Abstract
The cyclic AMP-responsive element-binding protein (CREB) is phosphorylated in response to a wide variety of signals, yet target gene transcription is only increased in a subset of cases. Recent studies indicate that CREB functions in concert with a family of latent cytoplasmic co-activators called cAMP-regulated transcriptional co-activators (CRTCs), which are activated through dephosphorylation. A dual requirement for CREB phosphorylation and CRTC dephosphorylation is likely to explain how these activator–co-activator cognates discriminate between different stimuli. Following their activation, CREB and CRTCs mediate the effects of fasting and feeding signals on the expression of metabolic programmes in insulin-sensitive tissues.
Phosphorylation provides a rapid but reversible mechanism by which extracellular signals regulate gene expression. Phosphorylation-dependent transcriptional activators are thought to be ‘first responders’ to such cues. They often promote genome-wide changes in gene expression through the induction of so-called immediate early genes, which typically encode other DNA-binding proteins; these in turn diversify the transcriptional response by triggering the expression of late genes. As was first demonstrated for the cyclic AMP-responsive element (CRE)-binding protein (CREB)1, phosphorylation modulates the activity of many eukaryotic transcription factors by altering their subcellular localization, DNA binding, or transactivation potential. CREB phosphorylation increases its activity by stimulating its association with the histone acetyl-transferase paralogues CREB-binding protein (CBP) and p3002–4. As its name suggests, CREB is phosphorylated in response to hormonal stimuli (for example, catecholamines) that increase intracellular cAMP production, but it can also be phosphorylated in response to a wide variety of extra-cellular signals, including growth factors, osmotic stress and ultraviolet irradiation5–11.
Like other second messengers, cAMP stimulates cellular gene expression with burst-attenuation kinetics. Transcription rates peak from 30 minutes to 1 hour after cAMP production is stimulated and they decrease progressively thereafter, paralleling changes in CREB phosphorylation at the regulatory Ser133 site12 as well as changes in CBP/p300-mediated histone acetylation over the promoter13. Binding of certain hormones to G protein-coupled receptors triggers the activation of adenyl cyclases, which catalyse the production of cAMP from ATP (FIG. 1). Binding of cAMP to regulatory subunits within the protein kinase A (PKA) heterotetramer liberates catalytic subunits, which migrate to the nucleus through a passive process. PKA phosphorylates CREB at Ser133 and the dynamics of CREB phosphorylation appear to be rate-limited by nuclear entry of the PKA catalytic subunit (FIG. 1), a passive process that requires from 30 minutes to 1 hour to reach maximal levels after stimulation with cAMP agonist14,15. Following its activation by cAMP and other signals, CREB is progressively dephosphorylated by the Ser/Thr phosphatases protein phosphatase 1 (PP1) and PP2A16,17, although the relative importance of these phosphatases in different tissues remains unclear.
Figure 1. cAMP stimulates creB phosphorylation.
The binding of ligand to G protein-coupled receptors (GPCR) that are linked to the stimulatory G proteins, which are comprised of α-, β- and γ-subunits, leads to the activation of adenylate cyclase (AC), which catalyses the synthesis of cyclic AMP. Increases in cellular cAMP stimulate protein kinase A (PKA) signalling. cAMP binds to the regulatory (R) subunits of PKA, thereby promoting their dissociation from the catalytic subunits. The liberated catalytic subunits enter the nucleus by passive diffusion and phosphorylate the cAMP-responsive element (CRE)-binding protein (CREB) at Ser133. Phosphorylated CREB promotes target gene expression at promoters containing CREs.
CREB and its family members — CRE modulator (CREM) and activating transcription factor 1 (ATF1) — stimulate target gene expression at promoters that contain CREs. These typically appear as either palindromic (TGACGTCA) or half-site (TGACG or CGTCA) sequences18–20, although a small number of functional variant sites21,22 have been described. Most of the 750,000 palindromic and half-site CREs in the human genome, a number of which are near silenced genes, are unoccupied in cells owing to disruptive cytosine methylation within the CREB-binding site23,24. Unmethylated, functional CREB-binding sites are primarily localized to promoter proximal regions within 250 base pairs of the transcription start site for about 5,000 genes, or roughly one quarter of the mammalian genome24,25. Indeed, like other promoter proximal elements, CREB-binding sites are most active when placed near the TATA box, and they become progressively weaker when moved further upstream26. Remarkably, CREB binds to, and undergoes Ser133 phosphorylation at, a majority of promoters with CREB-binding sites in cells exposed to a cAMP agonist. Despite this extensive profile of CREB activation, exposure to cAMP stimulates the expression of only about 100 CREB target genes, and the subset of cAMP-inducible genes appears to differ considerably between cell types24. Canonical TATA boxes — a requirement for transcriptional induction by cAMP27 — are absent from nearly two-thirds of CREB-occupied promoters, which might explain why so few CREB-occupied genes are upregulated. However, it seems likely that additional, as yet uncharacterized, epigenetic inputs exist, because only one in ten of the CREB-occupied promoters that contain a TATA-box seem to be upregulated in cells exposed to cAMP.
Additional effectors might be involved in modulating CREB target gene expression9,28. Indeed, the recent identification of the cAMP-regulated transcriptional co-activator (CRTC) family has provided insight into the mechanism underlying signal discrimination by the CREB pathway21,29. And parallel studies on the role of CREB and CRTCs in glucose and lipid metabolism have revealed how these proteins regulate cellular genes in response to circulating hormones and nutrients. Based on recent progress in the area of CREB and metabolism, this Review focuses on this aspect of signalling and transcription by CREB and the CRTC family of co-activators. Studies on the role of growth factors and stress signals in regulating CREB activity in the nervous system and other tissues have been reviewed extensively30 and will not be considered in this article.
Regulation of CREB activity
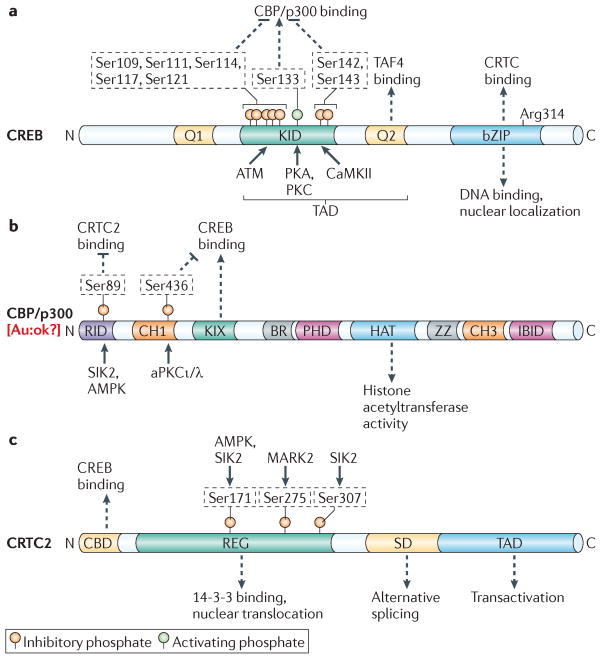
CREB and its family members share similar modular organization: they all contain an amino-terminal transactivation domain (TAD) and a carboxy-terminal basic Leu zipper (bZIP) DNA-binding and dimerization domain (FIG. 2a). The TAD is bipartite; it contains a central kinase-inducible domain (KID) and a Gln-rich constitutive activation (Q2) domain, which function cooperatively in response to cAMP31,32. The Q2 domain has been shown to enhance transcription by interacting with the TBP-associated factor 4 (TAF4), a component of basal transcription factor IID (TFIID)33,34. Supporting a functional role for this interaction, cells with a knockout of TAF4 are unable to stimulate CREB activity following exposure to cAMP agonist35. The CREB–TAF4 interaction is also blocked in certain neurodegenerative diseases with polyglutamine expansion, in which CREB target gene expression is downregulated due to the cytoplasmic sequestration of human TAF4 (REF. 36). The importance of TAF4 for transcriptional activation suggests that CREB mediates the recruitment of TFIID to relevant target genes in response to cAMP. Indeed, the ability of CREB to associate with TFIID could also explain why canonical TATA boxes are required for cAMP responsiveness.
Figure 2. Modular organization of creB and its co-activators.
a | Cyclic AMP-responsive element-binding protein (CREB) contains two Glu-rich domains (Q1 and Q2), a central kinase-inducible domain (KID) and a carboxy-terminal basic Leu zipper (bZIP) domain. The KID domain and the Q2 domain make up the amino-terminal transactivation domain (TAD). The Q2 domain binds to TBP-associated factor 4 (TAF4); phosphorylation of the KID domain at Ser133 promotes an interaction with CREB-binding protein (CBP) and its paralogue p300. Ser133 is phosphorylated by a number of basic directed kinases including protein kinase A (PKA) and PKC. Two clusters of phosphorylation sites flanking Ser133 inhibit CBP/p300 binding; these sites are phosphorylated by ataxia-telangiectasia mutated (ATM) and calcium- and calmodulin-dependent kinase II (CaMKII) as indicated. The bZIP domain promotes CREB DNA binding and dimerization; it also mediates CREB binding to cAMP-regulated transcriptional co-activators (CRTCs). Arg314 in the bZIP domain is critical for the CREB–CRTC interaction. b | Domain structure of CBP/p300, showing the nuclear receptor-interaction domain (RID), Cys and His-rich region 1 (CH1) and CH3, CREB-binding KIX domain, bromodomain (BR), plant homeodomain (PHD), histone acetyltransferase (HAT) domain, zinc-binding domain (ZZ) and interferon response factor-binding domain (IBID). Phosphorylation of both CBP and p300 at Ser89 by salt-inducible kinase 2 (SIK2) or AMP-activated protein kinase (AMPK) inhibits CRTC2 binding. Phosphorylation of CBP, but not p300, at Ser436 by atypical PKCι/λ (aPKCι/λ), within the CH1 region, inhibits binding to CREB. c | Domain structure of the CRTC family of CREB co-activators, as exemplified by CRTC2. CRTCs contain an N-terminal CREB binding domain (CBD), a central regulatory region (REG), a splicing domain (SD) and a C-terminal TAD. CRTC phosphorylation at Ser171 (by AMPK and SIK2), Ser275 (by microtubule affinity-regulating kinase 2 (MARK2)), and Ser307 (by SIK2) promotes 14-3-3 protein binding and the cytoplasmic sequestration of CRTC2. In contrast with CREB, CRTC2 phosphorylation in the TAD domain has not been described to date.
