of the quenched solution. Take enough long time points to accurately define the plateau cpm value (see Note 7).
At the end of the assay, remove two separate aliquots of 100 µL, and combine each with 20 µL Competitor Mix. Filter 100 µL of one of the 120 µL samples 10 seconds after mixing to determine the cpm value for this population of 100% CR complexes (cpm100% CR, used in determining filter efficiency; see section 4.1). Remove 100 µL from the other 120 µL sample and divide it into 20 µL aliquots. Spot these aliquots onto previously dried filters to determine the total cpm in each filtered aliquot (cpmTOT, used in determining the fraction of competitor-resistant filter retainable complexes, θ; see section 4.1).
3.3.2 Rapid-Mix Rapid-Quench Association Experiment at 0.5 M Urea
- Prepare solutions:
- DNA Mix (500µL total) and RNAP Mix (500 µL): Same proportions of reagents as in the corresponding mixes detailed in Section 3.3.1.1.
- Push Buffer for DNA and RNAP Mixes (10 mL total): Combine 833 µL 6 M Urea in 1× Transcription Buffer, 2290 µL 4× Transcription Buffer Salts, 10 µL 1 M DTT, 20 µL 50 mg/mL BSA, 1150 µL RNAP Storage Buffer, and 5700 µL deionized H2O. Remove 100 µL for determination of cpmbkgd and cpmTOT.
- Competitor Mix (5 mL total, for 50 µg/mL heparin in assay): Combine 417 µL 6 M Urea in 1× Transcription Buffer Salts, 1.15 mL 4× Transcription Buffer Salts, 5 µL 1 M DTT, 10µL 50 mg/mL BSA, 573 µL RNAP Storage Buffer, 15 µL 50 mg/mL heparin, and 2.83 mL deionized H2O.
Load Push Buffer into the left (A) and right (B) vertical drive syringes of a KinTek RQF-3 Rapid Quench-Flow Fast Mixer. Load Competitor Mix into the center (C) drive syringe.
Load the DNA Mix into the instrument’s left (A) sample port. Load Push Buffer (set aside in step 1c) into the right (B) sample port. Perform two reactions, mixing DNA Mix with Push Buffer in the reaction loop for 1 second. Filter one of the expelled samples to determine cpmbkgd. Apply the other expelled sample onto a set of dry filters to determine cpmTOT. Clean sample port B and load RNAP Mix.
Generate time points for the association of RNAP and DNA by allowing individual reactions to proceed for defined times prior to being quenched and expelled. Take enough long time points to accurately define the plateau cpm value (see Note 7). Collect the expelled volumes by inserting the exit line on the fast mixer into a 1.5 mL microcentrifuge tube into which a hole has been made in the top (see Note 5). Filter the entire expelled sample as described in Section 3.2.
3.4 Examples of Assays to Determine Solute Effects on Kinetics of Open Complex Dissociation
3.4.1 Manual Mixing Dissociation Experiment at 2 M urea
- Prepare solutions:
- RPo Mix (810 µL total): Combine 233 µL 4× Transcription Buffer Salts, 0.93 µL 1 M DTT, 1.9 µL 50 mg/mL BSA, X µL of radiolabeled DNA solution (final concentration 50–500 nM (see Note 8)), and (578 – X) µL deionized H2O (resulting in a volume of 814 µL). Remove 105 µL of this mix and combine with 15 µL Storage Buffer (resulting in 120 µL). To the remaining 709 µL, add 101 µL RNAP in Storage Buffer. Incubate at 37°C for 30–60 minutes to form open complexes.
- Urea/Competitor Mix (750 µL total, for 50 µg/mL heparin in assay): Combine 500 µL 6 M Urea in 1× Transcription Buffer Salts, 62.5 µL 4× Transcription Buffer Salts, 0.75 µL 1 M DTT, 1.5 µL 50 mg/mL BSA, 31.3 µL RNAP Storage Buffer, 1.5 µL 50 mg/mL heparin, and 153 µL deionized H2O.
Before initiation of dissociation reaction, remove 60 µL from RPo Mix and incubate at 37 °C to ensure the association reaction proceeds to completion (an additional 5–10 half-lives). Combine with 60 µL of 1× Transcription Buffer containing 100 µg/mL heparin, incubate for 10 seconds, and filter 100 µL to determine cpm100%CR. To the 120 µL sample from Section 1.a. above, add 120 µL Urea/Competitor Mix. Filter 100 µL of this to determine cpmbkgd. Remove 100 µL and spot onto a set of dry filters to determine cpmTOT.
To the remaining 750 µL of RPo Mix, add 750 µL of Urea/Competitor Mix at time 0. At various time points, remove 100 µL of the dissociation reaction and filter as described in Section 3.2.
