Abstract
Fungi are prolific producers of secondary metabolites (SMs) that show a variety of biological activities. Recent advances in genome sequencing have shown that fungal genomes harbor far more SM gene clusters than are expressed under conventional laboratory conditions. Activation of these “silent” gene clusters is a major challenge, and many approaches have been taken to attempt to activate them and, thus, unlock the vast treasure chest of fungal SMs. This review will cover recent advances in genome mining of SMs in Aspergillus nidulans. We will also discuss current updates in gene annotation of A. nidulans and recent developments in A. nidulans as a molecular genetic system, both of which are essential for rapid and efficient experimental verification of SM gene clusters on a genome-wide scale. Finally, we will describe advances in the use of A. nidulans as a heterologous expression system to aid in the analysis of SM gene clusters from other fungal species that do not have an established molecular genetic system.
Keywords: Aspergillus, secondary metabolite, polyketide synthase, nonribosomal peptide synthetase, gene cluster
Introduction
Filamentous fungi have played an important role in the history of drug discovery and development. The secondary metabolites (SMs) that these organisms produce have served as a source of low molecular weight molecules with a variety of biological activities. Examples of these are antibiotics such as penicillin, immunosuppressants such as cyclosporine, antifungals such as griseofulvin and the echinocandins, and antihypercholesterolemic drugs such as lovastatin [11, 24, 35]. Many of the bioactive SMs that are easily accessible under conventional laboratory conditions have already been isolated and patented for drug development. However, advances in genome sequencing [23, 32, 38, 40] revealed that fungal species harbor an abundance of SM gene clusters and these far exceed the number of known metabolites produced by the species [45]. This potential abundance of SMs may reflect their importance in nature as a chemical arsenal for niche security [41]. The carefully controlled growth conditions in laboratory culture settings prevent any competition or life-threatening circumstances that would trigger the production of SMs, thereby leaving many of the gene clusters dormant. Activating these silent gene clusters, revealing their biosynthetic pathways, and isolating the SMs produced by these pathways is a major challenge in the search for new SMs.
Various approaches have been taken in attempts to activate silent SM gene clusters [14], including fusing of regulatable promoters to a pathway-specific transcription factor [5, 17], removal of genes required for heterochromatin formation [7], genome-wide analysis of mutants of LaeA, a global regulator of SM [8], co-incubation with microorganisms to mimic conditions in nature [48], and the “one strain many compounds” (OSMAC) strategy [6]. Most of these approaches were developed in A. nidulans due to the availability of highly efficient gene-targeting systems in this model organism. The developed approaches are often subsequently applied to other filamentous fungi.
In this review we focus on recent advances in genome mining of secondary metabolism genes in A. nidulans. We also describe the current status of the annotation of the products of secondary metabolism genes in A. nidulans. We would also like to direct readers to the accompanying review in this issue by our collaborators Nancy Keller and Philipp Wiemann on general strategies for mining fungal natural products and to other recent reviews on this subject [10, 28, 53, 56, 61].
The status of annotating secondary metabolite genes in A. nidulans
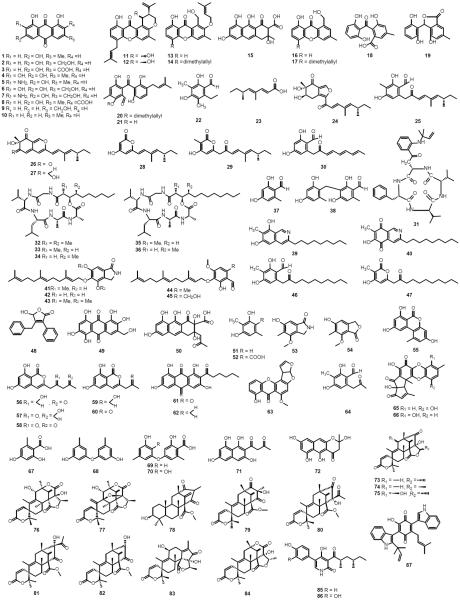
Among the Aspergillus species, A. nidulans has been used as a model organism, making it the most comprehensively studied and best characterized species in the genus with the largest body of literature. Most studies of secondary metabolite biosynthesis in A. nidulans have used strains derived from a common reference strain, A. nidulans FGSC A4. A. nidulans FGSC A4 was initially sequenced by Cereon Genomics (Monsanto) in 1998 to three-fold genome equivalent coverage and the sequence was publicly released in 2003. Shortly thereafter, additional sequencing was completed at the Whitehead Institute/MIT Center for Genomic Research to give a total of 13 genome-equivalent coverage. The seminal paper describing the A. nidulans genome was published in 2005 [23]. Access to this sequenced genome has allowed investigators to use sequence similarity to known genes from other species to mine for core genes that are involved in secondary metabolism in A. nidulans. Algorithms such as SMURF (Secondary Metabolite Unknown Regions Finder) [27] and antiSMASH (antibiotics and Secondary Metabolite Analysis Shell) [34] are extremely useful in predicting the core SM biosynthetic genes. Taking into consideration the most recent annotation and additional analysis of available genomic data, our group's most recent estimate is that the A. nidulans genome contains 56 putative secondary metabolism core genes including 27 polyketide synthase genes (PKS), 2 polyketide synthase-like genes (PKS-like), 11 nonribosomal peptide synthetase genes (NRPS), 15 NRPS-like genes, and 1 hybrid NRPS-PKS gene. Table 1 and Figure 1 show our current understanding of the products of these genes and the products from the pathways.
Table 1.
