SUMMARY
In HIV-1, the ability to mount antibody responses to conserved, neutralizing epitopes is critical for protection. Here we have studied the light chain usage of human and rhesus macaque antibodies targeted to a dominant region of the HIV-1 envelope second variable (V2) region involving lysine (K)169, the site of immune pressure in the RV144 vaccine efficacy trial. We found that humans and rhesus macaques used orthologous lambda variable gene segments encoding a glutamic acid-aspartic acid (ED) motif for K169 recognition. Structure determination of an unmutated ancestor antibody demonstrated that the V2 binding site was pre-configured for ED motif-mediated recognition prior to maturation. Thus, light chain usage for recognition of the site of immune pressure in the RV144 trial is highly conserved across species. These data indicate the HIV-1 K169-recognizing ED motif has persisted over the diversification between rhesus macaques and humans, suggesting an evolutionary advantage of this antibody recognition mode.
INTRODUCTION
The RV144 vaccine trial showed an estimated vaccine efficacy of 31% (Rerks-Ngarm et al., 2009), and a molecular sieve analysis of breakthrough infections demonstrated 48% vaccine efficacy when the second variable region (V2) of the infecting virus envelope (Env) matched the vaccine Env at lysine (K) at position 169 (Rolland et al., 2012). Isolation of V2 monoclonal antibodies (mAbs) from vaccinees demonstrated that in 4 V2 antibodies that recognized K169, all light chain second complementarity determining regions (LCDR2) contained a glutamic acid- aspartic acid (ED) motif, and crystal structures of two K169-reactive human mAbs, CH58 and CH59, demonstrated salt bridges formed with K169 by the E of CH58 and the D of CH59 (Liao et al., 2013). Of the four V2 antibodies initially isolated from RV144 vaccinees, mAb CH58 utilized lambda light chain V gene segment 6-57 (IGLV6-57) and mAb CH59 utilized IGLV3-10 both of which have a germline-encoded ED motif (Lefranc, 2001). Two additional V2 K169 antibodies, HG107 and HG120, independently isolated from RV144 vaccinees also expressed IGLV3-10 light chains and retained the LCDR2 ED motif (Liao et al., 2013). These observations raised the hypothesis that Vλ gene segments carrying the LCDR2 ED motif were required to recognize the HIV-1 V2 K169 epitope and exert the selective pressure apparent in the RV144 vaccine trial (Liao et al., 2013).
Rhesus macaques have been useful models of retrovirus infection pathogenesis as well as for evaluation of protective capacity of antibodies against simian immunodeficiency virus (SIV) and simian-human immunodeficiency virus (SHIV) infections. Study of the rhesus macaque genome has demonstrated that humans and rhesus macaques share ~93% of genome homology (Gibbs et al., 2007), with a common ancestor estimated at ~32 million years ago (Perelman et al., 2011). Thus, study of the rhesus antibody repertoire as a model for how humans will respond to pathogens and vaccines may be a useful approach for the prediction of human antibody responses.
One route to improvement on the results of the RV144 vaccine trial is to determine if any VL genes other than IGLV6-57 and IGLV3-10 can be used to target the K169 V2 region. To address this question, we immunized rhesus macaques with the RV144 vaccine Env, AE.A244 gp120, and isolated 27 antibodies that are dependent on V2 K169 for binding to HIV-1 Env gp120. We found that all 27 antibodies utilized the rhesus ortholog of human IGLV3-10, i.e. the rhesus IGLV3-17 gene segment that contains a germline-encoded LCDR2 ED motif. Two additional antibodies were isolated from rhesus immunizations with clade C Envs that recognized V2 K169 that were not IGLV3-17, but rather had ED motifs derived from LCDR2 somatic mutations. Thus, the only observed mechanism of recognition of K169 in primates is via antibodies with lambda light chains with a LCDR2 ED motif. That this germline-encoded rhesus IGLV3-17 and human IGLV3-10 light chain mode of recognition of V2 K169 has persisted through the divergence of humans and rhesus macaques suggests the Vλ LCDR2 ED motif confers a strong fitness advantage.
RESULTS
Restricted Utilization of Rhesus Macaque IGLV3-17 in Recognition of HIV-1 Env V2 K169
We immunized rhesus macaques with the RV144 vaccine trial Env AE.A244 gp120 in two regimens, one with the same immunogens used in the RV144 trial (Rerks-Ngarm et al., 2009) (NHP study 36; Fig S1A), and the other using a heterologous prime-boost strategy with RV144 AE.A244 gp120 Env glycoprotein immunogen as a prime, followed sequentially by AE.427299 gp120, B.9021 gp140, AE.A244 gp120, and then a final boost of AE.A244 and AE.427299 gp120s (NHP study 62.1; Fig S1B). Env proteins 92TH023 gp120 and CM244 gp120 in NHP study 36 and AE.A244 gp120 in NHP study 62.1 included K169 in their V2 loops while Env proteins MN.3 gp120 in NHP study 36 and AE.427299 gp120 and B.9021 gp140 in NHP study 62.1 did not (Gnanakaran et al., 2010; Liao et al., 2013).
We isolated antibodies from immunized monkeys utilizing antigen-specific memory B cell sorts using fluorophore-labeled AE.A244 gp120 (Bonsignori et al., 2012) or similarly labeled AE.A244 V1V2 tags protein (Liao et al., 2013) (Fig 1). A total of 39 antibodies were isolated that bound to the V2 region of Env (165LRDKKQKVHALFYKLDIVPIED186) in ELISA assays from NHP studies 36 and 62.1 (Tables S1 and S2). Of the 39 V2-targeted antibodies, 27 antibodies bound at K169 (the site of immune pressure in the RV144 vaccine efficacy trial) as established by alanine scanning binding ELISA assays (Table S1). All 27 V2 K169-dependent antibodies utilized IGLV3-17 genes with an ED motif in the LCDR2 (Table S2).
Figure 1. Antigen specific memory B-cell cell sorts on peripheral mononuclear blood cells (PMBCs) from three non-human primate (NHP) studies.
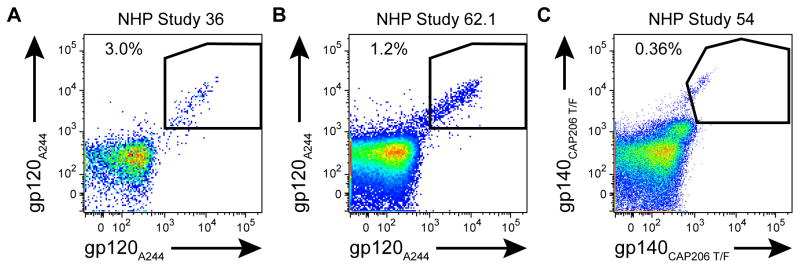
Live CD14−CD16−CD20+CD3−IgD−CD27+ cells were stained with antigens conjugated to two different fluorophores. Double positive antigen-specific cells were single-cell sorted into 96-well PCR plates for RT-PCR. AE.A244 gp120 specific memory B-cells were (A) 3.0% of NHP study #36 and (B) 1.2% of NHP study #62. (C) NHP study #53 was sorted with CAP206 T/F gp140 and 0.36% of cells were specific for this HIV-1 envelope. Data are representative results of at least 3 independent experiments.
Next, we selected a set of 18 V2-reactive antibodies with an ED motif for production in bulk and further characterization. Extensive epitope mapping by alanine scanning mutagenesis with a 19 residue AE.A244 V2 peptide (165LRDKKQKVHALFYKLDIVPIED186) confirmed that all 18 ED motif-containing antibodies demonstrated K169-dependent binding (Table 1, Fig S2). All antibodies also had H173 as an epitope component and additionally had combinations of K171, F176, Y177 and D180 amino acids in their epitopes. The epitopes recognized by the rhesus antibodies were most similar to the epitope utilized by the previously described human mAb, CH59, derived from an RV144 vaccinee (Liao et al., 2013).
Table 1.