3.4.2 Rapid-Mix Rapid-Quench Dissociation Experiment at 2 M urea
- Prepare solutions:
- RPo Mix (500 µL total): Combine 155 µL 4× Transcription Buffer Salts, 0.61 µL 1 M DTT, 1.24 µL 50 mg/mL BSA, X µL of radiolabeled DNA solution (final concentration 50–500 nM (see Note 10)), and (386 – X) µL deionized H2O (resulting in a volume of 543 µL). Remove 105 µL of this mix and combine with 15 µL Storage Buffer (resulting in 120 µL). To the remaining 438 µL, add 62.5 µL RNAP in Storage Buffer. Incubate at 37°C for 30–60 minutes to form open complexes.
- Urea/Competitor Mix (5 mL total): Same proportions of reagents as in the corresponding mixes detailed in Section 3.4.1.1.
- Push Buffer for RPo Mix (10 mL total): Combine 2.50 mL 4× Transcription Buffer Salts, 10 µL 1 M DTT, 20 µL 50 mg/mL BSA, 1.25 mL RNAP Storage Buffer, and 6.22 µL deionized H2O.
- Quench Buffer (10 mL total): Combine 2.5 mL 4× Transcription Buffer Salts, 10 µL 1 M DTT, 20 µL 50 mg/mL BSA, 1.25 mL RNAP Storage Buffer, 10 µL 50 mg/mL heparin, and 6.21 mL dH2O.
Load 5 mL Push Buffer for RPo Mix into the left (A) vertical drive syringe of a KinTek RQF-3 Rapid Quench-Flow Fast Mixer. (Set aside 5 mL Push Buffer for RPo Mix for later determination of cpm100%CR.) Load Urea/Competitor Mix into the right (B) drive syringe. Load Quench Buffer into the center (C) drive syringe.
Load mixture containing DNA in 1× Transcription Buffer (removed before addition of RNAP in production of RPo Mix) into the instrument’s left (A) sample port. Load Urea/Competitor Mix into the right (B) sample port. Perform two reactions, mixing DNA with Urea/Competitor Mix for 1 second. Filter one of the expelled samples to determine cpmbkgd. Apply the other expelled sample onto a set of dry filters to determine cpmTOT. Clean sample port A and load RPo Mix.
Generate time points for the dissociation of RPo by allowing individual reactions to proceed for defined times prior to being quenched and expelled (see Note 12). Collect and filter the expelled volumes as described in Section 3.3.2.
After all experimental time points are collected at the desired temperature, bring the temperature of the water bath connected to the fast mixer to 37 °C. Replace the urea solutions with Push Buffer for RPo. Mix and filter several samples with very short incubation times to determine cpm100%CR.
4. Data analysis
4.1 Determination of θt
The first step in the analysis of filter binding kinetic data is to calculate θt, the fraction of promoter DNA in competitor-resistant (CR) complexes at time t. CR complexes are operationally defined as those which survive a 10 s challenge with an inert (non-displacing) competitor like heparin (for λPR; see Note 2) or unlabeled promoter DNA. For λPR, because RPo is generally much more stable than I3 and I2, it is usually the dominant competitor-resistant open complex, though I3 may also be significant at lower temperatures. Values of θt are calculated by dividing the background-corrected counts per minute (cpm) for a time point filter by the total cpm per sample bound and correcting for filter efficiency (E), defined as the fraction of labeled promoter DNA retained on the filter under conditions where 100% of promoter DNA is bound in a CR complex in solution (E = cpm100%CR/cpmTOT; see Notes 13 and 14).
| (1) |
In RPo formation kinetics experiments, if the long-time plateau value of θt (called θt=∞) is significantly less than 1, then RPo formation is reversible and the kinetics are for the approach to an equilibrium distribution of RPo, intermediate complexes and free promoter DNA. If θt=∞ = 1, then RPo formation is irreversible.