Secondary metabolism gene clusters in A. tndulans
| No | AspGD Designation | Core Gene Name | Gene typea | Metabolites isolated from A. nidulans2 | References |
|---|---|---|---|---|---|
| 1 | AN0016 | pes1 | NRPS | ||
| 2 | AN0150 | mdpG | NR-PKS | emodin (1), emodin analogs (2–10), shamixanthone (11), epishamixanthone (12), variecoxanthone A (13), emericellin (14), afrochrysone carboxylic acid (15), 1-hydroxy-6-methyl-8-hydroxymethylxanthone (16), paeciloxanthone (17), monodictyphenone (18), 3-(2,6-dihydroxyphenyl)-4-hydroxy-6-methyl-1(3H)-isobenzofiiranone (19), arugosin A, H (20,21) | [7, 16, 36, 44, 46, 51] |
| 3 | AN0523 | pkdA | NR-PKS | 2-ethyl-4,6-dihydroxy-3,5-dimethylbenzaldehyde (22) | [1] |
| 4 | AN0607 | sidC | NRPS | ferricrocin | [20] |
| 5 | AN1034 | afoE | NR-PKS | (2Z,4Z)-4,6-dimethylocta-2,4-dienoic aicd (23), asperfiiranone (24), 6-[(3E,5E)-5,7-dimethyl-2-oxonona-3,5-dienyl-2,4-dihydr oxy-3-methylbenzaldehyde (25), preasperpyranone (26), asperpyranone (27), Proasperfuranone A, B (28, 29) | [17, 49] |
| 6 | AN1036 | afoG | HR-PKS | (2Z,4Z)-4,6-dimethylocta-2,4-dienoic aicd (23), asperfiiranone (24), 6-[(3E,5E)-5,7-dimethyl-2-oxonona-3,5-dienyl-2,4-dihydr oxy-3-methylbenzaldehyde (25), preasperpyranone (26), asperpyranone (27), Proasperfuranone A, B (28, 29) | [17, 49] |
| 7 | AN1242 | NRPS | nidulanin A (31) | [2] | |
| 8 | AN1680 | NRPS-like | |||
| 9 | AN1784 | pkjA | HR-PKS | ||
| 10 | AN2032 | pkhA | NR-PKS | 2,4-dihydroxy-6-[(3E,5E,7E)-2-oxonona-3,5,7-trienyl]benzaldehyde (30) | [1] |
| 11 | AN2035 | pkhB | HR-PKS | 2,4-dihydroxy-6-[(3E,5E,7E)-2-oxonona-3,5,7-trienyl]benzaldehyde (30) | [1] |
| 12 | AN2064 | NRPS-like | |||
| 13 | AN2545 | easA | NRPS | emericellamides (32–36) | [18] |
| 14 | AN2547 | easB | HR-PKS | emericellamides (32–36) | [18] |
| 15 | AN2621 | acvA | NRPS | penicillin | [31, 51] |
| 16 | AN2924 | NRPS-like | |||
| 17 | AN3230 | pkfA | NR-PKS | orsellinaldehyde (37), 3-(2,4-dihydroxy-6-methylbenzyl)-orsellinaldehyde (38), aspernidine A-E (41–45) | [1, 47, 58], |
| 18 | AN3386 | pkiA | NR-PKS | 7-methyl-3-nonylisoquinoline-6,8-diol (39), 6-hydroxy-7-methyl-3-nonylisoquinoline-5,8-dione (40), 2,4-dihydroxy-3-methyl-6-(2-oxoundecyl)benzaldehyde (46), 4-hydroxy-3-methyl-6-(2-oxoundecyl)-2-pyrone (47) | [1] |
| 19 | AN3396 | micA | NRPS-like | microperfuranone (48) | [59] |
| 20 | AN3495 | inpA | NRPS-like | ||
| 21 | AN3496 | inpB | NRPS | ||
| 22 | AN3612 | HR-PKS | |||
| 23 | AN4827 | NRPS-like | |||
| 24 | AN5318 | NRPS-like | |||
| 25 | AN6000 | aptA | NR-PKS | asperthecin (49), 2,3,6,8,9-pentahydroxy-1-oxo-3-(2-oxopropyl)-1,2,3,4-tetrahydroanthracene-2-carboxylic acid (50) | [1, 29, 52] |
| 26 | AN6236 | sidD | NRPS | ||
| 27 | AN6431 | HR-PKS | |||
| 28 | AN6444 | NRPS-like | |||
| 29 | AN6448 | pkbA | NR-PKS | 2,5-dimethylresorcinol (51), 3-methylorsellinic acid (52), cichorine (53), nidulol (54), | [1, 43] |
| 30 | AN6791 | HR-PKS | |||
| 31 | AN7071 | pkgA | NR-PKS | alternariol (55), citreoisocoumarin (56), analogs of citreoisocoumarin (57–60) | [1] |
| 32 | AN7084 | PKS-like | |||
| 33 | AN7489 | PKS-like | |||
| 34 | AN7825 | stcA (pksST) | NR-PKS | norsolorinic acid (61), norsolorinic acid anthrone (62), sterigmatocystin (63) | [1, 12, 54, 62] |
| 35 | AN7837+AN7838 | HR-PKS | |||
| 36 | AN7884 | NRPS | |||
| 37 | AN7903 | pkeA | NR-PKS | 2,4-dihydroxy-3-methyl-6-(2-oxopropyl)benzaldehyde (64) | [1] |
| 38 | AN7909 | orsA | NR-PKS | F9775A, B (65, 66), orsellinic acid (67), diorcinol (68), gerfelin (69), 10-deoxygerfelin (70) | [7, 42, 46, 48] |
| 39 | AN8105 | NRPS-like | |||
| 40 | AN8209 | wA | NR-PKS | 2-acetoacetyl T4HN (71), naphthopyrone YWA1 (72) | [1, 21, 55] |
| 41 | AN8383 | ausA | NR-PKS | isoaustinone (73), analogs of isoaustinone (74, 75), austinol (76), dehydroausitnol (77), protoaustinoid (78) preaustinoid A3–A5 (79–81), austinoneol A (82), neoaustinone (83), austinolide (84) | [1, 30, 36] |
| 42 | AN8412 | apdA | Hybrid | aspyridone A, B (85, 86) | [5] |
| 43 | AN8513 | tdiA | NRPS-like | terrequinone A (87) | [9, 51] |
| 44 | AN8910 | HR-PKS | |||
| 45 | AN9005 | HR-PKS | |||
| 46 | AN9129 | NRPS-like | |||
| 47 | AN9226 | nrpA | NRPS | ||
| 48 | AN9243 | NRPS-like | |||
| 49 | AN9244 | NRPS | |||
| 50 | AN9291 | NRPS-like | |||
| 51 | AN10297 | NRPS-like | |||
| 52 | AN10430 | HR-PKS | |||
| 53 | AN10486 | NRPS-like | |||
| 54 | AN10576 | ivoA | NRPS | ||
| 55 | AN11191 | pkkA | HR-PKS | ||
| 56 | AN12440 | NR-PKS |
Abbreviations: polyketide synthase (PKS), non ribosomal peptide synthetase (NRPS), hybrid PKS-NRPS (Hybrid), nonreduced polyketide synthase (NR-PKS), highly reduced polyketide synthase (HR-PKS)
Bold numbers correspond to chemical structures shown in Figure 1.