In contrast with the Q2 domain, the KID domain in CREB mediates its ability to be activated in response to cAMP and calcium, through a phosphorylation-dependent mechanism31,32. However, unlike the PKA-dependent effects of cAMP, intracellular calcium stimulates CREB phosphorylation through calcium- and calmodulin-dependent kinases30. Phosphorylation of CREB at Ser133 in the KID domain promotes an association with the KIX domain of CBP and p300 (REFs 37–39) (FIG. 2b). CBP/p300 are thought to enhance CREB target gene expression by acetylating nucleosomal histones40–42 and recruiting RNA polymerase II complexes43,44. In contrast with the profile of the promoter occupancy of phosphorylated CREB, recruitment of CBP/p300 to promoters is selective and predictive for transcriptional induction in response to cAMP24.
Structural studies of the CREB–CBP complex reveal that the KIX domain folds into a three-helix structure containing a shallow hydrophobic groove45. Phospho-Ser133 in the KID domain makes direct hydrogen bonds and salt bridge contacts with residues in the KIX domain; this association promotes a random coil-to-helix transition in the KID domain, which further stabilizes the complex through interactions with residues lining the hydrophobic groove in the KIX domain. The structure of the KID–KIX complex predicts that Ser133 phosphorylation is sufficient for CBP/p300 recruitment in response to extracellular signals. Supporting this idea, mutations in the KID and KIX domains that disrupt this interaction reduce target gene expression, whereas mutations that enhance the KID–KIX association increase expression46–49. Also, small molecules that block the formation of KID–KIX complexes also inhibit CREB target gene expression50.
However, despite its importance in stimulating CREB activity, Ser133 phosphorylation does not seem to be sufficient to recruit CBP/p300 or to increase target gene expression. Triggering of the phosphoinositol pathway in response to growth factor signals, for example, increases the PKC-mediated phosphorylation of CREB at Ser133 with comparable stoichiometry to cAMP agonists28; but, it does not promote CREB target gene expression or CBP/p300 recruitment51,52. In principle, the context-dependent induction of cellular genes could reflect the presence of other regulatory phosphorylation sites on CREB that modulate this interaction. Supporting this notion, the calcium- and calmodulin-dependent kinase II (CaMKII) has been found to block the expression of cAMP-responsive genes by phosphorylating CREB at Ser142 (REF. 53) within the KID domain and thereby disrupting its interaction with the KIX domain46. Similarly, ataxia-telangiectasia mutated (ATM)-mediated phosphorylation of CREB at Ser111 and Ser121 in response to DNA damage signals also inhibits CREB activity by blocking the CREB–CBP interaction54,55. Collectively, these studies demonstrate that accessory phosphorylation sites within the KID domain modulate the transcriptional response to signals that also promote Ser133 phosphorylation of CREB. But the biological contexts in which these accessory sites modulate CREB activity remain unclear. Future studies of mice with knock-in mutations at these sites should provide further insight into this process.
The CRTC family of CREB co-activators
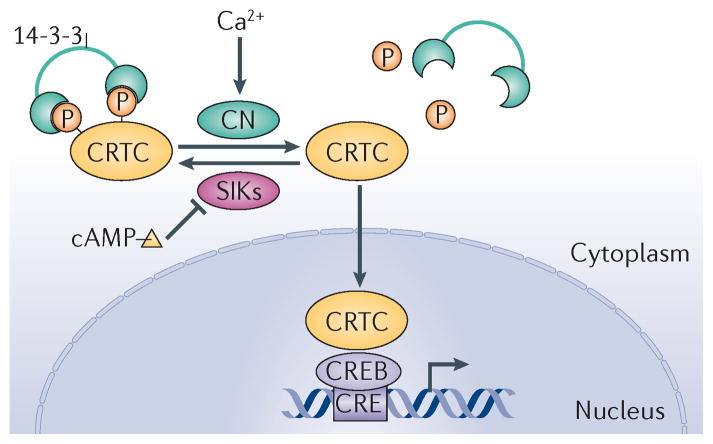
In addition to the regulatory effects of its constitutive and inducible domains, the bZIP domain in CREB also contributes to target gene activation in response to cAMP and calcium signals9. In high-throughput screens for modulators of a CRE–luciferase reporter, we and others have identified a family of CREB co-activators called CRTCs21,29,56,57, which increase CREB activity following their association with residues in the bZIP domain. The CRTC family consists of three members (CRTC1, CRTC2 and CRTC3) that have similar modular structures: they all contain an N-terminal CREB-binding domain (CBD), a central regulatory (REG) domain, a splicing domain (SD) and a C-terminal TAD (FIG. 2c). The CRTCs are also evolutionarily conserved, as functional homologues have been identified in both Drosophila melanogaster and Caenorhabditis elegans58,59. In the basal state, CRTCs are sequestered in the cytoplasm through phosphorylation-dependent interactions with 14-3-3 proteins (FIG. 3). Exposure to cAMP and calcium, but not other signals, triggers the calcineurin-mediated dephosphorylation and nuclear translocation of CRTCs, which then bind to CREB over relevant promoters. Binding of CRTCs to the bZIP domain of CREB leads to increased CREB occupancy over cognate binding sites60. Taken together, the importance of two regulatory events — the Ser133 phosphorylation of CREB and the dephosphorylation of CRTCs — for target gene activation has provided a two-hit model to explain why some signals that promote CREB phosphorylation have no effect on target gene expression.
Figure 3. crTc nuclear shuttling is regulated by phosphorylation.
Cyclic AMP and calcium signals regulate cAMP-responsive element (CRE)-binding protein (CREB) target genes by stimulating the nuclear translocation of cAMP-regulated transcriptional co-activators (CRTCs). Under basal conditions, CRTCs are phosphorylated and sequestered in the cytoplasm through interactions with 14-3-3 proteins. cAMP and calcium signals promote CRTC dephosphorylation through inhibition of the salt-inducible kinases (SIKs), which phosphorylate CRTCs, and through induction of the CRTC phosphatase calcineurin (CN), respectively. Dephosphorylated CRTC translocates to the nucleus where it binds to CREB and stimulates its activity. Image is modified, with permission, from REF. 57 © (2004) Elsevier.
Recent studies with cells deficient in both CBP and p300 point to perhaps more heterogeneous effects of the CBP/p300 and CRTC co-activators on cellular gene expression. Brindle and colleagues61 found that deletion of both CBP and p300 genes disrupted the expression of some CREB target genes but a subset of genes with multiple CREB binding sites were still induced by cAMP owing to compensatory recruitment of CRTCs to those promoters. Thus, CBP/p300 and CRTCs may provide alternative, rather than cooperative, pathways for target gene induction in certain cases. And the preferential recruitment of these co-activators to some target genes may depend on the distribution or sequence of CREB-binding sites over relevant promoters.
Whereas the CBP/p300 and CRTC co-activators seem critical for transcriptional induction in response to cAMP and calcium signals, the degree to which these co-activators modulate gene expression by increasing CREB occupancy has been controversial26,30. Although CREB appears to bind constitutively to many target genes24,25, Ginty and colleagues62 found that exposure of cells to cAMP increased CREB occupancy over binding sites on the FOS promoter. Arguing against a role for CBP/p300 in this process, cAMP promoted comparably the occupancy of both wild-type and Ser133 phosphorylation-defective CREB proteins. However, pointing to a potential role for CRTCs, another study found that CREB occupancy is reduced in CRTC2-knockout cells and that these effects are reversed when CRTC2 expression is restored60. By virtue of their ability to associate with the bZIP domain, CRTCs could increase CREB occupancy either by stabilizing the coiled-coil structure of the bZIP domain or by making accessory contacts with DNA. The consequent increase in CREB DNA binding affinity may also overcome potential inhibitory effects of other bZIP factors, which share overlapping DNA specificity with CREB. Structural studies of the CREB–CRTC complex should provide insight into this important question.
Beyond their role in transcription, CRTCs also seem to modulate the alternative splicing of certain CREB target genes via a conserved Pro-rich domain63,64. Consistent with this role, CRTC2 has been found to associate with the spliceosome factor non-POU domain-containing octamer-binding (NONO; also known as p54NRB)64. Indeed, the effects of CRTCs on splicing and transcription may be mutually exclusive, depending on promoter architecture. For example, although they have no effect on transcription from TATA-less promoters, CRTCs seem competent to promote alternative splicing of these genes. Future studies should reveal whether TATA-less CREB target genes, which are not transcriptionally upregulated by cAMP, are generally modulated by alternative splicing via CRTCs.