4.2 Determination of the Observed Rate Constant for RPo Formation
In excess RNAP, single-exponential kinetics of formation of CR complexes at λPR (primarily RPo) are observed under all conditions investigated. In these experiments, the observed rate constant for formation of these CR (open) complexes is called βCR, and is determined by fitting θt versus time to the first order rate equation
| (2) |
where θo is the fitted value of θt at t = 0 (typically θo less than 0.1) and θt=∞ is the long-time (plateau, equilibrium) value of θt (11, 12). Equivalently, βCR can be fit using the decay to equilibrium equation
| (3) |
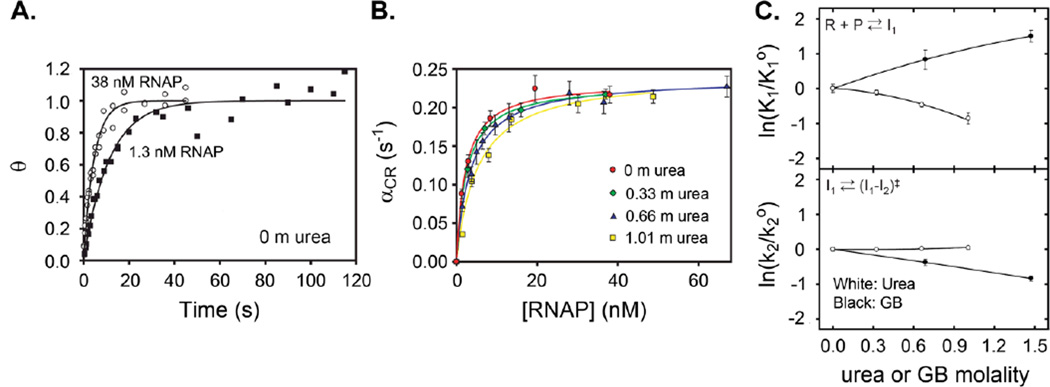
where Δθt = θt=∞ - θt and Δθo = θt=∞ - θo. Representative kinetic data for the formation of RPo using wild-type RNAP and λPR DNA are shown in Fig. 1.A., plotted as θt versus t.
Figure 1.
Data showing the determination of the urea dependences of K1 and k2. A. Rate constants of formation of filter-retainable open complexes (βCR) are determined at 25°C in BBassocurea at 0 m urea at 38 and 1.3 nM active RNAP using Equation 2. B. Data obtained for αCR are fit to Equation 5 as a function of active [RNAP] to obtain fits for K1 and k2 at varying concentrations of urea. C. Values of K1 and k2 are plotted as functions of urea and GB to determine solute concentration dependences, or “m-values”. Figure reproduced from (3) with permission (ACS Publications).
Where association is reversible (i.e., where θt=∞ is less than 1), then βCR is interpreted as the decay-to-equilibrium rate constant, while if association is irreversible (θt=∞ = 1), βCR is interpreted as the rate constant αCR for formation of competitor-resistant complexes. In general, βCR is related to αCR by
| (4) |
where kd is the dissociation rate constant. Where possible, αCR should be determined under conditions where RPo formation is irreversible (βCR >> kd, so that αCR = βCR). Where decay-to-equilibrium studies are necessary, the best way to determine αCR from βCR is to obtain an independent determination of kd. (see Note 15).
4.3 Determination of K1 and k2
The quantities K1 and k2 in Mechanism 1 are determined from analysis of the dependence of αCR on RNAP concentration in excess RNAP and at sufficiently high concentrations of DNA and RNAP that the kinetics of the initial diffusion-limited promoter binding step are not rate-determining. Single-exponential kinetics of CR complex formation indicates that all intermediates (I1 in Mechanism 1) formed before the rate-determining step (I1 → I2 in Mechanism 1) are in rapid equilibrium with each other and with free promoter DNA under the conditions investigated to date. For this situation, the rate constant αCR for RPo formation is a hyperbolic function of RNAP concentration (11–13):
| (5) |
where K1 is the equilibrium constant for I1 formation, k2 is the rate constant for the DNA opening step that converts I1 to I2, and [RNAP]tot is the total concentration of active RNAP (see Note 16). Nonlinear fitting of αCR versus [RNAP]tot is used to determine K1 and k2 (as in Fig. 1.B).
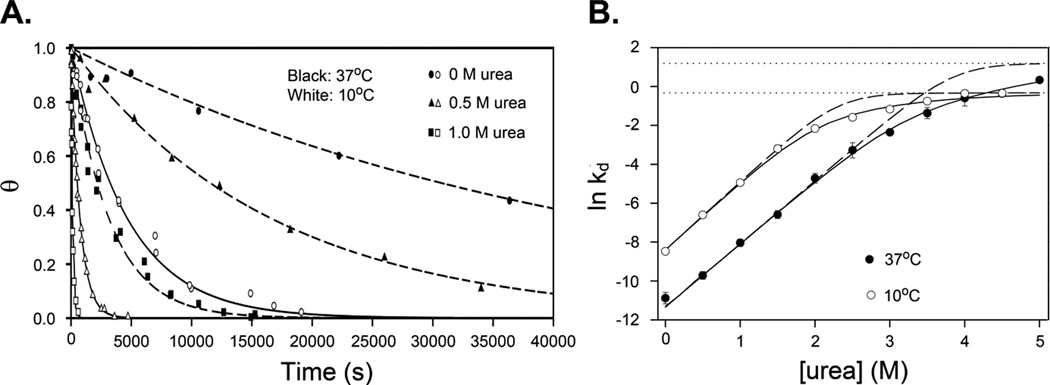
4.4 Determination of Observed Dissociation Rate Constant kd
The first order kinetics of dissociation of open complexes, made irreversible by the presence of an inert (non-displacing) competitor (see Note 2), are analyzed by the rate equation
| (6) |
to obtain the dissociation rate constant kd (see Fig. 2.A.): In Equation 6, θt=∞ is the fitted value of θt at completion of dissociation (typically θt=∞ less than 0.15), and θ0 is the fitted initial value of θ (typically θ0 ≈ 1).