Fig.1.
Structures of compounds isolated from A. nidulans
Bioinformatic advances
Since the original publication of the genome sequence data [23], A. nidulans gene annotations have been refined repeatedly to correct incomplete or inaccurate content [3, 4, 25, 39, 57]. The Aspergillus Genome Database (AspGD; http://www.aspgd.org/) provides gene and protein sequence data that are curated based on submitted information and published literature. Although the wealth of data and the availability of the algorithms mentioned previously have provided accurate predictions of core SM biosynthetic genes, it is still not possible to predict with accuracy the boundaries of secondary metabolite gene clusters or the functions of each member of the clusters based solely on genome sequence data. This is due to the fact that many of the genes surrounding the core SM biosynthetic genes often have unknown functions, making predictions of their involvement in the biosynthetic process of the SM almost impossible. Elucidation of biosynthetic gene clusters have thus been heavily dependent on experimental verification, a laborious process that involves single gene deletion of each gene with a suspected role in SM biosynthesis, followed by identification and characterization of SMs produced by the deletion strains. Improvements in “omics”-based methods for accurate prediction of SM gene cluster members and the availability of more precise annotations are desirable for a more rapid and efficient experimental verification of novel SM gene clusters.
Andersen et al. recently published a novel strategy for the accurate prediction of SM gene cluster boundaries [2] based on the fact that expression of genes of a given SM cluster is coordinately regulated. A DNA expression microarray was used to identify genes that were co-regulated with SM gene cluster backbone enzymes. A variety of culture media were selected that, based on SM profiling experiments, would elicit expression of as many gene clusters as possible. Samples were then taken from A. nidulans growing on the selected culture media for transcriptional profiling, and the generated data were combined with previously published data to form a superset of a total of 44 expression conditions for analysis. Andersen et al. developed clustering scores (CSs) that reflected the degree to which each gene was co-regulated with its neighbors. They developed statistical guidelines for identifying the extent of gene clusters, which were applied to the microarray data to generate cluster predictions. Comparisons with published data demonstrated that their algorithm predicted gene clusters with high accuracy and can even predict gene clusters that are scattered across different chromosomes. Using this algorithm, a list of 58 predicted SM gene clusters was generated.
These data have been curated at AspGD and applied as a criterion for the manual annotation of computationally predicted gene clusters as a part of a continued effort to improve and refine the prediction of SM gene cluster boundaries[25]. This updated gene cluster boundary annotation also incorporates published experimental data, synteny between clustered genes among different species, functional annotation of putative gene cluster members, and increase in the distance between predicted boundary genes and genes that are directly adjacent to it but not included in the cluster. This new and improved set of comprehensive SM gene cluster predictions will aid in facilitating the future investigation of novel Aspergillus SMs.
Genome-wide kinase knock-outs
The molecular genetic system of A. nidulans is powerful and technical advances in recent years have made genome-wide, systematic approaches more feasible. The Fungal Genetics Stock Center (FGSC) provides a systematic gene deletion construct collection, a valuable experimental resource for the A. nidulans research community. De Souza et al. have generated a set of gene deletion constructs for 9,851 genes, which represents 93.3% of the encoding genome [19]. Mutant strains generated with the cassettes are deposited with the FGSC after construction.
Using this deletion construct resource, a genome-wide kinase knock-out library consisting of deletion strains of most A. nidulans non-essential kinase genes was generated and deposited at the FGSC [19]. The kinase deletion strains were used for genome-wide functional analysis of kinases, resulting in identification of many previously unknown functions for kinases[19]. This kinase knock-out library was screened to test the hypothesis that manipulation of kinase expression has the potential to activate silent SM gene clusters [58]. This led to the discovery of an mpkA deletant that produced aspernidine A, a compound that had been discovered previously in A. nidulans [47] but the biosynthetic pathway remained unknown. The mpkA deletant produced a sufficient amount of aspernidine A to allow the identification and analysis of the gene cluster involved in its biosynthesis. From the chemical structure of aspernidine A combined with previous data [1], it was predicted that a nonreducing polyketide synthase (NR-PKS) gene, pkfA (AN3230) is involved in the biosynthesis of aspernidine A. Deletion of pkfA confirmed this, and the boundary of the gene cluster was identified through a series of gene deletions of the surrounding genes of pkfA. Analysis of the SMs produced by mpkA deletion strains resulted in isolation and characterization of novel intermediates that aided in generating a proposed pathway for aspernidine A.
A similar deletion set of 28 protein phosphatase genes was generated and used to identify four essential phosphatases and four required for normal growth [50]. The deposited deletion constructs were also used in a study that identified multiple kinases and phosphatases involved in the sensing of carbon and energetic status, and also contributed to the understanding of the signaling cascades that result in regulation of CreA derepression and hydrolytic enzyme production [13].