V2 Epitopes of Selected Antibodies
| Antibody | Species | VH | VL | V2 Epitope |
|---|---|---|---|---|
| 1678 | Rhesus | 3-SC11 | λ3-17 | K169, H173, F176, Y177 |
| 1697 | Rhesus | 3-SC11 | λ3-17 | K169, H173, F176, Y177 |
| 1811 | Rhesus | 3-SC11 | λ3-17 | K169, H173, D180 |
| 1410 | Rhesus | 4-48 | λ3-17 | K169, H173, D180 |
| 1825 | Rhesus | 4-48 | λ3-17 | K169, H173 |
| 2552 | Rhesus | 4-48 | λ3-17 | K169, H173 |
| 975 | Rhesus | 4-79 | λ3-17 | K169, K171, H173, Y177, D180 |
| 1447 | Rhesus | 4-79 | λ3-17 | K169, H173, Y177, D180 |
| 1601 | Rhesus | 4-79 | λ3-17 | K169, H173, D180 |
| 1671 | Rhesus | 4-79 | λ3-17 | K169, H173 |
| 1819 | Rhesus | 4-79 | λ3-17 | K169, H173, Y177 |
| 1823 | Rhesus | 4-79 | λ3-17 | K169, H173, Y177, D180 |
| 1824 | Rhesus | 4-79 | λ3-17 | K169, H173, Y177, D180 |
| 2553 | Rhesus | 4-79 | λ3-17 | K169, H173, D180 |
| 2536 | Rhesus | 4-96 | λ3-17 | K169, H173, D180 |
| 2547 | Rhesus | 4-96 | λ3-17 | K169, H173, F176 |
| 2653 | Rhesus | 4-96 | λ3-17 | K169, H173, D180 |
| 2548 | Rhesus | 4-SC6 | λ3-17 | K169, H173, D180 |
| 1056a | Rhesus | 3-SC11 | λ3-SC4 | K169, K171, H173, F176, Y177, D180 |
| 1534a | Rhesus | 3-SC11 | λ3-SC4 | K169, K171, H173, F176, Y177, D180 |
| CH58b | Human | 5-51 | λ6-57 | K168, K169, K171, H173, F176, Y177, K178, D180, P183 |
| CH59b | Human | 3-9 | λ3-10 | K169, H173, F176, Y177, D180 |
| HG107b | Human | 3-9 | λ3-10 | K169, K171, H173, F176, Y177, D180 |
| HG120b | Human | 3-23 | λ3-10 | K169, H173, F176, Y177, D180 |
V2 epitope residues were defined as residues where EC50 relative to WT for alanine mutations was reduced by >50% in ELISA assay. Epitope positions determined by ELISA were also confirmed by SPR for representative set of mutations (same as listed below). K169, the site of immune pressure in the RV144 trial, is shown in bold.
Antibody footprints mapped by SPR only for mutations K168A, K169A, K171A, H173A, F176A, Y177A, and D180A
Antibody footprints previously described (Liao Immunity 2013)
We then characterized the neutralization breadth of the 18 K169-recognizing, ED motif antibodies using both TZM-bl and A3R5 HIV-1 neutralization assays (McLinden et al., 2013; Sarzotti-Kelsoe et al., 2013). All 18 antibodies neutralized the easy-to-neutralize tier 1 virus AE.92TH023.6 in the TZM-bl assay (Table S3), but failed to neutralize the tier 1 HIV-1 isolate MN.3 (that does not have a K at position 169) and difficult-to-neutralize tier 2 HIV-1 isolate AE.CM244 in TZM-bl. All but one antibody failed to neutralize any HIV-1 isolates tested in the A3R5.7 cell assay (Table S3).
We next confirmed the ED motif as critical to V2 recognition by making single (D51A) and double (E50A+D51A) alanine mutations by site-directed mutagenesis to antibody 1518, a separate rhesus antibody from NHP 36 that was not included in the characterization above. Binding to AE.A244 gp120, V1V2 tags, and V2 peptide was reduced for the single alanine mutant and greatly reduced for the double alanine mutant (Table S1B). Additionally, neutralization of AE.92TH023 was lost for all three mutants (Table S3). Taken together these data demonstrate that the ED motif was essential for recognition of the V2 loop and mediation of neutralization.
The predominance of ED motif-containing antibodies in the V2 response suggested that the V2 is strongly antigenic for naïve B-cell receptors utilizing IGLV3-17. We computationally inferred the initial V(D)J rearrangements of three representative IGLV3-17, ED motif-containing antibody clonal lineages (Kepler, 2013) and recombinantly produced them for characterization. All three unmutated common ancestor (UCA) antibodies demonstrated binding to AE.A244 V1V2 tags protein with binding of two of the three UCAs reduced relative to the corresponding mutated antibodies (Table S1C). No neutralization was observed for the three UCAs (Table S3) demonstrating that the neutralization capacity of IGLV3-17 V2 antibodies is not attained until additional mutations are acquired from affinity maturation.
Acquisition of LCDR2 ED Motif Via Affinity Maturation
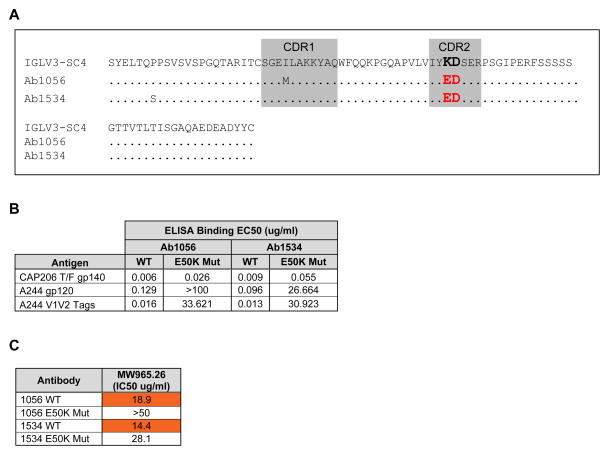
We next asked if we could observe examples of V2 K169 antibodies induced without the utilization of the CDRL2 ED motif encoding IGL3-17 gene from regimens from other NHP studies. For this analysis we isolated antibodies from a rhesus macaque that was immunized with three clade C (CAP206) Envs that each had a lysine at position 169 in V2 (NHP Study 54, Fig S1C). We isolated two antibodies (1056 and 1534) that bound AE.A244 V2 peptide that were dependent for their binding on K169 (Table 1 and Fig S2) and had an ED motif in LCDR2. However, these two antibodies did not use IGLV3-17, but rather used the IGLV3-SC4 gene segment that encodes a KD amino acid pair in the germline LCDR2 (Fig 2A). This K50E mutation in the 1056 and 1534 antibodies was likely the result of VJ hypermutation in germinal centers.
Figure 2. CDRL2 ED motif can be acquired by affinity maturation.
(A) Amino acid sequence alignment of VH region of antibodies 1056 and 1534 to their inferred germline VL gene segment. IGLV3-SC4 shows K50E somatic hypermutation confers the ED motif (red). Dots represent amino acid matches to germline. The ED motif is bolded for emphasis. (B) ELISA binding results for clonally-related WT antibodies 1056 and 1534 and reverted E50K mutants tested against CAP206 (autologous) and A244 antigens. (C) Neutralization of Clade C tier 1 MW965.26 in TZM-bl cell assay (orange denotes neutralization at <50 ug/ml). Data are from one experiment (B and C) with neutralization values (C) representing the average of duplicate measurements.
We next asked whether the K50E mutation resulted in improved Env binding. We used site-directed mutagenesis to revert E50 in the LCDR2s of antibodies 1056 and 1534 back to K. Indeed we found that binding to the CAP206 transmitted-founder gp140 improved 4–6 fold for the ED-bearing 1056 and 1534 antibodies compared to 1056 and 1534 E50K reversion mutants (Fig 2B). While wild-type E50 antibodies 1056 and 1534 bound well to AE.A244 gp120 and the AE.A244 V1V2 tag subunit protein, binding was lost or greatly diminished to these antigens in the E50K reverted antibodies (Fig 2B). Additionally, neutralization of the tier 1 HIV-1 strain C.MW965.26 was substantially reduced for 1534 E50K and lost for 1056 E50K (Fig 2C), demonstrating that acquisition of the ED motif resulted in the gain of heterologous tier 1 HIV-1 neutralization capacity.