Figure 2.
Data showing the determination of the urea dependences of k−2 and K3. A. Rates of dissociation of filter-retainable open complexes are determined at 10 and 37°C in DB at 0, 0.5, and 1 M urea using Equation 6. B. Data obtained for ln(kd) are fit as a function of urea concentration. Nonlinear fits are shown with (solid lines) and without (dashed lines, using Equation 7) consideration of the urea dependence of the term k−2/k−3. (For detailed analysis, see (5).) Horizontal dotted lines represent values of k−2, while the initial slopes of the fits determine the m-value -dlnK3/d[urea]. Figure reproduced from (5) with permission (Elsevier).
Single exponential kinetics of dissociation of RPo are observed after addition of an inert competitor because intermediates in dissociation formed before the rate-determining step (i.e. I3 and I2 in Mechanism 1) are unstable relative to RPo and rapidly equilibrate with one another and with RPo before the rate-determining conversion of I2 to I1. For this situation, kd is interpreted (12) as
| (7) |
where K3 is the overall equilibrium constant for the conversion of I2 to RPo. For most conditions investigated, K3 is much greater than 1 for competitor-initiated dissociation experiments for λPR and therefore kd ≈ k−2/K3. For dissociation initiated by a rapid shift to 1.1 M KCl (or 4 M urea), making K3 much less than 1, the observed dissociation rate constant kd ≈ k−2. Dissociation kinetics experiments initiated by upshifts to less extreme concentrations of urea have been used to determine rate constants k3 and k−3 and establish the existence of the intermediate I3 (5).
4.5 Interpreting Effects of Solutes
Effects of solutes on biopolymer processes arise from interactions of the solute with the biopolymer surface that is buried or exposed in the process (14). If the interaction of the solute with the biopolymer surface is favorable, relative to interactions with water, then addition of the solute will favor the direction of the process that exposes the biopolymer surface to water. Urea and Hofmeister salt ions like GuH+, I− and SCN− are protein denaturants because they interact favorably (or at least not unfavorably) with the amide and hydrocarbon surface of the protein that is exposed when the protein unfolds. On the other hand, osmolytes like GB and proline and Hofmeister salt ions like Glu−, F− and SO42− are protein stabilizers because their interactions with amide and aliphatic hydrocarbon surface of proteins are unfavorable relative to interactions with water. Effects of a solute on the rate constant for a step of a process are interpreted analogously in terms of interactions of the solute with the biopolymer surface that is buried or exposed in forming the high-free-energy transition state in that step (see (15)).
4.5.1 Interpreting Solute Effects on Equilibrium Constants
The effect of a solute on the equilibrium constant for a step is interpreted in terms of interactions of the solute with the biopolymer surface that is buried or exposed. For the conversion of reactant X to product Y, where the observed equilibrium constant Kobs = ([Y]/[X])eq at solute concentration [solute] and Kobs,o is the corresponding equilibrium constant in the absence of that solute,
| (8) |
where ΔASAX→Y is the change in water-accessible surface area in the conversion of X to Y and the proportionality constant αX→Y is the “interaction potential” quantifying the interaction of the solute with a unit area (i.e. 1 Å2) of a surface with the composition specified by ΔASAX→Y. Predicted values of αX→Y for interactions of urea with the predominantly hydrocarbon surfaces involved in various protein processes are generally in the range - (2.5 ± 0.5) × 10−4 M−1 (1). Therefore, as proposed originally by Myers et al. (16), urea can be used semiquantitatively as a probe of ΔASA in protein processes irrespective of the composition of the ΔASA. In particular, urea is an unambiguous probe for distinguishing steps with large changes in ASA (e.g. I2 → RPo, which has very large urea dependence of Kobs, indicating a very large reduction in ASA (see Fig. 1.C.)) from those with small or compensating changes in ASA (e.g., I1 → I2, which has no urea dependence of Kobs, indicating small or compensating changes in ASA (see Fig. 2.B.)).
Values of αX→Y for interactions of GB or proline with different surfaces can differ by as much as an order of magnitude, making them less easily interpretable and less useful as general probes of ΔASA in structurally uncharacterized processes (2). However, these solutes should prove very useful in conjunction with urea for beginning to characterize the composition of the ΔASA, in addition to the amount. For the surface exposed in local or global protein unfolding (approximately 70% hydrocarbon, 20% amide, 10% other polar or charged), effects of GB and proline are opposite in sign and slightly larger in magnitude than those of urea (2).