Collectively, these studies demonstrate that CRTCs are functional co-activators for cAMP- and calcium-responsive genes. However, whether CRTCs are dedicated CREB co-activators, remains uncertain, particularly as CRTC2 has been reported to bind other bZIP transcription factors and to enhance their transcriptional activities65–67. In the following section, we consider the role of CREB and CRTCs in modulating glucose and lipid metabolism in insulin-sensitive tissues, including the liver, pancreatic islets, adipose tissue, skeletal muscle and central nervous system (CNS). In particular, we examine the mechanisms by which hormonal and nutrient signals trigger the CREB–CRTC pathway, focusing on relevant target genes that seem to mediate cellular responses to these signals.
CREB in hepatic gluconeogenesis
During fasting, mammals maintain energy balance by shifting from glucose to fat burning. They also increase the levels of hepatic glycogenolysis and gluconeogenesis to provide fuel for glucose-dependent tissues, such as the brain and the red blood cell compartment, which lack the enzymes required to burn free fatty acids68–70. Fasting also triggers increases in skeletal muscle protein breakdown that lead to increases in circulating free amino acids, which are major precursors for hepatic gluconeogenesis. When fasting is prolonged, hepatic gluconeogenesis decreases as part of a protein-sparing process to protect against excessive muscle wasting, and liver-derived ketone bodies become the predominant fuel source for the brain. However, the mechanism underlying this shift from glucose to ketone production by the liver remains unclear.
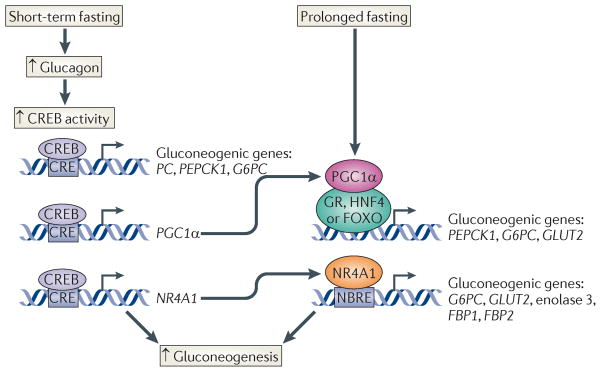
During short-term fasting, increases in circulating pancreatic glucagon stimulate the gluconeogenic programme through the activation of the cAMP pathway (FIG. 4). Activation of the cAMP-dependent Ser/Thr kinase PKA promotes CREB Ser133 phosphorylation, leading to the upregulation of gluconeogenic genes, such as phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase71. Hepatic expression of a dominant-negative CREB inhibitor called A-CREB72 causes fasting hypoglycaemia owing to impaired hepatic glucose output and to reduced gluconeogenic gene expression71. CREB seems to stimulate the gluconeogenic programme directly by binding to promoters for the PEPCK and glucose-6-phosphatase genes71,73,74. In parallel, decreases in insulin signalling also stimulate gluconeogenic gene expression through the dephosphorylation and nuclear translocation of the forkhead box (FOXO) domain proteins in the FOXO family75.
Figure 4. creB stimulates the gluconeogenic programme.
Cyclic AMP-responsive element (CRE)-binding protein (CREB) stimulates gluconeogenic gene expression through direct and feedforward mechanisms. During short-term fasting, CREB activity increases in response to glucagon and it directly stimulates the expression of the pyruvate carboxylase (PC), phosphoenolpyruvate carboxykinase 1 (PEPCK1) and glucose-6-phosphatase (G6PC) genes following its binding to CREs within their promoters. CREB activation also stimulates expression of peroxisome proliferator-activated receptor-γco-activator 1α (PGC1α) and members of the nuclear receptor subfamily 4 group A (NR4A) family (NR4A1, NR4A2 and NR4A3) of orphan nuclear receptors. PGC1α and NR4A1 further induce the expression of gluconeogenic genes as well as the glucose transporter 2 (GLUT2). PGC1α stimulates hepatic glucose production through its role as a co-activator for glucocorticoid receptor (GR), hepatocyte nuclear factor 4 (HNF4) and/or forkhead box (FOXO) transcription factor. NR4A1 stimulates hepatic fasting gene expression by binding to NGFIB-response elements (NBREs) within the promoters of the G6PC, GLUT2, enolase 3, and fructose-1,6-bisphosphatase 1 (FBP1) and FBP2 genes. Increases in PGC1α and NR4A1 expression increase gluconeogenic gene expression when fasting is prolonged.
CREB also increases the expression of the gluconeogenic programme through a feedforward mechanism involving the induction of peroxisome proliferator-activated receptor-γ(PPARγ) co-activator 1α (PGC1α), as well as members of the nuclear receptor subfamily 4 group A (NR4A) family of orphan nuclear hormone receptors71,76. The upregulation of PGC1α and NR4A genes by CREB seems to provide a mechanism to further amplify gluconeogenic gene expression in response to prolonged fasting (FIG. 4).
In addition to the effects of fasting and feeding hormones, hepatic gluconeogenesis is also regulated by the circadian clock, which coordinates hepatic metabolism with changes in the external environment77,78. The circadian clock is driven by E-box activators called circadian locomotor output cycles kaput (CLOCK) and brain and muscle ARNT-like (BMAL), which stimulate the expression of cryptochrome (CRY) and period (PER) family proteins. CRY and PER are transcriptional repressors that feedback on CLOCK and BMAL activity and generate the self-sustaining rhythm of the clock in the hypothalamus and peripheral tissues. Indeed, CRY1 and CRY2 are rhythmically expressed in liver; they accumulate during the night-to-day transition, when they inhibit fasting-induced CREB activity by blocking cAMP accumulation in response to glucagon, apparently by binding to the Gsα subunit of the heterotrimeric G protein79. Conversely, CREB activity and gluconeogenic gene expression are elevated during the day-to-night transition, when CRY protein levels are low and glucagon signalling is correspondingly elevated. Future studies should reveal the mechanism by which CRY proteins interfere with Gsα activity in hepatocytes and perhaps other cell types.
Opposing actions of glucagon and insulin
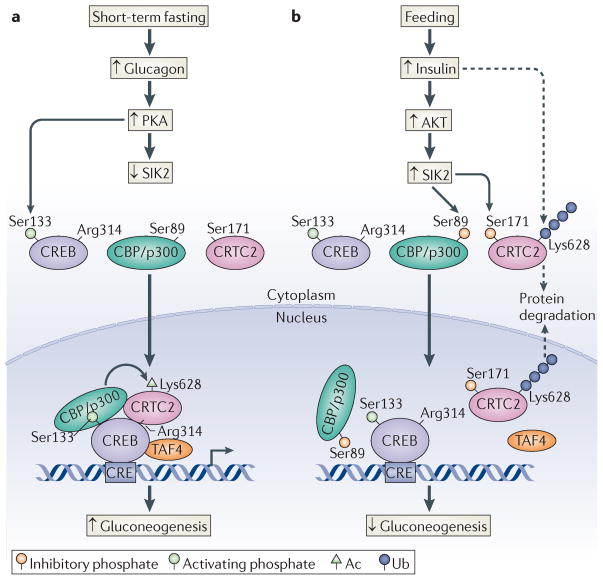
Consistent with their role in modulating CREB activity, CRTCs also seem to function importantly in the regulation of hepatic gluconeogenesis80,81. Under ad libitum feeding conditions, CRTC2, the most highly expressed member of this family in liver, is sequestered in the cytoplasm through phosphorylation-dependent interactions with 14-3-3 proteins (FIG. 5a). Hepatic CRTC2 is regulated through phosphorylation at Ser171 and Ser307 by salt-inducible kinases (SIKs) and other members of the AMP-activated protein kinase (AMPK) family of stress- and energy-sensing Ser/Thr kinases80,82. Increases in circulating glucagon during fasting promote the PKA-mediated phosphorylation and inhibition of SIK2, leading to CRTC2 dephosphorylation, nuclear translocation and recruitment to CREB-occupied genes. CRTC2 dephosphorylation seems to be sufficient for the upregulation of gluconeogenic genes, such as PEPCK and glucose-6-phosphatase, during fasting; overexpression of phosphorylation-defective (Ser171Ala) mutant CRTC2 increases gluconeogenic gene expression and hepatic glucose production. Cellular Ser/Thr phosphatases that promote hepatic CRTC2 activation in response to glucagon remain largely uncharacterized, although Koo and colleagues83 have recently found that SMEK1 and SMEK2, components of the Ser/Thr phosphatase PP4, are upregulated in livers from fasting mice, in which they may mediate CRTC2 dephosphorylation and activation.
Figure 5. Glucagon and insulin antagonism.
Opposing effects of glucagon and insulin on the cyclic AMP-responsive element (CRE)-binding protein (CREB) pathway. a | Glucagon stimulates the protein kinase A (PKA)-mediated phosphorylation of CREB at Ser133. PKA also stimulates cAMP-regulated transcriptional co-activator 2 (CRTC2) activity via the phosphorylation and inhibition of salt-inducible kinase 2 (SIK2), leading to the dephosphorylation of CRTC2. Dephosphorylated CRTC2 translocates to the nucleus, where it binds to CREB and promotes the recruitment of TBP-associated factor 4 (TAF4) and CREB-binding protein (CBP) and its paralogue p300. Nuclear CRTC2 is transiently stabilized by CBP/p300-mediated acetylation (Ac) at Lys628. b | Insulin signalling stimulates AKT, which phosphorylates and activates SIK2. Active SIK2 disrupts the CRTC2–CBP/p300 interaction by phosphorylating CBP/p300 at Ser89, leading to the deacetylation and ubiquitin (Ub)-dependent degradation of CRTC2. SIK2 also promotes the cytoplasmic translocation of CRTC2 through phosphorylation at Ser171.