Genome-wide analysis of all non-reduced polyketide synthases and NRPS-like enzymes in A. nidulans
Despite the success of various strategies to activate silent gene clusters, a large number of potential SM gene clusters remain untapped. To analyze clusters resistant to activation through existing approaches, a strategy was developed that completely bypasses normal regulation [1]. It takes advantage of recent advances in the construction of transforming fragments by fusion PCR and effective gene targeting to replace promoters of SM genes with the regulatable alcA promoter. It was applied to obtain a comprehensive understanding of the products of nonreducing polyketide synthase (NR-PKS) genes, a class of key genes of SM biosynthetic pathways [1]. The A. nidulans genome harbors 14 NR-PKS genes, and combined efforts by several groups over the years led to the identification of the chemical products of six of them [7, 12, 16, 17, 29, 42, 48, 52, 55, 62]. To determine the products of the remaining eight NR-PKS genes, the native promoters for each NR-PKS and other genes necessary for product formation or release were replaced with the alcA promoter. Induction of expression resulted in the production and release of compounds from each of the NR-PKS and allowed the completion of the determination of the products of NR-PKS genes of A. nidulans.
This approach can be applied to the discovery of other classes of SM biosynthetic gene clusters. This was demonstrated by systematically targeting nonribosomal peptide synthetase (NRPS)-like genes for promoter replacement, resulting in the discovery that one of the NRPS-like genes, micA, is the sole gene responsible for the biosynthesis of the metabolite microperfuranone [59].
In another strategy carried out by Nielsen et al., a genome-wide PKS deletion library was constructed by systematically deleting all 32 putative PKS genes [36]. A reference strain was cultured on an array of culture media to find conditions that would induce production of SMs that were not previously linked to a gene cluster, and this was followed by screening of the genome-wide PKS deletion library to establish the genetic link to the SMs. This approach provided novel links between PKS genes and SMs, demonstrating its strength and the potential usefulness of the deletion library as a resource for further PKS studies.
Use of A. nidulans as a host for heterologous expression of SM genes from other Aspergillus species
The highly advanced and established molecular genetic system of A. nidulans can be applied to the study of SM production of other fungal species that have poor or nonexistent molecular genetic systems [60]. Heterologous expression of fungal genes in other fungi has been used and with some success, but this approach is not without limitations including finding a suitable host and the difficulty of handling large genes and gene clusters. An advantage of fungal systems over bacterial for expressing fungal secondary metabolism genes is that fungi can correctly splice introns of secondary metabolism genes from other fungi resulting in successful expression [15, 22, 26]. Since many fungal SM genes are quite large and contain introns (often several introns) this is of considerable benefit.
Major advances have recently been made in establishing A. nidulans as a host for heterologous expression of fungal SMs. First, entire SM gene clusters have been deleted to eliminate production of unwanted A. nidulans SMs, resulting in reduced SM background and facilitating detection and isolation of compounds produced by the heterologously expressed genes [15].
Second, a system for transferring SM genes from other fungi while placing them under control of the alcA promoter has been developed [15, 33]. This system uses a strategy that involves 1) PCR amplification of each gene, 2) the use of fusion PCR to place each gene under control of the alcA promoter and to construct a transforming fragment, and 3) integration of the fragment into a target A. nidulans locus. For larger clusters several genes must be transferred into A. nidulans and, to avoid running out of selectable markers for transformation, a marker recycling strategy was developed [15]. Each time a new gene is introduced into A. nidulans a selectable marker is evicted and this marker can be used in the subsequent transformation. This strategy allows an unlimited number of genes to be transferred into and expressed in A. nidulans. The use of this approach resulted in the successful expression of all six genes of the gene cluster that encodes the production of asperfuranone, a cryptic gene cluster from A. terreus. Furthermore, various combinations of expression genes were tested, leading to clarification of the asperfuranone biosynthetic pathway.
Another recent approach to transfer members of entire SM gene clusters is to assemble the PCR amplified individual cluster fragments into a single large transforming fragment using USER fusion, followed by insertion into the integration vector by USER cloning [37]. Using this technique, a total of 13 genes of a putative gene cluster responsible for geodin biosynthesis from A. terreus were transferred into A. nidulans in a two step process, successfully enabling geodin biosynthesis in A. nidulans.
Conclusion
Advances in genome sequencing in fungi have provided us with a wealth of information that suggests that the number of SM gene clusters far exceeds the number of discovered compounds. A combination of bioinformatics and experimental verification is fundamental to elucidating the SM biosynthetic pathways that these SM gene clusters encode. Among the many species of Aspergillus, A. nidulans is used as a model organism and it is the species with the most abundant literature by far and the most advanced, highly efficient molecular genetic system. Recent advances in development of prediction algorithms in A. nidulans and updated curation by AspGD have given us access to improved SM gene cluster predictions, which we can use as a basis for subsequent experimental verification. Advances in transforming fragment construction techniques and effective gene targeting expedite the experimental verification process. These advances, in combination, have enabled quick and systematic approaches to uncover the potential of SM production by A. nidulans. The application of these advances is not limited to the SMs of A. nidulans. Combined efforts such as the “1000 Fungal Genomes Project (http://1000.fungalgenomes.org/home/)” by the DOE Joint Genome Institute (JGI) are dedicated to sequencing numerous different species of fungi and providing a database for the research community. Many of these fungi do not have good molecular genetic systems, which makes experimental verification a big challenge. Heterologous expression of fungal genes in other host fungi is one approach that is being used, and major advances have been made to establish A. nidulans as a host. Newly developed methods in constructing transforming fragments and improved transformation strategies have made it possible for large or multiple genes to be transformed into A. nidulans. These approaches will contribute greatly to uncovering the untapped resources of SMs that the fungal genomes encode.
Acknowledgements
C.C.C.W and B.R.O gratefully acknowledge the National Institutes of Health (GM084077) for supporting research on Aspergillus nidulans secondary metabolism. We also thank Elizabeth Oakley and Dr. James Sanchez for their editorial assistance.