4.5.2 Interpreting Solute Effects on Rate Constants
For the interconversion of reactant X and product Y via high free energy transition state TS, with single exponential kinetics and observed forward and back rate constants kf and kb:
| (9) |
Protein folding transition states (TS) have recently been characterized using effects of denaturants (urea or guanadinium chloride) on folding and unfolding rate constants (15). The slopes of linear plots of ln(k) versus [denaturant] (closely related to kinetic m-values) for folding and unfolding of thirteen globular proteins were analyzed together with data on the temperature dependences of these rate constants (activation heat capacities) to quantify the amounts of amide and hydrocarbon ASA buried in folding to and from TS. From these data, we conclude that folding to TS preferentially buries amide surface and that the amount of amide and hydrocarbon surface buried in folding to TS in most cases exceeds that buried in forming all the native secondary structure of the protein.
For RNAP, the only rate constants that have been characterized in this way are k2 and k−2 for the step that includes DNA opening (see Fig. 1.C. and Fig. 2.B.). For this step, neither k2 nor k−2 depends on urea concentration. Additionally, k2 is found to be independent of salt concentration and is unchanged when Cl− is replaced by Glu−, variables which greatly affect other steps in the mechanism of transcription initiation. We conclude that DNA opening occurs in the active site cleft, greatly reducing effects of solutes and salts from what would be observed for DNA melting in solution. Also, or alternatively, burial of RNAP surface in this step may compensate for the exposure of DNA surface upon melting. Investigation of effects of other solute probes on this step will help distinguish between these two scenarios.
4.5.3 Example Application of Solute Dependence: DME Assembly in Late Steps of RPo Formation
Recent work has shown the useful application of solute concentration dependence to the study of large conformational changes in the late steps of RPo formation. The slope of the initial linear region of a plot of ln(kd) versus urea for RPo complexes at λPR is approximately 3.0 M−1 (see Fig. 2 B); an increase in urea concentration from 0 to 1 M therefore increases kd by a factor of e3, about 20-fold. In this urea concentration range, kd ≈ k−2/K3 (Equation 7). Dissociation experiments after an upshift to high concentrations of urea in Fig. 2B reveal that k−2 is independent of urea concentration, so the entire effect of urea on αX→YΔASAX→Y is on the equilibrium constant K3 for the conversion I2 → RPo. Therefore, lnK3 decreases with increasing [urea] with a slope αI2→RPoΔASAI2→RPo = −3.0 M−1. Interpreted as a protein folding or a subunit assembly process involving the DME of RNAP, using αI2→ RPo = (2.5 ± 1) × 10−4 M−1 (see above) indicates that ΔASAI2 → RPo is about 1.2 × 104 A2. Folding buries 80–100 A2 per amino acid residue buried (ref?), so this ΔASAI2→RPo corresponds to 120–150 buried residues. The effect of GB on dissociation is similar in magnitude and opposite in sign to that of urea, consistent with this interpretation.
Many important questions remain about this folding transition in the late steps of the mechanism, including which regions of RNAP and promoter DNA are responsible for it, and its role in transcription initiation. Analysis of amino acid sequences of the beta and beta prime subunits with the program PONDR (Predictions of Natural Disordered Regions; (17, 18)) predicted regions of RNAP that are disordered in holoenzyme and that might fold in RPo formation, contributing to the massive urea dependence of the step I2→RPo (3). Several such unfolded regions, located at the downstream end of the cleft, are now designated downstream mobile elements (DME) (6). Variants of RNAP with deletions in these regions are being investigated to determine their effect on K3 and its urea dependence. For example, deletion of residues 1149–1190 of the β’ subunit of RNAP removes most of a DME called the β’ jaw (19, 20). This deletion (termed “ΔJAW RNAP”) reduces the urea dependence of K3 from −3.0 M−1 to −2.0 M−1, reducing the predicted number of residues folded in this step from 120–150 for WT RNAP to 80–100 for ΔJAW RNAP and suggesting that these 42 residues of the jaw are unstructured in the unbound WT RNAP holoenzyme and fold as part of the conformational changes that stabilize the initial open complex I2 (6). We find that the urea dependence of kd for RPo formed at the T7A1 promoter is similar to that seen for λPR (ER, unpublished data), indicating that a similar folding transition occurs to stabilize the initial open complex at this promoter. Solute studies performed at other promoters, as well as determinations of clamp width by FRET (21), RNAP-DNA crosslinking, or other methods will help to answer questions about the generality and physiological role of this massive conformational change late in the mechanism of transcription initiation.
Acknowledgments
Support of NIH grant GM 103061 (MTR) is gratefully acknowledged.