In addition to on–off effects of phosphorylation and dephosphorylation, cellular hepatic CRTC2 activity is further modulated by acetylation and ubiquitylation, modifications that have opposing effects on CRTC2 stability. Following its translocation to the nucleus in response to glucagon, dephosphorylated CRTC2 undergoes acetylation primarily at Lys628 by p300 and CBP52,84; this modification stabilizes CRTC2 in hepatocytes by blocking ubiquitylation at the same site by the nuclear-localized E3 ligase constitutive photomorphogenesis 1 (COP1)85 (FIG. 5a).
During refeeding, increases in circulating insulin reduce hepatic glucose output by downregulating the gluconeogenic programme. Following its insulin-dependent activation by the Ser/Thr kinase AKT, SIK2 phosphorylates nuclear CRTC2 and promotes its cytoplasmic translocation85. In parallel, refeeding also promotes CRTC2 deacetylation by inducing the SIK2-mediated phosphorylation of CBP/p300 at Ser89 (in p300), which disrupts the CRTC2–CBP/p300 interaction (FIG. 5b). Deacetylated CRTC2 is subsequently ubiquitylated at Lys628 and targeted for proteasomal degradation. The mechanism by which phosphorylation of CBP/p300 at Ser89 regulates its activity is unclear, although this site lies within a nuclear receptor-interaction domain (RID)86,87 that contains a canonical LXXLL motif for nuclear hormone receptor binding. Indeed, Ser89 phosphorylation has been found to disrupt binding of p300 to the nuclear hormone receptor PPARγ, although it is unclear whether the LXXLL motif in CBP/p300 also mediates an association with CRTC2.
Recently, insulin has also been found to inhibit gluconeogenesis by selectively disrupting the CREB–CBP interaction. Wondisford and colleagues88,89 found that refeeding triggers the phosphorylation of CBP at Ser436 within the Cys and His-rich region 1 (CH1) domain by the atypical PKC-ι/γ (aPKCι/γ). In turn, the Ser436 phosphorylation of CBP seems to block binding of CREB that is phosphorylated at Ser133. The regulatory Ser436 site in CBP is not conserved in p300, however, suggesting that hepatic CBP and p300 perform distinct roles in the liver. Supporting this model, CREB activity and hepatic gluconeogenesis are upregulated in knock-in mice expressing Ser436Ala phosphorylation-defective CBP88,89. The mechanism by which Ser436 phosphorylation blocks the CREB–CBP association is unclear because Ser436 is located within the conserved CH1 N-terminal to the KIX domain (FIG. 2). Future studies should reveal the structural basis for the phospho-Ser436-dependent regulation of CREB–CBP complexes.
CREB activity in nutrient sensing
During prolonged fasting or heavy exercise, decreases in cellular energy levels trigger the stress- and energy-sensing Ser/Thr kinase AMPK, which inhibits hepatic gluconeogenesis in part by phosphorylating and inactivating CRTC2 (REF. 80). Indeed, persistent elevations in hepatic gluconeogenesis seem to be sufficient to deplete hepatic energy stores and to increase AMPK activity90. Following its induction, AMPK has also been shown to increase hepatic β-oxidation and ketogenesis, probably contributing to a shift from glucose to ketone body production by the liver. In keeping with its ability to stimulate AMPK activity, the oral hypoglycaemic agent metformin has been found to reduce hepatic gluconeogenesis by promoting the AMPK-mediated phosphorylation of CRTC2 (REFs 80,91). However, the glucose-lowering effects of metformin appear to be largely preserved in mice even with a combined knockout of both AMPK catalytic subunits (α1 and α2), suggesting the presence of additional regulatory targets for this small molecule92. Future studies with insulin-resistant mouse models should reveal the extent to which metformin exerts its effects via AMPKs under hyperglycaemic conditions, in which the gluco-neogenic programme is abnormally upregulated.
In keeping with the characteristic burst-attenuation dynamics of cAMP-dependent transcription, the CREB–CRTC2 pathway seems to promote gluconeogenesis only transiently, during early fasting84. When fasting is prolonged, hepatic CRTC2 activity is decreased, in part through deacetylation by the NAD+-dependent deacetylase sirtuin 1 (SIRT1) and subsequent ubiquitin-dependent degradation. SIRT was first identified in yeast as a transcriptional silencer, the activity of which is upregulated by caloric restriction, and mammalian SIRT1 and other members of the sirtuin family are now recognized as important regulators of glucose and lipid metabolism93. SIRT1 seems to be activated during fasting and exercise, in part through an AMPK-dependent mechanism, at least in skeletal muscle94. Hepatic SIRT1 activity is also upregulated during prolonged fasting, when it promotes CRTC2 deacetylation at Lys628, leading to its COP1-mediated ubiquitylation and proteasome-dependent degradation84. In parallel, SIRT1 may support residual expression of the gluconeogenic programme during late fasting by deacetylating and enhancing the activity of other factors, such as the forkhead activator FOXO1 and the nuclear hormone receptor co-activator PGC1α95,96.
CREB and hyperglycaemia: a futile cycle
Hepatic glucose output is often elevated in insulin resistance owing to increases in gluconeogenic gene expression that promote the development of type II diabetes. Chronic hyperglycaemia also contributes independently to type II diabetes risk, in part through the O-glycosylation of intracellular proteins97. Increases in circulating glucose concentrations stimulate flux through the hexosamine biosynthetic pathway, leading to the enhanced O-glycosylation of relevant substrates at Ser/Thr residues. O-glycosylation is thought to alter protein function by blocking phosphorylation at the same site. Indeed, hepatic overexpression of rate-limiting enzymes in hexosamine biosynthesis and O-glycosylation pathways, such as O-GlcNAc transferase (OGT), seems to be sufficient to promote insulin resistance97.
OGT overexpression in the liver was found to increase gluconeogenic gene expression in mice through the O-glycosylation of CRTC2 at Ser171 (REF. 98). This modification increased CRTC2 activity by blocking its phosphorylation at the same site, which prevented 14-3-3 from binding and thus led to CRTC2 nuclear translocation and CREB activation. The O-glycosylation of CRTC2 may also contribute to glucose elevations in insulin resistance, as mice genetically pre-disposed to obesity have higher levels of O-glycosylated CRTC2 relative to lean controls. Reducing hepatic amounts of O-glycosylated CRTC2, through overexpression of the deglycosylating enzyme O-GlcNAcase, is sufficient to lower circulating glucose levels in this setting. Collectively, these studies indicate that the hyperglycaemia accompanying insulin resistance may paradoxically enhance hepatic glucose production through increases in the O-glycosylation of CRTC2 and other transcriptional effectors. Future studies should reveal the extent to which the hexosamine pathway also contributes to the physiologic regulation of gluconeogenesis and other metabolic pathways in insulin-sensitive tissues.
Based on its role as a gluconeogenic regulator, CRTC2 may provide a therapeutic target for the treatment of hyperglycaemia. Indeed, acute disruption of hepatic CREB or CRTC2 activity appears sufficient to lower circulating glucose and triglyceride levels in the setting of insulin resistance81,85,99. And mice with a knockout of CRTC2 exhibit improved insulin sensitivity with lowering of hepatic triglycerides under high-fat diet conditions60. In one study, knockout of the CRTC2 gene also promoted modest fasting hypoglycaemia under lean insulin-sensitive conditions60, whereas another found no change in a similar setting100. Although this discrepancy could reflect strain differences, the targeting vector used to disrupt CRTC2 gene in the study that found no change is predicted to generate an in-frame polypeptide with comparable activity to wild-type CRTC2. Nevertheless, both studies found an important role for CRTC2 in regulating hepatic gluconeogenic gene expression in cultured hepatocytes from these mice.
CREB pathway in pancreatic islets
In addition to its effects on hepatic glucose production, the CREB pathway regulates glucose and lipid metabolism in other insulin-sensitive tissues. For example, CREB and CRTC2 mediate the transcriptional effects of glucose and incretin hormones, such as glucagon-like peptide 1 (GLP1), in insulin-producing β-cells of pancreatic islets57. Transgenic mice expressing dominant-negative A-CREB in β-cells develop hyperglycaemia with decreased islet mass owing to decreases in β-cell proliferation and increases in β-cell apoptosis101,102. CREB was found to promote β-cell survival by stimulating the expression of insulin receptor substrate 2 (IRS2) — a key component of the insulin signalling pathway that binds to the insulin receptor — as well as the anti-apoptotic survival gene B-cell lymphoma 2 (BCL-2).