Limited availability of LCDR2 ED motif in the Antibody Repertoires of Humans and Rhesus Macaques
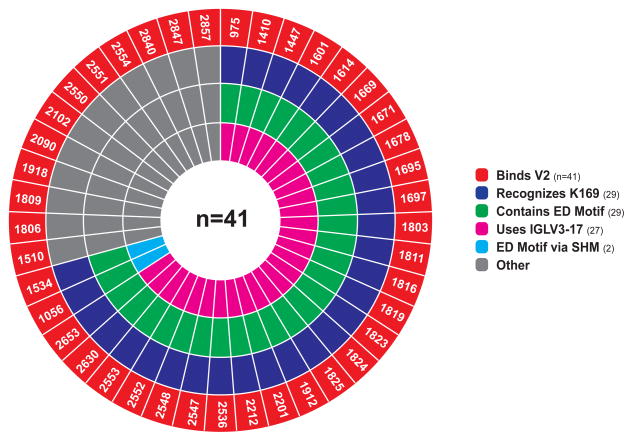
Thus, 71% (29/41) of the total number of V2 antibodies and 58% (15/26) of the V2-reactive clonal lineages isolated from 3 rhesus macaque Env immunization studies bound at K169, and all 29 K169-recognizing antibodies included the CDRL2 ED motif either by using IGLV3-17 in which ED is germline encoded (27 MAbs), or by acquiring ED through somatic mutations (2 MAbs) (Fig. 3). Given the remarkable usage of the ED motif for K169 recognition in both the human and rhesus immune response to HIV-1 Env immunization, we next asked how frequently the LCDR2 ED motif is encoded in the human and rhesus macaque light chain repertoires.
Figure 3. All K169-dependent V2 antibodies contain the CDRL2 ED motif.
Summary of characteristics of the rhesus antibodies. Antibody names are labeled in the segments of the outer ring for antibodies with V2-binding reactivity (red). Segments preceding inward towards center indicate additional characteristics of the antibody. The first inner ring specifies K169-dependent V2 recognition (blue) and second inner ring specifies presence of LCDR2 ED motif (green). The innermost ring specifies IGLV3-17 germline usage (magenta) or ED motif gained by somatic hypermutation (SHM) (cyan). Gray segments denote antibodies without the specified characteristic at that ring level. Data are representative of two independent experiments for purified antibodies and one experiment for antibodies from initial primary screening (see Table S1 for breakdown).
In humans, the LCDR2 ED motif is encoded in 3 Vλ gene segments: IGLV3-10, IGLV6-57 and IGLV3-22. IGLV3-22 has two alleles. IGLV3-22*02 is a pseudogene with a frame-shift mutation producing a premature stop codon and IGLV3-22*01 is potentially functional. (Frippiat and Lefranc, 1994; Lefranc, 2001). IGLV3-22 has not, however, been observed in lambda rearrangements in Genbank antibody sequences, suggesting the dominant allele is IGLV3-22*02 that effectively restricts germline encoding of the ED motif in the human repertoire to two light chain Vλ gene segments, IGLV6-57 and IGLV3-10. These two gene segments were precisely those utilized by all (n=4) of the human 4 V2 K169 antibodies isolated to date (Liao et al., 2013).
The known rhesus macaque light chain repertoire includes only two light chain V gene segments that encode for the LCDR2 ED motif in the germline, IGLV3-17 and IGLV3-30. Rhesus IGLV3-17 is an ortholog of human IGLV3-10 and the gene segments share 97% sequence similarity (Fig 4, Table S4). IGLV3-30 is an ortholog of human IGLV3-22. As with human IGLV3-22, no IGLV3-30 antibodies have been observed in Genbank. However, the incomplete characterization of rhesus Ig gene segments prevents us from establishing whether non-functional alleles for this gene segment exist. The rhesus ortholog of human IGLV6-57 is IGLV6-92, but IGLV6-92 encodes for a KD in its LCDR2. It is important to note that characterization of the rhesus Ig gene segment repertoire is incomplete, and it is possible that more gene segments or alleles that include an ED motif in the LCDR2 may be defined in the future.
Figure 4. Rhesus orthologs of human VLs encoding for CDRL2 ED motif in the germline.
Amino acid sequence alignments of the rhesus VL orthologs to human IGLV3-10 and IGLV6-57. Dots represent amino acid residue matches. The ED motif is bolded and colored red for emphasis.
Conservation of the IGLV3-10 V Gene Segment in Primate Phylogeny
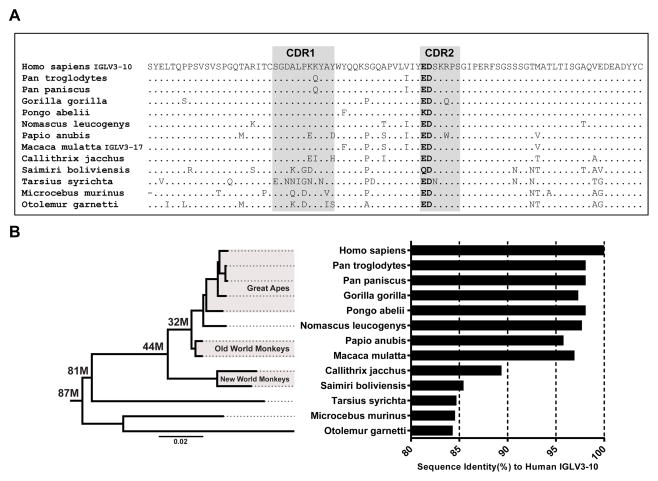
Since we have observed antibodies containing the LCDR2 ED motif both in human and rhesus antibody responses to HIV-1 V2 K169, and found that the ED motif is encoded in the germline Vλ segments of both species, we next asked if the ED motif is present in the Vλ gene segments of other primates. We searched for all human IGLV3-10 orthologs within 12 primate genomes. The genomes queried consisted of a diverse set of primates including representatives from the groups of great apes, old world monkeys, new world monkeys, tarsiers, and lemurs. All 12 primate genomes contained IGLV3-10 orthologs and in all but two (Pongo abelii and Saimiri boliviensis) the LCDR2 ED motif was present within their respective IGLV3-10 ortholog (Figure 5A). Within great apes, bonobos, and old world monkeys, the orthologs are highly similar to human IGLV3-10 with sequence identities greater than 95% (Figure 5B). Sequence identities of orthologs in the more distant extant primate groups of new world monkeys, tarsiers and lemurs are between 84 to 89%. Rhesus macaque and humans shared a common ancestor an estimated 32 million years ago (Perelman et al., 2011), demonstrating that the IGLV3-10 gene segment has not been altered substantially and has maintained the ED motif over that time period (Fig 5B). Given the presence of the ED motif in the mouse lemur (Microcebus murinus) light chain repertoire, the conservation of the ED motif in the primate lineage can be traced as far back as an estimated 87 million years ago (Perelman et al., 2011). The ED motifs in the IGLV3-10 primate orthologs are encoded by several different combinations of codon pairs (Fig. S3) with heterogeneity in codon usage observed in the more distant primate species from human, providing further evidence of a strong selective pressure for maintenance of the ED motif.
Figure 5. Conservation of IGLV3-10 and ED motif in primate phylogeny.
(A) Amino acid sequence alignment of human IGLV3-10 to orthologs from 12 primates with sequenced genomes. Dots represent amino acid residue matches and residues in the LCDR2 ED motif positions are bolded for emphasis. (B) Sequence identities of primate IGLV3-10 orthologs to human IGLV3-10 (right) juxtaposed to phylogenetic tree including respective primate species (left). Data on estimated time (in millions of years ago) to most common recent ancestor (shown in bold on tree nodes for relevant estimates) as well as for phylogenetic tree were taken from (Perelman et al., 2011). Branch length estimates units are substitution/site.