Footnotes
RNA polymerase binds well to nicked DNA and overhanging ends, so complete fill-in of the PCR fragment is essential. We use Vent DNAP (NEB) for PCR and a plasmid as our PCR template when possible, and we observe the best results with DNA fragments extending to at least −120 on the upstream end and +40 on the downstream end, relative to the transcription start site +1.
Possible competitors include unlabeled promoter DNA, single stranded DNA, or the polyanion heparin. No matter which competitor is used, it is necessary to test whether it is performing as intended. Heparin, typically used at concentrations in excess of 10 µg/mL, is an effective and inexpensive competitor that makes dissociation of WT RNAP from the λPR promoter irreversible. For WT RNAP and the λPR promoter, the dissociation rate constant kd is independent of heparin concentration above this threshold concentration, indicating that the mechanism of action of heparin as a competitor is to bind free RNAP and not to displace promoter DNA from open complexes. However, at some other promoters (e.g., T7 phage promoters (22)) and for some RNAP mutants (e.g., deletions of σ1.1 (Amanda Drennan, unpublished data), kd is observed to increase with increasing heparin concentration. In these situations, heparin must also bind to the open complex, displacing promoter DNA. Extrapolation of kd to zero heparin concentration provides an estimate of the unperturbed kd, which can also be determined using unlabeled full-length λPR promoter DNA (added in over 5–10-fold excess over RNAP) as a fast-binding, non-displacing competitor.
We use a 6 M urea stock here as a convenient example. Other concentrations can be used, up to the solubility limit of urea (approximately 10 M).
Dissolving urea is highly endothermic and occurs slowly. For high concentration stocks, it may be best accomplished by vortexing or stirring continuously in a water bath. Buffers containing urea can be stored at 4°C for approximately one week before the urea begins to hydrolyze significantly.
Using microcentrifuge tubes with holes punched in the top ensures that the expelled mixture does not splash out of the tube, as could be the case for a tube with an open cap. We find it convenient to produce a hole with an ideal diameter using a soldering iron.
Note that the protein-stabilizing solute glycerol, used at high concentration (50% v/v) in RNAP storage buffer, is of necessity present at moderately high concentration (typically 0.7 – 0.9 M) and must be held constant in series where concentrations of RNAP and other solutes are varied.
For accurate nonlinear fitting, at least half the time points on the time course should be before the first half life, and at least 3–4 time points should be used to define the plateau in counts as t approaches ∞. If the plateau cpm value is less than that expected for complete reaction (i.e. reversible, θt=∞ < 1 (see Section 4.1)), as is the case for WT RNAP- WT λPR experiments at low T, especially at low concentrations of excess RNAP, accurate plateau determination is particularly important.
Nitrocellulose filter binding is a highly quantitative kinetic assay on both long and short timescales which yields data readily compared to a large body of previous work. However, this method is not continuous, and not all promoter and RNAP variants form filter-retainable complexes. Use of a brief heparin challenge allows filter binding to distinguish between short-lived (competitor-sensitive, typically closed) complexes and long lived (competitor-resistant (CR), typically open) complexes. To test whether CR complexes are open, the kinetics of forming permanganate-reactive complexes are compared with the kinetics of forming CR complexes (Amanda Drennan and ER, unpublished data). These filter binding methods cannot determine individual rate constants k1 and k−1 for I1, as stated in Section 3. Alternative bulk kinetic methods include intrinsic tryptophan fluorescence (23) and the recently developed RNAP beacon assay (24). Both of these methods are still in development for quantitative use with long DNA fragments and are limited to relatively short assay times, but we are excited about their potential for fast collection of kinetic data. Single-molecule methods for analyzing kinetics of open complex formation and dissociation have also recently become available, including colocalization single-molecule spectroscopy (CoSMoS) (25).
We find it effective to dry filters by laying them flat onto a sheet of aluminum foil under a heat lamp. When quantifying filter cpm, we find that we obtain adequate signal detection by placing a dried filter into a standard borosilicate scintillation counter vial; however, one can also add scintillation fluid into the vial to amplify the signal.
The amount of DNA required will vary with the number of cpm filtered, the reversibility of association, etc. It should be adjusted empirically such that at equilibrium the filters retain enough cpm for easy detection and accurate data fitting, while the molar concentration of DNA does not exceed 1/5 the molar concentration of RNAP (so that RNAP is in sufficient excess for pseudo-first order kinetics).
RNAP should be added to buffer as close to the beginning of the experiment as possible to prevent damage to the enzyme from oxidation.