Reference list
- 1.Ahuja M, Chiang YM, Chang SL, Praseuth MB, Entwistle R, Sanchez JF, Lo HC, Yeh HH, Oakley BR, Wang CCC. Illuminating the diversity of aromatic polyketide synthases in Aspergillus nidulans. J Am Chem Soc. 2012;134(19):8212–8221. doi: 10.1021/ja3016395. doi:10.1021/ja3016395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen MR, Nielsen JB, Klitgaard A, Petersen LM, Zachariasen M, Hansen TJ, Blicher LH, Gotfredsen CH, Larsen TO, Nielsen KF, Mortensen UH. Accurate prediction of secondary metabolite gene clusters in filamentous fungi. Proc Natl Acad Sci U S A. 2013;110(1):E99–E107. doi: 10.1073/pnas.1205532110. doi: 10.1073/pnas.1205532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnaud MB, Cerqueira GC, Inglis DO, Skrzypek MS, Binkley J, Chibucos MC, Crabtree J, Howarth C, Orvis J, Shah P, Wymore F, Binkley G, Miyasato SR, Simison M, Sherlock G, Wortman JR. The Aspergillus Genome Database (AspGD): recent developments in comprehensive multispecies curation, comparative genomics and community resources. Nucleic Acids Res. 2012;40(D1):D653–D659. doi: 10.1093/nar/gkr875. doi:10.1093/nar/gkr875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnaud MB, Chibucos MC, Costanzo MC, Crabtree J, Inglis DO, Lotia A, Orvis J, Shah P, Skrzypek MS, Binkley G, Miyasato SR, Wortman JR, Sherlock G. The Aspergillus Genome Database, a curated comparative genomics resource for gene, protein and sequence information for the Aspergillus research community. Nucleic Acids Res. 2010;38:D420–D427. doi: 10.1093/nar/gkp751. doi:10.1093/nar/gkp751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol. 2007;3(4):213–217. doi: 10.1038/nchembio869. doi:10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- 6.Bode HB, Bethe B, Hofs R, Zeeck A. Big effects from small changes: Possible ways to explore nature's chemical diversity. Chembiochem. 2002;3(7):619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9. doi:10.1002/1439-7633(20020703)3:7<619::aid-cbic619>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 7.Bok JW, Chiang YM, Szewczyk E, Reyes-Domingez Y, Davidson AD, Sanchez JF, Lo H-C, Watanabe K, Strauss J, Oakley BR, Wang CCC, Keller NP. Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol. 2009;5(7):462–464. doi: 10.1038/nchembio.177. doi:10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bok JW, Hoffmeister D, Maggio-Hall LA, Murillo R, Glasner JD, Keller NP. Genomic mining for Aspergillus natural products. Chem Biol. 2006;13(1):31–37. doi: 10.1016/j.chembiol.2005.10.008. doi:10.1016/j.chembiol.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Bouhired S, Weber M, Kempf-Sontag A, Keller NP, Hoffmeister D. Accurate prediction of the Aspergillus nidulans terrequinone gene cluster boundaries using the transcriptional regulator LaeA. Fungal Genet and Biol. 2007;44(11):1134–1145. doi: 10.1016/j.fgb.2006.12.010. doi:10.1016/j.fgb.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 10.Brakhage AA. Regulation of fungal secondary metabolism. Nat Rev Microbiol. 2013;11(1):21–32. doi: 10.1038/nrmicro2916. doi:10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- 11.Brakhage AA, Schuemann J, Bergmann S, Scherlach K, Schroeckh V, Hertweck C. Activation of fungal silent gene clusters: A new avenue to drug discovery. Prog Drug Res. 2008;66:12. doi: 10.1007/978-3-7643-8595-8_1. [DOI] [PubMed] [Google Scholar]
- 12.Brown DW, Yu JH, Kelkar HS, Fernandes M, Nesbitt TC, Keller NP, Adams TH, Leonard TJ. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc Natl Acad Sci U S A. 1996;93(4):1418–1422. doi: 10.1073/pnas.93.4.1418. doi:10.1073/pnas.93.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown NA, de Gouvea PF, Krohn NG, Savoldi M, Goldman GH. Functional characterisation of the non-essential protein kinases and phosphatases regulating Aspergillus nidulans hydrolytic enzyme production. Biotechnol Biofuels. 2013;6 doi: 10.1186/1754-6834-6-91. doi:10.1186/1754-6834-6-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiang YM, Chang SL, Oakley BR, Wang CCC. Recent advances in awakening silent biosynthetic gene clusters and linking orphan clusters to natural products in microorganisms. Curr Opin Chem Biol. 2011;15(1):137–143. doi: 10.1016/j.cbpa.2010.10.011. doi:10.1016/j.cbpa.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiang YM, Oakley CE, Ahuja M, Entwistle R, Schultz A, Chang SL, Sung CT, Wang CCC, Oakley BR. An efficient system for heterologous expression of secondary metabolite genes in Aspergillus nidulans. J Am Chem Soc. 2013;135(20):7720–7731. doi: 10.1021/ja401945a. doi:10.1021/ja401945a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiang YM, Szewczyk E, Davidson AD, Entwistle R, Keller NP, Wang CCC, Oakley BR. Characterization of the Aspergillus nidulans monodictyphenone gene cluster. Appl Environ Microbiol. 2010;76(7):2067–2074. doi: 10.1128/AEM.02187-09. doi:10.1128/aem.02187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiang YM, Szewczyk E, Davidson AD, Keller N, Oakley BR, Wang CCC. A gene cluster containing two fungal polyketide synthases encodes the biosynthetic pathway for a polyketide, asperfuranone, in Aspergillus nidulans. J Am Chem Soc. 2009;131(8):2965–2970. doi: 10.1021/ja8088185. doi:10.1021/ja8088185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiang YM, Szewczyk E, Nayak T, Davidson AD, Sanchez JF, Lo HC, Ho WY, Simityan H, Kuo E, Praseuth A, Watanabe K, Oakley BR, Wang CCC. Molecular genetic mining of the Aspergillus secondary metabolome: Discovery of the emericellamide biosynthetic pathway. Chem Biol. 2008;15(6):527–532. doi: 10.1016/j.chembiol.2008.05.010. doi:10.1016/j.chembiol.