Structural Insights into Germline IGLV3-10 Usage and K169 Recognition
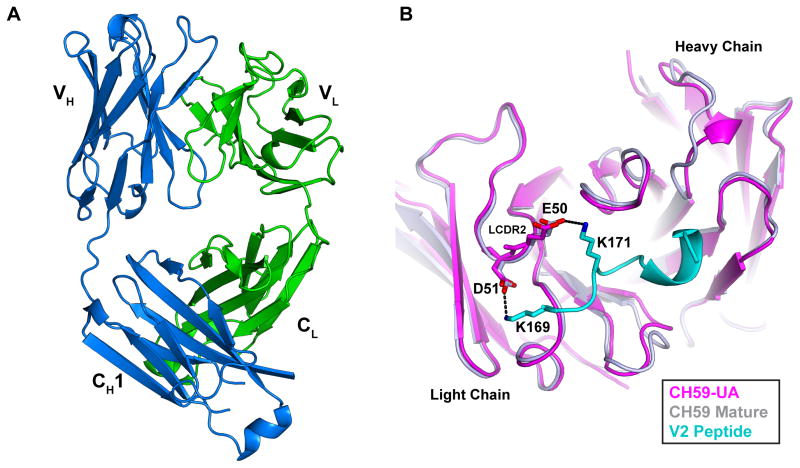
In order to gain structural insights into IGLV3-10 usage and V2 K169 recognition, we determined the crystal structure of the CH59 unmutated ancestor (CH59-UA) antibody that utilizes the germline IGLV3-10 segment (Fig 6A, Table S5). CH59-UA binds with micromolar affinity to the AE.A244 V2 peptide (Kd=0.5 μM), a value two orders of magnitude weaker than the affinity of the mutated CH59 antibody (Kd=3.1 nM). (Fig S4). Superposition of the CH59-UA structure onto the structure of mature CH59 in complex with a V2 peptide (Liao et al., 2013) revealed that the CH59 V2 K169 binding site is remarkably similar to the corresponding paratope of CH59-UA (Fig 6B). The average backbone root mean square deviation (RMSD) of the 6 CDRs was 1.26Å with the light chain CDRs aligning more closely (0.88Å RMSD) than the heavy chain CDRs (1.52Å RMSD). Particularly striking was the structural similarity between the respective ED motif conformations in the unliganded CH59-UA and the bound mature CH59. The CH59-UA E50 and D51 side-chains are positioned in nearly identical orientations as the bound conformations of the corresponding side-chains in the mature CH59, demonstrating in an unliganded and germline IGLV3-10 antibody the ED motif is in precisely the orientation used for recognizing K169 V2. Taken together, these data show that the CH59 binding site is largely pre-configured in the unliganded CH59-UA. Pre-configuration of an antibody binding site can mitigate the entropic penalty paid upon association (Schmidt et al., 2013) and consistent with this prediction, the CH59-UA has a very fast on-rate (Fig S4). The on-rate of the CH59-UA was observed to be similar to that of the mature CH59 on-rate; consequently affinity maturation in the CH59 lineage is driven by off-rate improvements. Given rhesus IGLV3-17 has 97% sequence identity to human IGLV3-10, we expect that the germline structure of rhesus IGLV3-17 is likely identical to the structure of human IGLV3-10. Consequently, the light chain recognition mode of rhesus IGLV3-17 antibodies to the HIV-1 V2 K169 epitope must closely resemble light chain recognition in the CH59 antibody lineage.
Figure 6. CH59-UA structure reveals pre-configured K169 recognition mode in IGLV3-10 germline.
(A) Crystal structure of the CH59-UA Fab with heavy chain colored blue and light chain colored green. See Table S5 for summary of structural data. (B) Superposition of crystal structures of CH59-UA (magenta) and mature CH59 (gray) in complex with a V2 peptide (cyan). The ED motif in CDRL2 of both antibodies and K169 and K171 of V2 peptide are shown in stick representation. Dashed lines indicate salt-bridges in mature CH59-V2 complex.
All rhesus macaque V2 antibodies isolated that contain the ED motif have LCDR3 lengths of 11 residues (Table S2), the same length as the LCDR3 of the human RV144 trial vaccine antibody, CH59. We next asked how compatible the ED antibody sequences were with the CH59 structure in complex with V2 (Liao et al., 2013). We threaded the sequence of a representative ED motif-containing rhesus antibody, Ab975, onto the CH59-V2 peptide complex structure, and the structural model of Ab975 demonstrated agreement with the CH59 mode of V2 recognition (Fig S4E). The ED motif of Ab975 is predicted to adopt a similar orientation as CH59 suggesting the ED motif of Ab975 could also form salt-bridges with K169 and K171 which is consistent with the epitope mapping of Ab975 (Table 1). The binding pocket opposite the C-terminal half of the V2 peptide in the structural model of Ab975 is shallower than CH59’s binding pocket, most likely reflecting differences in the heavy chain usage between Ab975 and CH59.
Analysis of Rhesus Macaque VH Pairing with IGLV3-17 ED Antibodies
All IGLV3-17 ED motif containing antibodies shared the amino acid H173 in their V2 epitopes (Table 1). In the crystal structure of the complex of CH59 with a V2 peptide, the heavy chain contacted residues C-terminal of K169 with H173 forming hydrogen bond contacts with Asp 95 (D95) in the heavy chain (Liao et al., 2013). While VH pairing with IGLV3-17 does not appear to be overly restricted with 4 distinct VH gene segments used among 15 IGLV3-17 clones (Table S2), all IGLV3-17 antibodies isolated contained D95 like CH59 (Fig S5) suggesting D95 may play a functional role in their V2 recognition. Due to the incomplete characterization at the 3′ end of VH gene segments in the rhesus repertoire we cannot be certain about the frequency in which D95 is encoded in heavy chains and whether this structural feature represents an additional restriction for recognition of the V2 epitope.
It is notable, however, that we have not observed the CH59 VH3-9 rhesus ortholog, IGHV3-78 (Table S4B), paired with the IGLV3-17 of the rhesus ED motif antibodies. We hypothesized that the lack of rhesus heavy and light chain pairs that were orthologous to the CH59 pairing could be due to incompatibly of the IGHV3-78 sequence in the CH59 V2 recognition mode. A structural model of IGHV3-78/IGLV3-17 in the CH59 recognition mode resulted in severe clashing between the Y59 residue in the HCDR2 and the V2 peptide (Figure S4F). In CH59, the VH3-9 germline gene does not encode a large amino acid such as tyrosine but instead the smallest amino acid residue, glycine, which allows for favorable paratope shape complementarity with the V2 peptide. Thus the steric hindrance of IGHV3-78 with the V2 peptide when structurally modeled into the CH59 recognition mode suggests a potential explanation for why we do not observe IGHV3-78/IGLV3-17 pairing in the rhesus V2 antibody response.
DISCUSSION
In this study, we demonstrate that the dominant antibody response in recognition of the V2 epitope surrounding lysine at amino acid position 169 in rhesus macaques utilizes the IGLV3-17 gene segment with a LCDR2 ED amino acid pair. We show that the VL ED motif can arise either by being encoded in the IGLV3-17 germline gene or arise in the course of somatic mutation from a germline-encoded KD LCDR2 motif. Remarkably, the rhesus IGLV3-17 gene is the ortholog of the human IGLV3-10 gene, which is utilized for recognition of the V2 K169 site in humans (Liao et al., 2013).
The rhesus IGLV3-17 ED motif antibodies had very similar neutralization profiles as human ED motif Abs CH58 and CH59, and rhesus ED motif Ab V2 footprints were most consistent with the V2 epitope of human V2 mAb CH59. The epitope footprints of human RV144 vaccinee V2 antibodies HG107 and HG120 were also similar to CH59, and all three antibodies used the IGLV3-10 gene segment, demonstrating that human IGLV3-10 and rhesus IGLV3-17 ortholog usage confers a highly consistent V2 recognition mode in both human and rhesus anti-V2 antibodies.