Combination of the dissociation reaction with the Quench Buffer at this point results in a dilution of the urea concentration, which slows down the rate of dissociation. The quench should be designed to slow down the rate of dissociation enough such that effectively no additional dissociation occurs while the solution is being collected and filtered. For λPR and wild-type RNAP, the quenched mixture should have concentrations of 0.08–0.12 M KCl and less than 1.5 M urea in order to effectively stop the dissociation reaction at room temperature. For other systems, the dilution of destabilizing solutes required should be empirically determined. It may be necessary to further dilute the reaction beyond what is achieved within the fast mixer itself. In such a case, samples can be further diluted with additional Quench Buffer already present in the microcentrifuge tube into which it is expelled. Where large volumes of Quench Buffer are required, we find it convenient to use 5 mL tubes.
For good quality DNA, background retention should be less than 5% of total counts.
Filter efficiency can vary considerably. Our standard of known high filter efficiency is 37°C in transcription buffer, where it varies from about 70% to about 100% depending on wash volume, vacuum pressure, pretreatment of filters, amount of free 32P label in the solution, etc. High concentrations of some solutes (e.g. over 0.24 M KCl or 1.5 M urea (5)) reduce filter efficiency, as does low salt concentration. Efficiency of retention of open complexes is not significantly temperature dependent (11). Under reversible conditions, it is best determined based in an irreversible assay performed side-by-side, or by removing 100 µL from the reaction mixture, incubating it at 37°C for one hour, and then subjecting to a heparin challenge.
The most accurate association kinetic data (and hence the most accurate values of K1, k2, and their solute dependences) are obtained under conditions where the process of open complex formation is irreversible. To counteract the reversibility potentially induced by destabilizing solutes such as urea, lower salt concentrations than those described here can be used.
We determine the active concentration of RNAP by titrating promoter DNA (at high concentration (typically over 1 nM) relative to the reciprocal of the overall equilibrium constant for RPo formation (typically less than 0.1 nM)) with RNAP so that all active RNAP added will bind to promoter DNA, as in (13). In general, we find that activity of WT holoenzyme varies from 30 to 100%, depending on enzyme preparation. Mutant polymerases, and especially mutants of the σ subunit, tend to be less active (10–30%). Activity can decrease in storage at −20 °C and should be determined regularly.
References
- 1.Guinn EJ, Pegram LM, Capp MW, Pollock MN, Record MT., Jr Quantifying why urea is a protein denaturant, whereas glycine betaine is a protein stabilizer. Procedings of the National Academy of Science USA. 2011;108:16932–16937. doi: 10.1073/pnas.1109372108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diehl RC, Guinn EJ, Capp MW, Tsodikov OV, Record MT., Jr Quantifying Additive Interactions of the Osmolyte Proline with Individual Functional Groups of Proteins: Comparisons with Urea and Glycine Betaine, Interpretation of m-Values. Biochemistry. 2013;52:5997–6010. doi: 10.1021/bi400683y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kontur WS, Saecker RM, Davis CA, Capp MW, Record MT., Jr Solute Probes of Conformational Changes in Open Complex (RPo) Formation by Escherichia coli RNA Polymerase at the lPR Promoter: Evidence for Unmasking of the Active Site in the Isomerization Step and for Large-Scale Coupled Folding in the Subsequent Conversion to RPo. Biochemistry. 2006;45:2161–2177. doi: 10.1021/bi051835v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kontur WS, Capp MW, Gries TJ, Saecker RM, Record MT., Jr Probing DNA Binding, DNA Opening, and Assembly of a Downstream Clamp/Jaw in Escherichia coli RNA Polymerase-lPR Promoter Complexes Using Salt and the Physiological Anion Glutamate. Biochemistry. 2010;49:4361–4373. doi: 10.1021/bi100092a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kontur WS, Saecker RM, Capp MW, Record MT., Jr Late steps in the formation of the E. coli RNA polymerase-lPR promoter open complexes: Characterization of conformational changes by rapid [perturbant] upshift experiments. Journal of Molecular Biology. 2008;376:1034–1047. doi: 10.1016/j.jmb.2007.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drennan AC, Kraemer MR, Capp MW, Gries TJ, Ruff EF, Sheppard C, et al. Key Roles of the Downstream Mobile Jaw of Escherichia coli RNA Polymerase in Transcription Initiation. Biochemistry. 2012;51:9447–9459. doi: 10.1021/bi301260u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang G-Q, Patel SS. Rapid Binding of T7 RNA Polymerase is Followed by Simultaneous Bending and Opening of the Promoter DNA. Biochemistry. 2006;45:4947–4956. doi: 10.1021/bi052292s. [DOI] [PubMed] [Google Scholar]