CRTC2 seems to function as a coincidence detector for incretin and glucose signals, which stimulate cAMP and calcium signalling pathways, respectively57. GLP1 promotes CRTC2 dephosphorylation at Ser171, in part via the PKA-mediated phosphorylation and inhibition of SIK2, whereas extracellular glucose promotes the dephosphorylation of CRTC2 at a different regulatory site (Ser275) through increases in intracellular calcium. Under basal conditions, Ser275 is phosphorylated by microtubule affinity-regulating kinase 2 (MARK2), a member of the AMPK family of Ser/Thr kinases103. Similar to the effects of Ser171 phosphorylation, phosphorylation at Ser275 promotes 14-3-3 protein binding and cytoplasmic sequestration of CRTC2. Ser275, and perhaps Ser171 as well, is dephosphorylated by the calcium-regulated Ser/Thr phosphatase calcineurin, which binds directly to CRTC2 through conserved so-called PIXIT motifs57,104. Thus, the cooperativity between GLP1 and glucose on β-cell specific gene expression appears to reflect, in part, the dephosphorylation of two independently-regulated residues, both of which mediate 14-3-3 protein binding and cytoplasmic sequestration of CRTC2. Future studies in CRTC2 knockout mice should reveal the extent to which this co-activator is required to mediate effects of incretin hormones on β-cell viability.
Role of CREB in adipose tissue
In addition to its role in glucose homeostasis, CREB also seems to modulate lipid metabolism in adipose tissue105; adipocyte CREB phosphorylation and activity are increased in response to high-fat diet feeding and obesity. Following its induction in adipocytes, CREB was found to enhance whole-body insulin resistance by stimulating the expression of the bZIP factor ATF3, which is a transcriptional repressor that decreases the expression of adiponectin, an insulin-sensitizing adipokine hormone. ATF3 also seems to mediate fasting- and obesity-associated decreases in the expression of insulin-sensitive glucose transporter 4 (GLUT4) in adipose tissue. Disrupting CREB activity through transgenic expression of A-CREB in mature adipocytes (F-ACREB) increased adiponectin and GLUT4 expression, and protected mice from the development of insulin resistance under conditions of diet-induced or genetically induced obesity105.
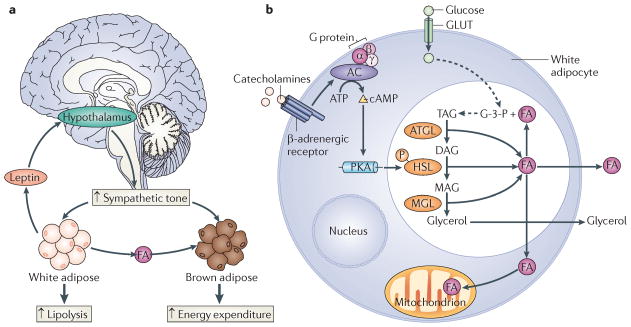
Consistent with the effects of CREB, CRTCs also seem to be important for lipid metabolism in adipose tissue. In particular, CRTC3 is highly expressed in white adipose tissue (WAT) and brown adipose tissue (BAT), in which it contributes to the development of insulin resistance106. Under lean conditions, the adipocyte-derived hormone leptin promotes energy expenditure by acting on hypothalamic centres that increase sympathetic outflow107 (FIG. 6). Increases in sympathetic nerve activity promote lipolysis in WAT and fat burning in BAT through the release of catecholamines, which in turn bind to adrenergic receptors and stimulate intracellular cAMP synthesis. Catecholamine signalling is disrupted in obesity, leading to decreases in lipid utilization that promote obesity. Mice with a knockout of CRTC3 remain lean even under high-fat diet feeding conditions; and they are insulin sensitive owing to increases in energy expenditure that protect them from the development of hepatic steatosis106. Fat burning is increased in CRTC3-mutant mice, reflecting increases in catecholamine signalling that enhance lipolysis in WAT and fatty acid oxidation in BAT. CRTC3 seems to attenuate catecholamine signalling in wild-type mice via induction of the regulator of G protein signalling 2 (RGS2) gene, which encodes a GTPase-activating protein that also binds and inhibits adenyl cyclase. In line with the increase in catecholamine signalling, brown adipocyte numbers are upregulated in CRTC3-knockout mice, contributing to their lean phenotype.
Figure 6. Leptin promotes lipolysis and energy expenditure.
a | The adipocyte-derived hormone leptin increases energy expenditure by acting on hypothalamic centres that increase sympathetic outflow. Increased sympathetic tone stimulates lipolysis and the release of fatty acids (FAs) from white adipose stores. Circulating FAs are taken up and oxidized by brown adipose tissue. b | In white adipocytes, catecholamines bind to β-adrenergic receptors and subsequently stimulate adenylate cyclase (AC) and cyclic AMP production. Increased cellular cAMP levels stimulate protein kinase A (PKA), which phosphorylates and activates hormone-sensitive lipase (HSL). During lipolysis, triacylglycerol (TAG) is hydrolysed by adipocyte triglyeride lipase (ATGL). The resultant diacylglycerol (DAG) is subsequently hydrolysed to monoacylglycerol (MAG) by HSL. MAG is further hydrolysed by MAG lipase (MGL) to generate glycerol, which enters the circulation. The FAs generated during the lipolysis of TAG may also enter the circulation. Alternatively, the FAs may undergo β-oxidation or may be re-esterified to TAG. Glycerol-3-phosphate (G-3-P) is utilized as the backbone for TAG synthesis and is generated from glucose, which is transported into the adipocyte via glucose transporters (GLUTs).
Similar to its role in mice, CRTC3 may also contribute to obesity in humans106. A common variant in the human CRTC3 gene, identified in the public database of single nucleotide polymorphisms (SNPs), was found to encode a gain-of-function protein (Ser72Asn) with increased transcriptional activity. Mexican-American individuals that are homozygous for the S72N variant seem to have increased adiposity, relative to individuals expressing only the wild-type CRTC3 gene. Future studies should reveal the relative extent to which changes in BAT or WAT function contribute to the effects of CRTC3 on obesity.
CREB function in skeletal muscle
In addition to its roles in other insulin-sensitive tissues, the CREB pathway also seems to function importantly in skeletal muscle: transgenic expression of dominant-negative A-CREB in skeletal muscle leads to a dystrophic phenotype characterized by progressive muscle wasting, muscle inflammation, and myonecrosis108. CREB was found to modulate expression of the myogenic programme by upregulating expression of the Ser/Thr kinase SIK1. In turn, SIK1 seems to regulate myogenic genes through the phosphorylation and cytoplasmic sequestration of class IIa histone deacetylases (HDACs), co-repressors that otherwise bind to myocyte-specific enhancer factor 2 (MEF2) and inhibit myogenic gene expression. The consequent inactivation of class IIa HDACs by SIK1 enhances the transcriptional activity of MEF2, leading to increases in myogenic gene expression.
Recently, CREB and CRTCs have also been found to increase mitochondrial oxidative capacity in muscle by upregulating the expression of the nuclear hormone receptor co-activator PGC1α109. Indeed, the CREB/CRTC pathway appears to be critical in promoting adaptive mitochondrial biogenesis. Depleting cellular ATP by exposure of cells to mitochondrial uncoupling agents was sufficient to upregulate CRTC–CREB activity and to promote increases in mitochondrial gene expression. Future studies should reveal whether exercise tolerance is correspondingly reduced in CRTC-deficient mice.
Metabolic role of hypothalamic CREB
Hypothalamic centres in the brain are thought to coordinate glucose and lipid metabolism in peripheral tissues after activation by hormonal and nutrient signals. Recent studies point to an important role for hypothalamic CREB and CRTC1 in this process. CRTC1 expression is almost exclusively confined to the CNS, where it mediates effects of hormonal and nutrient signals on energy balance, particularly in arcuate cells of the hypothalamus110. Mice with a knockout of the CRTC1 gene are hyperphagic, obese and leptin resistant. Although the mechanism remains unclear, leptin was found to stimulate the dephosphorylation and nuclear translocation of CRTC1 in arcuate cells. Following its induction, CRTC1 seemed to promote satiety by stimulating the CREB-dependent expression of the anorexigenic peptide cocaine- and amphetamine-regulated transcript (CART), a direct target for CREB action. In parallel, feeding-associated increases in circulating glucose also promoted the dephosphorylation of CRTC1, as well as CRTC2, in hypothalamic cells110,111, perhaps reprising the role of CRTCs as coincidence detectors for hormonal and nutrient signals in pancreatic islets. Future studies should reveal whether individual CRTC family members have overlapping or distinct effects on CREB target gene expression in the hypothalamus.
A role for CREB in longevity
Recent studies in lower organisms have revealed a broader role for CREB and CRTCs in regulating energy balance and lifespan. Disruption of the single CRTC homologue (called TORC) in D. melanogaster, for example, reduces adipose tissue stores and promotes starvation sensitivity in adult flies58. Similar to mammalian CRTC1, D. melanogaster TORC is expressed predominantly in the brain, where it also promotes energy balance and resistance to oxidative stress, in part through its role as a CREB co-activator. Re-expression of TORC in neurons is sufficient to rescue sensitivity to starvation and oxidative stress in TORC-mutant flies. Like its mammalian counterparts, D. melanogaster TORC is regulated through nucleo–cytoplasmic shuttling in response to phosphorylation by SIK. Indeed, SIK is also expressed primarily in the brain, where it modulates energy expenditure; SIK mutant flies have increased glycogen and lipid stores and they are starvation-resistant112. The effects of SIK appear to proceed via the CREB–TORC pathway because depletion of either CREB or TORC gene in neurons restores starvation sensitivity in SIK-mutant flies.
Based on their ability to regulate energy homeostasis, CRTCs might be expected to modulate lifespan. Indeed, recent work in C. elegans supports a role for CRTCs in this process59. C. elegans contains a single CRTC homologue, called CRTC-1, the activity of which is also regulated by AMPK phosphorylation and by calcineurin-mediated dephosphorylation. Increases in AMPK activity and decreases in calcineurin activity were found to promote longevity by downregulating worm CRTC-1 activity; reducing CRTC-1 expression also increased survival. Similar to its homologues in flies and mammals, worm CRTC-1 seems to exert its effects through an interaction with the worm CREB homologue CRH-1. Correspondingly, depletion of CRH-1 also increased lifespan. Future studies should reveal which CREB target genes modulate longevity in this and perhaps other organisms.