Of the four distinct VH gene segments observed to pair with IGLV3-17, two pairings, IGHV4-48:IGLV3-17 and IGHV4-79:IGLV3-17, occurred independently in two monkeys given two different immunization regimens. In addition, one VH:VL pair, IGHV3-SC11:IGLV3-17, was the orthologous pairing of the K169-recognizing ED motif antibody HG120 isolated from an RV144 subject. These data demonstrated that while there are many solutions to the problem of epitope recognition, the antibody response can enlist common modes of recognition through preferential VH:VL pairing. While VH:VL pairing is thought to be largely a stochastic process (de Wildt et al., 1999; Jayaram et al., 2012), the selection process itself is not random and those pairings that result in high affinity binding of antigen by the naïve BCR will be preferentially selected (Dal Porto et al., 1998; Shih et al., 2002). In HIV-1 infection, restricted pairing among different individuals has previously been reported for the VRC-01 CD4 binding site class of bnAbs where IGHV1-2 or IGHV1-46 must pair with light chain VJ rearrangements with a very short LCDR3 (West et al., 2012) in order to mimic CD4 in binding to the gp120 CD4 binding site at the correct angle for broad neutralization (Zhou et al., 2013). An additional example of restricted pairing in the HIV-1 antibody response is the VH:VL pairing of IGHV1-69:IGKV3-20 which has been observed for two gp41 neutralizing antibodies, 4E10 and CAP206-CH12 (Morris et al., 2011; Zwick et al., 2001), that recognize a similar MPER epitope. Unlike in broadly neutralizing HIV-1 antibodies, (Haynes et al., 2012b), the ED motif antibodies have low levels of somatic mutations. Therefore, while the ED motif-containing antibodies are associated to date with a low level of vaccine efficacy (Haynes et al., 2012a; Liao et al., 2013; Rerks-Ngarm et al., 2009), they are readily inducible, making the ED antibody response a desirable type of vaccine-induced response. One problem with the RV144 vaccine response was the limited breadth of the V2 response (Rolland et al., 2012). Efforts to broaden the V2 response with more V2 immunogen motifs will be one key to improving vaccine efficacy.
Several conformational V2 antibodies with limited neutralization have been elicited in HIV-1 infection and described previously (Gorny et al., 1994; Gorny et al., 2012; Mayr et al., 2013). Analysis of one of these antibodies, 697-D, demonstrated that unlike CH58 and CH59, V2 binding was not dependent on K169 (Liao et al., 2013). Additional analysis of published binding data of six additional infection-derived V2 antibodies showed that none of the conformational V2 antibodies were K169-sensitive (Gorny et al., 2012). Thus, while infection-derived V2 antibodies represent a class of V2 antibodies, they do not share the same recognition properties as the human or rhesus V2 ED-motif dependent antibodies elicited from vaccination.
It is important to note that ED motif-bearing antibodies target the V2 loop at K169 that is also a component of the V1V2-glycan epitope recognized by the PG9 class of broadly neutralizing antibodies comprised of PG9, PG16, CAP256-VRC26 and CH01 (Kwong and Mascola, 2012). This class of bnAbs is characterized by their extremely long HCDR3s (24–37 amino acids in length), sulfated tyrosines by post-translational modification, and recognition of glycans in the V2 loop. The pool of B-cells with extremely long CDRs is thought to be very small with <1% of B-cells of HCDR3s greater than 28 (Briney et al., 2012). In addition, a tyrosine within a larger amino acid motif is required for sulfation to occur (Stone et al., 2009). Taken together, the low likelihood of these bnAb requirements occurring simultaneously could help explain the difficulty of eliciting this type of bnAb response. The observation that ED motif antibodies target a linear V2 epitope and are easy to induce could also act to block elicitation of PG9-class bnAbs.
Finally, recent estimates have placed primate lentivirus infection occurring as long as 12 million years ago (Compton and Emerman, 2013) and lysine at position 169 is relatively well-conserved in Envs of simian immunodeficiency virus strains infecting many primate species (HIV/SIV Sequence Database: http://www.hiv.lanl.gov). The observation that IGLV3-10 has highly similar orthologs throughout the primate lineage that include the ED motif, suggests that the ED motif likely confers an evolutionary advantage within the antibody response. One hypothesis is that a recent evolutionary driving factor for ED motif maintenance could be primate retroviral defense.
EXPERIMENTAL PROCEDURES
Immunization of Rhesus Macaques
Rhesus macaque immunization regimens for NHP Studies 36, 54 and 62.1 are summarized in Figure S1. For NHP study 36, immunizations were given intramuscularly 5 times at weeks 0, 4, 12, 23 and 53 at 5×107 pfu per dose of ALVAC and 600μg per dose of AIDSVAX B/E gp120 (5 rhesus macaques). Blood, spleen, terminal ileum, as well as mesenteric, inguinal, anterior pelvic, posterior pelvic and periaortic lymph node samples were collected at 2 weeks after the 5th immunization. For NHP study 54, immunizations were given intramuscularly 7 times at 6 week intervals with 100 μg per dose (6 rhesus macaques). Blood samples were collected at 2 weeks after the 3rd immunization. For NHP study 62.1, immunizations were given intramuscularly 5 times at 6 weeks intervals with 100 μg per dose (6 rhesus macaques). Blood samples were collected at 2 weeks after the 5th immunization. All rhesus macaques were housed at Bioqual, Inc, Rockville, MD (NHP studies 54 and 62.1) and the New England Primate Research Center, Southborough, MA (NHP study 36). All rhesus macaques were maintained in accordance with the Association for Assessment and Accreditation of Laboratory Animals with the approval of the Animal Care and Use Committees of the National Institutes of Health and Harvard Medical School. Research was conducted in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, NRC Publication, 2011 edition.
Antibody Isolation
AE.A244 gp120D11 and AE.A244 V1V2 peptides were conjugated with either AF647 or BV421 (Invitrogen) dyes. Protein quality after conjugations was assessed by flow cytometry for binding to a panel of well-characterized HIV-1 specific antibodies. Peripheral blood mononuclear cells (PBMCs) of immunized rhesus macaques were stained with AquaVital dye, CD14 (BV570), CD16 (PE-Cy7), CD3 (PerCP Cy5.5), CD20 (FITC), CD27 (APC-Cy7), IgD (PE). Antigen-specific memory B-cells were single-cell sorted with a BD FACS Aria II into 96 well plates containing 20uL of reverse transcriptase buffer (RT) by gating on AquaVital dye-, CD14−, CD16−, CD3−, IgD−, CD20+, CD27+, BV421 and AF647 double positive cells. Immunoglobulin (Ig) heavy and light chain genes (VH and VL) were PCR amplified using a modified version of a previously described protocol (Liao et al., 2009), optimized for the amplification of rhesus antibodies (Zhang et al. unpublished data). The RT reaction was performed using 0.100 μM of constant region primers IgG, IgM, IgA, IgD, IgE, Igκ, Igλ (Table S6). Primers and RNA were first incubated at 65°C for 5 minutes. The PCR plate was chilled to 4°C and 0.75mM dNTPs (Qiagen), SSIII (U) (Invitrogen) and RNase Out (U) (Invitrogen) was added followed by an incubation at 50°C for 45 minutes, 55°C for 15 minutes. The cDNA was then amplified with two PCR steps. In the first step, 5μL of cDNA, 10X PCR Buffer (Qiagen), 10X Q Buffer (Qiagen), 0.20 mM dNTPs (Qiagen), MgCl2 (IgH-0.50mM, Igκ- 1.00mM, Igλ- 1.50mM) and 0.125 μM of either IgA1, IgA2, IgD, IgG, IgM, or Igκ, or Igλ with IgH, Igκ, or Igλ variable region primers (Table S6) were PCR amplified in a 50μL reaction at 95°C for 5 min, 94°C for 30 sec, 64°C (Igκ and Igλ) 62°C (IgH)for 45 sec, 72°C for 90 sec for 35 cycles followed by one cycle of 72°C for 7 min for IgH. Nested PCR was then performed in individual amplifications with 3.0μL of PCR product, 10X PCR Buffer (Qiagen), 10X Q Buffer (Qiagen), 0.20 mM dNTPs (Qiagen), MgCl2 (IgH-1.50mM, Igκ- 1.00mM, Igλ- 1.00mM), with 0.500 μM of IgH, or Igκ, or Igλ internal forward and reverse variable region primers (Table S6). The nested PCR conditions were: 95°C for 5 min, 94°C for 30 sec, 64°C (Igκ and Igλ) 62°C (IgH) for 45 sec, 72°C for 90 sec for 35 cycles followed by one cycle of 72°C for 7 min. PCR products were analyzed on 1.2% SYBER® Safe E-Gels® (Invitrogen). VH and VL PCR amplified genes were then purified, sequenced, sequences analyzed and VDJ arrangements inferred using previously described computational methods (Kepler, 2013; Munshaw and Kepler, 2010).