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Souza CP, Hashmi SB, Osmani AH, Andrews P, Ringelberg CS, Dunlap JC, Osmani SA. Functional analysis of the Aspergillus nidulans kinome. PLoS ONE. 2013;8(3) doi: 10.1371/journal.pone.0058008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisendle M, Oberegger H, Zadra I, Haas H. The siderophore system is essential for viability of Aspergillus nidulans: functional analysis of two genes encoding L-ornithine N-5-monooxygenase (sidA) and a non-ribosomal peptide synthetase (sidC) Mol Microbiol. 2003;49(2):359–375. doi: 10.1046/j.1365-2958.2003.03586.x. doi:10.1046/j.1365-2958.2003.03586.x. [DOI] [PubMed] [Google Scholar]
- 21.Fujii I, Watanabe A, Sankawa U, Ebizuka Y. Identification of Claisen cyclase domain in fungal polyketide synthase WA, a naphthopyrone synthase of Aspergillus nidulans. Chem Biol. 2001;8(2):189–197. doi: 10.1016/s1074-5521(00)90068-1. doi:10.1016/s1074-5521(00)90068-1. [DOI] [PubMed] [Google Scholar]
- 22.Fujii I, Yoshida N, Shimomaki S, Oikawa H, Ebizuka Y. An iterative type I polyketide synthase PKSN catalyzes synthesis of the decaketide alternapyrone with regio-specific octamethylation. Chem Biol. 2005;12(12):1301–1309. doi: 10.1016/j.chembiol.2005.09.015. doi: 10.1016/j.chembiol.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 23.Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D'Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Penalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW. Sequencing of Aspergillus nidulans and comparative analysis with A-fumigatus and A-oryzae. Nature. 2005;438(7071):1105–1115. doi: 10.1038/nature04341. doi:10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmeister D, Keller NP. Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat Prod Rep. 2007;24(2):393–416. doi: 10.1039/b603084j. doi:10.1039/b603084j. [DOI] [PubMed] [Google Scholar]
- 25.Inglis DO, Binkley J, Skrzypek MS, Arnaud MB, Cerqueira GC, Shah P, Wymore F, Wortman JR, Sherlock G. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiol. 2013;13 doi: 10.1186/1471-2180-13-91. doi:10.1186/1471-2180-13-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasahara K, Fujii I, Oikawa H, Ebizuka Y. Expression of Alternaria solani PKSF generates a set of complex reduced-type polyketides with different carbon-lengths and cyclization. Chembiochem. 2006;7(6):920–924. doi: 10.1002/cbic.200600034. doi: 10.1002/cbic.200600034. [DOI] [PubMed] [Google Scholar]
- 27.Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47(9):736–741. doi: 10.1016/j.fgb.2010.06.003. doi:10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klejnstrup ML, Frandsen RJN, Holm DK, Nielsen MT, Mortensen UH, Larsen TO, Nielsen JB. Genetics of polyketide metabolism in Aspergillus nidulans. Metabolites. 2012;2(1):100–133. doi: 10.3390/metabo2010100. doi: 10.3390/metabo2010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Chooi Y-H, Sheng Y, Valentine JS, Tang Y. Comparative characterization of fungal anthracenone and naphthacenedione biosynthetic pathways reveals an alpha-hydroxylation-dependent Claisen-like cyclization catalyzed by a dimanganese thioesterase. J Am Chem Soc. 2011;133(39):15773–15785. doi: 10.1021/ja206906d. doi:10.1021/ja206906d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo HC, Entwistle R, Guo CJ, Ahuja M, Szewczyk E, Hung JH, Chiang YM, Oakley BR, Wang CCC. Two separate gene clusters encode the biosynthetic pathway for the meroterpenoids austinol and dehydroaustinol in Aspergillus nidulans. J Am Chem Soc. 2012;134(10):4709–4720. doi: 10.1021/ja209809t. doi:10.1021/ja209809t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maccabe AP, Vanliempt H, Palissa H, Unkles SE, Riach MBR, Pfeifer E, Vondohren H, Kinghorn JR. Delta-(L-alpha-aminoadipyl)-L-cysteinyl-D-valine synthetase from Aspergillus nidulans: molecular characterization of the Acva gene encoding the 1st enzyme of the penicillin biosynthetic-pathway. J Biol Chem. 1991;266(19):12646–12654. [PubMed] [Google Scholar]
- 32.Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto KI, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu JJ, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Juvvadi PR, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, Wortman JR, Yamada O, Yamagata Y, Anazawa H, Hata Y, Koide Y, Komori T, Koyama Y, Minetoki T, Suharnan S, Tanaka A, Isono K, Kuhara S, Ogasawara N, Kikuchi H. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438(7071):1157–1161. doi: 10.1038/nature04300. doi:10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 33.Maiya S, Grundmann A, Li SM, Turner G. The fumitremorgin gene cluster of Aspergillus fumigatus: identification of a gene encoding brevianamide F synthetase. Chembiochem. 2006;7(7):1062–1069. doi: 10.1002/cbic.200600003. doi: 10.1002/cbic.200600003. [DOI] [PubMed] [Google Scholar]