Closing Remarks
Genome-wide studies indicate that the CREB pathway potentially modulates up to one-quarter of the mammalian genome. Surprisingly, only a subset of signals that promote the phosphorylation of CREB at Ser133 also stimulate target gene expression, indicating that CREB phosphorylation over target promoters does not invariably lead to target gene activation. In contrast with the wide array of signals leading to CREB phosphorylation, the CRTC co-activators are selectively activated by cAMP and calcium stimuli, and this selectivity is likely to underlie much of the signal discrimination by the CREB pathway. By virtue of their ability to associate with the bZIP domain of CREB, CRTCs may also direct CREB occupancy to a subset of potential target genes, depending on the sequence or spacing of CREB-binding sites. Although the structural basis for these changes is unknown, the predicted increase in CREB DNA-binding affinity may help to displace other factors in the bZIP family, which share overlapping specificity and could therefore interfere with CREB signalling. Finally, CREB–CRTC recruitment in response to cAMP may be modulated depending on nucleosome spacing or post-translational modification near CREB binding sites113.
Studies in insulin-sensitive tissues reveal a broad role for the CREB pathway in mediating effects of hormones and nutrients on energy homeostasis (TABLE 1). A key feature of the CREB pathway is its transient activation in response to hormonal and nutrient cues. Indeed, a collective analysis of CREB–CRTC function in peripheral tissues suggests that chronic activation of this pathway may underlie a number of pathologic changes that are associated with insulin resistance, including hyperglycaemia, hyperinsulinaemia and adipose tissue inflammation. Future efforts to find small molecules that effectively reduce CREB activity, particularly in liver and adipose tissue, may provide therapeutic benefits to affected individuals.
Table 1.
CREB–CRTC function in metabolic tissues
| Tissue | CREB target genes | Metabolic role |
|---|---|---|
| Liver |
|
Increased gluconeogenesis |
| Adipose |
|
Increased insulin resistance and obesity |
| β-cell |
|
Increased survival |
| Hypothalamus |
|
Decreased appetite |
| Skeletal muscle |
|
Mitochondrial biogenesis; increased myogenesis |
CREB, cyclic AMP-responsive element-binding protein; CRTC, cAMP-regulated transcriptional co-activator.
Glossary
- Second messenger
An intracellular molecule that mediates effects of extracellular signals on cellular function
- TATA box
An A/T-rich sequence usually located 30 nucleotides upstream from the transcriptional start site of RNA polymerase II-dependent promoters. TATA boxes specify transcriptional initiation; they are recognized by TATA-binding protein (TBP), the DNA-binding component of transcription factor IID (TFIID)
- KIX domain
A domain of three helices in CREB-binding protein (CBP) and p300 that mediates binding to phosphorylated cyclic AMP-responsive element-binding protein (CREB) and other transcription factors
- Spliceosome
A multi-protein complex that mediates splicing of nascent transcripts
- Glycogenolysis
The breakdown of glycogen into glucose monomers. This occurs in liver and muscle tissues following stimulation with glucagon or catecholamines
- Gluconeogenesis
The hepatic production of ‘new glucose’ from glycerol, pyruvate or Ala in response to glucagon, catecholamines and cortisol
- Heterotrimeric G protein
A membrane-associated protein complex, composed of α-, β- and γ-subunits, that associates with G protein-coupled receptors and mediates induction of intracellular signalling pathways in response to ligand binding
- β-oxidation
A fatty acid metabolic pathway that occurs in mitochondria and peroxisomes. Fatty acids are metabolized to acetyl-coA and then processed through the tricarboxylic acid (TCA) cycle
- Ketogenesis
A metabolic pathway in liver tissue that generates ketones (acetoacetate and β-hydroxybutyrate) using acetyl-coA from the β-oxidation pathway. Ketones provide an important fuel source for the brain and other tissues during long term fasting
- O-glycosylation
The enzymatic addition of a glycan to ser or Thr residues in proteins. O-glycosylation is thought to compete with phosphorylation in regulating protein function
- Hexosamine biosynthetic pathway
(HBP). An offshoot of the glycolytic pathway that normally accounts for 2–5% of glucose flux. The HBP generates UDP-glucosamine, which is used for O-glycosylation of proteins. Glucose flux through the HBP is increased in diabetes, in which it is thought to contribute to insulin resistance through the O-glycosylation of key proteins in the insulin signalling pathway
- Incretin hormones
A family of gastrointestinal hormones that are released into the circulation in response to oral feeding. They promote insulin release from β-cells of the pancreatic islets
- Sympathetic outflow
Activity of the sympathetic nervous system, which receives regulatory input from the hypothalamus. Among other functions, sympathetic nerve activity regulates heart rate, blood pressure and fat burning
- Hepatic steatosis
Pathological increases in hepatic lipid content that are often associated with obesity and insulin resistance. Also referred to as ‘fatty liver’
- Arcuate cell
One of a group of neurons in the hypothalamus that mediate effects of leptin and other signals on appetite through the expression of specific neuropeptides
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Marc Montminy’s homepage: http://www.salk.edu/labs/pbl-m
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at Serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 2.Chrivia JC, et al. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. Characterizes the role of CBP as a CREB co-activator. [DOI] [PubMed] [Google Scholar]
- 3.Kwok R, et al. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 4.Arias J, et al. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–228. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 5.Dash PK, Karl KA, Colicos MA, Prywes R, Kandel ER. cAMP response element-binding protein is activated by Ca2+/calmodulin- as well as cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 1991;88:5061–5065. doi: 10.1073/pnas.88.11.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheng M, Thompson MA, Greenberg ME. CREB: A Ca-regulated transcription factor phosphorylated by calmodulin-dependent kinases. Science. 1991;252:1427–1430. doi: 10.1126/science.1646483. [DOI] [PubMed] [Google Scholar]
- 7.Deak M, Clifton A, Lucocq J, Alessi D. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 1998;17:4426–4441. doi: 10.1093/emboj/17.15.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan Y, et al. FGF and stress regulate CREB and ATF-1 via a pathway involvin p38 MAP kinase and MAPKAP kinase-2. EMBO J. 1996;15:4629–4642. [PMC free article] [PubMed] [Google Scholar]
- 9.Bonni A, Ginty D, Dudek H, Greenberg M. Serine 133-phosphorylated CREB induces transcription via a cooperative mechanism that may confer specificity to neurotrophin signals. Mol Cell Neurosci. 1995;6:168–183. doi: 10.1006/mcne.1995.1015. [DOI] [PubMed] [Google Scholar]
- 10.Xing J, Ginty DD, Greenberg ME. Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science. 1996;273:959–963. doi: 10.1126/science.273.5277.959. [DOI] [PubMed] [Google Scholar]
- 11.Iordanov M, et al. CREB is activated by UVC through a p38/HOG-1-dependent protein kinase. EMBO J. 1997;16:1009–1022. doi: 10.1093/emboj/16.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michael LF, Asahara H, Shulman A, Kraus W, Montminy M. The phosphorylation status of a cyclic AMP-responsive activator is modulated via a chromatin-dependent mechanism. Mol Cell Biol. 2000;20:1596–1603. doi: 10.1128/mcb.20.5.1596-1603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canettieri G, et al. Attenuation of a phosphorylation-dependent activator by an HDAC–PP1 complex. Nature Struct Biol. 2003;10:175–181. doi: 10.1038/nsb895. [DOI] [PubMed] [Google Scholar]
- 14.Hagiwara M, et al. Coupling of hormonal stimulation and transcription via cyclic AMP-responsive factor CREB is rate limited by nuclear entry of protein kinase A. Mol Cell Biol. 1993;13:4852–4859. doi: 10.1128/mcb.13.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagiwara M, et al. Transcriptional attenuation following cAMP induction requires PP-1-mediated dephosphorylation of CREB. Cell. 1992;70:105–113. doi: 10.1016/0092-8674(92)90537-m. [DOI] [PubMed] [Google Scholar]
- 16.Alberts AS, Arias J, Hagiwara M, Montminy MR, Feramisco JR. Recombinant cyclic AMP response element binding protein (CREB) phosphorylated on Ser-133 is transcriptionally active upon its introduction into fibroblast nuclei. J Biol Chem. 1994;269:7623–7630. [PubMed] [Google Scholar]
- 17.Wadzinski B, et al. Nuclear protein phosphatase 2A dephosphorylates protein kinase A-phosphorylated CREB and regulates CREB transcriptional stimulation. Mol Cell Biol. 1993;13:2822–2834. doi: 10.1128/mcb.13.5.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montminy MR, Sevarino KA, Wagner JA, Mandel G, Goodman RH. Identification of a cyclic-AMP responsive element within the rat somatostatin gene. Proc Natl Acad Sci USA. 1986;83:6682–6686. doi: 10.1073/pnas.83.18.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Short JM, Wynshaw-Boris A, Short HP, Hanson RW. Characterization of the phosphoenolpyruvate carboxykinase (GTP) promoter-regulatory region. II. Identification of cAMP and glucocorticoid regulatory domains. J Biol Chem. 1986;261:9721–9726. [PubMed] [Google Scholar]