Transient Antibody Expression
Transient antibody expression was performed as previously described (Liao et al., 2009).
Recombinant Antibody Expression
Recombinant antibody expression was performed as previously described (Liao et al., 2011).
ELISA
ELISA assays were performed as previously described (Liao et al., 2011). K169 sensitivity in the initial screen of transient transfected V2 antibodies was established with ELISA by computing the area under the curve (AUC) for 3 serial dilutions (1:1, 1:3, and 1:9) for antibody binding to wildtype 171 V2 peptide and the K169A 171. Antibodies for which >50% reduction in K169A AUC relative to WT was observed were classified as K169 dependent. For epitope mapping of purified antibodies, ELISA half maximal effective concentrations (EC50s) were calculated by curve fitting using a five parameter logistic model and epitope positions defined by V2 171 peptide alanine scan mutations that reduced EC50s by >50% relative to wild type. Site-directed mutagenesis was performed as previous described (Gao et al., 2014).
Neutralization Assays
Neutralization activity of antibodies was measured in TMZ-bl cell-based (Sarzotti-Kelsoe et al., 2013) and A3R5.7 cell-based (McLinden et al., 2013) neutralization assays.
SPR Kinetics Measurements
Epitope mapping experiments in SPR were performed on a BIAcore 4000 (BIAcore Inc, Piscattaway, NJ) instrument at 25 C. Using a Series S CM5 chip, wild-type and ala-substituted peptides were amine-coupled directly on the chip surface. Data analyses were performed using the Biacore 4000 evaluation and BIAevaluation 4.1 software (BIAcore) as previously described (Alam et al., 2011; Alam et al., 2007). Binding responses of the irrelevant respiratory syncytial virus (RSV) antibody Synagis was used to subtract out responses due to non-specific interactions. Antibodies 1056 and 1534 failed to bind the AE.A244 171 peptide in ELISA assays but binding was detected with SPR and alanine scanning mutagenesis SPR was used to fine map the 1056 and 1534 epitopes using an epitope inclusion criteria of >50% reduction in SPR response units relative to wild type peptide.
Sequence analysis and Molecular modeling
Details of the sequence analysis and molecular modeling performed are provided in the Supplemental Experimental Procedures.
Biolayer interferometry (BLI) measurements
All BLI measurements were made using a ForteBio OctetRed 96 instrument and streptavidin sensors at 25°C and data analyses were performed using ForteBio data analysis 7 software. The gp120165-182 peptide sensors were prepared by dipping streptavidin sensors into wells containing biotinlyated gp120165-182 peptides (5μg/ml) for 300s. The peptide-loaded sensors were washed in PBS buffer (pH 7.4) for 60s before obtaining baseline. The affinity measurements of the mature and UA Fabs of CH59 to the gp120165-182 peptide were carried out by performing binding titrations (Fab concentrations ranged from 1–50μg/ml). The gp41 MPER specific 13H11 Fab (1–20μg/ml) binding to the WT peptide sensors was used in parallel to subtract out binding due to non-specific interactions with the sensors. The subtracted binding curves were fitted globally to a 1:1 binding model to obtain association (ka), dissociation (kd) rate constants and the apparent dissociation constant (Kd). The titrations were performed in triplicate.
Crystallographic Analysis of CH59-UA
Fab fragments of CH59-UA were produced recombinantly as previously described (Nicely et al., 2010) In short, Fab chains were generated by PCR using light and heavy chain genes as templates with appropriate primer pairs, and cloned into pcDNA3.1/hygro (Liao et al., 2006). Recombinant Fabs were produced in 293F cells by co-transfection with heavy chain and light chain gene plasmids, then purified using the methods described previously (Nicely et al., 2010). After affinity capture using CaptureSelect Fab lambda affinity matrix [BAC] under standard conditions, the Fab was further purified via gel filtration chromatography using a HiLoad 26/60 Superdex 200pg 26/60 column at 2 mL/min with a buffer of 10 mM Hepes pH 7.2, 50 mM NaCl, 0.02% NaN3. Fab peak elution fractions were pooled and exchanged into ddH2O via five dilute/concentrate cycles and brought to a final concentration of 15.0 mg/mL.
Unliganded Fab was tested in various crystallization screens in SBS format using reservoir volumes of 60 μL and drop volumes of 0.6 μL. Plates were incubated at 20°C. Crystals of CH59-UA were observed over a reservoir of 0.05 M calcium chloride dihydrate, 0.1 M MES monohydrate pH 6.0, 45% PEG 200 in drops composed of 0.3 μL protein + 0.3 μL reservoir with the protein solution at 15 mg/mL. These crystals were cryo-cooled directly from the drop.
Data on the crystal was collected at SER-CAT BM using an incident beam with wavelength of 1 Å. Data were reduced in HKL-2000 (Otwinowski and Minor, 1997) Matthews analysis suggested two Fabs in the asymmetric unit of the unliganded CH59-UA structure (Matthews, 1968) The structure was phased by molecular replacement in PHENIX (Terwilliger et al., 2008) using as the source model the mature CH59 Fab structure (Liao et al., 2013). Rebuilding and real-space refinements were done in Coot (Emsley et al., 2010) with reciprocal space refinements in PHENIX (Adams et al., 2010) and validations in MolProbity (Lovell et al., 2003). The crystal structure of unliganded CH59-UA Fab was refined to a resolution of 2.4 Å (Table S1).
Supplementary Material
Figure S1. Immunization regimens for the three NHP studies (Related to Fig. 1)
Regimens for (A) NHP #36 (B) NHP #54 and (C) NHP #62.1. Immunogens are listed in angled boxes above black horizontal arrows and corresponding times of immunization are listed below. Time of antibody isolation is indicated by a red box and line.
Figure S2. Epitope mapping of purified V2 antibodies (related to Table 1)
Purified antibodies were epitope mapped by alanine scanning mutations on the 171 V2 peptide. Red arrows indicate >50% reduction in binding (red line) of alanine scanning mutation at specified position relative to wild type peptide binding as measured by ELISA (EC50) (panels A-R) or SPR (Response Units) (panels S-T). Values listed are averages of measurements run in duplicate. For ease of comparison, scale of y-axis is set at 2.0 with values >2 listed above bars.
Figure S3. Codon usage in ED motif in IGLV3-10 primate orthologs (Related to Fig. 5)
The use of synonymous codons in primate orthologs of human IGLV3-10 encoding glutamic acid and aspartic acid (ED) at positions 50 and 51 suggests evidence of selective pressure for maintenance of LCDR2 ED motif in the primate lineage. Amino acids (uppercase) and DNA codons (lowercase) that differ from human are shown in red.
Figure S4. Structural models and binding kinetics of CH59-UA (Related to Fig. 6)
Representative bio-layer interferometry sensograms (red) and the best fit (black) are shown for the binding between wild type 171 V2 peptide and (A) CH59-UA and (B) CH59. (C) The binding kinetics parameters of CH59-UA and CH59 interactions with wild type 171 peptide are compared. The on-rate (ka), off-rate (kd) and dissociation constant (Kd) values were obtained by global fitting of specific binding sensograms to a 1:1 binding model and values listed are averages of triplicate measurements. (D) Epitope mapping of CH59-UA carried out using SPR measurements of alanine scans of AE.A244 gp120166-186 peptide binding of CH59-UA. Red line denotes 50% reduction in binding relative to WT peptide. Asterisk denotes alanine to glutamine mutation. To focus on epitope positions where binding decreased, the scale of y-axis is set at 2.0 with values >2 listed next to bars. Structural models of complexes of V2 peptide (green) and (E) rhesus antibody 975 and (F) a rhesus antibody with an unmutated IGHV3-78/IGLV3-17 pairing. In each model, the heavy chain (magenta) and light chain (gray) are shown in cartoon representation with a transparent surface overlayed to illustrate binding site shape. Predicted salt bridge interactions of the ED motif are shown by dashed black lines. The predicted steric clash between residue Y59 (yellow, stick representation) in the HCDR2 of IGHV3-78 and the V2 peptide is denoted by an asterisk.