- 34.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:W339–W346. doi: 10.1093/nar/gkr466. doi:10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75(3):311–335. doi: 10.1021/np200906s. doi:10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen ML, Nielsen JB, Rank C, Klejnstrup ML, Holm DK, Brogaard KH, Hansen BG, Frisvad JC, Larsen TO, Mortensen UH. A genome-wide polyketide synthase deletion library uncovers novel genetic links to polyketides and meroterpenoids in Aspergillus nidulans. FEMS Microbiol Lett. 2011;321(2):157–166. doi: 10.1111/j.1574-6968.2011.02327.x. doi:10.1111/j.1574-6968.2011.02327.x. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen MT, Nielsen JB, Anyaogu DC, Holm DK, Nielsen KF, Larsen TO, Nortensen UH. Heterologous reconstitution of the intact geodin gene cluster in Aspergillus nidulans through a simple and versatile PCR based approach. PLoS ONE. 2013;8(8):e72871. doi: 10.1371/journal.pone.0072871. doi:10.1371/journal.pone.0072871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Feldblyum TV, Fischer R, Fosker N, Fraser A, Garcia JL, Garcia MJ, Goble A, Goldman GH, Gomi K, Griffith-Jones S, Gwilliam R, Haas B, Haas H, Harris D, Horiuchi H, Huang J, Humphray S, Jimenez J, Keller N, Khouri H, Kitamoto K, Kobayashi T, Konzack S, Kulkarni R, Kumagai T, Lafton A, Latge JP, Li WX, Lord A, Majoros WH, May GS, Miller BL, Mohamoud Y, Molina M, Monod M, Mouyna I, Mulligan S, Murphy L, O'Neil S, Paulsen I, Penalva MA, Pertea M, Price C, Pritchard BL, Quail MA, Rabbinowitsch E, Rawlins N, Rajandream MA, Reichard U, Renauld H, Robson GD, de Cordoba SR, Rodriguez-Pena JM, Ronning CM, Rutter S, Salzberg SL, Sanchez M, Sanchez-Ferrero JC, Saunders D, Seeger K, Squares R, Squares S, Takeuchi M, Tekaia F, Turner G, de Aldana CRV, Weidman J, White O, Woodward J, Yu JH, Fraser C, Galagan JE, Asai K, Machida M, Hall N, Barrell B, Denning DW. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438(7071):1151–1156. doi: 10.1038/nature04332. doi:10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 39.Nitsche BM, Crabtree J, Cerqueira GC, Meyer V, Ram AFJ, Wortman JR. New resources for functional analysis of omics data for the genus Aspergillus. BMC Genomics. 2011;12 doi: 10.1186/1471-2164-12-486. doi:10.1186/1471-2164-12-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JAE, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EGJ, Debets AJM, Dekker P, van Dijck PWM, van Dijk A, Dijkhuizen L, Driessen AJM, d'Enfert C, Geysens S, Goosen C, Groot GSP, de Groot PWJ, Guillemette T, Henrissat B, Herweijer M, van den Hombergh J, van den Hondel C, van der Heijden R, van der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, van der Maarel M, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, van Peij N, Ram AFJ, Rinas U, Roubos JA, Sagt CMJ, Schmoll M, Sun JB, Ussery D, Varga J, Vervecken W, de Vondervoort P, Wedler H, Wosten HAB, Zeng AP, van Ooyen AJJ, Visser J, Stam H. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol. 2007;25(2):221–231. doi: 10.1038/nbt1282. doi:10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 41.Rohlfs M, Albert M, Keller NP, Kempken F. Secondary chemicals protect mould from fungivory. Biol Lett. 2007;3(5):523–525. doi: 10.1098/rsbl.2007.0338. doi:10.1098/rsbl.2007.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanchez JF, Chiang Y-M, Szewczyk E, Davidson AD, Ahuja M, Oakley CE, Bok JW, Keller N, Oakley BR, Wang CCC. Molecular genetic analysis of the orsellinic acid/F9775 gene cluster of Aspergillus nidulans. Mol Biosyst. 2010;6(3):587–593. doi: 10.1039/b904541d. doi:10.1039/b904541d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez JF, Entwistle R, Corcoran D, Oakley BR, Wang CCC. Identification and molecular genetic analysis of the cichorine gene cluster in Aspergillus nidulans. Medchemcomm. 2012;3(8):997–1002. doi: 10.1039/C2MD20055D. doi:10.1039/c2md20055d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanchez JF, Entwistle R, Hung J-H, Yaegashi J, Jain S, Chiang Y-M, Wang CCC, Oakley BR. Genome-based deletion analysis reveals the prenyl xanthone biosynthesis pathway in Aspergillus nidulans. J Am Chem Soc. 2011;133(11):4010–4017. doi: 10.1021/ja1096682. doi:10.1021/ja1096682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanchez JF, Somoza AD, Keller NP, Wang CCC. Advances in Aspergillus secondary metabolite research in the post-genomic era. Nat Prod Rep. 2012;29(3):351–371. doi: 10.1039/c2np00084a. doi:10.1039/c2np00084a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scherlach K, Sarkar A, Schroeckh V, Dahse H-M, Roth M, Brakhage AA, Horn U, Hertweck C. Two induced fungal polyketide pathways converge into antiproliferative spiroanthrones. Chembiochem. 2011;12(12):1836–1839. doi: 10.1002/cbic.201100132. doi:10.1002/cbic.201100132. [DOI] [PubMed] [Google Scholar]
- 47.Scherlach K, Schuemann J, Dahse HM, Hertweck C. Aspernidine A and B, prenylated isoindolinone alkaloids from the model fungus Aspergillus nidulans. J Antibiot. 2010;63(7):375–377. doi: 10.1038/ja.2010.46. doi:10.1038/ja.2010.46. [DOI] [PubMed] [Google Scholar]