- 20.Comb M, Birnberg NC, Seasholtz A, Herbert E, Goodman HM. A cyclic AMP- and phorbol ester-inducible DNA element. Nature. 1986;323:353–356. doi: 10.1038/323353a0. [DOI] [PubMed] [Google Scholar]
- 21.Iourgenko V, et al. Identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc Natl Acad Sci USA. 2003;100:12147–12152. doi: 10.1073/pnas.1932773100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hornbuckle LA, et al. Selective stimulation of G-6-Pase catalytic subunit but not G-6-P transporter gene expression by glucagon in vivo and cAMP in situ. Am J Physiol Endocrinol Metab. 2004;286:E795–E808. doi: 10.1152/ajpendo.00455.2003. [DOI] [PubMed] [Google Scholar]
- 23.Iguchi-Ariga SM, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci USA. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. Provides a genome-wide characterization of CREB occupancy and activity in different tissues. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Impey S, et al. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. Describes the genome-wide characterization of CREB target genes. [DOI] [PubMed] [Google Scholar]
- 26.Mayr B, Montminy M. Tanscriptional regulation by the phosphorylation dependent factor CREB. Nature Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 27.Conkright MD, et al. Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol Cell. 2003;11:1101–1108. doi: 10.1016/s1097-2765(03)00134-5. [DOI] [PubMed] [Google Scholar]
- 28.Brindle P, Nakajima T, Montminy M. Multiple protein kinase A-regulated events are required for transcriptional induction by cAMP. Proc Natl Acad Sci USA. 1995;92:10521–10525. doi: 10.1073/pnas.92.23.10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conkright MD, et al. TORCs: transducers of regulated CREB activity. Mol Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 30.Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 31.Quinn PG. Distinct activation domains within cAMP response element-binding protein (CREB) mediate basal and cAMP-stimulated transcription. J Biol Chem. 1993;268:16999–17009. [PubMed] [Google Scholar]
- 32.Brindle P, Linke S, Montminy M. Protein-kinase-A-dependent activator in CREB reveals a new role for the CREM family of repressers. Nature. 1993;364:821–824. doi: 10.1038/364821a0. [DOI] [PubMed] [Google Scholar]
- 33.Ferreri K, Gill G, Montminy M. The cAMP-regulated transcription factor CREB interacts with a component of the TFIID complex. Proc Natl Acad Sci USA. 1994;91:1210–1213. doi: 10.1073/pnas.91.4.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saluja D, Vassallo M, Tanese N. Distinct subdomains of human TAFII130 are required for interactions with glutamine-rich transcriptional activators. Mol Cell Biol. 1998;18:5734–5743. doi: 10.1128/mcb.18.10.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mengus G, et al. TAF4 inactivation in embryonic fibroblasts activates TGFβ signalling and autocrine growth. EMBO J. 2005;24:2753–2767. doi: 10.1038/sj.emboj.7600748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimohata T, et al. Expanded polyglutamine stretches interact with TAFII130, interfering with CREB-dependent transcription. Nature Genet. 2000;26:29–36. doi: 10.1038/79139. [DOI] [PubMed] [Google Scholar]
- 37.Lundblad JR, Kwok RP, Laurance ME, Harter ML, Goodman RH. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–8. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 38.Parker D, et al. Phosphorylation of CREB at Ser133 induces complex formation with CBP via a direct mechanism. Mol Cell Biol. 1996;16:694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 40.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 41.Ogryzko VV, Schiltz SR, Russanova V, Howard BH, Nakatani M. The transcriptional coactivators p300 and CBP are histone acetytransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 42.Asahara H, Santoso B, Du K, Cole P, Montminy M. Chromatin-dependent cooperativity between constitutive and inducible activation domains in CREB. Mol Cell Biol. 2001;21:7892–7900. doi: 10.1128/MCB.21.23.7892-7900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim T, Maniatis T. Efficient recruitment of TFIIB and CBP-RNA polymerase II holoenzyme by an interferon-β enhanceosome in vitro. Proc Natl Acad Sci USA. 1998;95:12191–12196. doi: 10.1073/pnas.95.21.12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kee B, Arias J, Montminy M. Adaptor-mediated recruitment of RNA polymerase II to a signal-dependent activator. J Biol Chem. 1996;271:2373–2375. doi: 10.1074/jbc.271.5.2373. [DOI] [PubMed] [Google Scholar]
- 45.Radhakrishnan I, et al. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. Describes the solution structure of the CREB–CBP complex and the role of CREB phosphorylation in promoting the CREB–CBP association. [DOI] [PubMed] [Google Scholar]
- 46.Parker D, et al. Analysis of an activator:coactivator complex reveals an essential role for secondary structure in transcriptional activation. Mol Cell. 1998;2:353–359. doi: 10.1016/s1097-2765(00)80279-8. [DOI] [PubMed] [Google Scholar]
- 47.Shaywitz AJ, Dove SL, Kornhauser JM, Hochschild A, Greenberg ME. Magnitude of the CREB-dependent transcriptional response is determined by the strength of the interaction between the kinase-inducible domain of CREB and the KIX domain of CREB-binding protein. Mol Cell Biol. 2000;20:9409–9422. doi: 10.1128/mcb.20.24.9409-9422.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kasper LH, et al. A transcription-factor-binding surface of coactivator p300 is required for haematopoiesis. Nature. 2002;419:738–743. doi: 10.1038/nature01062. [DOI] [PubMed] [Google Scholar]
- 49.Cardinaux JR, et al. Recruitment of CREB binding protein is sufficient for CREB-mediated gene activation. Mol Cell Biol. 2000;20:1546–1552. doi: 10.1128/mcb.20.5.1546-1552.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Best JL, et al. Identification of small-molecule antagonists that inhibit an activator:coactivator interaction. Proc Natl Acad Sci USA. 2004;101:17622–17627. doi: 10.1073/pnas.0406374101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner B, Bauer A, Schutz G, Montminy M. Stimulus-specific interaction between activator-coactivator cognates revealed with a novel complex-specific antiserum. J Biol Chem. 2000;275:8263–8266. doi: 10.1074/jbc.275.12.8263. [DOI] [PubMed] [Google Scholar]
- 52.Ravnskjaer K, et al. Cooperative interactions between CBP and TORC2 confer selectivity to CREB target gene expression. EMBO J. 2007;26:2880–2889. doi: 10.1038/sj.emboj.7601715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun P, Enslen H, Myung P, Maurer R. Differential activation of CREB by Ca2+/calmodulin-dependent protein kinase type II and type IV involves phosphorylation of a site that negatively regulates activity. Genes Dev. 1994;8:2527–2539. doi: 10.1101/gad.8.21.2527. [DOI] [PubMed] [Google Scholar]
- 54.Shi Y, et al. Direct regulation of CREB transcriptional activity by ATM in response to genotoxic stress. Proc Natl Acad Sci USA. 2004;101:5898–5903. doi: 10.1073/pnas.0307718101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shanware NP, Trinh AT, Williams LM, Tibbetts RS. Coregulated ataxia telangiectasia-mutated and casein kinase sites modulate cAMP-response element-binding protein-coactivator interactions in response to DNA damage. J Biol Chem. 2007;282:6283–6291. doi: 10.1074/jbc.M610674200. [DOI] [PubMed] [Google Scholar]
- 56.Bittinger MA, et al. Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr Biol. 2004;14:2156–2161. doi: 10.1016/j.cub.2004.11.002. Characterizes the mechanism by which calcium signals regulate the CRTC co-activators. [DOI] [PubMed] [Google Scholar]
- 57.Screaton RA, et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. Describes the role of AMPK family members and calcineurin in regulating CRTC activity. [DOI] [PubMed] [Google Scholar]
- 58.Wang B, et al. The insulin-regulated CREB coactivator TORC promotes stress resistance in Drosophila Cell. Metab. 2008;7:434–444. doi: 10.1016/j.cmet.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mair W, et al. AMPK and calcineurin induced longevity is mediated by CRTC-1 and CREB. Nature. 2011;470:404–408. doi: 10.1038/nature09706. Shows that CRTC-1 and CREB mediate effects of AMPK and calcineurin pathways on lifespan in C. elegans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y, et al. Targeted disruption of the CREB coactivator Crtc2 increases insulin sensitivity. Proc Natl Acad Sci USA. 2010;107:3087–3092. doi: 10.1073/pnas.0914897107. Demonstrates that CRTC expression in the brain regulates energy balance in D. melanogaster. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kasper LH, et al. CBP/p300 double null cells reveal effect of coactivator level and diversity on CREB transactivation. EMBO J. 2010;29:3660–3672. doi: 10.1038/emboj.2010.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Riccio A, et al. A nitric oxide signaling pathway controls CREB-mediated gene expression in neurons. Mol Cell. 2006;21:283–294. doi: 10.1016/j.molcel.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 63.Amelio AL, Caputi M, Conkright MD. Bipartite functions of the CREB co-activators selectively direct alternative splicing or transcriptional activation. EMBO J. 2009;28:2733–2747. doi: 10.1038/emboj.2009.216. Describes a novel role for the CRTC family in alternative splicing of CREB target genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Amelio AL, et al. A coactivator trap identifies NONO (p54nrb) as a component of the cAMP-signaling pathway. Proc Natl Acad Sci USA. 2007;104:20314–20319. doi: 10.1073/pnas.0707999105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee MW, et al. Regulation of hepatic gluconeogenesis by an ER-bound transcription factor, CREBH. Cell Metab. 2010;11:331–339. doi: 10.1016/j.cmet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 66.Canettieri G, et al. The coactivator CRTC1 promotes cell proliferation and transformation via AP-1. Proc Natl Acad Sci USA. 2009;106:1445–1450. doi: 10.1073/pnas.0808749106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Y, Vera L, Fischer WH, Montminy M. The CREB coactivator CRTC2 links hepatic ER stress and fasting gluconeogenesis. Nature. 2009;460:534–537. doi: 10.1038/nature08111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goodman MN, McElaney MA, Ruderman NB. Adaptation to prolonged starvation in the rat: curtailment of skeletal muscle proteolysis. Am J Physiol. 1981;241:E321–E327. doi: 10.1152/ajpendo.1981.241.4.E321. [DOI] [PubMed] [Google Scholar]