Figure S5. Multiple sequence alignments of rhesus antibodies using the IGLV3-17 gene segment (Related to Fig. 3)
Multiple sequence alignments for antibody (A) light chain amino acid sequences and (B) heavy chain amino acid sequences. Dots denote matches to reference sequence (gray fill). CDRs are shaded light blue and the LCDR2 ED motif (red) has been bolded for emphasis.
Table S1. ELISA Binding Data for V2-reactive Rhesus Antibodies, Unmutated Common Ancestors and ED Motif Alanine Mutations (Related to Fig 3)
Table S2. Characteristics of All V2 Antibodies in Study (Related to Fig. 1)
Table S3. Neutralization Data (Related to Table 1)
Table S4. Candidate Orthologs of Relevant V Gene Segments and Nomenclature Mapping (Related to Fig. 4)
Table S5. Data collection and refinement statistics for CH59-UA (Related to Fig 6)
Table S6. Rhesus PCR Primers (Related to Fig. 1)
Acknowledgments
This work was supported by Collaboration for AIDS Vaccine Discovery grants from the Bill & Melinda Gates Foundation and by the Center for HIV/AIDS Vaccine Immunology-Immuongen Discovery (CHAVI-ID; UMI-AI100645) grant from NIH/NIAID/DAIDS. This work was also supported by grant number 5T32-AI007392 from the NIH, NIAID. Flow cytometry work was additionally supported by The Duke University Center for AIDS Research Flow Cytometry core (NIH, NIAID, CFAR grant P30-AI-64518). Funding was also provided by Interagency Agreement Y1-AI-2642-12 between U.S. Army Medical Research and Material Command (USAMRMC) and the National Institutes of Allergy and Infectious Diseases through a cooperative agreement (W81XWH-07-2-0067) between the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., and the U.S. Department of Defense (DOD). The views expressed in this article are those of the authors and should not be construed as official or as representing the views of the Department of Defense or the Department of the Army. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. We thank A. Foulger for expert technical assistance in antibody production; D. Marshall and J. Whitesides for expert technical assistance in flow cytometry; A. Martelli for expert technical assistance in neutralization assays; and K. Soderberg and S. Bowen, for project management.
Footnotes
AUTHOR CONTRIBUTIONS
D.E., K.L. and T.B. isolated antibodies, designed assays and analyzed data. K.W. conducted structural and sequence analyses, data analyses and interpretation. K.W. and D.E. edited the manuscript. N.I.N. performed structural analysis of CH59. F.H.J., S.M.D, and S.M.A. performed protein purification and SPR assays. R.Z and H.X.L contributed to rhesus PCR and antibody production. K.E.L, C.S. and R.P. performed antibody binding assays. L.S., R.S. and S.S. performed rhesus immunizations. L.M. provided CAP206 Envs and sequences. J.K., S.N., P.P., S, R-N., N.L.M., and J.H.K. provided vaccine. G.K. and M. B. provided experimental design and interpreted data. D.C.M. performed neutralization assays. G.D.T. performed antibody binding assays. T.B.K. designed software and performed computational analyses of antibody sequences and inferred UCAs. M.A.M. produced fluorophor-labeled Env proteins for memory B cell cultures, and B.F.H. designed the study, oversaw all experiments and analyzed all data. K.W. and B.F.H wrote the paper.
ACCESSION NUMBERS
The Genbank accession numbers for VH and VL sequences of the rhesus antibodies reported in this paper are KJ956810 to KJ956891. Coordinates and structure factors for CH59-UA have been deposited into the Protein Data Bank with accession code 4QF1.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam SM, Liao HX, Dennison SM, Jaeger F, Parks R, Anasti K, Foulger A, Donathan M, Lucas J, Verkoczy L, et al. Differential reactivity of germ line allelic variants of a broadly neutralizing HIV-1 antibody to a gp41 fusion intermediate conformation. Journal of virology. 2011;85:11725–11731. doi: 10.1128/JVI.05680-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam SM, McAdams M, Boren D, Rak M, Scearce RM, Gao F, Camacho ZT, Gewirth D, Kelsoe G, Chen P, Haynes BF. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. Journal of immunology. 2007;178:4424–4435. doi: 10.4049/jimmunol.178.7.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Pollara J, Moody MA, Alpert MD, Chen X, Hwang KK, Gilbert PB, Huang Y, Gurley TC, Kozink DM, et al. Antibody-dependent cellular cytotoxicity-mediating antibodies from an HIV-1 vaccine efficacy trial target multiple epitopes and preferentially use the VH1 gene family. J Virol. 2012;86:11521–11532. doi: 10.1128/JVI.01023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briney BS, Willis JR, Crowe JE., Jr Human peripheral blood antibodies with long HCDR3s are established primarily at original recombination using a limited subset of germline genes. PLoS One. 2012;7:e36750. doi: 10.1371/journal.pone.0036750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton AA, Emerman M. Convergence and divergence in the evolution of the APOBEC3G-Vif interaction reveal ancient origins of simian immunodeficiency viruses. PLoS Pathog. 2013;9:e1003135. doi: 10.1371/journal.ppat.1003135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Porto JM, Haberman AM, Shlomchik MJ, Kelsoe G. Antigen drives very low affinity B cells to become plasmacytes and enter germinal centers. J Immunol. 1998;161:5373–5381. [PubMed] [Google Scholar]
- de Wildt RM, Hoet RM, van Venrooij WJ, Tomlinson IM, Winter G. Analysis of heavy and light chain pairings indicates that receptor editing shapes the human antibody repertoire. J Mol Biol. 1999;285:895–901. doi: 10.1006/jmbi.1998.2396. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frippiat JP, Lefranc MP. Genomic organisation of 34 kb of the human immunoglobulin lambda locus (IGLV): restriction map and sequences of new V lambda III genes. Mol Immunol. 1994;31:657–670. doi: 10.1016/0161-5890(94)90175-9. [DOI] [PubMed] [Google Scholar]
- Gao F, Bonsignori M, Liao HX, Kumar A, Xia SM, Lu X, Cai F, Hwang KK, Song H, Zhou T, et al. Cooperation of B Cell Lineages in Induction of HIV-1-Broadly Neutralizing Antibodies. Cell. 2014;158:481–491. doi: 10.1016/j.cell.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Gnanakaran S, Daniels MG, Bhattacharya T, Lapedes AS, Sethi A, Li M, Tang H, Greene K, Gao H, Haynes BF, et al. Genetic signatures in the envelope glycoproteins of HIV-1 that associate with broadly neutralizing antibodies. PLoS Comput Biol. 2010;6:e1000955. doi: 10.1371/journal.pcbi.1000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Moore JP, Conley AJ, Karwowska S, Sodroski J, Williams C, Burda S, Boots LJ, Zolla-Pazner S. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of human immunodeficiency virus type 1. J Virol. 1994;68:8312–8320. doi: 10.1128/jvi.68.12.8312-8320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Pan R, Williams C, Wang XH, Volsky B, O’Neal T, Spurrier B, Sampson JM, Li L, Seaman MS, et al. Functional and immunochemical cross-reactivity of V2-specific monoclonal antibodies from HIV-1-infected individuals. Virology. 2012;427:198–207. doi: 10.1016/j.virol.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012a;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nature Biotechnology. 2012b;30:423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram N, Bhowmick P, Martin AC. Germline VH/VL pairing in antibodies. Protein Eng Des Sel. 2012;25:523–529. doi: 10.1093/protein/gzs043. [DOI] [PubMed] [Google Scholar]