- 48.Schroeckh V, Scherlach K, Nuetzmann H-W, Shelest E, Schmidt-Heck W, Schuemann J, Martin K, Hertweck C, Brakhage AA. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci U S A. 2009;106(34):14558–14563. doi: 10.1073/pnas.0901870106. doi:10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Somoza AD, Lee KH, Chiang YM, Oakley BR, Wang CCC. Reengineering an azaphilone biosynthesis pathway in Aspergillus nidulans to create lipoxygenase inhibitors. Org Lett. 2012;14(4):972–975. doi: 10.1021/ol203094k. doi:10.1021/ol203094k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Son S, Osmani SA. Analysis of all protein phosphatase genes in Aspergillusnidulans identifies a new mitotic regulator, Fcp1. Eukaryot Cell. 2009;8(4):573–585. doi: 10.1128/EC.00346-08. doi:10.1128/ec.00346-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soukup AA, Chiang Y-M, Bok JW, Reyes-Dominguez Y, Oakley BR, Wang CCC, Strauss J, Keller NP. Overexpression of the Aspergillus nidulans histone 4 acetyltransferase EsaA increases activation of secondary metabolite production. Mol Microbiol. 2012;86(2):314–330. doi: 10.1111/j.1365-2958.2012.08195.x. doi:10.1111/j.1365-2958.2012.08195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szewczyk E, Chiang YM, Oakley CE, Davidson AD, Wang CCC, Oakley BR. Identification and characterization of the asperthecin gene cluster of Aspergillus nidulans. Appl Environ Microbiol. 2008;74(24):7607–7612. doi: 10.1128/AEM.01743-08. doi:10.1128/aem.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsunematsu Y, Ishiuchi Ki, Hotta K, Watanabe K. Yeast-based genome mining, production and mechanistic studies of the biosynthesis of fungal polyketide and peptide natural products. Nat Prod Rep. 2013;30(8):1139–1149. doi: 10.1039/c3np70037b. doi:10.1039/c3np70037b. [DOI] [PubMed] [Google Scholar]
- 54.Wang CCC, Chiang Y-M, Kuo P-L, Chang J-K, Hsu Y-L. Norsolorinic acid from Aspergillus nidulans inhibits the proliferation of human breast adenocarcinoma MCF-7 cells via fas-mediated pathway. Basic Clin Pharmacol Toxicol. 2008;102(6):491–497. doi: 10.1111/j.1742-7843.2008.00237.x. doi:10.1111/j.1742-7843.2008.00237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe A, Fujii I, Sankawa U, Mayorga ME, Timberlake WE, Ebizuka Y. Re-identification of Aspergillus nidulans wA gene to code for a polyketide synthase of naphthopyrone. Tetrahedron Lett. 1999;40(1):91–94. doi:10.1016/s0040-4039(98)80027-0. [Google Scholar]
- 56.Wiemann P, Keller NP. Strategies for mining fungal natural products. J Ind Microbiol Biotechnol. 2013;xxx:xxx–xxx. doi: 10.1007/s10295-013-1366-3. [DOI] [PubMed] [Google Scholar]
- 57.Wortman JR, Gilsenan JM, Joardar V, Deegan J, Clutterbuck J, Andersen MR, Archer D, Bencina M, Braus G, Coutinho P, von Dohren H, Doonan J, Driessen AJM, Durek P, Espeso E, Fekete E, Flipphi M, Estrada CG, Geysens S, Goldman G, de Groot PWJ, Hansen K, Harris SD, Heinekamp T, Helmstaedt K, Henrissat B, Hofmann G, Homan T, Horio T, Horiuchi H, James S, Jones M, Karaffa L, Karanyi Z, Kato M, Keller N, Kelly DE, Kiel J, Kim JM, van der Klei IJ, Klis FM, Kovalchuk A, Krasevec N, Kubicek CP, Liu B, MacCabe A, Meyer V, Mirabito P, Miskei M, Mos M, Mullins J, Nelson DR, Nielsen J, Oakley BR, Osmani SA, Pakula T, Paszewski A, Paulsen I, Pilsyk S, Pocsi I, Punt PJ, Ram AFJ, Ren QH, Robellet X, Robson G, Seiboth B, van Solingen P, Specht T, Sun JB, Taheri-Talesh N, Takeshita N, Ussery D, Vankuyk PA, Visser H, de Vondervoort P, de Vries RP, Walton J, Xiang X, Xiong Y, Zeng AP, Brandt BW, Cornell MJ, van den Hondel C, Visser J, Oliver SG, Turner G. The 2008 update of the Aspergillus nidulans genome annotation: A community effort. Fungal Genet Biol. 2009;46:S2–S13. doi: 10.1016/j.fgb.2008.12.003. doi:10.1016/j.fgb.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yaegashi J, Praseuth MB, Tyan S-W, Sanchez JF, Entwistle R, Chiang Y-M, Oakley BR, Wang CCC. Molecular genetic characterization of the biosynthesis cluster of a prenylated isoindolinone alkaloid aspernidine A in Aspergillus nidulans. OrgLett. 2013;15(11):2862–2865. doi: 10.1021/ol401187b. doi:10.1021/ol401187b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yeh HH, Chiang YM, Entwistle R, Ahuja M, Lee KH, Bruno KS, Wu TK, Oakley BR, Wang CCC. Molecular genetic analysis reveals that a nonribosomal peptide synthetase-like (NRPS-like) gene in Aspergillus nidulans is responsible for microperfuranone biosynthesis. Appl Microbiol Biotechnol. 2012;96(3):739–748. doi: 10.1007/s00253-012-4098-9. doi:10.1007/s00253-012-4098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yin WB, Chooi YH, Smith AR, Cacho RA, Hu Y, White TC, Tang Y. Discovery of cryptic polyketide metabolites from dermatophytes using heterologous expression in Aspergillus nidulans. ACS Synth Biol. 2013 doi: 10.1021/sb400048b. doi: 10.1021/sb400048b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yin WB, Keller NP. Transcriptional regulatory elements in fungal secondary metabolism. J Microbiol. 2011;49(3):329–339. doi: 10.1007/s12275-011-1009-1. doi:10.1007/s12275-011-1009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu JH, Leonard TJ. Sterigmatocystin biosynthesis in Aspergillus nidulans requires a novel type I polyketide synthase. J Bacteriol. 1995;177(16):4792–4800. doi: 10.1128/jb.177.16.4792-4800.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]