- 69.Cahill GF., Jr Fuel metabolism in starvation. Annu Rev Nutr. 2006;26:1–22. doi: 10.1146/annurev.nutr.26.061505.111258. [DOI] [PubMed] [Google Scholar]
- 70.Goodman MN, et al. Starvation in the rat. II. Effect of age and obesity on protein sparing and fuel metabolism. Am J Physiol. 1980;239:E277–E286. doi: 10.1152/ajpendo.1980.239.4.E277. [DOI] [PubMed] [Google Scholar]
- 71.Herzig S, et al. CREB regulates hepatic gluconeogenesis via the co-activator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 72.Ahn S, et al. A dominant-negative inhibitor of CREB reveals that it is a general mediator stimulus-dependent transcription of c- fos Mol Cell Biol. 1998;18:967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quinn PG, Granner DK. Cyclic AMP-dependent protein kinase regulates transcription of the phosphoenolpyruvate carboxykinase gene but not binding of nuclear factors to the cyclic AMP regulatory element. Mol Cell Biol. 1990;10:3357–3364. doi: 10.1128/mcb.10.7.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wynshaw-Boris A, Short JM, Loose DS, Hanson RW. Characterization of the phosphoenolpyruvate carboxykinase (GTP) promoter-regulatory region. I. Multiple hormone regulatory elements and the effects of enhancers. J Biol Chem. 1986;261:9714–9720. [PubMed] [Google Scholar]
- 75.Haeusler RA, Kaestner KH, Accili D. FoxOs function synergistically to promote glucose production. J Biol Chem. 2010;285:35245–35248. doi: 10.1074/jbc.C110.175851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pei L, et al. NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nature Med. 2006;12:1048–55. doi: 10.1038/nm1471. [DOI] [PubMed] [Google Scholar]
- 77.Ramsey KM, Marcheva B, Kohsaka A, Bass J. The clockwork of metabolism. Annu Rev Nutr. 2007;27:219–240. doi: 10.1146/annurev.nutr.27.061406.093546. [DOI] [PubMed] [Google Scholar]
- 78.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang E, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nature Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koo SH, et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437:1109–1111. doi: 10.1038/nature03967. Describes a role for CRTC2 and CREB in regulating hepatic gluconeogenesis. [DOI] [PubMed] [Google Scholar]
- 81.Saberi M, et al. Novel liver-specific TORC2 siRNA corrects hyperglycemia in rodent models of type 2 diabetes. Am J Physiol Endocrinol Metab. 2009;297:E1137–E1146. doi: 10.1152/ajpendo.00158.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Uebi T, Tamura M, Horike N, Hashimoto YK, Takemori H. Phosphorylation of the CREB-specific coactivator TORC2 at Ser307 regulates its intracellular localization in COS-7 cells and in the mouse liver. Am J Physiol Endocrinol Metab. 2010;299:E413–E425. doi: 10.1152/ajpendo.00525.2009. [DOI] [PubMed] [Google Scholar]
- 83.Yoon YS, et al. Suppressor of MEK null (SMEK)/protein phosphatase 4 catalytic subunit (PP4C) is a key regulator of hepatic gluconeogenesis. Proc Natl Acad Sci USA. 2010;107:17704–17709. doi: 10.1073/pnas.1012665107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu Y, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dentin R, et al. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature. 2007;449:366–369. doi: 10.1038/nature06128. [DOI] [PubMed] [Google Scholar]
- 86.Yuan LW, Gambee JE. Phosphorylation of p300 at serine 89 by protein kinase C. J Biol Chem. 2000;275:40946–40951. doi: 10.1074/jbc.M007832200. [DOI] [PubMed] [Google Scholar]
- 87.Yang W, et al. Regulation of transcription by AMP-activated protein kinase: phosphorylation of p300 blocks its interaction with nuclear receptors. J Biol Chem. 2001;276:38341–38344. doi: 10.1074/jbc.C100316200. [DOI] [PubMed] [Google Scholar]
- 88.He L, et al. Metformin and insulin suppress hepatic gluconeogenesis through phosphorylation of CREB binding protein. Cell. 2009;137:635–646. doi: 10.1016/j.cell.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou XY, et al. Insulin regulation of hepatic gluconeogenesis through phosphorylation of CREB-binding protein. Nature Med. 2004;10:633–637. doi: 10.1038/nm1050. [DOI] [PubMed] [Google Scholar]
- 90.Berglund ED, et al. Hepatic energy state is regulated by glucagon receptor signaling in mice. J Clin Invest. 2009;119:2412–2422. doi: 10.1172/JCI38650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shaw RJ, et al. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310:1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Foretz M, et al. Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. J Clin Invest. 2010;120:2355–2369. doi: 10.1172/JCI40671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Imai S, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Canto C, et al. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 2010;11:213–219. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Daitoku H, et al. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci USA. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 97.Veerababu G, et al. Overexpression of glutamine: fructose-6-phosphate amidotransferase in the liver of transgenic mice results in enhanced glycogen storage, hyperlipidemia, obesity, and impaired glucose tolerance. Diabetes. 2000;49:2070–2078. doi: 10.2337/diabetes.49.12.2070. [DOI] [PubMed] [Google Scholar]
- 98.Dentin R, Hedrick S, Xie J, Yates J, 3rd, Montminy M. Hepatic glucose sensing via the CREB coactivator CRTC2. Science. 2008;319:1402–1405. doi: 10.1126/science.1151363. [DOI] [PubMed] [Google Scholar]
- 99.Erion DM, et al. Prevention of hepatic steatosis and hepatic insulin resistance by knockdown of cAMP response element-binding protein. Cell Metab. 2009;10:499–506. doi: 10.1016/j.cmet.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Le Lay J, et al. CRTC2 (TORC2) contributes to the transcriptional response to fasting in the liver but is not required for the maintenance of glucose homeostasis. Cell Metab. 2009;10:55–62. doi: 10.1016/j.cmet.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Inada A, et al. Overexpression of inducible cyclic AMP early repressor inhibits transactivation of genes and cell proliferation in pancreatic β cells. Mol Cell Biol. 2004;24:2831–2841. doi: 10.1128/MCB.24.7.2831-2841.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jhala US, et al. cAMP promotes pancreatic β-cell survival via CREB-mediated induction of IRS2. Genes Dev. 2003;17:1575–1580. doi: 10.1101/gad.1097103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jansson D, et al. Glucose controls CREB activity in islet cells via regulated phosphorylation of TORC2. Proc Natl Acad Sci USA. 2008;105:10161–10166. doi: 10.1073/pnas.0800796105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aramburu J, et al. Selective inhibition of NFAT activation by a peptide spanning the calcineurin targeting site of NFAT. Mol Cell. 1998;1:627–637. doi: 10.1016/s1097-2765(00)80063-5. [DOI] [PubMed] [Google Scholar]
- 105.Qi L, et al. Adipocyte CREB promotes insulin resistance in obesity. Cell Metab. 2009;9:277–286. doi: 10.1016/j.cmet.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Song Y, et al. CRTC3 links catecholamine signaling to energy balance. Nature. 2010;468:933–939. doi: 10.1038/nature09564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Landsberg L. Feast or famine: the sympathetic nervous system response to nutrient intake. Cell Mol Neurobiol. 2006;26:497–508. doi: 10.1007/s10571-006-9010-7. [DOI] [PubMed] [Google Scholar]
- 108.Berdeaux R, et al. SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nature Med. 2007;13:597–603. doi: 10.1038/nm1573. [DOI] [PubMed] [Google Scholar]
- 109.Wu Z, et al. Transducer of regulated CREB-binding proteins (TORCs) induce PGC-1α transcription and mitochondrial biogenesis in muscle cells. Proc Natl Acad Sci USA. 2006;103:14379–14384. doi: 10.1073/pnas.0606714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Altarejos JY, et al. The Creb1 coactivator Crtc1 is required for energy balance and fertility. Nature Med. 2008;14:1112–1117. doi: 10.1038/nm.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lerner RG, Depatie C, Rutter GA, Screaton RA, Balthasar N. A role for the CREB co-activator CRTC2 in the hypothalamic mechanisms linking glucose sensing with gene regulation. EMBO Rep. 2009;10:1175–1181. doi: 10.1038/embor.2009.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Choi S, Kim W, Chung J. Drosophila salt-inducible kinase (SIK) regulates starvation resistance through CREB-regulated transcription coactivator (CRTC) J Biol Chem. 2010 doi: 10.1074/jbc.C110.119222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ramirez-Carrozzi VR, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schwartz MW, Porte D., Jr Diabetes, obesity, and the brain. Science. 2005;307:375–379. doi: 10.1126/science.1104344. [DOI] [PubMed] [Google Scholar]