- Kepler TB. Reconstructing a B-cell clonal lineage. I. Statistical inference of unobserved ancestors. F1000Res. 2013;2:103. doi: 10.12688/f1000research.2-103.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc MP. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 2001;29:207–209. doi: 10.1093/nar/29.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Bonsignori M, Alam SM, McLellan JS, Tomaras GD, Moody MA, Kozink DM, Hwang KK, Chen X, Tsao CY, et al. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity. 2013;38:176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Chen X, Munshaw S, Zhang R, Marshall DJ, Vandergrift N, Whitesides JF, Lu X, Yu JS, Hwang KK, et al. Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. J Exp Med. 2011;208:2237–2249. doi: 10.1084/jem.20110363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Levesque MC, Nagel A, Dixon A, Zhang R, Walter E, Parks R, Whitesides J, Marshall DJ, Hwang KK, et al. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J Virol Methods. 2009;158:171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Sutherland LL, Xia SM, Brock ME, Scearce RM, Vanleeuwen S, Alam SM, McAdams M, Weaver EA, Camacho Z, et al. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology. 2006;353:268–282. doi: 10.1016/j.virol.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- Mayr LM, Cohen S, Spurrier B, Kong XP, Zolla-Pazner S. Epitope mapping of conformational V2-specific anti-HIV human monoclonal antibodies reveals an immunodominant site in V2. PLoS One. 2013;8:e70859. doi: 10.1371/journal.pone.0070859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLinden RJ, Labranche CC, Chenine AL, Polonis VR, Eller MA, Wieczorek L, Ochsenbauer C, Kappes JC, Perfetto S, Montefiori DC, et al. Detection of HIV-1 neutralizing antibodies in a human CD4(+)/CXCR4(+)/CCR5(+) T-lymphoblastoid cell assay system. PLoS One. 2013;8:e77756. doi: 10.1371/journal.pone.0077756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris L, Chen X, Alam M, Tomaras G, Zhang R, Marshall DJ, Chen B, Parks R, Foulger A, Jaeger F, et al. Isolation of a human anti-HIV gp41 membrane proximal region neutralizing antibody by antigen-specific single B cell sorting. PLoS One. 2011;6:e23532. doi: 10.1371/journal.pone.0023532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshaw S, Kepler TB. SoDA2: a Hidden Markov Model approach for identification of immunoglobulin rearrangements. Bioinformatics. 2010;26:867–872. doi: 10.1093/bioinformatics/btq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicely NI, Dennison SM, Spicer L, Scearce RM, Kelsoe G, Ueda Y, Chen H, Liao HX, Alam SM, Haynes BF. Crystal structure of a non-neutralizing antibody to the HIV-1 gp41 membrane-proximal external region. Nat Struct Mol Biol. 2010;17:1492–1494. doi: 10.1038/nsmb.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Perelman P, Johnson WE, Roos C, Seuanez HN, Horvath JE, Moreira MA, Kessing B, Pontius J, Roelke M, Rumpler Y, et al. A molecular phylogeny of living primates. PLoS Genet. 2011;7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- Rolland M, Edlefsen PT, Larsen BB, Tovanabutra S, Sanders-Buell E, Hertz T, deCamp AC, Carrico C, Menis S, Magaret CA, et al. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature. 2012;490:417–420. doi: 10.1038/nature11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarzotti-Kelsoe M, Bailer RT, Turk E, Lin CL, Bilska M, Greene KM, Gao H, Todd CA, Ozaki DA, Seaman MS, et al. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J Immunol Methods. 2013 doi: 10.1016/j.jim.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AG, Xu H, Khan AR, O’Donnell T, Khurana S, King LR, Manischewitz J, Golding H, Suphaphiphat P, Carfi A, et al. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci U S A. 2013;110:264–269. doi: 10.1073/pnas.1218256109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih TA, Meffre E, Roederer M, Nussenzweig MC. Role of BCR affinity in T cell dependent antibody responses in vivo. Nat Immunol. 2002;3:570–575. doi: 10.1038/ni803. [DOI] [PubMed] [Google Scholar]
- Stone MJ, Chuang S, Hou X, Shoham M, Zhu JZ. Tyrosine sulfation: an increasingly recognised post-translational modification of secreted proteins. N Biotechnol. 2009;25:299–317. doi: 10.1016/j.nbt.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Jr, Diskin R, Nussenzweig MC, Bjorkman PJ. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc Natl Acad Sci U S A. 2012;109:E2083–2090. doi: 10.1073/pnas.1208984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev IS, Altae-Tran HR, Chuang GY, Joyce MG, et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity. 2013;39:245–258. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, Parren PW. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Immunization regimens for the three NHP studies (Related to Fig. 1)
Regimens for (A) NHP #36 (B) NHP #54 and (C) NHP #62.1. Immunogens are listed in angled boxes above black horizontal arrows and corresponding times of immunization are listed below. Time of antibody isolation is indicated by a red box and line.
Figure S2. Epitope mapping of purified V2 antibodies (related to Table 1)
Purified antibodies were epitope mapped by alanine scanning mutations on the 171 V2 peptide. Red arrows indicate >50% reduction in binding (red line) of alanine scanning mutation at specified position relative to wild type peptide binding as measured by ELISA (EC50) (panels A-R) or SPR (Response Units) (panels S-T). Values listed are averages of measurements run in duplicate. For ease of comparison, scale of y-axis is set at 2.0 with values >2 listed above bars.
Figure S3. Codon usage in ED motif in IGLV3-10 primate orthologs (Related to Fig. 5)
The use of synonymous codons in primate orthologs of human IGLV3-10 encoding glutamic acid and aspartic acid (ED) at positions 50 and 51 suggests evidence of selective pressure for maintenance of LCDR2 ED motif in the primate lineage. Amino acids (uppercase) and DNA codons (lowercase) that differ from human are shown in red.
Figure S4. Structural models and binding kinetics of CH59-UA (Related to Fig. 6)
Representative bio-layer interferometry sensograms (red) and the best fit (black) are shown for the binding between wild type 171 V2 peptide and (A) CH59-UA and (B) CH59. (C) The binding kinetics parameters of CH59-UA and CH59 interactions with wild type 171 peptide are compared. The on-rate (ka), off-rate (kd) and dissociation constant (Kd) values were obtained by global fitting of specific binding sensograms to a 1:1 binding model and values listed are averages of triplicate measurements. (D) Epitope mapping of CH59-UA carried out using SPR measurements of alanine scans of AE.A244 gp120166-186 peptide binding of CH59-UA. Red line denotes 50% reduction in binding relative to WT peptide. Asterisk denotes alanine to glutamine mutation. To focus on epitope positions where binding decreased, the scale of y-axis is set at 2.0 with values >2 listed next to bars. Structural models of complexes of V2 peptide (green) and (E) rhesus antibody 975 and (F) a rhesus antibody with an unmutated IGHV3-78/IGLV3-17 pairing. In each model, the heavy chain (magenta) and light chain (gray) are shown in cartoon representation with a transparent surface overlayed to illustrate binding site shape. Predicted salt bridge interactions of the ED motif are shown by dashed black lines. The predicted steric clash between residue Y59 (yellow, stick representation) in the HCDR2 of IGHV3-78 and the V2 peptide is denoted by an asterisk.
Figure S5. Multiple sequence alignments of rhesus antibodies using the IGLV3-17 gene segment (Related to Fig. 3)
Multiple sequence alignments for antibody (A) light chain amino acid sequences and (B) heavy chain amino acid sequences. Dots denote matches to reference sequence (gray fill). CDRs are shaded light blue and the LCDR2 ED motif (red) has been bolded for emphasis.
Table S1. ELISA Binding Data for V2-reactive Rhesus Antibodies, Unmutated Common Ancestors and ED Motif Alanine Mutations (Related to Fig 3)
Table S2. Characteristics of All V2 Antibodies in Study (Related to Fig. 1)
Table S3. Neutralization Data (Related to Table 1)
Table S4. Candidate Orthologs of Relevant V Gene Segments and Nomenclature Mapping (Related to Fig. 4)
Table S5. Data collection and refinement statistics for CH59-UA (Related to Fig 6)
Table S6. Rhesus PCR Primers (Related to Fig. 1)