- 8.Saecker RM, Record MT, Jr, deHaseth PL. Mechanism of Bacterial Transcription Initiation: RNA Polymerase-Promoter Binding, Isomerization to Initiation-Competent Open Complexes, and Initiation of RNA Synthesis. Journal of Molecular Biology. 2011;412:754–771. doi: 10.1016/j.jmb.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gries TJ, Kontur WS, Capp MW, Saecker RM, Record MT., Jr One-step DNA melting in the RNA polymerase cleft opens the initiation bubble to form an unstable open complex. Procedings of the National Academy of Science USA. 2010;107:10418–10423. doi: 10.1073/pnas.1000967107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis CA, Bingman CA, Landick R, Record MT, Jr, Saecker RM. Real-time footprinting of DNA in the first kinetically significant intermediate in open complex formation by Escherichia coli RNA polymerase. Procedings of the National Academy of Science USA. 2007;104:7833–7838. doi: 10.1073/pnas.0609888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saecker RM, Tsodikov OV, McQuade KL, Schlax PE, Jr, Capp MW, Record MT., Jr Kinetic studies and structural models of the association of E. coli s70 RNA polymerase with the lPR promoter: Large scale conformational changes in forming the kinetically significant intermediates. Journal of Molecular Biology. 2002;319:649–671. doi: 10.1016/S0022-2836(02)00293-0. [DOI] [PubMed] [Google Scholar]
- 12.Tsodikov OV, Record MT., Jr General method of analysis of kinetic equations for multistep reversible mechanisms in the single-exponential regime: application to kinetics of open complex formation between Es70 RNA polymerase and lambda (R) promoter DNA. Biophysical Journal. 1999;76:1320–1329. doi: 10.1016/S0006-3495(99)77294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McClure WR, Cech CL, Johnston DE. A Steady State Assay for the RNA Polymerase Initiation Reaction. Journal of Biological Chemistry. 1978;253(24):8941–8948. [PubMed] [Google Scholar]
- 14.Record MT, Jr, Guinn EJ, Pegram LM, Capp MW. Introductory Lecture: Interpreting and predicing Hofmeister salt ion and solute effects on biopolymer and model processes using the solute partitioning model. Faraday Discuss. 2013;160:9–44. doi: 10.1039/c2fd20128c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guinn EJ, Kontur WS, Tsodikov O, Shkel I, Record MT., Jr Probing the protein-folding mechanism using denaturant and temperature effects on rate constants. Procedings of the National Academy of Science USA. 2013 doi: 10.1073/pnas.1311948110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myers JK, Pace CM, Scholtz JM. Denaturant m-values and heat capacity changes: Relation to changes in accessible surface areas of protein unfolding. Protein Science. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Predicting protein disorder for N-, C-, and internal regions. Genome Informatics. 1999;10:30–40. [PubMed] [Google Scholar]
- 18.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42(38–48) doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 19.Ederth J, Artsimovitch I, Isaksson LA, Landick R. The Downstream DNA Jaw of Bacterial RNA Polymerase Facilitates Both Transcriptional Initiation and Pausing. Journal of Biological Chemistry. 2002;277:37456–37463. doi: 10.1074/jbc.M207038200. [DOI] [PubMed] [Google Scholar]
- 20.Ederth J, Isaksson LA, Abdulkarim F. Origin-specific reduction of ColE1 plasmid copy number due to mutations in a distinct region of the Escherichia coli RNA polymerase. Molecular Genetics and Genomics. 2002;267(5):587–592. doi: 10.1007/s00438-002-0689-y. [DOI] [PubMed] [Google Scholar]
- 21.Chakraborty A, Wang D, Ebright YW, Korlann Y, Kortkhonjia E, Kim T, et al. Opening and Closing of the Bacterial RNA Polymerase Clamp. Science. 2012;337:591–595. doi: 10.1126/science.1218716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cech CL, McClure WR. Characteriation of Ribonucleic Acid Polymerase-T7 Promoter Binary Complexes. Biochemistry. 1980;19:2440–2447. doi: 10.1021/bi00552a023. [DOI] [PubMed] [Google Scholar]
- 23.Johnson RS, Chester RE. Stopped-flow kinetic analysis of the interaction of Escherichia coli RNA polymerase with the bacteriophage T7 A1 promoter. Journal of Molecular Biology. 1998;283:353–370. doi: 10.1006/jmbi.1998.2101. [DOI] [PubMed] [Google Scholar]
- 24.Mekler V, Pavlova O, Severinov K. Interaction of Escherichia coli RNA Polymerase s70 Subunit with Promoter Elements in the Context of Free s70, RNA Polymerase Holoenzyme, and the b'-s70 Complex. Journal of Biological Chemistry. 2011;286:270–279. doi: 10.1074/jbc.M110.174102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedman LJ, Mumm JP, Gelles J. RNA polymerase approaches its promoter without sliding along DNA. Procedings of the National Academy of Science USA. 2013;110:9740–9745. doi: 10.1073/pnas.1300221110. [DOI] [PMC free article] [PubMed] [Google Scholar]